Short abstract
The voltage-gated sodium channel 1.5 (Nav1.5), encoded by the SCN5A gene, is responsible for the rising phase of the action potential of cardiomyocytes. The sodium current mediated by Nav1.5 consists of peak and late components (INa-P and INa-L). Mutant Nav1.5 causes alterations in the peak and late sodium current and is associated with an increasingly wide range of congenital arrhythmias. More than 400 mutations have been identified in the SCN5A gene. Although the mechanisms of SCN5A mutations leading to a variety of arrhythmias can be classified according to the alteration of INa-P and INa-L as gain-of-function, loss-of-function and both, few researchers have summarized the mechanisms in this way before. In this review article, we aim to review the mechanisms underlying dysfunctional Nav1.5 due to SCN5A mutations and to provide some new insights into further approaches in the treatment of arrhythmias.
Impact statement
The field of ion channelopathy caused by dysfunctional Nav1.5 due to SCN5A mutations is rapidly evolving as novel technologies of electrophysiology are introduced and our understanding of the mechanisms of various arrhythmias develops. In this review, we focus on the dysfunctional Nav1.5 related to arrhythmias and the underlying mechanisms. We update SCN5A mutations in a precise way since 2013 and presents novel classifications of SCN5A mutations responsible for the dysfunction of the peak (INa-P) and late (INa-L) sodium channels based on their phenotypes, including loss-, gain-, and coexistence of gain- and loss-of function mutations in INa-P, INa-L, respectively. We hope this review will provide a new comprehensive way to better understand the electrophysiological mechanisms underlying arrhythmias from cell to bedside, promoting the management of various arrhythmias in practice.
Keywords: Nav1.5, SCN5A, gain-of-function, loss-of-function, INa-P, INa-L
Introduction
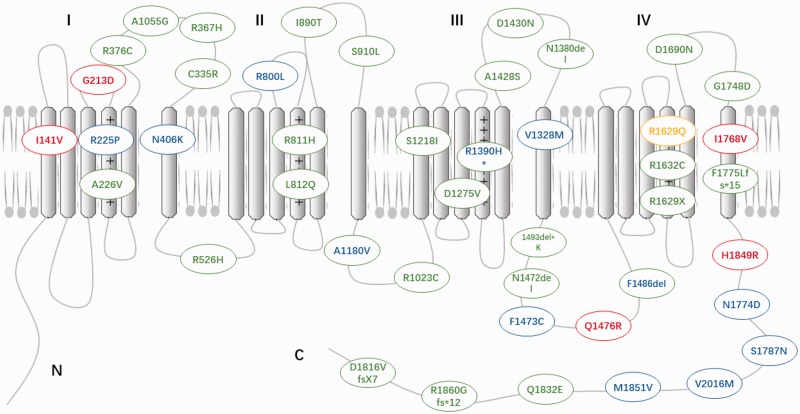
The α-subunit (Nav1.5) encoded by the SCN5A gene is the predominant element in heart tissue and plays a critical role in the excitability of cardiomyocytes. Nav1.5 channels mediate the inward sodium current (INa) and induce fast depolarization, thereby initiating the excitation–contraction coupling cascades in the cells. INa mediated by Nav1.5 can be classified into peak and late sodium currents (INa-P and INa-L). Mutations of SCN5A can impair Nav1.5 function and change the magnitude and duration of INa-P and INa-L, consequently leading to different types of fatal arrhythmias. Gain- or loss-of-function mutations are responsible for most of the pathogenesis of SCN5A mutation-induced cardiac disorders. More than 400 mutations have been identified in the SCN5A gene (updated SCN5A mutations from 2013 to 2018 are depicted in Figure 1). In this review, we will firstly introduce the biology of the Nav1.5 channel and then focus on the mechanisms underlying gain or loss of function and summarize arrhythmic consequences of mutant Nav1.5 and their clinical implications.
Figure 1.
Updated SCN5A mutations identified from 2013 to present. The red represents gain of INa-P, blue represents gain of INa-L, green represents loss of INa-P, yellow represents loss of INa-L, the mixed colors represent coexistence of gain- and loss-of-function mutations. (A color version of this figure is available in the online journal.)
Biology of the Nav1.5 channel
Sodium channels are hetero-multimeric proteins composed of a pore-forming α subunit and auxiliary β subunits. The α subunit consists of four homologous domains (DI–DIV). Each domain contains six transmembrane-spanning segments (S1–S6), of which the S4 segment functions as a voltage sensor and the S5 and S6 regions form the pore with the intermembrane P-loop.1 The α-subunit (Nav1.5) encoded by the SCN5A gene is the predominant element in the heart and plays a critical role in the excitability of cardiomyocytes. In terms of its biophysical properties, Nav1.5 channels can be observed at three states: closed at resting membrane potential (approximately −85 mV), activated during depolarization, and inactivated. Transition between these states depends primarily on the transmembrane potential, time, temperature, and pH value. Recovery from inactivation occurs within the repolarization phase during diastole under physiological conditions. The upstroke speed of the AP and conduction is determined by the numbers of Nav1.5 channels that are available for opening. The inactivation process is rapid and stable for most ion channels. However, sodium channels may inactivate incompletely, therefore generating a so-called INa-L throughout the plateau phase of the AP.2 In addition, some channels may reactivate during the repolarizing phase of the AP at a range of potentials in which inactivation is not complete and exhibits overlap with activation, resulting in the “window current.”3 Both INa-L and the window current can play critical roles in genetic and acquired cardiac diseases, as discussed below.
The SCN5A gene, which is expressed in a circadian pattern, is also expressed in extracardiac cells such as the excitable cells of the cerebral limbic system, and diverse subtypes of non-excitable cells, including microglia, astrocytes, T-lymphocytes, macrophages, fibroblasts, endothelial cells, and different type of cancer cells. Mounting evidence has demonstrated that sodium channels can take part in various effector functions and lead to non-classical effects in non-excitable cells. For instance, in cancer cells, Nav1.5 is related to enhanced invasiveness and metastasis. Nav1.5 affects Na+/H+ exchanger activity in breast cancer cells and causes local extracellular acidification, which results in activated cathepsin and consequently leads to the breakdown of the extracellular matrix. Na+ inflow is equally important in this process, as blocking Nav1.5 channels decreases the invasion of cancer cells. In addition, Nav1.5 in endosomes of macrophages from individuals with multiple sclerosis contributes to phagocytosis and pH regulation. It is suggested that targeting Nav1.5 could be a putative therapeutic approach in this disease.4
The Nav1.5 channel mediates the rapid entry of the sodium current (INa), a current that mainly contributes to the depolarization of the action potential (AP) in cardiac myocytes and the His-Purkinje system.5 INa mediated by Nav1.5 can be classified into peak and late sodium currents (INa-P and INa-L). INa-P occurs during phase 0 of the AP with a density of approximately 391 uA/uF and is quickly inactivated within 1–2 ms. The INa-L amplitude is much smaller than the INa-P amplitude in many species (approximately 0.1%–1%) and is inactivated more slowly during the plateau of the AP6 with the time constant ranging from 75 to 450 ms.7,8 The INa-P is mainly associated with the initiation of cardiac excitability and electrical conduction. The INa-P drives the rapid AP upstroke, resulting in further channel activation. This transient increase in intracellular sodium leads to calcium current (ICa) influx via L-type voltage-gated channels when the voltage upstroke reaches approximately −25 mV. The depolarization-activated ICa induces Ca2+ release from intracellular sarcoplasmic reticular Ca2+ stores and initiates myocardial mechanical activity.9
Pathogenesis of SCN5A mutations
SCN5A gene mutations impair Nav1.5 function and consequently change the magnitude and duration of INa-P and INa-L, which lead to different types of fatal arrhythmias.10 SCN5A mutations are responsible for various types of cardiac disorders, including Brudaga syndrome (BrS),11 long QT syndrome 3 (LQT3),12 cardiac conduction disease (CCD),13 sick sinus syndrome (SSS),14 atrial fibrillation (AF),15,16 progressive cardiac conduction defect (PCCD), dilated cardiomyopathy (DCM),17 multifocal ectopic Purkinje-related premature contraction (MEPPC),18 and the onset of a variety of non-cardiac diseases, including bowel syndrome,19 myotonic dystrophy,20 epilepsy,21 pain,22 and ataxia.23
SCN5A mutations result in the dysfunction of Nav1.5 due to defective protein trafficking, targeting, fixation to specific cellular compartments, post-translational protein processing, the modulation of biophysical properties and many unclear mechanisms.24 Genotype and phenotype vary significantly, as the phenotypic characterization ranges from asymptomatic phenotypes to sudden cardiac death (SCD) in individuals that carry the same mutations. In addition, specific SCN5A mutations cause an individual phenotype or compound phenotypes, indicating that a complex pathogenesis underlies SCN5A mutations.
Gain-of-function mutations and arrhythmias
Long QT syndrome (LQTS) is characterized by prolonged ventricular repolarization, which predisposes individuals to develop torsades de Pointes (TdP) and SCD. LQTS3 is caused by gain-of-function mutations of SCN5A. Approximately 8–10% of patients with SCN5A mutations are positively phenotypic as having LQTS.25,26 The first SCN5A mutation related to LQT3, the deletion of amino acids 1505–1507 (ΔKPQ), was identified by Wang et al.27 According to previous reports, cardiac events primarily occurred during sleep in LQT3 patients, and 18% died suddenly.28 The gain-of-function SCN5A mutation leads to enhanced INa-P and INa-L, which finally triggers life-threating arrhythmias primarily in LQT3 patients.
Gain-of-function mutations of INa-P
The underlying mechanisms of SCN5A mutations that lead to the gain-of-function of INa-P are mainly due to abnormalities in mutation-induced kinetic properties, including augmented INa-P amplitudes, negative shifts in the voltage-dependence of activation, and an increased speed of recovery from inactivation. The most recently identified SCN5A mutations over the last five years (from 2013 to 2018) are shown in Table 1.29–34
Table 1.
The newest identified gain of INa-P function of SCN5A mutations from 2013 to 2018 and their reported electrophysiological properties.
| Mutation | Protein domain | Biophysical properties of mutant protein | Cardiomyopathy and accompanied features | References |
|---|---|---|---|---|
| I141V | DI/S1 | Increased Iwindow: negative shift of activation | PVC, tachycardia | 33 |
| G213D | DI/S3-S4 | Increased INaP: negative shift of act, positive shift of inactivation | AA, VA, DCM | 34 |
| Q1476R | DIII-DIV | Increased INaP: positive shift of inactivation; increased INaL | LQT3 | 32 |
| G1748D | N-terminus DIV S6 | Increased INaP: positive shift of inactivation, accelerated recovery from inactivation | LQT3 | 29 |
| I1768V | DIV/S6 | Increased INaP, Iwindow: negative shift of the activation, faster recovery from inactivation | LQT3, SCD | 31 |
| H1849R | C-terminus | Increased INaP and INaL: negative shift of inactivation, slower inactivation | LQT, AF, VT, SCD | 30 |
AA: atrial arrhythmia; AF: atrial fibrillation; DCM: dilated cardiomyopathy; LQT: long QT syndrome; PVC: polymorphic ventricular complexes; SCD: sudden cardiac death; VA: ventricular arrhythmia; VT: ventricular tachycardia.
First, gains of channel function can be caused by variants that lead to augmented INa-P amplitudes. LQT3 mutations, such as I1748V31 and G1748D,29 exhibited greater INa-P than wild type, whereas variants A572D35 and G615E36 also showed a significant gain of function of INa-P, but with an unclear clinical phenotype. However, some SCN5A mutations identified in clinical LQTS patients, such as F1250L37 and N406K38 variants, showed no significant changes in INa-P amplitudes. These phenomena suggested that although altered INa-P amplitudes affect phenotypes of the diseases directly, there may be other unknown mechanisms that are related to certain environmental factors or unknown gene mutations that contribute to genotype–phenotype interactions.
Second, the gain of function of INa-P could be generated by a negative shift in voltage-dependent activation potentials. It was reported that Nav1.5 reached its maximal current at −20 mV, while the variants G1748D,29 H1849R,30 S216L,39 G983D, and F816Y40 showed peak inward currents at a more negative voltage. Patients with G1748D and H1849R showed typical LQT3 features. However, patients that carried the variants S216L, G983D, and F816Y showed an unclear phenotype. These cases indicate that not all negative shifts of activation result in a gain of function of SCN5A and manifest the LQT3 phenotype; there must be other unidentified mechanisms that underlie genotype–phenotype interactions.
Third, gain of function of INa-P can be induced by a faster recovery from inactivation. According to this underlying mechanism, variants A572D and G983D showed a faster resumption of inactivation due to a fast component of the recovery. Moreover, the dedication of the fast component to the recovery from inactivation was relatively augmented in G983D. The A572D variant manifested as atria tachycardia, and G983D manifested as an abnormal T-wave on the electrocardiography (ECG).41–43 The configuration of the T-wave pointed out the differences in the time course of ventricular repolarization. Morphologic changes in the T wave are sometimes more immediately remarkable than the mere prolongation of the QT interval; in some cases, the morphology of the T-wave is the only sensitive sign of ventricular repolarization disturbances.44,45
Gain-of-function mutations of INa-L
INa-L has also been called steady-state INa, slow inactivation, persistent current, and late current. Under physiological conditions, the amplitude of INa-L is larger in mid-myocardial cells (M cells) and Purkinje fibers than epicardial and endocardial cells.46 Although the magnitude of persistent INa-L is negligible compared to INa-P (0.1%–1%), the delay in inactivation breaks the delicate equilibrium of inward and outward currents, resulting in a prolongation of the action potential duration (APD), which manifests as a prolonged QT interval on ECG.7
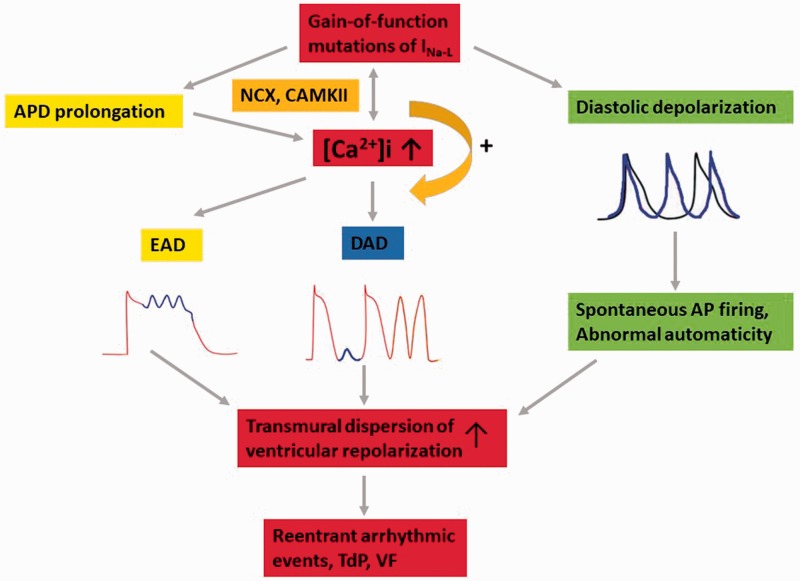
An increase in INa-L due to acquired conditions or inherited SCN5A mutations in favor of intracellular Ca2+ loading,47,48 the occurrence of early and delayed after depolarization (EAD and DAD),49,50 triggered activities,51 and spontaneous diastolic depolarization52 that promotes the spatial and temporal dispersion of ventricular repolarization can lead to reentrant arrhythmias (Figure 2).
Figure 2.
The ion mechanism of gain of function of INa-L leading to arrhythmia.
The detrimental effects of a pathological persistent INa-L contribute to the development of arrhythmic disorders. The mechanisms are as follows:
(i) During phase 2 of the AP plateau, membrane resistance is high while the ionic conductance is low,53 which caused marked APD prolongation. A prolonged APD helps L-type Ca2+ channels recover from inactivation and reactivate to form the upstroke of an EAD during the AP plateau.54 (ii) An increase in INa-L due to delayed inactivation increased intracellular Ca2+ entry via the Na+/Ca2+ exchanger (NCX, 3 Na+ out, 1 Ca2+ in)55,56 and interacted with calmodulin in a protein kinase II (CaMKII)-dependent manner,51,57 which had a positive feedback on Na+ loading. Additionally, it increased sarcoplasmic reticulum (SR) Ca2+ loading-induced Ca2+ release.47,58 The increases in INa-L, the activity of CaMKII and SR Ca2+ release contributed to a substrate precipitating DAD.59 (iii) Diastolic depolarization during phase 4 of the AP usually occurred in spontaneous pace-making cells of the sinoatrial and atrioventricular nodes.60 However, spontaneous diastolic depolarizations were often observed in Purkinje fibers and atrial tissue isolated from a diseased heart with a persistent INa-L. According to reports, the gain of function of SCN5A may lead to spontaneous AP firing and abnormal automaticity, especially in myocytes that were relatively depolarized and had low resting K+ conductance.61 After all, the formation of EAD and DAD due to the gain of function of INa-L occurred more frequently in M cells than endo and epi and increased the transmural dispersion of ventricular repolarization, which finally caused reentrant arrhythmic events, which manifested as TdP and ventricular fibrillation (VF).
Previous studies revealed several gain-of-function mechanisms of INa-L with an alteration in channel kinetics, including a slower speed of inactivation or a positive shift in the voltage-dependence of inactivation (Table 2).62–70
Table 2.
The newest identified gain of INa-L function of SCN5A mutations from 2013 to 2018 and their reported electrophysiological properties.
| Mutation | Protein domain | Biophysical properties of mutant protein | Cardiomyopathy and accompanied features | References |
|---|---|---|---|---|
| R225P | DI/S4 | Increased INaL and Iwindow: slower inactivation, shallower activation curve slope | Multifocal Ventricular Ectopy-associated Cardiomyopathy, LQT | 66 |
| N406Ka | DI/S6 | Decreased INaP: positive shift of activation; increased INaL | BrS, LQT | 65 |
| R800L | DII/S3-S4 | Increased INaL, Iwindow: incomplete inactivation and slowed decay of currents | LQTS, in compound with A261V-SNTA1 | 64 |
| A1180Va | DII-DIII | Decreased INaP: negative shift of inactivation; increased INaL | DCM, AVB | 63 |
| V1328M | DIV/S6 | Increased INaL: positive shift of inactivation | drug-induced BrS | 69 |
| F1473C | DIII-DIV | Increased INaL, Iwindow: negative shift of the inactivation | LQT3, TdP, VT, AVB | 62 |
| F1486dela | DIII-DIV | Decreased INaP; increased INaL: positive shift of inactivation, negative shift of activation, slower Na+ current decay | BrS, LQT | 65 |
| N1774D | C-terminus | Increased INaL: negative shift of activation, slower Na+ current decay; increased INaP | LQT | 65 |
| S1787N | C-terminus | Increased INaL due to splice variant and environmental factors | LQT3 | 67 |
| M1851V | C-terminus | Increased INaL: slower inactivation, faster recovery from inactivation, positive shift of inactivation | AF, VA | 70 |
| V2016Ma | C-terminus | Increased INaL: PKA activation; decreased INa | LQT, SND | 68 |
aBoth gain and loss of function mutations;
AF: atrial fibrillation; AVB: atrioventricular block; BrS: Brugada syndrome; DCM: dilated cardiomyopathy; LQT: long QT syndrome; SND: sudden nocturnal death; TdP: Torsade de pointes; VA: ventricular arrhythmia; VT: ventricular tachycardia
First, gain-of-function mutations of SCN5A resulted in slower inactivation kinetics and increased INa-L, which included the LQT3-causing variant A993T as well as A572D43 and K480N42 in patients with an unclear phenotype. In a patient with a mutation of T1526P,71 the cardiac examination was nearly normal, but T1526P showed a fractional gain-of-function property by a reduced speed of inactivation.
Second, gain of function can be caused by a positive shift in the voltage-dependence of inactivation, which also increased the magnitude and duration of INa-L. For example, the LQT3 mutation F1486L,72 which results in a gain of function, showed a positive shift in the voltage-dependence of inactivation. This effect of the N1325S73,74 variant with an unclear phenotype was also observed in patients.
Loss-of-function mutations and arrhythmias
SCN5A loss-of-function mutations often cause BrS which is characterized by ST-segment elevation in the right precordial leads (V1–V3). Over 300 SCN5A loss-of-function mutations have been identified in connection with BrS.75,76 Misfolded channels, trafficking defects, and negatively shifted steady-state inactivation curves contribute to a reduced availability of functional Nav1.5 channels on the plasma membrane.77
CCD mutations were also related to loss of function in Nav1.5.78 SCN5A loss of function reduced the AP upstroke velocity, which further delayed the rapid conduction of the electrical impulse through the highly specialized conduction system.79 Most SCN5A loss-of-function mutations prolonged the rising time of the AP and rendered it more difficult to reach the membrane potential, which is necessary for the fast AP upstroke. In addition, defects in channel gating kinetics, an inability to conduct sodium, and channel retention in the ER would further reduce the availability of channels. The SCN5A mutations that lead to a loss of channel functions can be classified as follows:
Loss-of-function mutations of INa-P
Mutations that lead to a loss of function of INa-P are related to an alteration in channel kinetics, including decreased INa-P amplitudes and retention in the ER, but the function of the channels was restored when they reached the membrane (Table 3).80–107
Table 3.
The newest identified loss of INa-P function of SCN5A mutations from 2013 to 2018 and their reported electrophysiological properties.
| Mutation | Protein domain | Biophysical properties of mutant protein | Cardiomyopathy and accompanied features | References |
|---|---|---|---|---|
| A226V | DI/S4 | Decreased INaP | BrS, in compound with p; R1629X | 102 |
| C335R | DI/S5-S6 | Decreased INaP | AF, BrS | 107 |
| D349N | No detected current | SSS | 89 | |
| R367G | Decreased INaP: trafficking defect | CCD | 40 | |
| R367H | DI/S5-S6 | Decreased INaP: positive shift of activation, negative shift of inactivation, faster recovery from inactivation | BrS, SUNDS | 106 |
| R376C | DI/S5-S6 | Decreased INaP: positive shift of activation | SSS, SCD | 91 |
| R526H | DI/DII | Decreased INaP: trafficking defect | BrS, SCD, RBBB | 94 |
| S528A | Phosphorylation site | Decreased INaP: trafficking defect | BrS, SCD, RBBB | 94 |
| R811H | DII/S4 | Decreased INaP: negative shift of inactivation, slower recovery from inactivation | BrS | 82 |
| L812Q | DII/S4 | Decreased INaP, Iwindow: trafficking defect, negative shift of inactivation | BrS | 96 |
| I890T | P-loop of DII | Decreased INaP: positive shift of the activation | BrS | 80 |
| S910L | DII/S5-S6 | Decreased INaP: trafficking defect, positive shift of activation | BrS, DCM | 20 |
| R1023C | DII-DIII | No detected current | ERS, VF, structural myocardial alteration | 84 |
| A1055G | DI/S5-S6 | Decreased INaP: negative shift of inactivation, degradation | ERS | 100 |
| W1095X | No detected current | BrS, epilepsy | 87 | |
| S1218I | DIII/S1 | Complete loss of INaP: trafficking defect | BrS | 82 |
| D1275V | DIII/S3 | Decreased INaP: positive shift of activation, enhanced degradation, trafficking defect | CCD, DCM, SND, AT, VT | 105 |
| N1380del | DIII/S5-S6 | No detected current | CCD, VT | 103 |
| R1390Ha | DIII/S4 | Decreased INaP: positive shift of activation, negative shift of inactivation, slower recovery from inactivation; increased INaL: slower deactivation | BrS, LQT, AA, VA | 99 |
| A1428S | DIII/S5-S6 | Decreased INaP | BrS | 97 |
| D1430N | DIII/S5-S6 | Complete loss of INaP: blockade of ion permeation | BrS, SCD | 86 |
| N1472del | DIII-DIV | Decreased INaP: positive shift of the activation and inactivation, slower recovery from inactivation | BrS, LQT, syncope, SCD, 2:1 AV block | 83 |
| 1493delKa | DIII-DIV | Decreased INaP: trafficking defect; enhanced recovery from inactivation | CCD, VA, SCD | 85 |
| R1629X | DIV/S4 | No detected current | BrS, in compound with p; A226V | 102 |
| R1632C | DIV/S4 | Decreased INaP: negative shift of inactivation, slower recovery from inactivation | BrS, sinus node dysfunction(SND) | 98 |
| D1690N | DIV/S5-S6 | Decreased INaP: positive shift of activation, slower recovery from inactivation; | BrS | 101 |
| G1748D | N-terminus of DIV/S6 | Decreased INaP: positive shift of activation curve, faster inactivation | BrS | 29 |
| F1775Lfs15a | DIV/S6 | Decreased INaP | overlap syndrome of SSS and BrS | 81 |
| L1786Qa | Decreased INaP: positive shift of activation, negative shift of inactivation; increased INaL | overlap syndrome of LQT3 and BrS | 92 | |
| D1790N | No detected current | SSS | 89 | |
| D1816VfsX7a | Truncation of C-terminus | Decreased INaP: trafficking defect, positive shift of activation; increased INaL: positive shift of inactivation, accelerated activation, faster recovery from inactivation | BrS, VF, bradycardia, AF | 90 |
| Q1832E | C-terminus | Decreased INaP: trafficking defect | BrS, SIDS, in compound with R1944△ | 104 |
| R1860Gfs12a | Truncation of C-terminus | Decreased INaP: negative shift of inactivation, degradation, positive shift of activation; increase INaL: delayed inactivation | SSS, AF, AVB | 93 |
| V2016M | SIV motif | Decreased INaP: trafficking defect, positive shift of activation | BrS | 95 |
| c; 4297G>C | DIII/S5-S6 | Decreased INaP: prolonged recovery from inactivation, positive shift of activation, affect translation process or degradation of the mutant protein | ERS | 88 |
aBoth gain and loss of function mutations;
AA: atrial arrhythmia; AF: atrial fibrillation; AT: atrial tachycardia; AVB: atrioventricular block; BrS: Brugada syndrome; CCD: cardiac conduction disease; DCM: dilated cardiomyopathy; ERS: early repolarization syndrome; LQT: long QT syndrome; RBBB: right bundle branch block; SCD: sudden cardiac death; SIDS: sudden infant death syndrome; SND: sudden nocturnal death; SSS: sick sinus syndrome; SUNDS: sudden unexpected nocturnal death syndrome; TdP: Torsade de pointes; VA: ventricular arrhythmia; VT: ventricular tachycardia.
Decreased current amplitudes cause loss of function of Nav1.5. As shown in the variants R222stop and R2012H,42 patients diagnosed with BrS matched the loss of function with a reduction of INa-P. In addition, there were a number of SCN5A mutations, such as E161K108 and P336L,109 that exhibited a dramatic reduction in INa-P density, while the kinetics and gating properties of the mutant channels were unaffected. These findings demonstrated that the decrease in INa-P density of the mutant channels was primarily caused by their retention in the ER, but the function of the channels was restored when they reached the membrane. Thus, we hypothesized that amino acid mutations might affect the protein structure, which would lead to misfolding and ER retention.
Loss-of-function mutations of INa-L
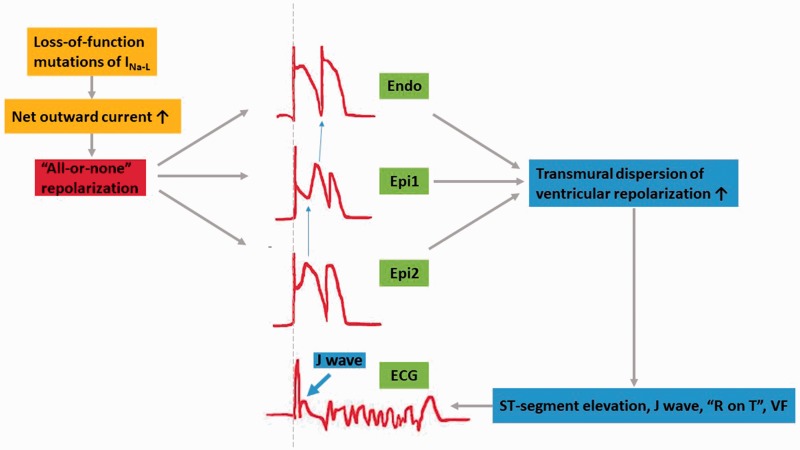
Loss of SCN5A channel function manifesting as ECG ST-segment elevation in right precordial leads was thought to contribute to an early repolarization of the right ventricular sub-epicardial myocardium that differentially altered the AP morphology of epicardial versus endocardial cells.11,110 Loss-of-function of SCN5A results in a reduction in INa-L; this alteration increases the relative amplitude of the fast, transient outward K+ current (Ito), which is the most prominent in epicardial cells of the right ventricle.111,112 Under normal conditions, Ito inhibited the depolarizing effect of the INa-L during the AP plateau, resulting in a marked AP notch in association with depolarizing Ca2+ currents in a “spike-and-dome” morphology.113 Consequently, the loss of INa-L and increase in Ito leads to a negative shift in the membrane potential, resulting in an “all-or-none” repolarization and causing an enhanced dispersion of repolarization in epicardial cells.114 Finally, this results in premature repolarization, phase 2 reentry, and significant AP shortening. In contrast, endocardial cells show a much smaller Ito and INa-L reduction, which do not significantly affect AP morphology and duration. The transmural heterogeneity of the cellular membrane voltage ultimately causes ST-segment elevation, a J wave, and even severe “R on T,” which leads to fatal VF on an ECG (Figure 3).115 Previous studies showed that the alteration of channel kinetics, such as a faster speed of inactivation or a negative shift in the voltage-dependence of inactivation, causes a loss of function of INa-L (Table 4).116
Figure 3.
The ion mechanism of loss of function of INa-L leading to arrhythmia.
Table 4.
The newest identified loss of INa-L function of SCN5A mutations from 2013 to 2018 and their reported electrophysiological properties.
| Mutation | Protien domain | Biophysical properties of mutant protein | Cardiomyopathy and accompanied features | References |
|---|---|---|---|---|
| R1629Q | DIV/S4 | Decreased INaL: negative shift of inactivation, enhanced intermediate inactivation, prolonged recovery from inactivation | BrS, SCD | 116 |
BrS: Brugada syndrome; SCD: sudden cardiac death.
First, faster inactivation kinetics contribute to a decreased INa-L. For example, V1591L was detected in a BrS patient with a decreased INa-L as a result of faster inactivation kinetics.117 However, an R568H variant identified in a patient42 diagnosed with QT prolongation also showed loss of function by faster inactivation kinetics. The mechanism of genotype-phenotype interactions remained unknown. Second, a negative shift of the voltage dependence of fast inactivation leading to a reduced INa-L caused loss of function of SCN5A. T1620K118,119 mutant channels in CCD patients, for instance, inactivate rapidly at less depolarized potentials, which may result in a significant reduction in INa-L and consequently lead to delayed AP upstroke in the Purkinje system. Similarly, the R2012H variant in BrS patients also resulted in a loss of function due to a negative shift of the voltage-dependence of inactivation.42
Loss-of-function mutations in both INa-P and INa-L
Studies pointed out that most of the mutant Nav1.5 associated with a conduction disorder displayed either a drastic current reduction or shifts of steady-state inactivation/activation or both,120,121 which may involve the alteration of INa-P and INa-L at the same time. As seen in G514C122,123 mutant channels, a positive shift of both steady-state activation and inactivation, leading to changes in both INa-P and INa-L, was observed and the shift of the activation curve predominated by only 3 mV, which was still sufficient to produce a reduced upstroke velocity. This type of mutation may cause overlap syndromes. It is reasonable to speculate that simultaneous alterations of inactivation/activation kinetics may lead to this type of phenotype.
Coexistence of gain- and loss-of function mutations
SCN5A mutations that present with an overlapped phenotype of LQT3 and BrS were also described.124 In vitro studies suggested that these uncommon SCN5A mutations cause a mixed phenotype by altering the amplitude of INa-P and INa-L through enhanced sodium channel inactivation, a negative shift in steady‐state sodium channel inactivation, and enhanced tonic block in response to sodium channel blockers.78,125
R1193Q,126,127 E1784K,128 and S216L variants in SCN5A are proposed to cause either BrS or LQTS. E1784K, originally described by Wei et al.129 and subsequently explored by several other groups, is the most common mutation that causes both LQT3 and BrS phenotypes. INa-P amplitude is significantly reduced, whereas the steady‐state inactivation is shifted to much more positive potentials that lead to an enhanced INa-L. The R1193Q mutation reveals a slower inactivation with a persistent INa-L, which accounts for the gains of channel function and is in line with the LQT3 manifestations. Additionally, the steady-state inactivation of this mutation was shifted to a more negative voltage that lowered INa-L, which explains the loss of function of sodium current and BrS clinical characteristics. S216L is proposed to be an LQTS3-causing mutation because of a significant increase in the persistent INa-L, as well as an acceleration of the recovery from inactivation. It was also identified in a BrS patient due to a significant reduction in the INa-P. Therefore, S216L is considered to cause a mixed BrS/LQT phenotype. The mechanisms of the coexistence of gain and loss of channel functions remain unclear; studies speculate that some environmental factors may play a vital role in the formation of the phenotype, and there might be other gene mutations to identify.
Perspectives
Since the first-generation gene sequencing technology was invented by Sanger in 1977, it has made tremendous progress. The brand-new third-generation sequencing technology has made the identification of gene mutations and genotypes more convenient. Functional analysis, including automatic patch clamp to study ion currents, in silico model simulation, and cryo-electronic microscopy to observe protein structure are advanced approaches to investigate the function of ion channels. New mutant models, such as iPSC-CM and transgenic animals, are more practical methods to investigate genotype–phenotype interactions. In clinical practice, we should also take more advantage of the current methods, such as ECG. ECG reflects the immediate manifestation of cardiac electrical activities on the body surface, provides the most direct evidence, and is a convenient approach for diagnosis. The alteration of T wave morphology and duration on the ECG is now used to distinguish different types of gene mutations. These discoveries stand out as notable landmarks in the progression of modern medical science that will allow further interpretations of the related pathogenesis. Additionally, in reference to these underlying mechanisms, many targeting therapies are desperately needed. Therapies that target these specific mutations with gene therapy, including RNA interference (RNAi) and CRISPER/Cas9, are under exploration, in addition to other traditional medicine therapies that affect the physiologic derangements of the mutations. The present findings enhance the general concept that the in vitro characterization of mutant ion channel functions is a key component for the generation of specific therapeutic strategies for patient management. Late sodium channel blockers, including mexiletine, ranolazine, flecainide, and a new compound, GS‐6615, have been used in patients with LQT3 to restore the gain of channel function. Mexiletine also rescues the membrane expression of the Nav1.5 channel that expresses the BrS mutation. Mexiletine may be able to rescue the retention of Na1.5 in the ER to increase the sodium current, but it can also block the INa-L, which manifests as an inhibitory effect.
In this review, we summarized the gain and loss of channel functions in the SCN5A gene and discussed the underlying mechanisms of its genotype–phenotype relationship. The detailed mechanisms that underlie dysfunctional Nav1.5 due to SCN5A mutations are described herein, and we also provide some new evidence for additional approaches in the treatment of arrhythmias due to mutant Nav1.5 channels.
Acknowledgements
We extend our gratitude to Dr. Ganxin Yan from Lankenau Hospital, Philadelphia, PA, USA, for editing this manuscript.
Authors’ contributions
Dan Han wrote the draft. Guoliang Li, Hui Tan and Chaofeng Sun revised it.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Chen-Izu Y, Shaw RM, Pitt GS, Yarov-Yarovoy V, Sack JT, Abriel H, Aldrich RW, Belardinelli L, Cannell MB, Catterall WA, Chazin WJ, Chiamvimonvat N, Deschenes I, Grandi E, Hund TJ, Izu LT, Maier LS, Maltsev VA, Marionneau C, Mohler PJ, Rajamani S, Rasmusson RL, Sobie EA, Clancy CE, Bers DM. Na+ channel function, regulation, structure, trafficking and sequestration. J Physiol 2015; 593:1347–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleber AG, Rudy Y. Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiol Rev 2004; 84:431–88 [DOI] [PubMed] [Google Scholar]
- 3.Attwell D, Cohen I, Eisner D, Ohba M, Ojeda C. The steady state TTX-sensitive (“window”) sodium current in cardiac Purkinje fibres. Pflugers Arch 1979; 379:137–42 [DOI] [PubMed] [Google Scholar]
- 4.Black JA, Waxman SG. Noncanonical roles of voltage-gated sodium channels. Neuron 2013; 80:280–91 [DOI] [PubMed] [Google Scholar]
- 5.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 2000; 26:13–25 [DOI] [PubMed] [Google Scholar]
- 6.Patlak JB, Ortiz M. Slow currents through single sodium channels of the adult rat heart. J Gen Physiol 1985; 86:89–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maltsev VA, Undrovinas AI. A multi-modal composition of the late Na+ current in human ventricular cardiomyocytes. Cardiovasc Res 2006; 69:116–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Undrovinas AI, Maltsev VA, Kyle JW, Silverman N, Sabbah HN. Gating of the late Na+ channel in normal and failing human myocardium. J Mol Cell Cardiol 2002; 34:1477–89 [DOI] [PubMed] [Google Scholar]
- 9.Fozzard HA. Cardiac sodium and calcium channels: a history of excitatory currents. Cardiovasc Res 2002; 55:1–8 [DOI] [PubMed] [Google Scholar]
- 10.Abriel H, Cabo C, Wehrens XH, Rivolta I, Motoike HK, Memmi M, Napolitano C, Priori SG, Kass RS. Novel arrhythmogenic mechanism revealed by a long-QT syndrome mutation in the cardiac Na(+) channel. Circ Res 2001; 88:740–5 [DOI] [PubMed] [Google Scholar]
- 11.Alings M, Wilde A. Brugada" syndrome: clinical data and suggested pathophysiological mechanism. Circulation 1999; 99:666–73 [DOI] [PubMed] [Google Scholar]
- 12.Berecki G, Zegers JG, Bhuiyan ZA, Verkerk AO, Wilders R, van Ginneken AC. Long-QT syndrome-related sodium channel mutations probed by the dynamic action potential clamp technique. J Physiol 2006; 570:237–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan BH, Iturralde-Torres P, Medeiros-Domingo A, Nava S, Tester DJ, Valdivia CR, Tusie-Luna T, Ackerman MJ, Makielski JC. A novel C-terminal truncation SCN5A mutation from a patient with sick sinus syndrome, conduction disorder and ventricular tachycardia. Cardiovasc Res 2007; 76:409–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL., Jr., Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J Clin Invest 2003; 112:1019–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther 2007; 81:35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benito B, Brugada R, Perich RM, Lizotte E, Cinca J, Mont L, Berruezo A, Tolosana JM, Freixa X, Brugada P, Brugada J. A mutation in the sodium channel is responsible for the association of long QT syndrome and familial atrial fibrillation. Heart Rhythm 2008; 5:1434–40 [DOI] [PubMed] [Google Scholar]
- 17.Hesse M, Kondo CS, Clark RB, Su L, Allen FL, Geary-Joo CT, Kunnathu S, Severson DL, Nygren A, Giles WR, Cross JC. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel Scn5a. Cardiovasc Res 2007; 75:498–509 [DOI] [PubMed] [Google Scholar]
- 18.Laurent G, Saal S, Amarouch MY, Beziau DM, Marsman RF, Faivre L, Barc J, Dina C, Bertaux G, Barthez O, Thauvin-Robinet C, Charron P, Fressart V, Maltret A, Villain E, Baron E, Merot J, Turpault R, Coudiere Y, Charpentier F, Schott JJ, Loussouarn G, Wilde AA, Wolf JE, Baro I, Kyndt F, Probst V. Multifocal ectopic Purkinje-related premature contractions: a new SCN5A-related cardiac channelopathy. J Am Coll Cardiol 2012; 60:144–56 [DOI] [PubMed] [Google Scholar]
- 19.Saito YA, Strege PR, Tester DJ, Locke GR, 3rd, Talley NJ, Bernard CE, Rae JL, Makielski JC, Ackerman MJ, Farrugia G. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol 2009; 296:G211–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pambrun T, Mercier A, Chatelier A, Patri S, Schott JJ, Le Scouarnec S, Chahine M, Degand B, Bois P. Myotonic dystrophy type 1 mimics and exacerbates Brugada phenotype induced by Nav1.5 sodium channel loss-of-function mutation. Heart Rhythm 2014; 11:1393–400 [DOI] [PubMed] [Google Scholar]
- 21.Noebels JL. Sodium channel gene expression and epilepsy. Novartis Found Symp 2002; 241:109–20 discussion 20–3, 226–32 [PubMed] [Google Scholar]
- 22.Priest BT, Garcia ML, Middleton RE, Brochu RM, Clark S, Dai G, Dick IE, Felix JP, Liu CJ, Reiseter BS, Schmalhofer WA, Shao PP, Tang YS, Chou MZ, Kohler MG, Smith MM, Warren VA, Williams BS, Cohen CJ, Martin WJ, Meinke PT, Parsons WH, Wafford KA, Kaczorowski GJ. A disubstituted succinamide is a potent sodium channel blocker with efficacy in a rat pain model. Biochemistry 2004; 43:9866–76 [DOI] [PubMed] [Google Scholar]
- 23.Vanmolkot KR, Babini E, de Vries B, Stam AH, Freilinger T, Terwindt GM, Norris L, Haan J, Frants RR, Ramadan NM, Ferrari MD, Pusch M, van den Maagdenberg AM, Dichgans M. The novel p.L1649Q mutation in the SCN1A epilepsy gene is associated with familial hemiplegic migraine: genetic and functional studies. Mutation in brief #957. Hum Mutat 2007; 28:522. [DOI] [PubMed] [Google Scholar]
- 24.Shy D, Gillet L, Abriel H. Cardiac sodium channel NaV1.5 distribution in myocytes via interacting proteins: the multiple pool model. Biochim Biophys Acta 2013; 1833:886–94 [DOI] [PubMed] [Google Scholar]
- 25.Priori SG, Schwartz PJ, Napolitano C, Bloise R, Ronchetti E, Grillo M, Vicentini A, Spazzolini C, Nastoli J, Bottelli G, Folli R, Cappelletti D. Risk stratification in the long-QT syndrome. N Engl J Med 2003; 348:1866–74 [DOI] [PubMed] [Google Scholar]
- 26.Napolitano C, Priori SG, Schwartz PJ, Bloise R, Ronchetti E, Nastoli J, Bottelli G, Cerrone M, Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA 2005; 294:2975–80 [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Shen J, Li Z, Timothy K, Vincent GM, Priori SG, Schwartz PJ, Keating MT. Cardiac sodium channel mutations in patients with long QT syndrome, an inherited cardiac arrhythmia. Hum Mol Genet 1995; 4:1603–7 [DOI] [PubMed] [Google Scholar]
- 28.Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation 2001; 103:89–95 [DOI] [PubMed] [Google Scholar]
- 29.Nunez L, Barana A, Amoros I, de la Fuente MG, Dolz-Gaiton P, Gomez R, Rodriguez-Garcia I, Mosquera I, Monserrat L, Delpon E, Caballero R, Castro-Beiras A, Tamargo J. p.D1690N Nav1.5 rescues p.G1748D mutation gating defects in a compound heterozygous Brugada syndrome patient. Heart Rhythm 2013; 10:264–72 [DOI] [PubMed] [Google Scholar]
- 30.Musa H, Kline CF, Sturm AC, Murphy N, Adelman S, Wang C, Yan H, Johnson BL, Csepe TA, Kilic A, Higgins RS, Janssen PM, Fedorov VV, Weiss R, Salazar C, Hund TJ, Pitt GS, Mohler PJ. SCN5A variant that blocks fibroblast growth factor homologous factor regulation causes human arrhythmia. Proc Natl Acad Sci U S A 2015; 112:12528–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauferstein S, Kiehne N, Peigneur S, Tytgat J, Bratzke H. Cardiac channelopathy causing sudden death as revealed by molecular autopsy. Int J Legal Med 2013; 127:145–51 [DOI] [PubMed] [Google Scholar]
- 32.Moreau A, Krahn AD, Gosselin-Badaroudine P, Klein GJ, Christe G, Vincent Y, Boutjdir M, Chahine M. Sodium overload due to a persistent current that attenuates the arrhythmogenic potential of a novel LQT3 mutation. Front Pharmacol 2013; 4:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swan H, Amarouch MY, Leinonen J, Marjamaa A, Kucera JP, Laitinen-Forsblom PJ, Lahtinen AM, Palotie A, Kontula K, Toivonen L, Abriel H, Widen E. Gain-of-function mutation of the SCN5A gene causes exercise-induced polymorphic ventricular arrhythmias. Circ Cardiovasc Genet 2014; 7:771–81 [DOI] [PubMed] [Google Scholar]
- 34.Calloe K, Broendberg AK, Christensen AH, Pedersen LN, Olesen MS, de Los Angeles Tejada M, Friis S, Thomsen MB, Bundgaard H, Jensen HK. Multifocal atrial and ventricular premature contractions with an increased risk of dilated cardiomyopathy caused by a Nav1.5 gain-of-function mutation (G213D). Int J Cardiol 2018; 257:160–7 [DOI] [PubMed] [Google Scholar]
- 35.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm 2005; 2:507–17 [DOI] [PubMed] [Google Scholar]
- 36.Albert CM, Nam EG, Rimm EB, Jin HW, Hajjar RJ, Hunter DJ, MacRae CA, Ellinor PT. Cardiac sodium channel gene variants and sudden cardiac death in women. Circulation 2008; 117:16–23 [DOI] [PubMed] [Google Scholar]
- 37.Yang P, Kanki H, Drolet B, Yang T, Wei J, Viswanathan PC, Hohnloser SH, Shimizu W, Schwartz PJ, Stanton M, Murray KT, Norris K, George AL, Jr, Roden DM. Allelic variants in long-QT disease genes in patients with drug-associated torsades de pointes. Circulation 2002; 105:1943–8 [DOI] [PubMed] [Google Scholar]
- 38.Xiao YF, Ke Q, Wang SY, Yang Y, Wang GK, Morgan JP, Leaf A. Point mutations in alpha-subunit of human cardiac Na+ channels alter Na+ current kinetics. Biochem Biophys Res Commun 2001; 281:45–52 [DOI] [PubMed] [Google Scholar]
- 39.Marangoni S, Di Resta C, Rocchetti M, Barile L, Rizzetto R, Summa A, Severi S, Sommariva E, Pappone C, Ferrari M, Benedetti S, Zaza A. A Brugada syndrome mutation (p.S216L) and its modulation by p.H558R polymorphism: standard and dynamic characterization. Cardiovasc Res 2011; 91:606–16 [DOI] [PubMed] [Google Scholar]
- 40.Yu R, Fan XF, Chen C, Liu ZH. Whole-exome sequencing identifies a novel mutation (R367G) in SCN5A to be associated with familial cardiac conduction disease. Mol Med Rep 2017; 16:410–4 [DOI] [PubMed] [Google Scholar]
- 41.Koval OM, Snyder JS, Wolf RM, Pavlovicz RE, Glynn P, Curran J, Leymaster ND, Dun W, Wright PJ, Cardona N, Qian L, Mitchell CC, Boyden PA, Binkley PF, Li C, Anderson ME, Mohler PJ, Hund TJ. Ca2+/calmodulin-dependent protein kinase II-based regulation of voltage-gated Na+ channel in cardiac disease. Circulation 2012; 126:2084–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortiz-Bonnin B, Rinne S, Moss R, Streit AK, Scharf M, Richter K, Stober A, Pfeufer A, Seemann G, Kaab S, Beckmann BM, Decher N. Electrophysiological characterization of a large set of novel variants in the SCN5A-gene: identification of novel LQTS3 and BrS mutations. Pflugers Arch 2016; 468:1375–87 [DOI] [PubMed] [Google Scholar]
- 43.Tester DJ, Valdivia C, Harris-Kerr C, Alders M, Salisbury BA, Wilde AA, Makielski JC, Ackerman MJ. Epidemiologic, molecular, and functional evidence suggest A572D-SCN5A should not be considered an independent LQT3-susceptibility mutation. Heart Rhythm 2010; 7:912–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iacoviello L, Bonaccio M, Di Castelnuovo A, Costanzo S, Rago L, De Curtis A, Assanelli D, Badilini F, Vaglio M, Persichillo M, Macfarlane PW, Cerletti C, Donati MB, de Gaetano G. Frontal plane T-wave axis orientation predicts coronary events: findings from the Moli-sani study. Ann Noninvasive Electrocardiol 2017; 264:51–7 [DOI] [PubMed] [Google Scholar]
- 45.Stocco FG, Evaristo E, Shah NR, Cheezum MK, Hainer J, Foster C, Nearing BD, Gervino E, Verrier RL. Marked exercise-induced T-wave heterogeneity in symptomatic diabetic patients with nonflow-limiting coronary artery stenosis. Ann Noninvasive Electrocardiol 2018; 23:e12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zygmunt AC, Eddlestone GT, Thomas GP, Nesterenko VV, Antzelevitch C. Larger late sodium conductance in M cells contributes to electrical heterogeneity in canine ventricle. Am J Physiol Heart Circ Physiol 2001; 281:H689–97 [DOI] [PubMed] [Google Scholar]
- 47.Schlotthauer K, Bers DM. Sarcoplasmic reticulum Ca(2+) release causes myocyte depolarization. Underlying mechanism and threshold for triggered action potentials. Circ Res 2000; 87:774–80 [DOI] [PubMed] [Google Scholar]
- 48.Katra RP, Laurita KR. Cellular mechanism of calcium-mediated triggered activity in the heart. Circ Res 2005; 96:535–42 [DOI] [PubMed] [Google Scholar]
- 49.Marban E, Robinson SW, Wier WG. Mechanisms of arrhythmogenic delayed and early afterdepolarizations in ferret ventricular muscle. J Clin Invest 1986; 78:1185–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss JN, Garfinkel A, Karagueuzian HS, Chen PS, Qu Z. Early afterdepolarizations and cardiac arrhythmias. Heart Rhythm 2010; 7:1891–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aiba T, Hesketh GG, Liu T, Carlisle R, Villa-Abrille MC, O'Rourke B, Akar FG, Tomaselli GF. Na+ channel regulation by Ca2+/calmodulin and Ca2+/calmodulin-dependent protein kinase II in guinea-pig ventricular myocytes. Cardiovasc Res 2010; 85:454–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah M, Akar FG, Tomaselli GF. Molecular basis of arrhythmias. Circulation 2005; 112:2517–29 [DOI] [PubMed] [Google Scholar]
- 53.Song Y, Shryock JC, Wu L, Belardinelli L. Antagonism by ranolazine of the pro-arrhythmic effects of increasing late INa in guinea pig ventricular myocytes. J Cardiovasc Pharmacol 2004; 44:192–9 [DOI] [PubMed] [Google Scholar]
- 54.January CT, Riddle JM. Early afterdepolarizations: mechanism of induction and block. A role for L-type Ca2+ current. Circ Res 1989; 64:977–90 [DOI] [PubMed] [Google Scholar]
- 55.Fujiwara K, Tanaka H, Mani H, Nakagami T, Takamatsu T. Burst emergence of intracellular Ca2+ waves evokes arrhythmogenic oscillatory depolarization via the Na+-Ca2+ exchanger: simultaneous confocal recording of membrane potential and intracellular Ca2+ in the heart. Circ Res 2008; 103:509–18 [DOI] [PubMed] [Google Scholar]
- 56.Armoundas AA, Hobai IA, Tomaselli GF, Winslow RL, O'Rourke B. Role of sodium-calcium exchanger in modulating the action potential of ventricular myocytes from normal and failing hearts. Circ Res 2003; 93:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie LH, Chen F, Karagueuzian HS, Weiss JN. Oxidative-stress-induced afterdepolarizations and calmodulin kinase II signaling. Circ Res 2009; 104:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoeker GS, Katra RP, Wilson LD, Plummer BN, Laurita KR. Spontaneous calcium release in tissue from the failing canine heart. Am J Physiol Heart Circ Physiol 2009; 297:H1235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joung B, Zhang H, Shinohara T, Maruyama M, Han S, Kim D, Choi EK, On YK, Lin SF, Chen PS. Delayed afterdepolarization in intact canine sinoatrial node as a novel mechanism for atrial arrhythmia. J Cardiovasc Electrophysiol 2011; 22:448–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyser RC, Miranda AM, Chen CM. Calcium handling precedes cardiac differentiation to initiate the first heartbeat. Elife 2016; 5:e17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mary-Rabine L, Hordof AJ, Danilo P, Jr., Malm JR, Rosen MR. Mechanisms for impulse initiation in isolated human atrial fibers. Circ Res 1980; 47:267–77 [DOI] [PubMed] [Google Scholar]
- 62.Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, Jean JC, Omari A, Sampson KJ, Kotton DN, Keller G, Kass RS. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol 2013; 141:61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen C, Xu L, Yang Z, Zou Y, Hu K, Fan Z, Ge J, Sun A. A1180V of cardiac sodium channel gene (SCN5A): is it a risk factor for dilated cardiomyopathy or just a common variant in Han Chinese? Dis Markers 2013; 35:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu RM, Tan BH, Orland KM, Valdivia CR, Peterson A, Pu J, Makielski JC. Digenic inheritance novel mutations in SCN5a and SNTA1 increase late I(Na) contributing to LQT syndrome. Am J Physiol Heart Circ Physiol 2013; 304:H994–h1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato K, Makiyama T, Wu J, Ding WG, Kimura H, Naiki N, Ohno S, Itoh H, Nakanishi T, Matsuura H, Horie M. Cardiac channelopathies associated with infantile fatal ventricular arrhythmias: from the cradle to the bench. J Cardiovasc Electrophysiol 2014; 25:66–73 [DOI] [PubMed] [Google Scholar]
- 66.Beckermann TM, McLeod K, Murday V, Potet F, George AL., Jr. Novel SCN5A mutation in amiodarone-responsive multifocal ventricular ectopy-associated cardiomyopathy. Heart Rhythm 2014; 11:1446–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu RM, Tan BH, Tester DJ, Song C, He Y, Dovat S, Peterson BZ, Ackerman MJ, Makielski JC. Arrhythmogenic biophysical phenotype for scn5a mutation s1787n depends upon splice variant background and intracellular acidosis. PLoS One 2015; 10:e0124921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Makiyama T, Wuriyanghai Y, Ohno S, Sasaki K, Hayano M, Harita T, Nishiuchi S, Yuta Y, Ueyama T, Shimizu A, Horie M, Kimura T. Cardiac sodium channel mutation associated with epinephrine-induced QT prolongation and sinus node dysfunction. Heart Rhythm 2016; 13:289–98 [DOI] [PubMed] [Google Scholar]
- 69.Turker I, Makiyama T, Vatta M, Itoh H, Ueyama T, Shimizu A, Ai T, Horie M. A novel SCN5A mutation associated with drug induced Brugada type ECG. PLoS One 2016; 11:e0161872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lieve KV, Verkerk AO, Podliesna S, van der Werf C, Tanck MW, Hofman N, van Bergen PF, Beekman L, Bezzina CR, Wilde AAM, Lodder EM. Gain-of-function mutation in SCN5A causes ventricular arrhythmias and early onset atrial fibrillation. Int J Cardiol 2017; 236:187–93 [DOI] [PubMed] [Google Scholar]
- 71.Kehl HG, Haverkamp W, Rellensmann G, Yelbuz TM, Krasemann T, Vogt J, Schulze-Bahr E. Images in cardiovascular medicine. Life-threatening neonatal arrhythmia: successful treatment and confirmation of clinically suspected extreme long QT-syndrome-3. Circulation 2004; 109:e205–6 [DOI] [PubMed] [Google Scholar]
- 72.Bankston JR, Yue M, Chung W, Spyres M, Pass RH, Silver E, Sampson KJ, Kass RS. A novel and lethal de novo LQT-3 mutation in a newborn with distinct molecular pharmacology and therapeutic response. PLoS One 2007; 2:e1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian XL, Yong SL, Wan X, Wu L, Chung MK, Tchou PJ, Rosenbaum DS, Van Wagoner DR, Kirsch GE, Wang Q. Mechanisms by which SCN5A mutation N1325S causes cardiac arrhythmias and sudden death in vivo. Cardiovasc Res 2004; 61:256–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao L, Fan P, Jiang Z, Viatchenko-Karpinski S, Wu Y, Kornyeyev D, Hirakawa R, Budas GR, Rajamani S, Shryock JC, Belardinelli L. Nav1.5-dependent persistent Na+ influx activates CaMKII in rat ventricular myocytes and N1325S mice. Am J Physiol Cell Physiol 2011; 301:C577–86 [DOI] [PubMed] [Google Scholar]
- 75.Schulze-Bahr E, Eckardt L, Breithardt G, Seidl K, Wichter T, Wolpert C, Borggrefe M, Haverkamp W. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat 2003; 21:651–2 [DOI] [PubMed] [Google Scholar]
- 76.Mizusawa Y, Wilde AA. Brugada syndrome. Circ Arrhythm Electrophysiol 2012; 5:606–16 [DOI] [PubMed] [Google Scholar]
- 77.Sommariva E, Pappone C, Martinelli Boneschi F, Di Resta C, Rosaria Carbone M, Salvi E, Vergara P, Sala S, Cusi D, Ferrari M, Benedetti S. Genetics can contribute to the prognosis of Brugada syndrome: a pilot model for risk stratification. Eur J Hum Genet 2013; 21:911–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Veltmann C, Barajas-Martinez H, Wolpert C, Borggrefe M, Schimpf R, Pfeiffer R, Caceres G, Burashnikov E, Antzelevitch C, Hu D. Further insights in the most common SCN5A mutation causing overlapping phenotype of long QT syndrome, Brugada syndrome, and conduction defect. J Am Heart Assoc 2016; 5:pii: e003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viswanathan PC, Balser JR. Inherited sodium channelopathies: a continuum of channel dysfunction. Trends Cardiovasc Med 2004; 14:28–35 [DOI] [PubMed] [Google Scholar]
- 80.Tarradas A, Selga E, Beltran-Alvarez P, Perez-Serra A, Riuro H, Pico F, Iglesias A, Campuzano O, Castro-Urda V, Fernandez-Lozano I, Perez GJ, Scornik FS, Brugada R. A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes Brugada syndrome. PLoS One 2013; 8:e53220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakajima S, Makiyama T, Hanazawa K, Kaitani K, Amano M, Hayama Y, Onishi N, Tamaki Y, Miyake M, Tamura T, Kondo H, Motooka M, Izumi C, Nakagawa Y, Horie M. A novel SCN5A mutation demonstrating a variety of clinical phenotypes in familial sick sinus syndrome. Intern Med 2013; 52:1805–8 [DOI] [PubMed] [Google Scholar]
- 82.Calloe K, Refaat MM, Grubb S, Wojciak J, Campagna J, Thomsen NM, Nussbaum RL, Scheinman MM, Schmitt N. Characterization and mechanisms of action of novel NaV1.5 channel mutations associated with Brugada syndrome. Circ Arrhythm Electrophysiol 2013; 6:177–84 [DOI] [PubMed] [Google Scholar]
- 83.Detta N, Frisso G, Zullo A, Sarubbi B, Cozzolino C, Romeo E, Wang DW, Calabro R, Salvatore F, George AL., Jr. Novel deletion mutation in the cardiac sodium channel inactivation gate causes long QT syndrome. Int J Cardiol 2013; 165:362–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watanabe H, Ohkubo K, Watanabe I, Matsuyama TA, Ishibashi-Ueda H, Yagihara N, Shimizu W, Horie M, Minamino T, Makita N. SCN5A mutation associated with ventricular fibrillation, early repolarization, and concealed myocardial abnormalities. Int J Cardiol 2013; 165:e21–3 [DOI] [PubMed] [Google Scholar]
- 85.Zumhagen S, Veldkamp MW, Stallmeyer B, Baartscheer A, Eckardt L, Paul M, Remme CA, Bhuiyan ZA, Bezzina CR, Schulze-Bahr E. A heterozygous deletion mutation in the cardiac sodium channel gene SCN5A with loss- and gain-of-function characteristics manifests as isolated conduction disease, without signs of Brugada or long QT syndrome. PLoS One 2013; 8:e67963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maury P, Moreau A, Hidden-Lucet F, Leenhardt A, Fressart V, Berthet M, Denjoy I, Bennamar N, Rollin A, Cardin C, Guicheney P, Chahine M. Novel SCN5A mutations in two families with "Brugada-like" ST elevation in the inferior leads and conduction disturbances. J Interv Card Electrophysiol 2013; 37:131–40 [DOI] [PubMed] [Google Scholar]
- 87.Parisi P, Oliva A, Coll Vidal M, Partemi S, Campuzano O, Iglesias A, Pisani D, Pascali VL, Paolino MC, Villa MP, Zara F, Tassinari CA, Striano P, Brugada R. Coexistence of epilepsy and Brugada syndrome in a family with SCN5A mutation. Epilepsy Res 2013; 105:415–8 [DOI] [PubMed] [Google Scholar]
- 88.Li N, Wang R, Hou C, Zhang Y, Teng S, Pu J. A heterozygous missense SCN5A mutation associated with early repolarization syndrome. Int J Mol Med 2013; 32:661–7 [DOI] [PubMed] [Google Scholar]
- 89.Kodama T, Serio A, Disertori M, Bronzetti G, Diegoli M, Narula N, Grasso M, Mazzola S, Arbustini E. Autosomal recessive paediatric sick sinus syndrome associated with novel compound mutations in SCN5A. Int J Cardiol 2013; 167:3078–80 [DOI] [PubMed] [Google Scholar]
- 90.Dolz-Gaiton P, Nunez M, Nunez L, Barana A, Amoros I, Matamoros M, Perez-Hernandez M, Gonzalez de la Fuente M, Alvarez-Lopez M, Macias-Ruiz R, Tercedor-Sanchez L, Jimenez-Jaimez J, Delpon E, Caballero R, Tamargo J. Functional characterization of a novel frameshift mutation in the C-terminus of the Nav1.5 channel underlying a Brugada syndrome with variable expression in a Spanish family. PLoS One 2013; 8:e81493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Detta N, Frisso G, Limongelli G, Marzullo M, Calabro R, Salvatore F. Genetic analysis in a family affected by sick sinus syndrome may reduce the sudden death risk in a young aspiring competitive athlete. Int J Cardiol 2014; 170:e63–5 [DOI] [PubMed] [Google Scholar]
- 92.Kanters JK, Yuan L, Hedley PL, Stoevring B, Jons C, Bloch Thomsen PE, Grunnet M, Christiansen M, Jespersen T. Flecainide provocation reveals concealed Brugada syndrome in a long QT syndrome family with a novel L1786Q mutation in SCN5A. Circ J 2014; 78:1136–43 [DOI] [PubMed] [Google Scholar]
- 93.Ziyadeh-Isleem A, Clatot J, Duchatelet S, Gandjbakhch E, Denjoy I, Hidden-Lucet F, Hatem S, Deschenes I, Coulombe A, Neyroud N, Guicheney P. A truncating SCN5A mutation combined with genetic variability causes sick sinus syndrome and early atrial fibrillation. Heart Rhythm 2014; 11:1015–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aiba T, Farinelli F, Kostecki G, Hesketh GG, Edwards D, Biswas S, Tung L, Tomaselli GF. A mutation causing Brugada syndrome identifies a mechanism for altered autonomic and oxidant regulation of cardiac sodium currents. Circ Cardiovasc Genet 2014; 7:249–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shy D, Gillet L, Ogrodnik J, Albesa M, Verkerk AO, Wolswinkel R, Rougier JS, Barc J, Essers MC, Syam N, Marsman RF, van Mil AM, Rotman S, Redon R, Bezzina CR, Remme CA, Abriel H. PDZ domain-binding motif regulates cardiomyocyte compartment-specific NaV1.5 channel expression and function. Circulation 2014; 130:147–60 [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Meng X, Yuchi Z, Zhao Z, Xu D, Fedida D, Wang Z, Huang C. De novo mutation in the SCN5A gene associated with Brugada syndrome. Cell Physiol Biochem 2015; 36:2250–62 [DOI] [PubMed] [Google Scholar]
- 97.Zhu JF, Du LL, Tian Y, Du YM, Zhang L, Zhou T, Tian LI. Novel heterozygous mutation c.4282G>T in the SCN5A gene in a family with Brugada syndrome. Exp Ther Med 2015; 9:1639–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakajima T, Kaneko Y, Saito A, Ota M, Iijima T, Kurabayashi M. Enhanced fast-inactivated state stability of cardiac sodium channels by a novel voltage sensor SCN5A mutation, R1632C, as a cause of atypical Brugada syndrome. Heart Rhythm 2015; 12:2296–304 [DOI] [PubMed] [Google Scholar]
- 99.Wang HG, Zhu W, Kanter RJ, Silva JR, Honeywell C, Gow RM, Pitt GS. A novel NaV1.5 voltage sensor mutation associated with severe atrial and ventricular arrhythmias. J Mol Cell Cardiol 2016; 92:52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo Q, Ren L, Chen X, Hou C, Chu J, Pu J, Zhang S. A novel mutation in the SCN5A gene contributes to arrhythmogenic characteristics of early repolarization syndrome. Int J Mol Med 2016; 37:727–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng Z, Xie Q, Huang Y, Zhao Y, Li W, Huang Z. p.D1690N sodium voltage-gated channel alpha subunit 5 mutation reduced sodium current density and is associated with Brugada syndrome. Mol Med Rep 2016; 13:5216–22 [DOI] [PubMed] [Google Scholar]
- 102.Tan BY, Yong RY, Barajas-Martinez H, Dumaine R, Chew YX, Wasan PS, Ching CK, Ho KL, Gan LS, Morin N, Chong AP, Yap SH, Neo JL, Yap EP, Moochhala S, Chong DT, Chow W, Seow SC, Hu D, Uttamchandani M, Teo WS. A Brugada syndrome proband with compound heterozygote SCN5A mutations identified from a Chinese family in Singapore. Europace 2016; 18:897–904 [DOI] [PubMed] [Google Scholar]
- 103.Yang Z, Lu D, Zhang L, Hu J, Nie Z, Xie C, Qiu F, Cheng H, Yan Y. p.N1380del mutation in the pore-forming region of SCN5A gene is associated with cardiac conduction disturbance and ventricular tachycardia. Acta Biochim Biophys Sin 2017; 49:270–6 [DOI] [PubMed] [Google Scholar]
- 104.Gando I, Morganstein J, Jana K, McDonald TV, Tang Y, Coetzee WA. Infant sudden death: mutations responsible for impaired Nav1.5 channel trafficking and function. Pacing Clin Electrophysiol 2017; 40:703–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayano M, Makiyama T, Kamakura T, Watanabe H, Sasaki K, Funakoshi S, Wuriyanghai Y, Nishiuchi S, Harita T, Yamamoto Y, Kohjitani H, Hirose S, Yokoi F, Chen J, Baba O, Horie T, Chonabayashi K, Ohno S, Toyoda F, Yoshida Y, Ono K, Horie M, Kimura T. Development of a patient-derived induced pluripotent stem cell model for the investigation of SCN5A-D1275N-related cardiac sodium channelopathy. Circ J 2017; 81:1783–91 [DOI] [PubMed] [Google Scholar]
- 106.Selga E, Sendfeld F, Martinez-Moreno R, Medine CN, Tura-Ceide O, Wilmut SI, Perez GJ, Scornik FS, Brugada R, Mills NL. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J Mol Cell Cardiol 2018; 114:10–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang H, Ding DB, Fan LL, Jin JY, Li JJ, Guo S, Chen YQ, Xiang R. Whole-exome sequencing identifies a Novel SCN5A mutation (C335R) in a Chinese family with arrhythmia. Cardiol Young 2018; 28:1–4 [DOI] [PubMed] [Google Scholar]
- 108.Smits JP, Koopmann TT, Wilders R, Veldkamp MW, Opthof T, Bhuiyan ZA, Mannens MM, Balser JR, Tan HL, Bezzina CR, Wilde AA. A mutation in the human cardiac sodium channel (E161K) contributes to sick sinus syndrome, conduction disease and Brugada syndrome in two families. J Mol Cell Cardiol 2005; 38:969–81 [DOI] [PubMed] [Google Scholar]
- 109.Cordeiro JM, Barajas-Martinez H, Hong K, Burashnikov E, Pfeiffer R, Orsino AM, Wu YS, Hu D, Brugada J, Brugada P, Antzelevitch C, Dumaine R, Brugada R. Compound heterozygous mutations P336L and I1660V in the human cardiac sodium channel associated with the Brugada syndrome. Circulation 2006; 114:2026–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan GX, Antzelevitch C. Cellular basis for the Brugada syndrome and other mechanisms of arrhythmogenesis associated with ST-segment elevation. Circulation 1999; 100:1660–6 [DOI] [PubMed] [Google Scholar]
- 111.Meregalli PG, Wilde AA, Tan HL. Pathophysiological mechanisms of Brugada syndrome: depolarization disorder, repolarization disorder, or more? Cardiovasc Res 2005; 67:367–78 [DOI] [PubMed] [Google Scholar]
- 112.Sieira J, Dendramis G, Brugada P. Pathogenesis and management of Brugada syndrome. Nat Rev Cardiol 2016; 13:744–56 [DOI] [PubMed] [Google Scholar]
- 113.Tan HL. Sodium channel variants in heart disease: expanding horizons. J Cardiovasc Electrophysiol 2006; 17:S151–s7 [DOI] [PubMed] [Google Scholar]
- 114.Trenor B, Cardona K, Saiz J, Noble D, Giles W. Cardiac action potential repolarization re-visited: early repolarization shows all-or-none behaviour. J Physiol 2017; 595:6599–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zimmer T, Surber R. SCN5A channelopathies–an update on mutations and mechanisms. Prog Biophys Mol Biol 2008; 98:120–36 [DOI] [PubMed] [Google Scholar]
- 116.Zeng Z, Zhou J, Hou Y, Liang X, Zhang Z, Xu X, Xie Q, Li W, Huang Z. Electrophysiological characteristics of a SCN5A voltage sensors mutation R1629Q associated with Brugada syndrome. PLoS One 2013; 8:e78382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang DW, Desai RR, Crotti L, Arnestad M, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Rognum T, Schwartz PJ, George AL., Jr. Cardiac sodium channel dysfunction in sudden infant death syndrome. Circulation 2007; 115:368–76 [DOI] [PubMed] [Google Scholar]
- 118.Surber R, Hensellek S, Prochnau D, Werner GS, Benndorf K, Figulla HR, Zimmer T. Combination of cardiac conduction disease and long QT syndrome caused by mutation T1620K in the cardiac sodium channel. Cardiovasc Res 2008; 77:740–8 [DOI] [PubMed] [Google Scholar]
- 119.Walzik S, Schroeter A, Benndorf K, Zimmer T. Alternative splicing of the cardiac sodium channel creates multiple variants of mutant T1620K channels. PLoS One 2011; 6:e19188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bezzina CR, Rook MB, Groenewegen WA, Herfst LJ, van der Wal AC, Lam J, Jongsma HJ, Wilde AA, Mannens MM. Compound heterozygosity for mutations (W156X and R225W) in SCN5A associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ Res 2003; 92:159–68 [DOI] [PubMed] [Google Scholar]
- 121.Niu DM, Hwang B, Hwang HW, Wang NH, Wu JY, Lee PC, Chien JC, Shieh RC, Chen YT. A common SCN5A polymorphism attenuates a severe cardiac phenotype caused by a nonsense SCN5A mutation in a Chinese family with an inherited cardiac conduction defect. J Med Genet 2006; 43:817–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan HL, Bink-Boelkens MT, Bezzina CR, Viswanathan PC, Beaufort-Krol GC, van Tintelen PJ, van den Berg MP, Wilde AA, Balser JR. A sodium-channel mutation causes isolated cardiac conduction disease. Nature 2001; 409:1043–7 [DOI] [PubMed] [Google Scholar]
- 123.Zhang ZS, Tranquillo J, Neplioueva V, Bursac N, Grant AO. Sodium channel kinetic changes that produce Brugada syndrome or progressive cardiac conduction system disease. Am J Physiol Heart Circ Physiol 2007; 292:H399–407 [DOI] [PubMed] [Google Scholar]
- 124.Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Brignole M, Giordano U, Giovannini T, Menozzi C, Bloise R, Crotti L, Terreni L, Schwartz PJ. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: a prospective evaluation of 52 families. Circulation 2000; 102:2509–15 [DOI] [PubMed] [Google Scholar]
- 125.Aoki H, Nakamura Y, Ohno S, Makiyama T, Horie M. Cardiac conduction defects and Brugada syndrome: a family with overlap syndrome carrying a nonsense SCN5A mutation. J Arrhythm 2017; 33:35–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin L, Takahashi-Igari M, Kato Y, Nozaki Y, Obata M, Hamada H, Horigome H. Prenatal diagnosis of atrioventricular block and QT interval prolongation by fetal magnetocardiography in a fetus with trisomy 18 and SCN5A R1193Q variant. Case Rep Pediatr 2017; 2017:6570465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sun A, Xu L, Wang S, Wang K, Huang W, Wang Y, Zou Y, Ge J. SCN5A R1193Q polymorphism associated with progressive cardiac conduction defects and long QT syndrome in a Chinese family. Case Rep Pediatr 2008; 45:127–8 [DOI] [PubMed] [Google Scholar]
- 128.Sumitomo N. E1784K mutation in SCN5A and overlap syndrome. Circ J 2014; 78:1839–40 [DOI] [PubMed] [Google Scholar]
- 129.Wei J, Wang DW, Alings M, Fish F, Wathen M, Roden DM, George AL., Jr. Congenital long-QT syndrome caused by a novel mutation in a conserved acidic domain of the cardiac Na+ channel. Circulation 1999; 99:3165–71 [DOI] [PubMed] [Google Scholar]