Short abstract
Congenital insensitivity to pain with anhidrosis (CIPA) is a rare autosomal recessive heterogeneous disorder mainly caused by mutations in the neurotrophic tyrosine receptor kinase 1 gene (NTRK1) and characterized by insensitivity to noxious stimuli, anhidrosis, and intellectual disability. We herein report the first north Han Chinese patient with CIPA who exhibited classic phenotypic features and severe intellectual disability caused by a homozygous c.851-33T>A mutation of NTRK1, resulting in aberrant splicing and an open reading frame shift. We reviewed the literature and performed in silico analysis to determine the association between mutations and intellectual disability in patients with CIPA. We found that intellectual disability was correlated with the specific Ntrk1 protein domain that a mutation jeopardized. Mutations located peripheral to the Ntrk1 protein do not influence important functional domains and tend to cause milder symptoms without intellectual disability. Mutations that involve critical amino acids in the protein are prone to cause severe symptoms, including intellectual disability.
Keywords: Congenital insensitivity to pain with anhidrosis, genetics, intellectual disability, neurotrophic tyrosine receptor kinase 1, north Han Chinese, in silico analysis
Introduction
Congenital insensitivity to pain with anhidrosis (CIPA, OMIM #256800), also known as hereditary sensory and autonomic neuropathy IV, is an autosomal recessive disorder caused by loss-of-function mutations in the neurotrophic tyrosine receptor kinase 1 gene (NTRK1, OMIM *191315),1 nerve growth factor beta gene (NGFβ, OMIM *162030),2,3 and necdin gene (NDN, OMIM *602117).4 The vast majority of mutations are observed in NTRK1.5 CIPA is characterized by insensitivity to noxious stimuli, anhidrosis, and intellectual disability.2,6 Classic symptoms of CIPA include recurrent unexplained fevers without sweating, self-mutilating behavior (e.g., biting of the tongue, lips, and distal extremities), recurrent bone fractures and Charcot arthropathy, nonhealing ulcers and injuries, and psychomotor retardation.5,7,8 The pain insensitivity in patients with CIPA is caused by an absence of small-diameter thinly myelinated primary afferent or unmyelinated neurons, and the anhidrosis is caused by a deficiency of sympathetic postganglionic neurons.5,9
Phenotypic heterogeneity in the occurrence of intellectual disability has been reported in patients with CIPA. Ntrk1-knockout mice lack basal forebrain cholinergic neurons and striatal cholinergic neurons.9 No autopsy data in humans with CIPA are available to validate these animal observations. Additionally, cranial magnetic resonance imaging has revealed no structural brain alterations in patients with CIPA.10 We herein present a case involving a patient with CIPA with classic phenotypic features and severe intellectual disability caused by a previously reported homozygous c.851-33T>A mutation of NTRK1 gene. We performed a literature review and in silico analysis to determine the association between mutations and intellectual disability in patients with CIPA.
Methods
Patient
This study was approved by the Ethical Review Board of the Peking Union Medical College Hospital. Written informed consent was obtained from the patient’s parents.
A 27-year-old Han Chinese woman was referred to our hospital in 2013 and diagnosed with CIPA. Her clinical history was reviewed and a physical examination was performed. Her intelligence quotient was measured with the Wechsler Adult Intelligence Scale (WAIS IV, 2008 version). A nerve biopsy was not conducted because of the minimal benefit to the patient.
Genetic analysis
After obtaining informed consent, we drew 4 ml of peripheral blood from the patient and her parents. Genomic DNA was extracted with the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol.
All 17 exons and intron–exon boundaries were amplified by polymerase chain reaction (PCR) with primers used in previous reports.6,11,12 Sanger sequencing was conducted by Berry Genomics Co., Ltd. (Beijing, China). The sequences were analyzed with Chromas software (version 2.1.1) and BLAT to UCSC Human Genome (Feb 2009, GRCh37/hg19, http://genome.ucsc.edu/) to call mutations.
The oligonucleotide-based comparative genomic hybridization (CGH) microarray is an efficient and reliable assay for copy number variation studies. The Agilent 1x1M Human Genome CGH microarray (Agilent, Santa Clara, CA, USA) was employed in this study. DNA processing, microarray handling, and data analysis were conducted by following the Agilent oligonucleotide array CGH protocol (version 6.0).
Literature review
The MeSH terms “congenital insensitivity to pain with anhidrosis” and “ntrk1 receptor” were used to search for articles pertinent to CIPA caused by NTRK1 mutation. Title and abstract screening was conducted to retrieve genetic studies, which elucidated genetic mutations causing CIPA. All mutations reported up to October 2017 were reviewed. Furthermore, using the known Ntrk1 protein structure (available at the RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do) and PyMOL software (Version 4.3.0), we attempted to interpret the heterogeneity of the intellectual disability in patients with CIPA.
Results
Clinical findings
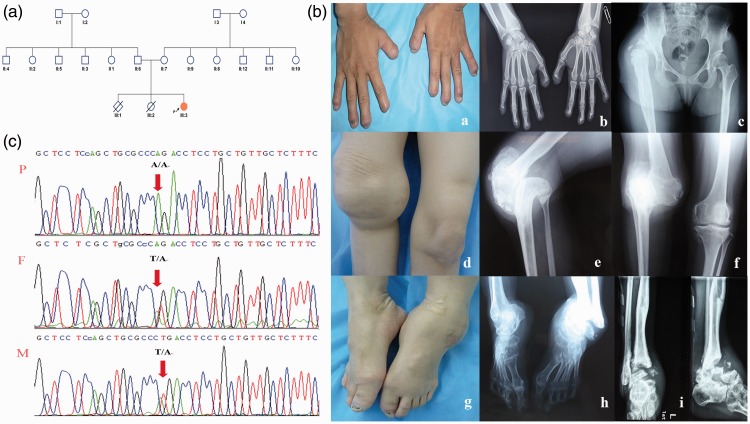
The patient in the present case was a 27-year-old Han Chinese woman (Figure 1(a), III:3) born to nonconsanguineous parents (Figure 1(a), II:6 and II:7). Her mother had a history of a terminated pregnancy (Figure 1(a), III:1) and fetal death at 7 months of gestation (Figure 1(a), III:2). The proband had a 27-year history of anhidrosis and insensitivity to pain and had experienced recurrent unexplained fevers (maximum body temperature, 42°C) during her first year of life. Bilateral dislocation of the hip joints had been present since age 2 years. Although her body development was normal, she showed cognitive impairment at a young age. Self-mutilating behavior was first observed at age 6 years and resulted in finger, ankle, tibia, hip, and femur fractures. She developed osteomyelitis due to poor healing following trauma or surgical treatment of fractures. She required a wheelchair for ambulation because of deformation of her right hip joint, right knee, and left ankle (Figure 1(b)). Her intelligence quotient was about 44 (WAIS IV, 2008 version). She showed emotional instability and communication difficulty. The clinical characteristics of patients with CIPA with the hotspot c.851-33T>A mutation are summarized in Table 1.
Figure 1.
Patient’s pedigree and clinical and imaging characteristics. Panel A: Pedigree of the patient (P), who is indicated by an arrow. Panel B: Clinical characteristics and imaging of the proband. (a, b) Hypoplasia of the distal phalanges of both hands due to self-mutilation and osteomyelitis. (c) Deformity of the hip joint. (d–f) Malunion of the right knee. (g–i) Deformity and multiple fractures of the ankle and tibia. Panel C: Sequencing of NTRK1 reveals the homozygous c.851-33T>A mutation in the patient (P) and heterozygous c.851-33T>A mutation in her father (F) and mother (M).
Table 1.
Clinical characteristics of patients with CIPA with the c.851-33T>A mutation
| Pt. | Genetic variants | Race | Sex | Age (y) | Insensitivity to pain | Charcot arthropathy | Recurrent bone fracture | Self-mutilation | Compromised proprioceptive sensation | Anhidrosis | Recurrent unexplained fever | Intellectual disability | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Homo. | Chinese | F | 27 | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Present report |
| 2 | Homo. | Korean | M | 28 | Yes | NA | Yes | Yes | NA | Yes | NA | Yes | Nam et al., 201713 |
| 3 | Co.Het. and IVS14 + 3A>T | Korean | F | 17 | Yes | NA | Yes | Yes | NA | Yes | NA | Yes | Nam et al., 201713 |
| 4 | Co.Het. and IVS14 + 3A>T | Korean | F | 16 | Yes | NA | Yes | Yes | NA | Yes | NA | Yes | Nam et al., 201713 |
| 5 | Co.Het. and IVS14 + 3A>T | Korean | M | 14 | Yes | NA | Yes | Yes | NA | Yes | NA | Yes | Nam et al., 201713 |
| 6 | Co.Het. and c.2303C>T | Korean | M | 16 | No | No | Yes | No | No | NA | No | No | Jung et al., 201314 |
| 7 | Co.Het. and c.1415delG | Chinese | F | 5 | Yes | NA | NA | Yes | No | Yes | Yes | Yes | Li et al., 201215 |
| 8 | Homo. | Korean | M | 2.7 | Yes | NA | No | Yes | NA | Yes | Yes | Yes | Lee et al., 200916 |
| 9 | Co.Het. and NA | Korean | M | 1.6 | Yes | NA | No | Yes | NA | Yes | Yes | Yes | Lee et al., 200916 |
| 10 | Co.Het. and NA | Korean | F | 2.4 | Yes | NA | No | Yes | NA | Yes | Yes | Yes | Lee et al., 200916 |
| 11 | Co.Het. and NA | Chinese | M | 20 | Yes | Yes | Yes | NA | Yes | Yes | Yes | Yes | Guo et al., 200417 |
| 12 | Co.Het. and NA | Chinese | M | 18 | Yes | Yes | Yes | NA | Yes | Yes | Yes | Yes | Guo et al., 200417 |
| 13 | Co.Het. and c.2281C>T | Chinese | M | 5.8 | Yes | Yes | Yes | NA | NA | Yes | Yes | NA | Lv et al., 201718 |
| 14 | Co.Het. and c.1652delA | Chinese | M | 20 | Yes | Yes | Yes | NA | NA | Yes | Yes | NA | Lv et al., 201718 |
| 15-19 | Co.Het. and NA | Japanese | NA | NA | Yes | NA | NA | NA | NA | Yes | Yes | NA | Miura et al., 200019 |
All patients had the c.851-33T>A mutation. Pt., patient; F, female; M, male; Homo., homozygous; Co.Het., compound heterozygous; NA ,not available.
Genetic analysis
PCR followed by Sanger sequencing of NTRK1 was completed. Because previous reports have indicated that point mutations in the intron cause abnormal splicing and resultant CIPA, we repeated the analysis and confirmed the presence of a homozygous c.851-33T>A mutation in intron 7 in the patient and a heterozygous c.851-33T>A mutation in both of her nonconsanguineous parents (Figure 1, Panel C). The CGH microarray showed no evidence of copy number variation in her genome (data not shown), validating the homozygosity of the c.851-33T>A mutation. This mutation was previously verified to result in aberrant splicing and a reading frame shift and can reportedly cause CIPA.13–19 To our knowledge, this is the first reported patient homozygous for the c.851-33T>A mutation in the north Han Chinese population.
Literature review
Structure of Ntrk1 protein
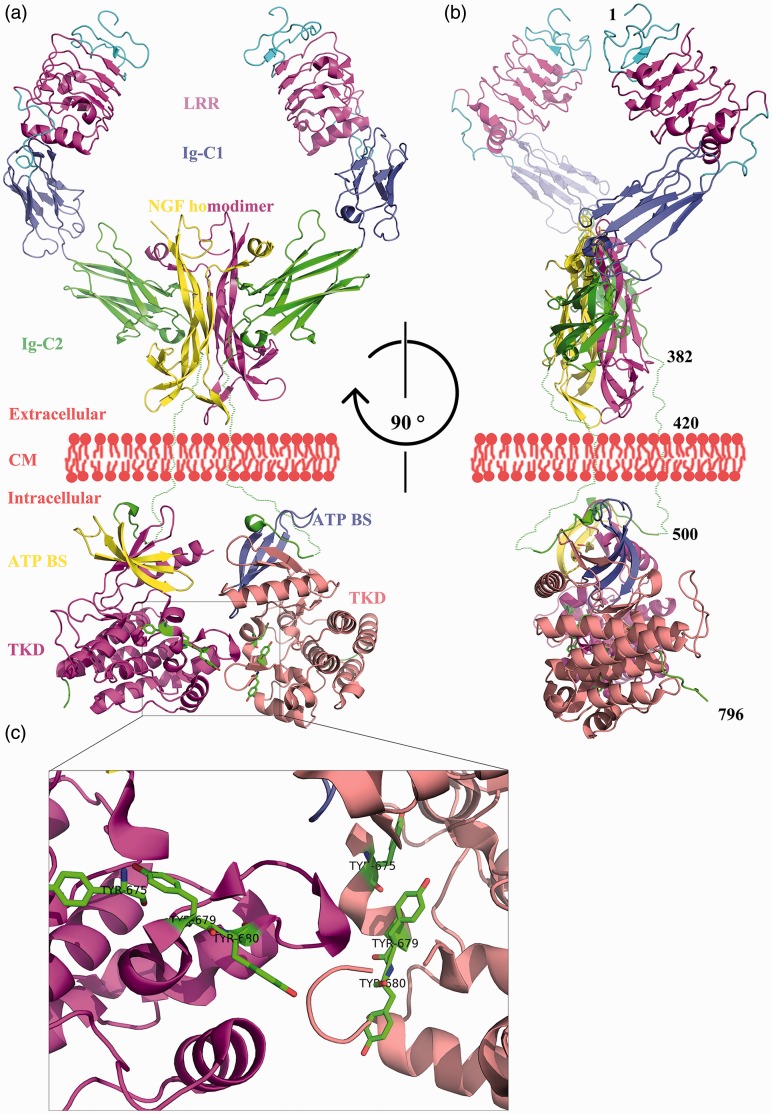
Crystal structures of the extracellular and intracellular parts of the Ntrk1 protein were identified by Wehrman et al.20 and Bertrand et al.,21, respectively (Figure 2). The Ntrk1 protein contains 796 amino acids and consists of three leucine-rich repeats, two immunoglobulin-like domains (Ig-C1 and Ig-C2), a transmembrane domain, an adenosine triphosphate (ATP) binding site, and a tyrosine kinase domain (TKD). Once the nerve growth factor (NGF) homodimer binds to Ntrk1, it causes Ntrk1 dimerization and autophosphorylation of tyrosine residues 675, 679, and 680.22 This initiates the cytoplasmic signaling cascade that is essential for the survival and growth of neurons.23
Figure 2.
Whole structure of Ntrk1 dimer binding with NGF homodimer. Panel A: Front view. Panel B: Lateral view. Panel C: Details of the interfaces of the cytoplasmic domains and three tyrosine residues phosphorylated during signaling transduction. Stereo visualization is realized by shading the further parts of the molecule. Residue numbers were labeled approximately in Panel B. Dotted lines were drawn manually to connect the extracellular and intracellular portions because no exact crystal structure for the transmembrane domain is available. LRR, leucine-rich repeats; Ig-C1 and Ig-C2, immunoglobulin-like domains; NGF, nerve growth factor; CM, cytomembrane; ATP BS, adenosine triphosphate binding site; TKD, tyrosine kinase domain.
Heterogeneity of intellectual disability in patients with CIPA
According to our literature review, 90% (99/110) of previously reported patients with CIPA had documented intellectual disability. However, about 10% (11/110) of the patients did not have intellectual disability (44 additional patients with CIPA whose intellectual disability status was not mentioned in the literature were excluded from the statistical analysis). We attempted to determine whether an association exists among CIPA mutation, its effect on Ntrk1 protein, and the presence of intellectual disability (Table 2).
Table 2.
NTRK1 mutations reported in patients with CIPA with or without intellectual disability
| Pt. | Mutations | Polyphen2 score and prediction* | SIFT score and prediction* | Patients’ information |
Reference | ||
|---|---|---|---|---|---|---|---|
| Race | Inheritance model | Intellectual disability | |||||
| 1 | c.722T>C; p.Leu213Proc.1556delG; p.G519fs28X | 1.000, Pr.D- | 0.00, D- | American | Co.Het. | No | Bonkowsky et al., 200324 |
| 2 | c.2347C>T; p.Arg760Trp#c.2337C>G; p.Tyr756X# | 0.653, Po.D- | 0.00, D- | Italy | Co.Het. | No | Indo, 200125 |
| 3 | c.2303C>T; p.Pro767Leu#c.1621G>C; p.Gly516Arg# | 1.000, Pr.D; 1.000, Pr.D | 0.03, D; 0.23, T | Japanese | Co.Het. | No | Ohto et al., 200426 |
| 4 | c.2303C>T; p.Pro767Leu#c.1660delC; p.Arg554Gly fs104X | 1.000, Pr.D- | 0.03, D- | Japanese | Co.Het. | No | Tanaka et al., 201227 |
| 5 | c.2303C>T; p. Pro767Leu#c.851-33T>A | 1.000, Pr.D- | 0.03, D- | Korean | Co.Het. | No | Jung et al., 201314 |
| 6 | c.574 + 1G>Ac.2206-2A>G | – | – | Spanish | Co.Het. | No | Sarasola et al., 201128 |
| 7 | c.1633–1G>T | - | - | Turkish | Homo. | No | Tuysuz et al., 200810 |
| 8 | c.1633–1G>T | - | - | Turkish | Homo. | Yes | Tuysuz et al., 200810 |
| 9 | c.G1247A; p.Glu388Lysc.C2429T; p.Gln782X | 0.883, Po.D- | 0.66, T- | Swedish | Co.Het. | Yes | Wieczorek et al., 200829 |
| 10 | c.1561T>C; p.Phe520Leu#c.2057G>A; p.Arg685His# | 0.993, Pr.D; 0.103, B | 0.00, D; 0.00, D | Chinese | Co.Het. | Yes | Gao et al., 201330 |
| 11 | c.1635G>C; p.Ala526Pro#c.2197G>A; p.Gly713Asp# | 0.999, Pr.D; 1.000, Pr.D | 0.13, T; 0.00, D | Chinese | Co.Het. | Yes | Wang et al., 201531 |
| 12 | c.1715T>G; p.Ile571Ser# | 1.000, Pr.D | 0.00, D | Turkish | Homo. | Yes | Li et al., 201215 |
| 13 | c.1825A>G; Met586Val# | 0.987, Pr.D | 0.01, D | Japanese | Homo. | Yes | Yotsumoto et al., 199932 |
| 14 | c.1945C>T; Arg648Trp#c.44G>A; Trp15X | 1.000, Pr.D- | 0.00, D- | Chinese | Co.Het. | Yes | Li et al., 201215 |
| 15 | c.2001C>T; p.Arg653Cys# | 0.989, Pr.D | 0.00, D | Turkish | Homo. | Yes | Guven et al., 201433 |
| 16 | c.2150C>T; Pro689Leu | 1.000, Pr.D | 0.00, D | Israeli | Homo. | Yes | Shatzky et al., 200034 |
| 17 | c.2209G>C; Val709Leu#c.2236G>A; Glu718Ser# | No Data; 0.997, Pr.D | 0.33, T; 0.01, D | Malaysia | Co. Het. | Yes | Shalimar et al., 200735 |
| 18 | c.2150T>G; p.Leu716Arg#c.1672C>T; p.Gln557X# | 0.999, Pr.D- | 0.00, D- | French | Co.Het. | Yes | Huehne et al., 200836 |
| 19 | c.2155G>A; p.Glu718Lys#c.287 + 2dupT | 0.995, Pr.D- | 0.00, D- | Korean | Co.Het. | Yes | Lee et al., 200916 |
| 20 | c.2206-11G>A; p.Glu734_Ala735insTrpProGln# | – | – | Turkish | Homo. | Yes | Yis et al., 201537 |
| 21 | c.783_785delGAA; p. 261delLys | - | - | Saudi | Homo. | Yes | Algahtani et al., 201638 |
| 22 | c.1970T>C; p.Leu657Pro | 0.786, Po.D | 0.00, D | Pakistani | Homo. | Yes | Shaikh et al., 201739 |
| 23 | c.2311C>T; p.Arg771Cys | 1.000, Pr.D | 0.00, D | Indian | Homo. | Yes | Shaikh et al., 201739 |
| 24 | c.963delG; p.Leu322Serfs*148c.851-33T>A | – | – | Chinese | Co.Het. | Yes | Wang et al., 201640 |
| 25 | c.1786G>A; p.Asp596Asnc.2350_2363del; p.Leu784Serfs*79 | 0.999, Pr.D- | 0.10, T- | Korean | Co.Het. | Yes | Nam et al., 201713 |
| 26 | c.704C>G; p.Ser235*c.2020G>T, p.Asp674Tyr | -0.879, Po.D | -0.00, D | Korean | Co.Het. | Yes | Nam et al., 201713 |
CIPA, congenital insensitivity to pain with anhidrosis; Pt., patient; Homo., homozygous; Co.Het., compound heterozygous; Pr.D, probably damaging; Po.D, possibly damaging; D, damaging; T, tolerated; B, benign
Notes: Patients who were Homo. or Co.Het. with frameshift mutations, premature stop mutations, and verified splice site mutations were not included in this table because of their highly deleterious effect with all patients exhibiting intellectual disability.
*Polyphen2 and SIFT scores and predictions (Pr.D, Po.D, D, T, and B) are given only for missense mutation. Discordances (patients without intellectual disability who had highly deleterious missense mutations or those with intellectual disability who had at least one possibly damaging or tolerable mutation) are in italics.
Because of the alternative splicing of NTRK1, different nomination systems were employed among the literature. Here, we mapped these mutations to the protein structure, in which the residue number may differ from that in the corresponding article.
-Data not available for frameshift or intron mutations.
In silico investigation of heterogeneity
We hypothesized that if the mutation on one of the two NTRK1 alleles is benign or tolerable, the patient may have less severe symptoms. We used the Polyphen2 (Harvard Medical School, Boston, MA, USA; http://genetics.bwh.harvard.edu/pph2/) and SIFT (J. Craig Venter Institute, La Jolla, CA, USA; http://sift.jcvi.org/) scores to predict the influence of a mutation. As shown in Table 2, these predictions were not correlated with the patients’ phenotype with respect to intellectual disability, as shown in italics. We then examined whether any observable association was present between the mutations and intellectual disability with an available Ntrk1 protein structure.
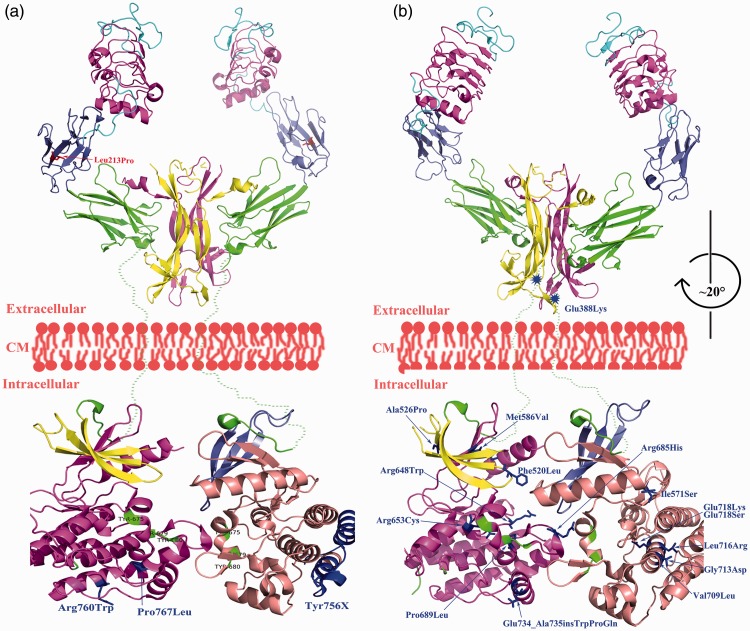
Figure 3(a) shows missense mutations (Table 2, Patients 1–5) in patients with CIPA without intellectual disability. As shown in the table, the point mutations p.Leu213Pro, Arg760Trp, and p.Pro767Leu were all located peripherally without apparent involvement in NGF binding, ATP binding, or interfaces of the TKD. In addition, leucine and proline are of similar structure with respect to their nonpolar side chains, which may have less effect on protein function. Because CIPA is a recessive disease, one copy of near fully functional genes will relieve the patient’s symptoms. The premature stop mutation p.Tyr756Ter caused Ntrk1 protein truncation of 40 amino acids at the C-terminal. This part of the Ntrk1 protein is located at a region distal to the kinase interface, which may be another reason that these children were not intellectually compromised.
Figure 3.
Observable association between mutations and the presence of intellectual disability. Panel A: At least one mutation reported in patients with CIPA without intellectual disability involved peripheral amino acids of the Ntrk1 protein. Panel B: Missense mutations in patients with CIPA with intellectual disability altered critical domains of the Ntrk1 protein. (Rotated by about 20º clockwise from the front view.)
Figure 3(b) shows missense mutations (Table 2, Patients 9–20) in patients with CIPA with apparent intellectual disability. All mutations were located in the functional domains; i.e., NFG binding site, ATP binding site, and interfaces of the TKD. Thus, we conclude that mutations located in functional domains of NTRK1 contribute to the observed intellectual disability.
In the present case, the c.851-33T>A mutation of NTRK1 caused a frame shift after the 283rd amino acid and a premature stop codon at the 319th codon,41 deleting the whole transmembrane domain, ATP binding site, and TKD. This mutation is highly deleterious and would be expected to cause intellectual disability in this patient.
Discussion
To our knowledge, this is the first case of CIPA in a north Han Chinese patient caused by a homozygous c.851-33T>A mutation that resulted in aberrant splicing and an open reading frame shift. We also reviewed all published Ntrk1 CIPA mutations in the literature and postulate that the presence of intellectual disability is determined by the location of the Ntrk1 mutation with respect to the functional protein domain. Missense mutations located peripheral to the Ntrk1 protein that do not jeopardize important domains (i.e., NGF binding site, ATP binding site, and TKD) tend to cause milder symptoms, usually without intellectual disability. Frame shift mutations, premature stop mutations, splice site mutations, and missense mutations that involve critical amino acids in the protein all cause severe intellectual disability.
However, several reports appear to be discordant. Sarasola et al.28 described a 6-year-old Spanish girl with c.574 + 1G>A and c.2206-2A>G compound heterozygous mutations (Table 2, Patient 6) who was cognitively normal. The author verified the influence of the two splice site mutations with reverse transcription PCR. The c.574 + 1G>A mutation caused exon 5 skipping and predicted Ntrk1 protein truncation after residue 164. The c.2206-2A>G mutation caused two alternative splicings, and the predicted proteins were Ala736_Gln742 interstitial deletion and truncation after residue 735, respectively. Tuysuz et al.10 reported three Turkish patients with CIPA with a homozygous c.1633–1G>T mutation, one 2.5-year-old boy without psychomotor retardation, and one 14-year-old boy and one 8-year-old girl with psychomotor retardation (Table 2, Patients 7 and 8). This discrepancy indicates that there may be other factors modifying the phenotypes of CIPA. The functional impact of the variant, including dimerization, autophosphorylation, PLCγ activity, and toxicity of the mutated protein to the cell should be validated to confirm the pathogenicity of the variant.
Conclusions
In this study, we observed that intellectual disability in patients with CIPA appears to be determined by which Ntrk1 protein domain a mutation has jeopardized. Although this observation method is qualitative, it provides another alternative to evaluate the function of a missense mutation in addition to the Polyphen2 and SIFT scores. We are awaiting the delineation of additional cases of CIPA to provide additional evidence supporting our observations.
Acknowledgments
The authors thank Dr. Uluc Yis from the Division of Child Neurology, Department of Pediatrics, School of Medicine, Dokuz Eylul University, Izmir, Turkey for sharing their full-text article, which added rationality to our analysis.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
Funding
This research was funded by the National Natural Science Foundation of China (81501852, 81472046, 81472045), Beijing Natural Science Foundation (7172175), Beijing Nova Program (Z161100004916123), Beijing Nova Program Interdisciplinary Collaborative Project (xxjc201717), 2016 Milstein Medical Asian American Partnership Foundation Fellowship Award in Translational Medicine, The Central Level Public Interest Program for Scientific Research Institute (2016ZX310177), PUMC Youth Fund & the Fundamental Research Funds for the Central Universities (3332016006), CAMS Initiative Fund for Medical Sciences (2016-I2M-3-003, 2016-I2M-2-006), the Distinguished Youth Foundation of Peking Union Medical College Hospital (JQ201506), and the 2016 PUMCH Science Fund for Junior Faculty (PUMCH-2016-1.1).
References
- 1.Axelrod FB, Chelimsky GG, Weese-Mayer DE. Pediatric autonomic disorders. Pediatrics 2006; 118: 309–321. [DOI] [PubMed] [Google Scholar]
- 2.Indo Y, Tsuruta M, Hayashida Y, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet 1996; 13: 485–488. [DOI] [PubMed] [Google Scholar]
- 3.Einarsdottir E, Carlsson A, Minde J, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet 2004; 13: 799–805. [DOI] [PubMed] [Google Scholar]
- 4.Kuwako K, Hosokawa A, Nishimura I, et al. Disruption of the paternal necdin gene diminishes TrkA signaling for sensory neuron survival. J Neurosci 2005; 25: 7090–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Indo Y. Nerve growth factor and the physiology of pain: lessons from congenital insensitivity to pain with anhidrosis. Clin Genet 2012; 82: 341–350. [DOI] [PubMed] [Google Scholar]
- 6.Mardy S, Miura Y, Endo F, et al. Congenital insensitivity to pain with anhidrosis: novel mutations in the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor. Am J Hum Genet 1999; 64: 1570–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-On E, Weigl D, Parvari R, et al. Congenital insensitivity to pain. Orthopaedic manifestations. J Bone Jt Surg Br 2002; 84: 252–257. [DOI] [PubMed] [Google Scholar]
- 8.Amano A, Akiyama S, Ikeda M, et al. Oral manifestations of hereditary sensory and autonomic neuropathy type IV. Congenital insensitivity to pain with anhidrosis . Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1998; 86: 425–431. [DOI] [PubMed] [Google Scholar]
- 9.Gabreels-Festin A. Hereditary neuropathies in childhood: morphologic hallmarks and pathophysiologic mechanisms. J Child Neurol 1999; 14: 52–53. [PubMed] [Google Scholar]
- 10.Tuysuz B, Bayrakli F, DiLuna ML, et al. Novel NTRK1 mutations cause hereditary sensory and autonomic neuropathy type IV: demonstration of a founder mutation in the Turkish population. Neurogenetics 2008; 9: 119–125. [DOI] [PubMed] [Google Scholar]
- 11.Verpoorten N, Claeys KG, Deprez L, et al. Novel frameshift and splice site mutations in the neurotrophic tyrosine kinase receptor type 1 gene (NTRK1) associated with hereditary sensory neuropathy type IV. Neuromuscul Disord 2006; 16: 19–25. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Wu N, Liu J, et al. Novel NTRK1 Frameshift Mutation in Congenital Insensitivity to Pain With Anhidrosis. J Child Neurol 2015; 30: 1357–1361. [DOI] [PubMed] [Google Scholar]
- 13.Nam TS, Li W, Yoon S, et al. Novel NTRK1 mutations associated with congenital insensitivity to pain with anhidrosis verified by functional studies. J Peripher Nerv Syst 2017; 22: 92–99. [DOI] [PubMed] [Google Scholar]
- 14.Jung CL, Ki CS, Kim BJ, et al. Atypical hereditary sensory and autonomic neuropathy type IV with neither mental retardation nor pain insensitivity. J Child Neurol 2013; 28: 1668–1672. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Liang JY, Sun ZH, et al. Novel nonsense and frameshift NTRK1 gene mutations in Chinese patients with congenital insensitivity to pain with anhidrosis. Genet Mol Res 2012; 11: 2156–2162. [DOI] [PubMed] [Google Scholar]
- 16.Lee ST, Lee J, Lee M, et al. Clinical and genetic analysis of Korean patients with congenital insensitivity to pain with anhidrosis. Muscle Nerve 2009; 40: 855–859. [DOI] [PubMed] [Google Scholar]
- 17.Guo YC, Liao KK, Soong BW, et al. Congenital insensitivity to pain with anhidrosis in Taiwan: a morphometric and genetic study. Eur Neurol 2004; 51: 206–214. [DOI] [PubMed] [Google Scholar]
- 18.Lv F, Xu X jie, Song Y wen, et al. Recurrent and novel mutations in the NTRK1 gene lead to rare congenital insensitivity to pain with anhidrosis in two Chinese patients. Clin Chim Acta 2017; 468: 39–45. [DOI] [PubMed] [Google Scholar]
- 19.Miura Y, Mardy S, Awaya Y, et al. Mutation and polymorphism analysis of the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor in congenital insensitivity to pain with anhidrosis (CIPA) families. Hum Genet 2000; 106: 116–124. [DOI] [PubMed] [Google Scholar]
- 20.Wehrman T, He X, Raab B, et al. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 2007; 53: 25–38. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand T, Kothe M, Liu J, et al. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol 2012; 423: 439–453. [DOI] [PubMed] [Google Scholar]
- 22.Stephens RM, Loeb DM, Copeland TD, et al. Trk receptors use redundant signal transduction pathways involving SHC and PLC-gamma 1 to mediate NGF responses. Neuron 1994; 12: 691–705. [DOI] [PubMed] [Google Scholar]
- 23.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc L B Biol Sci 2006; 361: 1545–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonkowsky JL, Johnson J, Carey JC, et al. An infant with primary tooth loss and palmar hyperkeratosis: a novel mutation in the NTRK1 gene causing congenital insensitivity to pain with anhidrosis. Pediatrics 2003; 112(3 Pt 2): e237–e241. [DOI] [PubMed] [Google Scholar]
- 25.Indo Y. Molecular basis of congenital insensitivity to pain with anhidrosis (CIPA): mutations and polymorphisms in TRKA (NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Hum Mutat 2001; 18: 462–471. [DOI] [PubMed] [Google Scholar]
- 26.Ohto T, Iwasaki N, Fujiwara J, et al. The evaluation of autonomic nervous function in a patient with hereditary sensory and autonomic neuropathy type IV with novel mutations of the TRKA gene. Neuropediatrics 2004; 35: 274–278. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Satoh T, Tanaka A, et al. Congenital insensitivity to pain with anhidrosis: a case with preserved itch sensation to histamine and partial pain sensation. Br J Dermatol 2012; 166: 888–891. [DOI] [PubMed] [Google Scholar]
- 28.Sarasola E, Rodriguez JA, Garrote E, et al. A short in-frame deletion in NTRK1 tyrosine kinase domain caused by a novel splice site mutation in a patient with congenital insensitivity to pain with anhidrosis. BMC Med Genet 2011; 12: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieczorek S, Bergstrom J, Saaf M, et al. Expanded HSAN4 phenotype associated with two novel mutations in NTRK1. Neuromuscul Disord 2008; 18: 681–684. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Guo H, Ye N, et al. Oral and craniofacial manifestations and two novel missense mutations of the NTRK1 gene identified in the patient with congenital insensitivity to pain with anhidrosis. PLoS One 2013; 8: e66863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Guo S, Duan G, et al. Novel and novel de novo mutations in NTRK1 associated with congenital insensitivity to pain with anhidrosis: a case report. Med 2015; 94: e871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yotsumoto S, Setoyama M, Hozumi H, et al. A novel point mutation affecting the tyrosine kinase domain of the TRKA gene in a family with congenital insensitivity to pain with anhidrosis. J Invest Dermatol 1999; 112: 810–814. [DOI] [PubMed] [Google Scholar]
- 33.Guven Y, Altunoglu U, Aktoren O, et al. Twins with hereditary sensory and autonomic neuropathy type IV with preserved periodontal sensation. Eur J Med Genet 2014; 57: 240–246. [DOI] [PubMed] [Google Scholar]
- 34.Shatzky S, Moses S, Levy J, et al. Congenital insensitivity to pain with anhidrosis (CIPA) in Israeli-Bedouins: genetic heterogeneity, novel mutations in the TRKA/NGF receptor gene, clinical findings, and results of nerve conduction studies. Am J Med Genet 2000; 92: 353–360. [DOI] [PubMed] [Google Scholar]
- 35.Shalimar A, Sharaf I, Farah WI, et al. Congenital insensitivity to pain with anhydrosis in a Malaysian family: a genetic analysis. J Orthop Surg (Hong Kong) 2007; 15: 357–360. [DOI] [PubMed] [Google Scholar]
- 36.Huehne K, Zweier C, Raab K, et al. Novel missense, insertion and deletion mutations in the neurotrophic tyrosine kinase receptor type 1 gene (NTRK1) associated with congenital insensitivity to pain with anhidrosis. Neuromuscul Disord 2008; 18: 159–166. [DOI] [PubMed] [Google Scholar]
- 37.Yis U, Mademan I, Kavukcu S, et al. A novel NTRK1 mutation in a patient with congenital insensitivity to pain with anhidrosis. Acta Neurol Belg 2015; 115: 509–511. [DOI] [PubMed] [Google Scholar]
- 38.Algahtani H, Naseer MI, Al-Qahtani M, et al. Congenital insensitivity to pain with anhidrosis: A report of two siblings with a novel mutation in (TrkA) NTRK1 gene in a Saudi family. J Neurol Sci 2016; 370: 35–38. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh SS, Chen YC, Halsall SA, et al. A Comprehensive Functional Analysis of NTRK1 Missense Mutations Causing Hereditary Sensory and Autonomic Neuropathy Type IV (HSAN IV). Hum Mutat 2017; 38: 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang QL, Guo S, Duan G, et al. Phenotypes and Genotypes in Five Children with Congenital Insensitivity to Pain with Anhidrosis. Pediatr Neurol 2016; 61: 63–69. [DOI] [PubMed] [Google Scholar]
- 41.Miura Y, Hiura M, Torigoe K, et al. Complete paternal uniparental isodisomy for chromosome 1 revealed by mutation analyses of the TRKA (NTRK1) gene encoding a receptor tyrosine kinase for nerve growth factor in a patient with congenital insensitivity to pain with anhidrosis. Hum Genet 2000; 107: 205–209. [DOI] [PubMed] [Google Scholar]