Abstract
To improve the diagnosis and treatment of Mycoplasma pneumoniae (Mp) infection and reduce the misuse of antibiotics, we sought to establish a loop-mediated isothermal amplification (LAMP) assay for rapid detection of Mp.
Six primers specific for the Mp P1 gene were designed, and the LAMP method was used to rapidly detect Mp. The sensitivity of the LAMP method was determined by serial dilution of the standard Mp strain FH (standard strains of Mycoplasma pneumoniae). Specificity was assessed with 17 common pathogenic microorganisms in the respiratory tract. Patient samples were collected from the Department of Respiratory and Critical Care Medicine at the 307th Hospital of Chinese People's Liberation Army from March 2016 to May 2017, examined prospectively, and compared with diagnosis by quantitative real-time polymerase chain reaction (qRT-PCR).
The LAMP assay for Mp detection can be completed within 60 minutes. The minimum detection limit was 39 pg/μL, and no cross-reaction was observed with 17 common respiratory tract pathogens. Of the 125 clinical specimens tested, 43 cases were positive by LAMP assay, and 40 cases were positive by qRT-PCR (P = .162). All 43 samples determined as positive by LAMP test were confirmed to be Mp by Mp P1 protein sequencing.
The LAMP assay is suitable for rapid detection of Mp. It has high sensitivity and specificity, and the detection results are not inferior to those of qRT-PCR.
Keywords: loop-mediated isothermal amplification, Mycoplasma pneumoniae, quantitative real-time polymerase chain reaction, rapid detection, respiratory infection
1. Introduction
Mycoplasma pneumoniae (Mp) is a common pathogen in respiratory infections.[1] It is the primary cause of community-acquired pneumonia in China,[2] and often leads to extra-pulmonary organ damage.[3] Due to its lack of a cell wall, Mp is not sensitive to common ß-lactam antibiotics; thus, early diagnosis of Mp infection can reduce the unnecessary use of these antibiotics. However, due to the slow growth of Mp, conventional culture takes 2 to 4 weeks.[4] Serological tests can only detect specific immunoglobulin M antibodies 5 to 7 days after the onset of the infection, and a repeated serum test is required 2 weeks later.[4] Detection based on quantitative real-time polymerase chain reaction (qRT-PCR) has high sensitivity and is rapid, taking only 2 to 3 hours to obtain results. However, due to the high cost and required technical expertise, qRT-PCR detection cannot be routinely performed in most areas of China, especially in grassroots laboratories. Therefore, a method for rapid and easy detection of Mp would have major clinical impact.
Loop-mediated isothermal amplification (LAMP) is a specific isothermal nucleic acid amplification technique.[5] Here we describe the development of an easy and rapid LAMP assay for detection of Mp. The method is fast and efficient, with high specificity and sensitivity. The results are easy to read, and the equipment is simple; the assay can be conducted using a small instrument at the bedside or in the field.
2. Materials and methods
2.1. Bacterial strains and clinical samples
The strains used in this study are shown in Table 1. Clinical specimens were collected from the throat swabs or alveolar lavage fluids of 125 patients (59 males, and 66 females with a mean age of 42.0 years; age range, 18–65 years) with acute respiratory tract infections highly suspected of Mp infection in the Department of Respiratory and Critical Care Medicine at the 307th Hospital of the Chinese People's Liberation Army between March 2016 and May 2017. The diagnosis of acute respiratory tract infections, including bronchitis (n = 75), bronchiolitis (n = 17), and pneumonia (n = 33), was made by clinical parameters (cough, productive sputum, dyspnea, chest pain, fever, or abnormal breath sounds), laboratory results (normal white blood cell count or mild elevation of C-reactive protein), and radiographic findings. Samples were stored in mycoplasma transport medium at −80°C. Informed consent was obtained from all the patients and the study protocol was approved by the Ethics Committee at the 307th Hospital. All the patients were blind to the study protocol.
Table 1.
Organisms used in this study.
2.2. Major materials and instruments
Mycoplasma broth base (CM0403, Oxoid, United Kingdom); Mycoplasma Supplement-G (SROO59C, Oxoid); Bst (Bacillus stearothermophilus) DNA polymerase, large fragment (New England Biolabs, Boston); Genomic DNA Purification Kit (Promega); real-time PCR kit (Da An Gene Co., Sun Yat-Sen University, China); dNTPs (Pharmacia); Chelex-100 and betaine (Sigma); magnesium sulfate, potassium chloride, sodium hydroxide, EDTA, and ammonium sulfate (Sinopharm Chemical Reagents, China); Tris-HCl (Shanghai Simco Biotech, Shanghai, China); Triton X-100 (Beijing Mylab Corporation, China); NP-40 (FLUKA); hydroxynaphthol blue (HNB; Regal Biotechnology, China); 2× Taq Mix (Tiangen Biotech, China); agarose (AMRESCO); thermal cycler, gel imager, and CFX96 real-time system (Bio-Rad, California); NanoDrop ND-1000 spectrophotometer (Thermo Scientific); Loopamp LA-320c Realtime Turbidimeter (Eiken Chemical Co., Tokyo, Japan); block heater (Hangzhou Bioer Technology, Hangzhou, China). Primers were synthesized by Beijing Sangon Biotech.
2.3. Preparation of bacterial DNA templates
Total bacterial genomic DNA was extracted by the Chelex method: a 1 mL throat swab culture was centrifuged at 12,500 rpm for 5 minutes at 4°C, the supernatant was discarded, and 100 μL double distilled water was added. Then, the same volume of Chelex DNA extraction solution (25 mmol/L NaOH, 10 mmol/L Tris-HCl, 1% Triton X-100, 1% NP-40, 0.1 mmol/L EDTA, 2% Chelex-100) was added and the mixture was boiled for 10 minutes, placed at 4° C for 5 minutes, and centrifuged at 14,000 rpm for 2 minutes at 4°C. The supernatant was used as the template for the LAMP assay and qRT-PCR reactions.
2.4. Primer design
The sequence of the Mp P1 gene (GenBank No. KC885956.1) was obtained from GenBank, and homology analysis was conducted with BLAST (National Center for Biotechnology Information) to identify specific conservative target sequences, for which primers were designed. The website http://primerexplorer.jp/e/ was used to design primers for the LAMP assay. The DNA sequences to be amplified were divided into 6 independent loci, and 5 pairs of primers were designed for the LAMP assay according to the 6 loci (internal primers forward inner primer (FIP) and backward inner primer (BIP), external primers F3 (forward outer primer) and B3 (backward outer primer), and loop primers LB (backward loop primer) and LF [forward loop primer]). All the designed primer sequences are listed in Table 2.
Table 2.
All the designed primers used for LAMP of the Mycoplasma pneumoniae P1 gene.
2.5. LAMP assay
LAMP reactions (25 μL total volume) were prepared, containing 20 mmol/L Tris-HCl (pH 8.8), 10 mmol/L KCl, 10 mmol/L (NH4)2SO4, 0.1% Tween-20, 0.8 mol/L betaine, 8 mmol/L MgSO4, 1.4 mmol/L dNTPs, 8 U of Bst DNA polymerase, 40 pmol of primers FIP and BIP, 20 pmol of primers LF and LB, and 5 pmol of primers F3 and B3. Finally, 2uL DNA template was added to the reaction. Reactions were incubated in a 63°C water bath for 50 minutes and then inactivated at 80°C for 5 minutes.
2.6. Detection of LAMP results
Realtime Turbidimeter detection: In the LAMP reaction, Bst DNA polymerase generates pyrophosphate, which reacts with divalent magnesium ions in the reaction solution to produce magnesium pyrophosphate, a white precipitate. A LA-320c Realtime Turbidimeter was used to measure the turbidity in the reaction tube every 6 seconds, which was plotted on a curve to determine positive and negative reactions.
Detection of calcein color change: Calcein is a metal ion indicator. It displays different colors according to the charge of magnesium ions in the reaction, with orange and green indicating negative and positive charges, respectively.
Determination of LAMP assay sensitivity: To evaluate the sensitivity, serial 10-fold dilutions of Mp FH (standard strains of Mycoplasma pneumoniae) genomic DNA extract from 0.039 pg/μL to 39 ng/μL were prepared, assayed in 3 replicates with LAMP, and compared to results obtained with qRT-PCR. LAMP turbidity measurements and colorimetric assays using calcein were performed.
2.7. qRT-PCR
The commercial qRT-PCR kit targeting the 16S rRNA gene of Mp was used as the benchmark comparison for LAMP. The total reaction volume was 45 μL, containing 2× Premix Ex Taq (Takara), 10 μM primers, 3 μM TaqMan probes, and 3 L of DNA template. The reaction conditions for qRT-PCR were as follows: 93°C for 2 minutes, then 10 cycles of 93°C for 45 seconds and 55°C for 60 seconds, then 30 cycles of 93°C for 30 seconds and 55°C for 45 seconds. Standard samples were used to plot a standard curve.
2.8. Statistic analysis
Comparison between the 2 methods of LAMP and qRT-PCR was analyzed by χ2test, and P < .05 was considered statistically significant. Significance was calculated using SPSS software, version 17 (SPSS Inc., San Francisco).
3. 3Results
3.1. Screen for optimal LAMP primers
Five pairs of primers were designed and tested for use in the Mp LAMP assay. The primer pair that displayed the earliest detection of the LAMP reaction, Mp16, was considered optimal (Fig. 1).
Figure 1.
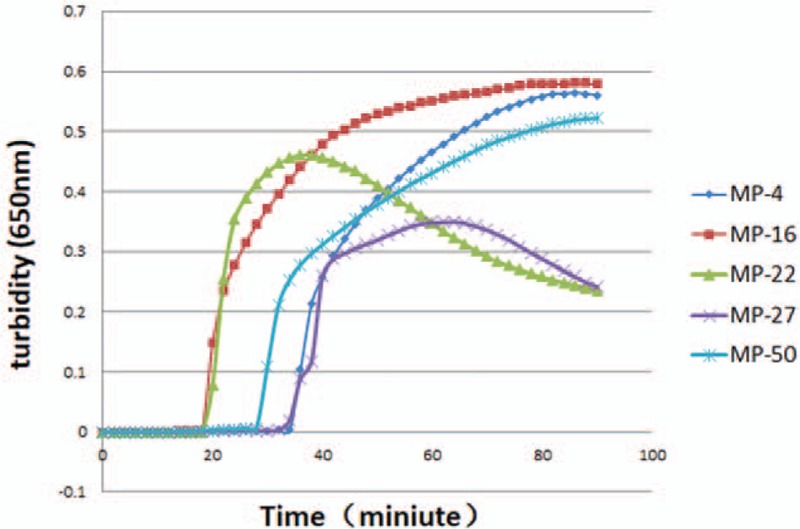
Screening of optimal LAMP primers for detection of Mycoplasma penumoniae FH. The number MP-4,16,22,27,50 means the 5 pairs of primers designed and tested for use in the Mp LAMP assay. LAMP = loop-mediated isothermal amplification.
3.2. Specificity test for LAMP detection of the P1 gene
The specificity of the test, or no cross-reactivity of the test, assayed in 3 replicates, was examined using DNA from 17 common pathogenic microorganisms in the respiratory tract as templates (Fig. 2). The primer pair only amplified the P1 gene of Mp, demonstrating the high specificity of the detection method. In addition, only the positive control was green, consistent with the turbidimetry results.
Figure 2.
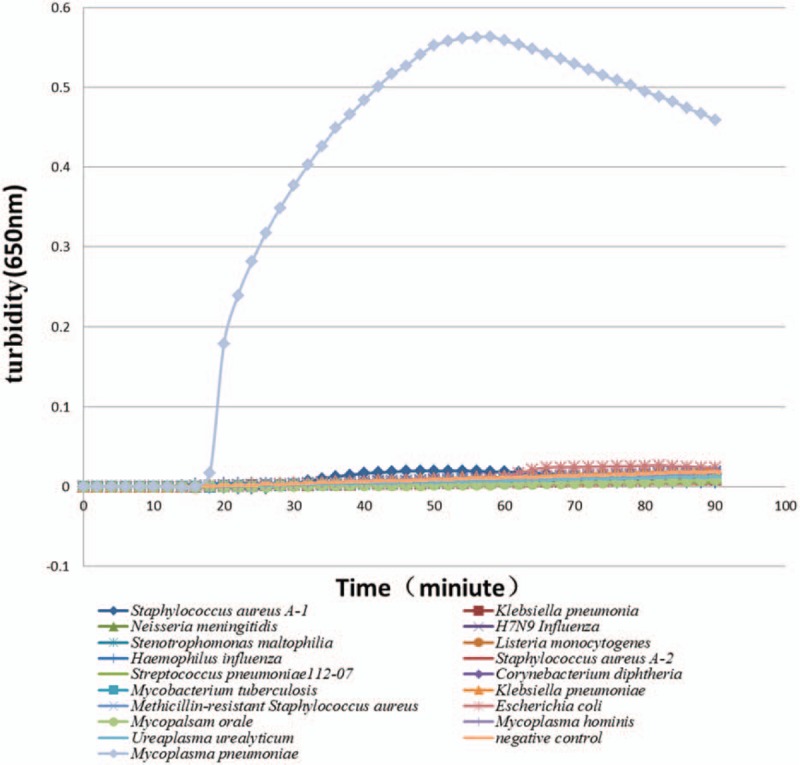
The specificity of LAMP for the detection of Mycoplasma pneumoniae FH. The line 1-17 refer to the DNA from 17 common pathogenic microorganisms in the respiratory tract as templates, the 18th line means the negative control group, and the 19th line means the positive control group. LAMP = loop-mediated isothermal amplification.
3.3. Sensitivity test for LAMP detection of P1
To examine the analytical sensitivity of the assay, or the limit of detection of the assay, assayed in 3 replicates, serially diluted Mp FH samples were used as templates in a LAMP assay for P1 gene detection. As shown in Figure 3, the concentration at which minimal detection occurred was 39 pg/μL, compared to the same result with qRT-PCR.
Figure 3.
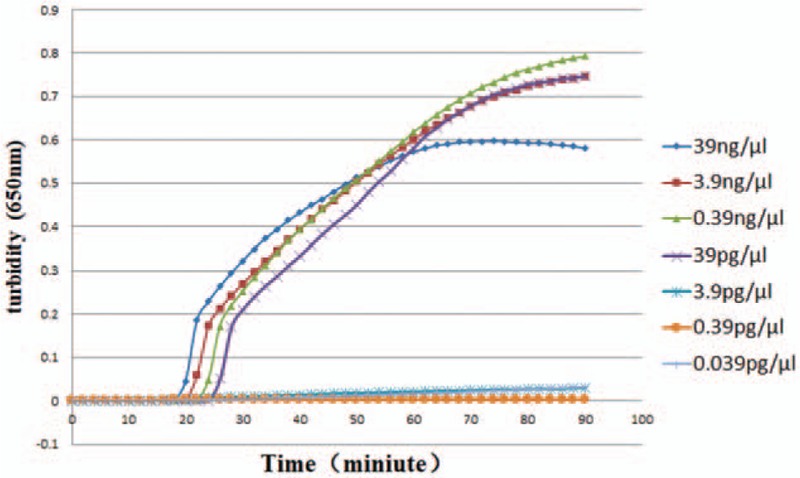
The sensitivity of LAMP for detection of Mycoplasma pneumoniae FH. The number in the right side of the figure means a serially diluted concentrations of Mp FH samples. LAMP = loop-mediated isothermal amplification.
3.4. Clinical sample test
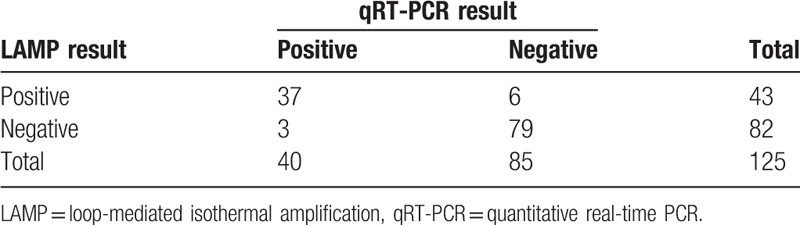
Clinical specimens were collected from 125 patients, of which 120 were throat swabs and 5 were alveolar lavage fluids. All the samples were tested in 3 replicates respectively with LAMP and qRT-PCR methods. The qRT-PCR method and the results of P1 gene sequencing with BLAST were taken as the “gold standard.” The Mp LAMP assay was positive in 43 cases and negative in 82 cases, whereas qRT-PCR analysis was positive in 40 cases and negative in 85 cases (Table 3). P1 gene sequencing was performed on the 43 LAMP-positive samples, and the results were analyzed with BLAST, which confirmed that all 43 samples were Mp. For qRT-PCR, 37 of the 40 positive samples were confirmed as Mp. Thus, the positive rate was 100% for the LAMP assay and 93% for qRT-PCR. There was no significant difference in Mp detection between the LAMP test and the qRT-PCR test (P = .162).
Table 3.
Correlation between LAMP and qRT-PCR for Mycoplasma pneumoniae detection from clinical specimens.
4. Discussion
We developed and evaluated a LAMP assay that targets the P1 gene of Mp. This assay showed good sensitivity and specificity.
LAMP is a specific isothermal nucleic acid amplification technique, in which 4 to 6 primers specific for 6 regions of the target gene are designed, and a DNA polymerase with strand displacement activity is used to amplify genes under isothermal conditions. The results can be directly determined by measuring the precipitation turbidity of the amplification byproduct magnesium pyrophosphate or judged by the color changes. The method has prominent advantages. First, it is rapid and efficient. The reaction can be completed within 30 to 60 minutes, and 109 to 1010 copies of target sequence can be obtained, which is an order of magnitude higher than traditional PCR. Second, it has high sensitivity and specificity. LAMP can detect a copy number 10 times lower than the minimal detection limit of PCR, and can amplify samples from miniscule numbers of cells. Four primers targeting 6 regions confer high specificity, and even in the presence of non-targeted DNA, specific amplification by LAMP is not affected. Third, the results are simple to interpret. The large amount of products generated in a LAMP reaction allows direct pyrophosphate turbidity measurement or fluorescence colorimetric detection for straightforward interpretation of results. Lastly, it requires only simple equipment (only a water bath is needed for DNA extraction and the LAMP reaction), enabling easy application. Due to its speed, sensitivity, specificity, simplicity, and low cost, the LAMP technique has attracted extensive attention from the World Health Organization, researchers, and relevant government agencies in the past decade, and has been successfully applied to fields including microbial pathogen detection, single nucleotide polymorphisms (SNP) detection, and surveillance of infectious diseases. At present, commercial products utilizing the LAMP assay for microorganisms have been approved in Japan and the United States.[6,7] In Japan, LAMP was capable of detecting Mp about 2×102 copies, corresponding to 2 to 20 colour changing units of Mp, and a concordance of 100% was observed between the LAMP and the qRT-PCRassays.[8] In the United States, the illumigene limit of detection of the LAMP assay for Mp was <88 CFU/reaction, the sensitivity was 100% and the specificity was 99% when compared to the qRT-PCR.[9] However, application of the commercial LAMP assay for detection of Mp is not available in China. In our study, the minimum detection limit of the LAMP assay was 39 pg/μL, and there was no significant difference in Mp detection between the LAMP and the qRT-PCR test. The results are almost consistent with the reported results.
A limitation of this study is the small number of samples collected. Additional clinical samples will be collected in the future to further assess the technique. However, our results demonstrate that LAMP assay provides comparable results to qRT-PCR analysis without requiring special equipment, and it can be routinely performed at the bedside or in the field with short detection time. This method is a promising option for rapid Mp detection in clinical practice.
Acknowledgments
We would like to express our warmest appreciation to Dr Jing Zheng, Zhizhi Heng, Fengjiang Li for their contributions in the collection of specimens.
Author contributions
Conceptualization: Xin Yuan, Qian Cui, Han Zhang, Jing Yuan, Yuzhong Feng, Xin Jin, Puyuan Li, Huiying Liu.
Data curation: Xin Yuan, Qian Cui, Han Zhang, Jing Yuan, Kaiwen Niu, Yuzhong Feng, Xin Jin, Puyuan Li, Huiying Liu.
Formal analysis: Xin Yuan, Yuzhong Feng, Puyuan Li.
Funding acquisition: Xin Yuan, Changqing Bai.
Investigation: Huiying Liu.
Methodology: Xin Yuan, Changqing Bai, Han Zhang, Kaiwen Niu, Yuzhong Feng, Huiying Liu.
Project administration: Xin Yuan, Changqing Bai, Jing Yuan, Yuzhong Feng.
Resources: Kaiwen Niu, Yuzhong Feng, Huiying Liu.
Software: Han Zhang, Kaiwen Niu.
Visualization: Qian Cui.
Writing – original draft: Qian Cui, Xin Jin.
Writing – review & editing: Xin Yuan, Changqing Bai.
Footnotes
Abbreviations: LAMP = loop-mediated isothermal amplification, Mp = Mycoplasma pneumoniae, qRT-PCR = quantitative real-time polymerase chain reaction, SNP = single nucleotide polymorphisms.
XY, CB, QC, and HZ should be considered co-first authors.
The study protocol was approved by the Ethics Committee at the 307th Hospital.
This study was supported by the Capital clinical application research fund (Grant: Z141107002514182) and National Natural Science Foundation of China (Grant: 81400009). The authors declare no conflict of interest.
References
- [1].Waites KB, Xiao L, Liu Y, et al. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin Microbiol Rev 2017;30:747–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu YN, Chen MJ, Zhao TM, et al. A multicentre study on the pathogenic agents in 665 adult patients with community:acquired pneumonia in cities of China. Zhonghua Jie He He Hu Xi Za Zhi 2006;29:3–8. Chinease. [PubMed] [Google Scholar]
- [3].Waites KB, Balish MF, Atkinson TP. New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol 2008;3:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 2004;17:697–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Parida M, Sannarangaiah S, Dash PK, et al. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 2008;18:407–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aizawa Y, Oishi T, Tsukano S, et al. Clinical utility of loop-mediated isothermal amplification for rapid diagnosis of Mycoplasma pneumoniae in children. J Med Microbiol 2014;63:248–51. [DOI] [PubMed] [Google Scholar]
- [7].Petrone BL, Wolff BJ, DeLaney AA, et al. Isothermal detection of Mycoplasma pneumoniae directly from respiratory clinical specimens. J Clin Microbiol 2015;53:2970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saito R, Misawa Y, Moriya K, et al. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Mycoplasma pneumoniae. J Med Microbiol 2005;54:1037–41. [DOI] [PubMed] [Google Scholar]
- [9].Ratliff AE, Duffy LB, Waites KB. Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J Clin Microbiol 2014;52:1060–3. [DOI] [PMC free article] [PubMed] [Google Scholar]


