Abstract
Physical activity is associated with a lower risk of breast, colon, and endometrial cancer. Epigenetic mechanisms such as changes in DNA methylation may help to explain these protective effects. We assessed the impact of a one year aerobic exercise intervention on DNA methylation biomarkers believed to play a role in carcinogenesis. The Alberta Physical Activity and Breast Cancer Prevention (ALPHA) Trial was a two-armed randomized controlled trial in 320 healthy, inactive, postmenopausal women with no history of cancer. In an ancillary analysis, frozen blood samples (n = 256) were reassessed for levels of DNA methylation within LINE-1 and Alu repeats as well as within the promoter regions of APC, BRCA1, RASSF1, and hTERT genes. Differences between the exercise and control arm at 12-months, after adjusting for baseline values, were estimated within an intent-to-treat and per-protocol analysis using linear regression. No significant differences in DNA methylation between the exercise and control arms were observed. In an exploratory analysis, we found that the prospective change in estimated VO2max was negatively associated with RASSF1 methylation in a dose-response manner (p-trend = 0.04). A year-long aerobic exercise intervention does not affect LINE-1, Alu, APC, BRCA1, RASSF1, or hTERT methylation in healthy, inactive, postmenopausal women. Changes in DNA methylation within these genomic regions may not mediate the association between physical activity and cancer in healthy postmenopausal women. Additional research is needed to validate our findings with RASSF1 methylation.
Trial Registration: ClinicalTrials.gov NCT00522262.
Introduction
Regular physical activity has been shown to protect against a multitude of cancers in a wide variety of study populations and settings. It is well established that being more physically active lowers the risk of colorectal, breast, and endometrial cancer [1, 2]. Emerging evidence suggests that exercise plays an important etiologic role in the prevention of other types of cancer as well. A recent pooled analysis of 1.44 million individuals suggested that the protective effects of physical activity also extend to head and neck, esophageal, lung, kidney, blood, and bladder cancers independent of body mass index [3]. The burden of cancer attributable to physical inactivity is considerable. It was recently estimated that roughly 7% of colorectal and breast cancer cases worldwide were attributable to physical inactivity in 2013, representing a total healthcare cost of $5.2 billion for these two cancer sites alone [4].
The underlying biologic mechanisms whereby physical activity influences cancer risk has been investigated in recent randomized controlled trials in healthy populations [5–9]. One recently identified but poorly understood mechanism whereby physical activity may prevent carcinogenesis is DNA methylation [10]. Patterns of DNA methylation can become dysregulated in response to ageing and exposure to carcinogenic agents which can lead to genomic instability and abnormal gene expression [11–13]. Such dysregulation is widely thought to be a molecular state that predisposes an individual to cancer [14, 15].
We invested the impact of a year-long exercise intervention on levels of DNA methylation within two major types of repetitive elements (LINE-1 and Alu) and within the promoter regions of four candidate genes (APC, BRCA1, RASSF1, and hTERT) in a group of healthy, inactive postmenopausal women. High levels of LINE-1 and Alu methylation are associated with chromosomal stability [16–18]. Although the evidence is conflicting and additional research is needed, some studies have found that individuals with lower levels of LINE-1 and Alu methylation in tissue or in blood have a higher risk of developing cancer [19–22]. We selected four genes based on evidence of an association between genetic mutations and epigenetic alterations in these genes and the risk of cancer. APC, BRCA1, and RASSF1 are tumour suppressor genes which play important etiologic roles in carcinogenesis. High levels of methylation within the promoter regions of these genes is associated with gene expression silencing and is a common occurrence in several types of cancer tissue [23]. Recent epidemiologic studies have reported an increased risk of cancer among individuals with higher levels of APC, BRCA1, and RASSF1 promoter methylation in tissue or in blood [24–28]. In contrast, elevated levels of methylation within the promoter region of hTERT is potentially associated with a protective carcinogenic effect via increased telomerase expression [29]. The hTERT gene encodes for the catalytic subunit of telomerase which regulates telomeric DNA length and plays a vital role in the cellular immortalization of cancers [30]. In healthy populations, shorter telomeres have been associated with an increased risk of cancer [31].
The objective of the current study was to determine if a year-long aerobic exercise intervention could impact blood-based measures of DNA methylation with LINE-1 and Alu regions as well as within the promoter regions of the APC, BRCA1, RASSF1, and hTERT genes. Although it has yet to be proven, we hypothesized that physical activity would have a systemic effect on levels of DNA methylation across all tissues such that any changes in DNA methylation detected within blood would also reflect changes in breast tissue–the target tissue of interest. We also hypothesized that higher levels of DNA methylation within LINE-1 and Alu regions and within the promoter region of the hTERT gene and that lower levels of DNA methylation within the promoter regions of the APC, BRCA1, and RASSF1 genes would be associated with a reduced risk of cancer. As such, we hypothesized that women randomized to the exercise intervention would have significantly higher levels of LINE-1, Alu, and hTERT methylation and significantly lower levels of APC, BRCA1, and RASSF1 methylation after adjusting for baseline differences.
Methods
Study population
Participant recruitment and eligibility have been detailed elsewhere [7]. To be eligible, participants had to be physically inactive postmenopausal women between the ages of 50 and 75 and non-smokers or excessive alcohol consumers. In addition, eligible participants had no prior history of cancer, had physician clearance to participate, and had to provide written informed consent. Ethics approval was obtained from the Alberta Cancer Research Ethics Committee, the University of Calgary, and the University of Alberta.
Study design, intervention, and covariate information
The Alberta Physical Activity and Breast Cancer Prevention (ALPHA) Trial was a two-armed randomized year-long exercise intervention that assessed the effects of aerobic activity on biomarkers associated with breast cancer risk [7]. There were 320 participants who were randomized in a 1:1 ratio to either the intervention or control arm. The intervention began with 15–20 minutes of aerobic activity at 50–60% of the maximum heart rate three times/week. This exercise protocol was increased over the course of three months to 45 minutes of activity at 70–80% of the maximum heart rate five times/week which was sustained for the remaining nine months. A minimum of three weekly sessions were supervised at designated facilities while the remaining sessions were unsupervised.
Baseline and follow-up information on several covariates was collected. A baseline health questionnaire captured the participants’ age, ethnicity, and prior smoking history. Weight and height were objectively measured. Dietary folate intake, alcohol intake, and levels of physical activity in the year before study entry were estimated using validated questionnaires [32, 33]. At both baseline and follow-up, a submaximal exercise test was used to estimate maximal oxygen uptake (VO2max) as described previously [7].
Blood collection and DNA methylation assays
Blood samples were collected from each participant at baseline (n = 320) and at 12-months (n = 310) after a ten hour fast using standardized collection, processing, and storage protocols [7]. In the Translational Laboratory at the Tom Baker Cancer Center (Calgary, Alberta), peripheral blood mononuclear DNA from 630 samples was purified and extracted using the Hamilton STARlet liquid handling instrument (Hamilton Robotics Inc., Reno, USA) and the Machery-Nagel NucleoMag Blood 200 μl kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany). As determined by the Qubit dsDNA HS Assay Kit (ThermoFisher Scientific Inc., Waltham, USA), a total of 33 samples had insufficient DNA for further analysis. The remaining 597 samples were plated on seven 96-well plates. At the McGill University Génome Québec Innovation Centre (Montréal, Canada), the plated samples underwent sodium bisulfite conversion which was carried out with the EZ-96 DNA Methylation-Gold Kit (Zymo Research, Irvine, USA, Catalog No. D5007). DNA methylation within successfully treated samples (n = 573) was assessed using two methods. A pyrosequencing assay involving the HotStarTaq DNA Polymerase kit (Qiagen, Hilden, Germay) and PyroMark Q24 (Qiagen, Hilden, Germay) was used to measure the degree of LINE-1 and Alu methylation. The primers used in our assessment of LINE-1 and Alu methylation were developed in a prior investigation [34]. Methylation analyses for APC, BRCA1, RASSF1 and hTERT were performed using Genome Quebec’s Sequenom® EpiTYPER platform and standard EpiPanel (Agena Bioscience, San Diego, USA). This mass spectrometry-based platform enables accurate and quantitative measurement of DNA methylation levels at multiple CpG genomic regions [35].The targeted regions and the corresponding CpG sites are described in S1 Table. Target Regions for Gene-Specific Outcomes.
DNA methylation data processing
A total of 23, 43, 20, and 26 CpG sites were assessed within the promoter regions of the APC, BRCA1, RASSF1, and hTERT genes respectively. As done in previous studies [35, 36], we removed CpG sites deemed unreliable by any one of the following criteria: 1) the mass was beyond the limit of detection; 2) overlapping signals or silent peaks were detected; 3) there were multiple CpG units with the same mass; 4) the CpG unit had a low success rate which we defined as a failure rate of at least 10% across samples. After removing these unreliable CpG sites, there remained 7, 23, 13, and 21 CpG sites for the APC, BRCA1, RASSF1, and hTERT genes respectively (S1 Table. Spearman’s Correlation between Repetitive Element and Gene-Specific Baseline DNA Methylation Measures).
For each outcome, we excluded observations that were missing more than 15% of data within the remaining CpG sites. This step resulted in the exclusion of the following number of observations: LINE1 (n = 3), Alu (n = 3), APC (n = 28), BRCA1 (n = 10), RASSF1 (n = 3), and hTERT (n = 12). For both LINE1 and Alu methylation, we also excluded observations that failed to meet quality control standards specified by the PyroMark Q24 software (n = 3). Among the remaining observations, there were no missing methylation values for any of the CpG sites assessed within LINE1, Alu, or APC. Missingness within the other outcomes was addressed using a plate-specific mean imputation. For any given individual, no more than 3 CpG sites were imputed for the hTERT and BRCA1 genes and no more than 1 CpG site was imputed for the RASSF1 gene.
The overall level of DNA methylation within each genomic region of interest was defined as the average percent methylation across the remaining CpG sites. These raw values were adjusted for batch effects using the following mean centering approach: where yi is the adjusted value for the ith observation, xi is the raw value for the ith observation, is the mean specific to the jth batch, and is the overall mean [37]. These batch adjusted methylation values were used in all subsequent analyses.
Reliability assessment
We evaluated the inter-batch reliability for each DNA methylation measure. The samples were processed in seven batches. We had enough DNA to duplicate the laboratory analyses for eight samples. One of the seven batches was used as a reference batch. For each of the eight duplicated samples, we placed one of the samples in this reference batch. We then placed the paired duplicate samples in one of the other six remaining batches, in random order, such that each of the six remaining batches had at least one of the eight duplicate samples. Based on a visual examination of the Bland-Altman plots, one outlier was removed from the BRCA1 and APC analyses. Inter-batch reliability was quantified using the coefficient of variation (CV) and limits of agreement (LOA) as follows: LINE1 (CV = 1.4%;LOA = -0.6–3.4), Alu (CV = 2.2%;LOA = -0.2–1.3), APC (CV = 8.6%;LOA = -0.1–0.6), BRCA1 (CV = 9.8%;LOA = -0.2–0.7), RASSF1 (CV = 15.0%;LOA = 0.0–1.3), and hTERT (CV = 10.8%;LOA = -2.3–8.1).
Data analysis
The outcome of interest was the absolute mean difference in percent DNA methylation between the exercise arm and the control arm at follow-up after adjusting for baseline values. This effect was estimated using linear regression whereby follow-up methylation was modeled as a function of baseline methylation and treatment arm. APC methylation was log transformed to address non-normality. Intent-to-treat and per-protocol analyses were conducted. In the per-protocol analysis, individuals randomized to the intervention arm were excluded if they did not adhere to at least 90% of the target exercise time. Within the intent-to-treat framework, subgroup analyses were conducted by testing the significance of the interaction term between treatment arm and the following covariates: age (<60/≥60 years), BMI (normal:18.5–24.9 kg/m2; overweight:25.0–29.9 kg/m2; obese:30.0+ kg/m2), baseline VO2max (above/below the median), and family history of breast cancer (yes/no).
As an exploratory analysis, we also investigated the association between the amount of physical activity, weight loss, and change in aerobic fitness that occurred during the intervention and differences in DNA methylation at 12-months.
Individuals with incomplete DNA methylation data were not included in the foregoing analyses. We assumed that these data were missing completely at random since any missingness would be due to shortcomings in laboratory methods. To assess the robustness of our results, we conducted a sensitivity analysis whereby the analyses were repeated after imputing DNA methylation values among individuals with partial outcome information. A multiple imputation procedure was used to impute ten datasets using DNA methylation and baseline covariate data as auxiliary variables. SAS v.9.4 was used for all analyses.
Results
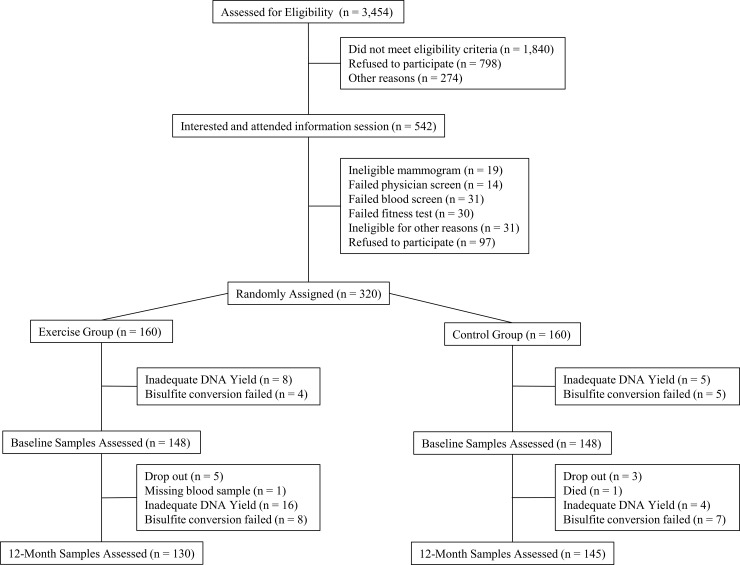
The flow of participants through the study and the numbers available for analysis are summarized in Fig 1. The two randomized groups were balanced with respect to baseline covariates (Table 1). The women included in these analyses had a mean age of 61.1 years (range = 50.6–74.9), a BMI of 29.2 kg/m2 (range = 20.4–43.9), and VO2max of 26.7 ml/kg/min (range = 11.5–52.2). Most of these participants (91.8%) were Caucasian.
Fig 1. CONSORT diagram of ALPHA trial ancillary study.
This Consolidated Standards of Reporting Trials (CONSORT) diagram describes the inclusion and exclusion of participants included in the current trial.
Table 1. Baseline characteristics of ALPHA trial participants included in intent-to-treat analysis.
| Baseline Characteristic | Exercise (N = 122) | Control (N = 134) |
|---|---|---|
| Age (years), mean (SD) | 61.5 (5.4) | 60.7 (5.6) |
| Caucasian, n (%) | 109 (90.1)a | 125 (93.3) |
| Never smoker, n (%) | 88 (75.9)b | 82 (63.1)c |
| Body mass index (kg / m2), mean (SD) | 28.8 (4.2) | 29.6 (4.3) |
| Weight (kg), mean (SD) | 74.7 (12.3) | 77.4 (12.9) |
| Total activity (MET-hours/week), mean (SD) | 116.7 (60.7) | 134.4 (81.9) |
| Physical fitness (VO2max, ml/kg/min), mean (SD) | 26.8 (6.2) | 26.6 (6.1) |
| Alcohol (g/day), median (IQR) | 4.6 (6.0)d | 5.1 (7.9)a |
| Folate (mcg DFE/day), median (IQR)e | 405.7 (175.7)d | 425.6 (191.2)a |
a N miss = 1
b N miss = 6
c N miss = 4
d N miss = 3
e Dietary folate equivalents (DFE)
A weak positive correlation was found between baseline LINE1 and Alu methylation (Spearman’s rho = 0.15, p<0.01). We did not detect a statistically significant correlation between any of the other DNA methylation measures at baseline (S2 Table).
The results of the intent-to-treat and per-protocol analyses are summarized in Tables 2 and 3 respectively. In both analyses, we did not detect a statistically significant difference in DNA methylation between the exercise and the control arms at follow-up after adjusting for baseline methylation values. The magnitude of the observed effect in these analyses was small–the absolute mean difference in estimated percent methylation between the two groups was less than 0.5% for all outcomes.
Table 2. Intent-to-treat analysis comparing changes in the DNA methylation outcomes between the exercise and control groups.
| N | Baselinea | 12-Monthsa | Difference at 12-Monthsb |
P-value | |
|---|---|---|---|---|---|
| LINE1 | |||||
| Control | 131 | 72.85 (72.59 to 73.10) | 73.01 (72.72 to 73.30) | Ref. | |
| Exercise | 120 | 72.83 (72.54 to 73.12) | 72.78 (72.54 to 73.02) | -0.23 (-0.60 to 0.14) | 0.23 |
| Alu | |||||
| Control | 131 | 19.11 (19.03 to 19.19) | 19.09 (19.00 to 19.17) | Ref. | |
| Exercise | 120 | 19.09 (19.01 to 19.18) | 19.12 (19.05 to 19.19) | +0.03 (-0.08 to 0.14) | 0.57 |
| APCc | |||||
| Control | 124 | 1.76 (1.65 to 1.88) | 1.73 (1.61 to 1.85) | Ref. | |
| Exercise | 109 | 1.79 (1.68 to 1.91) | 1.68 (1.58 to 1.78) | 0.97 (0.88 to 1.06)c | 0.52 |
| BRCA1d | |||||
| Control | 128 | 1.76 (1.68 to 1.85) | 1.79 (1.71 to 1.87) | Ref. | |
| Exercise | 117 | 1.77 (1.69 to 1.84) | 1.74 (1.66 to 1.81) | -0.02 (-0.09 to 0.05) | 0.60 |
| RASSF1 | |||||
| Control | 133 | 3.33 (3.24 to 3.42) | 3.22 (3.14 to 3.31) | Ref. | |
| Exercise | 121 | 3.26 (3.17 to 3.35) | 3.21 (3.11 to 3.30) | -0.01 (-0.14 to 0.11) | 0.84 |
| hTERT | |||||
| Control | 132 | 17.32 (16.73 to 17.90) | 17.58 (16.96 to 18.20) | Ref. | |
| Exercise | 117 | 16.64 (16.08 to 17.21) | 17.55 (16.88 to 18.23) | +0.20 (-0.67 to 1.07) | 0.65 |
a Estimated mean (95% C.I.) percent methylation
b Estimated mean difference (95% C.I.) in percent methylation between the exercise arm and control arm at follow-up after adjusting for baseline methylation
c The data were log transformed to address non-normality. Presented are the geometric means (95% C.I.) at baseline and 12-months. The estimated difference is the ratio of the geometric means of the exercise arm and control arm at follow-up after adjusting for baseline methylation.
d Four influential observations excluded from analysis (n exercise = 2; n control = 2)
Table 3. Per-protocol analysis comparing changes in the DNA methylation outcomes between the exercise and control groups.
| N | Baselinea | 12-Monthsa | Difference at 12-Monthsb |
P-value | |
|---|---|---|---|---|---|
| LINE1 | |||||
| Control | 131 | 72.85 (72.59 to 73.10) | 73.01 (72.72 to 73.30) | Ref. | |
| Exercisec | 79 | 72.86 (72.48 to 73.23) | 72.83 (72.52 to 73.14) | -0.18 (-0.62 to 0.26) | 0.42 |
| Alu | |||||
| Control | 131 | 19.11 (19.03 to 19.19) | 19.09 (19.00 to 19.17) | Ref. | |
| Exercisec | 79 | 19.13 (19.02 to 19.23) | 19.13 (19.03 to 19.23) | +0.04 (-0.09 to 0.18) | 0.51 |
| APCd | |||||
| Control | 124 | 1.76 (1.65 to 1.88) | 1.73 (1.61 to 1.85) | Ref. | |
| Exercisec | 72 | 1.80 (1.66 to 1.96) | 1.68 (1.56 to 1.81) | 0.97 (0.87 to 1.08)d | 0.59 |
| BRCA1e | |||||
| Control | 128 | 1.76 (1.68 to 1.85) | 1.79 (1.71 to 1.87) | Ref. | |
| Exercisec | 76 | 1.69 (1.60 to 1.77) | 1.75 (1.67 to 1.83) | +0.04 (-0.04 to 0.12) | 0.36 |
| RASSF1 | |||||
| Control | 133 | 3.33 (3.24 to 3.42) | 3.22 (3.14 to 3.31) | Ref. | |
| Exercisec | 79 | 3.28 (3.17 to 3.39) | 3.18 (3.07 to 3.29) | -0.04 (-0.18 to 0.09) | 0.55 |
| hTERT | |||||
| Control | 132 | 17.32 (16.73 to 17.90) | 17.58 (16.96 to 18.20) | Ref. | |
| Exercisec | 77 | 16.79 (16.06 to 17.52) | 17.67 (16.78 to 18.57) | +0.29 (-0.71 to 1.29) | 0.57 |
a Estimated mean (95% C.I.) percent methylation
b Estimated mean difference (95% C.I.) in percent methylation between the exercise arm and control arm at follow-up after adjusting for baseline methylation
c Excluded participants who did not adhere to 90% of target exercise time (i.e. an average of 180 min/week of moderate to vigorous aerobic activity over 12-months)
d The data were log transformed to address non-normality. Presented are the geometric means (95% C.I.) at baseline and 12-months. The estimated difference is the ratio of the geometric means of the exercise arm and control arm at follow-up and after adjusting for baseline methylation.
e Four influential observations excluded from analysis (n exercise = 2; n control = 2)
In subgroup analyses, no significant modification of the intervention effect by baseline age, BMI, estimated VO2max, or family history of breast cancer was observed for any outcome (p-interaction>0.05;data not shown).
The exploratory analyses examining the association between estimated change in VO2max and DNA methylation (Table 4) revealed a statistically significant negative dose-response association between change in physical fitness and RASSF1 methylation at 12-months after adjusting for age and baseline values (p-trend = 0.04). Weight loss and time spent exercising were not associated with any of the DNA methylation outcomes (Table 4).
Table 4. The association between the amount of exercise, weight loss, change in VO2 max and DNA methylation during a year-long aerobic physical activity intervention.
| Estimated Mean Difference at 12-months Adjusted for Age and Baseline Values (95% CI) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | LINE-1 | N | Alu | N | APCa | N | BRCA1b | N | RASSF1 | N | hTERT | |
| Time Exercisingc | ||||||||||||
| Controls | 131 | Ref. | 131 | Ref. | 124 | Ref. | 129 | Ref. | 133 | Ref. | 132 | Ref. |
| <150 min/wk | 30 | -0.41 (-1.01 to 0.19) p = 0.18 |
30 | 0.05 (-0.13 to 0.23) p = 0.61 |
25 | 0.90 (0.77 to 1.05) p = 0.16 |
29 | -0.15 (-0.27 to -0.02) p = 0.02 |
30 | 0.09 (-0.11 to 0.30) p = 0.36 |
29 | 0.14 (-1.27 to 1.55) p = 0.84 |
| ≥150 to <225 min/wk | 50 | -0.07 (-0.56 to 0.43) p = 0.79 |
50 | -0.02 (-0.17 to 0.13) p = 0.80 |
49 | 0.95 (0.85 to 1.07) p = 0.43 |
50 | -0.01 (-0.12 to 0.09) p = 0.81 |
51 | -0.10 (-0.26 to 0.07) p = 0.26 |
49 | -0.02 (-1.18 to 1.13) p = 0.97 |
| ≥225 min/wk | 40 | -0.27 (-0.81 to 0.26) p = 0.36 |
40 | 0.09 (-0.07 to 0.25) p = 0.29 |
35 | 1.04 (0.90 to 1.19) p = 0.60 |
39 | 0.08 (-0.04 to 0.19) 0.18 |
40 | 0.02 (-0.17 to 0.20) p = 0.87 |
39 | 0.39 (-0.86 to 1.64) p = 0.54 |
| p-trendd | 0.33 | 0.48 | 0.99 | 0.38 | 0.70 | 0.64 | ||||||
| Weight Losse | ||||||||||||
| Gain (>3%) | 26 | Ref. | 26 | Ref. | 25 | Ref. | 25 | Ref. | 26 | Ref. | 26 | Ref. |
| Maintenance (+/-3%) | 142 | -0.09 (-0.73 to 0.54) p = 0.78 |
142 | -0.05 (-0.24 to 0.14) p = 0.63 |
132 | 1.08 (0.92 to 1.25) p = 0.35 |
140 | 0.04 (-0.09 to 0.17) p = 0.57 |
141 | 0.01 (-0.21 to 0.22) p = 0.96 |
142 | -0.72 (-2.18 to 0.73) p = 0.33 |
| Minor loss(>3% to <5%) | 27 | -0.17 (-0.99 to 0.65) p = 0.69 |
27 | -0.02 (-0.27 to 0.23) p = 0.87 |
26 | 1.03 (0.85 to 1.25) p = 0.77 |
27 | 0.03 (-0.14 to 0.19) p = 0.75 |
27 | 0.04 (-0.24 to 0.31) p = 0.80 |
27 | -0.03 (-1.90 to 1.85) p = 0.98 |
| Meaningful loss (≥5%) | 54 | 0.00 (-0.71 to 0.71) p = 0.99 |
54 | -0.10 (-0.31 to 0.11) p = 0.36 |
47 | 1.09 (0.92 to 1.29) p = 0.34 |
54 | -0.02 (-0.17 to 0.12) p = 0.75 |
55 | 0.02 (-0.22 to 0.26) p = 0.86 |
53 | 0.21 (-1.42 to 1.85) 0.83 |
| p-trendf | 0.64 | 0.90 | 0.75 | 0.68 | 0.80 | 0.95 | ||||||
| Physical Fitnessg | ||||||||||||
| Meaningful decrease (≥10%) | 40 | Ref. | 40 | Ref. | 41 | Ref. | 40 | Ref. | 42 | Ref. | 41 | Ref. |
| Moderate to no change (+/-9.9%) | 100 | +0.02 (-0.55 to 0.58) p = 0.95 |
100 | +0.06 (-0.11 to 0.23) p = 0.47 |
92 | 0.90 (0.79 to 1.02) p = 0.10 |
99 | -0.13 (-0.24 to -0.02) p = 0.02 |
99 | -0.06 (-0.24 to 0.13) p = 0.56 |
99 | +0.85 (-0.44 to 2.14) p = 0.20 |
| Meaningful increase (≥10%) | 101 | +0.32 (-0.24 to 0.88) p = 0.26 |
101 | +0.07 (-0.10 to 0.23) p = 0.44 |
90 | 0.93 (0.81 to 1.05) p = 0.24 |
98 | -0.10 (-0.21 to 0.01) p = 0.09 |
102 | -0.17 (-0.35 to 0.02); p = 0.07 |
100 | +0.39 (-0.89 to 1.67) p = 0.65 |
| p-trendh | 0.15 | 0.53 | 0.46 | 0.29 | 0.04 | 0.90 | ||||||
a APC methylation was log-transformed to address non-normality. Presented are the estimated ratio (95% CI) of the geometric means at 12-months after adjusting for baseline differences
b Two influential observations were excluded from the analysis
c The average amount of moderate to vigorous aerobic activity completed during the intervention
d Assessed by modeling the rank of the categories as a measured variable
e The amount of weight lost during the intervention as a percentage of the participant’s baseline weight
f Assessed by modeling the median cut-points (gain = 4.51%; maintenance = 0.16%; minor loss = 4.23%; meaningful loss = 7.31%) as a measured variable
g The change in VO2 max that occurred during the intervention as a percentage of the participant’s baseline VO2 max
h Assessed by modeling the median cut-points (decrease = -16.80%; no = +1.62%; increase = 24.34%) as a measured variable
After imputing values for individuals with incomplete DNA methylation data, similar results for the intent-to-treat and per-protocol analyses were obtained (data not shown). In the exploratory analysis, however, the dose-response association between change in VO2max and RASSF1 methylation was attenuated and no longer statistically significant (p-trend = 0.16). Compared to those who had a meaningful decrease in VO2max, the estimated mean difference at 12-months in RASSF1 percent methylation was -0.04 (95% CI:-0.23–0.14) and -0.12 (95% CI:-0.31–0.07) for individuals who had moderate to no change and for those who had a meaningful increase respectively. No other noteworthy differences between the complete-case and multiple imputation analyses were observed.
Discussion
In this group of healthy, inactive, postmenopausal women, a year-long exercise intervention did not have a significant impact on levels of DNA methylation within LINE-1 and Alu repeats or within the promoter regions of APC, BRCA1, RASSF1, or hTERT. The current state of epidemiologic evidence pertaining to the epigenetic effects of physical activity is disparate and has been previously reviewed [10, 38–40]. Few candidate gene studies targeting APC, BRCA1, RASSF1, or hTERT have been conducted [40]. Our null findings with respect to APC, BRCA1, and RASSF1 are consistent with results from two previous cross-sectional candidate gene studies in healthy women [41, 42] and several epigenome-wide association studies [40]. These null results might suggest that DNA methylation within these genes is tightly regulated in healthy individuals and is resistant to environmental influence.
In contrast to our gene-specific measures, a greater number of investigations on the epigenetic effects of physical activity have focused on repetitive element DNA methylation, particularly LINE-1 methylation [40]. Consistent with our findings, a year-long randomized weight loss trial found that a dietary and exercise intervention did not impact LINE-1 methylation in a group of overweight sedentary women [43]. Similar null findings have also been reported in various observational settings [44, 45]. However, White et al. conducted a large cross-sectional study on 647 middle aged Caucasian women with a family history of breast cancer and found that childhood, teenage, and adulthood levels of physical activity were positively associated with LINE-1 methylation [46]. One possible explanation for this disparity may be the duration and timing of the exposure. The studies reporting null findings have assessed levels of physical activity over relatively short periods of time, ranging from four-day accelerometer assessments to one-year exercise interventions or past-year levels of physical activity. In contrast, White et. Al (2013) assessed lifetime levels of physical activity. These findings may suggest that LINE-1 methylation is relatively impervious to short term changes in physical activity and is more largely determined by levels of physical activity sustained over long periods of time.
In an exploratory analysis, we found a significant positive dose-response association between changes in VO2max and RASSF1 methylation. The direction of this finding is consistent with a cancer prevention mechanism. However, the magnitude of the estimated mean difference in RASSF1 methylation at 12-months between the highest and lowest categories of change in VO2max was 0.17% which may not be clinically meaningful. In addition, this analysis was post-hoc, the dose-response association was no longer statistically significant after imputing DNA methylation values for individuals with incomplete data, and multiple comparisons were made. As such, the finding of an association between VO2max and RASSF1 methylation should strictly be considered hypothesis-generating.
The weaknesses of this study should be acknowledged. The current study was an ancillary analysis of the ALPHA trial which was not originally designed to explore changes in DNA methylation. As such, we were forced to rely on the use of blood samples in our analyses which is a major limitation of this study. Specifically, the lack of an association between exercise and DNA methylation that we observed within blood may not reflect a lack of an association within breast, colon, endometrial, and other tissues of greater etiologic interest [47]. We were unable to estimate the distribution of white blood cell types in our analysis and measurement error arising from failure to adjust for cell-type distribution could partially account for our null findings [48]. Finally, the exclusion criteria used in the current study limits the generalizability of these results.
The strengths of this investigation should also be noted. The risk of bias due to confounding has been minimized through randomization. There was minimal attrition in this study since only nine (2.8%) participants were lost to follow-up for reasons unrelated to the intervention. Lastly, the intervention was also highly successful with respect to increasing physical activity levels. Among the individuals included in our intent-to-treat analysis, 70.5% completed 150+ min/week of exercise over the course of the intervention.
Future research should consider assessing levels of physical activity over extended periods of time. Studies may also wish to target more heterogeneous study populations to allow for the exploration of modification by cancer risk profile. DNA methylation investigations should ideally be carried out within the target tissue of interest and those conducted in blood should estimate the distribution of blood cells directly or should consider indirect estimation methods when available [49]. In addition, future studies could explore the association between physical activity and DNA methylation within genomic regions other than those investigated in the current analysis. Rather than focusing on cancer prevention, researchers could also consider exploring the mediating role of DNA methylation with respect to physical activity and cancer survival as previously done. [50]
Conclusion
A year-long aerobic exercise intervention did not impact levels of DNA methylation within LINE-1 and Alu repeats or within the promotor regions of the APC, BRCA1, RASSF1, and hTERT genes in the white blood cells of healthy, inactive, postmenopausal women. An exploratory analysis suggested that changes in physical fitness may be negatively associated with changes in RASSF1. Future large-scale studies examining changes in physical activity over longer periods of time in diverse study populations are required to validate these findings.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
The ALPHA Trial study set-up was performed by Kim van der Hoek and Marla Orenstein. The Study Coordinators were Rosemary Crosby, Ame-Lia Tamburrini, and Sarah MacLaughlin. Fitness Centre Managers were Ben Wilson, Lisa Workman and Diane Cook. Exercise Trainers were Shannon Hutchins, Kathy Traptow, Shannon Brown, Susan Daniel, Parissa Gillani, Stephanie Sanden, Karen Mackay, and Sandra Olsen. Data preparation for this ancillary study was done by Qinggang Wang. DNA extraction and quantification was performed by Angela Chan and Michelle Dean in the Translational Labratory. Dr. Nigel Brockton provided advice on the DNA extraction protocols developed for this study. DNA isolation and extraction was carried out by Angela Chan of the Tom Baker Cancer Centre Translational Laboratories. Rosalie Fréchette from the McGill University Génome Québec Innovation Centre supervised the DNA methylation assays.
Data Availability
There are ethical restrictions on sharing a de-identified dataset imposed by Alberta Health Services since the data contains sensitive and potentially identifying patient information. De-identified data from this study can be shared with other investigators by applying to the Cancer Committee of the Health Research Ethics Board of Alberta (cancer@hreba.ca) for access to the data. A data sharing agreement needs to be created between Alberta Health Services and the requesting investigator’s host institution. This data sharing agreement must be reviewed and approved by the Vice President of Research Administration of Alberta Health Services.
Funding Statement
The ALPHA Trial was originally funded by a research grant from the Canadian Breast Cancer Research Alliance (No. 13576 and No. 017468) and the Alberta Cancer Foundation (No. 22170) to Christine M Friedenreich. The ALPHA ancillary study was funded by grants from Canadian Institutes for Health Research and the Canadian Cancer Society (No. MOP-130238) to Christine M Friedenreich. Funding for the DNA extraction was provided by an Alberta Cancer Foundation grant for the Biospecimen Processing Unit within the Translational Laboratories at the Tom Baker Cancer Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Devon J Boyne was supported by funding from the Queen's University Terry Fox Foundation Training Program in Transdisciplinary Cancer Research in partnership with the Canadian Institutes for Health Research (CIHR) and an Ontario Graduate Scholarship and is currently supported by the University of Calgary Ruby Doctoral Recruitment Scholarship and the Faculty of Graduate Studies Doctoral Scholarship. Christine M Friedenreich was supported by an Alberta Innovates Health Solutions Health Senior Scholar Award and by the Alberta Cancer Foundation Weekend to End Women's Cancers Breast Cancer Chair. Kerry S Courneya is supported by the Canada Research Chairs Program.
References
- 1.Friedenreich CM, Neilson HK, Lynch BM. State of the epidemiological evidence on physical activity and cancer prevention. Eur J Cancer. 2010;46(14): 2593–604. doi: 10.1016/j.ejca.2010.07.028 [DOI] [PubMed] [Google Scholar]
- 2.Kyu HH, Bachman VF, Alexander LT, Mumford JE, Afshin A, Estep K, et al. Physical activity and risk of breast cancer, colon cancer, diabetes, ischemic heart disease, and ischemic stroke events: systematic review and dose-response meta-analysis for the Global Burden of Disease Study 2013. BMJ. 2016;354: i3857 doi: 10.1136/bmj.i3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore SC, Lee IM, Weiderpass E, Campbell PT, Sampson JN, Kitahara CM, et al. Association of Leisure-Time Physical Activity With Risk of 26 Types of Cancer in 1.44 Million Adults. JAMA Intern Med. 2016;176(6): 816–25. doi: 10.1001/jamainternmed.2016.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding D, Lawson KD, Kolbe-Alexander TL, Finkelstein EA, Katzmarzyk PT, van Mechelen W, et al. The economic burden of physical inactivity: a global analysis of major non-communicable diseases. Lancet. 2016;388(10051): 1311–24. doi: 10.1016/S0140-6736(16)30383-X [DOI] [PubMed] [Google Scholar]
- 5.Campbell KL, Foster-Schubert KE, Alfano CM, Wang CC, Wang CY, Duggan CR, et al. Reduced-calorie dietary weight loss, exercise, and sex hormones in postmenopausal women: randomized controlled trial. J Clin Oncol. 2012;30(19): 2314–26. doi: 10.1200/JCO.2011.37.9792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedenreich CM, MacLaughlin S, Neilson HK, Stanczyk FZ, Yasui Y, Duha A, et al. Study design and methods for the Breast Cancer and Exercise Trial in Alberta (BETA). BMC cancer. 2014;14: 919 doi: 10.1186/1471-2407-14-919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedenreich CM, Woolcott CG, McTiernan A, Ballard-Barbash R, Brant RF, Stanczyk FZ, et al. Alberta physical activity and breast cancer prevention trial: sex hormone changes in a year-long exercise intervention among postmenopausal women. J Clin Oncol. 2010;28(9): 1458–66. doi: 10.1200/JCO.2009.24.9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monninkhof EM, Peeters PH, Schuit AJ. Design of the sex hormones and physical exercise (SHAPE) study. BMC public health. 2007;7: 232 doi: 10.1186/1471-2458-7-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Gemert WA, Iestra JI, Schuit AJ, May AM, Takken T, Veldhuis WB, et al. Design of the SHAPE-2 study: the effect of physical activity, in addition to weight loss, on biomarkers of postmenopausal breast cancer risk. BMC cancer. 2013;13: 395 doi: 10.1186/1471-2407-13-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Irwin ML. Effects of Physical Activity on DNA Methylation and Associations with Breast Cancer In: Epigenetics, Energy Balance, and Cancer: New York: Springer; 2016. pp. 251–64. [Google Scholar]
- 11.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5): 698–711. doi: 10.1016/j.devcel.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 12.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging cell. 2015;14(6): 924–32. doi: 10.1111/acel.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochmanski J, Montrose L, Goodrich JM, Dolinoy DC. Environmental Deflection: The Impact of Toxicant Exposures on the Aging Epigenome. Toxicol Sci. 2017;156(2): 325–35. doi: 10.1093/toxsci/kfx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1): 50–5. doi: 10.1016/j.gde.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 15.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1): 27–36. doi: 10.1093/carcin/bgp220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ade C, Roy-Engel AM, Deininger PL. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 2013;3(6): 639–45. doi: 10.1016/j.coviro.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124(1): 81–7. doi: 10.1002/ijc.23849 [DOI] [PubMed] [Google Scholar]
- 18.Kemp JR, Longworth MS. Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Front Chem. 2015;3:68 doi: 10.3389/fchem.2015.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M, Agodi A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PloS one. 2014;9(10): e109478 doi: 10.1371/journal.pone.0109478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan K, Flanagan JM. Is there a link between genome-wide hypomethylation in blood and cancer risk? Cancer Prev Res (Phila). 2012;5(12): 1345–57. [DOI] [PubMed] [Google Scholar]
- 21.Joyce BT, Gao T, Zheng Y, Liu L, Zhang W, Dai Q, et al. Prospective changes in global DNA methylation and cancer incidence and mortality. Br J Cancer. 2016;115(4): 465–72. doi: 10.1038/bjc.2016.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PloS one. 2012;7(4): e34615 doi: 10.1371/journal.pone.0034615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garinis GA, Patrinos GP, Spanakis NE, Menounos PG. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002;111(2): 115–27. doi: 10.1007/s00439-002-0783-6 [DOI] [PubMed] [Google Scholar]
- 24.Ding Z, Jiang T, Piao Y, Han T, Han Y, Xie X. Meta-analysis of the association between APC promoter methylation and colorectal cancer. Onco Targets Ther. 2015;8: 211–22. doi: 10.2147/OTT.S75827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen C, Sheng Q, Zhang X, Fu Y, Zhu K. Hypermethylated APC in serous carcinoma based on a meta-analysis of ovarian cancer. Ovarian Res. 2016;9(1): 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi DT, Han M, Gao N, Tian W, Chen W. Association of RASSF1A promoter methylation with gastric cancer risk: a meta-analysis. Tumour Biol. 2014;35(2): 943–8. doi: 10.1007/s13277-013-1123-2 [DOI] [PubMed] [Google Scholar]
- 27.Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin epigenetics. 2016;8: 115 doi: 10.1186/s13148-016-0282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Long X. Association of BRCA1 promoter methylation with sporadic breast cancers: Evidence from 40 studies. Sci Rep. 2015;5: 17869 doi: 10.1038/srep17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis KA, Tollefsbol TO. Regulation of the Telomerase Reverse Transcriptase Subunit through Epigenetic Mechanisms. Front Genet. 2016;7: 83 doi: 10.3389/fgene.2016.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6(6): 584–93. doi: 10.1158/2159-8290.CD-16-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H, Zhou Z, Wei S, Liu Z, Pooley KA, Dunning AM, et al. Shortened telomere length is associated with increased risk of cancer: a meta-analysis. PloS one. 2011;6(6): e20466 doi: 10.1371/journal.pone.0020466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csizmadi I, Kahle L, Ullman R, Dawe U, Zimmerman TP, Friedenreich CM, et al. Adaptation and evaluation of the National Cancer Institute's Diet History Questionnaire and nutrient database for Canadian populations. Public Health Nutr. 2007;10(1): 88–96. doi: 10.1017/S1368980007184287 [DOI] [PubMed] [Google Scholar]
- 33.Friedenreich CM, Courneya KS, Neilson HK, Matthews CE, Willis G, Irwin M, et al. Reliability and validity of the Past Year Total Physical Activity Questionnaire. Am J Epidemiol. 2006;163(10): 959–70. doi: 10.1093/aje/kwj112 [DOI] [PubMed] [Google Scholar]
- 34.Kile ML, Baccarelli A, Tarantini L, Hoffman E, Wright RO, Christiani DC. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PloS one. 2010;5(10): e13730 doi: 10.1371/journal.pone.0013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suchiman HE, Slieker RC, Kremer D, Slagboom PE, Heijmans BT, Tobi EW. Design, measurement and processing of region-specific DNA methylation assays: the mass spectrometry-based method EpiTYPER. Frontiers Genet. 2015;6: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho V, Ashbury JE, Taylor S, Vanner S, King WD. Quantification of gene-specific methylation of DNMT3B and MTHFR using sequenom EpiTYPER(R). Data Brief. 2016;6: 39–46. doi: 10.1016/j.dib.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nygaard V, Rodland EA, Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17(1): 29–39. doi: 10.1093/biostatistics/kxv027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denham J, Marques FZ, O'Brien BJ, Charchar FJ. Exercise: putting action into our epigenome. Sports Med. 2014;44(2): 189–209. doi: 10.1007/s40279-013-0114-1 [DOI] [PubMed] [Google Scholar]
- 39.Horsburgh S, Robson-Ansley P, Adams R, Smith C. Exercise and inflammation-related epigenetic modifications: focus on DNA methylation. Exerc Immunol Rev. 2015;21: 26–41. [PubMed] [Google Scholar]
- 40.Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiol (Oxf). 2015;213(1): 39–59. [DOI] [PubMed] [Google Scholar]
- 41.Bryan AD, Magnan RE, Hooper AE, Harlaar N, Hutchison KE. Physical activity and differential methylation of breast cancer genes assayed from saliva: a preliminary investigation. Ann Behav Med. 2013;45(1): 89–98. doi: 10.1007/s12160-012-9411-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coyle YM, Xie XJ, Lewis CM, Bu D, Milchgrub S, Euhus DM. Role of physical activity in modulating breast cancer risk as defined by APC and RASSF1A promoter hypermethylation in nonmalignant breast tissue. Cancer Epidemiol Biomarkers Prev. 2007;16(2):192–6. doi: 10.1158/1055-9965.EPI-06-0700 [DOI] [PubMed] [Google Scholar]
- 43.Duggan C, Xiao L, Terry MB, McTiernan A. No effect of weight loss on LINE-1 methylation levels in peripheral blood leukocytes from postmenopausal overweight women. Obesity (Silver Spring). 2014;22(9): 2091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morabia A, Zhang FF, Kappil MA, Flory J, Mirer FE, Santella RM, et al. Biologic and epigenetic impact of commuting to work by car or using public transportation: a case-control study. Prev Med. 2012;54(3–4): 229–33. doi: 10.1016/j.ypmed.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics. 2011;6(3): 293–9. doi: 10.4161/epi.6.3.14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White AJ, Sandler DP, Bolick SC, Xu Z, Taylor JA, DeRoo LA. Recreational and household physical activity at different time points and DNA global methylation. Eur J Cancer. 2013;49(9):2199–206. doi: 10.1016/j.ejca.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15(4): r54 doi: 10.1186/gb-2014-15-4-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adalsteinsson BT, Gudnason H, Aspelund T, Harris TB, Launer LJ, Eiriksdottir G, et al. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PloS one. 2012;7(10): e46705 doi: 10.1371/journal.pone.0046705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics. 2017;18(1): 105 doi: 10.1186/s12859-017-1511-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng H, Irwin ML, Lu L, Risch H, Mayne S, Mu L, Deng Q, Scarampi L, Mitidieri M, Katsaros D, Yu H. Physical activity and breast cancer survival: an epigenetic link through reduced methylation of a tumor suppressor gene L3MBTL1. Breast Cancer Res Treat. 2012;133(1):127–35. doi: 10.1007/s10549-011-1716-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
There are ethical restrictions on sharing a de-identified dataset imposed by Alberta Health Services since the data contains sensitive and potentially identifying patient information. De-identified data from this study can be shared with other investigators by applying to the Cancer Committee of the Health Research Ethics Board of Alberta (cancer@hreba.ca) for access to the data. A data sharing agreement needs to be created between Alberta Health Services and the requesting investigator’s host institution. This data sharing agreement must be reviewed and approved by the Vice President of Research Administration of Alberta Health Services.