Short abstract
Oral mucositis is still one of the most painful side effects of chemotherapeutic treatment and a mounting body of evidence suggests a key role for the oral microbiome in mucositis development. However, the underlying mechanisms remain elusive. In this work, we have investigated the interactions between the host, the microbiome, and chemotherapeutic treatments in more detail. The effect of 5-fluorouracil, commonly inducing mucositis, was assessed on a co-culture model that consists of an epithelial cell layer and a biofilm derived from oral microbiota from different types of samples (saliva, buccal swabs and tongue swabs) and donors (healthy individuals and patients suffering from mucositis). After 24 h co-incubation, all oral microbial samples were found to reduce wound healing capacity with 26 ± 15% as compared with untreated condition. Compared with saliva and tongue samples, buccal samples were characterized by lower bacterial cell counts and hence higher wound healing capacity. For samples from healthy individuals, an inverse correlation was observed between bacterial cell counts and wound healing capacity, whereas for patients suffering from mucositis no correlation was observed. Moreover, patient-derived samples had a less diverse microbial community and higher abundances of pathogenic genera. No major impact of 5-fluorouracil on wound healing capacity or the composition of the microbiome was seen at physiologically relevant concentrations in the mouth. In conclusion, bacterial cell count is inversely correlated with wound healing capacity, which emphasizes the importance of oral hygiene during oral wound healing in healthy individuals. However, future research on extra measures besides oral hygiene is needed to assure a good wound healing during mucositis, as for patients the bacterial composition seems also crucial. The direct effect of 5-fluorouracil on both the microbiome and wound healing is minimal, pointing to the importance of the host and its immune system in chemotherapy-induced microbial shifts.
Impact statement
Chemotherapy-induced oral mucositis has a major impact on the quality of life of patients. The additional costs and treatment time associated with this pathology are significant. Although the pathology of the disease is well understood, the role and importance of oral microbiota currently are less clear. In this study, we focused on the effect of oral microbiota on wound healing, the final phase of oral mucositis, during 5-FU exposure. We show that the bacterial load and composition have a major impact on the healing process in contrast to 5-FU which only marginally slows down healing. This emphasizes the importance of good oral health care during oral mucositis to minimize bacterial load around the oral lesions. However, since we show that also the composition of the oral microbiome plays a role in wound recovery, the identification of specific pathogenic species or their metabolites might be worthwhile to allow proper treatment.
Keywords: Bacteria, microbiome, chemotherapy, host–microbe interactions, in vitro model, oral mucositis
Introduction
Oral mucositis is a painful and debilitating complication of cancer treatment with a major impact on the quality of life of the patient. Its frequency is high but varies depending on the type of treatment with around 20–40% incidence in conventional chemotherapeutic treatment of solid tumors, to almost 100% for high-dose chemotherapy prior to hematopoietic stem cell transplantation or radiotherapy for head and neck cancer.1–3 Although it is one of the most studied toxicities of cancer treatment, only few therapeutic agents are available for oral mucositis.4 One of the chemotherapeutic agents with high risk of developing mucositis is 5-fluorouracil (5-FU),2 an antimetabolite that inhibits thymidylate synthase (TS) and is incorporated in DNA and RNA.5,6 The incidence of developing grade 3–4 oral mucositis (i.e. confluent ulcers and unable to eat solids) in case of 5-FU treatment is more than 15%.7 During continuous infusion (22 h), plasma levels of 5-FU range from 3 to 10 µM and saliva levels from 0.08 to 0.8 µM.8,9 Previous research has indicated that some oral species are sensitive to 5-FU starting from 0.4 µM.10
The pathogenesis of mucositis is described by the 5-stage model of Sonis. Briefly, reactive oxygen species (ROS) are generated in the initiation phase, followed by the activation of transcription factors, such as nuclear factor-kappa B (NF-κB). These induce the production of pro-inflammatory cytokines and activate other signaling pathways. Feedback-loops induce more inflammation and apoptosis which lead to the ulceration phase, in which bacteria colonize the ulcers and can penetrate to the submucosa. In most cases, spontaneous healing takes place within two to three weeks after completion of the treatment. Although this last phase is of great importance in terms of recovery and further continuation of the cancer treatment, it is also the least understood.1
More and more evidence is emerging on the role of the oral microbiome in the pathogenesis of oral mucositis.11–13 Microbiota can play a negative role in mucositis and induce infection of the ulcers which encourages the use of antimicrobial agents. However, no clinical guidelines have been formulated regarding the use of antimicrobial agents due to insufficient and conflicting scientific data.12,14 Microbiota may also be involved in phases other than the ulceration phase, and this role can be both positive and negative.15 Microbiota are for example able to influence the activation of toll like receptors (TLRs), NF-κB, and mitogen-activated protein kinase (MAPK), which are all proteins involved in important signaling pathways regulating mucositis. This way, microbiota might contribute to a higher tissue inflammation level and therefore increase apoptosis rate.13
Clinical studies have shown shifts in the oral microbial profile of patients, both after chemo and radiotherapy. However, the great variability in patient population, sample collection and technical methods to analyze the microbiota makes it difficult to generalize conclusions.12 It seems that for blood cultures and oral swabs taken during chemotherapy, the most frequently isolated Gram-negative species are Enterobacteriaceae spp., Pseudomonas spp., and E. coli, whereas Staphylococcus spp. and Streptococcus spp. are the most frequently isolated Gram-positive species.12,16 Not only microbial composition, but also functional factors such as the mucus layer and microbial adhesion can be affected by the cancer treatment.12,17 Moreover, oral microbiota may regulate wound recovery, with positive or negative effects depending on the species and the bacterial density.18–20 These factors will depend on both the donor and on the specific site in the oral cavity, as they each have their own microbial community.21 For example, the saliva microbiome resembles the tongue microbiome but is distinct from the buccal mucosal microbiome.21
In this study, we further investigated the role of oral microbiota on wound healing capacity and the effect of chemotherapy on both the microbiota and wound healing in an in vitro co-culture model that was previously optimized.22 First, the toxicity of 5-FU towards oral epithelial cells was determined using the MTT/SRB cytotoxicity tests. Next, the impact of oral microbiota and 5-FU, and the combination thereof, on epithelial wound healing were studied in the co-culture model for 24 h, with a special focus on the potential impact of the type of oral sample and donor variability.
Material and methods
Cell culture
The TR146 cell line, obtained from the Laboratory of Experimental Cancer Research (Ghent University Hospital), is an oral squamous cell carcinoma cell line isolated from a local lymph node metastasis23 and is often used as a model for human buccal epithelium24,25 and in wound healing assays.22 Cells were cultured at 37°C, 10% CO2, and 90% relative humidity in Dulbecco’s modified Eagle’s Medium (DMEM) (Gibco, Merelbeke, Belgium) with 10% heat inactivated fetal bovine serum (Greiner Bio-one, Vilvoorde, Belgium), 100 IU/ml penicillin (Gibco, Merelbeke, Belgium), 100 µg/ml streptomycin (Gibco, Merelbeke, Belgium), and 2.5 µg/ml amphotericin B (Gibco, Merelbeke, Belgium).
Oral samples
Oral samples were obtained from healthy children or patients suffering from oral mucositis (Ethical approval from Ghent University hospital, Belgian Registration number B670201112526), all aged 6–14 years. All patients were treated for hematological malignancies. Three types of samples were collected: saliva, buccal swab, and tongue swab. All samples were collected at least 2 h after eating or brushing teeth and before sampling, the oral cavity of the individuals was flushed with drinking water. For the buccal and tongue samples, a sterile cotton swab was gently wiped 10 times along the inner cheek or on the dorsal side of the tongue and subsequently dissolved in 1 ml of phosphate-buffered solution (PBS).
Chemicals
A filter-sterilized stock solution of 100 mM 5-FU (Sigma Aldrich, Overijse, Belgium) was prepared in dimethylsulfoxide (DMSO) and further diluted to 75, 50, 20, 10, 5, 1, 0.1, 0.01 mM in DMSO. Stock solutions were further diluted (1:1000) in culture medium for the experiments.
MTT/SRB test
To test the cytotoxicity of 5-FU, an MTT/SRB test was performed. The MTT assay26 was used to measure the mitochondrial activity and the SRB assay27 to measure cellular protein content. TR146 cells were seeded in 96-well plates at a density of 40,000 cells/well (100 µl DMEM with serum/well). After 24 h, medium was discarded and 100 µl serum-free, antibiotic-free DMEM was added together with different 5-FU concentrations (0.01–100 µM). DMSO (1:1000) was used as a control. All plates were incubated at 37°C and 5% CO2. After 24 h, 48 h, and five days, an MTT and SRB test was performed. Six biological replicates were included for each 5-FU concentration and for each time point. For the MTT-assay, 20 µl MTT (3–(4,5-demethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (5 mg/ml in PBSD−) was added and incubated for 2 h at 37°C. All mediums were removed and formazan crystals were resuspended in 100 µl DMSO. The absorbance was measured at 570 nm. For the SRB (sulforhodamine B) assay, cells were fixated by adding 25 µl 50% trichloroacetic acid (TCA) and incubated for 1 h at 4°C. After removal of the TCA, the plate was rinsed with water and dried. Next, 75 µl SRB solution (0.4% in 1% glacial acetic acid) was added and the plate was incubated for 30 min at 4°C. The plate was then rinsed with 1% glacial acetic acid and dried. The stained cells were resuspended in 200 µl 10 mM Tris buffer and the absorbance was measured at 490 nm.
Co-culture model
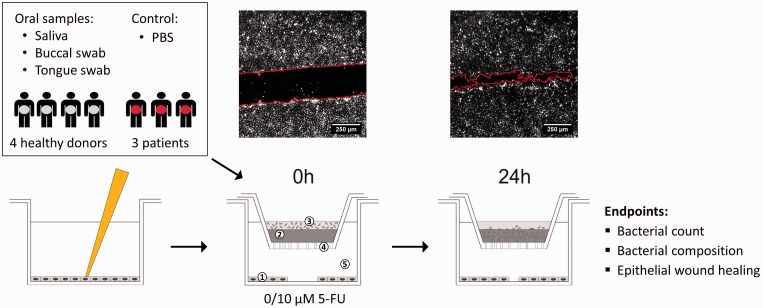
To investigate the interactions of oral microbiota and oral epithelial cells without direct contact, we used an oral in vitro model described by De Ryck et al22 (Figure 1). Briefly, the model consists of a 24-well Transwell® plate with removable inserts with a polycarbonate membrane of pore size 0.4 µm (Corning Incorporated, New York, USA). In the apical part, 20 µl of the bacterial suspension was brought on top of a solidified agar/mucin solution (75 µl, 5% porcin mucin Type II, 0.8% agar). PBS was used as a control. In the basolateral side, epithelial cells were seeded at a density of 250,000 cells/well and at confluency, a wound healing assay was performed (see below). During co-culture, the inserts with the microbiota were transferred to the wells containing the epithelial cells and incubated at 37°C, 10% CO2 in serum-free, antibiotic-free DMEM with 5-FU (10 µM) or DMSO as a control (1:1000). After 24 h of co-culture, inserts were removed and 100 µl PBS was added to collect the bacteria for further analysis. For each of the seven donors (four healthy individuals and three patients suffering from mucositis), a buccal sample, a saliva sample, and a tongue sample as well as a blank (without microbiota) were tested in this co-culture model, each with and without 10 µM 5-FU (Figure 1). Each condition was tested in triplicate or quadruplicate.
Figure 1.
Experimental set-up of the co-culture model with (1) oral epithelial TR146 cells stained with DiI, (2) agar/mucin layer, (3) microbial biofilm or PBS as a control, (4) polycarbonate membrane with 0.4 µm pores, (5) DMEM with 0 µM or 10 µM 5-FU. Fluorescent images show examples of wounds (red line) at 0 h and 24 h. (based on De Ryck et al.22). (A color version of this figure is available in the online journal.)
Wound healing assay
During co-incubation, a wound healing assay was performed based on the protocol by De Ryck et al.22 (Figure 1). TR146 cells were stained with Vybrant DiI cell labeling solution (Life Technologies, Merelbeke, Belgium) before seeding in 24 well Transwell® plates at 250,000 cells/well. At the start of the experiment, two scratches were made in the confluent monolayer using a sterile 100 µl pipette tip. Cell medium was discarded to remove cellular debris and 1 ml of new serum-free, antibiotic-free DMEM was added to the cells. At four selected fields per well and at each time point, images of the wound were acquired using a fully automated widefield fluorescent microscope (Nikon Ti, Nikon Instruments, Amsterdam, The Netherlands), equipped with a 4×/0.15 Plan Achromat objective and EM-CCD camera (Andor Ixon+, Andor Technology, Belfast, UK). The surface area of the wound was calculated for each time point using a home-written script for FIJI freeware (http://fiji.sc) that is available upon reasonable request (www.uantwerpen.be/Cell-group/scripts). In brief, the DiI counterstained time-lapse images are first pre-processed by background subtraction and local contrast enhancement, after which the non-damaged part of the cell monolayer is detected by a combination of variance, maximum and Gaussian blur filtering, and segmented using a user-defined or automatic threshold. The inverse of this mask is selected as wounded area. The relative wound size was calculated by normalizing to the wound area at 0 h.
At the end of the wound healing experiment, metabolic activity and viability of the epithelial cells were evaluated with an MTT-assay. To each well, 1 ml of serum-free, antibiotic-free DMEM, and 200 µl MTT (5 mg/ml in PBSD−) were added and incubated for 2 h at 37°C. After removal of the medium, the formazan crystals were dissolved in 1 ml DMSO. Absorbance was measured at 540 nm (200 µl) (Infinite F50 Tecan, Tecan, Mechelen, Belgium). Percentage of viability, compared to the control, was calculated.
Colony-forming units
To measure the number of viable cells present in the insert, the oral samples (saliva, oral swab, tongue swab) were plated using brain heart infusion (BHI)-agar plates. A dilution series was made and 10 µl of bacterial suspension was plated and incubated aerobically at 37°C in triplicate.
Flow cytometry
The number of intact and damaged bacterial cells in the insert after 24 h was measured by flow cytometry as described by Van Nevel et al.28 The samples were diluted in a filter sterile (0.22 µm) PBS to obtain cell numbers within the detection range (104–106 cells/ml). Next, the samples were stained with SYBR Green I (10,000× diluted from stock, Invitrogen, Merelbeke, Belgium) and propidium iodide (final concentration 4 µM, Invitrogen, Merelbeke, Belgium) and incubated for 13 min at 37°C before measurement. The flow cytometer (BD Accuri C6 flow cytometer, BD, Erembodegem, Belgium) was equipped with a 488 nm solid-state laser and Milli-Q was used as sheath fluid. Signals were detected in fluorescent channels FL1 (green) and FL3 (red), respectively, equipped with a 518–548 nm and 670 nm bandpass filter. Cells were counted by measuring the number of particles in a set volume after gating on green vs. red fluorescence plots in the BD CSampler software. Quality control of absolute cell counting was done with standardized beads. Background was monitored by measuring a filtered sample, equally diluted as the test samples.
Microbial community analysis
All protocols concerning microbial community analysis are further described in Supplementary Information. Briefly, DNA extraction was performed based on Vilchez-Vargas et al.,29 and the quality of the DNA samples was analyzed by gel electrophoresis. On all samples, denaturing gel electrophoresis was performed using the PRBA338F-GC and 518R primers targeting the V3 region.30,31 Illumina sequencing was performed on one replicate of each condition for the saliva samples of all individuals by LGC Genomics (Berlin, Germany) on the MiSeq platform. The Illumina sequencing data were deposited to the European Nucleotide Archive (SRA) via GFBio32 with study number PRJEB20819.
Statistical analysis
All statistical analyses were performed in R (version 3.3.2). Mixed-model regression of MTT and SRB data was performed for each time point with the concentration as categorical predictor. A random intercept effect was incorporated for each replicate measurement. In order to make correct statistical inference, all models were evaluated for normal distributed residuals with homogenous variance, by Shapiro–Wilk tests (P > 0.05) and visually by Q–Q plots. Model parameters were estimated by maximum likelihood. When a significant concentration effect was present (ANOVA, P < 0.01), the categories were compared pair-wise by post hoc analysis using Tukey’s honestly significant difference (HSD) method. All tested concentrations were compared with the control condition (0 µM) and differences were considered significant at P < 0.05.
For all other basic statistics, linear models were built using forward selection of parameters (fixed factors and interactions) on the scaled and centered data. All models were evaluated for normal distributed residuals with homogenous variance. When a significant effect was present, the post hoc analysis was performed using multiple comparisons with Benjamini Hochberg correction. When interactions were present, data were split in subgroups to define significant differences. Differences were considered significant at P < 0.05.
The packages phyloseq33 and vegan34 were used for microbial community analysis. Heatmaps were generated with the pheatmap package and order-based Hill’s numbers35 were calculated. Non-metric distance scaling (NMDS) plots of the bacterial community data were created based on the Bray–Curtis distance measures. Significant differences were identified by means of Permutational ANOVA (PERMANOVA) using the adonis function (vegan). For the comparison of relative abundances between two groups, the Wilcoxon Rank Sum test was used.
Results
5-FU toxicity to oral epithelial TR146 cells
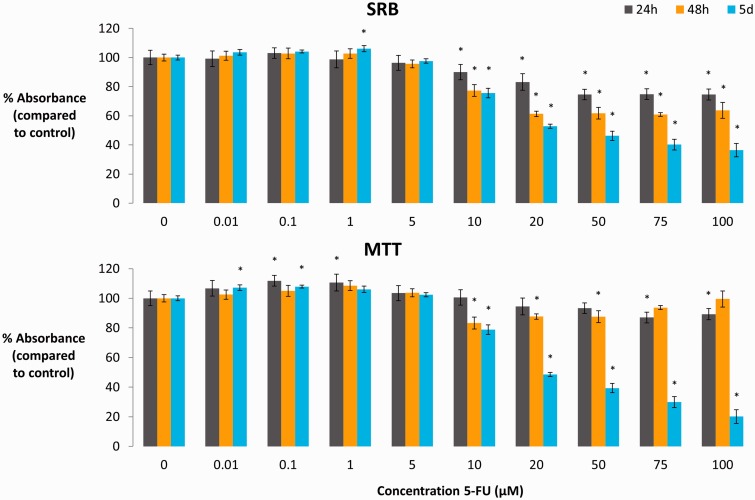
To assess the direct toxicity of 5-FU towards TR146 oral epithelial cells, an MTT/SRB test was performed after 24 h, 48 h, and five days of treatment (Figure 2). The SRB test showed a significant decrease (P < 0.05) in protein content starting from 10 µM for all time points. These decreases ranged from a drop with 10% for 10 µM after 24 h to 63.6% for 100 µM after five days. The MTT test showed a small but significant (P < 0.05) increase in mitochondrial activity for some time points at low concentrations of 5-FU (0.01–1 µM). At higher concentrations (starting from 10 µM), small decreases were observed after 24 h and 48 h. Viability dropped to less than 50% after five days of treatment with 5-FU at levels higher than 20 µM (P < 0.05). Together, these data show that 5-FU was toxic for TR146 cells starting from 20 µM after 24 h and starting from 10 µM after 48 h or five days.
Figure 2.
MTT and SRB toxicity test of 5-FU (0.01–100 µM) on oral epithelial TR146 cells (AV ± SD, n = 6). Significant deviations from the control condition (0 µM) are indicated by the asterisks (P < 0.05). (A color version of this figure is available in the online journal.)
Bacterial cell counts are determined by sample and donor type
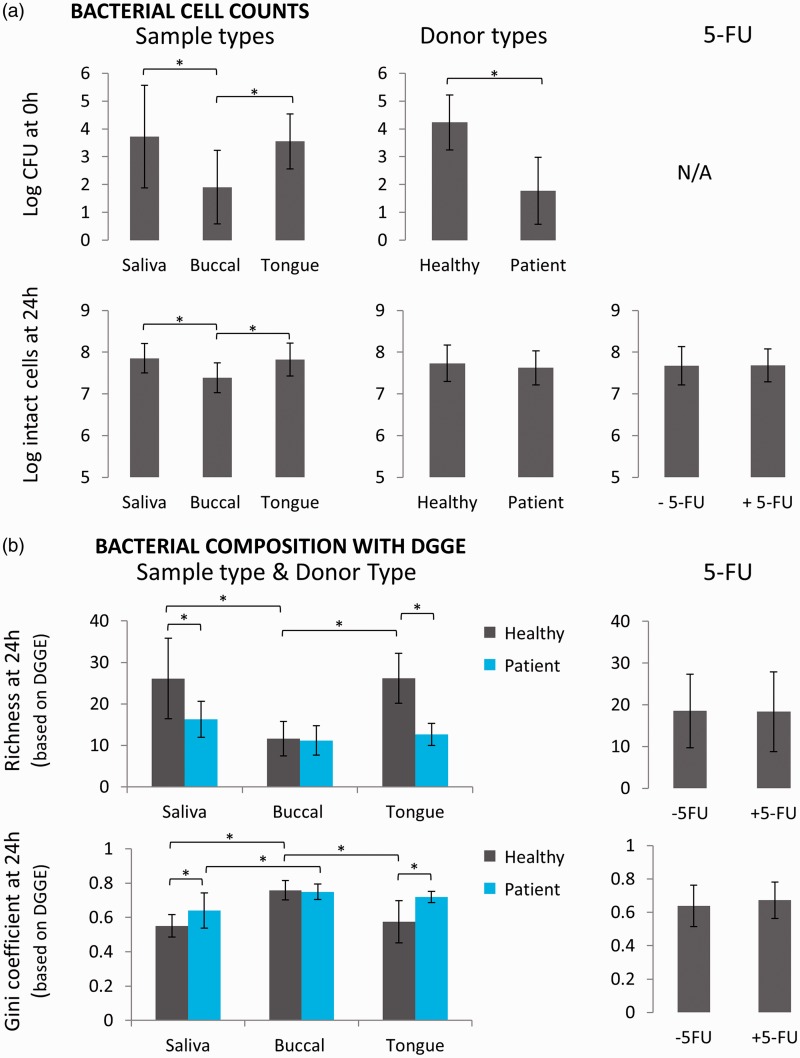
For seven donors (four healthy individuals and three patients suffering from mucositis), three types of samples (saliva, buccal swabs, and tongue swabs) were investigated in a co-culture model (Figure 1). In this model, the microbial sample was incubated for 24 h on an agar/mucin layer in indirect contact with oral epithelial cells. Each sample was tested in absence or presence of 10 µM 5-FU in the basolateral compartment. Both the initial (directly after taking the sample, t = 0 h) and final (after 24 h of co-culture, t = 24 h) bacterial cell counts were evaluated (Figure 3(a)). Depending on the type of sample and the type of donor, the initial bacterial concentration ranged between 1 and 5 log CFU. With regard to the different oral sample types, a clear distinction was observed between buccal swabs on the one hand and saliva and tongue swabs on the other. The initial (t = 0 h) bacterial concentration in buccal swabs (1.9 ± 1.3 log CFU) was significantly lower compared to saliva (3.7 ± 1.8 log CFU, P < 0.001) and tongue swabs (3.6 ± 1.0 log CFU, P < 0.001). Despite this variation in initial number, all samples were able to grow up to a concentration of 7–8 log CFU after 24 h of co-culture in the in vitro model. The difference in concentration, depending on the sample type, was still present after 24 h, with slightly lower bacterial cell counts for the buccal swab amended wells (7.4 ± 0.4 log cells) compared to saliva (7.9 ± 0.4 log cells, P < 0.001) and tongue swab amended wells (7.8 ± 0.4 log cells, P < 0.001). Also, the type of donor affected the bacterial cell counts. While patient samples displayed a 2–3 log lower initial bacterial concentration compared to healthy individuals (P < 0.001), no significant differences were noted after 24 h in the co-culture model (P = 0.12). Surprisingly, treatment with 5-FU did not alter bacterial cell counts at 24 h (P = 0.60). Thus, bacterial cell counts are determined by both sample type and donor type, but are not affected by 5-FU.
Figure 3.
Bacterial cell counts and composition of microbiota derived from different sample and donor types cultured in the oral co-culture model in presence or absence of 5-FU. Sample type data represent pooled data of all donor types in presence and absence of 5-FU and similar pooled data are used for graphs concerning donor type and 5-FU. (a) Bacterial cell counts at t = 0 h and t = 24 h (AV ± SD); (b) Richness and Gini coefficient as measure for bacterial diversity by DGGE (AV ± SD). Significant differences between groups are indicated by the asterisks (P < 0.05). (A color version of this figure is available in the online journal.)
Buccal-derived samples have lower microbial diversity, compared to saliva and tongue amended samples
DGGE analysis (Figure 3(b) and Figure S1) showed that differences in microbial diversity between sample types were dependent on the type of donor, as a significant interaction between donor type and sample type was seen. For healthy individuals, the microbial community of the buccal swab amended wells was lower in richness and evenness, compared to saliva (for both P < 0.001) and tongue swab amended wells (for both P < 0.001) at 24 h. For patients, only a significant increase in Gini coefficient was seen for buccal amended wells, compared to saliva (P = 0.0055). For each donor, Bray–Curtis analysis of DGGE profiles also showed significant differences between the different sample types (Table S1).
Patient-derived samples are less diverse and enriched in pathogenic genera as compared to healthy donor samples
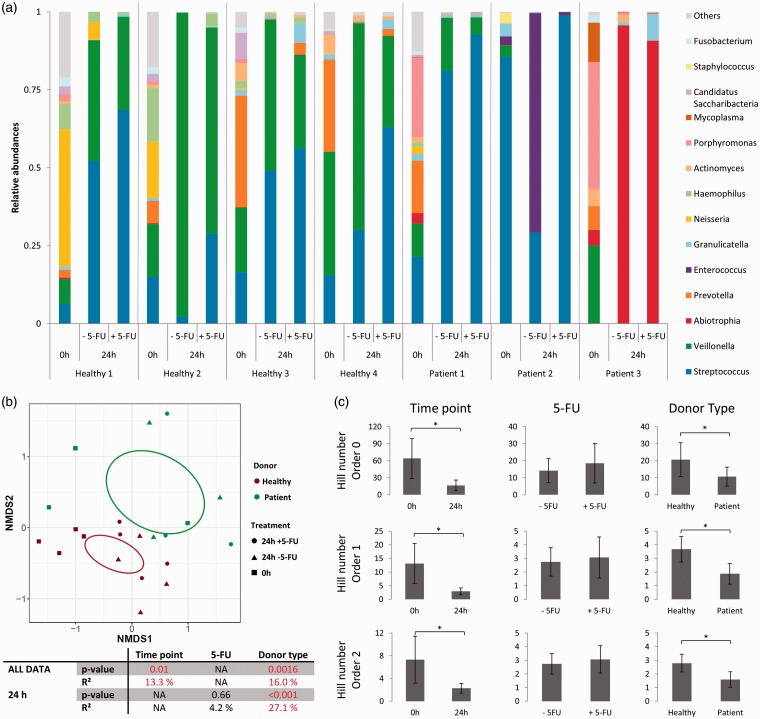
With regard to donor type, DGGE analysis showed different responses depending on the type of sample. For saliva and tongue swab amended wells, a lower richness (P < 0.001 for both) and evenness (P = 0.0084 and P < 0.001) was observed for patient-derived samples at 24 h, compared to wells with samples from healthy individuals (Figure 3(b)). However, no differences were seen for buccal swab amended wells (richness P = 0.91 and evenness P = 0.77). The high cell density in saliva samples allowed for performing Illumina sequencing (guaranteeing high-quality data acquisition). As could be expected, the results showed clear differences between donor types (Figure 4(a) and Figure S2). Bray–Curtis analysis at OTU level revealed that 16.0% of the variation in the composition of the saliva samples could be attributed to the type of donor (P = 0.0016). Visualization by NMDS plots confirmed the major impact of donor type as all patient-derived samples cluster to one side of the plot (Figure 4(b)). In correspondence with DGGE results, diversity parameters were lower for patient-derived samples, compared to samples from healthy individuals (Hill number order 0, P = 0.0067; order 1, P = 0.028; order 2, P = 0.026) (Figure 4(c)). At 24 h, patient-derived saliva samples were more dominated by Lactobacillales (containing Streptococcus, Abiotrophia, and Enterococcus) (95.3 ± 6.9%) compared to samples derived from healthy individuals (45.3 ± 23.0%) (P < 0.001) (Figure 4(a)). In contrast, Veillonella is more abundant in samples derived from healthy individuals at 24 h (50.8 ± 24.3%) in comparison with patient-derived samples (3.8 ± 6.4) (P < 0.0023). The initial (t = 0 h) samples from patients contained also more pathogenic genera, for example 25.5% of Porphyromonas for patient 1, 2.8% of Enterococcus and 3.3% of Staphylococcus for patient 2, and 40.8% of Porphyromonas and 12.7% of Mycoplasma for patient 3. These genera were not (Enterococcus and Mycoplasma) or at much lower abundances (Staphylococcus 0–0.04%, Porphyromonas 0.2–2.3%) detected in the samples derived from healthy individuals. Interestingly, the initial microbial composition of the saliva samples of healthy individuals 1 and 2 on the one hand, and 3 and 4 on the other hand was very similar. This can be explained by the fact that these were samples from siblings, living in the same environment and having similar eating habits. In brief, patient samples had lower microbial diversity and higher abundance of pathogenic genera.
Figure 4.
Illumina sequencing of the 16S rRNA gene of the microbiota in the saliva samples. (a) Bar plot representing the 14 most abundant genera; (b) NMDS plot with 95% confidence ellipsoid, P-values and R2 for different confounding factors based on Bray–Curtis dissimilarities; (c) Hill numbers order 0, 1, and 2 representing richness, evenness, and diversity, respectively (AV ± SD). Significant differences between groups are indicated by the asterisks (P < 0.05).
5-FU had no major impact on bacterial composition
DGGE showed that 5-FU did not affect richness (P = 0.83) nor evenness (P = 0.069) of the bacteria. Bray–Curtis analysis showed that only for patient 2, a significant effect of 5-FU on the microbial profile could be detected based on the DGGE profile (P = 0.0014) (Table S1). For all sample types of this patient, two dominant bands clearly disappeared following 5-FU treatment (Figure S6). Similar to DGGE, Illumina sequencing showed that 5-FU treatment did not significantly affect the bacterial diversity (Figure 4(c)). However, following 5-FU treatment, a general trend in increased Streptococcus abundance (from 40.6 ± 26.7% to 68.1 ± 25.5%; P = 0.099) and of decreased Veillonella abundance (from 44.7 ± 34.8% to 26.8 ± 23.4%; P = 0.32) was observed (Figure 4(a)). In contrast to the other individuals, wells derived from patient 3 were dominated by Abiotrophia after 24 h of co-culture both with and without 5-FU. More specifically, Prevotella abundance increased following 5-FU treatment for samples derived from healthy individuals 3 and 4 (0.4% to 3.8% and 0.4% to 2.2%, respectively). For patient 2, Enterococcus and Streptococcus were the most abundant genera in the untreated wells (70.4% and 29.2%, respectively), whereas in presence of 5-FU, Streptococcus dominated with 98.9%. This result confirmed the changed DGGE profiles of patient 2 following 5-FU treatment (Figure S6). Altogether, these results indicate small yet non-significant changes in the composition of the biofilm following 5-FU treatment (based on Bray–Curtis dissimilarities on OTU level, P = 0.66).
Bacterial composition changes after 24 h of co-culture
Finally, a significant change in bacterial composition was observed with Illumina sequencing attributed between sampling time points (t = 0 h vs. t = 24 h) (P = 0.01), which explained 13.3% of the variation in all samples (based on Bray–Curtis dissimilarities on OTU level). This difference was also visible in the NMDS plot (Figure 4(b)). Moreover, all Hill numbers showed a decrease in diversity at 24 h compared to the initial samples (Hill number order 0, P < 0.001; order 1, P < 0.001; order 2, P < 0.001) (Figure 4(c)). Streptococcus and Veillonella were the dominating genera in the saliva samples after 24 h in the in vitro model (together 95.6 ± 4.2%), apart from the control sample derived from patient 2, which was dominated by Enterococcus and the samples derived from patient 3, which were dominated by Abiotrophia (Figure 4(a)). Next to Streptococcus and Veillonella, the initial saliva samples were also populated by Prevotella, Neisseria, Granulicatella, Haemophilus, Actinomyces, Porphyromonas, Fusobacterium, and Megasphaera of which levels depended on the donor. A lot of this diversity was lost during the 24 h incubation in the co-culture model. For some donors, most genera were still present albeit at relatively low abundances.
Epithelial wound healing is reduced by oral microbiota, irrespective of the presence of 5-FU
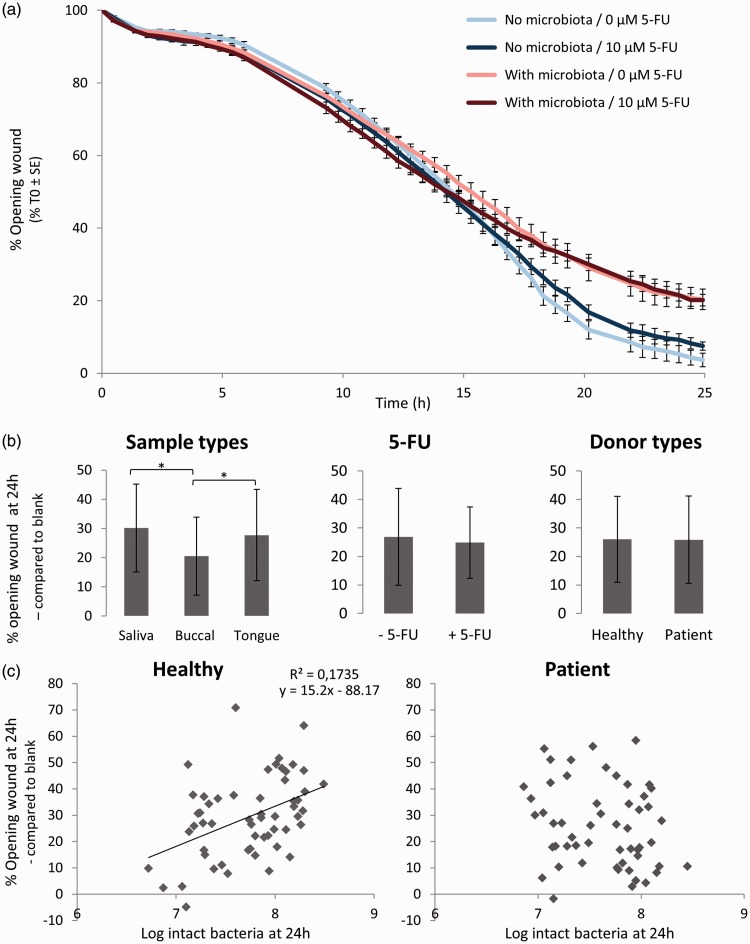
To investigate the closure of artificially induced wounds in an epithelial monolayer over time, we followed wound healing in a separate set-up with and without microbiota derived from a buccal swab from a healthy individual in presence or absence of 5-FU (Figure 1). Compared to the (unchallenged) control wells, epithelial cell wound healing slowed down in presence of microbiota starting from 16 h, eventually resulting in a 16% lower wound healing after 25 h (Figure 5(a)). The presence of 5-FU had no effect on the wound healing capacity and this was independent of microbial presence.
Figure 5.
(a) Oral microbiota derived from a buccal swab reduce wound healing capacity of oral epithelial cells in an in vitro mucosa model, irrespective of the treatment with 5-FU (10 µM) (AV ± SE); (b) Sample type affect wound healing capacity, whereas donor type and presence of 5-FU do not (AV ± SD); (c) For healthy individuals, a positive correlation between the opening of the wound and bacterial cell count at 24 h was observed, but for patients no such trend was noticed. (A color version of this figure is available in the online journal.)
This experiment showed that co-culture with microbiota reduces wound healing of oral epithelial cells. This effect might, however, be caused by different bacterial cell counts and composition, which have been shown to depend on the type of donor and the sample type. Indeed, although a general reduction (25.9 ± 15.1%) of wound healing capacity was observed by the addition of oral microbiota, different trends could be noticed for sample and donor types (Figure 5).
First, addition of microbiota derived from saliva and tongue swabs had a more detrimental effect on wound healing in comparison with buccal-derived microbiota (P = 0.0051 for saliva; P = 0.041 for tongue) (Figure 5(b)). Regarding the type of donor, no difference in wound healing capacity was noticed (P = 0.95). However, plotting the wound opening at 24 h as a function of the bacterial cell counts revealed two different trends between healthy and patient samples (Figure 5(c)). Microbial samples from healthy individuals displayed a linear relationship with each additional log CFU of bacterial cells resulting in a 15.2% increase in wound opening (P = 0.00082). Independent of microbiome composition, this is indicative (adjusted R2 = 0.17) of a higher wound healing capacity at lower bacterial loads. However, no such trend could be observed for patient samples (P = 0.13). Again, no modulating effect of 5-FU on wound healing was observed after 24 h in the presence (P = 0.49) and absence (P= 0.21) of microbiota. An MTT assay performed after 24 h of co-culture showed no effect of sample type (P = 0.26) or type of donor (P = 0.23) on the cell viability of TR146 cells (Figure S3). A small but significant increase in epithelial cell viability was observed following 5-FU treatment in presence of microbiota (89.5 ± 11.0% to 95.3 ± 11.4%, P = 0.004), whereas no effect was observed in absence of microbiota (P = 0.94). Together, these data indicate that wound healing potential is determined by both bacterial cell count and bacterial composition.
Discussion
Oral mucositis is a debilitating side effect of chemotherapeutic treatment in which microbiota are more and more shown to play an important role. In this study, we investigated the interactions between the oral microbiome, oral epithelial cells, and a chemotherapeutic (5-FU) using an in vitro co-culture model. As wound healing is crucial in recovering from mucositis, this was one of the functional endpoints in the model apart from microbial numbers and composition.
Our data showed that oral microbiota reduced wound healing capacity for all seven donors with 25.9 ± 15.1%. Previous research using the same in vitro model showed that oral microbiota had similar negative effects on wound healing.22 However, this reduction appeared to be species- and concentration-dependent.18,19 It has been shown for chronic wounds that low amounts of microbiota can improve wound healing, whereas in infectious conditions with high bacterial loads, wound healing capacity is significantly reduced.19 Our data confirmed that for healthy individuals, lower bacterial cell counts correlated with higher wound healing capacity. This encourages the use of good oral hygiene during mucositis, shown previously to be of high importance in oral mucositis, as colonization of the ulcers by microbiota may prolong the healing phase.4,36 However, for patients that are in the acute mucositis phase, more measures might be needed, as we have shown that for such patients wound healing capacity was independent of the bacterial cell counts. This indicates that also bacterial composition might be important in acute mucositis patients. De Ryck et al.18 indeed showed that wound healing capacity seems to be species-dependent with Klebsiella oxytoca having a deleterious effect on wound healing, whereas Streptococcus mitis and Streptococcus oralis stimulated wound healing.
Further, we observed differences in the composition and diversity of oral microbiota derived from patients suffering from mucositis compared to healthy individuals. The abundance of Lactobacillalles was higher in patient samples in comparison with healthy individuals and the diversity of samples derived from patients was lower. Our results are in accordance with a prospective study with 454-sequencing of mucosal samples also showing a lower diversity in patient samples compared to reference individuals.37 Moreover, the Illumina data from our study revealed the presence of larger numbers of genera containing pathogenic species, like Porphyromonas, Enterococcus, and Staphylococcus, in the patient-derived samples, which could lead to a higher infection risk. Porphyromonas gingivalis was shown previously to be predictive for the development of oral ulcerations in hematopoietic stem cell transplantation patients.38
Different sites in the oral cavity are colonized with distinct microbial communities.21 In our study, buccal swabs had lower bacterial cell counts, compared to saliva and tongue swabs, leading to a higher wound healing capacity, which is in line with the previous results. DGGE also indicated lower richness and evenness in the buccal samples. This lower diversity of buccal microbiome compared to saliva and tongue samples has already been explained by extensive data derived from The Human Microbiome Project by the dominance of Streptococcus in buccal samples.21 All these differences may impact the microbiome of the ulcers, depending on the location of the ulcer.
We also investigated the effect of 5-FU on different endpoints in the co-culture model. We chose to work with a dose of 10 µM, as this was the highest non-toxic concentration for TR146 cells after 24 h. Similar toxicity profiles have been recorded for other cell lines such as for Caco-2 cells.39 In vivo concentrations range from 3 to 10 µM in plasma and 0.08–0.8 µM in saliva following continuous treatment,8,9 but significantly increase in case of dihydropyrimidine dehydrogenase (DPD) deficiency.40 Previous research showed a variable sensitivity among oral species towards 5-FU.10 However, in our system which comprises a plethora of oral species cultured in a biofilm, we did not see an impact of 5-FU on both bacterial cell counts or wound healing. Further, 5-FU had only a minor impact on bacterial composition with an increasing trend in Streptococcus and a decreasing trend in Veillonella. S. oralis, S. mitis, and Streptococcus salivarius have been shown in a previous study to be resistant to 5-FU at 10 µM,10 which might explain their ability to increase in abundance. No data are available on the sensitivity of Veillonella to 5-FU; however, our data suggest that Veillonella is sensitive towards 5-FU. The results for patient 2 indicated high sensitivity of Enterococcus towards 5-FU, confirming previous research.17 Moreover, patient 2 was the only donor for which a significant effect of 5-FU on the microbial composition was shown. This indicates a donor-specific effect of 5-FU which encourages the use of a personalized approach.
At 24 h, the biofilm formed in the model was mainly dominated by Streptococcus and Veillonella for the saliva samples. Although this indicates a loss of diversity of the original saliva sample when cultured in the in vitro model, this loss might be due to biofilm formation. In vivo growth of an oral biofilm on enamel-dentin slabs in the mouth of healthy volunteers also showed a dominance of Streptococcus (62%) and Veillonella (27%) after 48 h.41 Although we used saliva samples, the high abundance of Streptococcus is more similar to buccal samples.21 We hypothesize that the use of an agar/mucin layer as a substrate promotes biofilm formation of a buccal community, despite the use of a saliva sample as a microbial source. This immature biofilm is formed by Streptococci, known to be initial colonizers of the oral biofilm.42 With respect to Veillonella, dependency on the lactic acid produced by Streptococci has been shown43 and therefore these species are likely to co-occur.
In conclusion, oral microbiota reduce wound healing capacity of epithelial cells with higher bacterial cell counts linked to lower wound healing capacity in healthy individuals. However, for patients suffering from mucositis, the mechanism of wound healing is more related to microbial composition, rather than microbial load as their oral samples are characterized by a disturbed microbial community with higher abundances of pathogenic genera. More research on the link between oral microbial composition and wound healing capacity is needed to fully understand their role in the wound healing process in patients suffering from mucositis.
Supplementary Material
Acknowledgements
The authors would like to thank Charlotte Grootaert for the use of cell culture equipment for part of the work and Charlotte De Rudder and Massimo Marzorati for their review of the manuscript.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; EV, BV, TS, JB and AB conducted the experiments, BDM supplied the oral samples from patients with oral mucositis, WDV supplied the microscopy facilities and analysis tool for wound healing results, and EV wrote the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was supported by the Bijzonder Onderzoeksfonds (Ghent University grant numbers BOF13/DOC/280, BOF17/GOA/032 and BOF/11267/09; University of Antwerp grant number TTBOF29267; and the Seventh Framework Programme (FP7/2011) (grant number 299169).
References
- 1.Sonis S. Pathobiology of oral mucositis: novel insights and opportunities. J Support Oncol 2007; 5:3–11 [PubMed] [Google Scholar]
- 2.Villa A, Sonis ST. Mucositis: pathobiology and management. Curr Opin Oncol 2015; 27:159–64 [DOI] [PubMed] [Google Scholar]
- 3.Lalla RV, Sonis ST, Peterson DE. Management of oral mucositis in patients with cancer. Denta Clin N Am 2008; 52:61–viii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villa A, Sonis ST. Pharmacotherapy for the management of cancer regimen-related oral mucositis. Expert Opin Pharmacother 2016; 17:1801–7 [DOI] [PubMed] [Google Scholar]
- 5.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003; 3:330–8 [DOI] [PubMed] [Google Scholar]
- 6.Grem JL. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs 2000; 18:299–313 [DOI] [PubMed] [Google Scholar]
- 7.Sonis ST, Elting LS, Keefe D, Peterson DE, Schubert M, Hauer-Jensen M, Bekele BN, Raber-Durlacher J, Donnelly JP, Rubenstein EB. Perspectives on cancer therapy-induced mucosal injury – pathogenesis, measurement, epidemiology, and consequences for patients. Cancer 2004; 100:1995–2025 [DOI] [PubMed] [Google Scholar]
- 8.Joulia JM, Pinguet F, Ychou M, Duffour J, Astre C, Bressolle F. Plasma and salivary pharmacokinetics of 5-fluorouracil (5-FU) in patients with metastatic colorectal cancer receiving 5-FU bolus plus continuous infusion with high-dose folinic acid. Eur J Cancer 1999; 35:296–301 [DOI] [PubMed] [Google Scholar]
- 9.Takimoto CH, Yee LK, Venzon DJ, Schuler B, Grollman F, Chabuk C, Hamilton JM, Chen AP, Allegra CJ, Green JL. High inter- and intrapatient variation in 5-fluorouracil plasma concentrations during a prolonged drug infusion. Clin Cancer Res 1999; 5:1347–52 [PubMed] [Google Scholar]
- 10.Vanlancker E, Vanhoecke B, Smet R, Props R, Van de Wiele T. 5-Fluorouracil sensitivity varies among oral micro-organisms. J Med Microbiol 2016; 65:775–83 [DOI] [PubMed] [Google Scholar]
- 11.Vasconcelos RM, Sanfilippo N, Paster BJ, Kerr AR, Li Y, Ramalho L, Queiroz EL, Smith B, Sonis ST, Corby PM. Host-microbiome cross-talk in oral mucositis. J Dent Res 2016; 95:725–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanhoecke B, De Ryck T, Stringer A, Van de Wiele T, Keefe D. Microbiota and their role in the pathogenesis of oral mucositis. Oral Dis 2015; 21:17–30 [DOI] [PubMed] [Google Scholar]
- 13.Stringer AM, Logan RM. The role of oral flora in the development of chemotherapy-induced oral mucositis. J Oral Pathol Med 2015; 44:81–7 [DOI] [PubMed] [Google Scholar]
- 14.Saunders DP, Epstein JB, Elad S, Allemano J, Bossi P, van de Wetering MD, Rao NG, Potting C, Cheng KK, Freidank A, Brennan MT, Bowen J, Dennis K, Lalla RV, Mascc I. Systematic review of antimicrobials, mucosal coating agents, anesthetics, and analgesics for the management of oral mucositis in cancer patients. Support Care Cancer 2013; 21:3191–207 [DOI] [PubMed] [Google Scholar]
- 15.van Vliet MJ, Harmsen HJM, de Bont E, Tissing WJE. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog 2010; 6:e1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Napenas JJ, Brennan MT, Bahrani-Mougeot FK, Fox PC, Lockhart PB. Relationship between mucositis and changes in oral microflora during cancer chemotherapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2007; 103:48–59 [DOI] [PubMed] [Google Scholar]
- 17.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh ASJ, Hamilton J, Keefe DMK. Gastrointestinal microflora and mucins may play a critical role in the development of 5-fluorouracil-induced gastrointestinal mucositis. Exp Biol Med 2009; 234:430–41 [DOI] [PubMed] [Google Scholar]
- 18.De Ryck T, Vanlancker E, Grootaert C, Roman BI, De Coen LM, Vandenberghe I, Stevens CV, Bracke M, de Wiele TV, Vanhoecke B. Microbial inhibition of oral epithelial wound recovery: potential role for quorum sensing molecules? AMB Exp 2015; 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis 2004; 17:91. [DOI] [PubMed] [Google Scholar]
- 20.Laheij A, de Soet JJ, Veerman ECI, Bolscher JGM, van Loveren C. The influence of oral bacteria on epithelial cell migration in vitro. Mediat Inflamm 2013; 2013:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012; 13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Ryck T, Grootaert C, Jaspaert L, Kerckhof F-M, Van Gele M, De Schrijver J, Van den Abbeele P, Swift S, Bracke M, Van de Wiele T, Vanhoecke B. Development of an oral mucosa model to study host-microbiome interactions during wound healing. Appl Microbiol Biotechnol 2014; 98:6831–46 [DOI] [PubMed] [Google Scholar]
- 23.Rupniak HT, Rowlatt C, Lane EB, Steele JG, Trejdosiewicz LK, Laskiewicz B, Povey S, Hill BT. Characteristics of 4 new human cell-lines derived from squamous-cell carcinomas of the head and neck. J Natl Cancer Inst 1985; 75:621–35 [PubMed] [Google Scholar]
- 24.Jacobsen J, Pedersen M, Rassing MR. TR146 cells as a model for human buccal epithelium: II. Optimisation and use of a cellular sensitivity MTS/PMS assay. Int J Pharm 1996; 141:217–25 [Google Scholar]
- 25.Naglik J, Moyes D. Epithelial cell innate response to Candida albicans. Adv Dent Res 2011; 23:50–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth 1983; 65:55–63 [DOI] [PubMed] [Google Scholar]
- 27.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006; 1:1112–6 [DOI] [PubMed] [Google Scholar]
- 28.Van Nevel S, Koetzsch S, Weilenmann HU, Boon N, Hammes F. Routine bacterial analysis with automated flow cytometry. J Microbiol Meth 2013; 94:73–6 [DOI] [PubMed] [Google Scholar]
- 29.Vilchez-Vargas R, Geffers R, Suarez-Diez M, Conte I, Waliczek A, Kaser VS, Kralova M, Junca H, Pieper DH. Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ Microbiol 2013; 15:1016–39 [DOI] [PubMed] [Google Scholar]
- 30.Ovreas L, Forney L, Daae FL, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 1997; 63:3367–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer G, Dewaal EC, Uitterlinden AG. Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal-RNA. Appl Environ Microbiol 1993; 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diepenbroek M, Glöckner FO, Grobe P, Güntsch A, Huber R, König-Ries B, Kostadinov I, Nieschulze J, Seeger B, Tolksdorf R, editors. Towards an integrated biodiversity and ecological research data management and archiving platform: the german federation for the curation of biological data (GFBio). In: Plödereder E, Grunske L, Schneider E, Ull D (eds), Informatik 2014 – Big Data Komplexität meistern. GI-Edition: Lecture Notes in Informatics (LNI) – Proceedings 232: 1711-1724. Köllen Verlag: Bonn, 2014.
- 33.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. R package version 2.3-4, 2016.
- 35.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology 1973; 54:427–32 [Google Scholar]
- 36.Keefe DM, Schubert MM, Elting LS, Sonis ST, Epstein JB, Raber-Durlacher JE, Migliorati CA, McGuire DB, Hutchins RD, Peterson DE, Mucositis Study Section M. Updated clinical practice guidelines for the prevention and treatment of mucositis. Cancer 2007; 109:820–31 [DOI] [PubMed] [Google Scholar]
- 37.Ye Y, Carlsson G, Agholme MB, Wilson JAL, Roos A, Henriques-Normark B, Engstrand L, Modeer T, Putsep K. Oral bacterial community dynamics in paediatric patients with malignancies in relation to chemotherapy-related oral mucositis: a prospective study. Clin Microbiol Infect 2013; 19:E559–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laheij A, de Soet JJ, von Dem Borne PA, Kuijper EJ, Kraneveld EA, van Loveren C, Raber-Durlacher JE. Oral bacteria and yeasts in relationship to oral ulcerations in hematopoietic stem cell transplant recipients. Support Care Cancer 2012; 20:3231–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang SB, Shih HY, Huang CH, Li LT, Chen CC, Fang HW. Live and heat-killed Lactobacillus rhamnosus GG upregulate gene expression of pro-inflammatory cytokines in 5-fluorouracil-pretreated Caco-2 cells. Support Care Cancer 2014; 22:1647–54 [DOI] [PubMed] [Google Scholar]
- 40.Saif MW, Choma A, Salamone SJ, Chu E. Pharmacokinetically guided dose adjustment of 5-fluorouracil: a rational approach to improving therapeutic outcomes. J Natl Cancer Inst. 2009; 101:1543–52. [DOI] [PubMed] [Google Scholar]
- 41.Klug B, Santigli E, Westendorf C, Tangl S, Wimmer G, Grube M. From mouth to model: combining in vivo and in vitro oral biofilm growth. Front Microbiol 2016; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Micro 2010; 8:471–80 [DOI] [PubMed] [Google Scholar]
- 43.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 2000; 54:413–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.