Abstract
Tuberculosis is the deadliest infectious disease worldwide. Although the BCG vaccine is widely used, it does not efficiently protect against pulmonary tuberculosis and an improved tuberculosis vaccine is therefore urgently needed. Mycobacterium tuberculosis uses different ESX/Type VII secretion (T7S) systems to transport proteins important for virulence and host immune responses. We recently reported that secretion of T7S substrates belonging to the mycobacteria-specific Pro-Glu (PE) and Pro-Pro-Glu (PPE) proteins of the PGRS (polymorphic GC-rich sequences) and MPTR (major polymorphic tandem repeat) subfamilies required both a functional ESX-5 system and a functional PPE38/71 protein for secretion. Inactivation of ppe38/71 and the resulting loss of PE_PGRS/PPE-MPTR secretion were linked to increased virulence of M. tuberculosis strains. Here, we show that a predicted total of 89 PE_PGRS/PPE-MPTR surface proteins are not exported by certain animal-adapted strains of the M. tuberculosis complex including M. bovis. This Δppe38/71-associated secretion defect therefore also occurs in the M. bovis-derived tuberculosis vaccine BCG and could be partially restored by introduction of the M. tuberculosis ppe38-locus. Epitope mapping of the PPE-MPTR protein PPE10, further allowed us to monitor T-cell responses in splenocytes from BCG/M. tuberculosis immunized mice, confirming the dependence of PPE10-specific immune-induction on ESX-5/PPE38-mediated secretion. Restoration of PE_PGRS/PPE-MPTR secretion in recombinant BCG neither altered global antigenic presentation or activation of innate immune cells, nor protective efficacy in two different mouse vaccination-infection models. This unexpected finding stimulates a reassessment of the immunomodulatory properties of PE_PGRS/PPE-MPTR proteins, some of which are contained in vaccine formulations currently in clinical evaluation.
Author summary
One of the major findings of the pioneering Mycobacterium tuberculosis H37Rv genome sequencing project was the identification of the highly abundant PE and PPE proteins, named after their N-terminal motifs Pro–Glu (PE) or Pro–Pro–Glu (PPE). Within the 20 years of research since then, many claims were made that PE/PPE proteins, including the two large subgroups encoded by repetitive sequences with very high GC content (PE_PGRS and PPE-MPTR families), are exported to the bacterial surface or beyond, and show broad immunomodulatory impact on host-pathogen interaction. We thus screened strains from different branches of the M. tuberculosis complex, including the attenuated Mycobacterium bovis BCG vaccine strains, for their capacity to export PE_PGRS/PPE-MPTR proteins. Strikingly, we found that BCG strains were unable to export the plethora of PE_PGRS/PPE-MPTR proteins due to the absence of the region of difference RD5, which in M. tuberculosis encodes PPE38, required for PE_PGRS/PPE-MPTR export. Surprisingly, the restoration of PE_PGRS/PPE-MPTR export by genetic complementation in recombinant BCG did not result in immunomodulatory changes or altered protection in mouse models. Our results thus put into perspective the numerous reports on virulence-associated immunomodulatory impact of individual PE_PGRS and PPE-MPTR proteins and open novel questions on their biological function(s).
Introduction
Tuberculosis is the deadliest infectious disease worldwide and is responsible for more than 1.7 million deaths per year [1]. Its causative agent, Mycobacterium tuberculosis, is a slow growing bacterium inherently resistant to many antibiotics. This problem is further exacerbated by rising levels of acquired drug resistance, resulting in multi-drug-resistant (MDR) and extensively-drug-resistant (XDR) strains of M. tuberculosis, which require treatment regimens of two years with low treatment success rates and severe side effects [1–3]. These worrying developments highlight the need for a successful vaccine, halting the transmission of tuberculosis [4]. The currently used vaccine is based on Mycobacterium bovis, attenuated through serial culture by Calmette and Guérin and therefore known as Bacille Calmette-Guérin (BCG) [5–7]. BCG is generally believed to protect relatively well against severe forms of disseminated tuberculosis in children, but is unable to induce full protection or halt transmission of M. tuberculosis in adolescents and adults [4,8,9]. Furthermore, even these protective traits are subject to controversy, which may be caused by the plethora of genomic mutations and recombination events that have accrued during the worldwide sub-culturing of the original BCG strain [5,6,10,11].
One possible reason for sub-optimal protection by BCG and other candidate vaccines is the absence or secretion defect of certain immunogenic proteins. M. tuberculosis secretes many proteins through its different secretion systems, including Sec-translocation (Sec), Twin-arginine-translocation (Tat), or Type VII secretion (T7S) systems [12,13]. M. tuberculosis possesses five different T7S systems called ESX-1 to ESX-5 [14]. The first T7S system to be discovered was ESX-1, identified by the Region of Difference (RD)1 deletion in BCG [15], responsible for the loss of ESX-1-mediated secretion in this vaccine strain [16,17]. Substrates of the ESX-1 system are responsible for the rupture of mycobacterium-containing phagosomes and represent a major virulence factor of pathogenic mycobacteria [18–21]. Corresponding to this information, the expression of the ESX-1 secretion system in BCG increased protective activity, but was also associated with increased pathogenesis [22]. Interestingly, a recently developed recombinant BCG strain expressing ESX-1 of Mycobacterium marinum was able to induce cytosolic pattern recognition and better protective responses, without a significant increase in virulence [23]. Similarly, the vaccine candidate MTBVAC was recently shown to induce immune responses to selected ESX-1 substrates and this ability was found to be the major determinant of improved protective efficacy as compared to BCG [24].
While the ESX-1 system is the best studied T7S system in mycobacteria, the ESX-5 system has the largest repertoire of substrates [25–27]. The ESX-5 system is essential for slow-growing mycobacteria, because of its role in outer membrane permeability [26,28]. Therefore, this system is present and considered functional in BCG. The coding sequences of the potential substrates of the ESX-5 system together form almost 8% of the coding potential of the M. tuberculosis genome [29]. Most notable amongst the ESX-5 substrates are the PE and PPE proteins, named for the proline and glutamic acid residues in their N-terminal domains. Defined functions have been described for some PE-PPE proteins, such as the lipase LipY [30,31] and PPE10, the latter of which is important for capsular integrity of M. marinum [32]. Furthermore, many studies have ascribed immunomodulatory functions to PE-PPE proteins, such as altering host cytokine responses by interaction with Toll-like receptors or inhibition of antigenic presentation [33–36]. However, most PE and PPE proteins have no known functions and their high degree of homology makes them difficult to study. The latter is especially true for the two most-recently evolved subgroups of ESX-5 substrates, i.e. the PE_PGRS and PPE-MPTR proteins. Both these sub-groups are characterized by their GC-rich DNA sequences, repetitive glycine-rich amino acid motifs and high molecular weight ranging up to ~365 kDa [27,29].
We recently identified the PPE protein PPE38 and its highly similar, duplicated variant PPE71, as essential factors in the secretion of both the PE_PGRS and PPE-MPTR proteins, in both M. marinum and M. tuberculosis [37]. The genes encoding PPE38 and PPE71 are organized in a 4-gene locus that also includes the esxX and esxY genes (Fig 1A), which however are not required for PE_PGRS secretion in M. tuberculosis strain CDC1551 [37]. Strains with naturally occurring, or engineered, loss-of-function mutations of the ppe38-locus were unable to secrete both PE_PGRS and PPE-MPTR proteins and were more virulent in a mouse infection model [37]. Indeed, deletion of the ppe38-locus occurred at the branching point of modern Beijing (Lineage 2) strains and may have aided in their global dispersal [37]. Moreover, the ppe38-locus was previously shown to be a hypervariable genetic region and many strains within the M. tuberculosis complex (MTBC) have polymorphisms in this locus [38]. Such polymorphisms are often caused by recombination events involving IS6110 elements [38,39]. Insertion, homologous recombination, deletion of IS6110 copies and/or deletion of intervening sequences between two IS6110 elements can lead to overexpression or gene deletion/truncation events with possible effects on transmission or virulence [37,40–43]. The most well-known of the polymorphisms affecting the ppe38-locus is the deletion of the RD5 region from BCG and several other animal-adapted strains of the MTBC [38,44]. The biological impact of the RD5 deletion has been a controversial subject of research and has focused solely on the phospholipase C encoding genes plcABC. Deletion of plcABC was reported to either attenuate [45] or increase virulence of M. tuberculosis [46]. However, a more recent study of the plc-genes in different mouse and cellular models showed no relevant contribution of these genes to the virulence of M. tuberculosis [47].
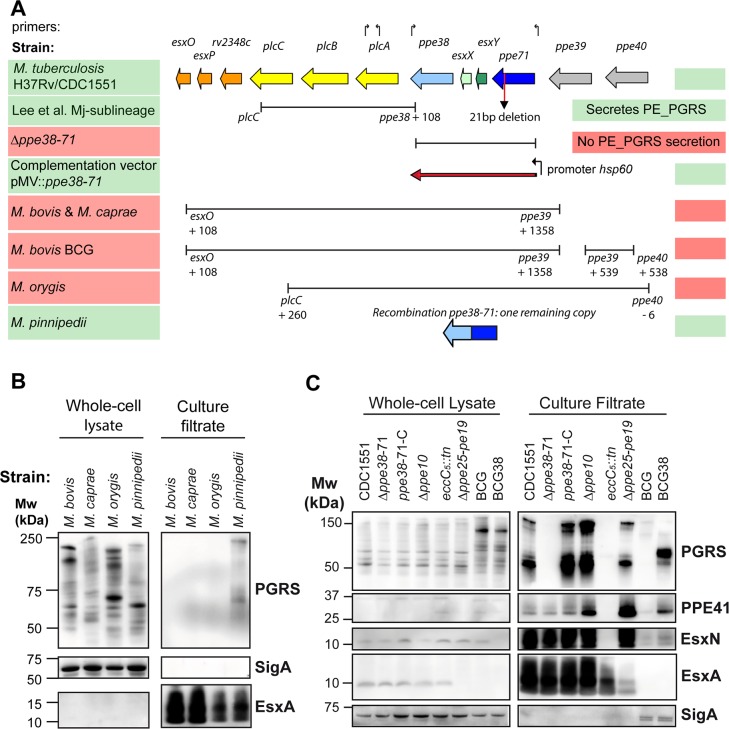
Fig 1. RD5-like genetic deletions in the M. tuberculosis complex and their effect on PE_PGRS secretion.
A) The genetic organization of the RD5 locus in M. tuberculosis strains CDC1551 and H37Rv is depicted in colored arrows. Bars below the genes indicate the size and location of different RD5-like and ppe38-deletions examined in this work. Arrows above the genes indicate primers used in this study to verify the presence of RD5 associated genes, sequences can be found in S4 Table. Functional PE_PGRS secretion is indicated by shading of the strain name in green, while red shading represents strains in which PE_PGRS secretion is not functional (based on immunoblot analysis). Figure adapted from Mc Evoy et al. 2009 with permission [38]. B) Immunoblot secretion analysis of animal-adapted MTBC strains verifies that strains with RD5 deletions do not secrete PE_PGRS proteins. Samples were prepared as described in materials and methods section. C) Immunoblot secretion analysis reveals PE_PGRS secretion defect in BCG, comparable to the M. tuberculosis ppe38-71-deletion strain or a general ESX-5 secretion mutant (eccC5::tn). SigA was used as a loading and lysis control. Some lysis could be found in both BCG and BCG38, but was not markedly different between strains. Please note that these immunoblots correspond to the same pre-cultures as those that were used in the immunogenicity experiment depicted in Fig 5 and therefore also include the Δppe10 and Δppe25-pe19 isolates. Full western blots corresponding to panels depicted in B-C are depicted in S5 Fig and S7 Fig, respectively.
Here, we investigated the effect of RD5-like polymorphisms of the ppe38-locus in a number of MTBC-branches and discovered that the RD5 deletions in animal-adapted strains and the BCG vaccine strains have profound effects on the repertoire of secreted substrates in these strains. Restoration of PPE38-dependent secretion results in a wider antigenic repertoire of BCG, whereby the identification of two immunogenic epitopes in one of the substrates, i.e. the PPE-MPTR protein PPE10, has allowed us to monitor the immunological impact of the corresponding secretion characteristics on host immune responses.
Results
Variation in PE_PGRS secretion in MTBC lineages and outgroups reveal genome sequence assembly problems
The genetically most-distant tubercle bacilli are represented by the Mycobacterium canettii clade. This outgroup mirrors the genomic diversity likely present within the ancestor of M. tuberculosis before branching and clonal expansion of the MTBC [48]. Recent studies of M. canettii have improved our understanding of adaptations that have shaped the transition from an M. canettii-like ancestor into extant M. tuberculosis, such as the gain of surface hydrophobicity through loss of lipooligosaccharide production [49] and the apparent loss of the capacity to exchange chromosomal DNA in the MTBC [50]. Interestingly, the available genome sequence information of five M. canettii isolates revealed potential polymorphisms in the ppe38-locus [48]. While strains D, K and L all possessed copies of the ppe38 and ppe71 genes, the sequence of strain J in the database indicated the potential absence of ppe38 and ppe71 from the strain. Such a deletion would be expected to affect PE_PGRS secretion [37]. However, secretion analysis revealed that all 5 isolates secreted PE_PGRS proteins (S1A Fig). Subsequent PCR analysis confirmed the presence of a complete ppe38-71 locus, similar to M. tuberculosis H37Rv, for all tested M. canettii strains, including strain J (S1B Fig). It is likely that the sequence polymorphisms in the previously deposited dataset may have arisen due to automated sequence assembly-associated bio-informatic artefact, which is a known problem for this region [37,38].
Another interesting group of strains, which were reported to have major polymorphisms in the RD5/ppe38-locus, was recently described by Lee et al. [51]. The Inuit population of the Nunavik region in Canada is affected by high levels of tuberculosis incidence. The majority of all cases in this cohort were shown to have resulted from the introduction of a single, particular M. tuberculosis strain, about one century ago. This sublineage was defined by genomic deletions, two of which affect the RD5/ppe38 locus. A 5,759 bp RD5-like deletion (CDC1551-D17) removed the three phospholipase C genes plcABC and truncated ppe38 (Fig 1A). The other ppe gene in this locus, ppe71 (mt2422), was reported to be affected by 22 bp frameshift deletion (Fig 1A)[51]. Reinvestigation of the sequence of ppe71 by inspection of the whole genome sequence data, and by PCR and Sanger sequence analysis revealed that this deletion was in fact a 21 bp deletion causing a 7 amino-acid deletion (Amino acids 354-MGGAGAG-361) relative to PPE71 of M. tuberculosis H37Rv, but not a frameshift. This deletion has been previously described to occur also in other strains of M. tuberculosis, including CDC1551 (MT2422 - http://www.genome.jp/dbget-bin/www_bget?mtc:MT2422) [38]. To test whether the RD5-like polymorphism negatively affects PE_PGRS secretion, five strains with and one strain without this deletion were subjected to secretion analysis by immunoblotting. All strains exhibited similar secretion levels of both PE_PGRS proteins and the ESX-1 substrate EsxA, as compared to reference strain CDC1551 (S1B Fig). These data show that the PPE71 variant carrying the MGGAGAG-deletion is able to sustain PE_PGRS secretion levels in M. tuberculosis, independently of truncation of PPE38. Furthermore, there is no apparent phenotypic difference when M. tuberculosis has one or two functional copies of PPE38/71.
RD5 deletions in animal-adapted strains and in M. bovis BCG block PE_PGRS secretion
A striking amount of different RD5-like polymorphisms are present in the animal-adapted lineages/ecotypes of M. tuberculosis complex. These strains share their most recent common ancestor with M. africanum Lineage 6 [52,53], which is reported to have two copies of ppe38/ppe71 [54]. Mycobacterium pinnipedi, a pathogen for seals and sea lions, has one intact copy of the ppe38 gene, but no esxXY-genes (Fig 1A). M. bovis and Mycobacterium caprae share an identical RD5 deletion, while Mycobacterium orygis possesses a unique RD5 deletion (Fig 1A) [38,55,56]. To investigate the effect of RD5 deletions on PE_PGRS secretion in animal-adapted strains, we performed secretion analysis of M. bovis, M. caprae, M. orygis and M. pinnipedi (Fig 1B). As expected, M. pinnipedi was the only tested species able to secrete PE_PGRS proteins in concordance with the presence of one functional copy of ppe38 (Fig 1B). In contrast, M. bovis, M. caprae and M. orygis were deficient in PE_PGRS secretion, while EsxA secretion was not affected and no marked cell lysis occurred (Fig 1B). Intracellular PE_PGRS expression was detected in strains with a secretion defect and was strikingly different between isolates (Fig 1B).
Since M. bovis and M. caprae share the same RD5 deletion with M. bovis BCG, we hypothesized that this vaccine strain is also deficient in PE_PGRS secretion (Fig 1A). Indeed, BCG did not secrete PE_PGRS proteins comparable to M. tuberculosis-Δppe38-71 and the ESX-5 deficient strain eccC5::tn (Fig 1C) [37,57]. This PE_PGRS secretion defect was at least partially restored in the recombinant BCG strain complemented with the M. tuberculosis ppe38-71-locus, which we have called BCG38. As expected, BCG and BCG38 were deficient in secretion of the ESX-1 substrate EsxA (ESAT-6) and exhibited only low levels of PPE41 and EsxN. The increase of PPE41 secretion in BCG38 compared to the parental strains (Fig 1C) was consistent in this experiment and other replicates (S1C and S1D Fig). Furthermore, five different M. bovis BCG isolates, which were selected for their relative genetic distance [6,10], were all deficient in PE_PGRS secretion (S1D Fig), emphasizing that all BCG strains are likely unable to secrete PE_PGRS proteins. It is of interest to note that M. bovis BCG Tice secretes higher levels of the ESX-5 substrates PPE41 and EsxN (S1D Fig), likely because of its genetic duplication of the esx-5 genetic locus [10]. However, despite this ESX-5 duplication, BCG Tice is unable to secrete PE_PGRS proteins. The PE_PGRS secretion defect of BCG was not restored in a previously constructed BCG strain with a cosmid containing the complete RD5 region of M. tuberculosis H37Rv (S1C Fig) [16]. In contrast, introduction of the ppe38-71 locus from M. tuberculosis on an integrative plasmid constitutively expressing these genes under control of the hsp60 promoter [37], partially restored PE_PGRS secretion of recombinant M. bovis BCG (Fig 1C, S1C and S1D Fig). This finding was especially surprising since emergence of RD5-deleted M. bovis/M. caprae progenitor strains likely dates back thousands of years [52].
Taken together, our data show that the different BCG vaccines are all deficient for the secretion of PE_PGRS proteins and that this is at least partially revertible by complementation with the ppe38-71 locus of M. tuberculosis. Based on our previous work, this secretion defect is expected to affect up to 89 proteins classified as PE_PGRS or PPE-MPTR [27,37].
Secretion of PE_PGRS/PPE-MPTR proteins in M. tuberculosis or BCG does not alter phenotypic and functional maturation of host innate immune cells, or antigenic presentation
The ability to restore PPE38-dependent secretion in M. bovis BCG allowed us to investigate to what extent this secretion defect affects properties of the BCG vaccine. Many of the 89 members of the PE_PGRS and PPE-MPTR proteins have been suggested to perform biological roles in virulence and immune modulation, although the molecular mechanisms and biological relevance remain unestablished for most of these [14,27,33,34]. Increasing the repertoire of immunogenic proteins secreted by BCG could lead to increased protection, since protein secretion by mycobacteria is essential for the efficient induction of protective CD4+ T-cell responses [22,58–61]. However, restoring secretion of proteins that have been proposed to exhibit immunomodulatory functions could also decrease efficacy of the vaccine strain. In particular, recent reports suggest that PPE38 itself downregulates Major Histocompatibility Complex class-I (MHC-I) expression in murine macrophages [62] and that PE_PGRS47 inhibits autophagy and is responsible for reducing MHC-II-restricted antigen presentation during in vivo infection of mice [35].
We set out to establish whether presence of PPE38 and the ability to secrete PE_PGRS and PPE-MPTR proteins, affected phenotypic and functional maturation of infected murine innate immune cells. Bone marrow-derived dendritic cells (BM-DCs) of C57BL/6 mice were infected (MOI = 0.5) with isogenic M. tuberculosis [37] or BCG strains, with or without the ppe38-locus. All the infected BM-DCs exhibited a clear upregulation of co-stimulatory markers CD40, CD80 and CD86, as well as modulation of MHC-I (H-2Kb) and MHC-II (I-Ab) expression, compared to uninfected controls. However, no differences in the induction of any such phenotypic maturation markers could be observed for the different isogenic WT and recombinant strains (Fig 2, S1 Table). Quantification of several inflammatory cytokines in the culture supernatants of the infected BM-DCs showed highly similar levels of TNFα, IL-12p40/70 and IL-6 production induced by the isogenic strains of BCG and M. tuberculosis (Fig 2B). These results indicate that PPE38-dependent secretion defects are unlikely to have a major effect on the phenotypic or functional maturation of DCs, even though many PE_PGRS and PPE-MPTR proteins have previously been suggested to perform such biological roles [33,35].
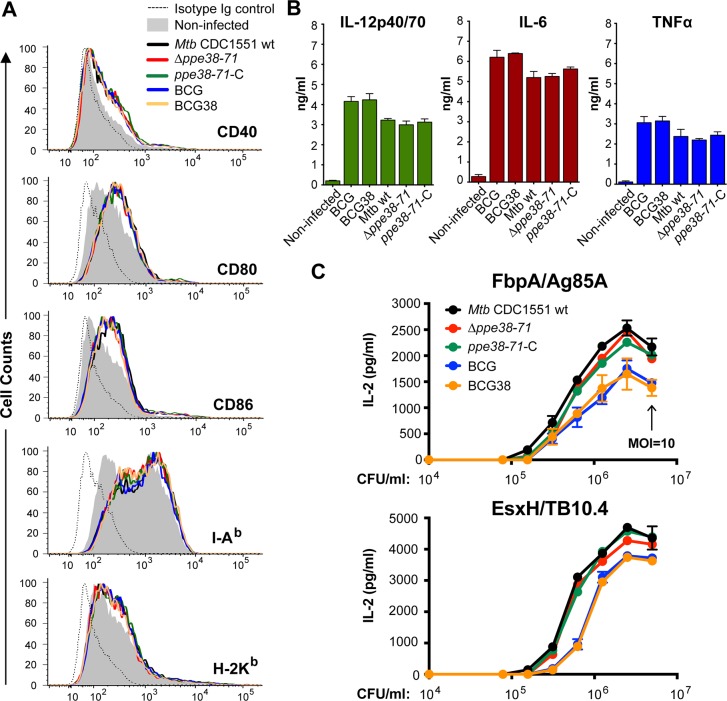
Fig 2. Secretion of PPE38 and PE_PGRS/PPE-MPTR proteins in BCG or M. tuberculosis does not alter phenotypic and functional maturation, or antigen presentation by innate immune cells.
A) BM-DCs (C57BL/6, H-2b) infected with the indicated mycobacterial strains were stained for surface expression of co-stimulation markers CD40, CD80 and CD86, or MHC components I-Ab and H-2Kb. Depicted are the cell counts (Y-axis) and fluorescent intensity (X-axis) as quantified by flow cytometric analyses. Quantification of mean fluorescent intensity and quantification of cell survival can be found in S1B Table) Culture supernatant of the experiment described in A was assessed for the presence of cytokines IL-12p40/70, IL-6 and TNF-α. No differences were detected between cells infected with the isogenic BCG or M. tuberculosis isolates. C) Antigenic presentation by infected DCs is not affected by disruption or restoration of PPE38–dependent protein secretion in M. tuberculosis or BCG. BM-DCs (BALB/c, H-2d) were infected with two-fold dilutions (data points in graph) of the indicated M. tuberculosis or BCG strains starting at MOI = 10 (indicated by black arrow). IL-2 production was quantified by ELISA after overnight co-culture with I-Ed-restricted T-cell hybridoma specific for FbpA (Ag85A101-120 (2A1), upper panel) or with I-Ad-restricted T-cell hybridoma specific for EsxH (TB10.474−88 (1G1), lower panel). Data are representative of biological duplicates.
In addition, we assessed whether PPE38-dependent protein secretion influences MHC-II-restricted presentation of other mycobacterial antigens. Such a phenotype might possibly be caused by a direct effect on the host phagocytes due to restored PE_PGRS secretion [35,36], or by competition in the hosts antigen presentation machinery upon secretion of the large number of PPE38-dependent substrates. To test this hypothesis, BM-DCs were infected with serial two-fold dilutions of M. tuberculosis or BCG strains with and without the ppe38-locus. IL-2 secretion in culture medium by MHC-II restricted T-cell hybridomas specific to FbpA (Ag85A101-120 –Fig 2C, upper panel) or EsxH (TB10.474−88 –lower panel) was quantified by ELISA as a measure of antigen presentation and hybridoma T-cell activation. Higher levels of IL-2 were detected in response to M. tuberculosis strains compared to BCG strains, but no differences were observed between isogenic strains with, or without, functional PPE38-dependent PE_PGRS/PPE-MPTR secretion. These data show that PPE38-dependent PE_PGRS/PPE-MPTR secretion does not reduce MHC-II-restricted antigen presentation of other mycobacterial antigens by the host DCs.
Together, these results suggest that introduction of PPE38 and restoration of PE_PGRS secretion do not negatively affect phenotypic and functional maturation of innate immune cells, or their capacity to present antigen to CD4+ T cells.
Restoration of PPE38-dependent PE_PGRS/PPE-MPTR protein secretion in BCG does not impact protection potential against M. tuberculosis in mice
Since we found no evidence suggesting that antigen presentation of mycobacterial antigens by DCs is negatively affected by restoration of PPE38-dependent secretion, we hypothesized that the enlarged repertoire of secreted proteins in BCG38 could increase its vaccine potential compared to the parental BCG. In parallel, we hypothesized that the capsule of BCG could be altered upon restoration of PPE38-dependent secretion. We recently reported that transposon insertions in the gene encoding an ESX-5 associated chaperone (espG5), or in the PPE-MPTR encoding gene ppe10 (mmar_0761), reduce capsule integrity of M. marinum [32]. Similarly, an eccC5::tn mutant in the M. tuberculosis strain CDC1551, completely deficient in ESX-5 secretion, also exhibited reduced capsule integrity [32,57]. Since PPE10 is dependent on PPE38 for its secretion [37], we hypothesized that restoration of PPE10 secretion might positively affect capsule integrity. The presence of an intact capsule on BCG, achieved by culturing in detergent-free growth medium, has recently been shown to be important for a more potent immune response and could therefore be relevant for the protective efficacy of BCG38 [63].
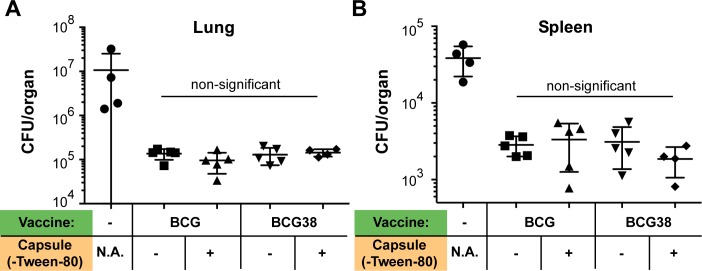
To test both hypotheses, C57BL/6 mice were subcutaneously (s.c.) immunized with 1 million CFU of either BCG, or BCG38, cultured either in shaking condition in the presence of 0.025% Tween-80, or in unperturbed conditions without detergent. Four weeks post-immunization, mice were challenged by an aerosol infection of M. tuberculosis H37Rv (bacterial load: 680 CFU/lung at Day 1, prepared without detergent). Mice were killed four weeks post infection, at which time lungs and spleens were harvested and assessed for bacterial burdens by CFU counting. An approximate 100-fold reduction in bacterial lung burdens was achieved by all conditions of vaccination irrespective of the presence of detergent, or the BCG vs BCG38 vaccine strains (Fig 3A). This reduction of bacterial lung burden coincided with improved macroscopic state of the lungs (S2A Fig). Similarly, an approximately 10-fold reduction in spleen CFUs and reduction in splenomegaly was detected in the vaccinated mice irrespective of the method of vaccine preparation (Fig 3A, S2B Fig). No significant (p<0.05) differences in bacterial burdens were observed between any of the four tested conditions in either the spleens or lungs. Together, these results show that restoration of PPE38-dependent PE_PGRS/PPE-MPTR secretion in BCG does not significantly improve protection against M. tuberculosis in the murine model used. Moreover, we did not find a significant difference in protective efficacy between conventional and detergent-free preparation of either BCG or BCG38, suggesting that capsular integrity is not altered or does not affect protection in this model.
Fig 3. Restoring PPE38-dependent protein secretion of BCG does not increase protection against M. tuberculosis in C57BL/6 mice.
Lung (A) or spleen (B) bacterial burdens of C57BL/6 mice infected with M. tuberculosis H37Rv via aerosol administration. Mice were vaccinated s.c. four weeks before the challenge, with 1 x 106 CFU/mouse of either BCG or BCG38 (indicated in green). Both strains were prepared, either in standard culture conditions in medium containing 0.025% Tween-80 considered as no capsule (indicated with (-)), or in culture allowing capsule formation/retention in detergent free condition (indicated with (+)). Photographs of the assessed organs are depicted in S2A and S2B Fig. Each data point represents the CFU/organ of one single mouse counted and averaged from two technical duplicates. Error bars depict the standard deviation. Differences between different vaccination conditions were non-significant (p>0.05), but all vaccination conditions were statistically different from the unimmunized control group (p<0.01). For simplicity, this latter information is not depicted in the figure. Significance was calculated with Prism software using ordinary one-way ANOVA followed by Tukey’s test for multiple comparisons.
Identification of immunogenic T-cell epitopes of the PPE-MPTR protein PPE10
Secretion of T7S-mediated mycobacterial proteins is essential to induce host CD4+ T-cell responses and the great majority of immunogenic and protective antigens of M. tuberculosis are secreted proteins [64]. Many of the known immunodominant antigens are PE and PPE proteins and these form an integral part in a number of subunit or recombinant vaccines [58,65–68]. Therefore, the finding that restoration of PPE38-dependent PE_PGRS/PPE-MPTR secretion in BCG did not significantly affect protective efficacy was surprising, particularly as up to 89 individual proteins are predicted to be concerned. In order to explain these unexpected data, we reflected on our hypotheses and found additional variables that could affect the assumptions on which they are based. In particular, while PPE-MPTR secretion was shown to be strictly dependent on PPE38 in both M. marinum and M. tuberculosis, we had no direct evidence of PPE-MPTR secretion in BCG38. In contrast to PPE-MPTR proteins, PE_PGRS proteins may not contain immunodominant epitopes or be protective antigens [69–72]. Furthermore, although previous studies have found a strict correlation between in vitro secretion and the capability to induce CD4+ T-cell responses [23,58,68], it is conceivable that the PPE38-dependant substrates are still membrane, or surface, associated in ppe38-71-deficient strains and thereby remain able to induce T-cell responses.
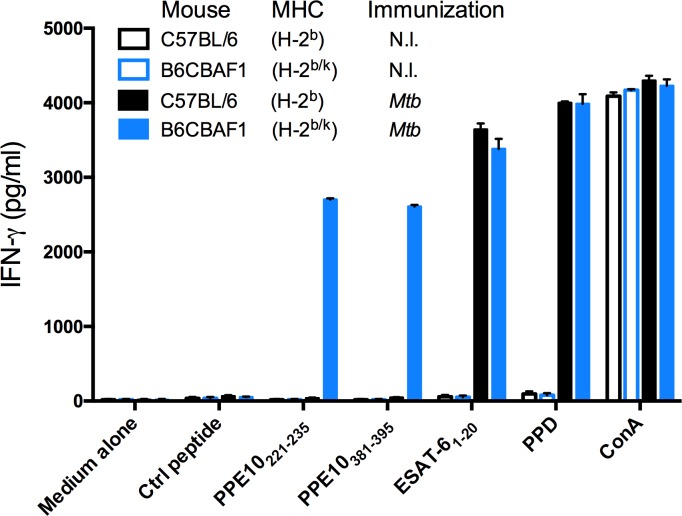
Since tools to study PPE-MPTR proteins are scarce and currently insufficient to answer the questions above, we set out to develop an immunological approach to study PPE-MPTR secretion and their immunogenicity in more detail. We selected PPE10 as a model MPTR-protein, because PPE10 is predicted to be the most ancestral MPTR protein in mycobacteria [27]. The PPE domain covers the N-terminal 181 residues of PPE10 and is highly similar to other PPE proteins. The middle of the protein contains a typical MPTR repeat domain, which is very similar to other MPTR proteins. The C-terminus contains a domain unique to PPE10, which is secreted in vitro [25,32,37]. PPE10 is also of biological interest, since it is detected in vivo in guinea pig lungs and this protein is required for capsular integrity of M. marinum [32,73]. We set out to assess whether PPE10 has the potential to induce CD4+ T-cell mediated immune responses in mice. To increase the likelihood of identifying immunogenic epitopes, we immunized not only C57BL/6 mice, but also C57BL/6 x CBA (H-2b/k) F1 mice, which express a more diverse repertoire of MHC restricting elements (S2 Table). Mice were s.c. immunized with wild-type M. tuberculosis H37Rv and were killed three weeks later. Splenocytes were isolated and stimulated in vitro with a peptide library consisting of sixty 15-mers with a 5-amino acid shifting frame spanning PPE10181-487 of M. tuberculosis H37Rv [29,74]. None of the sixty peptides were able to induce specific T-cell mediated IFN-γ responses by splenocytes from unimmunized mice or immunized C57BL/6 mice (S3 Fig). However, two peptides were immunogenic in the C57BL/6 x CBA (H-2b/k) F1 mice and induced high levels of IFN-γ, similar to the positive control peptide ESAT-61−20 (Fig 4, S3 Fig). Interestingly, one of these immunogenic peptides (PPE10221-235: GSGNTGSGNLGLGNL) was situated in the MPTR domain of PPE10, while the other (PPE10381-395: NVLNSGLTNTPVAAP) was derived from the PPE10-specific C-terminal domain. The MPTR peptide PPE10221-235 has 17 close homologues within the M. tuberculosis genome (identity > 65%, but no 100% homologues), while this was not the case for PPE10381-395 (S3 Table). These results show that immunization with M. tuberculosis induces immune responses against PPE10 and that this response can be elicited both against the PPE10-specific C-terminal domain or the MPTR domain.
Fig 4. Epitope mapping of PPE10 identifies two novel immunogenic T-cell epitopes.
C57BL/6 H-2b (black) or C57BL/6 x CBA (H-2b/k) F1 mice (B6CBAF1, blue) were immunized s.c. with 1 x 106 CFU/mouse of M. tuberculosis H37Rv (Mtb, filled bars), or were left non-immunized (N.I. empty bars). Three weeks post-immunization, splenocytes were stimulated with control peptides or a library of 15-mers spanning PPE10 excluding the PPE domain. T-cell mediated IFN-γ responses were quantified buy ELISA as a measure of immunogenicity. Two immunogenic PPE10-peptides were identified (PPE10221-235 & PPE10381-395) in B6CBAF1 mice. Error bars depict standard deviation over two technical replicates. This figure depicts only newly identified epitopes and controls. Full results of the pep-scan epitope mapping can be found in S2 Fig.
Deletion of ppe10 does not significantly alter protein secretion of other Type VII secretion substrates in M. tuberculosis
The newly identified immunogenic peptides derived from PPE10 are a tool that allowed us to answer different questions regarding the PPE-MPTR proteins. First, to determine the specificity and cross-reactivity of the epitopes, we constructed a deletion mutant of ppe10 (Rv0442c) in the M. tuberculosis CDC1551 background by homologous recombination and phage transduction (S4 Fig) [75]. In contrast to M. marinum-ppe10::tn [32], no altered colony morphology or other growth phenotype was observed in M. tuberculosis-Δppe10. This finding is in concordance with the absence of such a phenotype in ESX-5 mutants of M. tuberculosis and highlights this as a species-specific difference between M. marinum and M. tuberculosis [32,57,76].
We performed biochemical secretion analysis on the Δppe10 strain in parallel with the strains that were examined (see below) for their immunogenic potential (Fig 1C). In contrast to a previous report, we found that M. tuberculosis Δppe25-pe19 did secrete PPE41 and EsxN, which may be due to differences in bacterial growth conditions and/or methods in protein extraction and detection [76]. This strain harbors intact genes coding for the ESX-5-membrane complex [57,77] and is able to induce in vivo CD4+ T-cell responses against PE and PPE proteins, in contrast to the general ESX-5 deficient strain ΔeccD5 in the same background [58,68]. The Δppe10 strain showed no difference in PE_PGRS secretion. Similarly, secretion of EsxA and EsxN was not affected by deletion of ppe10. Although slightly elevated levels of PPE41 secretion were observed, we concluded from these combined data that M. tuberculosis-Δppe10 does not have a general supersecretion phenotype as was previously reported for M. marinum-ppe10::tn [32].
BCG and M. tuberculosis-Δppe38-71 are unable to induce immune responses against PPE10
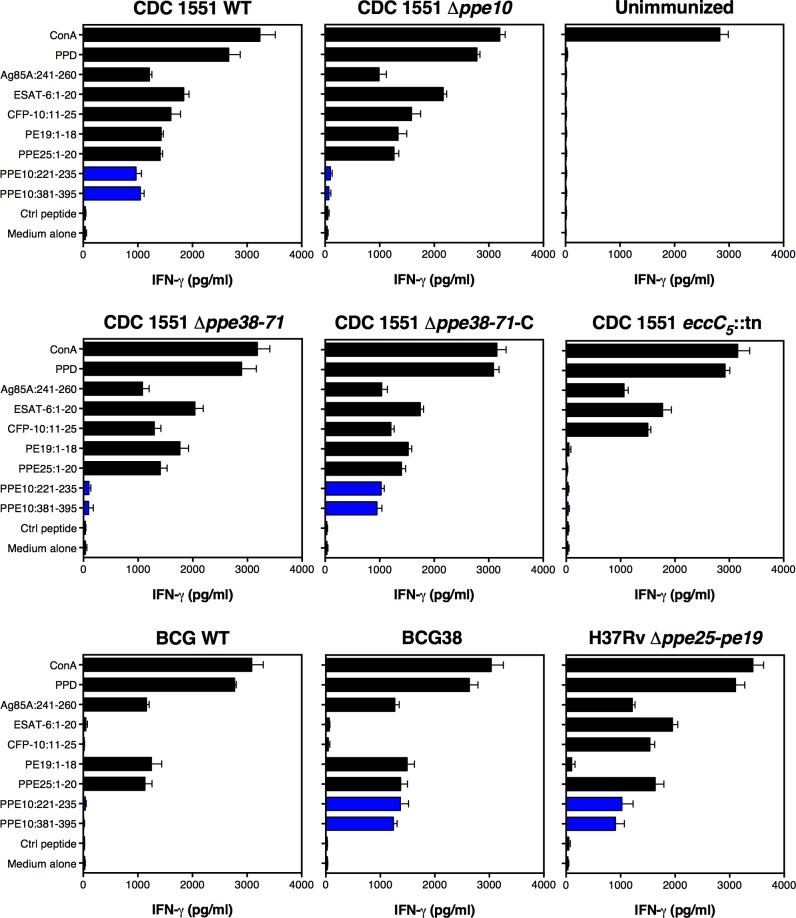
To assess the specificity of the newly identified PPE10 epitopes and to better understand the effect of the ppe38-dependent secretion on immunogenicity, we immunized C57BL/6 x CBA F1 mice with the different M. tuberculosis and BCG strains for which the secretion phenotype was characterized (Fig 1C). Three weeks post-immunization, splenocytes were collected and stimulated with the PPE10221-235 and PPE10381-395 peptides, as well as purified protein derivate (PPD—a positive control for immunization by Mycobacteria) and a number of known antigenic peptides derived from proteins secreted via ESX-1 (EsxA1-20 [78] and CFP-1011−25 [79]), ESX-5 (PE191-18 and PPE251-20 [58]) or the twin-arginine-translocation (TAT) pathway (Ag85A241-260) [80,81]. As expected, splenocytes of mice immunized with M. tuberculosis CDC1551 produced high levels of IFN-γ after stimulation with PPE10221-235, PPE10381-395 or all other immunogenic peptides, but not when incubated with a negative control peptide (E. coli MalE100-114), or the medium control (Fig 5). The Δppe10 deletion strain did not induce IFN-γ production in response to either PPE10221-235, or PPE10381-395, whereas responses against the other peptides were unaffected (Fig 5). Unexpectedly, this result shows that both of the newly identified PPE10 peptides are highly specific, even though we hypothesized cross-reactivity to occur for PPE10221-235, because of the high similarity to other MPTR domains (S3 Table). As expected, the ESX-5 secretion mutant eccC5::tn did not induce T-cell responses against the ESX-5 substrates PE19, PPE25 and PPE10, further confirming that the export of these antigens by the ESX-5 secretion system is indispensable for the induction of T-cell immune responses [58,68,82]. Importantly, Δppe38-71 was not able to induce immunogenicity against either of the PPE10 epitopes, a phenotype that was fully reverted in the complemented strain ppe38-71-C. This confirms that secretion and in vivo immunogenicity of PPE10 as a model PPE-MPTR protein are dependent on PPE38 in the M. tuberculosis CDC1551 background, which we were previously unable to assess. Similar to Δppe38-71, also BCG was completely unable to induce immune responses against either of the PPE10 epitopes. In contrast, BCG38 induced immunogenicity against both PPE10 epitopes at similar levels to the M. tuberculosis isolates. Together, these results clearly confirm that the secretion and in vivo immunogenicity of the ancestral PPE-MPTR protein PPE10 is strictly dependent on PPE38. These data also provide evidence that the in vitro observed PPE38-dependence of PE_PGRS and PPE-MPTR proteins is a phenotype that can be directly translated to the in vivo situation. Here, we show that the vaccine strain BCG is unable to induce T-cell responses against the ancestral PPE-MPTR protein PPE10, because of the deletion of its ppe38-71-locus as part of RD5.
Fig 5. Ability of mycobacteria to induce T-cell responses against PPE-MPTR protein PPE10 is dependent on functional ESX-5- and PPE38-dependent secretion.
C57BL/6 x CBA F1 mice were immunized with the indicated mycobacterial strains. Three weeks post-immunization, splenocytes were stimulated with the indicated peptides and IFN-γ production was measured by ELISA. Responses to the newly identified PPE10-derived immunogenic peptides are depicted in blue. Error bars represent the standard deviation over two technical duplicates. The results are representative of two biological replicates performed on different timepoints.
Finally, we compared the results obtained for the different WT and recombinant BCG strains with a recently developed attenuated M. tuberculosis strain, deleted for 5 pe/ppe genes in the esx-5 locus, named MtbΔppe25-pe19 [76]. Genes encoding the ESX-5 secretion core machinery [57,77] are intact in this strain, as is the ppe38 gene, a finding which is confirmed by the fact that this strain induced T-cell responses against both PPE10 epitopes. This result highlights that attenuated M. tuberculosis vaccine strains may avoid certain M. bovis related secretion differences that result in immunogenic properties.
Prime-boost vaccination regimen to improve PPE10-specific immune responses does not increase protection against M. tuberculosis
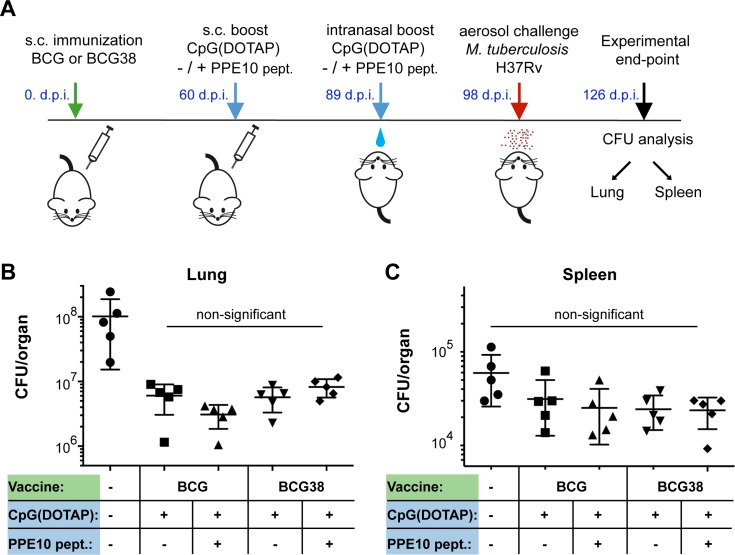
The results of our epitope mapping analysis showed that C57BL/6 mice were unable to develop T-cell responses against PPE10, which could provide an explanation for the lack of improved protection conferred by BCG38 compared to BCG. Therefore, we performed a similar experiment in these C57BL/6 x CBA F1 mice, designed to maximize any potential increase in PPE-MPTR-specific immune responses, by boosting vaccination of BCG or BCG38 with the immunogenic PPE10- peptides (Fig 6A). Sixty days after s.c. vaccination with BCG strains, a CpG(DOTAP)-formulated peptide booster, or the adjuvant alone, was administered s.c., followed by an intranasal booster twenty-nine days later. Mice were challenged by an aerosol challenge of M. tuberculosis H37Rv nine days after the final booster and were killed 28 days later to assess lung and spleen bacterial burdens (Fig 6B and 6C). No significant differences were observed among the groups of vaccinated animals. Only a modest decrease in spleen CFUs was achieved by any of the vaccination regimens. This reduction was not significant (p<0.05) for BCG-vaccinated mice and injected with the adjuvant alone, but was significant for the three other groups. However, no significant differences between any of the vaccinated groups was detected. Vaccination with all regimens reduced lung CFU values at least 10-fold. In fact, vaccination with BCG38, boosted with PPE10-derived immunogenic peptides, had the highest average bacterial lung burden of the four different vaccination regimens. These data clearly oppose our hypothesis, that restoring the lack of PPE-MPTR immune responses in BCG increases its protective efficacy.
Fig 6. Boosting PPE10 specific immune responses does not increase protection against M. tuberculosis.
A) Graphical representation of the prime-boost vaccination protocol. Mice were immunized with either BCG or BCG38 (Green). 60 days post-infection (d.p.i.) C57BL/6 x CBA F1 mice were injected s.c. with a booster consisting of adjuvant CpG(DOTAP), alone or in combination with a mix of PPE10221-235 and PPE10381-395 peptides (blue). The same formulation was intranasally administered four weeks later. Nine days after the intranasal boost, mice were exposed to M. tuberculosis H37Rv aerosol infection (220 CFU/lung 1 d.p.i). Bacterial lung (B) and Spleen (C) burdens were assessed by dilution and counting 4 weeks post-infection (experimental end-point) after being photographed for macroscopic investigation (S2C and S2D Fig). Each data point represents the CFU value of one organ from a single mouse, error bars depict the standard deviation. No significant differences between the vaccination conditions were detected by ordinary one-way ANOVA followed by Tukey’s test of multiple comparisons. All vaccination conditions resulted in a significant (p<0.01) reduction in lung burden compared to unimmunized controls (Ordinary one way ANOVA; Dunnett’s test of multiple comparisons against a single control). For simplicity, this latter information is not depicted in the figure. Reduction in spleen CFUs was not significant for any of the vaccination conditions. Statistical analyses were performed using PRISM software.
Together, we could find no evidence of an immunomodulatory effect of PPE38-dependent proteins. Inversely, restoration of BCG’s capacity to secrete PE_PGRS and PPE-MPTR proteins and thereby enlarging the PE_PGRS/PPE-MPTR antigenic repertoire of BCG, did not result in improved vaccine protection in two mouse models.
Discussion
We previously demonstrated that loss-of-function mutations in the ppe38-locus of M. tuberculosis block PE_PGRS and PPE-MPTR secretion and increase virulence in a mouse model [37]. In this work, we examined the correlation of known ppe38 deletions in other lineages of the MTBC with a PE_PGRS/PPE-MPTR secretion defect. We hypothesized that the success of certain clinical isolates of Lineage 4 could perhaps be explained by their RD5-like deletion, which includes ppe38 [51,83]. However, secretion analysis of these Lineage 4 strains revealed that a single copy of PPE71 carrying a 21 bp deletion (corresponding to the loss of amino acids MGGAGAG), seems to be functional and sufficient to support PE_PGRS secretion. Similarly, although intriguing differences in protein secretion levels were observed between M. canettii strains, we found that all analyzed strains secreted PE_PGRS proteins. The anticipated polymorphisms in the ppe38-locus of selected M. canettii strains [48] were likely caused by a sequence assembly problem of repetitive sequences. These results highlight the difficulties of bio-informatic analyses of this locus, which is hampered by the high sequence similarity between ppe38 and ppe71, that seem to cause already some discrepancies between the reference genomes of M. tuberculosis H37Rv and CDC1551 [29,37,38,84].
In contrast, our investigation of RD5-like polymorphisms did reveal that multiple members of the animal adapted lineage of the MTBC are completely devoid of PE_PGRS secretion because of their RD5 deletion. It should be emphasized that the RD5-like deletion of M. orygis occurred independently of that of M. bovis and M. caprae. Furthermore, even more members of the animal adapted lineage, such as M. microti, M. suricattae and the Dassie Bacillus, are reported to have independent RD5-like deletions, which we hypothesize to also block PE_PGRS and PPE-MPTR secretion [38,85–87]. Together, these findings suggest a specific selective advantage associated to loss of the ppe38-locus and its associated secretion phenotype in certain animal adapted strains. The modern Beijing strains, also defective in PPE38-dependent secretion, have expanded concurrently with increased human population densities and mobility [88]. These changes in the host-population alter the optimal balance between virulence/infectivity and lower the advantage to stay dormant or subclinical in the host [89]. It is tempting to speculate that the loss of PPE38 and its associated secretion and virulence phenotype has helped ancestral M. tuberculosis strains derived from human hosts, to adapt towards survival and transmission in a new host niches.
We were surprised that we were able to restore the secretion defect of BCG by introducing the ppe38-locus from M. tuberculosis. Since the RD5 deletion of BCG already occurred in the most-recent common ancestor of M. bovis and M. caprae, this deletion likely dates back millennia [52]. Furthermore, the 13 years of in vitro culturing by Calmette and Guérin to create BCG and the ensuing decades of culturing while it was distributed worldwide has caused accumulation of even more mutations [6,10,11]. Still, introduction of the integrative vector constitutively expressing the ppe38-locus was clearly able to at least partially restore both PE_PGRS and PPE-MPTR secretion in BCG.
Our newly identified immunogenic epitopes in the PPE-MPTR protein PPE10, provide a tool to gain more understanding about this group of proteins. Firstly, although previous work only definitively detected the C-terminal domain of PPE10 to be secreted [25,32,37], immunization with M. tuberculosis also clearly induced immune responses against the MPTR-associated epitope. This provides evidence that the MPTR domain is accessible to the host antigen presentation machinery and that these repetitive domains have the potential to contain functional T-cell epitopes. Furthermore, wild-type BCG and M. tuberculosis with impaired PPE38-dependent secretion were completely unable to induce immune responses against PPE10, similar to a general ESX-5 secretion mutant. We recently reported that processing, presentation on MHC-II and recognition of immunogenic proteins by CD4+ T-cells occurs only when the protein is transported over the inner membrane by its cognate TypeVII secretion system [82]. Secreted proteins, but also the cell-wall associated protein PE19 induced strong CD4+ T-cell responses, whereas this was not the case in an ESX-5 deficient isolate, where PE19 is only present in the cytoplasm or associated to the plasma membrane [82]. Together with the current work, this is important evidence that PPE38 is essential for the translocation of PPE-MPTR proteins through the ESX-5 secretion machinery in vivo and that without PPE38, these proteins are not surface associated or otherwise accessible to the immune system.
It is perhaps striking that the PE_PGRS and PPE-MPTR secretion defect of BCG has not been previously reported, considering the amount of research done on this vaccine. Based on the available literature on PE_PGRS and PPE-MPTR proteins, it is logical to hypothesize that a vaccine strain that does not secrete these proteins might in fact be a relatively effective vaccine. Many immunomodulatory properties have been attributed to PE_PGRS and PPE-MPTR proteins [14,33,34,67]. Perhaps the most relevant of these, is the reported function of certain PE_PGRS proteins to inhibit antigen presentation [35,36]. If PE_PGRS proteins indeed inhibit antigen presentation, it would be highly detrimental to introduce a vaccine that secretes these proteins. Notably, this is an urgent question since a number of novel tuberculosis vaccine candidates based on attenuated M. tuberculosis are currently in clinical or pre-clinical development. We showed for one of these candidate vaccines (i.e. M. tuberculosis-Δppe25-pe19), that PE_PGRS and PPE-MPTR secretion is indeed fully functional [58,68,76]. Our isogenic Δppe38-71 strains of M. tuberculosis and the BCG38 strain form an ideal tool to answer such questions and to understand more about these proteins as a group. In this work, we did not find any evidence of inhibition of antigen presentation in strains secreting PPE38-dependent substrates, or lack thereof in strains without PPE38. Similarly, and in contrast to many reports of immunomodulatory effects of PE_PGRS proteins, we did not find any evidence of differential immune modulation by strains with, or without, functional PPE38-dependent secretion. More specifically, no differences were observed in DC maturation [90], MHC-I or -II expression [62] or cytokine production [91–93]. Finally, PE_PGRS and PPE-MPTR proteins have often been implicated as mycobacterial virulence factors [14,34,35,94,95]. The previously described increased virulence in strains lacking PPE38-dependent secretion, including the hypervirulent Beijing isolates, put this work in perspective [37]. Here, we bolster our previously published evidence that strains without PPE38, including a number of animal adapted species and the BCG vaccine, are truly unable to translocate these proteins. Although many of these animal adapted strains have reduced virulence in humans compared to M. tuberculosis, they are clearly pathogenic for their natural host and should not be seen as attenuated [96]. This is in line with a role for PPE38-dependent substrates as virulence attenuating factors [37]. The biological roles of the PE_PGRS and PPE-MPTR proteins that are reported to be required for virulence, may not require secretion of these effector proteins or might in certain cases be due to indirect effects on other proteins. This hypothesis is further supported by the fact that many of the studies that attribute virulence traits to PE_PGRS and PPE-MPTR proteins, are performed in M. smegmatis, which lacks an ESX-5 secretion system and is unable to secrete these proteins [27,33,77]. Further work on the biological function of PE_PGRS and PPE-MPTR proteins, either on an individual basis or grouped, will have to take into account these findings and critically assess the impact of localization on effector function. Whether complementation of the ppe38-locus in BCG might further attenuate BCG strains or otherwise affect persistence in the host was not studied in further detail, in part because BCG38 did not confer superior protection to M. tuberculosis infection. However, we do not expect significant virulence differences between BCG and BCG38, since PPE38-dependent virulence effects generally occur in the chronic infection stage [37], when BCG is already expected to be cleared.
Perhaps the most relevant finding of this work is that BCG is unable to secrete PE_PGRS and PPE-MPTR proteins and therefore does not raise T-cell responses against these proteins. Previous studies have shown that antibodies can be raised against PE_PGRS proteins, suggesting that it could be a beneficial property of a vaccine to secrete these proteins [25,97,98]. Here we provide evidence that PPE-MPTR proteins can be immunogenic in mice, which is further supported by a recent publication investigating immunogenicity of the PPE-MPTR protein PPE39 [72]. Kim et al. identified two immunogenic epitopes of which one (MTBK_2482085−102) is located in the PPE-domain and has high homology to non-MPTR PPE proteins, while the other (MTBK_24820217−234) was located in the MPTR domain of this protein. Interestingly, the authors reported that vaccination with the recombinant PPE39 protein induced a higher level of protection against M. tuberculosis Erdman, compared to a hypervirulent Beijing isolate [72]. This difference might be explained by our data, which suggest that immune responses against the MPTR epitope would not be helpful against a PPE38-deficient Beijing isolate. A related issue that requires further work is whether the PPE38-dependent secretion effect in modern Beijing isolates is somehow related to that of the BCG vaccine and whether their respective secretion defects affect vaccine efficacy.
There is strong evidence for the importance of PPE-MPTR proteins in human immune responses, because the PPE-MPTR protein PPE42 (Rv2608) is an integral part of the subunit fusion-protein vaccine candidate ID93 [65,71]. The fusion protein ID93 consists of four different proteins and has been tested as a vaccine candidate in both a Phase 1 and Phase 2A clinical trial [99,100]. Bertholet et al. 2008 demonstrated that PBMCs isolated from PPD+ healthy subjects produced IFN-γ in response to PPE42 and that almost 70% of subjects showed a reaction against the recombinant protein in a recall experiment [71]. Interestingly, 100% of PPD+ subjects exhibited recall responses against the other (non-MPTR) PPE proteins that were tested, which could possibly be explained by exposure to modern Beijing, or other PPE38-deficient strains, in the subject cohort. PPE42 was selected as part of the ID93 vaccine due to its excellent ability to induce both humoral and cellular immune responses and immunization with PPE42 provided protection in mice almost comparable to BCG [65,71]. In Guinea pigs, ID93 significantly boosted the protection induced by BCG, which was interpreted as an ability to boost immune responses elicited by BCG [65]. However, based on our work it should be assumed that BCG does not induce immune responses against the PPE-MPTR protein PPE42 and that boosting with ID93 may in fact broaden antigenic repertoire of the combined vaccination. Similarly, ID93 is able to induce protective immune responses to the M. tuberculosis Beijing isolate HN878, but it is unclear what the role of PPE42 is in this response. The analyses performed in Bertholet et al. 2010 and Baldwin et al. 2015 were performed with the four-gene fusion protein ID93 and not with the individual PPE42 subunit, which makes it impossible to assess these questions more thoroughly. What remains clear however, is that the PPE-MPTR protein PPE42 is an important part of a vaccine currently in clinical trials. The finding that ID93 includes a protein to which parental BCG is likely not able to induce immune responses, may actually put the proven booster qualities of this vaccine candidate in a different light and lead to optimal strategies to employ it.
The question whether immune responses against PPE38-dependent proteins are important for a vaccine to be protective against tuberculosis, needs an urgent answer, especially since it concerns a total of 89 proteins. There are multiple vaccine candidates in clinical, or pre-clinical, development that are based on attenuated M. tuberculosis strains and which likely secrete PE_PGRS and PPE-MPTR proteins [24,58,67,68,76,101]. Should we knock-out ppe38-71 in these vaccine candidates to avoid immune modulation by the secreted substrates, or should we prioritize these vaccine candidates, because they have a broader potential repertoire of epitopes? Should BCG vaccination be boosted by vaccine candidates including PPE-MPTR proteins such as ID93, or should this be avoided? Are there differences between designing vaccine candidates against strains secreting PE_PGRS/PPE-MPTR proteins and those with a PPE38-dependent secretion defect, such as the modern Beijing isolates? Are murine or other small animal infection models appropriate to predict PE_PGRS and PPE-MPTR-mediated impact on vaccine efficacy? These are questions that we are not yet able to answer in this work, but they reveal the need to increase our understanding of PE_PGRS and PPE-MPTR proteins. Better knowledge on PE_PGRS/PPE-MPTR proteins is not just an intellectual goal, but may also help to make more informed decisions in the design of novel vaccines against tuberculosis.
Methods
Strains and growth conditions
All strains used in the study and the sources they are derived from can be found in S5 Table. Unless otherwise specified, all mycobacterial strains were grown on Middlebrook 7H11 solid medium (Difco) supplemented with OADC (BD Biosciences), or liquid 7H9 medium supplemented with ADC supplement and 0.05% Tween-80. Antibiotics were added where opportune at a concentration of 50μg/ml for Hygromycin (Euromedex), or 25μg/ml for Kanamycin (Sigma). Strains were incubated at 37ºC. Liquid cultures were grown in shaking conditions at 80 rotations per minute. For animal-adapted strains M. bovis, M. caprae, M. orygis and M. pinnipedii, 0.2% w/v of Pyruvate (Sigma) was added to the growth medium [102]. Infection stocks of M. tuberculosis H37Rv used for aerosol infection experiments and BCG or BCG38 vaccination stocks without Tween-80 were prepared by inoculating 0.1 OD/ml bacteria in 100ml liquid culture without Tween-80. This culture was incubated for 7 days, after which it was washed with phosphate buffered saline (PBS) and sonicated (5x (100 pulses of 0.1s)) and left to rest for at least one hour before collecting the cell suspension considered to obtain a single-cell solution of encapsulated mycobacteria. Standard vaccination stocks were prepared in Dubos medium containing 0.025% Tween-80 in standing conditions and were harvested at an optical density between 0.4 and 0.7 OD600/ml.
PCR verification of RD5 deletions
RD5 deletions were PCR verified by previously published primers specific for plcA (rv2351c, S4 Table), which produce a product of approximately 500bp when this gene is present [52]. Primers amplifying the ppe38-71-locus (S4 Table) produce a 3378bp product when the complete ppe38-71 locus is present [38]. This includes two copies of ppe38/71 (mt2419/mt2422) flanking the esxX (mt2420) and esxY (mt2421) in between in CDC1551. When only one copy of ppe38 and no esxX/esxY are present this PCR produces a product of approximately 1500 bp [38].
Recombinant strains and mutant construction
The complementation plasmid containing the ppe38-locus from CDC1551 (mt2419-22) under expression of hsp60 promoter was previously described [37]. The cosmid containing the RD5 region (pYUB::RD5) was part of the library described by Bange et al. 1999 and contains the genetic region spanning 2,611 kb– 2,645 kb of the M. tuberculosis H37Rv reference genome [29,103].
M. tuberculosis-Δppe10 was constructed as described by Bardarov et al. [75]. The homologous recombination construct was created by a PCR combining primers PPE10 KO LF & LR to amplify the 3’ end of rv0442c and another PCR with primers PPE10 KO RF & PPE10 KO RR to amplify the 5’ end of rv0442c (See S4 Table for primer sequences). After phage packaging and infection, seven transformed colonies were tested by PCR with either primer PPE10(mtb) flank F & p0004s-HR, or PPE10(mtb) flank R & p0004s-HL (S4A and S4B Fig). All colonies were found to have the correct deletion spanning from 152bp to 1133bp after the 5’ of rv0442c. We attempted to complement the Δppe10 mutant with a previously published plasmid (p19kPro::rv0442c-HA) overexpressing HA-tagged PPE10 under control of the lpqH promotor [25]. Although clones expressing the HA-tag on this plasmid were obtained, these had a considerable in vitro growth defect, which would conflict with in vivo and in vitro studies and therefore this complemented strain was not analyzed further.
Secretion analysis
Strains were pre-cultured until mid-logarithmic phase under normal growth conditions (described above). Cultures were washed two times in 7H9 medium without ADC, supplemented with 0.2% Dextrose and 0.05% Tween and were incubated in this medium for 48 hours. Cultures were centrifuged to separate cells and the supernatant was filtered through a 0.02μm filter, after which it was TCA-precipitated to concentrate. Cellular material was washed with PBS, resuspended in solubilisation/denaturation buffer and boiled for 10 min at 95°C. After sterilisation by heating for 2 hours at 80°C, samples were sonicated to disrupt cells and boiled at 95°C during 10 minutes.
Samples were loaded on 12% or 4–12% SDS-Page gels (NuPage, Novex, Life technologies) and transferred to nitrocellulose filters by dry western blotting (iBlot, Invitrogen). Proteins were stained by primary antibodies: Anti-PGRS 7C4.1F7 [25] (Clone 7C4.1F7 was a kind gift from Michael J. Brennan, USA), polyclonal anti-SigA (Kind gift from I. Rosenkrands, Denmark), Rabbit polyclonal anti-EsxN (rMTb9.9A) [104], monoclonal ESAT-6 (hyb76-8), or anti PPE41 [105].
Cell infection, ELISA and flow cytometry
BM-DCs derived from C57BL/6 (H-2b) female mice were generated directly in 6-well plates and infected at day 6 of culture with different mycobacterial strains at M.O.I of 0.5 in RPMI 1640-GlutaMax medium (Invitrogen) containing 10% FBS (4 x 106 cells/well in 4 ml volume). After over-night of infection at 37°C and 5% CO2, IL-6 (clone MP5-20F3 for coating and clone MP5-32C11 for detection, BD Pharmingen), IL12p40/70 (clone C17.8 RUO, BD Pharmingen) and TNF-α (clone 1F3F3D4 for coating and clone clone XT3/XT22 for detection, eBioscience) cytokine production was quantified in the culture supernatants by ELISA.
For viability and phenotypic maturation evaluation, infected DCs were washed with PBS and incubated first with Live/Dead-Pacific Blue reagent (Invitrogen) during 35 minutes at 10°C in the dark. Cells were then washed twice and incubated with appropriate dilution of anti-CD16/CD32 (2.4G2 mAb, BD Pharmingen) during 20 minutes followed by surface staining by 30 minutes of incubation with appropriate dilutions of APC-anti-CD11b (BD Pharmingen), PE-Cy7-anti-CD11c (BD Pharmingen), FITC-anti-CD40 (clone HM40-3, SONY), FITC-anti-CD80 (B7-1) (clone 16-10A1 Biolegend), FITC-anti-CD86 (B7-2) (clone PO3, SONY), FITC-anti-MHC-II (I-A/I-E) (clone MS/114.15.2, eBioscience), FITC-anti-MHC-I (H-2kb) (clone AF6-88-5-5-3, eBioscience) or FITC-anti-IgG1k isotype control. The stained cells were washed twice with FACS buffer (PBS containing 3% fetal bovine serum (FBS) and 0.1% NaN3) and then fixed with 4% paraformaldehyde during 18h at 10°C prior to sample acquisition by a LSR Fortessa flow cytometer system (BD Bioscience) and BD FACSDiva software. The obtained data were analyzed using FlowJo software (Treestar, OR, USA).
Antigen presentation assay
BM-DCs derived from BALB/c (H-2d) female mice were used at day 6 of culture as antigen presenting cells. Cells were seeded in 96-well plates at 5 x 104 cells/well and loaded with 1 μg/ml of homologous or negative control synthetic peptides, or infected with different mycobacterial strains with serial two-fold dilutions of M.O.I., starting at M.O.I. = 10, in RPMI 1640-GlutaMax medium (Invitrogen) containing 10% FBS. After 18h of infection at 37°C and 5% CO2, cells were washed twice with RPMI medium to eliminate the IL-2 possibly produced by the infected DCs and then co-cultured with 1 x 105 cells/well of T-cell hybridoma specific to EsxH/TB10.474−88 (1G1) or Ag85A101-120 (2A1), respectively restricted by I-Ad or I-Ed. After over-night of co-culture at 37°C and 5% CO2, the IL-2 secretion was quantified in the culture supernatants by ELISA (clone JES6-1A12 for coating and clone JES6-5H4 for detection, BD Pharmingen).
Epitope mapping of PPE10 and T-cell assay
A peptide library of sixty 15-mers with a 5-amino acid shifting frame, spanning amino acids 181–487 of PPE10 (Rv0442c), was constructed commercially (Mimotopes Europe, United Kingdom). Epitope screening of PPE10 and immunogenicity assays were performed as previously described [58], with some modifications. Briefly, 6-8-week-old female C57BL/6 (H-2b) or C57BL/6 x CBA F1 (H-2b/k) mice were immunized s.c. with 1 x 106 CFU/mouse of different mycobacterial strains obtained from exponential culture in Dubos medium. Epitope mapping was performed with mice immunized with M. tuberculosis H37Rv. Three to four weeks post-immunization, mice were sacrificed and pool of total splenocytes (n = 2 mice per group) were restimulated in 96-well flat-bottom plates (TPP, Den- mark) at 5 x 105cells per well in HL-1 medium (Biowhittaker, Lonza, France), complemented with 2 mM GlutaMax (Invitrogen, Life Technologies, France), 5 x 10−5 M β-mercaptoethanol, 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma-Aldrich, France) in the presence of 10–20 μg/ml of individual peptides. IFN-γ production in the supernatant was quantified by ELISA after 72h of culture at 37°C and 5% CO2 (clone AN-18 for coating and clone R46A2 for detection), BD Pharmingen.
Protection assays
BCG and BCG38 were grown in 10ml Dubos medium or in 100ml 7H9-medium with ADC-supplement without Tween-80. M. tuberculosis H37Rv and BCG-strains cultured without Tween-80 were sonicated (5 X 100 pulses; 0.1 seconds/pulse; 0.9 seconds’ rest; amplitude 30%) to disrupt clumps and were frozen at -80°C. Frozen stocks were counted for CFU’s before immunization to assess dose while the dose of Dubos-grown strains was estimated based on optical density.
Eight-week-old C57BL/6 mice (n = 5 mice/group), were immunized with 1 x 106 CFU/mouse of BCG Danish (cultured—or + Tween-80), or BCG38 (cultured—or + Tween-80) in 200 μl PBS. Eight mice were concurrently injected with sterile PBS. Thirty days after vaccination, mice were challenged with aerosolized WT M. tuberculosis H37Rv strain. Three mice were sacrificed to assess bacterial lung burdens 1 day post challenge (assessed at 680 CFU/lung). All other mice were killed four weeks post-challenge due to human end-point criteria of unvaccinated mice. Lungs and spleens were homogenized by beadbeating, serially diluted in PBS and plated on 7H11 plates with (Lungs) or without (spleens) BBL MGIT PANTA (Beckton Dickinson, Ireland).
The Prime-boost vaccination and challenge experiment was performed similar as above, with the following modifications. BCG or BCG38 were precultured in Dubos medium and five first generation C57BL/6 x CBA crossover mice were left unvaccinated or s.c. immunized (n = 5 mice/group). Eight weeks post-immunization, a subcutaneous boost was administered. This boost consisted of 200 μl/mouse of formulation containing 50 μl of each PPE10-derived peptide (PPE10221-235 and PPE10381-395) ProteoGenix, France, 30 μg of CpG 1826 oligodeoxynucleotides as adjuvant (Sigma-Aldrich, France) at 1 μl/mL concentration, 60 μl of liposomal transfection reagent DOTAP (N-[1-(2,3-DioleOyloxy)]-N,N,N-Trimethyl Ammonium Propane methylsulfate, Roche, France) and 10 μl Opti-MEM (Life Technologies, France) as described in Sayes et al., 2016 [68]). Four weeks later, an intranasal boost was given to mice via intra-nasal route, under anesthesia as described in Sayes et al., 2016, 25 μl/mouse contained 10 μg of PPE10 peptides, 2 μg of CpG at 10 μl/mL concentration, 10 μl of DOTAP and 3 μl Opti-MEM contained in 20 μl/mouse [68]. Ten days after the intranasal boost, mice were aerosol challenged with WT M. tuberculosis H37Rv strain. Three non-immunized mice were killed one day post challenge to assess infectious dose administered, which was calculated at 220 CFU/lung. Four weeks later all other mice were killed and one lung and the spleen were homogenized with a MillMixer organ homogenizer (Qiagen, Courtaboeuf, France) and plated to assess bacterial burdens on 7H11 Agar medium supplemented with ADC (Difco, Becton Dickinson). The CFU were counted after 3–4 weeks of incubation at 37°C.
All immunized and infected mice for immunogenicity and protection experiments were placed and manipulated in isolator in BSL-III protection-level animal facilities at the Institut Pasteur.
To determine the statistical significance of the data, analyses were performed by use of GraphPad Prism software (GraphPad Software, La Jolla, CA, USA), using ordinary one-way ANOVA followed by Tukey’s test for multiple comparisons.
Ethics statement
All animal experiments were performed in animal facilities that meet all legal requirements in France and by qualified personnel in such a way to minimize discomfort for the animals. All procedures including animal studies were conducted in strict accordance with European and French regulations (Directive 86/609/CEE and Decree 87–848 of 19 October 1987). All protocols were reviewed and approved by the Institut Pasteur Safety and Animal Care and Use Committee (Protocol 11.245) and the local ethical committee CETEA “Comité d'Ethique en Expérimentation Animale” (approved protocols CETEA 2012–0005 and CETEA 2013–0036).
Supporting information
Immunoblots of whole-cell lysates or culture filtrates of the indicated M. canetti (A), M. tuberculosis (B) or BCG (C, D) isolates [48,51]. A) Although differences in protein secretion could be observed between different M. canetti isolates (A-J), all isolates exhibited PE_PGRS secretion. B) PE_PGRS secretion of Mj-sublineage strains with a deletion affecting ppe38, but not ppe71 (Lanes 4–8) was not discernible from Lineage 4 control isolate CDC1551 or an isolate from the same cohort without this deletion (MT13848). C) Introduction of plasmid pMV::ppe38-71 in BCG complemented PE_PGRS secretion (BCG38), while complementation was not observed when performed with pYUB::RD5, even though presence of genetic presence of RD5 was PCR-confirmed with primers RD5B-plcA.int.F/R [52]. D) Immunoblot secretion analysis of five genetically divergent BCG isolates confirms the PE_PGRS secretion defect in all BCG isolates. Cop. 38 indicates the strain M. bovis BCG Copenhagen, transformed with vector pMV::ppe38-71 and is hereafter referred to as BCG38. Anti-SigA staining is uses as a lysis control in A and D, while anti-GroEL2 is used in B and C. Strain details can be found in S5 Table. Full blots of panels A-D are depicted in S5 Fig and S6 Fig.
(TIF)
Organs depicted in A and B correspond to the experiment depicted in Fig 3. Organs depicted in C and D correspond to the experiment depicted in Fig 6. After photography of the lungs (A, C), a single lung lobe was used for lung CFU quantification. Splenomegaly (B, D) was reduced, by all vaccination conditions, but did not differ markedly between vaccination conditions.
(TIF)
IFN-γ production in response to peptides covering the indicated amino acid positions of PPE10 (Rv0442c) in C57BL/6 (grey/green) or C57BL/6 x CBA F1 (B6CBAF1, blue/brown) mice. Mice were immunized with M. tuberculosis H37Rv (left) or unimmunized (right).
(TIF)
A) Schematic representation of deletion strategy and primers. The genetic region around PPE10, as taken from tuberculist, is depicted in colored arrows [74]. Flanking fragments used for homologous recombination are depicted in black bars. Left (PPE10KO-LF and PPE10KO-LR) and right (PPE10KO-RF and PPE10KO-RR) flanking regions were amplified by primers depicted in black. Primers used to verify successful homologous recombination are depicted in dark blue. All primer sequences can be found in S4 Table. B) PCR verification of successful homologous recombination in seven different colonies that grew on hygromycin selection plates. Colony 1 was taken for further analyses. Full gels used to create B are depicted in S7 Fig.
(TIF)
(TIF)
As expected, the percentage of live cells was higher for BCG-infected cells than for cells infected with M. tuberculosis strains, but did not vary significantly between isogenic strains (≤ 1.0% difference between isogenic strain). These values are derived from the experiment depicted in Fig 2A.
(PDF)
A promoter mutation disrupts production of I-Eαb in C57BL/6 mice (Grey font), which are therefore unable to produce MHC-II I-E (Grey). In contrast, H-2k mice can produce both I-Aαk I-Aαk and I-Eαk I-Eαk. C57BL/6 x CBA F1 mice have an even bigger repertoire of possible functional MHC-II isoforms available due to heterodimerization of α and β subunits from H-2b and H-2k haplotypes.
(PDF)
Black letters indicate identical amino acids. Red letters indicate non-identical amino acids. Top: homologues of the MPTR-containing peptide PPE10221-235 ordered by percentage of sequence identity. Bottom: Homologues of the peptide PPE10381-395, which is part of the C-terminal secreted domain of PPE10.
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors would like to thank Alexandre Pawlik and Fiona McIntosh for help. We also thank Michael J. Brennan for initially sharing a clone producing PE_PGRS antibody and acknowledge the BEI resources for the Mtb CDC1551 Transposon Mutant 1291 (MT1844, Rv1795) (BEI ID NR-14751). We further thank Robyn Lee and Anzaan Dippenaar for data analysis and James Gallant, Maroeska Burggraaf and Edith NG Houben for insightful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
LSA and RB acknowledge the support in part by grants of the Agence national de la Recherche ANR-14-JAMR-001-02, ANR-10-LABX-62-IBEID, and ANR-16-CE35-0009, the Fondation pour la Recherche Médicale FRM (DEQ20130326471) and the European Union's Horizon 2020 Research and Innovation Program grant TBVAC2020 643381. LSA and JWJvH are supported by the Netherlands Organisation for Scientific Research (Vidi grant 91717305 to JWJvH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Global tuberculosis report 2017. WHO. World Health Organization; 2017; Available: http://www.who.int/tb/publications/global_report/en/
- 2.Arnold A, Witney AA, Vergnano S, Roche A, Cosgrove CA, Houston A, et al. XDR-TB transmission in London: Case management and contact tracing investigation assisted by early whole genome sequencing. J Infect. 2016;73(3):210–8. doi: 10.1016/j.jinf.2016.04.037 [DOI] [PubMed] [Google Scholar]
- 3.Dheda K, Limberis JD, Pietersen E, Phelan J, Esmail A, Lesosky M, et al. Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study. Lancet Respir Med. 2017;5(4):269–281. doi: 10.1016/S2213-2600(16)30433-7 [DOI] [PubMed] [Google Scholar]
- 4.Fletcher HA, Schrager L. TB vaccine development and the End TB Strategy: importance and current status. Trans R Soc Trop Med Hyg. 2016;110: 212–218. doi: 10.1093/trstmh/trw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr M., Small P. A historical and molecular phylogeny of BCG strains. Vaccine. 1999;17: 915–922. doi: 10.1016/S0264-410X(98)00277-1 [DOI] [PubMed] [Google Scholar]
- 6.Brosch R, Gordon S V., Garnier T, Eiglmeier K, Frigui W, Valenti P, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci U S A. 2007;104: 5596–601. doi: 10.1073/pnas.0700869104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calmette A. Preventive Vaccination Against Tuberculosis with BCG. Proc R Soc Med. 1931;24: 1481–90. [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufmann SH. Fact and fiction in tuberculosis vaccine research: 10 years later. Lancet Infect Dis. 2011;11: 633–640. doi: 10.1016/S1473-3099(11)70146-3 [DOI] [PubMed] [Google Scholar]
- 9.Dockrell HM, Smith SG. What Have We Learnt about BCG Vaccination in the Last 20 Years? Front Immunol. 2017;8: 1134 doi: 10.3389/fimmu.2017.01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdallah AM, Hill-Cawthorne GA, Otto TD, Coll F, Guerra-Assunção JA, Gao G, et al. Genomic expression catalogue of a global collection of BCG vaccine strains show evidence for highly diverged metabolic and cell-wall adaptations. Sci Rep. 2015;5: 15443 doi: 10.1038/srep15443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran V, Behr MA, Liu J. BCG Vaccines Molecular Genetics of Mycobacteria, Second Edition American Society of Microbiology; 2014. pp. 49–59. doi: 10.1128/microbiolspec.MGM2-0028-2013 [Google Scholar]
- 12.van der Woude AD, Luirink J, Bitter W. Getting Across the Cell Envelope: Mycobacterial Protein Secretion. Current topics in microbiology and immunology. 2012. pp. 109–134. doi: 10.1007/82_2012_298 [DOI] [PubMed] [Google Scholar]
- 13.Perkowski EF, Zulauf KE, Weerakoon D, Hayden JD, Ioerger TR, Oreper D, et al. The EXIT Strategy: an Approach for Identifying Bacterial Proteins Exported during Host Infection. MBio. 2017;8: e00333–17. doi: 10.1128/mBio.00333-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol. 2016;14: 677–691. doi: 10.1038/nrmicro.2016.131 [DOI] [PubMed] [Google Scholar]
- 15.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178: 1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46: 709–17. [DOI] [PubMed] [Google Scholar]
- 17.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100: 12420–5. doi: 10.1073/pnas.1635213100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129: 1287–98. doi: 10.1016/j.cell.2007.05.059 [DOI] [PubMed] [Google Scholar]
- 19.Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14: 1287–98. doi: 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- 20.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8: e1002507 doi: 10.1371/journal.ppat.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simeone R, Sayes F, Song O, Gröschel MI, Brodin P, Brosch R, et al. Cytosolic Access of Mycobacterium tuberculosis: Critical Impact of Phagosomal Acidification Control and Demonstration of Occurrence In Vivo. PLoS Pathog. 2015;11: e1004650 doi: 10.1371/journal.ppat.1004650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, et al. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9: 533–9. doi: 10.1038/nm859 [DOI] [PubMed] [Google Scholar]
- 23.Gröschel MI, Sayes F, Shin SJ, Frigui W, Pawlik A, Orgeur M, et al. Recombinant BCG Expressing ESX-1 of Mycobacterium marinum Combines Low Virulence with Cytosolic Immune Signaling and Improved TB Protection. Cell Rep. 2017;18: 2752–2765. doi: 10.1016/j.celrep.2017.02.057 [DOI] [PubMed] [Google Scholar]
- 24.Aguilo N, Gonzalo-Asensio J, Alvarez-Arguedas S, Marinova D, Gomez AB, Uranga S, et al. Reactogenicity to major tuberculosis antigens absent in BCG is linked to improved protection against Mycobacterium tuberculosis. Nat Commun. 2017;8: 16085 doi: 10.1038/ncomms16085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jiménez C, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73: 329–40. doi: 10.1111/j.1365-2958.2009.06783.x [DOI] [PubMed] [Google Scholar]
- 26.Ates LS, Ummels R, Commandeur S, van der Weerd R, Sparrius M, Weerdenburg E, et al. Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria. PLOS Genet. 2015;11: e1005190 doi: 10.1371/journal.pgen.1005190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6: 95 doi: 10.1186/1471-2148-6-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Luca M, Bottai D, Batoni G, Orgeur M, Aulicino A, Counoupas C, et al. The ESX-5 associated eccB-EccC locus is essential for Mycobacterium tuberculosis viability. PLoS One. 2012;7: e52059 doi: 10.1371/journal.pone.0052059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393: 537–44. doi: 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 30.Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281: 3866–75. doi: 10.1074/jbc.M505556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, et al. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J Biol Chem. 2011;286: 19024–34. doi: 10.1074/jbc.M110.204966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJP, Garcia-Vallejo JJ, et al. The ESX-5 System of Pathogenic Mycobacteria Is Involved In Capsule Integrity and Virulence through Its Substrate PPE10. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol. 2011;2011: 497203 doi: 10.1155/2011/497203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ates LS, Houben ENG, Bitter W. Type VII Secretion: A Highly Versatile Secretion System. Virulence Mechanisms of Bacterial Pathogens, Fifth Edition American Society of Microbiology; 2016. pp. 357–384. doi: 10.1128/microbiolspec.VMBF-0011-2015 [DOI] [PubMed] [Google Scholar]
- 35.Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, et al. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol. 2016;1: 16133 doi: 10.1038/nmicrobiol.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan MJ, Delogu G. The PE multigene family: a “molecular mantra” for mycobacteria. Trends Microbiol. 2002;10: 246–249. doi: 10.1016/S0966-842X(02)02335-1 [DOI] [PubMed] [Google Scholar]
- 37.Ates LS, Dippenaar A, Ummels R, Piersma SR, van der Woude AD, van der Kuij K, et al. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol. 2018;3: 181–188. doi: 10.1038/s41564-017-0090-6 [DOI] [PubMed] [Google Scholar]
- 38.McEvoy CRE, van Helden PD, Warren RM, Gey van Pittius NC. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol Biol. 2009;9: 237 doi: 10.1186/1471-2148-9-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEvoy CRE, Warren RM, van Helden PD, Gey van Pittius NC. Multiple, independent, identical IS6110 insertions in Mycobacterium tuberculosis PPE genes. Tuberculosis (Edinb). 2009;89: 439–42. doi: 10.1016/j.tube.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 40.Ho TB, Robertson BD, Taylor GM, Shaw RJ, Young DB. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates.Yeast. 2000;17(4):272–82. doi: 10.1002/1097-0061(200012)17:4<272::AID-YEA48>3.0.CO;2-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalo-Asensio J, Pérez I, Aguiló N, Uranga S, Picó A, Lampreave C, et al. New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis Complex lineages. PLOS Genet. 2018;14: e1007282 doi: 10.1371/journal.pgen.1007282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soto CY, Menéndez MC, Pérez E, Samper S, Gómez AB, García MJ, et al. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J Clin Microbiol. 2004;42: 212–9. doi: 10.1128/JCM.42.1.212-219.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso H, Aguilo JI, Samper S, Caminero JA, Campos-Herrero MI, Gicquel B, et al. Deciphering the role of IS6110 in a highly transmissible Mycobacterium tuberculosis Beijing strain, GC1237. Tuberculosis (Edinb). 2011;91: 117–26. doi: 10.1016/j.tube.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 44.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32: 643–55. [DOI] [PubMed] [Google Scholar]
- 45.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, Manganelli R, et al. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2002;45: 203–17. [DOI] [PubMed] [Google Scholar]
- 46.Smith D. A., Parish T., Smith S. M., Dockrell H. M., Stoker N. G. & Bancroft GJ. Deletion of mycobacterial phospholipases C and haemolysin alters virulence and inhibits T cell recognition of Mycobacterium tuberculosis H37Rv. Fifth Int Conf Pathog Mycobact Infect Stock Sweden, Abstr book. 2002; 11. [Google Scholar]
- 47.Le Chevalier F, Cascioferro A, Frigui W, Pawlik A, Boritsch EC, Bottai D, et al. Revisiting the role of phospholipases C in virulence and the lifecycle of Mycobacterium tuberculosis. Sci Rep. 2015;5: 16918 doi: 10.1038/srep16918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45: 172–9. doi: 10.1038/ng.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boritsch EC, Frigui W, Cascioferro A, Malaga W, Etienne G, Laval F, et al. pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat Microbiol. 2016;1: 15019 doi: 10.1038/nmicrobiol.2015.19 [DOI] [PubMed] [Google Scholar]
- 50.Boritsch EC, Khanna V, Pawlik A, Honoré N, Navas VH, Ma L, et al. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proc Natl Acad Sci U S A. 2016;113: 9876–9881. doi: 10.1073/pnas.1604921113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee RS, Radomski N, Proulx J-F, Levade I, Shapiro BJ, McIntosh F, et al. Population genomics of Mycobacterium tuberculosis in the Inuit. Proc Natl Acad Sci U S A. 2015;2000: 201507071. doi: 10.1073/pnas.1507071112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brosch R, Gordon S V., Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99: 3684–3689. doi: 10.1073/pnas.052548299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coscolla M, Lewin A, Metzger S, Maetz-Rennsing K, Calvignac-Spencer S, Nitsche A, et al. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg Infect Dis. 2013;19: 969–76. doi: 10.3201/eid1906.121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu L, Zhong J, Jia X, Liu G, Kang Y, Dong M, et al. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 2016;44: 730–743. doi: 10.1093/nar/gkv1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mostowy S, Inwald J, Gordon S, Martin C, Warren R, Kremer K, et al. Revisiting the evolution of Mycobacterium bovis. J Bacteriol. 2005;187: 6386–95. doi: 10.1128/JB.187.18.6386-6395.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, et al. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis. 2012;18: 653–5. doi: 10.3201/eid1804.110888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houben ENG, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jiménez CR, et al. Composition of the type VII secretion system membrane complex. Mol Microbiol. 2012;86: 472–84. doi: 10.1111/j.1365-2958.2012.08206.x [DOI] [PubMed] [Google Scholar]
- 58.Sayes F, Sun L, Di Luca M, Simeone R, Degaiffier N, Fiette L, et al. Strong immunogenicity and cross-reactivity of Mycobacterium tuberculosis ESX-5 type VII secretion: encoded PE-PPE proteins predicts vaccine potential. Cell Host Microbe. 2012;11: 352–63. doi: 10.1016/j.chom.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 59.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, et al. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 2008;4: e33 doi: 10.1371/journal.ppat.0040033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majlessi L, Brodin P, Brosch R, Rojas M-J, Khun H, Huerre M, et al. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J Immunol. 2005;174: 3570–9. [DOI] [PubMed] [Google Scholar]
- 61.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74: 88–98. doi: 10.1128/IAI.74.1.88-98.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng L, Tong J, Wang H, Tao C, Wang Q, Niu C, et al. PPE38 Protein of Mycobacterium tuberculosis Inhibits Macrophage MHC Class I Expression and Dampens CD8+ T Cell Responses. Front Cell Infect Microbiol. 2017;7: 68 doi: 10.3389/fcimb.2017.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prados-Rosales R, Carreno LJ, Weinrick B, Batista-Gonzalez A, Xu J, Chan J, et al. The presence of a mycobacterial capsule is associated with differences in the protective efficacy of BCG vaccination against Mycobacterium tuberculosis in mice. J Infect Dis. 2016; jiw153-. doi: 10.1093/infdis/jiw153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, et al. Memory T Cells in Latent Mycobacterium tuberculosis Infection Are Directed against Three Antigenic Islands and Largely Contained in a CXCR3+CCR6+ Th1 Subset. PLoS Pathog. 2013;9: e1003130 doi: 10.1371/journal.ppat.1003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertholet S, Ireton GC, Ordway DJ, Windish HP, Pine SO, Kahn M, et al. A Defined Tuberculosis Vaccine Candidate Boosts BCG and Protects Against Multidrug-Resistant Mycobacterium tuberculosis. Sci Transl Med. 2010; 2(53):53ra74 doi: 10.1126/scitranslmed.3001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baldwin SL, Reese VA, Huang PD, Beebe EA, Podell BK, Reed SG, et al. Protection and long-lived immunity induced by the ID93/GLA-SE vaccine candidate against a clinical Mycobacterium tuberculosis isolate. Clin Vaccine Immunol. 2015;23(2):137–47. doi: 10.1128/CVI.00458-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brennan MJ. The enigmatic PE/PPE Multi-gene Family of Mycobacteria and TB Vaccination. Infect Immun. 2017; 85: e00969–16. doi: 10.1128/IAI.00969-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayes F, Pawlik A, Frigui W, Gröschel MI, Crommelynck S, Fayolle C, et al. CD4+ T Cells Recognizing PE/PPE Antigens Directly or via Cross Reactivity Are Protective against Pulmonary Mycobacterium tuberculosis Infection. PLOS Pathog. 2016;12: e1005770 doi: 10.1371/journal.ppat.1005770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McEvoy CRE, Cloete R, Müller B, Schürch AC, Helden PD van, Gagneux S, et al. Comparative Analysis of Mycobacterium tuberculosis pe and ppe Genes Reveals High Sequence Variation and an Apparent Absence of Selective Constraints. PLoS One. 2012;7: e30593 doi: 10.1371/journal.pone.0030593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Copin R, Coscollá M, Seiffert SN, Bothamley G, Sutherland J, Mbayo G, et al. Sequence diversity in the pe_pgrs genes of Mycobacterium tuberculosis is independent of human T cell recognition. MBio. 2014;5: e00960–13. doi: 10.1128/mBio.00960-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bertholet S, Ireton GC, Kahn M, Guderian J, Mohamath R, Stride N, et al. Identification of Human T Cell Antigens for the Development of Vaccines against Mycobacterium tuberculosis. J Immunol. 2008; 181(11):7948–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim A, Hur Y-G, Gu S, Cho S-N. Protective vaccine efficacy of the complete form of PPE39 protein from Mycobacterium tuberculosis Beijing/K strain in mice. Clin Vaccine Immunol. 2017;24(11):e00219–17. doi: 10.1128/CVI.00219-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One. 2010;5: e13938 doi: 10.1371/journal.pone.0013938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kapopoulou A, Lew JM, Cole ST. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb). 2011;91: 8–13. doi: 10.1016/j.tube.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 75.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, et al. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94: 10961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, et al. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol. 2012;83: 1195–209. doi: 10.1111/j.1365-2958.2012.08001.x [DOI] [PubMed] [Google Scholar]
- 77.Beckham KSH, Ciccarelli L, Bunduc CM, Mertens HDT, Ummels R, Lugmayr W, et al. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single particle analysis. Nat Microbiol. 2017;2: 1–8. doi: 10.1038/nmicrobiol.2017.47 [DOI] [PubMed] [Google Scholar]
- 78.Brandt L, Oettinger T, Holm A, Andersen AB, Andersen P. Key epitopes on the ESAT-6 antigen recognized in mice during the recall of protective immunity to Mycobacterium tuberculosis. J Immunol. 1996;157: 3527–33 [PubMed] [Google Scholar]
- 79.Wu Y, Woodworth JS, Shin DS, Morris S, Behar SM. Vaccine-elicited 10-kilodalton culture filtrate protein-specific CD8+ T cells are sufficient to mediate protection against Mycobacterium tuberculosis infection. Infect Immun. 2008;76: 2249–55. doi: 10.1128/IAI.00024-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marrichi M, Camacho L, Russell DG, DeLisa MP. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J Biol Chem. 2008;283: 35223–35. doi: 10.1074/jbc.M802660200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.D’Souza S, Rosseels V, Romano M, Tanghe A, Denis O, Jurion F, et al. Mapping of murine Th1 helper T-Cell epitopes of mycolyl transferases Ag85A, Ag85B, and Ag85C from Mycobacterium tuberculosis. Infect Immun. 2003;71: 483–93. doi: 10.1128/IAI.71.1.483-493.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sayes F, Blanc C, Ates LS, Deboosere N, Orgeur M, Le Chevalier F, et al. Multiplexed Quantitation of Intraphagocyte Mycobacterium tuberculosis Secreted Protein Effectors. Cell Rep. 2018;23(4):1072–1084. doi: 10.1016/j.celrep.2018.03.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kato-Maeda M, Rhee JT, Gingeras TR, Salamon H, Drenkow J, Smittipat N, et al. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 2001;11: 547–54. doi: 10.1101/gr166401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alland D, Whittam TS, Murray MB, Cave MD, Hazbon MH, Dix K, et al. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J Bacteriol. 2003;185: 3392–9. doi: 10.1128/JB.185.11.3392-3399.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brodin P, Eiglmeier K, Marmiesse M, Billault A, Garnier T, Niemann S, et al. Bacterial Artificial Chromosome-Based Comparative Genomic Analysis Identifies Mycobacterium microti as a Natural ESAT-6 Deletion Mutant. Infect Immun. 2002;70: 5568–5578. doi: 10.1128/IAI.70.10.5568-5578.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mostowy S, Cousins D, Behr MA. Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J Bacteriol. 2004;186: 104–9. doi: 10.1128/JB.186.1.104-109.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dippenaar A, Parsons SDC, Sampson SL, van der Merwe RG, Drewe JA, Abdallah AM, et al. Whole genome sequence analysis of Mycobacterium suricattae. Tuberculosis (Edinb). 2015;95: 682–8. doi: 10.1016/j.tube.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 88.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47: 242–9. doi: 10.1038/ng.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brites D, Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev. 2015;264: 6–24. doi: 10.1111/imr.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bansal K, Elluru SR, Narayana Y, Chaturvedi R, Patil SA, Kaveri S V., et al. PE_PGRS Antigens of Mycobacterium tuberculosis Induce Maturation and Activation of Human Dendritic Cells. J Immunol. 2010;184: 3495–3504. doi: 10.4049/jimmunol.0903299 [DOI] [PubMed] [Google Scholar]
- 91.Deng W, Long Q, Zeng J, Li P, Yang W, Chen X, et al. Mycobacterium tuberculosis PE_PGRS41 Enhances the Intracellular Survival of M. smegmatis within Macrophages Via Blocking Innate Immunity and Inhibition of Host Defense. Sci Rep. 2017;7: 46716 doi: 10.1038/srep46716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chatrath S, Gupta VK, Dixit A, Garg LC. PE_PGRS30 of Mycobacterium tuberculosis mediates suppression of proinflammatory immune response in macrophages through its PGRS and PE domains. Microbes Infect. 2016;18: 536–42. doi: 10.1016/j.micinf.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 93.Zumbo A, Palucci I, Cascioferro A, Sali M, Ventura M, D’Alfonso P, et al. Functional dissection of protein domains involved in the immunomodulatory properties of PE_PGRS33 of Mycobacterium tuberculosis. Pathog Dis. 2013;69: 232–239. doi: 10.1111/2049-632X.12096 [DOI] [PubMed] [Google Scholar]
- 94.Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288: 1436–9. [DOI] [PubMed] [Google Scholar]
- 95.Fishbein S, Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96: 901–916. doi: 10.1111/mmi.12981 [DOI] [PubMed] [Google Scholar]
- 96.Malone KM, Gordon S V. Mycobacterium tuberculosis Complex Members Adapted to Wild and Domestic Animals. Adv Exp Med Biol. 2017;1019:135–154. doi: 10.1007/978-3-319-64371-7_7 [DOI] [PubMed] [Google Scholar]
- 97.Delogu G, Brennan MJ. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun. 2001;69: 5606–11. doi: 10.1128/IAI.69.9.5606-5611.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koh KW, Soh SE, Seah GT. Strong antibody responses to Mycobacterium tuberculosis PE-PGRS62 protein are associated with latent and active tuberculosis. Infect Immun. 2009;77: 3337–43. doi: 10.1128/IAI.01175-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phase 1 ID93 + GLA-SE Vaccine Trial in Healthy Adult Volunteers—Full Text View—ClinicalTrials.gov [Internet]. [cited 21 Sep 2017]. Available: https://clinicaltrials.gov/ct2/show/NCT01599897
- 100.Phase 2a ID93 + GLA-SE Vaccine Trial in TB Patients After Treatment Completion—Full Text View—ClinicalTrials.gov [Internet]. [cited 21 Sep 2017]. Available: https://clinicaltrials.gov/ct2/show/NCT02465216
- 101.Arbues A, Aguilo JI, Gonzalo-Asensio J, Marinova D, Uranga S, Puentes E, et al. Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated M. tuberculosis-based vaccine to enter clinical trials. Vaccine. 2013;31: 4867–73. doi: 10.1016/j.vaccine.2013.07.051 [DOI] [PubMed] [Google Scholar]
- 102.Keating LA, Wheeler PR, Mansoor H, Inwald JK, Dale J, Hewinson RG, et al. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol Microbiol. 2005;56: 163–174. doi: 10.1111/j.1365-2958.2005.04524.x [DOI] [PubMed] [Google Scholar]
- 103.Bange F-C, Collins FM, Jacobs WR. Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79: 171–180. doi: 10.1054/tuld.1998.0201 [DOI] [PubMed] [Google Scholar]
- 104.Alderson MR, Bement T, Day CH, Zhu L, Molesh D, Skeiky YA, et al. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4(+) T cells. J Exp Med. 2000;191: 551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62: 667–79. doi: 10.1111/j.1365-2958.2006.05409.x [DOI] [PubMed] [Google Scholar]
- 106.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36: W5–W9. doi: 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblots of whole-cell lysates or culture filtrates of the indicated M. canetti (A), M. tuberculosis (B) or BCG (C, D) isolates [48,51]. A) Although differences in protein secretion could be observed between different M. canetti isolates (A-J), all isolates exhibited PE_PGRS secretion. B) PE_PGRS secretion of Mj-sublineage strains with a deletion affecting ppe38, but not ppe71 (Lanes 4–8) was not discernible from Lineage 4 control isolate CDC1551 or an isolate from the same cohort without this deletion (MT13848). C) Introduction of plasmid pMV::ppe38-71 in BCG complemented PE_PGRS secretion (BCG38), while complementation was not observed when performed with pYUB::RD5, even though presence of genetic presence of RD5 was PCR-confirmed with primers RD5B-plcA.int.F/R [52]. D) Immunoblot secretion analysis of five genetically divergent BCG isolates confirms the PE_PGRS secretion defect in all BCG isolates. Cop. 38 indicates the strain M. bovis BCG Copenhagen, transformed with vector pMV::ppe38-71 and is hereafter referred to as BCG38. Anti-SigA staining is uses as a lysis control in A and D, while anti-GroEL2 is used in B and C. Strain details can be found in S5 Table. Full blots of panels A-D are depicted in S5 Fig and S6 Fig.
(TIF)
Organs depicted in A and B correspond to the experiment depicted in Fig 3. Organs depicted in C and D correspond to the experiment depicted in Fig 6. After photography of the lungs (A, C), a single lung lobe was used for lung CFU quantification. Splenomegaly (B, D) was reduced, by all vaccination conditions, but did not differ markedly between vaccination conditions.
(TIF)
IFN-γ production in response to peptides covering the indicated amino acid positions of PPE10 (Rv0442c) in C57BL/6 (grey/green) or C57BL/6 x CBA F1 (B6CBAF1, blue/brown) mice. Mice were immunized with M. tuberculosis H37Rv (left) or unimmunized (right).
(TIF)
A) Schematic representation of deletion strategy and primers. The genetic region around PPE10, as taken from tuberculist, is depicted in colored arrows [74]. Flanking fragments used for homologous recombination are depicted in black bars. Left (PPE10KO-LF and PPE10KO-LR) and right (PPE10KO-RF and PPE10KO-RR) flanking regions were amplified by primers depicted in black. Primers used to verify successful homologous recombination are depicted in dark blue. All primer sequences can be found in S4 Table. B) PCR verification of successful homologous recombination in seven different colonies that grew on hygromycin selection plates. Colony 1 was taken for further analyses. Full gels used to create B are depicted in S7 Fig.
(TIF)
(TIF)
As expected, the percentage of live cells was higher for BCG-infected cells than for cells infected with M. tuberculosis strains, but did not vary significantly between isogenic strains (≤ 1.0% difference between isogenic strain). These values are derived from the experiment depicted in Fig 2A.
(PDF)
A promoter mutation disrupts production of I-Eαb in C57BL/6 mice (Grey font), which are therefore unable to produce MHC-II I-E (Grey). In contrast, H-2k mice can produce both I-Aαk I-Aαk and I-Eαk I-Eαk. C57BL/6 x CBA F1 mice have an even bigger repertoire of possible functional MHC-II isoforms available due to heterodimerization of α and β subunits from H-2b and H-2k haplotypes.
(PDF)
Black letters indicate identical amino acids. Red letters indicate non-identical amino acids. Top: homologues of the MPTR-containing peptide PPE10221-235 ordered by percentage of sequence identity. Bottom: Homologues of the peptide PPE10381-395, which is part of the C-terminal secreted domain of PPE10.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.