Abstract
Because the epidemiology of Clostridium difficile infection (CDI) is region-specific, the present study was undertaken to examine the epidemiology of C difficile outbreaks in Beijing, China.
Eighty nonduplicate isolates were collected from March, 2016 to December, 2016. The molecular type and phylogenetic analysis were evaluated by multilocus sequence typing (MLST). The minimum inhibitory concentrations (MICs) for 11 antibiotics and the resistance mechanisms were investigated.
Sixty-five toxigenic strains (81.25%), including 22 tcdA-B+CDT- strains (27.5%) and 43 tcdA+B+CDT- strains (53.75%), and also 15 nontoxigenic strains (tcdA-B-CDT-; 18.75%) were detected. MLST identified 21 different sequence types (STs), including 2 novel types (ST409 and ST416). All isolates were susceptible to metronidazole, vancomycin, fidaxomicin, piperacillin/tazobactam, and meropenem, and all were effectively inhibited by emodin (MICs 4–8 μg/mL). The resistance rates to rifaximin, ceftriaxone, clindamycin, erythromycin, and ciprofloxacin were 8.75%, 51.25%, 96.25%, 81.25%, and 96.25%, respectively; 81.25% (65/80) of isolates were multidrug-resistant. Amino acid mutations in GyrA and/or GyrB conferred quinolone resistance. One novel amino acid substitution, F86Y in GyrA, was found in 1 CIP-intermediate strain. The erm(B) gene played a key role in mediating macrolide-lincosamide-streptogramin B (MLSB) resistance. Erm(G) was also found in erm(B)-negative strains that were resistant to both erythromycin and clindamycin. RpoB mutations were associated with rifampin resistance, and 2 new amino mutations were identified in 1 intermediate strain (E573A and E603N).
Regional diversity and gene heterogeneity exist in both the ST type and resistant patterns of clinical C difficile isolates in Northern China.
Keywords: antibiotic resistance, Beijing, Clostridium difficile, epidemiology
1. Introduction
Clostridium difficile is a Gram-positive, spore-forming anaerobic bacillus that causes a range of gastrointestinal syndromes, from mild diarrhea to severe pseudomembranous colitis, lethal toxic megacolon, and sepsis.[1] Due, in part, to the emergence and subsequent spread of the hypervirulent NAP1/BI/027 strain,[1]C difficile infection (CDI) has become a major cause of healthcare antibiotic-associated diarrhea, accounting for up to 20% of cases.[2,3] Treatment with antimicrobials, including the macrolide-lincosamide-streptogramin B (MLSB) group of antibiotics (eg, clindamycin and erythromycin), fluoroquinolones, and cephalosporins, is a high risk factor for CDI.[4,5]
The major virulence factors of C difficile include enterotoxin A (TcdA) and cytotoxin B (TcdB).[6] On the basis of the tcdA and or tcdB genes located within the pathogenicity locus (PaLoc) and potentially an additional binary toxin, clinical isolates are classified into several types. Most isolates are positive for both tcdA and tcdB (tcdA+B+); the toxin variant isolates are only positive for tcdB (tcdA−B+). Nontoxigenic strains (tcdA−B−) are also isolated from clinical patients. Moreover, the frequency of hypervirulent strains, known as North American pulsed field electrophoresis type 1 or ribotype027 (NAP1/027) encoding cdtA and cdtB, has increased in recent years, resulting in nosocomial outbreaks in North America, Canada, and Europe.[7,8]
Analysis of antimicrobial susceptibility and mechanisms of resistance are required for surveillance of the emergence and distribution of C difficile, and guiding public health measures. In addition to the classical antimicrobials used for CDI, emodin can inhibit the growth of anaerobic bacteria, including Propoinibacterium, Eubacterium, and Clostridium[9]; however, few isolates were analyzed. Therefore, investigating the minimum inhibitory concentrations (MICs) of 11 antimicrobials for C difficile strains and the related mechanism of resistance is important to effectively understand, prevent, and control CDI outbreaks.
Molecular epidemiology analyses of C difficile are crucial for identifying and controlling outbreaks of CDI. Using multilocus sequence typing (MLST) analysis, a nucleotide sequence-based characterization of allelic polymorphisms in 7 housekeeping genes was used to analyze the evolutionary genetics and global epidemiology of C difficile.[10] Similar analyses in China indicate that CDI epidemiology is region-specific.[11–14] Specifically, analysis of 432 CDI specimens from Eastern China revealed that the sequence type (ST) ST37 was predominant,[11] whereas ST3 and ST54 were predominant in other studies in Northeast China.[12,13] Moreover, genetic diversity of C difficile within a region changes over time.[15] Thus, the present study was undertaken to examine the epidemiology of C difficile outbreaks in a single institution in Northern China.
2. Materials and methods
2.1. Clostridium difficile isolate collection and culture
Clostridium difficile isolates were obtained from loose stool specimens of patients with suspected CDI at the Beijing Friendship Hospital of Capital Medical University. In total, 80 nonduplicate isolates were collected from 475 in-patients with diarrhea from March, 2016 to December, 2016. This study was approved by the Institutional Review Board of the Beijing Friendship Hospital of Capital Medical University.
Stool samples were cultured on C difficile agar selective D-cycloserine–cefoxitin–fructose agar plates (CM0601, Oxoid, Basingstoke, UK) in an anaerobic atmosphere at 37°C for 48 hours. Presumptive C difficile isolates were identified by Gram stain, odor, and typical colony morphology, and confirmed by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight mass spectrometry (MALDI-TOF-MS) (VITEK MS, bioMerireux, Lyon, France). Isolates were maintained in 20% skimmed milk at −80°C for further studies.
2.2. MLST analysis and detection of toxin genes by PCR
Bacterial genomic DNA was extracted using a Bacterial Genomic DNA Extraction Kit (D1600, Solarbio, Beijing, China), and the endotoxin genes, cdtA and cdtB, were detected as described previously.[16] Seven housekeeping loci (adk, atpA, dxr, glyA, recA, soda, and tpi) were selected for MLST analysis.[10] PCR was performed, and amplification products were sequenced with a forward primer. DNA sequences were submitted to PubMLST (http://pubmlst.org/clostridium difficile) to obtain the ST; new ST or alleles were given a novel number by C difficile database curators.
Genetic diversity was analyzed using MEGA version 7 software. For MLST homology analysis, forward sequences of all the stains were trimmed to the same length, aligned, and analyzed using the MEGA 7.0 software package. The phylogenetic trees from the concatenated sequences were constructed by the neighbor joining method with a Kimura 2-parameter distance model using MEGA 7.0 software, and Bootstrap analysis with 1000 replicates was performed. The evolutionary distance was computed using the maximum composite likelihood method, according to the units of the number of base substitutions per site.[17]
2.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the reference agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines.[18] Briefly, frozen C difficile isolates were thawed and subcultured on Brucella agar plates supplemented with vitamin K1 (1 μg/mL), hemin (5 μg/mL), and 5% laked sheep red blood twice. Individual colonies were subsequently suspended to an equivalent of 0.5 McFarland, and 1 μL of the suspension was inoculated on 100-mm Brucella blood agar plates with a gradient antimicrobial concentration for 48 hours at 37°C in anaerobic conditions. The following 11 antibiotics were tested: metronidazole, vancomycin, fidaxomicin, piperacillin/tazobactam, meropenem, rifaximin, ceftriaxone, clindamycin, erythromycin, ciprofloxacin, and emodin. MICs of the antimicrobials were determined using the broth micro dilution method and were interpreted according to the 2017 CLSI recommendations. For agents without referenced standards, the breakpoints were as follows: rifaximin, ≥32 μ g/mL; vancomycin, ≥32 μ g/mL; erythromycin, ≥8 μ g/mL; and ciprofloxacin, ≥8 μ g/mL.[19,20] There was no previously published reference standard for emodin. C difficile ATCC700057 and Bacteroides fragilis ATCC 25285 were used as quality control strains.
2.4. Detection of resistance genes and sequencing
The fluoroquinolone resistance-determining regions (QRDRs) of GyrA and GyrB, RpoB, and erm(B), and also the 23S rRNA gene, were amplified using the related primers as previously described and shown in Table 1.[21–24]
Table 1.
Primers for analysis of resistance genes used in this study.

Erythromycin resistance determinants were identified via a set of primers by overlapping PCR.[25] Other classes of erm genes (D, F, G, and Q) were also detected.[26–29] For analysis of erm(B)-negative, clindamycin and/or erythromycin-resistant isolates, analysis of 23S genes were analyzed by PCR mapping using 6 primer pairs as described by Schmidt et al.[21] The DNA products were purified and sequenced on an ABI 3700 sequencer using a forward PCR primer. Mutations in the resistance genes were aligned with the reference sequence of C difficile 630 (GenBank accession no. NC 009089.1) using ORF Finder and DNAMAN alignment software.
2.5. Statistical analysis
Data analysis was performed using SPSS 20.0 Version 20.0 (IBM. Armonk, NY). The resistance rates (R%) were summarized as toxigenic and nontoxigenic isolate strains. Differences in the resistance rates between toxigenic and nontoxigenic isolate strains were compared using the Person chi-square test or Fisher exact test if any number of strains was less than 5. The statistical assessments were 2-tailed, and a value of P < .05 was considered statistically significant.
3. Results
3.1. Toxin typing and molecular epidemiology of the C difficile isolates
On the basis of the toxin genes patterns identified by PCR, the 80 isolates were divided into 65 toxigenic strains (81.25%) and 15 nontoxigenic strains (18.75%). The 65 toxigenic strains were classified into 2 toxin types, including 22 tcdA−B+CDT− strains (27.5%) and 43 tcdA+B+CDT− strains (53.75%). The A+B+CDT+ strain was not detected.
The MLST analysis identified 21 different ST types, among which ST409 and ST416 were novel (Table 2). ST409 was a new combination of 7 existing allelic genes, and ST416 contained 5 new alleles (adk33, atpA47, glyA65, recA36, and tpi62). ST81 was the prevalent type identified in 20 isolates (25%), followed by ST8 (13 isolates, 16.25%), ST42 (9 isolates, 11.25%), and ST39, ST3, and ST2 (6 isolates for each ST type, 7.54% individually). Eleven ST types were found in 1 single isolate.
Table 2.
Phenotypic and genotypic characteristics of Clostridium difficile isolates.
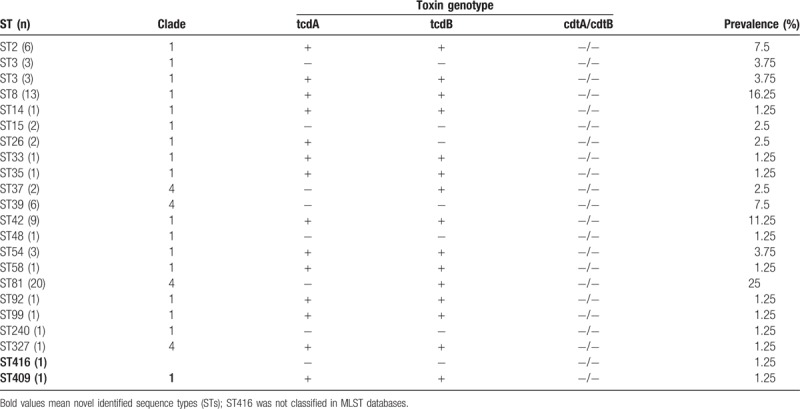
There was an interesting correlation between ST types and toxin types. All ST42, ST8, and ST2 isolates were toxin type A+B+CDT− strains, and all ST39 isolates were nontoxigenic strains. Moreover, ST81 was the most dominant type, all of which were A−B+CDT− strains. Thus, it is possible that the CDI outbreaks occurring during the study period at our institution was caused by ST81.
3.2. Phylogenetic analysis
The genetic diversity and phylogenetic relationships of the 80 isolates were analyzed using MEGA software. As shown in Fig. 1, a neighbor-joining tree was generated from concatenated sequences of 7 loci. Most ST types were classified into clade 1; ST81, ST37, ST39, and ST327 belonged to clade 4. ST416 was not closely related to any of the other isolates.
Figure 1.
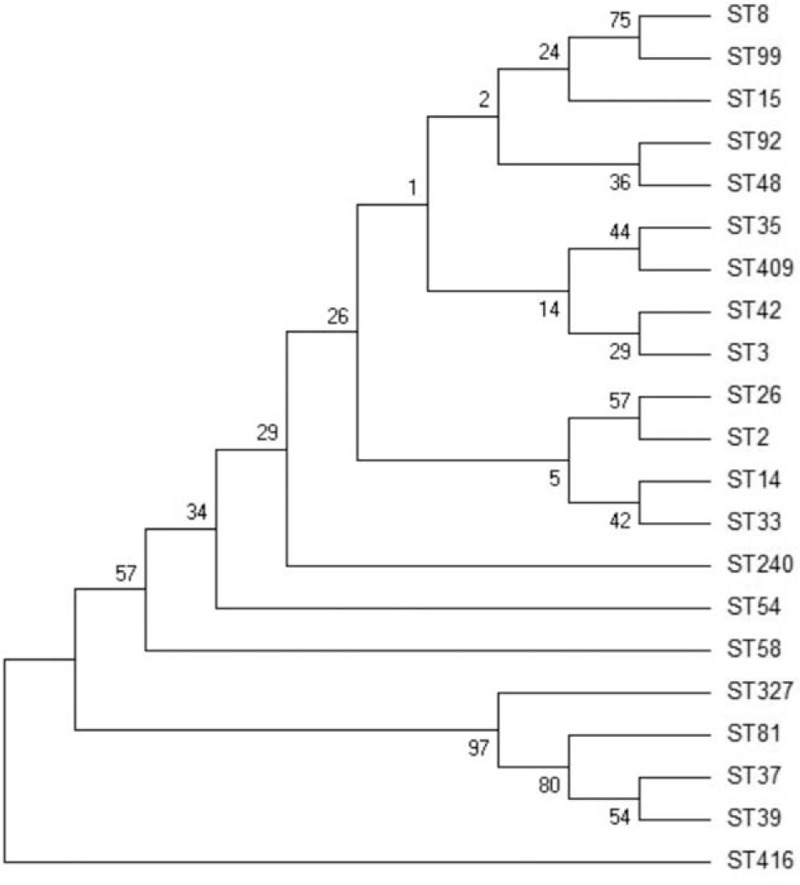
Phylogenetic tree reconstructed using the neighbor-joining method based on composite sequences of seven housekeeping gene fragments.
3.3. Antimicrobial susceptibility analysis
As shown in Table 3, all 80 isolates were susceptible to metronidazole, vancomycin, and fidaxomicin with MIC90 values of 0.5, 0.25, and 0.25 μg/mL, respectively. All isolates were also susceptible to meropenem and piperacillin/tazobactam. Emodin also exhibited effective in vitro antibacterial activities against the C difficile isolates with MICs ranging from 4 to 8 μg/mL. Seventy (87.5%) isolates were susceptible to rifaximin (MIC ≤0.0078 μg/mL), 7 (8.75%) isolates were resistant (MIC ≥32 μg/mL), and 3 (3.75%) strains showed intermediate (MIC range 0.25–2 μg/mL) susceptibility to rifaximin. In contrast, 96.25% of the isolates were equally resistant to clindamycin and ciprofloxacin, with MIC90 of 256 and 64 μg/mL, respectively (Table 3). The resistance rates to erythromycin and ceftriaxone were 81.25% and 51.25%, respectively.
Table 3.
Toxin genotypes and antimicrobial susceptibilities of Clostridium difficile isolates.
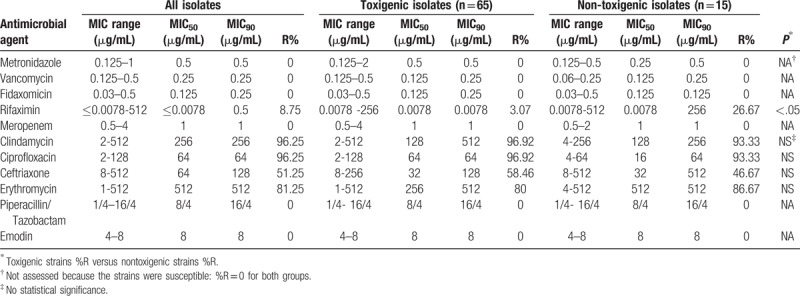
There was no significant difference in the resistance rates to ceftriaxone, clindamycin, ciprofloxacin, and erythromycin between the toxigenic and nontoxigenic isolates (Table 3). Interestingly, the nontoxigenic strains showed higher resistance to rifaximin as compared with the toxigenic strains (P < .05).
Analysis of multidrug-resistant strains defined as those resistant to at least 3 antibiotics revealed that 81.25% (65/80) of isolates were multidrug-resistant strains in this study. Of note, all ST81 strains were multidrug-resistant strains with high resistance to erythromycin (MIC ≥256 μg/mL), clindamycin (MIC ≥32 μg/mL), ciprofloxacin (MIC ≥32 μg/mL), and ceftriaxone (MIC ≥128 μg/mL) (data not shown). All control results were in standard reference ranges.
3.4. Molecular analysis of the mechanisms of resistance
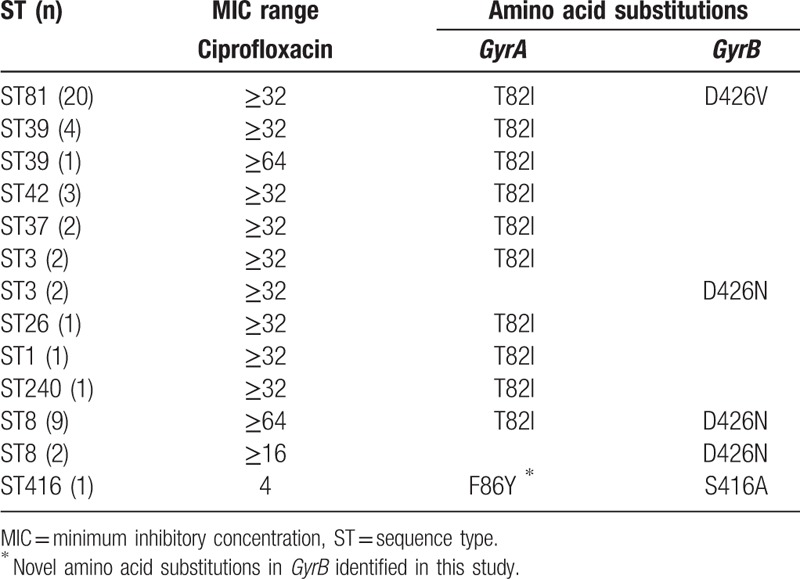
The quinolone resistance-determining regions of GyrA and GyrB were individually sequenced from the nucleotide codons 48 to 152 and 371 to 479. The results indicated that 18.75% (15/80) of the C difficile isolates had 1 single amino acid substitution in GyrA, including T82I in 14 strains and T82V in 1 strain. In addition, 36.25% (29/80) had 1 amino acid substitution in both GyrA and GyrB, and 5% (4/80) had a single amino acid change in GyrB D426N (Table 4). Moreover, 1 intermediate nontoxigenic isolate, ST416 (MIC 4 μg/mL), showed 1 novel amino acid substitution, F86Y, in GyrA. No amino acid substitutions in GyrA and GyrB were found in the C difficile-susceptible strains (data not shown). T82I was the most frequent amino acid change in the highly resistant C difficile isolates (ie, those with a MIC ≥32 μg/mL) that was found in various ST types. C difficile isolates with amino acid substitutions in both GyrA (T82I) and GyrB (D426V) belonged to the ST81 type. In addition, strains with amino acid substitutions in both GyrA (T82I) and GyrB (D426N) were the ST8 type.
Table 4.
Amino acid substitution in gryA and gryB associated with resistance to quinolones in the Clostridium difficile isolates.
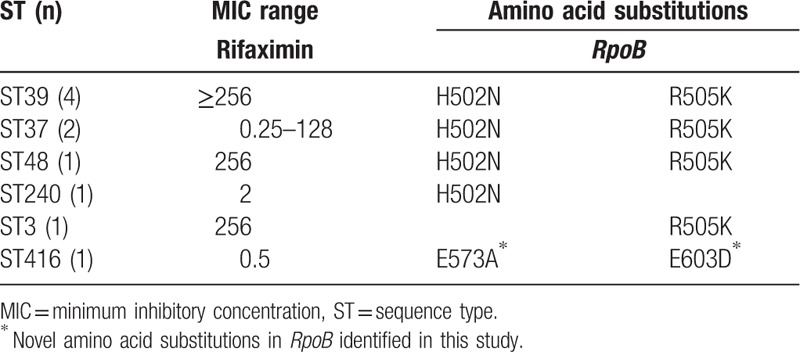
We next analyzed the RpoB gene (nucleotide codons 480–616) in all isolates for possible amino acid substitutions. As shown in Table 5, there were 5 different RpoB polymorphisms identified in the rifaximin-intermediate and rifaximin-resistant C difficile isolates. Three amino acid substitutions had been previously reported.[20,30] However, the amino acid substitutions E573A combined with G603D in the ST416 strain were both novel. The amino acid substitutions H502N and R505K in RpoB were the most frequent mutations in rifaximin-resistant strains (Table 5). However, there was no relationship between ST type and the mechanism of resistance with respect to the RpoB gene. All 60 C difficile-susceptible strains had no amino acid substitution in RpoB (data not shown).
Table 5.
Amino acid substitutions in RpoB associated with rifaximin resistance in Clostridium difficile isolates.
Among all 80 C difficile isolates, 60 strains (75%) were erm(B)-positive, and these isolates were resistant to both erythromycin and clindamycin. However, the other 20 erm(B)-negative isolates contained 12 clindamycin-resistant strains (≥8 μg/mL), and 6 were resistant to both erythromycin and clindamycin, including some that were highly resistant (erythromycin, MIC ≥64 μg/mL and clindamycin, MIC ≥64 μg/mL).
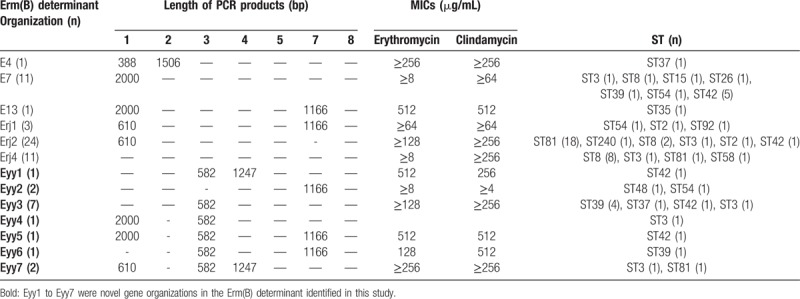
We further analyzed the genetic organizations of the Erm(B) determinant region of 66 isolates resistant to both erythromycin and clindamycin, including 60 erm(B)-positive isolates and 6 erm(B)-negative isolates. In total, 13 different genetic organizations were identified, and 7 arrangements (designated Eyy1–Eyy7) were new (Table 6). E4, E7, E13, Erj1, Erj2, and Erj4 were reported previously.[19,25] The majority of isolates were Erj2 (36.36%), followed equally by Erj4 and E7 (both 16.67%).
Table 6.
Characterization of erm(B) determinant organization of the Clostridium difficile strains resistant to macrolide-lincosamide-streptogramin B.
All erm(B)-negative strains were also negative for other erm class genes found in anaerobes (eg, D, F, G, and Q). However, erm(G) was found in 1 erm(B)-positive isolate with resistance to both erythromycin and clindamycin, and also 5 erm(B)-negative strains, including 2 isolates with resistance to both erythromycin and clindamycin and 3 clindamycin-resistant isolates. Although erm(G) nucleotide sequences were not in alignment with the reference isolate, C difficile 630, they had 98% sequence identity with erm(G) of Bacteroides thetaiotaomicron conjugal transposon Tcr Emr 7853 (GenBank accession no. L42817). This suggests that erm(G) may mediate resistance to the macrolide antibiotics in C difficile isolates.
We next analyzed the 23S rDNA gene of 20 erm(B)-negative strains and 5 erm(B)-positive strains resistant to both erythromycin and clindamycin. No mutation of the nucleotide in position 656 of the 23S rDNA gene was found in 12 of 20 isolates that were erm(B)-negative, including 2 strains resistant to both erythromycin and clindamycin and 10 clindamycin-resistant only strains. In addition, PCR amplification of the 23S rDNA gene was low in 2 strains; therefore, they were not sequenced successfully. In addition, this mutation was also not identified in 4 erm(B)-positive strains resistant to both erythromycin and clindamycin. In contrast, a nucleotide C656T substitution within the 23S rDNA gene was found in 6 strains, including 4 strains—3 erm(B)-negative and 1 erm(B)-positive strains—resistant to both erythromycin and clindamycin, 1 strain resistant to only clindamycin, and 1 susceptible strain.
4. Discussion
Clostridium difficile is recognized as a major source of nosocomial antibiotic-associated diarrhea. In recent years, the incidence of CDI has increased in the United States, Canada, and Europe, and a recent study in Eastern China found a 10% prevalence of CDI in hospitalized patients with diarrhea.[11] Because the epidemiology of CDI is region-specific, the present study was undertaken to examine the epidemiology of C difficile outbreaks in Northern China. Of the 80 isolates analyzed, 65 were toxigenic strains (81.25%), including 22 tcdA−B+CDT− strains (27.5%) and 43 tcdA+B+CDT− strains (53.75%), and 15 were nontoxigenic strains (tcdA−B−CDT−; 18.75%). Twenty-one different STs were detected, including 2 novel types (ST409 and ST416). All isolates were susceptible to metronidazole, vancomycin, fidaxomicin, piperacillin/tazobactam, and meropenem, and all were effectively inhibited by emodin. Resistance to rifaximin, ceftriaxone, clindamycin, erythromycin, and/or ciprofloxacin was detected in some isolates, and 81.25% were multidrug-resistant. Amino acid mutations in GyrA and/or GyrB conferred quinolone resistance. The erm(B) gene played a key role in mediating macrolide-lincosamide-streptogramin B (MLSB) resistance, and RpoB mutations were associated with rifampin resistance.
The prevalence of CDI in the present study was 13.7%, which is higher than that (9.8%) reported in another study conducted in China[31] and lower than that reported in a similar epidemiological study in Indonesia.[32] TcdA−B+CDT− strains accounted for a large proportion (27.5%) of the collected isolates, which was similar to a previous report in China.[33] However, we did not identify any tcdA+B+CDT+ stains, indicating that the hypervirulent strain, NAP1/027, was not prevalent in our hospital. Although ST54 and ST37 were the major types reported in other studies in China,[31,34] ST81 was the dominant type observed in the present study, followed by ST8 and ST42. Moreover, ST81 carried a single allelic variant (atpA). Thus, C difficile isolates likely have genetic variances that differ between regions as a result of genetic shift over the years. Interestingly, 90.9% of the tcdA−B+ strains belonged to the prevalent type ST81, which implied that the CDI outbreaks were caused by ST81 in our hospital during the study period. Both of the toxigenic strains (classified into 14 STs) and nontoxigenic strains (divided into 7 STs) had great genetic diversity, which was in accordance with data published by Kuwata et al,[35] and also other studies conducted in China.[11–14]
Metronidazole is considered the first choice for mild-to-moderate CDI, and vancomycin is the first-line antibiotic for moderate-to-severe CDI. Although the emergence of strains with reduced susceptibility to metronidazole and vancomycin has been reported,[19,36–38] antimicrobial susceptibility testing of both toxigenic strains and nontoxigenic strains did not identify any strain resistant to metronidazole and vancomycin in the present study. Fidaxomicin was also approved by the US Food and Drug Administration for the treatment of CDI in 2011; it has potent antibiotic activity in vitro with minimal impact on intestinal microbiota composition in vivo.[39–41] Although fidaxomicin has not been licensed for the treatment of CDI in China, we analyzed the susceptibility of the C difficile isolates to this antibiotic, which revealed a MIC50 of 0.125 μg/mL and MIC90 of 0.125 μg/mL, which was identical to a previous report in Taiwan.[36] Both meropenem and piperacillin/tazobactam showed activity against all strains.
In contrast to a study conducted in Hebei Province in Northeast China,[13] some of the isolates in the present study were resistant to rifaximin—a gastrointestinal-selective antibiotic that inhibits the gene transcription of bacteria via binding RNA bacteria polymerase target sites. Although it holds promise as an alternative for the treatment of relapsing CDI,[42] sufficient clinical data worldwide are lacking. We observed that the MICs for rifaximin were either very high or very low, which was consistent with previous findings,[20] with a resistance rate of 8.7%, which is lower than the 29.8% resistance rate reported in Shanghai,[43] but similar to another report (10.9%) in Taiwan.[36] In addition, we found that nontoxigenic strains had a higher resistance rate to rifaximin than toxigenic strains, which was likely a result of the high-level resistance observed with 5 isolates (≥256 μg/mL). This result also demonstrated that although nontoxigenic strains were part of the normal intestinal tract flora, they may mediate acquired drug resistance of toxigenic strains by horizontal transmission.
Wang et al[9] first described that emodin inhibited the growth of Clostridia in 1990; however, only 5 C difficile isolates were analyzed. Using the broth microdilution method to determine the MICs of emodin, we found that all isolates were effectively inhibited. However, the antibacterial action and gastrointestinal selectivity of emodin remain to be examined.
In the present study, 81.25% of the isolates were multidrug-resistant strains, which was higher than the 73.33% reported by Dong et al.[19] ST81 strains showed high-level resistance to erythromycin, ciprofloxacin, and clindamycin, which was identical to another study described in Japan.[35] Amino acid substitutions in both GyrA and/or GyrB have been associated with fluoroquinolone resistance.[44] In the present study, the T82I mutation was the most frequently observed in high-level ciprofloxacin-resistant strains, which is similar to previous reports, respectively.[39,40,45,46] Liao et al[36] detected 1 isolate (MIC 8 μg/mL) with only amino acid mutation, S416A in GyrB or T82I in GyrA. In the present study, we identified a ciprofloxacin-intermediate strain ST416 (MIC 4 μg/mL) with a novel amino acid substitution, F86Y, in GyrA combined with the S416A mutation in GyrB. We also observed different MICs in isolates with identical amino acid substitutions in GyrB, suggesting that GryB mutations alone or combination with those in GyrA result in diverse resistance to CIP.
We also identified H502N and/or R505K mutations in the RpoB gene in all of the rifaximin-resistant and intermediate strains. In addition, we identified 2 previously unreported amino acid substitutions in 1 intermediate strain: E573A and E603N with a MIC of 0.5 μg/mL. Further studies are required to examine how these novel substitutions reduced the susceptibility to rifaximin.
Erm(B) carried by the mobile element Tn5398 plays a major role in resistance to the MLSB group of antibiotics. A detailed analysis of the genetic organization of the Erm(B)-determinant region revealed the E4, E7, E13, Erj1, Erj2, and Erj3 genetic arrangements with Erj2 as the most prevalent form, which is consistent with a previous study in China.[19] However, we identified 7 genetic arrangements, Eyy1 to Eyy7, that were novel. PCR fragment 3 with a new product of 582 bp may be a result of a DNA fragment deletion between ORF298 and the erm(2B) gene, suggesting that genetic exchange and recombination frequently occur in clinical strains. Therefore, the genetic arrangement of the Erm(B)-determinant region diverges between different countries.[19,47] Although other Erm classes (ie, D, F, and Q) were also negative in Erm(B)-negative strains, we demonstrated the presence of erm(G) in ERY-resistant strains, including those that were erm(B)-negative and positive. Although the nucleotide sequence of erm(G) was not aligned with the reference strain 630, high sequence identity was observed with the B thetaiotaomicron conjugal transposon, Tcr Emr 7853. There were only 2 amino acids change (T201M and A238T) in the nucleotide codons of erm(G), suggesting that the dissemination of erythromycin resistance genes between Bacteroides and Clostridia may occur horizontally via conjugative transposons, demonstrating the importance of mobile genetic elements in the spread of resistance in clinical C difficile isolates.
We also identified a C656T mutation in the 23S rDNA gene in both clindamycin-resistant and susceptible strains, suggesting that this mutation does not impact resistance to MLSB antibiotics. Moreover, all ST2 isolates were susceptible to ceftriaxone, indicating a possible relationship between ST type and ceftriaxone resistance. It is possible that the C difficile isolates were not consistently resistant to ceftriaxone, and this divergence of resistance to ceftriaxone depended is strain-specific. Further studies are necessary to characterize the mechanisms by which the isolates were resistant to ceftriaxone, including the presence of antibiotic-degrading enzymes, ß-lactamases, and penicillin-binding proteins (PBPs), and also modification of target sites.
In this study, ciprofloxacin was 1 of the antibiotics studied. However, it is known that C difficile resistance to ciprofloxacin is relatively common. Because of its low price and its excellent antibacterial effect, ciprofloxacin is regarded as the drug of first choice for treating nosocomial infectious diarrhea caused by pathogenic bacteria, such as Escherichia coli and Salmonella, in the Infectious Diseases Department in our hospital. In addition, ciprofloxacin is a drug that is important in the treatment of upper and lower respiratory tract infections, urinary tract infections, and other common infections seen in general practice.[48–51] In this study, the resistant rate of isolated C difficile strains to ciprofloxacin was high (96.25%), which suggests that use of ciprofloxacin may be a risk factor for the development of CDI in our hospital. We believe that investigating the resistance of C difficile strains to CIP will help clinicians to rationally choose the antibiotics and prevent secondary C difficile infections in our hospital.
The present study is limited in the small number of specimens characterized from a single institution. Thus, additional larger studies are required to confirm our results in this specific region, especially given that transmission between healthcare facilities may occur.[52] Furthermore, the mechanisms by which some of the isolates were resistant to specific antimicrobials were not determined in detail. In addition, the mechanism underlying emodin suppression of C difficile growth was not analyzed. Finally, the association between the genotypes identified and severity of CDI was not assessed.
In conclusion, this study described the genetic diversity and resistance patterns of toxigenic and nontoxigenic strains of C difficile isolates collected from Beijing Friendship Hospital in China. No strain was resistant to metronidazole, vancomycin, and fidaxomicin, and emodin effectively inhibited the growth of all strains in vitro. ST81 was the most prevalent type with high-level resistance to erythromycin, clindamycin, ciprofloxacin, and ceftriaxone. In addition, several novel resistance genes were identified, which may affect the patterns of resistance and increase their dissemination between C difficile isolates. Given the high degree of resistance and diversity observed in the present study, it is necessary to continually examine the epidemiological distribution of C difficile in China. Further studies analyzing the resistance mechanisms of C difficile in detail will be of potential benefit in effectively preventing and controlling CDI in hospitals.
Author contributions
Conceptualization: Baoya Wang, Jianrong Su.
Data curation: Baoya Wang, Zhi Lv, Pingping Zhang.
Formal analysis: Baoya Wang, Pingping Zhang.
Methodology: Zhi Lv.
Supervision: Baoya Wang.
Writing – original draft: Baoya Wang, Jianrong Su.
Writing – review & editing: Jianrong Su.
Footnotes
Abbreviations: CDI = Clostridium difficile infection, CLSI = Clinical and Laboratory Standards Institute (CLSI), MIC = minimum inhibitory concentration, MLSB = macrolide-lincosamide-streptogramin B, MLST = multilocus sequence typing, PBPs = penicillin-binding proteins, PCR = polymerase chain reaction, ST = sequence type.
The authors of this work have nothing to disclose.
References
- [1].Elliott B, Androga GO, Knight DR, et al. Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect Genet Evol 2017;49:1–1. [DOI] [PubMed] [Google Scholar]
- [2].To KB, Napolitano LM. Clostridium difficile infection: update on diagnosis, epidemiology, and treatment strategies. Surg Infect 2014;15:490–502. [DOI] [PubMed] [Google Scholar]
- [3].Miller BA, Chen LF, Sexton DJ, et al. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 2011;32:387–90. [DOI] [PubMed] [Google Scholar]
- [4].Owens RC, Jr, Donskey CJ, Gaynes RP, et al. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis 2008;46:S19–31. [DOI] [PubMed] [Google Scholar]
- [5].Loo VG, Bourgault AM, Poirier L, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med 2011;365:1693–703. [DOI] [PubMed] [Google Scholar]
- [6].Kuehne SA, Cartman ST, Heap JT, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010;467:711–3. [DOI] [PubMed] [Google Scholar]
- [7].Kuijper EJ, Coignard B, Tüll P. ESCMID Study Group for Clostridium difficile; EU Member States; European Centre for Disease Prevention and Control. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 2006;12:2–18. [DOI] [PubMed] [Google Scholar]
- [8].Pépin J, Valiquette L, Cossette B. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 2005;173:1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang WF, Chen CM, Wang SC, et al. A series of studies on anti-anaerobic bacteria of traditional Chinese medicine-III. Experimental study on anaerobic bacteria of rhubarb. Chin J Microecol 1990;2:17–23. [Google Scholar]
- [10].Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 2010;48:770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jin D, Luo Y, Huang C, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol 2017;55:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheng JW, Xiao M, Kudinha T, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolates from a university teaching hospital in China. Front Microbiol 2016;7:1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tian TT, Zhao JH, Yang J, et al. Molecular characterization of Clostridium difficile isolates from human subjects and the environment. PLoS One 2016;11:e0151964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hawkey PM, Marriott C, Liu WE, et al. Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an under recognized problem in Asia? J Clin Microbiol 2013;51:3308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Collins DA, Gasem MH, Habibie TH, et al. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect 2017;18:34–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Persson S, Torpdahl M, Olsen KE. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 2008;14:1057–64. [DOI] [PubMed] [Google Scholar]
- [17].Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 2016;33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- [19].Dong D, Zhang L, Chen X, et al. Antimicrobial susceptibility and resistance mechanisms of clinical Clostridium difficile from a Chinese tertiary hospital. Int J Antimicrob Agents 2013;41:80–4. [DOI] [PubMed] [Google Scholar]
- [20].O’Connor JR, Galang MA, Sambol SP, et al. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 2008;52:2813–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schmidt C, Löffler B, Ackermann G. Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn Microbiol Infect Dis 2007;59:1–5. [DOI] [PubMed] [Google Scholar]
- [22].Dridi L, Tankovic J, Burghoffer B, et al. gyrA and gyrB mutations are implicated in cross-resistance to Ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother 2002;46:3418–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Curry SR, Marsh JW, Shutt KA, et al. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis 2009;48:425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 2006;38:779–86. [DOI] [PubMed] [Google Scholar]
- [25].Farrow KA, Lyras D, Rood JI. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 2001;147:2717–28. [DOI] [PubMed] [Google Scholar]
- [26].Kim HS, Choi EC, Kim BK, et al. A macrolide-lincosamide-streptogramin B resistance determinant from Bacillus anthracis 590: cloning and expression of ermJ. J General Microbiol 1993;139:601–7. [DOI] [PubMed] [Google Scholar]
- [27].Comstock LE, Coyne MJ, Tzianabos AO, et al. Analysis of a capsular polysaccharide biosynthesis locus of Bacteroides fragilis. Infection & Immunity 1999;67:3525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cooper AJ, Shoemaker NB, Salyers AA, et al. The erythromycin resistance gene from the Bacteroides conjugal transposon Tcr Emr 7853 is nearly identical to ermG from Bacillus sphaericus. Antimicrob Agents Chemother 1996;40:506–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Berryman DI, Lyristis M, Rood JI, et al. Cloning and sequence analysis of ermQ, the predominant macrolide-lincosamide-streptogramin B resistance gene in Clostridium perfringens. Antimicrob Agents Chemother 1994;38:1041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pecavar V, Blaschitz M, Hufnagl P, et al. High-resolution melting analysis of the single nucleotide polymorphism hot-spot region in the rpoB gene as an indicator of reduced susceptibility to rifaximin in Clostridium difficile. J Med Microbiol 2012;61:780–5. [DOI] [PubMed] [Google Scholar]
- [31].Chen YB, Gu SL, Wei ZQ, et al. Molecular epidemiology of Clostridium difficile in a tertiary hospital of China. J Med Microbiol 2014;63:562–9. [DOI] [PubMed] [Google Scholar]
- [32].Collins DA, Putsathit P, Elliott B, et al. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology 2017;49:309–13. [DOI] [PubMed] [Google Scholar]
- [33].Huang H, Weintraub A, Fang H, et al. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe 2010;16:633–5. [DOI] [PubMed] [Google Scholar]
- [34].Yan Q, Zhang J, Chen C, et al. Multilocus sequence typing (MLST) analysis of 104 Clostridium difficile strains isolated from China. Epidemiol Infect 2013;141:195–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kuwata Y, Tanimoto S, Sawabe E, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis 2015;34:763–72. [DOI] [PubMed] [Google Scholar]
- [36].Liao CH, Ko WC, Lu JJ, et al. Characterizations of clinical isolates of Costridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother 2012;56:3943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Baines SD, O’Connor R, Freeman J, et al. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother 2008;62:1046–52. [DOI] [PubMed] [Google Scholar]
- [38].Peláez T, Cercenado E, Alcalá L, et al. Metronidazole resistance in Clostridium difficile is heterogeneous. J Clin Microbiol 2008;46:3028–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Louie TJ, Miller MA, Mullane KM, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 2011;364:422–31. [DOI] [PubMed] [Google Scholar]
- [40].Lee C, Louie TJ, Weiss K, et al. Fidaxomicin versus vancomycin in the treatment of Clostridium difficile infection: Canadian outcomes. Can J Infect Dis Med Microbiol 2016;2016:8048757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Poxton IR. Fidaxomicin: a new macrocyclic, RNA polymerase-inhibiting antibiotic for the treatment of Clostridium difficile infections. Future Microbiol 2010;5:539–48. [DOI] [PubMed] [Google Scholar]
- [42].Iv EC, Iii EC, Johnson DA. Clinical update for the diagnosis and treatment of Clostridium difficile infection. World J Gastrointest Pharmacol Ther 2014;5:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huang H, Wu S, Wang M, et al. Molecular and clinical characteristics of Clostridium difficile infection in a University Hospital in Shanghai, China. Clin Infect Dis 2008;47:1606–8. [DOI] [PubMed] [Google Scholar]
- [44].Drudy D, Kyne L, O’Mahony R, et al. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg Infect Dis 2007;13:504–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huang H, Wu S, Wang M, et al. Clostridium difficile infections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes. Int J Antimicrob Agents 2009;33:339–42. [DOI] [PubMed] [Google Scholar]
- [46].Spigaglia P, Barbanti F, Mastrantonio P, et al. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol 2008;57:784–9. [DOI] [PubMed] [Google Scholar]
- [47].Spigaglia P, Carucci V, Barbanti F, et al. ErmB determinants and Tn916-Like elements in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 2005;49:2550–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cao X, Cavaco LM, Lv Y, et al. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol 2011;49:2496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang HL, Tan M, Qiu AM, et al. Antibiotics for treatment of acute exacerbation of chronic obstructive pulmonary disease: a network meta-analysis. BMC Pulm Med 2017;17:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lu B, Zhou H, Zhang X, et al. Molecular characterization of Klebsiella pneumoniae isolates from stool specimens of outpatients in sentinel hospitals Beijing, China, 2010-2015. Gut Pathog 2017;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gurov AV, Kriukov AI, Kunelskaya VY, et al. Evaluation of the efficacy and tolerability of oral ciprofloxacin used in the comprehensive treatment of external bacterial otitis: an observational prospective study. Int Arch Otorhinolaryngol 2017;21:329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Beran V, Kuijper EJ, Harmanus C, et al. Molecular typing and antimicrobial susceptibility testing to six antimicrobials of Clostridium difficile isolates from three Czech hospitals in Eastern Bohemia in 2011-2012. Folia Microbiol 2017;62:1–7. [DOI] [PubMed] [Google Scholar]


