Abstract
The DNA repair protein O6-alkylguanine alkyltransferase (AGT) is responsible for removing promutagenic alkyl lesions from exocyclic oxygens located in the major groove of DNA, i.e. the O6 and O4 positions of guanine and thymine. The protein carries out this repair reaction by transferring the alkyl group to an active site cysteine and in doing so directly repairs the premutagenic lesion in a reaction that inactivates the protein. In order to trap a covalent AGT–DNA complex, oligodeoxyribonucleotides containing the novel nucleoside N1,O6-ethanoxanthosine (eX) have been prepared. The eX nucleoside was prepared by deamination of 3′,5′-protected O6-hydroxyethyl-2′-deoxyguanosine followed by cyclization to produce 3′,5′-protected N1,O6-ethano-2′-deoxyxanthosine, which was converted to the nucleoside phosphoramidite and used in the preparation of oligodeoxyribonucleotides. Incubation of human AGT with a DNA duplex containing eX resulted in the formation of a covalent protein–DNA complex. Formation of this complex was dependent on both active human AGT and eX and could be prevented by chemical inactivation of the AGT with O6-benzylguanine. The crosslinking of AGT to DNA using eX occurs with high yield and the resulting complex appears to be well suited for further biochemical and biophysical characterization.
INTRODUCTION
The alkylation of exocyclic oxygens located in the major groove of DNA, i.e. the O6 position of guanine and to a lesser extent the O4 position of thymine, is promutagenic and can be carcinogenic. While alkylation at the O6 position of guanine typically constitutes a small fraction of the damage produced by exogenous alkylators it is principally responsible for the GC→AT transition mutations associated with mutagenic and carcinogenic effects of many alkylating agents (1). O6-alkylguanine-DNA alkyltransferase (AGT) is an evolutionarily conserved DNA repair protein that is the principle cellular mechanism responsible for repair of O6-alkylguanine (see 2–6 and references therein). AGT carries out the repair reaction by transferring the alkyl group from the exocyclic oxygen to an active site cysteine in an SN2-like transfer. This alkyl transfer results in the formation of an alkyl cysteine and inactivation of the protein. AGT therefore is a stoichiometric DNA repair molecule rather than a true enzyme. These alkyltransferases include the Escherichia coli proteins Ada (the C-terminal domain) and Ogt, as well as homologs identified from eukaryotic, prokaryotic and eubacterial sources (7).
First identified as a component of the adaptive response to alkylation damage in E.coli, alkyltransferase activity presumably evolved to protect cellular DNA from the effects of endogenous sources of alkylation. It is, however, the ability of AGT to attenuate the effect of antitumor compounds like methylating agents (e.g. dacarbazine, procarbazine and temozolomide) and chloroethylating agents [e.g. bis-chloroethylnitrosourea (BCNU), clomesone and fotemustine] that has prompted much of the recent interest in AGT (8–10). Despite detailed biochemical characterization, recent structural data and extensive screening of AGT inhibitors, the inability to prepare AGT–DNA complexes suitable for structural characterization has hampered our understanding of how these DNA repair proteins recognize and repair damaged DNA.
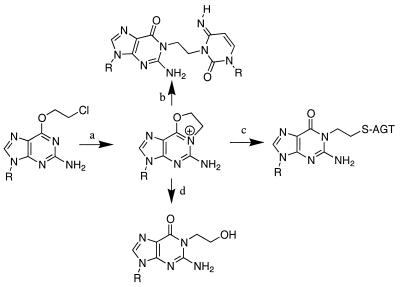
We here describe a strategy for covalently trapping a post-repair AGT–DNA complex. This strategy is based on the observation that covalent protein–DNA complexes form when chloroethylnitrosourea (CENU)-treated DNA is incubated with AGT (11). The CENUs are a class of bifunctional crosslinking agents that produce interstrand crosslinks via N1,O6-ethanoguanine (eG), an activated compound that results from intramolecular cyclization of the initial chloroethylation at the O6 position of guanine (12). As shown in Figure 1, intramolecular cyclization of the initial CENU alkylation product O6-chloroethylguanine (pathway a) gives rise to eG. The positively charged eG is highly reactive and can undergo nucleophilic attack by the cytosine located on the opposite strand of DNA (pathway b) to produce a G-C interstrand crosslink (12). This G-C crosslink is believed to be the principal cytotoxic lesion produced by CENU. The observation that DNA treated with BCNU in the presence of AGT produced a small amount of covalently trapped AGT–DNA suggested that eG might be a substrate for AGT (13–15). The ‘repair’ of eG (pathway c) results in a covalently trapped protein–DNA complex. Characterization of the crosslinked species confirmed the structure of the covalent protein–DNA crosslink and suggested that crosslinking occurred as a result of nucleophilic attack by the active site cysteine on the O6-CH2- portion of the ethano bridge (11). The hyper-reactivity of eG also results in its hydrolysis (pathway d), producing N1-hydroxyethylguanine (16).
Figure 1.
Formation and reactions of N1,O6-ethanoguanine. Intramolecular cyclization of the initial chloroethylnitrosourea adduct (a) generates the reactive ethano intermediate which can crosslink DNA (b), be ‘repaired’ by AGT (c) or react with water (d).
The ability of the N1,O6-ethano bridge to be both a substrate for AGT and a covalent tether is the basis of our crosslinking strategy. Unfortunately, the hydrolytic sensitivity of eG as well as its incompatibility with the methods associated with automated synthesis of oligodeoxyribonucleotides greatly reduce its potential as a mechanism-based crosslinker. Therefore, we set out to design a nucleoside analog that would retain the major groove structure of eG but be stable enough to withstand both chemical and enzymatic manipulation. Replacement of the exocyclic amino group of guanine with oxygen to give N1,O6-ethanoxanthine (eX) (Fig. 2) satisfied these conditions. This xanthine derivative maintains the major groove structure while taking advantage of the uncharged tautomeric form accessible to xanthine.
Figure 2.
Structures of N1,O6-ethanoguanine (eG) and N1,O6-ethanoxanthine (eX).
Below we report the synthesis of the nucleoside phosphoramidite of eX, the preparation of oligodeoxyribonucleotides containing eX and the formation of covalent protein–DNA complexes between human AGT and an oligodeoxyribonucleotide containing a single eX.
MATERIALS AND METHODS
Materials
2′-Deoxyguanosine was purchased from US Biochemicals (Cleveland, OH). All other reagents were purchased from Aldrich Chemical Co. (Milwaukee, WI). Anhydrous solvents were used as supplied. TLC was conducted on EM plastic-backed sheets (silica gel 60 F254, 0.2 mm). The 5′-O-dimethoxytrityl-2′-deoxyribonuceoside-3′-O-(β-cyanoethyl-N,N-diisopropyl)phosphoramidites of N-acetyl-2′-deoxycytidine, N-phenoxyacetyl-2′-deoxyadenosine and N-(4-isopropyl-phenoxyacetyl)-2′-deoxyguanosine and thymidine as well as the appropriately protected deoxyribonucleoside controlled pore glass supports were purchased from Glen Research Inc. (Sterling, VA). Anhydrous acetonitrile for use in the oligodeoxyribonucleotide synthesis was stored over calcium hydride. The enzymes shrimp alkaline phosphatase (SAP) and T4 polynucleotide kinase (PNK) were purchased from New England Biolabs (Beverly, MA); snake venom phosphodiesterase (SVP) from Crotalus durissus was purchased from Boehringer Mannheim (Indianapolis, IN). Human AGT was a gift from Dr Phil Potter (St Jude).
General procedures
Flash column chromatography was performed on Merck silica gel 60, 230–400 mesh. Absorption spectra were recorded using a Varian DMS100S UV/Vis spectrophotometer. HPLC separations and analyses were conducted on a Varian 9010/50 system. 1H, 13C and 31P NMR were performed on a Bruker AMX300 spectrometer; chemical shifts are in p.p.m. and proton chemical shifts are reported relative to internal tetramethylsilane (CDCl3) or to residual solvent peaks (DMSO); 31P spectra were collected with broadband proton noise decoupling and referenced to an external 85% H3PO4 standard. High resolution mass spectrometry (MALDI-FTMS) was performed by the Scripps Research Institute Mass Spectrometry Facility.
Synthesis of 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-2′-deoxyguanosine (3)
The 3′- and 5′-hydroxyls of commercially available 2′-deoxyguanosine were protected using t-butyldimethylsilylchloride (17). 3′,5′-TBDMSi-dG (2) (3 g, 6 mmol) was dried by rotary evaporation (in vacuo) with anhydrous pyridine, dissolved in 60 ml of anhydrous pyridine and cooled in an ice bath. To this stirred mixture, trifluoroacetic anhydride (3.0 ml, 21 mmol) was added drop-wise. After stirring for 30 min, 150 ml of a freshly prepared solution of sodium in ethylene glycol (12.5 g sodium in 250 ml of anhydrous ethylene glycol dissolved in 3 g batches over a 24 h period) was added. The reaction was stirred overnight at room temperature. The resulting reddish-brown mixture was poured into 500 ml of water and extracted with four 200 ml portions of chloroform. The combined organic layers were dried over sodium sulfate and, following filtration, the solvent was removed by rotary evaporation (in vacuo). Traces of pyridine were removed by co-evaporation with two 50 ml portions of toluene. The resulting oil was dissolved in 10 ml of chloroform/methanol (95:5) and purified by flash column chromatography on a silica gel column using chloroform/methanol (95:5). Fractions containing product were pooled and the solvent removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-2′-deoxyguanosine (3) as a yellow foam (2.75 g, 5.4 mmol; yield 90%). UVmax 279, 247 in CH3OH; Rf 0.15 in CHCl3/CH3OH (95:5); 1H NMR (d6-DMSO) δ (p.p.m.) 8.05 [s, 1.0H, H8], 6.42 [s, 1.9H, N2H2], 6.21 [t, 1.0H, H1′], 4.89 [t, 0.5H, O6CH2CH2OH], 4.52 [m, 1.0H, H3′], 4.41 [t, 1.9H, O6CH2CH2OH], 3.82 [m, 1.0H, H4′], 3.62–3.77 [m, 3.9H, H5′H5′′/O6CH2CH2OH], 2.72 [m, 1.0, H2′], 2.27 [m, 1.0H, H2′], 0.89–0.87 [s&s, 17.7H, (CH3)3C(CH3)2Si], 0.10 [s, 5.8H, (CH3)3C(CH3)2Si], 0.04 [s, 5.8H, (CH3)3C(CH3)2Si].
Synthesis of 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-2′-deoxyxanthosine (4)
Deamination was accomplished by dissolving 3 (2.75 g, 5.4 mmol) in 10 ml of acetone, which was transferred to a reaction vessel containing a solution of NaNO3 (10 g) in 30 ml of water. To this mixture 15 ml of glacial acetic acid was added and the solution was vigorously agitated until the evolution of nitrogen ended, ∼2 h. The reaction was neutralized by adding sodium bicarbonate in batches, poured into 500 ml of water and extracted with two 200 ml portions of chloroform. The combined organic layers were dried over sodium sulfate and, after filtration, the solvent removed by rotary evaporation (in vacuo). The resulting foam was dissolved in 5 ml of chloroform/methanol (95:5) and purified by flash column chromatography on a silica gel column using chloroform/methanol (95:5). Fractions containing product were pooled and solvent removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-2′-deoxyxanthosine (4) as a tan foam (1.76 g, 3.5 mmol; yield 65%). UVmax 266, 237 in CH3OH; Rf 0.38 in CHCl3/CH3OH (90:10); 1H NMR (d6-DMSO) δ (p.p.m.) 11.57 [bs, 0.4H, N3H], 8.22 [s, 1.0H, H8], 6.25 [t, 1.0H, H1′], 4.91 [bs, 0.3H, O6CH2CH2OH], 4.60 [m, 1.0H, H3′], 4.47 [t, 2.0H, O6CH2CH2OH], 3.82 [m, 1.0H, H4′], 3.78–3.62 [m, 3.9H, H5′H5″/O6CH2CH2OH], 2.77 [m, 1.0, H2′], 2.30 [m, 1.0H, H2′], 0.89–0.86 [s&s, 17.7H, (CH3)3C(CH3)2Si], 0.11 [s, 5.8H, (CH3)3C(CH3)2Si], 0.02 [d, 5.8H, (CH3)3C(CH3)2Si].
Synthesis of 3′,5′-t-butyldimethylsilyl-O6-(dimethoxytrityl-O-ethyl)-2′-deoxyxanthosine (5)
4 (1.7 g, 3.4 mmol) was dried by rotary evaporation (in vacuo) with anhydrous pyridine and dissolved in 20 ml of anhydrous pyridine. Dimethoxytrityl chloride (1.5 equiv., 1.7 g) was added and the reaction stirred overnight at room temperature. The reaction was quenched by addition of 3 ml of 95% ethanol and allowed to stir for 15 min. The reaction mixture was concentrated by rotary evaporation (in vacuo), dissolved in 200 ml of ethyl acetate and extracted with two 200 ml portions of 5% sodium bicarbonate and one 200 ml portion of a saturated NaCl solution. The resulting organic layer was dried over sodium sulfate and, after filtration, the solvent removed by rotary evaporation (in vacuo). The resulting foam was dissolved in 5 ml chloroform/methanol (99:1) with a trace of triethylamine and purified by flash column chromatography using chloroform/methanol (99:1) in the presence of trace triethylamine. Fractions containing product were pooled and the solvent was removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-O6-(dimethoxytrityl-O-ethyl)-2′-deoxyxanthosine (5) (1.9 g, 2.35 mmol; yield 70%). 1H NMR (d6-DMSO) δ (p.p.m.) 11.61 [bs, 0.6H, N3H], 8.29 [s, 0.8H, H8], 7.39–6.81 [m, 12.9, Ph-H], 6.27 [t, 1.0H, H1′], 4.68 [bs, 1.9H, O6CH2CH2OH], 4.57 [m, 1.0H, H3′], 3.77 [m, 1.0H, H4′], 3.76–3.63 [m, 7.9H, H5′H5′′/Ph-OCH3], 3.3 [bs, 1.5H, O6CH2CH2OH], 2.80 [m, 1.0, H2′], 2.32 [m, 1.0H, H2′], 0.96–0.80 [s&s, 18.1H, (CH3)3C(CH3)2Si], 0.12 [s, 5.1H, (CH3)3C(CH3)2Si], 0.02 [d, 5.0H, (CH3)3C(CH3)2Si].
Synthesis of 3′,5′-t-butyldimethylsilyl-O6-(dimethoxytrityl-O-ethyl)-O2-nitrophenethyl-2′-deoxyxanthosine (6)
5 (1.9 g, 2.35 mmol) was dissolved in 25 ml of anhydrous 1,4-dioxane and stirred at room temperature. To the reaction vessel 3 equiv. of triphenylphosphine (1.85 g), 3 equiv. of diethyl azodicarboxylate (1.11 ml) was added, followed by 3 equiv. of nitrophenylethanol (1.17 g). Following a rapid evolution of heat, the reaction vessel was stirred overnight at room temperature. The reaction was quenched by the addition of 5 ml of 95% ethanol and stirred for 5 min. The solvents were removed by rotary evaporation (in vacuo). The resulting oil was dissolved in 5 ml of chloroform and purified using flash column chromatography on silica gel using chloroform. Pure fractions were pooled; fractions containing product and contaminating reaction components were pooled, dried and re-chromatographed. Fractions containing product were pooled and the solvent was removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-O6-(dimethoxytrityl-O-ethyl)-O2-nitrophenethyl-2′-deoxyxanthosine (6) as a white foam (1.4 g, 1.46 mmol; yield 62%). 1H NMR (d6-DMSO) δ (p.p.m.) 8.36 [s, 1.0H, H8], 8.14 [d, 1.7H, Ph-H], 7.74–6.78 [m, 19.4, Ph-H], 6.30 [t, 1.0H, H1′], 4.71 [bs, 2.1H, O6CH2CH2OH], 4.56 [m, 3.0H, H3′, O2-CH2CH2-Ar], 3.83 [m, 1.1H, H4′], 3.73–3.63 [m, 8.8H, H5′H5″/Ph-OCH3/O2-CH2CH2-Ar], 3.3 [t, 2.5H, O6CH2CH2OH], 2.91 [m, 1.3, H2′], 2.31 [m, 1.1H, H2′], 0.95–0.80 [s&s, 20.6H, (CH3)3C(CH3)2Si], 0.12 [s, 5.8H, (CH3)3C(CH3)2Si], –0.02 [d, 5.6H, (CH3)3C(CH3)2Si].
Synthesis of 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-O2-nitrophenethyl-2′-deoxyxanthosine (7)
6 (1.4 g, 1.46 mmol) was dissolved in 3 ml of dichloromethane and transferred to a separation funnel. To the separation funnel was added 50 ml of 2.5% dichloroacetic acid in dichloromethane. The solution turned bright orange and was allowed to stand for 5 min. Water (50 ml) was added to the separation funnel followed by small batches of solid sodium bicarbonate. Following agitation the pH of the aqueous layer was checked and sodium bicarbonate was added until the solution was neutralized. The organic layer was collected and washed with two 50 ml portions of 5% sodium bicarbonate and one 50 ml portion of a saturated NaCl solution. The organic layer was dried over sodium sulfate and, after filtration, the solvent removed by rotary evaporation (in vacuo). The resulting foam was dissolved in 5 ml of chloroform and purified by flash column chromatography on a silica gel column using chloroform/methanol (99:1). Fractions containing product were pooled and solvent removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-O6-hydroxyethyl-O2-nitrophenethyl-2′-deoxyxanthosine (7) (700 mg, 1.07 mmol; yield 73%). 1H NMR (d6-DMSO) δ (p.p.m.) 8.31 [s, 1.3H, H8], 8.17 [d, 1.6H, Ph-H], 6.28 [t, 1.0H, H1′], 4.93 [t, 0.7H, O6CH2CH2OH], 4.58 [m, 3H, H3′, O2-CH2CH2-Ar], 4.49 [t, 2.0H, O6CH2CH2OH], 3.83–3.64 [m, 5.3H, H4′, H5′/H5″], 2.89 [m, 1.2, H2′], 2.31 [m, 1.1H, H2′], 0.88–0.84 [s&s, 19.0H, (CH3)3C(CH3)2Si], 0.09 [s, 5.5H, (CH3)3C(CH3)2Si], –0.01 [d, 5.2H, (CH3)3C(CH3)2Si].
Synthesis of 3′,5′-t-butyldimethylsilyl-N1,O6-ethano-2′-deoxyxanthosine (9)
7 (700 mg, 1.07 mmol) was dried by rotary evaporation (in vacuo) with anhydrous pyridine and dissolved in 20 ml of anhydrous pyridine and 2,4,6-triisopropylbenzenesulphonyl chloride (4.5 equiv., 1.5 g) was added. The reaction was allowed to stir at room temperature for 2 h; an additional equivalent of 2,4,6-triisopropylbenzenesulphonyl chloride was required to get complete conversion of the starting material as measured by TLC. The reaction was quenched by the addition of 20 ml of absolute ethanol. The resulting solution was concentrated by rotary evaporation (in vacuo) to give an oil. The oil was dissolved in 5 ml of pyridine and 1,8-diazabicyclo[5.4.0]-undec-7-ene (DBU) was added to give a final concentration of 0.5 M. The reaction was stirred for 3 h. The reaction was concentrated by rotary evaporation (in vacuo). The resulting oil was dissolved in 50 ml of ethyl acetate and washed with two 50 ml portions of a 5% sodium bicarbonate solution and one 50 ml portion of a saturated NaCl solution. The organic layer was dried over sodium sulfate and, after filtration, the solvent was removed by rotary evaporation. The resulting oil was dissolved in chloroform/methanol and purified by flash column chromatography on a silica gel column using chloroform/methanol (95:5). Fractions containing product were pooled and solvent removed by rotary evaporation (in vacuo) to give 3′,5′-t-butyldimethylsilyl-N1,O6-ethano-2′-deoxyxanthosine (9) as a foam (350 mg, 0.71 mmol; yield 67%). 1H NMR (d6-DMSO) δ (p.p.m.) 8.07 [s, 1.0H, H8], 6.15 [t, 1.0H, H1′], 4.77 [t, 1.9H, O6-CH2CH2-N1], 4.57 [m, 1.0H, H3′], 4.21 [t, 2.0H, O6-CH2CH2-N1], 3.85–3.61 [m, 3.2H, H4′, H5′/H5″], 2.72 [m, 1.1H, H2′], 2.29 [m, 1.1H, H2′], 0.89–0.85 [s&s, 19.0H, (CH3)3C(CH3)2Si], 0.11 [s, 6.3H, (CH3)3C(CH3)2Si], 0.02 [d, 6.2H, (CH3)3C(CH3)2Si]; MALDI-FTMS, expected for C24H42N4O5Si2 522.79, found 523 [H+], 545 [Na+].
Synthesis of N1,O6-ethano-2′-deoxyxanthosine (10)
9 (350 mg, 0.71 mmol) was dissolved in 2 ml of anhydrous tetrahydrofuran. One milliliter of a 1 M solution of tetrabutylammonium fluoride in tetrahydrofuran was added and the reaction stirred at room temperature for 45 min. The solvent was removed by rotary evaporation (in vacuo). The resulting oil was dissolved in 2 ml of chloroform/methanol (95:5) and loaded onto a silica gel column. The column was washed with two column volumes of chloroform/methanol (95:5) and product eluted with chloroform/methanol (80:20). Fractions containing product were pooled and solvent removed by rotary evaporation (in vacuo) to give N1,O6-ethano-2′-deoxyxanthosine (10) as a white solid (160 mg, 0.54 mmol; yield 77%). UVmax 246 in CH3OH; Rf 0.24 in CHCl3/CH3OH (80:20); 1H NMR (d6-DMSO) δ (p.p.m.) 8.11 [s, 0.93H, H8], 6.16 [t, 1.0H, H1′], 5.30 [d, 0.21H, 3′-OH], 4.93 [t, 0.19H, 5′-OH], 4.77 [t, 1.9H, O6-CH2CH2-N1], 4.35 [m, 1.0H, H3′], 4.21 [t, 2.1H, O6-CH2CH2-N1], 3.84 [m, 1.0H, H4′] 3.59–3.46 [m, 2.1H, H5′/H5″], 2.23 [m, 1.2H, H2′]; MALDI-FTMS, expected for C12H14N4O5 317.0856 [Na+], found 317.0859 [Na+].
Synthesis of N1,O6-ethano-5′-O-dimethoxytrityl-2′-deoxyxanthosine (11)
10 (160 mg, 0.54 mmol) was dried by rotary evaporation (in vacuo) with anhydrous pyridine and dissolved in 5 ml of anhydrous pyridine. Dimethoxytrityl chloride (1.1 equiv., 200 mg) was added. The reaction was stirred overnight at room temperature; an additional 100 mg dimethoxytrityl chloride and 10 mg dimethylaminopyridine were required to get complete conversion of the starting material as measured by TLC. The reaction was quenched by addition of 0.5 ml of 95% ethanol and stirred for 5 min. The reaction mixture was concentrated by rotary evaporation (in vacuo). The resulting oil was dissolved in 25 ml of ethyl acetate and the organic layer washed with two 25 ml portions of a 5% sodium bicarbonate solution and one 25 ml portion of a saturated NaCl solution. The organic layer was dried over sodium sulfate and, after filtration, the solvent was removed by rotary evaporation. The resulting foam was dissolved in 1.5 ml of chloroform/methanol/triethylamine (95:4.5:0.5) and purified by flash column chromatography on a silica gel column using chloroform/methanol/triethylamine (95:4.5:0.5). Fractions containing product were pooled and solvent removed by rotary evaporation (in vacuo) to give N1,O6-ethano-5′-O-dimethoxytrityl-2′-deoxyxanthosine (11) as a white foam (190 mg, 0.32 mmol; yield 60%). 1H NMR (CDCl3) δ (p.p.m.) 8.96 [s, 1.0H, H8], 7.42–6.76 [m, 14.8H, Ar-H], 6.25 [t, 1.0H, H1′], 4.83 [t, 2.2H, O6-CH2CH2-N1], 4.56 [m, 1.1H, H3′], 4.36 [t, 2.3H, O6-CH2CH2-N1], 4.07 [m, 1.1H, H4′] 3.88–3.71 [m, 8.1H, H5′/H5′′/Ar-OCH3], 2.66 [m, 1.1H, H2′], 2.37 [m, 1.1H, H2′].
Synthesis of N1,O6-ethano-5′-O-dimethoxytrityl-3′-O- (β-cyanoethyl-N,N-diisopropyl)phosphoramidite-2′-deoxyxanthosine (12)
11 (172 mg; 0.29 mmol) was dissolved in 4 ml of anhydrous dichloromethane. To the stirred solution 500 µl of diisopropylethylamine was added, followed by drop-wise addition of 150 µl of 2-cyanoethyl diisopropylchlorophosphoramidite. The reaction was allowed to proceed for 1 h, at which time the reaction was judged complete by TLC (chloroform/ethyl acetate/triethylamine 45:45:10). The reaction was quenched with 1 ml of anhydrous methanol and the solvents were removed by rotary evaporation (in vacuo). The resulting oil was dissolved in 1 ml of chloroform and purified by flash column chromatography on silica gel using chloroform/ethyl acetate/triethylamine (45:45:10). Fractions containing product were pooled and the solvent removed by rotary evaporation (in vacuo). The resulting oil was dissolved in 1 ml of ethyl acetate and the product precipitated from rapidly stirring petroleum ether at –40°C. The resulting precipitate was collected by centrifugation and dried under vacuum to give N1,O6-ethano-5′-O-dimethoxytrityl-3′-O-(β-cyanoethyl-N,N-diisopropyl)phosphoramidite-2′-deoxyxanthosine (12) as a white powder (190 mg, 0.24 mmol; yield 84%). 31P NMR (CD3OD) δ (p.p.m.) 157.50, 157.23.
Oligodeoxyribonucleotide synthesis, purification and characterization
Three oligodeoxyribonucleotides, NC-test (d-ACeXGT), NC4 (d-CAGACTGCCGeXCGCTGCAGGT) and NC1 (d-ACCTGCAGCGCCGGCAGTCTG), were prepared on an Applied Biosystems model 392 DNA synthesizer using standard phosphoramidite chemistry. Phosphoramidite solutions of N-acetyl-protected 2′-deoxycytidine, N-phenoxyacetyl-protected 2′-deoxyadenosine and N-(4-isopropyl-phenoxyacetyl)-protected 2′-deoxyguanosine were 0.1 M in concentration. A 0.15 M solution of 12 was used with an extended coupling period of 120 s. Coupling efficiency of 12 was >97% as measured by the trityl cation. The 5′-terminal dimethoxytrityl group was removed by the synthesizer and the oligomer-derivatized support transferred to a screw cap glass vial. The support was treated with 500 µl of concentrated ammonium hydroxide for 2 h at 37°C. The supernatant was removed from the support and the support washed with two 400 µl aliquots of 50% aqueous acetonitrile. The combined supernatant and washings were evaporated to dryness under vacuum at 37°C. Oligodeoxyribonucleotides were purified by strong anion exchange (SAX) HPLC on a Rainin Dynamax II column (0.46 × 15 cm) using a linear gradient of 0.0–0.8 M ammonium sulfate in a buffer that contained 1 mM ammonium acetate, pH 6.2, in 20% acetonitrile at a flow rate 0.6 ml/min. The column was monitored at 290 nm for preparative runs. The oligomers were desalted on a C-18 SEP PAK cartridge. The SEP PAK was pre-equilibrated by washing with 10 ml aliquots of acetonitrile, 50% aqueous acetonitrile and 2% acetonitrile in 50 mM sodium phosphate, pH 5.8 (buffer A). The oligodeoxyribonucleotide solution was diluted in buffer A to a final acetonitrile concentration of 3% and the solution applied to the cartridge. The cartridge was washed with 10 ml of water and the oligomer was eluted with 3 ml of 50% aqueous acetonitrile. Oligomers containing eX were subjected to enzymatic digestion with a combination of SVP and SAP by dissolving 0.1 A260 units in 10 µl of a solution containing 10 mM Tris, pH 8.1, 2 mM magnesium chloride, 1 µl of SVP and 0.5 µl of SAP. After 16 h at 37°C digests were analyzed by C-18 reversed phase HPLC on a Rainin Microsorb-C-18 column (0.46 × 15 cm) using a gradient of 2–3% acetonitrile over 12 min followed by a gradient of 3–20% acetonitrile over 8 min in buffer A.
Crosslinking experiments
Oligomer NC4 (4 × 10–10 mol) was radiolabeled in 9.5 µl of a solution containing 70 mM Tris, pH 7.6, 10 mM magnesium chloride, 5 mM dithiothreitol (DTT) and 63 µM [γ-32P]ATP (sp. act. 17 µCi/mmol). The reaction was initiated by addition of 0.5 µl (5 U) of T4 PNK and incubated at 37°C for 16 h. End-labeling reactions were heated to 65°C for 20 min in order to inactivate T4 PNK. Excess radiolabeled ATP was not removed. Radiolabeled NC4 (5 µl) was added to a 20 µM solution of cold duplex (45 µl) containing a 10% excess of NC1 to produce a 22 µM NC1/NC4* solution in crosslinking buffer (20 mM Tris, pH 7.0, 100 mM NaCl, 5 mM MgCl2, 10 mM DTT and 10% glycerol). Annealing was carried out by placing samples in a heat block at 95°C for 5 min, which was then allowed to gradually cool to room temperature and finally placed on ice for 5 min.
Crosslinking was initiated by addition of 4 µl of the 22 µM NC1/NC4* solution (final concentration 4 µM) to a reaction mixture containing 50 µM human AGT (a gift from Phil Potter) in crosslinking buffer. The reaction was incubated at 37°C and quenched by addition of 6× SDS loading buffer, followed by heating at 100°C for 5 min. The reaction volume was increased 10-fold for crosslinking time courses and the reaction mixture was centrifuged at 14 000 r.p.m. in a microcentrifuge prior to removal at each time point. Inactivation of hAGT was accomplished by heating the sample to 95°C for 2 min and then equilibrating at 37°C prior to addition of radiolabeled duplex or by incubating hAGT with 100 µM O6-benzylguanine [i.e. 1 µl of 2 mM O6-benzylguanine dissolved in dimethylsulfoxide (DMSO)] prior to addition of the radiolabeled duplex. O6-Benzylguanine was prepared as previously described (18). In order to control for the effect that DMSO has on crosslinking efficiency, a mock inactivation sample was adjusted to 10% DMSO prior to addition of hAGT.
RESULTS
Preparation of N1,O6-ethano-2′-deoxyxanthosine
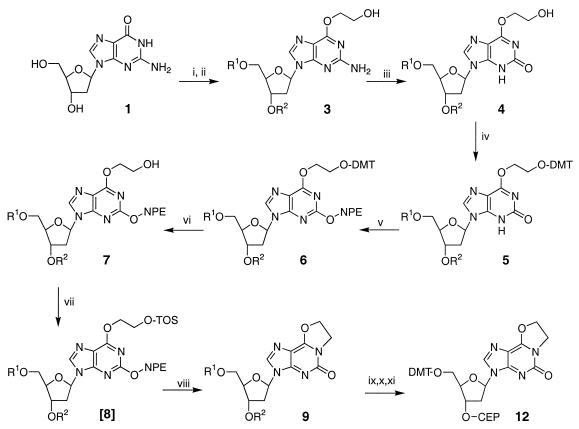
The synthetic route used to prepare the 5′-dimethoxytrityl-3′-cyanoethylphosphoramidite of eX is shown in Figure 3. The 5′- and 3′-hydroxyl groups of commercially available 2′-deoxyguanosine were protected with t-butyldimethylsilyl groups (17). A one-pot double displacement procedure (19) allowed the introduction of a hydroxyethyl group at the O6 position and nitrous acid-mediated deamination (20) of O6-hydroxyethylguanine gave the protected O6-hydroxyethylxanthosine, 4. The direct cyclization of 4 to produce the ethano bridged compound resulted in low (<2%) yields. To increase overall yields a step-wise approach was pursued. The newly introduced hydroxyethyl group was protected using dimethoxytritylchloride to give 5, followed by protection of the carbonyl O2 with a nitrophenethyl group via the Mitsunobo reaction (21) to give 6. The DMT group was then removed with acid to give 7. Activation of the exposed hydroxyl was carried out by converting it to a benzene sulphonyl derivative, 8. Attempts to purify 8 were problematical but in situ treatment of 8 (no purification) with 0.5 M DBU resulted in production of the desired eX derivative. The ethano bridge presumably results via an intramolecular cyclization in which N1 displacement of the sulphonyl derivative is initiated by base-catalyzed removal of the NPE protecting group. The protected nucleoside was converted to the phosphoramidite of N1,O6-ethano-2′-deoxyxanthosine, 12, using conventional strategies.
Figure 3.
Synthetic route used to prepare the nucleoside phosphoramidite of N1,O6-ethano-2′-deoxyxanthosine: (i) TBDMSiCl, imidazole, dimethylformamide, 1 h; (ii) trifluoroacetic anhydride, pyridine, 0°C, followed by Na/ethylene glycol; (iii) NaNO3, aqueous acetic acid, acetone, 2 h with agitation; (iv) 4,4′-dimethoxytritylchloride (DMTrCl), pryidine; (v) p-nitrophenylethanol (NPE), triphenylphosphine, diethyl azodicarboxylate, dioxane; (vi) 2% dichloroacetic acid/CH2Cl2, followed by NaHCO3 neutralization; (vii) 2,4,6-triisopropylbenzenesulfonylchloride, pyridine; (viii) 0.1 M DBU in pyridine; (ix) 0.5 M t-butylammonium fluoride in tetrahydrofuran; (x) DMTrCl, pyridine; (xi) N,N-diisopropylethylamine, 2-cyanoethyl diisopropylchlorophosphoramidite, CH2Cl2. R1, R2, TBDMSi; DMT, dimethoxytrityl; NPE, p-nitrophenylethane; TOS, 2,4,6-triisopropylbenzenesulfonyl; CEP, cyanoethyl diisopropylphosphoramidite.
Preparation of oligodeoxyribonucleotides containing eX
Oligodeoxyribonucleotides NC4 and NC-test, each containing a single eX, were prepared as described in Materials and Methods. Initial attempts to deprotect oligodeoxyribonucleotides containing eX using concentrated NH4OH at elevated temperatures (65°C for 3 h) resulted in complete degradation of eX. In order to determine the exact nature of the degradation an aliquot of 10 was treated with concentrated NH4OH at 65°C for 3 h and analyzed by reverse phase HPLC and mass spectrometry. Three degradation products whose retention times differed from 10 were observed. The two major species (50 and 40%) were identified from their molecular weight as the nucleoside and free base which correspond to ammonia displacement of the O6-ethyl portion of the ethano bridge (data not shown). Efforts to identify the third species were unsuccessful. This type of degradation of O6-alkylguanine analogs has been noted in the literature (22) and can be easily circumvented by using nucleoside phosphoramidites with protecting groups that can be removed under milder conditions (e.g. acetyl, PAC and iPR-PAC groups).
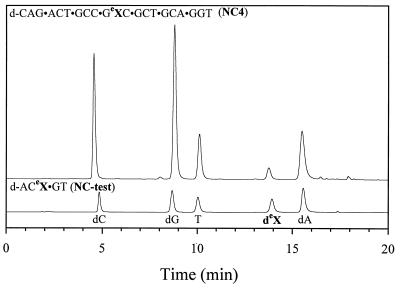
Oligodeoxyribonucleotides prepared in this manner were cleaved from the support and deprotected using concentrated NH4OH at 37°C for 2 h, purified using strong anion exchange chromatography and shown to be >98% pure as measured by analytical reverse phase HPLC and denaturing PAGE. Figure 4 shows the nucleoside composition of oligodeoxyribonucleotides containing eX following digestion of the oligodeoxyribonucleotides with SVP and SAP followed by analysis by reverse phase HPLC on a C18 column. Co-injection of 10 with oligodeoxyribonucleotide digests confirmed that the peak labeled deX is the expected product, indicating that the ethano bridge survived the mild deprotection procedure (data not shown). Estimated degradation of the ethano portion of eX under these conditions was <2%.
Figure 4.
Reversed phase HPLC analysis of oligomers NC4 and NC-test after digestion with SVP and calf intestinal phosphatase. The oligodeoxyribonucleotide sequences are shown above the chromatogram and the nucleoside peaks are labeled at the bottom.
Formation of covalent hAGT–DNA complexes
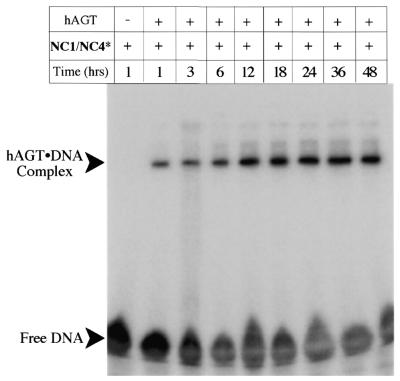
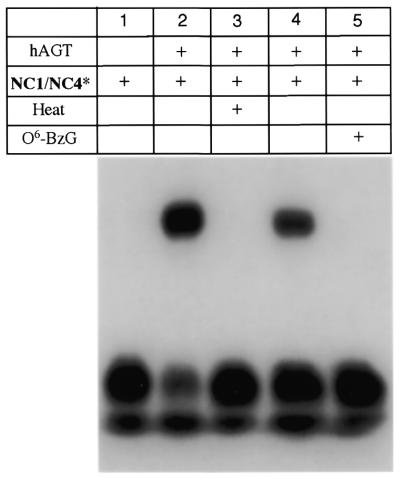
The ability of eX to covalently trap hAGT in a post-repair protein–DNA complex is shown in Figure 5. Covalent crosslinking of the hAGT to radiolabeled DNA oligomers containing eX can be monitored by following the formation of a protein–DNA complex that is insensitive to thermal and chemical denaturation. Thus aliquots from a solution containing 4 µM oligodeoxyribonucleotide and 50 µM hAGT in crosslinking buffer were quenched with SDS gel loading buffer and boiled for 5 min at the indicated times. Prior to electrophoresis on a 12% SDS–polyacrylamide gel the samples were boiled for 5 min and following electrophoresis complexes were visualized by autoradiography. Lane 1 shows the electrophoretic mobility of the radiolabeled DNA duplex NC1/NC4* in the absence of protein. On SDS–polyacrylamide gels the oligodeoxyribonucleotide NC4* runs near the gel front and appears as a poorly resolved band. Unincorporated radiolabeled ATP is responsible for a slightly faster moving band (see Fig. 6 for clearer resolution of the bands); this material does not participate in crosslinking. Lane 2 shows formation of a slowly migrating radiolabeled species that is insensitive to denaturation. Visualization of the gel by staining with Coomassie Blue showed a new protein-containing band that runs coincident with the radioactive species (data not shown), consistent with a covalent protein–DNA complex.
Figure 5.
Autoradiograph of an SDS–polyacrylamide gel showing covalent crosslinking of N1,O6-ethano-2′-deoxyxanthosine-containing oligodeoxyribonucleotides to hAGT as a function of time. Radiolabeled oligodeoxyribonucleotide duplexes were incubated at 37°C in the absence of hAGT (lane 1) or in the presence of hAGT (lanes 2–8) for up to 48 h.
Figure 6.
Autoradiograph of an SDS–polyacrylamide gel showing covalent crosslinking of N1,O6-ethano-2′-deoxyxanthosine-containing oligodeoxyribonucleotides to hAGT. Radiolabeled oligodeoxyribonucleotide duplexes (4 µM) were incubated for 50 h at 37°C in the absence of hAGT (lane 1), in the presence of 50 µM hAGT (lane 2), in the presence of heat-inactivated hAGT (lane 3), in the presence of hAGT pretreated with 10% DMSO (lane 4) and in the presence of hAGT pretreated with 10% DMSO and 100 µM O6-benzylguanine.
It was observed over these prolonged incubations that a precipitate of free hAGT formed. This precipitation depleted the reaction mixture of soluble hAGT within 24 h and led to the apparent leveling off of crosslinking seen in Figure 5, lanes 2–8. Complete conversion of radiolabeled NC4 to the protein–DNA complex can be achieved by addition of more hAGT (data not shown). We also note that while precipitated protein was cleared from the solution by centrifugation the covalent complex formed remained soluble even after 48 h at 37°C.
Figure 6 shows the effect of thermal and chemical inactivation of hAGT on formation of the protein–DNA complex. Lanes 1 and 2 show the electrophoretic mobility of the eX-containing oligodeoxyribonucleotide with and without hAGT. Thermal denaturation (90°C for 2 min) of hAGT prior to incubation with the oligodeoxyribonucleotide abolishes formation of the covalent complex (lane 3), as does incubation with O6-benzylguanine (18), a mechanism-based inhibitor of hAGT (lane5). Lane 4 is a mock inactivation of hAGT by DMSO (the solvent used to add the O6-benzylguanine). Crosslinking could also be abolished if NC4 was treated with concentrated NH4OH for 3 h at 65°C prior to annealing and incubation with hAGT (data not shown), presumably due to degradation of the ethano bridge.
DISCUSSION
In the past several years the proposed model for the recognition and repair of alkylated guanine by AGT has evolved. Early models, based in part on the crystal structure of the C-terminal domain of the E.coli Ada protein, postulated a conformational change that would bring the buried active site cysteine into proximity to an alkylated base in an unperturbed helix (23). The realization that some DNA-modifying proteins (e.g. cytosine 5-methyltransferases and DNA glycosylases) carry out the chemical modification of DNA by extruding the target base from the double helix into a buried active site led to the suggestion that AGT may use this mechanism. Recent structures of hAGT in the unalkylated and alkylated forms (24,25) and models of E.coli (26) and human AGT (24) bound to duplex DNA suggest that base flipping could result in productive repair complexes. Furthermore, NMR studies of the C-terminal domain of the Ada protein show that DNA binding does not involve significant conformational changes by the protein, an observation consistent with a base flipping mechanism (27).
In spite of the growing consensus that AGT recognition and repair of alkylated DNA involves base flipping there is little direct evidence. Efforts to produce complexes of AGT with DNA suitable for structural and biophysical characterization have been unsuccessful. Isolation of pre-repair complexes is complicated by a lack of appropriate cofactors which can be omitted in order to trap complexes and while active site mutants which retain binding specificity for DNA containing methylated guanine have been reported (28), it is not clear how useful these mutants will be for structural work. Post-repair complexes also pose a significant technical challenge. Alkyl transfer results in a significant destabilization of the protein leading to an increased Stokes radius (29), increased proteolytic sensitivity (30) and rapid degradation in HT29 cells and cell-free extracts (31). The structure of alkylated AGT suggests that alkyltransfer causes the disruption of an extensive hydrogen bonding network, promotes helix displacement and results in dramatic destabilization of alkylated AGT (24).
The strategy pursued by our laboratory to prepare AGT–DNA complexes has been to use modified nucleosides to trap covalent complexes. To that end we previously demonstrated that it was possible to crosslink hAGT to a synthetic oligodeoxyribonucleotide containing a thiol-derivatized guanine (32). The N6-thioethyl derivative of 2,6-diaminopurine was shown to produce a covalent disulfide crosslink between the hAGT active site cysteine and the modified nucleoside. Unfortunately, the crosslinking reaction was inefficient and required the use of very large excesses of protein to achieve low levels of crosslinking.
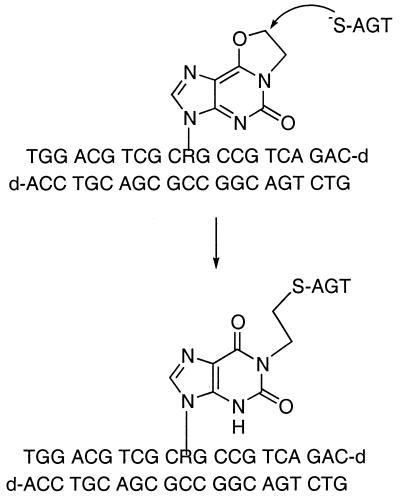
The eX analog described here was designed to circumvent many of the limitations of the earlier thiol-based crosslinker and is based on the observation that DNA treated with CENUs produced covalently trapped AGT–DNA complexes. Early work done by Brent and co-workers (11,13–15) elucidated the mechanism by which a CENU-derived intermediate could serve as a substrate for and crosslink AGT to DNA. This intermediate is the reactive N1,O6-ethanoguanine (eG), which results from intramolecular cyclization of the original alkylation product, O6-chlorethylguanine. AGT can recognize eG in a repair reaction that involves nucleophilic attack by the active site cysteine on the O6-methylene group to form a covalent complex between AGT and DNA via an N1-ethylcysteine crosslink (11).
Unfortunately, eG is not well suited to the preparation of homogeneous site-specific AGT–DNA complexes. In situ generation of eG by treatment with CENU results in the formation of a variety of alkylation products (33) and the eG adduct itself undergoes both hydrolysis (16) and interstrand crosslink formation (12), significantly dampening our enthusiasm for its utility as a useful crosslinker. Rather, we set out to design a nucleoside analog that maintained the major groove structure of eG, in order to facilitate recognition and crosslinking with AGT, but which removed the positive charge in order to ameliorate the reactivity of eG. The most straightforward and least perturbing route to preparing such a nucleoside analog was to replace the exocyclic amino group of eG with a carbonyl oxygen. The deamination of guanine to xanthine allows the normally protonated N3 of xanthine to lose its proton and undergo a tautomeric shift in which the positive charge at N1 can be ameliorated.
The strategy presented here for the preparation of eX was one of several under consideration. The availability of 2′-deoxyguanosine and a method for introduction of the ethano portion of the molecule were the principal advantages to starting with 2′-deoxyguanosine rather than 2′-deoxyxanthosine. A convenient procedure for the introduction of a variety of alkyl groups at the O6 position of deoxyguanosine had been reported (19) and appeared to be adaptable to the introduction of a hydroxyethyl group. This one-pot double displacement procedure relies upon trifluoroacetic anhydride to activate the O6, which when carried out in pyridine results in in situ generation of the pyridinium derivative. Quenching the reaction with an alcohol in the presence of a base, a solution of sodium dissolved in ethylene glycol in this case, results in the displacement of pyridine to produce the desired O6-alkyl derivative. A large excess of freshly prepared ethylene glycol/sodium solution was required to prevent the decreases in yield noted in the original description of this procedure (19). With the introduction of the 2-carbon bridge the conversion to a deoxyxanthosine derivative was pursued. Deamination of 2′-deoxyguanosine using nitrous acid has been reported (20) and was used to convert O6-hydroxyethyl-dG 3 to the 2′-deoxyxanthosine derivative 4. Attempts at deamination using n-pentylnitrite as reported by Steinbrecher et al. (34) for O6-nitrophenethyl-protected dG were unsuccessful.
A direct cyclization of 4 to produce eX was initially envisioned, however, in a non-productive competing reaction the O2 carbonyl oxygen reacted with the benzenesulphonyl chlorides used to activate the hydroxyethyl group. This competing reaction decreased the yield of the desired cyclized compound and necessitated masking the O2 position. The nitrophenethyl group is a convenient protecting group that has been used to protect the exocyclic oxygens of guanine (35), xanthine (19) and uric acid (36) and could be easily introduced via the Mitsunobo reaction (21). Thus the hydroxyethyl group was first masked with dimethoxytrityl chloride, the O2 protected with an NPE group and finally the DMT was removed to afford the desired O2-protected hydroxyethyl derivative, 7. The exposed hydroxyl group could then be selectively activated using 2,4,6-triisopropylbenzenesulphonyl chloride to produce the tosylate, 8. This tosylate was poorly behaved during purification attempts and had to be converted to the desired ethano compound without purification. Thus after quenching the tosylation reaction and evaporation of solvents, the resulting oil was dissolved in pyridine and, following the addition of a strong base, DBU, the desired O6-CH2CH2-N1 bridge-containing compound was produced. Cyclization presumably occurs following base-catalyzed β elimination of the NPE group from the O2 position, which in turn increases the nucleophilicity of N1.
Conversion of 9 to the nucleoside phosphoramidite and its incorporation into oligodeoxyribonucleotides was accomplished using standard procedures, with one important exception. Due to the anticipated sensitivity of eX to ammonia-catalyzed displacement of the O6-alkyl moiety, protecting groups for the dA, dC and dG amidites were selected that could be removed under mild conditions. The sensitivity of O6-alkylguanine to displacement by ammonia has been observed (22) and while there have been no such reports for alkylxanthine derivatives, it seemed likely that similar reactivity might exist. Indeed, when the NC-test sequence was prepared with traditional protecting groups (i.e. N-benzoyl-dC and dA) deprotection using concentrated NH4OH at 65°C for 3 h resulted in complete degradation of eX. When acetyl- and phenoxyacetyl-protected monomers were substituted the preparation of oligodeoxyribonucleotides containing intact eX was easily accomplished.
Oligodeoxyribonucleotides containing eX produce covalent protein–DNA complexes when incubated with hAGT, presumably via the proposed mechanism shown in Figure 7. Nucleophilic attack by the active site cysteine on the O6-methylene carbon in a repair-like reaction results in alkyl transfer and covalent capture of a protein–DNA complex. The ability of O6-benzylguanine, a mechanism-based inhibitor of hAGT (18), to abolish crosslink formation is consistent with the proposed mechanism-based crosslinking scheme. While nucleophilic attack at the N1-methylene carbon would be expected to produce a covalent complex indistinguishable at the current level of characterization it seems unlikely that the active site architecture would accommodate such an attack.
Figure 7.
Scheme showing the proposed mechanism of crosslink formation between hAGT and eX.
Though designed to mimic eG, there are several important differences between the crosslinking reported with eG-containing DNA (11) and the crosslinking described here. First, the rate of crosslink formation with eX-containing oligomers appears to be slower than that of eG-containing oligomers. Crosslinking with in situ generated eG (i.e. DNA treated with CENU) is complete within 1 h, while crosslinking with eX occurred more slowly (∼12 h to give 50% crosslinking). While direct comparisons are complicated, it is not unreasonable to assume that the eX analog would be less reactive than eG. The presence of a positive charge on eG can stabilize the negative charge that forms on the leaving group in an SN2 type displacement (i.e. the O6 of eG). Removing the positive charge would then be expected to decrease the rate of crosslinking. It is also possible that substitution of oxygen for the exocyclic amino group of guanine has an effect on binding and/or recognition of the substrate, also contributing to slower crosslinking. The second difference between eG and eX is the extent of crosslink formation. Maximal crosslinking efficiencies for eG-containing DNA were ∼50% compared to the near quantitative crosslinking efficiency achievable with oligomers containing eX. The inability to achieve complete crosslinking using eG is likely a result of several competing reactions: (i) removal of the O6-chloroethyl group by AGT prior to cyclization; (ii) hydrolysis of eG producing the unreactive N1-hydroxyethyl derivative; (iii) formation of the G-C interstrand crosslink. The third difference achieved with eX is the formation of only a single protein–DNA species, whereas DNA treated with CENU and incubated with hAGT resulted in the formation of multiple crosslinked protein–DNA species.
The differences observed between crosslinking with eG and the nucleoside analog eX are consistent with the hypothesis that amelioration of the positive charge results in a better behaved, albeit slower, crosslinker. The crosslinking reactions described here required only a 10-fold excess of protein over DNA and resulted in high levels of covalent protein–DNA complexes. Furthermore, the persistent solubility of the hAGT–DNA complex versus the free protein suggests that these complexes may be amenable to further biophysical characterization.
CONCLUSIONS
The modified nucleoside N1,O6-ethano-2′-deoxyxanthosine has been prepared, incorporated into oligodeoxyribonucleotides and used to generate covalent complexes with hAGT. Mechanism-based covalent trapping of the hAGT–DNA complex occurs with high yield and the resulting complexes should facilitate structural studies of the recognition and repair activity of this important class of DNA repair proteins. Further characterization of the covalent protein–DNA complexes involving both hAGT and the AGT domain of E.coli Ada protein is underway.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Patterson Trust and the Alexander and Margaret Stewart Trust. We thank Dr Phil Potter for the hAGT and Dr Paul Miller for space, equipment and helpful discussions regarding the synthesis of eX.
References
- 1.Bignami M., O’Driscoll,M., Aquilina,G. and Karran,P. (2000) Unmasking a killer: DNA O(6)-methylguanine and the cytotoxicity of methylating agents. Mutat. Res., 462, 71–82. [DOI] [PubMed] [Google Scholar]
- 2.Samson L. (1992) The suicidal DNA repair methyltransferases of microbes. Mol. Microbiol., 6, 825–831. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T., Sedgwick,B., Sekiguchi,M. and Nakabeppu,Y. (1988) Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem., 57, 133–157. [DOI] [PubMed] [Google Scholar]
- 4.Mitra S. and Kaina,B. (1993) Regulation of repair of alkylation damage in mammalian genomes. Prog. Nucleic Acid Res. Mol. Biol., 44, 109–142. [DOI] [PubMed] [Google Scholar]
- 5.Pegg A.E., Dolan,M.E. and Moschel,R.C. (1995) Structure, function and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol., 51, 167–223. [DOI] [PubMed] [Google Scholar]
- 6.Pegg A.E. (2000) Repair of O(6)-alkylguanine by alkyltransferases. Mutat. Res., 462, 83–100. [DOI] [PubMed] [Google Scholar]
- 7.Daniels D.S. and Tainer,J.A. (2000) Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O6-alkylguanine-DNA alkyltransferase. Mutat. Res., 460, 151–163. [DOI] [PubMed] [Google Scholar]
- 8.Encell L.P., Landis,D.M. and Loeb,L.A. (1999) Improving enzymes for cancer gene therapy. Nat. Biotechnol., 17, 143–147. [DOI] [PubMed] [Google Scholar]
- 9.Dolan M.E. and Pegg,A.E. (1997) O6-benzylguanine and its role in chemotherapy. Clin. Cancer Res., 3, 837–847. [PubMed] [Google Scholar]
- 10.Glassner B.J., Weeda,G., Allan,J.M., Broekhof,J.L., Carls,N.H., Donker,I., Engelward,B.P., Hampson,R.J., Hersmus,R., Hickman,M.J., Roth,R.B., Warren,H.B., Wu,M.M., Hoeijmakers,J.H. and Samson,L.D. (1999) DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis, 14, 339–347. [DOI] [PubMed] [Google Scholar]
- 11.Gonzaga P.E., Potter,P.M., Nui,T.-Q., Yu,D., Ludlum,D.B., Rafferty,J.A., Margison,G.P. and Brent,T.P. (1992) Identification of the cross-link between the human O6-methylguanine-DNA methyltransferase and chloroethynitrosourea-treated DNA. Cancer Res., 52, 6052–6058. [PubMed] [Google Scholar]
- 12.Fishhaber P.L., Gall,A.S., Duncan,J.A. and Hopkins,P.B. (1999) Direct demonstration in synthetic oligonucleotides that N,N′-bis(2-chloroethyl)-nitrosourea cross-links N1 of deoxyguanosine to N3 of deoxycytidine on opposite strands of duplex DNA. Cancer Res., 59, 4363–4368. [PubMed] [Google Scholar]
- 13.Brent T.S. and Remack,J.S. (1988) Formation of covalent complexes between human O6-alkylguanine-DNA alkytransferase and BCNU-treated defined length synthetic oligodeoxynucleotides. Nucleic Acids Res., 16, 6779–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brent T.P., Remack,J.S. and Smith,D.G. (1987) Characterization of a novel reaction by human O6-alkylguanine-DNA alkyltransferase with 1,3-bis(2-chloroethyl)-1-nitrosourea-treated DNA. Cancer Res., 47, 6185–6188. [PubMed] [Google Scholar]
- 15.Gonzaga P.E., Harris,L., Margison,G.P. and Brent,T.P. (1990) Evidence that covalent complex formation between BCNU-treated oligonucleotides and E. coli alkyltransferase requires the O6-alkylguanine function. Nucleic Acids Res., 18, 3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker S., Kirk,M.C. and Ludlum,D.B. (1987) Synthesis and characterization of O6-(2-chloroethyl)guanine: a putative intermediate in the cytotoxic reaction of chloroethylnitrosoureas with DNA. Biochem. Biophys. Res. Commun., 148, 1124–1128. [DOI] [PubMed] [Google Scholar]
- 17.Ogilve K.K. (1973) The tert-butyldimethylsilyl group as a protecting group in deoxynucleosides. Can. J. Chem., 51, 3799–3807. [Google Scholar]
- 18.Dolan M.E., Moschel,R.C. and Pegg,A.E. (1990) Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and thereputic alkylating agents. Proc. Natl Acad. Sci. USA, 87, 5368–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fathi R., Goswami,B., Kung,P.-P., Gaffney,B.L. and Jones,R.A. (1990) Synthesis of 6-substituted 2′-deoxyguanosine derivatives using trifluoroacetic anhydride in pyridine. Tetrahedron Lett., 31, 319–322. [Google Scholar]
- 20.Eritja R., Horowitz,D.M., Walker,P.A., Ziehler-Martin,J.P., Boosalis,M.S., Goodman,M.F., Itakura,K. and Kaplan,B.E. (1986) Synthesis and properties of oligonucleotides containing 2′-deoxynebularine and 2′-deoxyxanthosine. Nucleic Acids Res., 14, 8135–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsunobo O. (1981) The use of diethyl azodicarboxylate and triphenylphosphine in synthesis and transformation of natural products. Synthesis, 1, 1–28. [Google Scholar]
- 22.Wang L., Spratt,T.E., Pegg,A.E. and Peterson,L.A. (1999) Synthesis of DNA oligonucleotides containing site-specifically incorporated O6-[4-oxo-4-(3-pyridyl)butyl]guanine and their reaction with O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol., 12, 127–131. [DOI] [PubMed] [Google Scholar]
- 23.Moore M.H., Gulbis,J.M., Dodson,E.J., Demple,B. and Moody,P.C.E. (1992) Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E.coli. EMBO J., 13, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniels D.S., Mol,C.D., Arvai,A.S., Kanugula,S., Pegg,A.E. and Tainer,J.A. (2000) Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding. EMBO J., 19, 1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wibley J.E., Pegg,A.E. and Moody,P.C. (2000) Crystal structure of the human O(6)-alkylguanine-DNA alkyltransferase. Nucleic Acids Res., 28, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vora R.A., Pegg,A.E. and Ealick,S.E. (1998) A new model for how O6-methylguanine-DNA methyltransferase binds DNA. Proteins, 32, 3–6. [PubMed] [Google Scholar]
- 27.Verdemato P.E., Brannigan,J.A., Damblon,C., Zuccotto,F., Moody,P.C. and Lian,L.Y. (2000) DNA-binding mechanism of the Escherichia coli Ada O(6)-alkylguanine-DNA alkyltransferase. Nucleic Acids Res., 28, 3710–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazra T.K., Roy,R., Biswas,T., Grabowski,D.T., Pegg,A.E. and Mitra,S. (1997) Specific recognition of O6-methylguanine in DNA by active site mutants of human O6-methylguanine-DNA methyltransferase. Biochemistry, 36, 5769–5776. [DOI] [PubMed] [Google Scholar]
- 29.Hora J.F., Eastman,A. and Bresnick,E. (1983) O6-methylguanine methyltransferase in rat liver. Biochemistry, 22, 3759–3763. [DOI] [PubMed] [Google Scholar]
- 30.Kanugula S., Goodtzova,K. and Pegg,A.E. (1998) Probing of conformational changes in human O6-alkylguanine-DNA alkyl transferase protein in its alkylated and DNA-bound states by limited proteolysis. Biochem. J., 329, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pegg A.E., Wiest,L., Mummert,C., Stine,L., Moschel,R.C. and Dolan,M.E. (1991) Use of antibodies to human O6-alkylguanine-DNA alkyltransferase to study the content of this protein in cells treated with O6-benzylguanine or N-methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis, 12, 1679–1683. [DOI] [PubMed] [Google Scholar]
- 32.Paalman S.R., Noll,D.M. and Clarke,N.D. (1997) Formation of a covalent complex between methylguanine methyltransferase and DNA via disulfide bond formation between the active site cysteine and a thiol-containing analog of guanine. Nucleic Acids Res., 25, 1795–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodell W.J. and Pongracz,K. (1993) Chemical synthesis and detection of the cross-link 1-[N3-(2′-deoxycytidyl)]-2-[N1-(2′-deoxyguanosinyl)]ethane in DNA reacted with 1-(2-chloroethyl)-1-nitrosourea. Chem. Res. Toxicol., 6, 434–438. [DOI] [PubMed] [Google Scholar]
- 34.Steinbrecher T., Wameling,C., Oesch,F. and Seidel,A. (1993) Activation of the C2 position of purine by the trifluoromethanesulfonate group: synthesis of N2-alkylated deoxyguanosines. Angew. Chem. Int. Ed. Engl., 32, 404–406. [Google Scholar]
- 35.Trichtinger T., Charubala,R. and Pleiderer,W. (1983) Syntheis of O6-p-nitrophenylethyl guanosine and 2′-deoxyguanosine derivatives. Tetrahedron Lett., 24, 711–714. [Google Scholar]
- 36.Schulz B.S. and Pfleiderer,W. (1985) A new synthesis of 9-β-d-ribofuranosyluric acid and its 5′-monophosphate. Tetrahedron Lett., 26, 5421–5424. [PubMed] [Google Scholar]