Supplemental Digital Content is available in the text.
Keywords: antisense, Elavl1, endothelium, RNA, zebrafish
Abstract
Objective—
Tie1 (tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1), an endothelial and hematopoietic cell–specific receptor tyrosine kinase, is an important regulator of angiogenesis and critical for maintaining vascular integrity. The post-transcriptional regulation of tie1 mRNA expression is not understood, but it might partly explain Tie1’s differential expression pattern in endothelium. Following up on our previous work that identified natural antisense transcripts from the tie1 locus—tie1 antisense (tie1AS), which regulates tie1 mRNA levels in zebrafish—we attempted to identify the mechanism of this regulation.
Approach and Results—
Through in vitro and in vivo ribonucleoprotein binding studies, we demonstrated that tie1AS long noncoding RNA interacts with an RNA binding protein—embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1)—that regulates tie1 mRNA levels. When we disrupted the interaction between tie1AS and Elavl1 by using constitutively active antisense morpholino oligonucleotides or photoactivatable morpholino oligonucleotides, tie1 mRNA levels increased between 26 and 31 hours post-fertilization, particularly in the head. This increase correlated with dilation of primordial midbrain channels, smaller eyes, and reduced ventricular space. We also observed these phenotypes when we used CRISPR (clustered regularly interspaced short palindromic repeats)–mediated CRISPRi (CRISPR-mediated interference) to knock down tie1AS. Treatment of the morpholino oligonucleotide–injected embryos with a small molecule that decreased tie1 mRNA levels rescued all 3 abnormal phenotypes.
Conclusions—
We identified a novel mode of temporal and spatial post-transcriptional regulation of tie1 mRNA. It involves long noncoding RNA, tie1AS, and Elavl1 (an interactor of tie1AS).
Long noncoding RNAs (lncRNAs), loosely defined as RNA molecules longer than 200 nucleotides that lack the general properties of protein-coding RNAs, have emerged as important regulators of gene expression at both the transcriptional and post-transcriptional levels.1–3 LncRNAs are often classified by their origins in the genome and their relationships to protein-coding regions.4 One subclass of lncRNA, natural antisense transcripts (NATs), is abundant in eukaryotic genomes.5 NATs share complementarity with protein-coding transcripts but are transcribed in the opposite orientation. We previously identified NATs from the tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 (tie1) loci in zebrafish (2 NATs), mice (1 NAT), and humans (3 NATs).6 None of those tie1 antisense (tie1AS) transcripts have been shown to encode proteins, and all are >200 nucleotides long. By current definition, they are therefore lncRNAs.
Tie1 is an essential gene in development, and deletion of the murine tie1 gene leads to local hemorrhage and edema, causing embryonic lethality.7,8 TIE1 protein, encoded by the sense strand in the tie1 locus, is a receptor tyrosine kinase expressed on the surfaces of endothelial cells and hematopoietic cells,9 and most of its function is context dependent.10 TIE1 protein is required for vessel integrity in the late phases of embryonic angiogenesis and for survival of endothelial cells in mature vessels.7,8,11,12 Further, Tie1 is differentially expressed across vascular beds.13–15 The critical role of Tie1 and its nonuniform expression in different types of endothelia during embryogenesis and in adult tissues suggest tight regulation and control of its expression at the post-transcriptional and post-translational level. Although a large body of work describes post-translational regulation of Tie1, very little is known on the context-dependent post-transcriptional regulatory mechanisms for tie1 mRNA. Here, we describe a molecular mechanism used by tie1AS and the RNA-binding protein embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1) to regulate tie1 mRNA levels post-transcriptionally both spatially and temporally in the vascular bed of the developing zebrafish brain.
Materials and Methods
Data available on request from the authors.
Animal Research
All zebrafish (Danio rerio) studies were performed according to the Medical College of Wisconsin (MCW) animal protocol guidelines under protocol AUA320. The MCW Institutional Animal Care and Use Committee approved the studies conducted here. Transgenic lines used in this study included: Tg(kdrl:eGFP), Tg(kdrl:eGFP)roya9; mitfaw2, Tg(kdrl:NLS-EGFP)/Tg(−6.5kdrl:mCherry), and Tg(kdrl:NLS-EGFP). Additional details on the transgenic lines used are given in Major Resources Table in the online-only Data Supplement.
Wild-type or transgenic adult zebrafish males and females were crossed to obtain embryos. Only zebrafish embryos (1-cell stage to 48 hours post-fertilization [hpf]) were used in our experiments. To obtain freshly fertilized wild-type and transgenic zebrafish embryos, we crossed adult fish at a ratio of 1 male to 2 females, housing them overnight in off-system breeding tanks (1 L volume) at a density of ≤5 fish per tank. The fish were separated by a divider and were maintained in a crossing tank containing a breeding chamber and a lower collection chamber. Embryos were collected in the morning when the separator was removed. The embryos were placed in an incubator with an ambient temperature of 28.5°C. The embryo medium (E3) consisted of 5 mmol/L NaCl, 0.17 mmol/L KCl, 0.33 mmol/L CaCl2, and 0.33 mmol/L MgSO4.
For imaging studies, we anesthetized the embryos by incubating them in E3 medium containing a final concentration of 0.003% tricaine (3-amino benzoic acid ethyl ester; Sigma) at pH 7.
RNA Binding Ultraviolet Cross-Linking Assays
Ultraviolet (UV) cross-linking assays were performed as described previously.16 In brief, probes were synthesized with the T7 bacteriophage RNA polymerase (Ambion) and α-[P32]-rUTP (ribonucleoside uridine triphosphate; Perkin Elmer, Boston, MA) from linearized, sequence-verified plasmids in which double-stranded oligos were ligated into the EcoR I and Hind III sites of pGEM3. 100 000 cpm of each probe was combined with 5 µL 2× binding buffer (1× binding buffer is 20 mmol/L HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], 3 mmol/L MgCl2, 40 mmol/L KCl, and 1 mmol/L DTT [dithiothreitol], pH 7.6). Sequence-specific complexes were generated upon addition of 2 μL of total zebrafish 28 hpf extract (25–50 μg/μl) and 1-μL E.coli tRNA (5 mg/mL). The mixture was incubated for 30 minutes, UV cross-linked for 12 minutes at 254 nm, and RNA digested with 5-μg RNase I (Thermo Fisher; EN0602) for 10 minutes at 37°C. The samples were resolved on SDS-polyacrylamide gels and analyzed with a GE Storm 820 PhosphorImager.
RNA Pull-Down and Mass Spectrometry
Overlapping sense and antisense single-stranded DNA oligos were purchased from Eurofins MWG Operon LLC (Louisville, KY) and hybridized to obtain double-stranded DNA. All double-stranded DNA had the T7 promoter region at the 5′ end. The MaxiScript T7 (Ambion; AM1312) kit was used to transcribe RNA from double-stranded DNA in vitro. The RNA was biotinylated at its 3′ end, using a Pierce Desthiobiotinylation Kit (Thermo Scientific; 20163). Desthiobiotinylated RNA probes were incubated with streptavidin magnetic beads from a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific; 20164) as per kit protocol. After washes with Wash Buffer (Thermo Scientific; 20164), 200 µg of zebrafish total lysate (26–28 hpf) was added to the RNA-bound streptavidin magnetic beads, and the complex was incubated at 4°C with gentle agitation for 90 minutes. After three washes with Wash Buffer (Thermo Scientific; 20164), the RNA-bound proteins were eluted in SDS sample buffer and resolved by SDS-PAGE on 4% to 20% acrylamide gel (Bio-Rad, Hercules, CA). RNA-bound proteins were visualized by silver staining. Protein bands of interest were excised and sequenced by mass spectrometry (MS) at the Taplin Mass Spectrometry Facility (Harvard Medical School, Boston, MA). MS hits were analyzed for the number of unique and total peptides obtained and by determining the area under the curve. Each hit was digested in silico with trypsin, and the number of peptides obtained was compared with the number of peptides in the MS results. Vitellogenin, a yolk protein known to contaminate zebrafish embryo protein lysate,17 was excluded from the analysis.
RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction
Wild-type and transgenic zebrafish embryos were manually dechorionated with forceps. Whole embryo, head, and tail/trunk were manually dissected and homogenized in Trizol with a motorized pestle (Sigma Aldrich; Z359971). Heads were obtained by cutting behind the hindbrain ventricle, and tails/trunks were obtained by cutting at the start of the yolk sac extension with microscissors (World Precision Instruments). After Trizol treatment and DNase I digestion, RNA was purified with the help of RNeasy columns (Qiagen; 74034) as per the manufacturer’s instructions. Three hundred nanograms to 500 ng of RNA was incubated with 50 mmol/L oligo(dT)20 primer (Invitrogen; 18418) and 10 mmol/L dNTPs (deoxynucleotide triphosphates), and heated to 65°C for 5 minutes. After being cooled on ice, the RNA was reverse transcribed to cDNA with Superscript III (Invitrogen; 18080044) as per the manufacturer’s protocol. We performed quantitative real-time polymerase chain reaction (PCR) by mixing 1 μL cDNA with 10 μL 2× qPCR Buffer Mastermix iCycler (MidSci BEQPCR-IC) and 5 pmol of each forward and reverse tie1, tie1AS, or gapdh primer. Quantitative real-time PCR was also performed with VE (vascular endothelial)-cadherin and p53 forward and reverse primers. The sequence of the primers is listed in Table II in the online-only Data Supplement. Fluorescence emitted during each cycle was monitored using the iQ5 Multicolor Real-Time PCR Detection System (BioRad, Hercules, CA). The absence of nonspecific products was determined by analyzing dissociation curves. End point PCR was performed by combining 1 μL cDNA with 5 pmol of each forward and reverse tie1, tie1AS, or gapdh primer, 10 μL 5× high-fidelity buffer, and 2 U of Phusion DNA polymerase (Thermo Scientific; F530L).
Western Blotting After RNA Pull-Down
Western blotting was performed after the RNA pull-down to confirm the interaction between tie1AS and Elavl1. RNA pull-down was performed as mentioned above and resolved by SDS-PAGE on a 12% acrylamide gel (Bio-Rad, Hercules, CA). Proteins were electrophoretically transferred onto PVDF (polyvinylidene fluoride) membrane overnight at 12 V. After washes in 1× TBST (Tris-buffered saline tween20; 1× TBS with 1% Tween-20), the PVDF membrane was blocked in blocking solution (5% wt/vol BSA in 1× TBST) for 1 hour at room temperature. Anti-Elav antibody 1:500 (Invitrogen; A21271) was added and incubated overnight at 4°C. After washes in 1× TBST, the membrane was incubated for 1 hour with anti-mouse IgG (Cell Signaling; 7076S) in blocking solution (1:2000). The membrane was washed again in 1× TBST and developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher; 34078) on X-ray films (MidSci; EBNU2).
Whole Fish In Situ Hybridization, Fluorescence In Situ Hybridization, and Immunostaining
Whole fish in situ hybridization was performed as described previously.6 Fluorescence in situ hybridization and immunofluorescence were performed with an RNAscope kit from Advanced Cell Diagnostics, Inc, following the protocol described by Gross-Thebing et al.18 Fluorescence in situ hybridization probes to detect tie1 mRNA and tie1AS lncRNA were designed by Advanced Cell Diagnostics, Inc. Probe details are mentioned in the Major Resource Table in the online-only Data Supplement. Anti-Elav antibody (Invitrogen; A21271) was used as the primary antibody to detect Elavl1, and Alexa Fluor 488 goat anti-mouse antibody (Invitrogen A11001) was used as the secondary. Control immunofluorescence staining was performed with anti-mouse IgG (Cell Signaling; 7076) as the primary antibody and Alexa Fluor 488 goat anti-mouse antibody (Invitrogen A11001) as the secondary.
tie1AS:Elavl1 Interaction Studies In Vivo
The interaction between tie1AS lncRNA and Elavl1 in vivo was detected with a Magna RIP kit (Millipore 17–700) according to the manufacturer’s protocol with the following modifications: zebrafish wild-type embryos (26–28 hpf) were dechorionated and washed in 1× PBS. Embryos were homogenized in 1× PBS with a motorized pestle (Sigma Aldrich; Z359971) and dissolved in lysis buffer from the Magna RIP kit. Anti-Elav antibody (Invitrogen; A21271), anti-tyrosine hydroxylase antibody (Pel Freeze; P-40101), or IgG from the kit was added to the kit’s magnetic beads as per the manufacturer’s protocol. After being washed with Wash Buffer (also from the kit), the antibody-bound magnetic beads were incubated with zebrafish lysate overnight at 4°C. Protein-bound RNAs were isolated according to the kit’s protocol and reverse transcribed to cDNA. They were used in end point PCR and quantitative PCR, as described earlier.
CRISPRi Knockdown of tie1AS
We knocked down tie1AS by following the CRISPRi (clustered regularly interspaced short palindromic repeats–mediated interference) method described in Long et al19 2015. In brief, 3 guide RNAs (gRNAs) targeting the transcription start site of tie1AS (250 pg/embryo each) were coinjected with dCas9-KRAB (dead cas9 enzyme-kruppel-associated box domain) mRNA (catalytically inactive Cas9, 250 pg/embryo) into 1-cell stage zebrafish embryos: wild-type, Tg(kdrl:eGFP), or Tg(kdrl:NLS-EGFP)/Tg(-6.5kdrl:mCherry). As a negative control, scrambled gRNAs (250 pg/embryo) were coinjected with dCas9-KRAB (250 pg/embryo). The dCas9-KRAB plasmids (pXT7-dCas9-KRAB) were generously donated by Dr Dong Liu of Peking University, China. The plasmids were linearized, and mRNA was synthesized in vitro for injections. gRNAs were generated as per Varshney et al20 2015. RNA from at least 50 embryos or embryo heads was collected between 28 and 31 hpf and pooled for RT-qPCR analysis as described earlier. The average RT-qPCR value for tie1AS RNA (relative to gapdh) from 3 biological replicates was compared between the control knockdown group and the tie1AS knockdown group to determine the efficacy of CRISPRi-mediated knockdown of tie1AS. tie1 mRNA levels in the control CRIPSRi group and the tie1AS CRISPRi group were determined by the method used for tie1AS lncRNA. The difference in tie1AS levels between the control CRISPRi group and the tie1AS CRISPRi group was analyzed for statistical significance as described in Imaging, Quantification, and Statistical Analysis.
CRISPRi gRNA Sequences
Sequences of the T7 promoter gRNAs cassettes used in this study were as follows, with the T7 promoter italicized:
TTAATACGACTC ACTATAGA TGAGTTGTTTTC TT CAG TGT TT TAGAGCT AGAAATAGCAAGTTAAAA TA AGGC TAGTCCGTTA TCAACTTGAA AAAGTGGCACCGAGTCGGTGCTTTT
TTAATAC GACT CACTATAG AG G TTGAC TTG CGTTTTCTGT TTTAG A GC TAGAAATAGCAAG TTAAAA TAAGGCTA GTCCGTTA TCAACTTGAA AAAGTGGCA CCGAGTCGGTGCTTTT
TTAATACG ACTCA CTATA GACAAAAG CTCAA TTAGCATGTT TTAGAGC TAGAAATAGCAA GTTAAAA TAAGGCTA GTCCGT TA TCAAC TTGAAAA AGTGGCACCGAG TCGGTGCTTTT
The scrambled gRNA used in the study had the following sequence, with the T7 promoter italicized:
TT AAT ACGA CTCACTATA GTTTTAG AGCTA GAAAT AGCA AGTTAAA ATAA GGCTA GT CCGTTATC AACTTGAAA AAGTGGCACCGA GTCG GTGCTTTT
Morpholino Studies
Gene Tools Inc designed the morpholinos that blocked the Elavl1-binding site (tie1AS-Elavl-bMOs) and the control morpholino oligonucleotides (cMOs). MO injections were performed as described previously.6 Photo-cleavable morpholinos obtained from Gene Tools Inc were incubated with cMO or tie1AS:Elavl-bMO1 at a ratio of 1.2:1 to form a photo-cleavable MO:MO complex (pMO complexes) that cages MOs to render them inactive. Two nanograms of pMO complexes was injected into 1-cell zebrafish embryos, which were developed to 14 to 16 somite (som) stages in petri dishes at 28.5°C in the dark before being exposed to 365 nm UV light in a light box for 8 minutes. After the MO was photo-activated, the embryos were returned to the 28.5°C incubator. Between 28 and 31 hpf, their phenotypes were analyzed. Then their RNA was extracted, processed, and quantified by RT-qPCR, as described earlier. RT-qPCR values for tie1 mRNA and tie1AS RNA (relative to gapdh) were obtained from at least 3 biological replicates for embryos injected with cMO/control pMO or tie1AS-Elavl-bMOs/tie1AS-Elavl-pMO. The average RT-qPCR values, along with SDs, were plotted in Figures 3 and 5 and Figures IX, XI, XVI, XVIII, and XIX in the online-only Data Supplement. We determined the significance of the difference between the RT-qPCR values of the control group versus the experimental groups (modulation of the Elavl1-tie1AS interaction or tieAS knockdown) as described below.
Figure 3.

Analysis of tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 (tie1) mRNA and tie1 antisense (tie1AS) RNA levels in zebrafish embryos injected with tie1AS-Elavl-bMOcomb. One-cell wild-type or Tg(Kdrl:eGFP) zebrafish embryos were injected with control morpholino oligonucleotides (cMOs) or morpholinos targeting the embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1)–binding sites in tie1AS (tie1AS-Elavl-bMOcomb). The embryos were grown at 28.5°C, and at least 50 were collected at time points shown on the x axis. RNA was extracted from whole embryos (A and D), embryo heads (B and E), or trunk/tail (C and F) and used in RT-qPCR. Levels of tie1 and tie1AS RNA relative to gapdh mRNA expression are shown for embryos injected with cMO (blue open squares) or tie1AS-Elavl-bMOcomb (red open squares). Error bars represent SDs. For each specified time point, the difference was analyzed for statistical significance. The asterisk denotes adj P<0.05 after multiple testing corrections. In A and D, n=≈50 embryos were analyzed per time point, with a total of 3 biological replicates. In B, C, E, and F, n=50 to 60 embryo heads or trunks/tails were analyzed per time point, with a total of 3 biological replicates.
Figure 5.
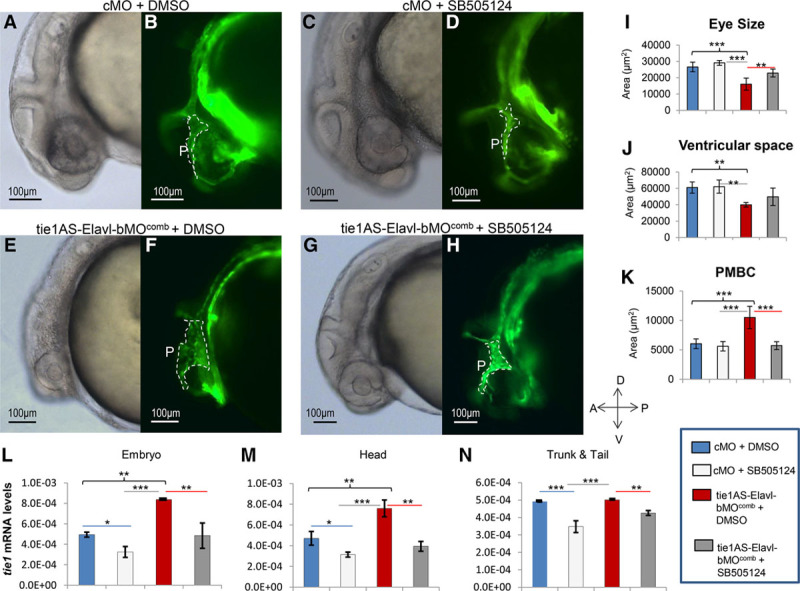
Phenotypic evaluation and analysis of tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 (tie1) mRNA levels in embryos treated with morpholino or dimethyl sulfoxide (DMSO)/SB-505124. One-cell Tg(kdrl:eGFP) zebrafish embryos were injected with control morpholino oligonucleotides (cMOs) or morpholinos targeting embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1)–binding sites in tie1AS (tie1AS-Elavl-bMOcomb). They were then grown at 28.5°C in the dark and treated with 100 μm DMSO or 100 μm SB-505124 between somite stages 18 and 20. Around 28 hours post-fertilization (hpf), the embryos were imaged (A–H), and areas of eyes, ventricular space, and primordial midbrain channels (PMBCs) were quantified from 6 lateral images (I–K). Also, tie1 mRNA levels in some embryos collected between 28 and 31 hpf were determined (L–N). A, A representative (156/180) phase-contrast image of a 28 hpf embryo injected with cMO and treated with DMSO (cMO+DMSO). B, A fluorescence image corresponding to A. C, A representative (165/216) phase-contrast image of a 28 hpf embryo injected with cMO and treated with SB-505124 (cMO+SB-505124). D, A fluorescence image corresponding to C. E, A representative (98/200) phase-contrast image of a 28 hpf embryo injected with tie1AS-Elavl-bMOcomb and treated with DMSO (tie1AS-Elavl-bMOcomb+DMSO). F, A fluorescence image corresponding to E. G, A representative (152/200) phase-contrast image of a 28 hpf embryo injected with tie1AS-Elavl-bMOcomb and treated with SB-505124 (tie1AS-Elavl-bMOcomb+SB-505124). H, A fluorescence image corresponding to G. White dotted lines outline PMBCs (also denoted by P). Outlines of eyes, ventricular spaces, and PMBCs were traced in lateral images (phase-contrast or fluorescence) with Axiovision 4.8.2, and areas within the outlines were deduced with the Measure tool from Axiovision 4.8.2. Average areas of eyes (I), ventricular spaces (J), and PMBCs (K) were deduced for the cMO+DMSO group (blue bars), the cMO+SB-505124 group (light gray bars), the tie1AS-Elavl-bMOcomb+DMSO group (red bars), and the tie1AS-Elavl-bMOcomb+SB-505124 group (dark grey bars). Error bars represent SDs (n=6). cMO+DMSO embryos (blue bars), cMO+SB-505124 embryos (light gray bars), tie1AS-Elavl-bMOcomb+DMSO embryos (red bars), and tie1AS-Elavl-bMOcomb+SB-505124 embryos (dark gray bars) were collected between 28 and 31 hpf. Their RNA was extracted from whole embryo (L), embryo head (M), and trunk/tail (N), and used in RT-qPCR. Average tie1 mRNA levels (ratio to β-actin) were plotted in L, M. Error bars represent SDs. n=50 embryos or heads or trunks/tails per group, with a total of 3 biological replicates. Multi-group differences in I–N were analyzed using ANOVA followed by Tukey test. *Adj (adjusted after multiple testing corrections) P<0.5; **Adj P<0.01; and ***Adj P<0.001. We found significant differences between the cMO+DMSO group vs the tie1AS-Elavl-bMOcomb+DMSO group (black braces), the cMO+DMSO group vs the cMO+SB-505124 group (blue lines), the cMO+SB-505124 group vs the tie1AS-Elavl-bMOcomb+DMSO group (gray lines), and the tie1AS-Elavl-bMOcomb+DMSO group vs the tie1AS-Elavl-bMOcomb+SB-505124 group (red lines).
Imaging, Quantification, and Statistical Analysis
Details related to this section are available in the Supplemental Materials section in the online-only Data Supplement.
Drug Treatment
SB-505124, a selective inhibitor of TGF (transforming growth factor)-βR, was purchased from Millipore Sigma (catalogue number S4696) and dissolved in dimethyl sulfoxide (DMSO) as per the manufacturer’s instructions. One-cell-stage Tg(kdrl:eGFP) embryos were injected with cMO or tie1AS-Elavl-bMOcomb and allowed to grow in the dark at 28.5°C. Between 18 and 20 somite stages, we treated 20 to 25 embryos per petri dish (60×15 mm) with either 100 μm DMSO or 100 μm SB-505124 (diluted in E3 solution). Embryos were grown until 28 to 31 hpf, dechorionated, washed 3× in E3 solution and imaged (phase contrast, fluorescence, and confocal) or processed for RNA extraction as described earlier.
Results
Elavl1 Protein Interacts With tie1AS
Previously, we demonstrated that an 804-nucleotide zebrafish tie1AS lncRNA forms a duplex with tie1 mRNA in the cytoplasm and that overexpression of the tie1AS lncRNA leads to specific degradation of tie1 mRNA.6 tie1AS lncRNA is complementary in sequence to tie1 mRNA apart from a 148 nucleotide-long region in the 5′ end, referred to as a unique sequence (Figure 1A). We hypothesized that tie1AS uses the unique sequences to interact with protein(s) involved in the regulation of tie1 mRNA (resembling the way that mouse ubiquitin carboxyl-terminal hydrolase L1 [Uchl1] antisense lncRNA regulates Uchl1 mRNA21). To test our hypothesis, we performed RNA affinity chromatography (RNA pull-down assay) to detect any RNA-binding proteins that interact with the unique sequences in tie1AS lncRNA. We divided the 148 nucleotide-long unique sequence into 3 overlapping fragments: tie1AS(1–70), tie1AS(45–105), and tie1AS(88–151) (Figure 1A) and generated biotinylated RNA probes. We then incubated those probes with zebrafish embryo lysate (26 to 28 hpf). This time point was chosen because we previously observed that tie1AS and tie1 mRNA levels peak around 24 hpf. Zebrafish proteins interacting with biotinylated RNA probes were pulled down with streptavidin magnetic beads, separated on SDS-PAGE gel, and visualized with silver stain (Figure 1B). Interestingly, only tie1AS(88–151) showed a specific complex—at 37 kDa (Figure 1B, lane labeled tie1AS(88–151), arrow)—though all 3 probes and scrambled RNA (our negative control) produced nonspecific complexes at ≈40 kDa (Figure 1B, asterisk). We excised the tie1AS(88–151)–protein complex and analyzed its proteins through MS (Table I in the online-only Data Supplement). Spectral counts and a comparison of average precursor intensities identified Embryonic lethal abnormal vision Drosophila-like 1 (Elavl1) as the most abundant protein in the complex. This method of quantifying the relative abundance of proteins in MS samples was previously used in Cullinane et al22 in 2015. We focused on the interaction between Elavl1 and tie1 because both have been implicated in angiogenesis.23,24
Figure 1.

Identification of embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1) interaction with unique sequences in tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 antisense (tie1AS) long noncoding RNA (lncRNA). A, Diagram of the convergent tie1 mRNA and tie1AS lncRNA hybrid. tie1AS lncRNA sequences noncomplementary to tie1 mRNA are shown as unique sequences. The unique sequences in tie1AS were subdivided into 3 probes with overlapping regions: tie1AS(1–70) [purple], tie1AS(45–105) [cyan], and tie1AS(88–151) [red]. These probes were used in RNA pull-down assays and ultraviolet (UV) cross-linking assays. Elavl1-binding sites in tie1AS are underlined in red. B, The 3 RNA probes described in A and a scrambled RNA were transcribed in vitro, biotinylated at the 3′ end, and used in RNA pull-down assays as described in Methods in the online-only Data Supplement. From left to right, scrambled RNA (negative control), tie1AS(1–70), tie1AS(45–105), and tie1AS(88–151). The asterisk indicates the nonspecific ribonucleoprotein complex. The black arrow points to the tie1 AS-specific RNA-protein complex, which was excised and analyzed by mass spectrometry. C, Biotinylated RNAs (B) were incubated with zebrafish lysate (26–28 hours post-fertilization [hpf]), resolved by SDS-PAGE, and blotted onto a PVDF membrane. The membrane was probed with anti-Elav antibody (primary) and anti-mouse antibody (secondary), and developed by chemiluminescence. D, Zebrafish 26 to 28 hpf lysate was incubated with protein A-bound anti-Elav antibody, protein A-bound Tyrosine Hydroxylase (TH) antibody, or IgG, and immobilized on magnetic beads. The beads were isolated and washed, and the RNA was extracted. After reverse transcription, the cDNA obtained was used in quantitative PCR. The x axis shows the antibodies used in immunoprecipitation. The y axis shows the percentages of tie1AS lncRNA (dark gray) or tie1 mRNA (light gray) obtained relative to the input. E, UV cross-linking of RNA and zebrafish lysate. RNA labeled with [α]P32-UTP was incubated with zebrafish lysate (26–28 hpf) and cross-linked with UV for 12 minutes. After digestion with RNase I, the complexes were boiled in 2× SDS sample buffer and resolved by SDS-PAGE. The gel was visualized using a phosphorimager. RNA probes from left to right, tie1AS(1–70), tie1AS(40–105), wild-type tie1AS(88–151) in 2 successive lanes, and mutated tie1AS(88–151), tie1ASmut,UAAACUAACAGCAAGCUGCCUGGUCAGAGCAGAACAGAUGACUUUCAGUAAGGAUAGGAUAAUGCAUCGUUU (mutated sequences are underlined). The black arrow indicates that mutations within the adenosine uridine-rich element prevent the formation of the ribonucleoprotein complex.
We repeated the RNA pull-down assay and performed Western blotting with an anti-Elav antibody, which confirmed that Elavl1 binds to tie1AS(88–151) (Figure 1C, lane 4) but not to tie1AS(1–70), tie1AS(45–105), or the negative control (Figure 1C, lanes 1–3). The anti-Elav antibody was raised against the human Elav orthologues HuB (human antigen B), HuC, and HuD and was previously used to detect zebrafish Elavl125 and Elavl3.26 Indeed, the antibody cross-reacts with all 4 members of the zebrafish Elav family of proteins: Elavl1 (HuR, HuA), Elavl2 (HuB), Elavl3 (HuC), and Elavl4 (HuD; Figure I in the online-only Data Supplement), which differ in size: 37 kDa, 42 kDa, 39 kDa, and 52 kDa, respectively. We therefore concluded that the 37 kDa protein that interacts with tie1AS (88–151) is Elavl1. However, we observed 2 bands of almost similar size in the Western blot (Figure 1C), and 2 isoforms of Elavl1 (Elavl1a and Elavl1b) have been reported in zebrafish, with Elavl1a accounting for 90% of Elavl1 proteins.27 To confirm that tie1AS lncRNA interacts with Elavl1 in vivo, we used the anti-Elav antibody to immunoprecipitate Elav-like proteins from 26 to 28 hpf zebrafish lysates. Then, we reverse transcribed the RNA and used quantitative PCR for tie1AS or tie1 mRNA. Indeed, we found that tie1AS—but not tie1—coprecipitated with Elav-like proteins (Figure 1D). The negative control immunoprecipitated with anti-IgG, and anti-tyrosine hydroxylase antibodies failed to pull-down tie1AS (Figure 1D). Also, abundant RNAs, such as gapdh, did not coprecipitate with Elavl1 nonspecifically (Figure II in the online-only Data Supplement). Thus, our in vitro and in vivo results together show that tie1AS lncRNA and Elavl1 form a complex together.
To narrow down the Elavl1-binding site within tie1AS, we performed UV cross-linking assays by incubating body-labeled RNA probes [tie1AS(1–70), tie1AS(45–105), and tie1AS(88–151)] with zebrafish total lysate. We resolved the RNA–protein complexes on SDS-PAGE gel and visualized them by autoradiography. As in the RNA pull-down assays described above, only tie1AS(88–151) (Figure 1E, lane 3, and lane 4) and not tie1AS(1–70) or tie1AS(45–105) (Figure 1E, lanes 1–2) produced 2 ribonucleoprotein complexes at ≈37 kDa. Because Elav proteins are known to bind adenosine uridine (AU)-rich elements within RNA28—and given that tie1AS lncRNA harbors an AU-rich element, UUAAUUUUAU, between nucleotides 105 and 115—we investigated whether this element is required for binding Elavl1. Mutation of the AU-rich element within tie1AS(88–151) (5′UUAAUUUUAU3′ to GCUGCCUGGC3′) prevented the specific ribonucleoprotein complex from forming at 37 kDa (Figure 1E, compare lane 4 to lane 5, arrow). Thus, a 10 nucleotide element, 5′UUAAUUUUAU3′, within tie1AS(88–151) is necessary for tie1AS to interact with Elavl1.
Next, we investigated the spatial expression of tie1 mRNA, tie1AS lncRNA, and Elavl-like proteins in 26 to 28 hpf zebrafish embryos. We used double fluorescence in situ hybridization to detect tie1 mRNA (Figure 2A and 2D; Figures III, V, VI, and VII in the online-only Data Supplement; Movie I in the online-only Data Supplement) and tie1AS lncRNA (Figure 2B and 2D; Figures III, IV, VI, and VII in the online-only Data Supplement; Movie M1 in the online-only Data Supplement) followed by immunofluorescence staining for Elav-like proteins (Figure 2C and 2D; Figures III, IV, VI, and VII in the online-only Data Supplement; Movie I in the online-only Data Supplement) in 26 to 28 hpf embryos. Our results show that both tie1AS and Elav-like proteins have broad regions of expression in the head (Figure 2; Figures IV and VI in the online-only Data Supplement) but not in the tail or trunk (Figure III in the online-only Data Supplement). In the 26 to 28 hpf head, we observed close apposition of tie1 mRNA expression (Figure 2A; Figure V in the online-only Data Supplement) with head vessels, and that expression region was sharply demarcated. In contrast, tie1AS lncRNA (Figure 2B) was expressed almost exclusively in regions of head where tie1 mRNA was absent. In the merged image (Figure 2D), note the regions of exclusivity for tie1 mRNA and tie1AS lncRNA along carotid arteries and primordial midbrain channels (PMBCs). Expression of Elav-like proteins (Figure 2C), was abundant in the forebrain (f), hindbrain (hb), and developing eyes (e). To investigate any overlap in expression, especially between tie1AS and Elav-like proteins, we performed a detailed analysis (Methods in the online-only Data Supplement) across all the z-stack images that contributed to the merged image in Figure 2D, using the colocalization module of LSM510 software (Figures IV and VI in the online-only Data Supplement).
Figure 2.

Spatial expression of tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 (tie1) mRNA, tie1 antisense (tie1AS) long noncoding RNA (lncRNA), and embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1) protein in the head of 26 to 28 hours post-fertilization (hpf) zebrafish embryo. A–D, Following double fluorescence in situ hybridization (RNAscope) to detect tie1 mRNA and tie1AS lncRNA expression, we used immunofluorescence to detect Elav-like protein expression in the same embryo. Representative (6/6) confocal photomicrographs of zebrafish embryo head (lateral) between 26 and 28 hpf captured with a 20× objective lens and processed as z-projections show tie1 mRNA expression in magenta (A), tie1AS lncRNA expression in yellow (B), and Elav-like protein expression in cyan (C). A–C, Merged in D. E, Diagram of 28 hpf zebrafish embryo. Inset: Blow-up of head vasculature. Veins are shown in blue, arteries in red. Vessels indicated are cranial division of the internal carotid artery (CrDI), primordial midbrain channel (PMBC), middle cerebral vein (MCeV), primordial hind-brain channel (PHBC), anterior cerebral vein (ACeV), lateral dorsal aortae (LDA), aortic arch 1 (AA1), primitive internal carotid arteries (PICA), caudal division of the internal carotid artery (CaDI), and primitive mesencephalic artery (PMsA). e indicates eye; f, forebrain; and hb, hindbrain.
This analysis was necessary because the signal associated with Elav-like proteins was overwhelmingly strong across most 1 micron z-stack sections, whereas the signal for tie1AS was very strong in some z-stack sections at submicron level. The imaging analysis showed strong overlap between the expression of tie1AS lncRNA and Elav-like proteins in the forebrain, hindbrain, and eye (Figures IV and VIC in the online-only Data Supplement). When tie1 mRNA expression was included in this analysis, overlap between the 3 molecules was limited to only a few cells in the eye (Figure VI in the online-only Data Supplement). We concluded from this spatial analysis that there is considerable overlap between the expression of tie1AS and Elavl1 at 26 to 28 hpf but that those 2 molecules overlap with tie1 mRNA only in a small number of cells.
Functional Analysis of tie1AS:Elavl1
We used 3 distinct approaches to explore the functional relevance of the interaction between tie1AS and Elavl1 (Figure VIII in the online-only Data Supplement): (1) morpholino antisense oligonucleotides29(MOs), (2) photoactivatable caged MOs (pMOs),30 and (3) CRISPR/Cas9 technology-based transcriptional repression/knockdown31 of tie1AS lncRNA (CRISPRi).
Using 2 different MOs—tie1AS-Elavl-bMO1 and tie1AS-Elavl-bMO2—we blocked Elavl1-binding sites in tie1AS (Table II and Figure VIII in the online-only Data Supplement). When the 2 MOs were injected separately (1 ng each), levels of tie1 mRNA and tie1AS lncRNA were not significantly different from those in embryos injected with cMO (Figure IX in the online-only Data Supplement). However, when we injected 2 ng of tie1AS-Elavl-bMO1 or 2 ng of tie1AS-Elavl-bMO2 separately or coinjected the 2 MOs (1 ng each; tie1AS-Elavl-bMOcomb), we observed a 2-fold increase in tie1 mRNA levels in whole embryo (Figure 3A) and a 2-fold increase in the head (Figure 3B). In contrast, tie1AS levels were 0.4-fold lower in the head (Figure 3E) at 28 to 31 hpf than when we injected the cMOs. We validated the ability of tie1AS-Elavl-bMOcomb to disrupt the interaction between tie1AS and Elavl1 through an in vitro ribonucleoprotein complex formation assay (Figure X in the online-only Data Supplement). We also did not observe changes in levels of p53 mRNA, a gene known to be nonspecifically induced by MO injection,32 or in levels of VE-cadherin mRNA, an important regulator of angiogenesis (Figure XI in the online-only Data Supplement). When we compared the embryos injected with cMO (Figure 4A and 4B) with those injected with tie1AS-Elavl-bMOcomb, we saw a 0.5-fold decrease in eye size (Figure 4C, red arrow and E), a 0.4-fold reduction in ventricular space (Figure 4C, red asterisk and V), and a 2-fold increase (dilation) in the size of PMBCs (Figure 4D, white outline and P). The defects coincided with changes in tie1 and tie1AS lncRNA RNA levels in the head (Figure 3). By performing chromogenic in situ hybridization on embryos injected with 28 hpf tie1AS-Elavl1-bMOcomb, we found a strong correlation between the change in the spatial expression of tie1 mRNA in the head and abnormalities in the head vasculature (compare Figure 4M with Figure 4N, white arrows; also compare Figure XIIA and XIIB in the online-only Data Supplement). We subclassified those embryos into 4 phenotypes: (1) normal/unaffected (38.3%, n=300), (2) mild (32%, n=300), (3) moderate (21%, n=300), and (4) severe (11.7%, n=300; Figure XIII in the online-only Data Supplement). (Descriptions of each phenotype are given in Figure XIII in the online-only Data Supplement.)
Figure 4.
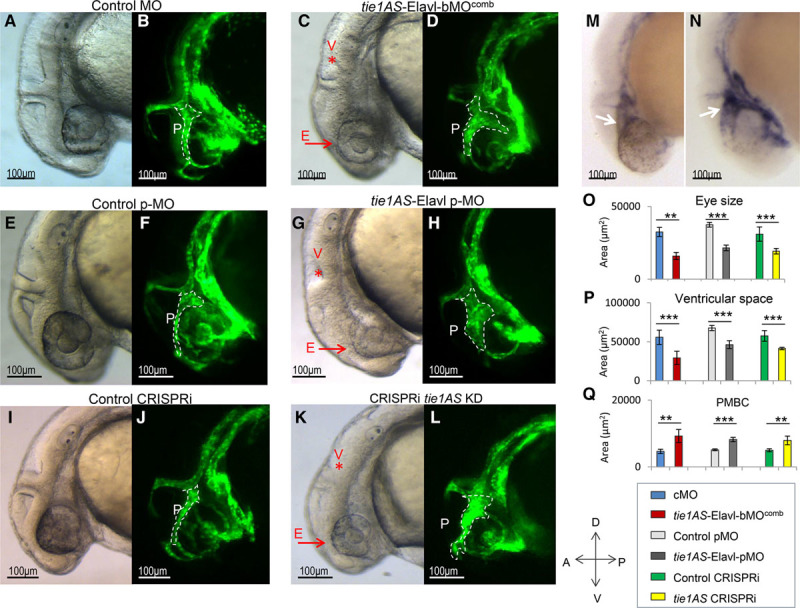
Phenotypic evaluation of CRISPRi (clustered regularly interspaced short palindromic repeats–mediated interference) embryos and embryos injected with morpholino (MO) or photoactivatable morpholino (pMO). One-cell stage Tg(kdrl:eGFP) zebrafish embryos that expressed eGFP(enhanced green fluorescent protein) in the blood vasculature were injected with control morpholinos (cMOs) or morpholinos targeting the embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1)–binding sites in tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 antisense (tie1AS; tie1AS-Elavl-bMOcomb). The embryos were grown at 28.5°C and analyzed between 28 and 31 hours post-fertilization (hpf) for phenotype (A–D). A, A lateral phase-contrast image of a 28 hpf embryo injected with cMO. B, The corresponding fluorescence image. C, A lateral phase-contrast image of a 28 hpf embryo injected with tie1AS-Elavl-bMOcomb. D, The corresponding fluorescence image. A, B, Representative of 102 of the 114 embryos observed. C, D, Representative of 159 of 300 embryos. Two nanograms of photoactivatable control morpholinos (control pMO) or photoactivatable caged morpholinos targeting Elavl1-binding sites in tie1AS (tie1AS-Elavl-pMO) were injected into 1-cell Tg(kdrl:eGFP) embryos, allowed to develop in the dark at 28.5°C to the 14 to 16 somite stage and exposed to 365 nm ultraviolet to activate the pMOs. The embryos were imaged between 28 and 31 hpf. E, A representative (170/200) phase-contrast image of an activated control pMO embryo and (F) and its corresponding fluorescence image. G, Representative (123/300) phase-contrast image of an activated tie1AS-Elavl-pMO embryo and (H) its corresponding fluorescence image. Tg(kdrl:eGFP) embryos were injected at the 1-cell stage with 250 pg of scrambled guide RNAs (gRNAs) along with 250 pg of dCas9-KRAB (dead cas9 enzyme-kruppel-associated box domain; scgRNA+dCas9) or 250 pg of guide RNAs targeting tie1AS transcription start site (TSS), along with dCas9-KRAB (tie1AS gRNAs+dCas9). They were allowed to develop at 28.5°C, and were imaged between 28 and 31 hpf. I, A representative (88/100) phase-contrast image of scgRNA+dCas9 and (J) its corresponding fluorescence image. K, A representative (90/200) phase-contrast image of a tie1AS gRNAs+dCas9 embryo and (L) its corresponding fluorescence image. M, Embryos injected with control morpholinos (cMO) or (N) tie1AS-Elavl-bMOcomb were fixed between 28 and 31 hpf, and chromogenic in situ hybridization was used to detect the expression of tie1 mRNA. Purple staining in M and N shows tie1 mRNA expression. M, Representative of 18 of 20 embryos analyzed, and N is representative of 10 of 25 embryos analyzed. White dotted lines in B, D, F, H, J, and L outline primordial midbrain channels (PMBCs; denoted by P). Red arrows next to E denote smaller eyes, and red asterisks under the V in C, G, and K denote reduced ventricular space. White arrows in M and N indicate tie1 mRNA expression along PMBC. Outlines of eyes, ventricular space, and PMBCs were traced in lateral images (phase-contrast or fluorescence) with Axiovision 4.8.2, and areas within the outlines were deduced with the Measure tool from Axiovision 4.8.2. We then plotted the average areas of eyes (O), ventricular spaces (P), and PMBCs (Q) for the embryos injected with cMO (blue bars), tie1AS-Elavl-bMOcomb (red bars), control pMO (light gray bars), or tie1AS-Elavl-pMO (dark gray bars) and for the control clustered regularly interspaced short palindromic repeats interference (CRISPRi; green bars) and tie1AS CRISPRi embryos (yellow bars). Error bars represent SDs (n=6). *Adj P<0.05, **Adj P<0.01, and ***Adj P<0.001 (P adjusted after multiple testing corrections). The horizontal lines under the asterisks denote the groups compared.
To understand how tie1 mRNA is regulated by tie1AS–Elav-like protein in space and time, we determined whether the 2 molecules colocalize in the head at 48 hpf (Figure XIV in the online-only Data Supplement), a time point at which disrupting the interaction between tie1AS and Elavl1 has no effect on tie1 mRNA levels (Figure 3C). Our data show that, at 48 hpf, Elav-like proteins are expressed in a limited number of cells in the eye and the hind brain, where tie1AS and tie1 mRNA are not expressed (Figure XIIIA in the online-only Data Supplement). Thus, in contrast to the 26 to 28 hpf embryonic stages (Figure 2), there was no overlap between the spatial expression of tie1AS and Elav-like proteins in the 48 hpf head. However, the expression patterns of Elav-like proteins in the trunk and tail are similar at 26 to 28 hpf and 48 hpf (Figures III versus XIIIB in the online-only Data Supplement). From this constitutively active MO approach, we conclude that interaction between tie1AS and Elavl1 around 28 hpf is indeed required to regulate tie1 mRNA levels in the head of the zebrafish embryo.
To further investigate temporal aspects of tie1AS-mediated regulation of tie1 mRNA, we used a pMO. Photo-cleavable MOs have been shown to be nontoxic to embryonic zebrafish,33 as has exposure to UV for >10 minutes post-gastrulation (5 hpf).33 The photo-cleavable MO (pMO1) complementary to the tie1AS-Elavl-bMO1 we used previously caged tie1AS-Elavl-bMO1 (Figure VIII in the online-only Data Supplement). On exposure to UV light, the photo-cleavable MO in the tie1AS-Elavl-bMO1:pMO1 complex (tie1AS-Elavl-pMO) was cleaved, facilitating the activation of tie1AS-Elavl-bMO1 and its binding to its target (additional details in Methods and Figure VIII in the online-only Data Supplement). This strategy allowed us to explore the temporal activation of MOs. We injected control pMO or tie1AS-Elavl-bMO1:pMO mix (1:1.2) (tie1AS-Elavl-pMO) into 1-cell zebrafish embryos that were then incubated in the dark at 28.5°C. Between the 14 and 16 somite stages, the embryos were exposed to 365 nm UV light for 8 minutes to activate the MO. They were then developed at 28.5°C and analyzed for RNA levels and morphological defects between 28 and 31 hpf. Compared with the embryos activated with control pMO (Figure 4E and 4F; Figure XVE and XVF in the online-only Data Supplement), the embryos activated by tie1AS-Elavl-pMO (Figure 4G and 4H; Figure XVG and XVH in the online-only Data Supplement) showed a 0.3-fold reduction in eye size (Figure 4G, red arrow and E), a 0.3-fold reduction in ventricular space (Figure 4G, red asterisk and V), a 1.6-fold increase in the size of PMBCs (Figure 4H, white outline and P), and an increase in tie1 mRNA levels in the head (Figure XVI in the online-only Data Supplement). Dilated PMBCs, reduced ventricular space, and smaller eyes were noticed in ≈40% of the embryos activated with tie1AS-Elavl-pMO (n=300). Thus, our pMO approach confirmed the constitutive MO results and also showed that Elavl1-bound tie1AS downregulates tie1 mRNA levels in the head post-segmentation between 28 and 31 hpf.
Using a third independent approach, we downregulated tie1AS transcription with CRISPR interference (CRISPRi; Figure VIII in the online-only Data Supplement), following a protocol described in Long et al19 2015. We preferred CRISPRi to the more conventional CRISPR because of the proximity of the Elavl1-binding sites to the tie1-coding sequences and because of the scarcity of available canonical protospacer adjacent motif sites (NGG) near Elavl1-binding sites in tie1AS. We injected 3 gRNAs that target the transcription start site of tie1AS into zebrafish embryos together with a catalytically inactive form of Cas9 fused to the repressive Krüppel-associated box domain (dCas9-KRAB). As a control, we coinjected scrambled gRNAs with dCas9-KRAB. The embryos were developed until 28 to 31 hpf at 28.5°C and analyzed for tie1AS levels. Compared with the control (scrambled gRNA+dCas9-KRAB), there was a 5-fold decrease in tie1AS levels in whole embryos and a 2.3-fold decrease in the heads of embryos coinjected with the gRNAs that targeted the transcription start site of tie1AS and dCas9-KRAB (Figure XVIIIA and XVIIIB in the online-only Data Supplement). As in the case of the MO and pMO-based approaches, CRISPRi-mediated knockdown of tie1AS caused an increase (≈2-fold) in tie1 mRNA levels (Figure XVIIIB and XVIIID in the online-only Data Supplement), a 0.4-fold reduction in eye size (Figure 4K, red arrow and E), a 0.3-fold reduction in ventricular space (Figure 4K, red asterisk and V), and dilated PMBCs (Figure 4L, white outline and P) compared with the control embryos (Figure 4I and 4J; Figure XVIIA and XVIIB in the online-only Data Supplement). The defects were apparent in ≈50% of the tie1AS knockdown embryos (n=200). Thus, with all 3 approaches, we noticed improper brain vessel patterning and morphological defects (smaller eyes and reduced ventricular space) that coincided with elevated tie1 mRNA levels in the head.
We used the MO approach to further analyze the correlation between increased tie1 mRNA levels and changes in eye size, ventricular space, and PMBCs. We injected 1-cell transgenic zebrafish embryos with cMO or tie1AS-Elavl-bMOcomb, developed the embryos at 28.5°C, and treated them with either 100 µm DMSO or 100 µm SB-505124 between somite stages 18 and 20. SB-505124 is a commercially available selective inhibitor of TGFβ type I receptors that was previously shown to reduce tie1 mRNA expression in embryonic zebrafish.34 Using this reagent, we determined the phenotypic effects of decreasing tie1 mRNA in embryos injected with tie1AS-Elavl1-bMOcomb. We did not observe any statistically significant difference in eye size, ventricular space, or PMBCs between the cMO embryos treated with DMSO or SB-505124 (Figure 5A and 5B versus 5C and 5D, and blue bars versus light gray bars in Figure 5I through 5K). When we compared the cMO+DMSO group with the tie1AS-Elavl-bMOcomb+DMSO group, however, we observed a statistically significant decrease in eye size (Figure 5A and 5B versus 5E and 5F, and 5I, dark blue versus red bars) and ventricular space (Figure 5A and 5B versus 5E and 5F, and 5J, dark blue versus red bars) and an increase in PMBCs (Figure 5A and 5B versus 5E and 5F, and 5K, dark blue versus red bars), which is consistent with our previous findings (Figure 4). Moreover, when we compared the tie1AS-Elavl-bMOcomb+DMSO embryos with the tie1AS-Elavl-bMOcomb+SB-505124 embryos (Figure 5E and 5F versus 5G and 5H; red bars versus dark grey bars in Figure 5I through 5K), we saw statistically significant increases in eye size (0.5-fold) and ventricular space and a decrease (1.8-fold) in PMBCs. Furthermore, the cMO+SB-505124 and tie1AS-Elavl-bMOcomb+SB-505124 embryos (Figure 5C and 5D versus 5G and 5H and 5I through 5K, compare light gray bars to dark gray bars) were similar in eye size, ventricular space, and PMBCs, resembling the cMO+DMSO group. These results collectively imply that suppressing tie1 mRNA levels in embryos injected with tie1AS-Elavl-bMOcomb rescues the 3 phenotypes.
To correlate phenotypic changes with tie1 mRNA levels, we performed RT-qPCR on the groups outlined above. Compared with the cMO+DMSO group, the cMO+SB-505124 group showed the expected decrease (≈0.3-fold) in tie1 mRNA levels across all tissues (whole embryo, head, trunk, and tail; Figure 5L through 5N, compare blue bars versus light gray bars). When we compared the cMO+DMSO group with the tie1AS-Elavl-bMOcomb+DMSO group, we observed the same pattern of tie1 mRNA levels as reported in Figure 3 (Figure 5L through 5N, compare blue bars versus red bars). When we compared tie1 mRNA levels in the tie1AS-Elavl-bMOcomb+DMSO embryos with those in tie1AS-Elavl-bMOcomb+SB-505124 embryos, we observed a decrease in tie1 mRNA levels in whole embryo, head, tail, and trunk (Figure 5L through 5N, compare red bars versus dark gray bars). The cMO+DMSO group and the tie1AS-Elavl-bMOcomb+SB-505124 group had comparable tie1 mRNA levels across all tissues (Figure 5L through 5N, compare blue bars versus dark gray bars). Thus, administering SB-505124 to embryos that had been injected with tie1AS-Elavl-bMOcomb suppressed the increase in tie1 mRNA levels reported in Figure 3.
In both cMO+SB-505124 and tie1AS-Elavl-bMOcomb+SB-505124 groups, we noticed defects in trunk and tail vasculature, as reported previously.34 From our studies of drug treatment after cMO or tie1AS-Elavl-bMOcomb morpholino injections, we conclude that there is strong correlation between an increase in tie1 mRNA levels, particularly in the head, and the occurrence of dilated PMBCs, reduced ventricular space, and smaller eye size.
Next, we counted endothelial cells in the dilated PMBCs in the CRISPRi/MO embryos. First, we injected MOs, pMOs, and CRISPRi into the transgenic embryos Tg(kdrl: mCherry; NLS-EGFP), which express mCherry in the vasculature and eGFP (enhanced green fluorescent protein) in endothelial cell nuclei. Then we counted endothelial cell nuclei in PMBCs. Compared with the embryos injected with cMO, those injected with tie1AS-Elavl-bMOcomb showed a 1.5-fold increase in the number of endothelial cells within the PMBCs (embryos with severe phenotypes were excluded from the analysis; Figure 6A through 6C). Similarly, the number of endothelial cells was higher in the tie1AS-Elavl-pMO embryos (Figure 6E and 6F) and the tie1AS knockdown embryos (Figure 6H and 6I) than in the control pMO embryos (Figure 6D and 6F) or the control knockdown embryos (Figure 6G and 6I). To further examine the correlation between increased tie1 mRNA levels in the head and higher endothelial cell numbers within PMBCs, we counted endothelial cell numbers in embryos that were injected with tie1AS-Elavl-bMOcomb or cMO after being treated with SB-505124 or DMSO (treatments performed as in Figure 5). The tie1AS-Elavl-bMOcomb+DMSO group showed the expected higher number (1.8-fold) of endothelial cells in PMBCs around 28 hpf compared with the cMO+DMSO embryos (Figure 6J versus 6L; and Figure 6L and 6N, compare blue bar versus red bar). The cMO+DMSO group and the cMO+SB-505124 group (Figure 6J versus 6K; Figure 6N, blue bar versus light gray bar) had similar numbers of endothelial cells along their PMBCs. However, the tie1AS-Elavl-bMOcomb+SB-505124 embryos had only half as many endothelial cells along their PMBCs as the tie1AS-Elavl-bMOcomb+DMSO (Figure 6L versus 6M; also, Figure 6N, compare red bar versus dark gray bar). Endothelial cell numbers along PMBCs in the tie1AS-Elavl-bMOcomb+SB-505124 group and the cMO+DMSO/SB-505124 group were similar (Figure 6M versus 6J and 6K; also, Figure 6N, compare dark gray bar versus blue bar or light gray bar). Thus, suppressing the increase in tie1 mRNA levels in the tie1AS-Elavl-bMOcomb embryos with SB-505124 rescued the endothelial cell number (within PMBCs) phenotype.
Figure 6.
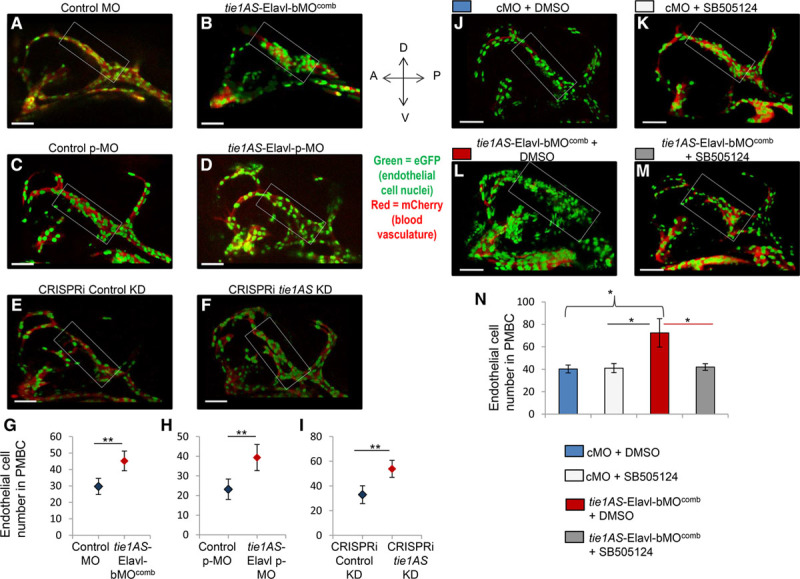
Endothelial cell number in Tg(kdrl:mCherry; NLS-eGFP zebrafish embryos that expressed eGFP (enhanced green fluorescent protein) in endothelial cell nuclei and red fluorescent protein (mCherry) in blood vessels. A representative confocal image (z-stack) of a 28 hours post-fertilization (hpf) transgenic zebrafish embryo injected at the 1-cell stage with (A) Control morpholino oligonucleotide (cMO; 7 of 7 images), (B) tie1AS-Elavl-bMOcomb (8/10), (C) control pMO that was activated between somite stages14 and 16 by exposure to 365 nm ultraviolet light for 8 minutes (6/6), (D) tie1AS-Elavl-bMO:pMO mix (tie1AS-Elavl-pMO) that was photo-activated between 14 and 16 somite stages (6/6), (E) scrambled guide RNA (gRNA) and dCas9-KRAB (dead cas9 enzyme-kruppel-associated box domain; CRISPRi [clustered regularly interspaced short palindromic repeats–mediated interference] negative control; 6/6), (F) 3 gRNAs targeting transcription start site (TSS) of tyrosine kinase containing immunoglobulin and epidermal growth factor homology 1 antisense (tie1AS) along with dCas9-KRAB (tie1AS knockdown; 5/7). The average number of endothelial cells in the primordial midbrain channel (PMBC) of 28 hpf embryos injected with (G) Control morpholinos (cMO; blue diamond) or tie1AS-Elavl-bMOcomb (red diamond); (H) control photoactivatable MO (pMO; blue diamond) or tie1AS-Elavl-pMO (red diamond), which were photo-cleaved between somite stages 14 and 16. The other 2 groups were (I) the control knockdown (blue diamond) and the tie1AS knockdown (red diamond) embryos. The endothelial cells were counted manually on 6 merged z-stack images on Velocity 3D imaging software (Perkin Elmer). J–M, One-cell stage transgenic zebrafish embryos, Tg(kdrl:mCherry; NLS-eGFP, were injected with cMO or tie1AS-Elavl-bMOcomb, grown at 28.5°C in the dark, and treated with 100 µm dimethyl sulfoxide (DMSO) or 100 µm SB-505124 between somite stages 18 and 20. Representative (6/6) confocal images (z-stack) of 28 hpf embryos are shown for (J) cMO+DMSO, (I) cMO+SB-505124, (L) tie1AS-Elavl-bMOcomb+DMSO, and (M) tie1AS-Elavl-bMOcomb+SB-505124. N, Average number of endothelial cells in cMO+DMSO embryos (blue bars), cMO+SB-505124 embryos (light gray bars), tie1AS-Elavl-bMOcomb+DMSO embryos (red bars), and tie1AS-Elavl-bMOcomb+SB-505124 embryos (dark gray bars). White rectangular boxes around PMBC in A–F and J–M denote regions where endothelial cell nuclei were counted. *Adj P<0.05 and **adj P<0.01 (P values were adjusted after multiple testing corrections). Error bars represent SDs (n=6). White horizontal bars at the left bottom corners of A–F and J–M are scale bars (50 µm).
In summary, increased tie1 mRNA levels in the head between 28 and 31 hpf strongly correlated with dilated and distended PMBCs and with a higher number of endothelial cells in the PMBCs. Collectively, our data suggest that the tie1AS-Elavl1 complex plays an important role in regulating tie1 mRNA levels in both space and time to achieve proper PMBC patterning in the developing zebrafish embryo.
Discussion
These studies provide a mechanistic explanation for our previous observation that tie1AS mediates the degradation tie1 mRNA.6 We show that binding of Elavl1 to a 10 nucleotide AU-rich element in tie1AS sustains tie1AS levels, facilitating the degradation of tie1 mRNA in the cytoplasm. Blocking the interaction between Elavl1 and tie1AS with MOs and pMOs caused a ≈2-fold decrease in tie1AS levels in the head and a >2-fold increase in tie1 mRNA levels. CRISPRi confirmed this result, showing a 2.5-fold decrease in tie1AS in the head and a corresponding ≈2-fold increase in tie1 mRNA levels. On the basis of these results, we hypothesize that Elavl1 plays an active role in the eventual degradation of tie1 mRNA, using tie1AS as an intermediary. Our current model (Figure XX in the online-only Data Supplement) is that the interaction of Elavl1 with tie1AS brings Elavl1 within proximity to the poly(A) tail of tie1 mRNA, allowing Elavl1 to interfere with the interaction of poly(A) binding protein with tie1 mRNA. This interaction also recruits proteins involved in the degradation of tie1 mRNA. Indeed, Elav-like proteins have been shown to bind to AU-rich elements and poly(A) sequences simultaneously in vitro.35 Also, the Elavl1 binding site identified in zebrafish tie1AS is present in 1 of the 3 TIE1AS lncRNAs identified in humans6 and in mouse Tie1AS lncRNA (Figures XXI and XXII in the online-only Data Supplement). Coincidently, the human TIE1AS variant that was previously shown to regulate TIE1 mRNA and protein levels contains the Elavl1 binding site, and its position relative to the length of the lncRNA is also conserved (Figure XXI in the online-only Data Supplement). Thus, the mechanism proposed here could be evolutionarily conserved. This mechanism was implicated previously for an Elavl1 orthologue, HuR, which recruits proteins involved in the degradation of nucleophosmin mRNA.36
Coupling fluorescence in situ hybridization with immunofluorescence, we observed that tie1 mRNA expression is restricted within head vessels (Figure 2A; Figure V in the online-only Data Supplement), whereas tie1AS lncRNA (Figure 2B) and Elavl1 (Figure 2C) show broader expression in the head. In the 26 to 28 hpf head, we observed strong overlap in the expression of tie1AS lncRNA and Elav-like proteins in the forebrain, hindbrain, and eye (Figure IV in the online-only Data Supplement). Consistent with that expression pattern, interrupting the tie1AS-Elavl1 interaction (with MOs) restricted the abnormal phenotype primarily to the head (smaller eyes, dilated PMBCs, and reduced ventricular space), and few or no changes were observed in the trunk (26–28 hpf), where no overlap between tie1AS lncRNA and Elav-like proteins was evident (Figure III in the online-only Data Supplement). Thus, we hypothesize that the head phenotype (smaller eyes, dilated PMBCs, and reduced ventricular space) associates with the interaction between tie1AS and Elavl1. In addition, when we used CRISPRi to knock down tie1AS, we observed trunk intersomitic vessel defects in ≈35% of the embryos (data not shown). In this approach, we downregulated tie1AS levels without influencing the interaction between tie1AS and Elavl1. Thus, the intersomitic vessel defects observed in the CRISPRi embryos likely associate with tie1AS via an Elavl1-independent mechanism. Another possibility that cannot be excluded is that tie1AS may possess additional functions (other than regulating tie1 mRNA) that influence intersomitic vessel development. These questions need further evaluation.
From a vascular phenotype perspective, the PMBCs in the head are influenced by modulating the tie1AS-Elavl1 interaction. During vasculogenesis, PMBCs give rise to all the superficial zebrafish ocular vessels.37 Expression of tie1, tie1AS, and Elavl1 in PMBCs at 26 to 28 hpf indicates some overlap between Elavl1 and tie1 (Figure VIIB in the online-only Data Supplement) but no overlap between Elavl1 and tie1AS (Figure VIIA in the online-only Data Supplement) or between tie1 and tie1AS (Figure VIIC in the online-only Data Supplement). The overlap observed between Elavl1 and tie1 in fluorescence in situ hybridization and immunofluorescence does not necessarily imply interaction because in vivo pull-down assays showed no interaction between tie1 mRNA and Elavl1 (Figure 1D).
If the colocalization of tie1AS and Elavl1 in PMBCs is minimal at 26 to 28 hpf, how can we understand the PMBC dilation phenotype? Our spatial expression analysis of tie1, tie1AS, and Elavl1 provides an explanation. We hypothesize that the interaction between tie1AS and Elavl1 occurs in cells surrounding PMBCs, preventing those cells from expressing tie1. Thus, the interaction creates tie1-negative boundaries in the head at 26 to 28 hpf. If functional approaches are used to influence the tie1AS-Elavl1 interaction or decrease tie1AS levels (CRISPRi), tie1 mRNA levels increase abruptly in cells originally destined for tie1-negative status. Support for this idea emerged from our expression analysis involving chromogenic in situ hybridization, in which PMBCs in embryos injected with tie1AS-Elavl-bMOcomb showed spatial expansion of tie1 mRNA expression (Figure 4M versus 4N; Figure XII in the online-only Data Supplement).
Indeed, increased tie1 mRNA levels are linked to PMBC dilation, as shown by our small molecule approach. SB-505124, a small molecule that selectively inhibits activin receptor-like kinases, was previously shown to decrease tie1 mRNA expression in zebrafish.34 When we injected embryos with cMO or tie1AS-Elavl-bMOcomb and treated them with DMSO, vehicle control, or SB-505124, we observed the expected decrease in tie1 mRNA in the heads of the cMO+SB-505124 embryos (Figure 5L through 5N). The tie1AS-Elavl-bMOcomb+DMSO embryos showed the expected increase in tie1 mRNA in the head, which was brought down to control levels by SB-505124 treatment (Figure 5L and 5M). Importantly, the eye size, ventricular space, and PMBC dilation phenotypes were all rescued when the tie1AS-Elavl-bMOcomb embryos were treated with SB-505124 (Figure 5I through 5K). SB-505124 treatment of embryos injected with cMO or tie1AS-Elavl-bMOcomb upregulated tie1AS expression in the head and trunk/tail (Figure XIX in the online-only Data Supplement), which was unexpected but does coincide with the reduction in tie1 mRNA levels and defects in the trunk and tail. We hypothesize that defects in the trunk and tail could arise from either suboptimal levels of tie1 mRNA that are independent of tie1AS or are caused by an Elavl1-independent mechanism of tie1AS or by many genes, including tie1, that are affected by SB-505124.
In conclusion, our findings show that vascular patterning in the brain associated with tie1 is fine-tuned by a low-abundance transcript, tie1AS, which interacts with an abundant RNA-binding protein, Elavl1, in a temporal and spatial manner. Thus, regulation of tie1 mRNA mediated by the tie1AS:Elavl1 complex is critical during development.
Acknowledgments
T.A. Chowdhury designed and performed experiments, interpreted data, and wrote and edited the article. C. Koceja designed and performed experiments and interpreted data. S. Eisa-Beygi designed and performed experiments and edited parts of the article. B.P. Kleinstiver provided intellectual input and designed reagents. S.N. Kumar contributed to imaging and interpreted data. C.W. Lin performed statistical analyses, reviewed and interpreted data, and edited parts of the article. K. Li designed and performed experiments and interpreted data. S. Prabhudesai designed and performed experiments and interpreted data. J.K. Joung provided intellectual input and reviewed data. R. Ramchandran provided intellectual input, gave direction in experimental design, interpreted data, provided resources, and wrote the article. We thank Mark T. McNally and Lisa M. McNally for UV cross-linking studies, Developmental Vascular Biology Program members for their input and suggestions, and Luis Monterroso (Rice University) and Nazmul Hasan (University of Southern California) for participating in this project during the summer of 2015 and 2017, respectively. We also thank Dr Dong Liu for the generous gift of pXT7-dCas9-KRAB.
Sources of Funding
This work was supported by grants from the National Institutes of Health (1R01HL112639 and 1R01HL123338; R. Ramchandran). T.A. Chowdhury is a recipient of an American Heart Association postdoctoral fellowship. R. Ramchandran and K. Li are partly supported by funds from the Women’s Health Research Program at MCW. R. Ramchandran, C. Koceja, and S. Prabhudesai are partly supported by funds from the Department of Pediatrics and the Children’s Research Institute at MCW. S. Eisa-Beygi is supported by funds from Kelleigh’s Cause Foundation.
Disclosures
J.K. Joung has financial interests in Monitor Biotechnologies, Beam Therapeutics, Editas Medicine, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K. Joung interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. The other authors report no conflicts.
Nonstandard Abbreviations and Acronyms
- cMO
- control morpholino
- CRISPRi
- clustered regularly interspaced short palindromic repeats–mediated interference
- DMSO
- dimethyl sulfoxide
- eGFP
- enhanced green fluorescent protein
- Elavl1
- embryonic lethal and abnormal vision Drosophila-like 1
- gRNA
- guide RNA
- hpf
- hours post-fertilization
- lncRNA
- long noncoding RNA
- MO
- morpholino antisense oligonucleotides
- MS
- mass spectrometry
- NAT
- natural antisense transcript
- PMBC
- primordial midbrain channel
- pMO
- photoactivatable morpholino antisense oligonucleotides
- TGF
- transforming growth factor
- Tie1
- tyrosine kinase containing immunoglobulin and epidermal growth factor homology-1
- tie1AS
- tyrosine kinase containing Immunoglobulin and epidermal growth factor homology-1 antisense
The online-only Data Supplement is available with this article at http://atvb.ahajournals.org/lookup/suppl/doi:10.1161/ATVBAHA.118.310848/-/DC1.
Highlights
Tie1 antisense (tie1AS) long noncoding RNA binds embryonic lethal and abnormal vision Drosophila-like 1 (Elavl1) in vitro and in vivo.
The interaction between tie1AS and Elavl1 fine-tunes tissue-specific tie1 mRNA expression.
An increase in tie1 mRNA levels because of perturbation of tie1AS-mediated regulation of tie1 mRNA coincides with reduced ventricular space, smaller eye size, and dilated primordial midbrain channels.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama S, Tomaru Y, Kasukawa T, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Li K, Blum Y, Verma A, et al. A noncoding antisense RNA in Tie-1 locus regulates Tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puri MC, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. EMBO J. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato TN, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–74. doi: 10.1038/376070a0. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong E, Korhonen J, Silvennoinen O, Cleveland JL, Lieberman MA, Alitalo R. Expression of Tie receptor tyrosine kinase in leukemia cell lines. Leukemia. 1993;7:1585–1591. [PubMed] [Google Scholar]
- 10.Mueller SB, Kontos CD. Tie1: an orphan receptor provides context for angiopoietin-2/Tie2 signaling. J Clin Invest. 2016;126:3188–3191. doi: 10.1172/JCI89963. doi: 10.1172/JCI89963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puri MC, Partanen J, Rossant J, Bernstein A. Interaction of the TEK and TIE receptor tyrosine kinases during cardiovascular development. Development. 1999;126:4569–4580. doi: 10.1242/dev.126.20.4569. [DOI] [PubMed] [Google Scholar]
- 12.Gjini E, Hekking LH, Küchler A, Saharinen P, Wienholds E, Post JA, Alitalo K, Schulte-Merker S. Zebrafish Tie-2 shares a redundant role with Tie-1 in heart development and regulates vessel integrity. Dis Model Mech. 2011;4:57–66. doi: 10.1242/dmm.005033. doi: 10.1242/dmm.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savant S, La Porta S, Budnik A, et al. The orphan receptor Tie1 controls angiogenesis and vascular remodeling by differentially regulating Tie2 in Tip and stalk cells. Cell Rep. 2015;12:1761–1773. doi: 10.1016/j.celrep.2015.08.024. doi: 10.1016/j.celrep.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korhonen J, Partanen J, Armstrong E, Vaahtokari A, Elenius K, Jalkanen M, Alitalo K. Enhanced expression of the Tie receptor tyrosine kinase in endothelial cells during neovascularization. Blood. 1992;80:2548–2555. [PubMed] [Google Scholar]
- 15.Boutet SC, Quertermous T, Fadel BM. Identification of an octamer element required for in vivo expression of the TIE1 gene in endothelial cells. Biochem J. 2001;360(pt 1):23–29. doi: 10.1042/0264-6021:3600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury TA, Kleene KC. Identification of potential regulatory elements in the 5’ and 3’ UTRs of 12 translationally regulated mRNAs in mammalian spermatids by comparative genomics. J Androl. 2012;33:244–256. doi: 10.2164/jandrol.110.012492. doi: 10.2164/jandrol.110.012492. [DOI] [PubMed] [Google Scholar]
- 17.Link V, Shevchenko A, Heisenberg CP. Proteomics of early zebrafish embryos. BMC Dev Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross-Thebing T, Paksa A, Raz E. Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol. 2014;12:55. doi: 10.1186/s12915-014-0055-7. doi: 10.1186/s12915-014-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long L, Guo H, Yao D, Xiong K, Li Y, Liu P, Zhu Z, Liu D. Regulation of transcriptionally active genes via the catalytically inactive Cas9 in C. Elegans and D. Rerio. Cell Res. 2015;25:638–641. doi: 10.1038/cr.2015.35. doi: 10.1038/cr.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshney GK, Pei W, LaFave MC, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25:1030–1042. doi: 10.1101/gr.186379.114. doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 22.Cullinane DL, Chowdhury TA, Kleene KC. Mechanisms of translational repression of the Smcp mRNA in round spermatids. Reproduction. 2015;149:43–54. doi: 10.1530/REP-14-0394. doi: 10.1530/REP-14-0394. [DOI] [PubMed] [Google Scholar]
- 23.Chang SH, Hla T. Post-transcriptional gene regulation by HuR and microRNAs in angiogenesis. Curr Opin Hematol. 2014;21:235–240. doi: 10.1097/MOH.0000000000000040. doi: 10.1097/MOH.0000000000000040. [DOI] [PubMed] [Google Scholar]
- 24.Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J Biol Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leigh NR, Schupp MO, Li K, Padmanabhan V, Gastonguay A, Wang L, Chun CZ, Wilkinson GA, Ramchandran R. Mmp17b is essential for proper neural crest cell migration in vivo. PLoS One. 2013;8:e76484. doi: 10.1371/journal.pone.0076484. doi: 10.1371/journal.pone.0076484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veldman MB, Rios-Galdamez Y, Lu XH, Gu X, Qin W, Li S, Yang XW, Lin S. The N17 domain mitigates nuclear toxicity in a novel zebrafish Huntington’s disease model. Mol Neurodegener. 2015;10:67. doi: 10.1186/s13024-015-0063-2. doi: 10.1186/s13024-015-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Lu YC, Dai K, Torregroza I, Hla T, Evans T. Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood. 2014;123:1384–1392. doi: 10.1182/blood-2013-09-526962. doi: 10.1182/blood-2013-09-526962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekker SC. Morphants: a new systematic vertebrate functional genomics approach. Yeast. 2000;17:302–306. doi: 10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. doi: 10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 31.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulton JD. Making a morpholino experiment work: controls, favoring specificity, improving efficacy, storage, and dose. Methods Mol Biol. 2017;1565:17–29. doi: 10.1007/978-1-4939-6817-6_2. doi: 10.1007/978-1-4939-6817-6_2. [DOI] [PubMed] [Google Scholar]
- 33.Tallafuss A, Gibson D, Morcos P, Li Y, Seredick S, Eisen J, Washbourne P. Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development. 2012;139:1691–1699. doi: 10.1242/dev.072702. doi: 10.1242/dev.072702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrial M, Paffett-Lugassy N, Jeffrey S, Jordan D, O’Loughlin E, Frederick CJ, III, Burns CG, Burns CE. TGF-β signaling is necessary and sufficient for pharyngeal arch artery angioblast formation. Cell Rep. 2017;20:973–983. doi: 10.1016/j.celrep.2017.07.002. doi: 10.1016/j.celrep.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma WJ, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cammas A, Sanchez BJ, Lian XJ, Dormoy-Raclet V, van der Giessen K, López de Silanes I, Ma J, Wilusz C, Richardson J, Gorospe M, Millevoi S, Giovarelli M, Gherzi R, Di Marco S, Gallouzi IE. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat Commun. 2014;5:4190. doi: 10.1038/ncomms5190. doi: 10.1038/ncomms5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman R, Weiss O, Sebbagh M, Ravid R, Gibbs-Bar L, Yaniv K, Inbal A. Development and origins of zebrafish ocular vasculature. BMC Dev Biol. 2015;15:18. doi: 10.1186/s12861-015-0066-9. doi: 10.1186/s12861-015-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


