Abstract
Phospholipids, sphingolipids, and cholesterol are integral components of eukaryotic cell organelles, including the nucleus. Recent evidence shows characteristic features of nuclear lipid composition and signaling, which are distinct from that of the cytoplasm and plasma membrane. While the nuclear phosphoinositol lipid signaling in cell cycle regulation and differentiation has been well described, there is a paucity on the role of nuclear sphingolipids and sphingolipid signaling in different physiological and pathophysiological human conditions. In this prospective, we describe the role of sphingolipids and specifically focus on the sphingoid bases, such as sphingosine, ceramide, and sphingosine-1-phosphate (S1P) generation and catabolism in nuclear signaling and function. Particularly, S1P generated in the nucleus by phosphorylation of SphK2 modulates HDAC activity either by direct binding or through activation of nuclear reactive oxygen species and regulates cell cycle and pro-inflammatory gene expression. Potential implication of association of SphK2 with the co-repressor complexes and generation of S1P in the nucleus on chromatin remodeling under normal and pathological conditions is discussed. A better understanding of sphingolipid signaling in the nucleus will facilitate the design and development of new and novel therapeutic approaches to modulate expression of pro-inflammatory and cell cycle dependent genes in human pathologies such as cancer, bacterial lung infection, neurodegeneration and cystic fibrosis.
Keywords: Nuclear Signaling, Lipids, Phosphoinositides, Epigenetics, Sphingolipids, Sphingosine-1-Phosphate, Sphingosine kinase 2, Inflammation, Histone acetylation and deacetylation
INTRODUCTION
The nucleus of a prokaryotic cell is a highly compartmentalized and a functionally distinct organelle; within its well-defined nuclear envelope (inner and outer membrane) are several distinct endonuclear domains including nuclear matrix, chromatin, and nucleolus. Lipids comprising of phospholipids, sphingolipids, and cholesterol (CHOL) are integral components of all eukaryotic cellular organelle membranes. While the composition and role of plasma membrane and cytosolic organelle membrane lipids have been extensively investigated, only in the last two decades, studies related to eukaryotic nuclear lipids and its role in nuclear signaling and function have gained momentum. The phospho- and sphingo-lipids within the nucleus are distinct from those present in the nuclear envelope. The nuclear envelope is a lipid bilayer that exhibits unique lipid composition between the outer and the inner membranes. The outer membrane of the nuclear envelope is contiguous with the endoplasmic reticulum (ER) and shares some lipidomics characteristics with the ER, while the inner membrane is associated with the nuclear lamina and chromatin architecture and has certain unique lipoid signatures. Several of the phospho- and sphingo-lipid metabolizing enzymes have been identified in the nucleus, suggesting its ability to generate and metabolize lipids independent of other cellular organelles. The nuclear membranes also express a number of receptors including those for platelet activating factor (PAF), prostaglandin (PG) E2, thromboxane (TX) 2, inositol (1,4,5) trisphosphate (IP3), PPARγ, retinoic acid, and G-protein coupled receptors (GPCRs) that transduce nuclear signaling and initiate potential cross-talk between the nucleus and other organelles in the cell. The lipid signaling cross-talk between the nucleus and other organelles under normal and pathological conditions can influence the inter- and intra-cellular functions and cellular physiology. While several studies have elucidated the potential importance of nuclear phospholipids and in particular inositol phospholipid signaling in gene expression, only limited information is available on nuclear sphingolipids, sphingolipid signaling in the nucleus, and its role in regulation of gene expression and chromatin modifications. This perspective focuses on recent revelations in nuclear sphingosine-1-phosphate (S1P) metabolism and activities of S1P metabolizing enzymes that regulate cell cycle and pro-inflammatory gene expression via chromatin modifications.
PHOSPHOLIPIDS AND PHOSPHOINOSITIDES IN NUCLEAR SIGNALING
Similar to other cellular organelles, the nucleus is rich in phospholipids, sphingolipids, and cholesterol (Irvine, 2003; Farooqui, 2009; Ledeen and Wu, 2008; Lucki and Sewer, 2012). Majority of the lipids are localized in the nuclear envelope and provide structural support, as well as participate in signal transduction. Nuclei isolated from different cell types and tissues contain high levels of phosphatidylcholine (PC), sphingomyelin (SM), and CHOL that interact with nuclear proteins to form lipid-protein complexes ( Albi et al., 1994; Albi et al., 2003b; Albi and Magni, 2004;. Recent studies suggest that organization of nuclear PC, SM, CHOL in lipid microdomains could regulate nuclear processes such as transcription, proliferation, differentiation, and apoptosis (Cascianelli et al., 2008). In addition to the major lipids, the nucleus also contains bioactive lipids that are locally generated by enzymes specifically localized in nuclear compartments such as nuclear matrix, chromatin, and nuclear speckles ( Ledeen and Wu, 2006; Ledeen and Wu, 2008; Farooqui, 2009; Lucki and Sewer, 2012). Nuclear lipid levels and lipid composition are in a dynamic state, and are regulated by altered metabolic fluxes in response to plasma membrane/cytoplasmic and nuclear signaling cascades.
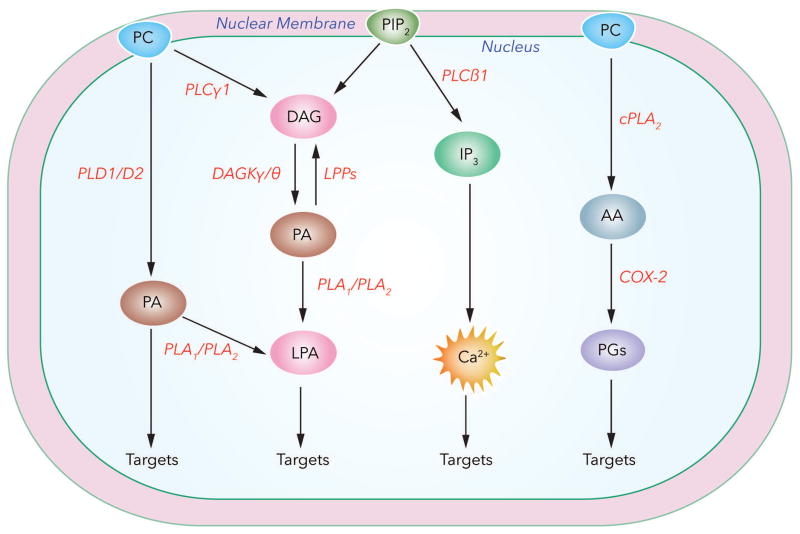
A well-studied example of modulation of phospholipid signal transduction cascade in the nucleus is turnover of inositol phospholipids, especially phosphatidylinositol-4,5-bisphosphate (PIP2). Extracellular stimulation of hepatocytes and other cell types by insulin growth factor (IGF) results in enhanced nuclear diacylglycerol (DAG) production via hydrolysis of nuclear PIP2 by phospholipase C (PLC) β (Payrastre et al., 1992) and modulation of nuclear processes such as proliferation and gene expression (Alessenko and Burlakova, 2002). Interestingly, generation of nuclear DAG also promotes PKC translocation to the nucleus (Divecha et al., 1991) and initiates DNA synthesis (Martelli et al., 1991) (Raben and Baldassare, 2000). In the nucleus, DAG is also generated by a second pathway involving hydrolysis of PC or phosphatidylethanolamine (PE) by nuclear phospholipase D (PLD) to form phosphatidic acid (PA) that is subsequently converted to DAG by a nuclear lipid phosphate phosphatase (LPP) (Gaveglio et al., 2011). Nuclear DAG has a short half-life as it is rapidly converted to PA by nuclear DAG kinase θ ;Tu-Sekine et al., 2016) that is stimulated by a variety of agonists (Martelli et al., 2000). Further, PA produced in the nucleus can function as a second messenger. PA interacts with a variety of cellular and cytoskeletal proteins and these interactions may modulate enzyme activities, spatio-temporal localization of the protein, and cellular functions such as migration, vesicular trafficking, cytoskeletal organization, secretion, and proliferation (Cummings et al., 2002; Liu et al., 2013). PA in the nucleus can also generate another bioactive signaling lipid mediator, lysophosphatidic acid (LPA), by the action of PA specific phospholipase A1/A2 (Fourcade et al., 1995; Sonoda et al., 2002; Aoki et al., 2008). LPA mediates cellular functions by signaling extracellularly via G-protein coupled LPA1–6 receptors and intracellularly by binding to target proteins such as PPARγ (McIntyre et al., 2003; Tsukahara et al., 2013). No definitive measurements of LPA levels in nuclear preparations have been reported; however, LPA1 is known to be constitutively expressed in the nucleus of porcine cerebral microvascular endothelial, rat hepatoma HTC4, (Gobeil et al., 2003) and human bronchial epithelial and neuronal PC12 cells (Waters et al., 2006). Further, in human bronchial epithelial cells (HBECs) and PC12 cells, trafficking of LPA1 to the nucleus was influenced by cell-matrix interactions via integrin signaling, and nuclear LPA1 may be involved in regulating intra-nuclear protein phosphorylation (Waters et al., 2006). PLC-β mediated hydrolysis of PIP2 in the nucleus, in addition to DAG, also generates the second messenger, inositol IP3 that binds to its cognate receptor present on the inner nuclear membrane to release calcium that is essential for transcriptional regulation of calcium dependent enzymes such as calmodulin kinase (Bootman et al., 2009). In the nucleus, IP3 is phosphorylated by inositol kinases localized in the nucleus (Nalaskowski et al., 2002; Seeds and York, 2007; Kim et al., 2017) resulting in the generation of inositol polyphosphates that regulate chromatin remodeling, histone modifications, transcription and mRNA export (Monserrate and York, 2010; Burton et al., 2013). In addition to PIP2, phosphatidylinositol-5 phosphate (PI5P) was shown to be necessary for the binding of Ubiquitin-like with PHD and RING finger domain 1 (UHRF1) to tri-methylated residues of histone3 (H3K9me3) (Arita et al., 2008). UHRF1 is involved in DNA replication and repair and also associated with targeting HDACs to actively transcribed chromatin via interaction with PI5P. Nuclear PIP2 also mediates interaction between BASP-1 and WILMS’TUMOR 1 protein in association with HDAC1 (Toska et al., 2012) further implicating a role for polyphosphoinositides and other phospholipid-derived mediators in gene regulation and chromatin structure. Thus, there is compelling evidence that mammalian cells have the ability to synthesize and metabolize phospholipids and generate lipid mediators within the nucleus that signal both intra- and extra-nuclear for cellular processes (Fig. 1).
Fig. 1.
SIGNALING NUCLEAR SPHINGOLIPIDS
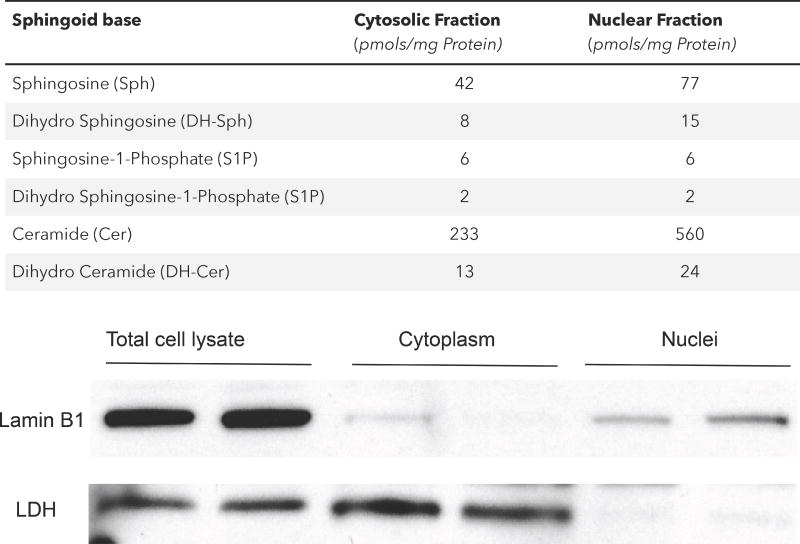
Sphingolipids comprising of sphingophospholipids, sphingoglycolipids and neutral sphingolipids play important roles in several cellular functions. A common feature of all sphingolipids is the long chain sphingosine base (usually C18) that is modified by long-chain fatty acids linked to the amino group and phosphate, phophosphorylcholine, or sugar moieties such as glucose and/or galactose attached to the primary hydroxyl group. Nuclear sphingolipid levels have been partially quantified and different sphingolipid species have been identified in different nuclear domains including nuclear envelope, matrix and chromatin (Lucki and Sewer, 2012; Ledeen and Wu, 2008). While SM is the major sphingolipid in the nucleus and associated with the nuclear matrix and chromatin (Neitcheva and Peeva, 1995; Albi et al., 1994; Lucki and Sewer, 2012), other sphingoid bases such as sphingosine, ceramide and sphingosine-1-phosphate (S1P) are present in the nuclear and cytosolic preparations from lung epithelial cells (Table 1). Many of the nuclear sphingolipids are in a dynamic state and exhibit turnover suggesting localization of sphingolipid metabolizing enzymes such as SM synthase, sphingomyelinase (SMase) ceramidase and sphingosine kinases (SphKs) in the nucleus and its sub-compartments. The nucleus is also rich in glycosphingolipids such as GM1, GM3, GD1, and GD3 gangliosides (Ledeen and Wu, 2008). Nuclear GM1 is enriched in the inner membrane of NE and in tight association with Na+/Ca2+ exchanger (Xie et al., 2002), and was shown to modulate nuclear calcium levels (Gerasimenko and Gerasimenko, 2004; Ledeen and Wu, 2007). Sphingolipid (Table 2) and its metabolizing enzymes (Table 3) are components of chromatin and recent evidence points to interaction between sphingolipids/sphingolipid metabolizing enzymes and HDACs in co-repressor complexes such as Sin3 and NuRD (Hait et al., 2009; Ebenezer et al., 2017). Thus, nuclear sphingolipids function as membrane components, signaling lipid mediators, and modulators of calcium and double stranded RNA (Ledeen and Wu, 2008; Micheli et al., 1998).
Table 1.
Sphingoid bases Levels in Epithelial Cell Nuclear Fraction
Cytosol and nuclear fractions were isolated from mouse lung alveolar epithelial cell (MLE-12) lysate using differential sucrose density gradient-centrifugation. Lipids were isolated by Bligh and Dyer extraction and sphingoid based were quantified by LC/MS/MS and normalized to mg protein in the nuclear fraction. The purity of nuclear fraction was verified by western blotting for Lamin B1 and Lactose dehydrogenase.
Table 2.
Nuclear localization and function of Sphingolipids
| Sphingolipid | Generated From | Nuclear Localization | Function | References |
|---|---|---|---|---|
| Sphingomyelin | Ceramide + Phosphatidylcholine | Nuclear Envelope | DNA synthesis, Chromatin Assembly, RNA Stability |
Ledeen & Wu., 2011 Lucki & Sewer., 2012 Albi et al., 2003b Chocian et al., 2010 |
| Ceramide | Dihydro Ceramide | Chromatin/Nucleosome | Apoptosis Protein degradation |
Albi et al., 2003a Albi & Magni. 2004 Tsugane et al., 1999 |
| Ceramide-1-P | Ceramide | Perinuclear | cPLA2 activation |
Simanshu et al., 2013 Lamour & Chalfant, 2005 |
| Sphingosine | Ceramide | Chromatin/Nucleosome | SF-1 gene transcript, Host-pathogen Interaction |
Tsugane et al., 1999 Lucki & Sewer., 2012 LaBauve & Wargo., 2014 |
| S1P | Sphingosine | Chromatin/Nucleosome | Histone Acetylation, ROS Generation |
Hait et al., 2009 Gardner et al., 2016a Ebenezer at al., 2016a, 2017a,b Fu et al., 2014 Matsushima et al., 2012 |
| Dihydro S1P | Dihydro-Sphingosine | Chromatin/Nucleosome | Hisone Acetylation, ROS Generation | Gardener et al., 2016b |
| Ganglioside M1 | Ceramide + Hexoses + Mono-Sialic acid | Nuclear Envelope | Nuclear Ca2+ Homeostasis |
Keenan et al., 1972 Matyas & Moore., 1987 Ledeen & Wu., 2006, 2008, 2011 Lucki & Sewer, 2012 Xie stal., 2002 Gerasimenko et al., 1995 Wu et al., 2009 |
| Ganglioside D1a | Ceramide + Hexoses + Di-Sialic acid | Nuclear Envelope | Storage Precursor of GM1 |
Ledeen & Wu., 2008, 2011 Wang et al., 2009 Xie et al., 2002 |
| ▵2-Hexadecenal | S1P | ? | Modification of HDACs | Ebenezer at al., 2015., 2016a, 2017a,b |
Table 3.
Summary of Sphingolipid Metabolizing enzymes in Cell Nucleus
| Metabolizing Enzyme | Sphingolipid Subtrate ▶ Sphingolipid Product | References |
|---|---|---|
| Sphingomyelin Synthase | Ceramide + Phosphatidylcholine▶ Sphingomyelin |
Albi et al., 2003a Farooqui, 2009 Lucki & Sewer, 2012 |
| Reverse Sphingomyelin Synthase | Sphingomyelin + Diacylglycerol ▶ Ceramide |
Albi et al., 2003a Albi & Magni, 2004 |
| Ceramidase | Ceramide ▶ Sphingosine | Shiraishi et al., 2003 |
| Sphingomyelinase | Sphingomyelin ▶Ceramide |
Alessenko & Chatterjee, 1995 Tsugane et al., 1999 Albi et al., 2006a |
| Sphingosine Kinase 1 | Sphingosine ▶ Sphingosine-1-Phosphate Dihydro Sphingosine ▶ Dihydro Sphingosine-1-Phosphate |
Ohotski et al., 2013 |
| Sphingosine Kinase 2 | Sphingosine ▶ Sphingosine-1-Phosphate FTY720 ▶ FTY720-Phosphate |
Hait et al., 2009 Fu et al; 2014 Ebenezer et al., 2016b Gardner et a.l., 2016a |
| Sphingosine-1-Phosphatases 1 & 2 | Sphingosine-1-Phosphate ▶ Sphingosine | Not shown |
| Ceramide Kinase | Ceramide ▶ Ceramide-1-Phosphate | Rovina et al., 2009 |
| Sphingosine-1-Phosphate Lyase | Sphingosine-1-Phosphate ▶ Δ2-Hexadecenal Dihydro Sphingosine-1-Phosphate ▶ Hexadecanal + ethanolamine-P |
Ebenezer et al., 2015, 2016a Ebenezer et al., 2017b |
| Neuraminidase | Ganglioside GD1a ▶ Ganglioside GM1 |
Wang et al., 2009 Ledeen & Wu., 2011 |
Sphingomyelin in Nuclear Signaling and Function
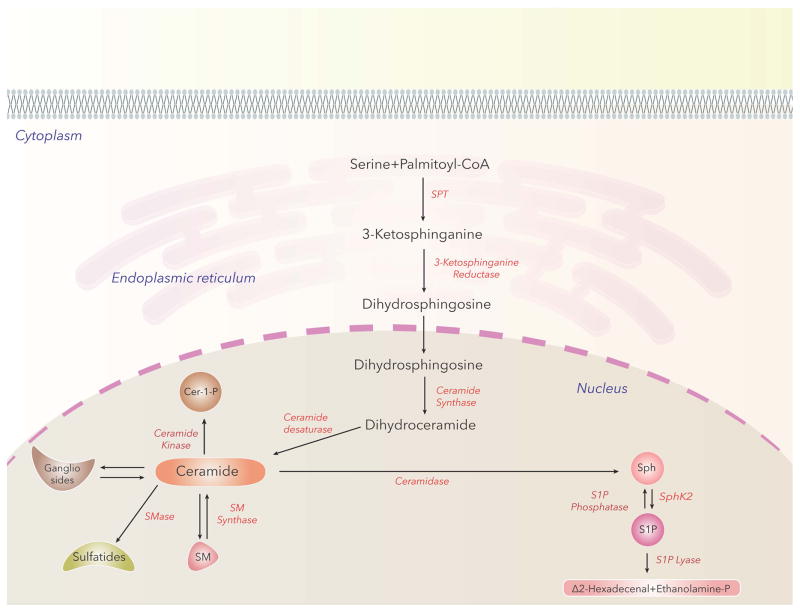
SM is the most abundant sphingophospholipid in the nucleus and a major component of chromatin (Ledeen and Wu, 2008; Lucki and Sewer, 2012). Additionally, SM is also enriched in nuclear matrix (Albi et al., 2003b). Nuclear SM metabolism is independent from the Golgi/ER; its levels are in a dynamic state in the nucleus and modulated by high fat diet, stress, cell cycle and tissue regeneration. Rats on high fat diet revealed elevated SM and ceramide levels in both liver homogenates and hepatocellular nuclei with a shift towards greater fatty acid saturation status in SM and ceramide (Cer) (Chocian et al., 2010). Enzymes regulating SM biosynthesis and catabolism have been identified in the nucleus and nuclear fractions. In the nucleus, the primary pathway involved in generation of SM requires de novo biosynthesis of the Cer intermediate from palmitoyl-CoA and L-serine. Condensation of palmitoyl CoA and L-serine to 3-ketosphinganine is mediated by serine palmitoyl transferase, the rate limiting enzyme in SM biosynthesis, while other enzymes involved in Cer generation are 3-ketosphinganine reductase, Cer synthase and Cer desaturase (Fig. 2). Conversion of Cer to D-erythro-N-palmitoyl-sphingosine-1-phosphocholine (SM) requires SM synthase 1 or 2, which utilize phosphocholine derived from nuclear PC, which is present in both the chromatin and NE (Albi et al., 2003a). SM generated in the nucleus is metabolized by neutral sphingomyelinase (nSMase), first detected in the nuclear matrix of rat ascites hepatoma cells (Tamiya-Koizumi et al., 1989) and subsequently in NE (Alessenko and Chatterjee, 1995), chromatin (Albi and Magni, 1997), and nuclear matrix (Neitcheva and Peeva, 1995). Another SM metabolizing enzyme that carries out the reverse reaction of SM-synthase, namely transfer of phosphocholine from SM to DAG with the formation of PC was detected in rat liver chromatin (Albi et al., 2003a). The role of SM-synthase and reverse SM-synthase in chromatin is unclear; however these two enzymes may modulate Cer/DAG ratio in the chromatin and thus regulate proliferation or apoptosis (Albi and Magni, 2004). Recent studies on intra-nuclear SM localization in various cell types were carried out using nSMase conjugated to colloidal gold particles and transmission electron microscopy, which revealed preferential localization of SM within the peri-chromatin region (Scassellati et al., 2010). Further, microinjection of SMase into living cell nucleus led to deterioration of nuclear structures supporting the intra-nuclear SM association with transcriptionally active ribosomal genes within the dense fibrillar component of the nucleolus and possible involvement of SM in internal nuclear architecture and mRNA processing (Scassellati et al., 2010). However, it is unclear if the resultant loss of SM or accumulation of Cer is responsible for the rapid degradation of intra-nuclear structure. It is not only SM, but also nSMase and SM synthase that appear to be associated with intra-nuclear complexes containing STAT3, a transcriptional factor, showing SM and its metabolizing enzymes to be part of the newly synthesized double stranded RNA in active chromatin (Rossi et al., 2007). Nuclear SM also interacts with CHOL and PC in nuclear lipid microdomains (NLMs) (Slotte, 2013; Cascianelli et al., 2008) and the ratio of CHOL/SM/PC is higher in nuclear matrix during rat liver regeneration suggesting an inverse relationship between nuclear matrix fluidity and DNA synthesis (Albi et al., 2003b). Thus, NLMs may facilitate cross-talk between glycerolipids, sphingolipids, CHOL and lipid mediators generated by metabolic enzymes, which regulate cellular processes such as proliferation, apoptosis and cell cycle in normal and pathological situations. Interestingly, NLMs isolated from hepatocytes and hepatoma cells showed a shift in the ratio between very long chain fatty acid (C24:0) and long chain fatty acid (C16:0). In hepatoma cells, NLMs had a higher proportion of C16:0 fatty acid compared to C24:0 in SM fraction, an increase in the expression of STAT3, Raf1 and PKC δ and a decrease in relative levels of vitamin D receptors (Lazzarini et al., 2015) indicating a potential link between enhanced dynamic properties of NLMs with increasing shuttling of signaling proteins in the nucleus of cancer cells. Increased expression of PKC δ and SM synthase and decreased SMase activities in murine melanoma cell line compared to the wild type resulted in elevated DAG and decreased ceramide pools in the nucleus, (Albi et al., 2005). The inhibition of SMase, in turn, may increase nuclear SM-synthase activity and enrichment of nuclear SM pool or alter the ratio of Cer to DAG levels in the nucleus to modulate nuclear signaling. Interestingly, exposure of non-Hodgkin lymphoblastic lymphoma cells to gentamycin, an aminoglycoside antibiotic, stimulated cytosolic SMase, inhibited nuclear SMase, increased nuclear SM-synthase activity with elevated nuclear SM levels and induced cell death (Codini et al., 2015). Further analysis of nuclear fractions and NLMs using lipidomics and proteomics should allow identification, quantification and association between SM and its metabolites with proteins in the nucleus, chromatin and nuclear matrix.
Fig. 2.
Nuclear Ceramide and Ceramide-1-Phosphate Metabolism and Signaling
Cer is central to sphingolipid metabolism as it is not only the building block for all the major sphingophospholipids and sphingoglycolipids in the biosynthetic pathway but also serves as an immediate intermediate of SM degradation by SMase in cells including the nucleus (Fig. 2). In the nucleus, Cer is converted to sphingosine (Sph) by ceramidase (Shiraishi et al., 2003). Cells generate ceramide-1-phosphate (C1P) from Cer in endoplasmic reticulum/golgi organelles through ceramide kinase (CERK) (Sugiura et al., 2002), and there is evidence for CERK to be associated in the nucleus that could generate nuclear C1P (Rovina et al., 2009). In addition to Sph and Cer, nucleus also has dihydro (DH) Sph and DH Cer. Lipidomics of sphingoid bases from mouse lung epithelial cell line, MLE-12, revealed high levels of Cer in the nuclear fraction (Table 3). Further, nuclear Cer were enriched in C16:0, C22:0, C24:0 and C24:1 and C28:0 fatty acids linked to the amino group of Cer (Ebenezer, Fu and Natarajan unpublished data). Nuclear Cer levels are modulated by extracellular agonists, bacterial infections, high fat diet, serum-deprivation and apoptosis inducing agent such as Fas ligand (Watanabe et al., 2004). Hepatic vein ligation in rats enhanced Cer accumulation in hepatocyte nucleus through activation of neutral SMase (Tsugane et al., 1999) that correlated with DNA fragmentation and cell death. Serum-deprivation increased nuclear Cer levels during the early phase of apoptosis while late phase apoptosis correlated with extranuclear stimulation of SMase activity and accumulation of ceramide in the cytoplasmic compartments (Albi et al., 2006). High fat diet increased Cer levels in rat liver nuclei from 500 nmoles/gm dry tissue to 1600 nmoles/gm dry tissue (~3 fold change) and a significant increase in saturated fatty acid species (C14:0, C16:0 and C18:0) (Chocian et al., 2010). Although Cer has been implicated in apoptosis, no specific nuclear target of Cer has been identified. Cer localized in NLM may be involved in 1,25-dihydroxy vitamin D3 mediated differentiation of embryonic hippocampal cells (Marini et al., 2010), as exposure of the cells to exogenous SMase or serum deprivation increased SMase activity, changed the lipid composition of NLM and impaired the localization of vitamin D3 receptor in the NLM. It is unclear if Cer traffics out from the nucleus into the cytoplasm or vice-versa via lipid protein(s) such as Cer transport protein CERT, and FAPP2 that have been identified (Schroeder et al., 2008; Yamaji et al., 2008). However, there is evidence in support of the blood-borne fatty acids to rapidly enter nuclear lipid pools in hepatocytes (Bucki et al., 1997). Conversion of Cer to C1P catalyzed by Cer kinase (CERK) takes place at the Golgi and C1P-transfer protein (CPTP) transfers C1P to the plasma membrane and other organelles including the nucleus (Simanshu et al., 2013). In addition to CERK’s primary localization in Golgi, it is also associated with the nucleus through the nuclear import signal at the N-terminal and exported to the cytosol with its nuclear export signal at the C-terminal region (Rovina et al., 2009). The C1P generated in the nucleus may have specific signaling function(s) that are yet to be described. C1P is an activator of group IVA cytosolic phospholipase A2, the rate-limiting step in eicosanoid generation (Lamour and Chalfant, 2005). Most of the cellular effects of C1P have been described to take place in intracellular membrane compartments; however, C1P also stimulated cell migration that was blocked by pertussis toxin suggesting signaling of C1P via PTx-sensitive G-protein coupled receptor(s) on the plasma membrane of the cell (Arana et al., 2013). Further studies are necessary to determine the role of nuclear C1P in nuclear eicosanoid synthesis and cellular functions such as proliferation, apoptosis and survival.
Nuclear Sphingosine Signaling and Regulation of Gene Transcription
Total cellular and nuclear free sphingosine (Sph) levels are much lower compared to Cer and nuclear Sph levels are regulated by ceramidases that hydrolyze ceramide to Sph and ceramide synthase, a reversible enzyme that can convert Sph to Cer (Tsugane et al., 1999; Lucki and Sewer, 2012). Sph is a ligand for steroidogenic factor 1 (SF-1) and regulates the transcription of CYP17 (Urs et al., 2006). Under basal conditions, Sph is bound to SF-1 in the nucleus with Sin3A, HDAC and other co-repressor proteins and stimulation with adrenocorticotropin hormone (ACTH) activates protein kinase A that facilitates release of Sph from SF-1 receptor’s ligand binding pocket and stimulates transcription of genes involved in steroid hormones synthesis from cholesterol via binding of PA to SF-1 (Lucki and Sewer, 2012). This study implicates an important role for nuclear Sph in regulating gene transcription. Sphingolipids play an important role in host-pathogen interaction and detection of host-derived Sph by Pseudomonas aeruginosa (PA) is essential for survival of the Gram-positive bacterium in the murine lung. Sph, present in the pulmonary surfactant is generated via metabolism of SM ▶ ceramide ▶ S1P, induced a transcript sphA (PA5325) that was regulated by an AraC-family transcription factor, PA5324 (SphR) (LaBauve and Wargo, 2014). Deletion of sphR resulted in reduced bacterial survival during P. aeruginosa infection of mouse lung. Sph is bactericidal and deletion of sphR increased sensitivity of the antimicrobial effects of Sph suggesting that Sph detection by the AraC-family transcription factor SphR may be more prevalent in other bacteria and not limited to P. aeruginosa.
P. aeruginosa infection of mouse lung significantly reduced free Sph levels in BAL fluids from 40 pmoles/ml to 10 pmoles/ml 6–48 h post-infection; however, the Sph levels in ling tissue was elevated at 48 h post-infection (Ebenezer, Fu and Natarajan, unpublished data) indicating modulations in Sph levels in BALF and lung tissue after bacterial infection. The pathophysiological significance of Sph reduction in BAL fluids is yet to be correlated to P. aeruginosa infection of the lung and resolution but there is evidence for sphingoid long chain bases to prevent lung infection by P. aeruginosa Jung and Tabazavareh, 2014). The contradictory role of Sph in P. aeruginosa survival and infection of mouse lung needs further interrogation.
Nuclear Sphingosine-1-Phosphate Metabolism and Signaling
S1P, the simplest bioactive sphingophospholipid of the human sphingolipidome, is one of most extensively investigated lipid that is known to play a key role in various cellular functions. S1P is enriched in plasma (0.1–1.0 μM) compared to tissues and interstitial fluids(Olivera et al., 2013). Cellular sources of plasma S1P are unclear; however recent studies suggest that platelets (Yatomi et al., 2004), red blood cells (Ito et al., 2007) and non-hematopoietic sources such as liver sinusoidal endothelial cells (Venkataraman et al., 2008) may be the predominant sources (Książek et al., 2015). S1P is biosynthesized in mammalian cells by phosphorylation of Sph, catalyzed by sphingosine kinases (SphKs) 1 & 2 ( Neubauer and Pitson, 2013; Gault et al., 2010). Although both the isoforms of SphK catalyze S1P formation from Sph, the sub-cellular localization and expression vary in different cells that dictate spatio-temporal S1P production and function. SphK1 expression is higher in the lung and heart, and SphK2 in the liver and spleen (Melendez et al., 2000; Siow et al., 2011). Two functional nuclear export signal sequences (NES) direct SphK1 localization to the cytosol (Inagaki et al., 2003); in contrast, SphK2 has nuclear import and export sequences, and is found predominantly in the nucleus in many cell types (Igarashi et al., 2003). Both SphK1 and SphK2, when activated by growth and survival factors, undergo translocation, post-translational modifications, protein-lipid, and protein-protein interactions that ultimately result in elevated S1P levels in the cell (Alemany et al., 2007), which regulates various biological responses. Cytosolic S1P produced by SphK1 enhances cell growth, whereas Sphk2 generated S1P in ER and/or membranes promote apoptosis, as evident from studies using different model systems ( Liu et al., 2003; Hait et al., 2006). Despite their differences, in vivo studies have shown that SphK1 and SphK2 can compensate for each other, as neither SphK1 nor SphK2 knockout mice models reveal any ostensible phenotype in adulthood; however the double knockouts are embryonically lethal. Additionally, S1P could be also be generated from C1P by the action of an amidase to cleave the long chain fatty acid or sphingosylphosphorylcholine by autotaxin (ATX)/lysophospholipase D (Lyso PLD) (Aoki et al., 2008). These pathways are yet to be established and may not represent the major pathway of S1P formation in the cell or the nucleus.
S1P signals via G-protein coupled S1P1–5 receptors present on the cell surface (“inside-out” signaling) and initiate downstream cellular responses (Pyne and Pyne, 2010; ; Natarajan et al., 2013; Ebenezer et al., 2016a). However, the possibility of intracellular action of S1P via intracellularly localized S1PRs is understudied and cannot be ruled out. Recent evidences suggest localization and differential expression of S1PRs in normal and malignant human tissues. All the five S1PRs are expressed in both the cytoplasm and nucleus of benign and malignant tissues from multiple human organs as evidenced by IHC and ICC (Wang et al., 2014). Additionally, in estrogen receptor-positive breast cancer, high cytoplasmic S1P1 and nuclear S1P2 and S1P3 expression and association of these receptors with signaling proteins such as ERK1/2, AKT or SphK1 was reported to be associated with survival or recurrence of estrogen receptor-positive breast cancer (Ohotski et al., 2013). S1P, generated within cells is catabolized to sphingosine by S1P phosphatases (SPPs) 1 & 2 (Pyne et al., 2009; Lépine et al., 2011). Additionally, S1P is also terminally hydrolyzed by S1P lyase (S1PL) to ethanolamine phosphate and hexadecenal in cells (Van Veldhoven et al., 2000; Saba and Hla, 2004). Thus, S1P accumulation is the result of a balance between synthesis, catalyzed by SphKs, and degradation mediated by LPPs, SPPs, and S1PL. In addition to inside-out signaling, S1P can also signal inside the cell by increasing free Ca2+ levels (Birchwood et al., 2001; Itagaki and Hauser, 2003); however, the intracellular target(s) is yet to be identified. Recent studies suggest binding of S1P to human telomerase (Panneer Selvam et al., 2015) and HDACs 1& 2 in normal and cancerous epithelial cells (Hait et al., 2009; Ebenezer et al., 2016b), suggesting a role for nuclear S1P and signaling in normal physiology and pathophysiology (Fig. 2).
Nuclear S1P or S1P analog(s) Generation Requires SphK2 Phosphorylation and Activation in the Nucleus
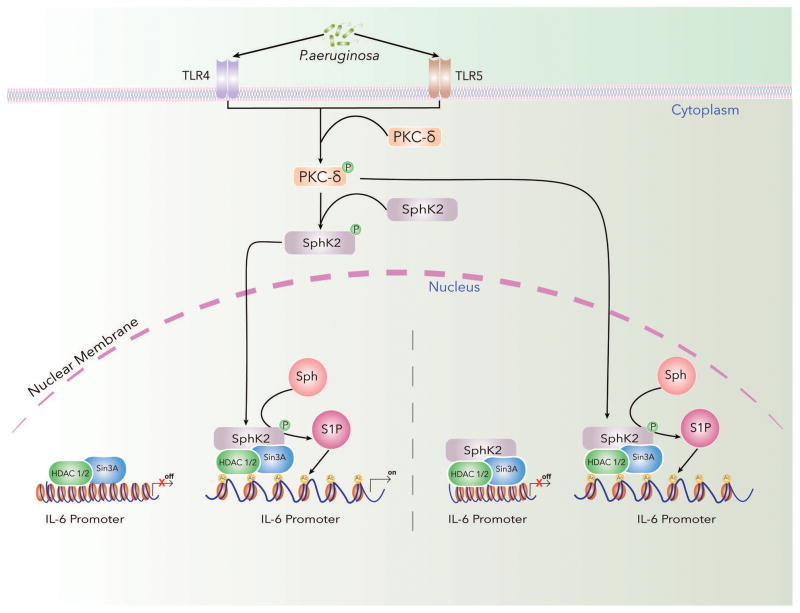
SphK2/S1P signaling axis in the nucleus and its role in gene regulation via epigenetic mechanism(s) is an emerging paradigm. In many cells, SphK2 is localized in the nucleus; however, its role in S1P production and nuclear signaling is yet to be fully defined. S1P levels in the nucleus of epithelial (<10 pmoles/mg protein) (Fu et al., 2014; Ebenezer et al., 2017a) and breast cancer cell line MCF-7 (~2 pmol) (Hait et al., 2009) is low as compared to Sph and ceramide. However, infection of epithelial cells with P. aeruginosa or overexpression of SphK2 in MCF-7 cells resulted in enhanced nuclear S1P levels by several fold (Ebenezer et al., 2016; Hait et al., 2009). Similarly, treatment of mouse embryonic fibroblasts with FTY720, an analog of Sph, resulted in significant accumulation of FTY720-P in both the cytoplasm and nuclear fractions (Gardner et al., 2016a) and blocking SphK2 attenuated nuclear S1P levels in cells (Hait et al., 2009; Ebenezer et al., 2017a) as well as FTY720-P (Gardner et al., 2016a). Administration of FTY720 in wild type mice resulted in accumulation of FTY720-P in the hippocampal region and mice with genetic deletion of SphK2 showed reduced FTY720-P (Hait et al., 2014). A critical step in nuclear S1P or FTY720-P production is phosphorylation of SphK2 and its nuclear localization in response to stimuli. Stimulation of cells with phorbol myristoyl acetate (PMA) (Hait et al., 2009) or infection of mouse lung or lung epithelial cells with P. aeruginosa induced SphK2 phosphorylation at ser/thr 614 and nuclear localization (Ebenezer et al., 2017). Inhibition of SphK2 activity with ABC294640, a specific inhibitor of SphK2, reduced the amount of S1P (Ebenezer et al., 2017a) accumulation in the nucleus suggesting a role for nuclear SphK2 in cell functions. Similarly, SH-SY5Y neuroblastoma cells treated with FTY720 exhibited elevated levels of FTY720-P in the nucleus compared to the cytoplasm, which required SphK2 activity as transfection of the catalytically inactive SphK2G212E plasmid attenuated FTY720-P accumulation (Hait et al., 2014). As SphK2 is localized in the cytoplasm as well as nucleus of many cell types including lung epithelial cells (Ebenezer et al., unpublished data), it will be important to determine if SphK2 is phosphorylated in the cytoplasm and translocated to the nucleus or nuclear SphK2 is phosphorylated preferentially compared to cytoplasmic SphK2 or if both the steps are operative in response to an exogenous stimulus. It appears that both the cytoplasmic and nuclear SphK2 are phosphorylated in lung alveolar and bronchial epithelial cells after infection with heat-inactivated P. aeruginosa and further characterization of the phosphorylation status is warranted (Ebenezer et al., 2017a; Ebenezer et al., 2017b). Both SphK1 and SphK2 are phosphorylated at ser/thr residues by ERK1/2 (Pitson et al., 2003; Hait et al., 2007), and other protein kinases such as PKC. P. aeruginosa infection of alveolar and bronchial epithelial cells stimulated phosphorylation of PKC δ, ε, and ζ and blocking PKC δ, but not PKC ε or ζ, attenuated SphK2 phosphorylation in the nucleus and S1P production suggesting an important role for this PKC isoform in nuclear S1P accumulation after bacterial infection of epithelial cells (Fu et al., 2014; Ebenezer et al., 2017a). Further, the phosphorylated PKC δ is co-localized with SphK2/p-SphK2 in the nucleus of the epithelial cells after P. aeruginosa infection. The role of ERK1/2 or other protein kinases in phosphorylation of SphK2 needs to be evaluated. Additionally, total cellular and nuclear S1P, dihydro S1P or FTY720-P levels could be elevated by inhibiting S1P lyase, dihydroceramide synthase and ceramide synthase or overexpression of either SphK1 or SphK2 in vivo or in vitro. Genetic deletion of sgpl1 in mouse embryonic fibroblasts (Ihlefeld et al., 2012) or inhibition of S1PL by chemical agent such as 2-acetyl-5-tetrahydroxybutl imidazole (THI) increased total lung (Zhao et al., 2011) and nuclear S1P (Nguyen-Tran et al., 2014) levels. Further, treatment of the mouse embryonic fibroblast with fumonisin B1 (an inhibitor of both dihydroceramide synthase and ceramide synthase) resulted in elevated levels of cytoplasmic and nuclear dihydro S1P that was significantly reduced by Sphk1 inhibitor PF-543 and SphK2 inhibitor, ABC294640 (Gardner et al., 2016b) however, the underlying mechanism of enhanced accumulation of S1P or dihydro S1P is not known. Recent evidences suggest that elevated nuclear S1P and S1P analogs are linked to increased risk for neural tube defects (Gardner et al., 2016a; Gardner et al., 2016b) neurodegeneration in neuron models of Huntington disease (Moruno-Manchon et al., 2017) and bacterial lung inflammation (Fu et al., 2014; Ebenezer et al., 2016a; Ebenezer et al., 2017a; Ebenezer et al., 2017b). Further studies are necessary to define the role of elevated nuclear S1P or S1P analogs in vivo and in vitro to understand the role of nuclear S1P signaling pathway(s) in the development human pathologies for potential therapeutic intervention.
Nuclear S1P and FTY720-P are Co-Epigenetic Regulator of Chromatin Remodeling, Gene Transcription and Inflammatory responses via association with HDACs
The role of nuclear S1P and S1P analogs mediated regulation of chromatin remodeling and transcriptional regulation of pro-inflammatory and cell cycle genes is a new and novel emerging area. Nuclear SphK2/p-Sphk2 and S1P co-immunoprecipitated with HDAC1/2; S1P and FTY720-P inhibited HDAC activity and increased histone acetylation at H3K9, H4K8, H3K18 and H3K23 in MCF-7, MLE-12 and MEFs (Hait et al., 2009; Ebenezer et al., 2016a; 2017a; Hait et al., 2014; 2015; Gardner et al., 2016a). Blocking SphK2 activity with ABC294640 or down-regulation of SphK2 siRNA reduced PMA or P aeruginosa mediated H3 and H4 histone acetylation in MCF-7 (Hait et al., 2009) and MLE-12 cells (Ebenezer et al., 2017a), respectively. Transcription regulation through chromatin compaction and decompaction is regulated through two major co-repressor complexes in mammalian cells, namely Sin3a, and NuRD/Mi2 (Vermeulen et al., 2006; Baymaz et al., 2015). Both the Sin3 and NuRD co-repressor complexes are large multi-protein complexes in which all the components and their functions have not been identified; however, two of the key components include HDAC1 and 2, and methyltransferases (including methyl-CpG-binding proteins) (Delcuve et al., 2012; Chen et al., 2015; Li and Seto, 2016). SphK2/p-SphK2 was associated with HDAC1/HDAC2 as part of the Sin3 and NuRD co-repressor complexes and was selectively enriched at the promoters of genes encoding pro-inflammatory cytokines such as IL-6 and TNF-α (Ebenezer et al., 2017a), and cyclin-dependent kinase inhibitor p21 or transcriptional regulator of c-fos (Hait et al., 2009). More recently, down-regulation of SphK2 with siRNA or inhibiting SphK2 activity with a selective inhibitor K145 reduced hypoxia-mediated nuclear S1P, histone acetylation and nuclear HIF-1α synthesis in breast cancer cell lines (Hait and Maiti, 2017). Thus, HDAC1/2 are direct nuclear targets of S1P and link nuclear S1P production to epigenetic regulation of pro-inflammatory, transcriptional factor and cell cycle genes. The role of SphK2/S1P signaling in histone acetylation was not restricted to cells in culture but was observed in vivo in animal models of inflammation and injury. Genetic deletion of SphK2, but not SphK1, protected mouse against P. aeruginosa mediated lung inflammation and injury and SphK2 knockdown resulted in reduced H3 and H4 histone acetylation and secretion of pro-inflammatory IL-6 and TNF-αsecretion in bronchoalveolar lavage (BAL) fluids (Fu et al., 2014; Ebenezer et al., 2015; Ebenezer et al., 2016b). Administration of SphK2 inhibitor, ABC294640 to mice reduced lung inflammation, pro-inflammatory cytokines in BAL fluids, and histone acetylation both pre- and post-challenge of P aeruginosa implicating a therapeutic role of blocking SphK2 in reducing bacterial lung inflammation (Ebenezer et al., 2016b) (Fig. 3). In the dystrophic mouse (mdx), increasing nuclear S1P levels by inhibiting S1PL with THI decreased HDAC activity, and increased histone acetylation that resulted in up-regulation of muscle metabolic genes and key micro RNAs miR-29 and miR-1 (Nguyen-Tran et al., 2014). These effects observed in mdx mouse muscle after blocking S1PL lyase seem to be beneficial for the maintenance of energy metabolism in mice while S1P generated by SphK2 activation in epithelial cells seem to regulate the pro-inflammatory cytokine secretion via modulation of HDAC1/2. Partial deletion of sgpl1 in mice (sgpl1+/− heterozygous) offered protection against P.aeruginosa mediated lung inflammation and injury; however, the involvement of cytosolic and/or nuclear S1P in chromatin remodeling and mechanism of this protective response to bacterial inflammation needs to be established (Ebenezer, Fu, and Natarajan unpublished data). Recent studies with the Sph analog, FTY720 showed that FTY720-P generated in the nucleus by SphK2 mediated phosphorylation of FTY720 reactivated expression of silenced estrogen receptor-α (ERα) and sensitized the breast cancer cells to tamoxifen that required inhibition of HDAC activity (Hait et al., 2015) suggesting an important role for SphK2/FTY720-P nuclear signaling for effective treatment of therapy-resistant breast cancer. The SphK2/FTY720-P signaling axis also seems to play a key role in ameliorating lipid accumulation and trafficking defects in Niemann-Pick type C (NPC) syndrome, a fatal neurodegenerative disorder caused by mutations in the NPC1 and NPC2 genes leading to cholesterol and sphingolipids in late endosomes and lysosomes (Loftus et al., 1997) (Vanier, 2013). FTY720 administration in mice increased hippocampal and cerebellar NPC1 and NPC2 expression that required SphK2 (Newton:2017d) Further, FTY720 mediated expression of NPC1 and NPC2 in NPC1 mutant fibroblasts correlated with generation of FTY720-P and decreased accumulation of cholesterol and glycosphingolipids in NCP1 mutant fibroblasts (Newton et al., 2017). FTY720-P akin to S1P is a potent HDAC inhibitor and hence the FDA approved drug, Fingolimod (FTY720), for treatment of multiple sclerosis may be useful for the treatment of lipid storage disorders such as NPC in humans.
Fig. 3.
Mechanisms of Nuclear S1P Mediated Regulation of HDAC1/2 and Chromatin Remodeling
(i) S1P binding to HDAC1/2 and modulation of activity
Based on the ability of S1P to bind to HDAC1/2 and inhibit its activity in vitro, modeling of S1P into the active site of HDAC1 homologue from Aquifex aeolicus indicated that it docked well in the pocket with the highly conserved Arg27 with a predicted binding energy of −6.85 kcal/mol comparable to −7.09 kcal/mol of the HDAC1/2 inhibitor suberoylanilide hydroxamic acid (SAHA) (Hait et al., 2009). Further, the bound [32P] S1P was totally displaced by inhibitors of HDAC1/2, SAHA and trichostatin A, and unlabeled S1P suggesting competitive binding. While these studies support that S1P is an endogenous ligand of HDAC1/2, the mechanism(s) of S1P mediated inhibition of HDAC1/2 activity remains to be investigated.
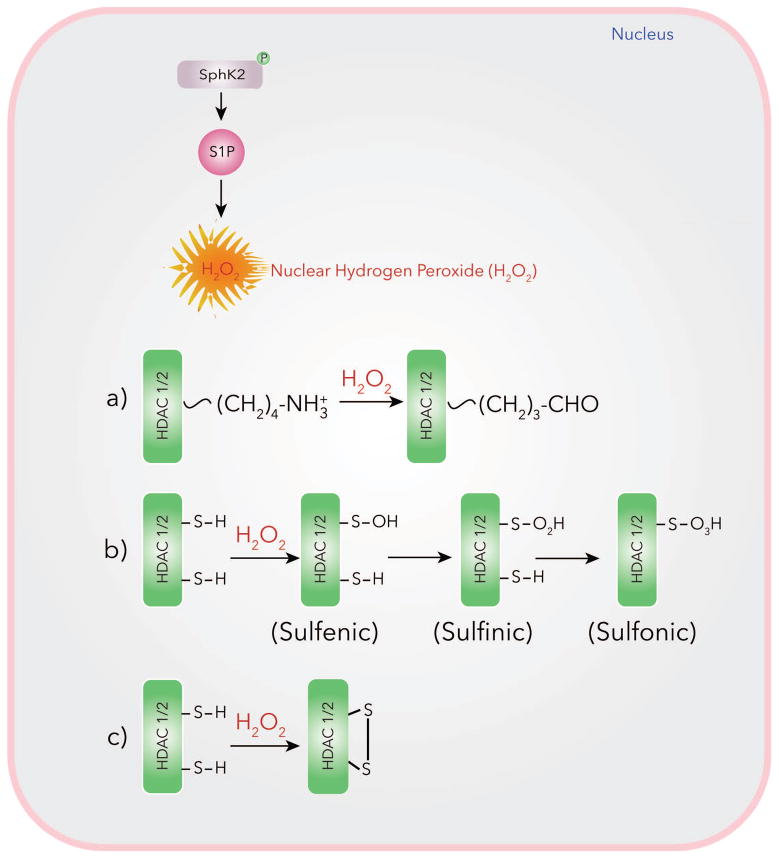
(ii) ROS-mediated oxidative modification of HDAC1/2
Inhibition of HDAC1/2 and enhanced histone acetylation by SphK2/S1P signaling axis in the nucleus may involve S1P mediated reactive oxygen species (ROS) generation in the nucleus that modulates HDAC1/2 oxidation of lysine residues as well as oxidation of SH groups to sulfenic acid and formation of disulfide linkage in HDAC1/2. In vitro, S1P stimulated p47phox dependent ROS generation in human lung endothelial cells and genetic deletion of SphK1 in mice attenuated hyperoxia-induced ROS generation in BAL fluids (Harijith et al., 2016). P. aeruginosa infection of mouse lung increased S1P and hydrogen peroxide levels in lungs and BAL fluids while genetic deletion of SphK2, but not SphK1, in mice also attenuated accumulation of hydrogen peroxide after bacterial infection (Fu et al., 2014). Blocking SphK2 activity with ABC294640 or knockdown of PKC δ with siRNA reduced nuclear ROS generation, H3/H4 histone acetylation and IL-6 secretion. Inhibition of SphK2 with ABC294640 had no effect on cytoplasmic or mitochondrial ROS production as determined by p-Hyper redox sensor measurement (Ebenezer et al., 2017a; Ebenezer et al., 2017b). ROS production in mammalian cells is regulated by NOX1–5 and DUOX1 &2 (Altenhofer et al., 2012; Brandes et al., 2014). P. aeruginosa activation of mouse and human lung epithelial cells stimulated ROS production via NOX2 and NOX4 (Fu et al., 2013) and NOX 4 was predominantly localized in the nucleus compared to NOX2 (Fu P et al., 2014; Ebenezer et al., 2016a &b). The P. aeruginosa mediated H3 and H4 histone acetylation and IL-6 production was reduced by NOX4 siRNA and blocking SphK2 with ABC294640 attenuated NOX4 mediated hydrogen peroxide production (Fu et al., 2014; Ebenezer et al., 2017a & b). Thus, P. aeruginosa infection of lung epithelial cells drives SphK2-dependent S1P generation that stimulates nuclear hydrogen peroxide via NOX4 modulating HDAC activity, and histone acetylation pattern. Increased NOX4 dependent oxidative stress in the nucleus of cardiac myocytes induced NOX4 dependent cysteine oxidation of HDAC4 and nuclear exit of HDAC4, resulting in cardiac hypertrophy response to phenylephrine and pressure overload (Matsushima et al., 2012). These studies suggest a role for oxidative stress in modulation of chromatin remodeling. The mechanism of regulation of HDAC1/2 activity by P. aeruginosa mediated nuclear hydrogen peroxide seems to involve oxidation of HDAC1/2. Further, P. aeruginosa induced oxidation of HDACs was attenuated by PKC δ, SphK2 and Nox4 siRNAs (Ebenezer et al., 2017a) (Fig. 4).
Fig. 4.
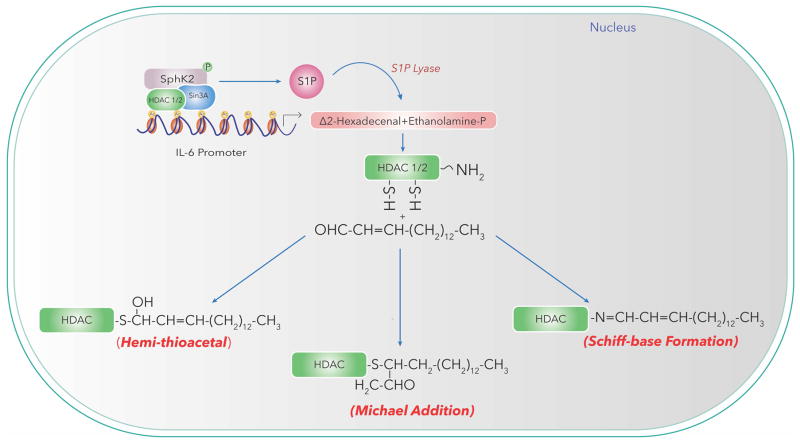
(iii) Modulation of HDAC1/2 by Aldehydes Generated in the Nucleus
Short-chain and long-chain fatty aldehydes released in cell nucleus could interact with ε-amino groups of lysine residues or SH groups of HDAC1/2 to generate adduct resulting in modulation of HDAC activity and histone acetylation/methylation patterns, and cellular functions. Δ2-hexadecenal is one of the products generated by S1PL action on S1P in cells (Van Veldhoven, 2000; Saba and Hla, 2004; Natarajan et al., 2013). Although S1PL is predominantly localized in the endoplasmic reticulum, nuclear fractions isolated from human bronchial and mouse lung epithelial cells exhibited immunostaining for S1PL and activity as measured by ability to generate Δ2-hexadecenal by mass spectrometry (Ebenezer et al., 2015; Ebenezer et al., 2016a). The nuclear fractions isolated from mouse lung epithelial MLE-12 cells were enriched in lamin B (marker for nuclear membrane) with least contamination of cytosol (LDH marker); however, showed positive staining for ER markers suggesting potential ER contamination (Ebenezer et al., 2015). It is noteworthy to point out that nuclear outer membrane is contiguous to ER and use of mild detergents to release the ER and nuclear outer membrane may be necessary to further characterize S1PL localization in the nucleus. Inhibition of S1PL by 4-deoxypyridoxine in mouse lung epithelial cells not only elevated P. aeruginosa mediated S1P levels but also reduced H3/H4 histone acetylation in the nuclear fraction suggesting a potential role for long-chain fatty aldehyde(s) released from S1P to modulate HDAC activity and histone acetylation (Ebenezer et al., 2015; Ebenezer et al., 2017a; Ebenezer et al., 2017b). Further, exogenous addition of Δ2-hexadecenal to nuclear preparations from MLE-12 cells stimulated H3K9 histone acetylation in a dose- and time-dependent manner (Ebenezer et al., 2015) suggesting a role for S1PL released fatty aldehyde in chromatin remodeling. Trapping of the aldehydes released by agents such as phloretin, a polyphenol, also reduced P. aeruginosa stimulated histone acetylation. These studies suggest a potential role for reactive aldehydes such as 4-hydroxynonenal and Δ2-hexadecenal generated by lipid peroxidation and S1P catabolism, respectively as potential modulators of HDAC1/2 and could serve as epigenetic co-regulators of pro-inflammatory gene expression. In the nucleus, long chain fatty aldehydes such as hexadecanal (C16:0 fatty aldehyde) could also be generated from ethanolamine or choline plasmalogens through plasmalogenase activity (Ansell and Spanner, 1968; McMaster et al., 1992). Further studies are necessary to quantify hexadecenal and hexadecanal generated in the nucleus and delineate molecular mechanism(s) of short and long-chain fatty aldehyde mediated modulation of nuclear HDAC1/2 in lung pathologies (Fig. 5).
Fig. 5.
Nuclear Glycosphingolipids
Complex glycosphingolipids, comprised of oligosaccharide chain containing hexose and N-acetylneuraminic acid (sialic acid) residues, are intrinsic components of cell organelles including the nucleus. Gangliosides are sialic acid containing glycosphingolipids that have been identified to occur both in the outer and inner membranes of the nucleus (Keenan et al., 1972; Matyas and Morré, 1987; Ledeen and Wu, 2006; Lucki and Sewer, 2012). Two major species of Gangliosides, GM1, GD1a, were found to be present in both membranes of NE (Ledeen and Wu, 2008; Ledeen and Wu, 2011) along with two isoforms of neuraminidase (Neu), which convert GD1a to GM1 (Wang et al., 2009). GM1 in the inner membrane interacts with Na+/Ca2+ exchanger (NCX) (Xie et al., 2002) and conversion of GD1a to GM1 by Neu1 or Neu3 potentiates the NCX activity. The nuclear NCX/GM1 complex situated in the inner membrane of NE mediates Ca2+ transfer from cytosol to endoplasmic reticulum (Gerasimenko et al., 1995; Wu et al., 2009). Furthermore, nuclear NCX/GM1 complex may provide cytoprotection as GM1 knockout mice exhibited temporal lobe seizures of greater severity and duration to kainic acid compared to normal mice, and administration of LIGA-20, a membrane permeable analog of GM1 reduced the duration of kainic acid induced seizures by enhanced penetration to the nucleus of hippocampal neurons and activate the NCX of the NE (Wu et al., 2005). Thus increasing GM1 and other gangliosides in the nucleus may offer a therapeutic protective application in neurodegenerative disorders such as Parkinson’s disease.
CONCLUSION AND FUTURE PRESPECTIVES
This perspective highlights the role of nuclear sphingolipids signaling in cellular responses in normal and pathological conditions. Nuclear sphingolipids is an area that has been actively investigated for more than a decade; however, only in the past 5 years considerable progress has been made in delineating the link between sphingolipid metabolism and epigenetic regulation of cellular functions such as gene expression, cell proliferation, and inflammation. Among the nuclear sphingolipids, S1P generation in the nucleus and signaling have been implicated in several human pathologies including cancer, pulmonary inflammation, and neurodegenerative disorders. There is strong evidence for S1P generation in the nucleus mediated by activation/phosphorylation of SphK2 catalyzed by PKC δ or ERK 1/2 and modulation of H3 and H4 histone acetylation in vivo and in vitro. In addition to S1P, the S1P analog FTY720-P is also emerging as a modulator of nuclear HDAC activity and gene transcription. SphK2 or p-SphK2 in the nucleus is associated with HDACs 1 & 2 as part of the co-repressor complex of Sin3, NuRD or CoREST and S1P generated by SphK2 activation affects HDAC 1/2 activity, and H3 and H4 histone acetylation pattern in the chromatin. At least three potential mechanisms have been identified for S1P dependent regulation of HDAC1/2 activity. The first proposed mechanism involves docking of S1P in the pocket of HDAC1/2 inhibitor SAHA binding site while the second mechanism points out to the role of nuclear S1P signaling in nuclear ROS generation and oxidation of HDACs1/2 leading to inhibition of HDAC activity, increased acetylation of H3 and H4 histones, chromatin remodeling and altered gene expression. A third potential pathway might involve short and long fatty aldehydes such as 4-hydroxynonenal and Δ2-hexadecenal released in the nucleus by lipid peroxidation and S1P degradation, respectively to modulate HDAC activity. Nuclear S1P levels are regulated by synthesis and catabolism. Very little is known regarding breakdown of S1P in the nucleus either by S1P phosphatases or S1P lyase, which require further investigation. Similarly, histone acetylation is a balance between HDACs and histone acetylases (HATs); however, S1P modulation of HATs is unclear. Limited information is available on the role of Sph and ceramides in the nucleus. Further interrogation is necessary to identify the role of these two sphingoid bases in nuclear signaling and function. To-date only a few genes have been identified to be epigenetically regulated by nuclear S1P and use of ChIP-Sequencing (ChIP-Seq) will provide a power tool to identify genome-wide DNA binding sites for co-repressor complexes such as Sin3, NuRD and CoREST that contain HDACs, SphK2 and other target proteins. Finally, detailed lipidomics analysis of nuclear sphingolipids in different cells types will provide a better understanding of these lipids in nuclear signaling involved in various human pathologies.
Acknowledgments
This study was supported by the National Institutes of Health grant HLBI P01 098050 to VN. We are grateful to Ms. Kavya Chowdary Yarlagadda for technical assistance and Dr. Prasad Kanteti for valuable suggestions and editing of the manuscript.
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interests relating to this manuscript.
References
- Albi E, Magni MP. Chromatin neutral sphingomyelinase and its role in hepatic regeneration. Biochem Biophys Res Commun. 1997;236:29–33. doi: 10.1006/bbrc.1997.6803. [DOI] [PubMed] [Google Scholar]
- Albi E, Magni MPV. The role of intranuclear lipids. Biology of the Cell. 2004;96:657–667. doi: 10.1016/j.biolcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Albi E, La Porta CAM, Cataldi S, Magni MV. Nuclear sphingomyelin-synthase and protein kinase C delta in melanoma cells. Archives of Biochemistry and Biophysics. 2005;438:156–161. doi: 10.1016/j.abb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Albi E, Mersel M, Leray C, Tomassoni ML, Viola-Magni MP. Rat liver chromatin phospholipids. Lipids. 1994;29:715–719. doi: 10.1007/BF02538916. [DOI] [PubMed] [Google Scholar]
- Albi E, Lazzarini R, Magni MV. Reverse sphingomyelin-synthase in rat liver chromatin. FEBS Lett. 2003a;549:152–156. doi: 10.1016/s0014-5793(03)00810-x. [DOI] [PubMed] [Google Scholar]
- Albi E, Cataldi S, Bartoccini E, Magni MV, Marini F, Mazzoni F, Rainaldi G, Evangelista M, Garcia Gil M. Nuclear sphingomyelin pathway in serum deprivation-induced apoptosis of embryonic hippocampal cells. J Cell Physiol. 2006;206:189–195. doi: 10.1002/jcp.20448. [DOI] [PubMed] [Google Scholar]
- Albi E, Cataldi S, Rossi G, Magni MV. A possible role of cholesterol-sphingomyelin/phosphatidylcholine in nuclear matrix during rat liver regeneration. Journal of Hepatology. 2003b;38:623–628. doi: 10.1016/s0168-8278(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Alemany R, van Koppen CJ, Danneberg K, Ter Braak M, Meyer zu Heringdorf D. Regulation and functional roles of sphingosine kinases. Naunyn Schmiedebergs Arch Pharmacol. 2007;374:413–428. doi: 10.1007/s00210-007-0132-3. [DOI] [PubMed] [Google Scholar]
- Alessenko A, Chatterjee S. Neutral sphingomyelinase: localization in rat liver nuclei and involvement in regeneration/proliferation. Mol Cell Biochem. 1995;143:169–174. doi: 10.1007/BF01816950. [DOI] [PubMed] [Google Scholar]
- Alessenko AV, Burlakova EB. Functional role of phospholipids in the nuclear events. Bioelectrochemistry. 2002;58:13–21. doi: 10.1016/S1567-5394(02)00135-4. [DOI] [PubMed] [Google Scholar]
- Altenhofer S, Kleikers PW, Radermacher KA, Scheurer P, Rob Hermans JJ, Schiffers P, Ho H, Wingler K, Schmidt HH. The Nox toolbox: validating the role of NADPH oxidase in physiology and disease. Cell Mol Life Sci. 2012;69:2327–2343. doi: 10.1007/s00018-012-1010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell GB, Spanner S. Plasmalogenase activity in normal and demyelinating tissue of the central nervous system. Biochem J. 1968;108:207–209. doi: 10.1042/bj1080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781:513–518. doi: 10.1016/j.bbalip.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Arana L, Ordoñez M, Ouro A, Rivera I-G, Gangoiti P, Trueba M, Gomez-Muñoz A. Ceramide 1-phosphate induces macrophage chemoattractant protein-1 release: involvement in ceramide 1-phosphate-stimulated cell migration. American Journal of Physiology - Endocrinology and Metabolism. 2013;304:E1213–E1226. doi: 10.1152/ajpendo.00480.2012. [DOI] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Baymaz HI, Karemaker ID, Vermeulen M. Perspective on unraveling the versatility of “co-repressor” complexes. Biochim Biophys Acta. 2015;1849:1051–1056. doi: 10.1016/j.bbagrm.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium Influx and Signaling in Yeast Stimulated by Intracellular Sphingosine 1-Phosphate Accumulation. J Biol Chem. 2001;276:11712–11718. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. An update on nuclear calcium signalling. J Cell Sci. 2009;122:2337–2350. doi: 10.1242/jcs.028100. [DOI] [PubMed] [Google Scholar]
- Brandes RP, Weissman N, schroder K. Nox family NADPH oxidases: Molecular mechanisms of activation. Free Radic Biol Med. 2014;76:208–226. doi: 10.1016/j.freeradbiomed.2014.07.046. [DOI] [PubMed] [Google Scholar]
- Bucki R, Żendzian-Piotrowska M, Nawrocki A, Górski J. Effect of increased uptake of plasma fatty acids by the liver on lipid metabolism in the hepatocellular nuclei. Prostaglandins, Leukotrienes and Essential Fatty Acids. 1997;57:27–31. doi: 10.1016/S0952-3278(97)90489-0. [DOI] [PubMed] [Google Scholar]
- Burton A, Azevedo C, Andreassi C, Riccio A, Saiardi A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc Natl Acad Sci USA. 2013;110:18970–18975. doi: 10.1073/pnas.1309699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascianelli G, Villani M, Tosti M, Marini F, Bartoccini E, Magni MV, Albi E. Lipid Microdomains in Cell Nucleus. Mol Biol Cell. 2008;19:5289–5295. doi: 10.1091/mbc.E08-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 2015;20:35–47. doi: 10.1615/critrevoncog.2015012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chocian G, Chabowski A, Zendzian-Piotrowska M, Harasim E, Łukaszuk B, Górski J. High fat diet induces ceramide and sphingomyelin formation in rat’s liver nuclei. Mol Cell Biochem. 2010;340:125–131. doi: 10.1007/s11010-010-0409-6. [DOI] [PubMed] [Google Scholar]
- Codini M, Cataldi S, Ambesi-Impiombato F, Lazzarini A, Floridi A, Lazzarini R, Curcio F, Beccari T, Albi E. Gentamicin Arrests Cancer Cell Growth: The Intriguing Involvement of Nuclear Sphingomyelin Metabolism. International Journal of Molecular Sciences. 2015;16:2307–2319. doi: 10.3390/ijms16022307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R, Parinandi N, Wang L, Usatyuk P, Natarajan V. Phospholipase D/phosphatidic acid signal transduction: role and physiological significance in lung. Mol Cell Biochem. 2002;234–235:99–109. [PubMed] [Google Scholar]
- Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4(1):5. doi: 10.1186/1868-7083-4-5. doi:10:1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfić H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991;10:3207. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P, Ebenezer DL, Huang LS, Usatyuk PV, Harijith AK, Berdyshev EV, Natarajan V. Epigenetic regulation of P. aeruginosa mediated acute lung injury by sphingosine kinase 2. Am J Respir Critical Care Medicine. 2014;189:A1061. [Google Scholar]
- Ebenezer DL, Fu P, Berdyshev E, Natarajan V. Nuclear S1P lyase regulates histone acetylation in Pseudomonas aeruginosa induced lung inflammation. The FASEB J. 2015;29 Supplement 863.26. [Google Scholar]
- Ebenezer DL, Fu P, Natarajan V. Targeting sphingosine-1-phosphate signaling in lung diseases. Pharmacology & Therapeutics. 2016a;168:143–157. doi: 10.1016/j.pharmthera.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer DL, Fu P, Berdyshev E, Natarajan V. Sphingosine kinase 2 generated nuclear S1P is an epigenetic co-regulator of Pseudomonas aeruginosa-induced lung inflammation. The FASEB J. 2016b;30 Supplement 1053.6. [Google Scholar]
- Ebenezer DL, Fu P, Ha AW, Natarajan V. PKC-delta mediated SphK2 activation and histone acetylation in Pseudomonas aeruginosa induced lung inflammation. The FASEB J. 2017a;31 Supplement 1629.27. [Google Scholar]
- Ebenezer DL, Fu P, Suryadevara V, Zhao Y, Natarajan V. Epigenetic regulation of pro-inflammatory cytokine secretion by sphingosine 1-phosphate (S1P) in acute lung injury: Role of S1P lyase. Adv Biol Regul. 2017b;63:156–166. doi: 10.1016/j.jbior.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui AA. Lipid Mediators in the Neural Cell Nucleus: Their Metabolism, Signaling, and Association with Neurological Disorders. The Neuroscientist. 2009 doi: 10.1177/1073858409337035. [DOI] [PubMed] [Google Scholar]
- Fourcade O, Simon MF, Viodé C, Rugani N, Leballe F, Ragab A, Fournié B, Sarda L, Chap H. Secretory phospholipase A2 generates the novel lipid mediator lysophosphatidic acid in membrane microvesicles shed from activated cells. Cell. 1995;80:919–927. doi: 10.1016/0092-8674(95)90295-3. [DOI] [PubMed] [Google Scholar]
- Fu P, Mohan V, Mansoor S, Tiruppathi C, Sadikot RT, Natarajan V. Role of Nicotinamide Adenine Dinucleotide Phosphate–Reduced Oxidase Proteins in Pseudomonas aeruginosa–Induced Lung Inflammation and Permeability. Am J Respir Cell Mol Biol. 2013 doi: 10.1165/rcmb.2012-0242OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner NM, Riley RT, Showker JL, Voss KA, Sachs AJ, Maddox JR, Gelineau-van Waes JB. Elevated Nuclear and Cytoplasmic FTY720-Phosphate in Mouse Embryonic Fibroblasts Suggests the Potential for Multiple Mechanisms in FTY720-Induced Neural Tube Defects. Toxicol Sci. 2016a;150:161–168. doi: 10.1093/toxsci/kfv321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner NM, Riley RT, Showker JL, Voss KA, Sachs AJ, Maddox JR, Gelineau-van Waes JB. Elevated nuclear sphingoid base-1-phosphates and decreased histone deacetylase activity after fumonisin B1 treatment in mouse embryonic fibroblasts. Toxicol Appl Pharmacol. 2016b;298:56–65. doi: 10.1016/j.taap.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Gault CR, Obeid LM, Hannun YA. Sphingolipids as Signaling and Regulatory Molecules. Springer; New York, NY, New York, NY: 2010. An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaveglio VL, Pasquaré SJ, Giusto NM. Metabolic pathways for the degradation of phosphatidic acid in isolated nuclei from cerebellar cells. Archives of Biochemistry and Biophysics. 2011;507:271–280. doi: 10.1016/j.abb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Gerasimenko O, Gerasimenko J. New aspects of nuclear calcium signalling. J Cell Sci. 2004;117:3087–3094. doi: 10.1242/jcs.01295. [DOI] [PubMed] [Google Scholar]
- Gerasimenko OV, Gerasimenko JV, Tepikin AV. ATP-dependent accumulation and inositol trisphosphate-or cyclic ADP-ribose-mediated release of Ca2+ from the nuclear envelope. Cell. 1995 doi: 10.1016/0092-8674(95)90494-8. [DOI] [PubMed] [Google Scholar]
- Gobeil F, Bernier SG, Vazquez-Tello A, Brault S, Beauchamp MH, Quiniou C, Marrache AM, Checchin D, Sennlaub F, Hou X, Nader M, Bkaily G, Ribeiro-da-Silva A, Goetzl EJ, Chemtob S. Modulation of pro-inflammatory gene expression by nuclear lysophosphatidic acid receptor type-1. J Biol Chem. 2003;278:38875–38883. doi: 10.1074/jbc.M212481200. [DOI] [PubMed] [Google Scholar]
- Hait NC, Bellamy A, Milstien S, Kordula T, Spiegel S. Sphingosine kinase type 2 activation by ERK-mediated phosphorylation. J Biol Chem. 2007;282:12058–12065. doi: 10.1074/jbc.M609559200. [DOI] [PubMed] [Google Scholar]
- Hait NC, Wise LE, Allegood JC, O’Brien M, Avni D, Reeves TM, et al. Active, phosphorylated fingolimod inhibits histone deacetylases and facilitates fear extinction memory. Nature Neuroscience. 2014;17(7):971–980. doi: 10.1038/nn.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Maiti A. Abstract 1388: SphK2/S1P axis regulates hypoxia-mediated HIF-α synthesis in breast cancer cells. Cancer Res. 2017;77:1388–1388. doi: 10.1158/1538-7445.AM2017-1388. [DOI] [Google Scholar]
- Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochimica Et Biophysica Acta-Biomembranes. 2006;1758:2016–2026. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of Histone Acetylation in the Nucleus by Sphingosine-1-Phosphate. Science. 2009;325:1254–pe1. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Avni D, Yamada A, Nagahashi M, Aoyagi T, Aoki H, et al. The phosphorylated prodrug FTY720 is a histone deacetylase inhibitor that reactivates ERα expression and enhances hormonal therapy for breast cancer. Oncogenesis. 2015;4(6):e156. doi: 10.1038/oncsis.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harijith A, Pendyala S, Ebenezer DL, Ha AW, Fu P, Wang Y-T, Ma K, Toth PT, Berdyshev EV, Kanteti P, Natarajan V. Hyperoxia-induced p47phox activation and ROS generation is mediated through S1P transporter Spns2, and S1P/S1P1&2 signaling axis in lung endothelium. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2016;311:L337–L351. doi: 10.1152/ajplung.00447.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura S-I. Sphingosine Kinase 2 Is a Nuclear Protein and Inhibits DNA Synthesis. J Biol Chem. 2003;278:46832–46839. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- Ihlefeld K, Claas RF, Koch A, Pfeilschifter JM, Meyer zu Heringdorf D. Evidence for a link between histone deacetylation and Ca 2+homoeostasis in sphingosine-1-phosphate lyase-deficient fibroblasts. Biochem J. 2012;447:457–464. doi: 10.1101/cshperspect.a004168. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Li P-Y, Wada A, Mitsutake S, Igarashi Y. Identification of functional nuclear export sequences in human sphingosine kinase 1. Biochem Biophys Res Commun. 2003;311:168–173. doi: 10.1016/j.bbrc.2003.09.194. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–361. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Itagaki K, Hauser CJ. Sphingosine 1-Phosphate, a Diffusible Calcium Influx Factor Mediating Store-operated Calcium Entry. J Biol Chem. 2003;278:27540–27547. doi: 10.1074/jbc.M301763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Anada Y, Tani M, Ikeda M, Sano T, Kihara A, Igarashi Y. Lack of sphingosine 1-phosphate-degrading enzymes in erythrocytes. Biochem Biophys Res Commun. 2007;357:212–217. doi: 10.1016/j.bbrc.2007.03.123. [DOI] [PubMed] [Google Scholar]
- Jung YP, Tabazavareh ST. Sphingoid long chain bases prevent lung infection by Pseudomonas aeruginosa. EMBO molecular …. 2014 doi: 10.15252/emmm.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan TW, Morré DJ, Huang CM. Distribution of gangliosides among subcellular fractions from rat liver and bovine mammary gland. FEBS Lett. 1972 doi: 10.1016/0014-5793(72)80768-3. [DOI] [PubMed] [Google Scholar]
- Kim E, Ahn H, Kim MG, Lee H, Kim S. The Expanding Significance of Inositol Polyphosphate Multikinase as a Signaling Hub. Mol Cells. 2017;40:315–321. doi: 10.14348/molcells.2017.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Książek M, Chacińska M, Chabowski A, Baranowski M. Sources, metabolism, and regulation of circulating sphingosine-1-phosphate. The Journal of Lipid Research. 2015;56:1271–1281. doi: 10.1194/jlr.R059543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GT, Maceyka M, Milstien S, Spiegel S. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nature Reviews Drug Discovery. 2013;12:688–702. doi: 10.1038/nrd4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBauve AE, Wargo MJ. Detection of Host-Derived Sphingosine by Pseudomonas aeruginosa Is Important for Survival in the Murine Lung. PLOS Pathogens. 2014;10:e1003889. doi: 10.1371/journal.ppat.1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour NF, Chalfant CE. Ceramide-1-phosphate: The “missing” link in eicosanoid biosynthesis and inflammation. Molecular Interventions. 2005;5:358–367. doi: 10.1124/mi.5.6.8. [DOI] [PubMed] [Google Scholar]
- Lazzarini A, Macchiarulo A, Floridi A, Coletti A, Cataldi S, Codini M, Lazzarini R, Bartoccini E, Cascianelli G, Ambesi-Impiombato FS, Beccari T, Curcio F, Albi E. Very-long-chain fatty acid sphingomyelin in nuclear lipid microdomains of hepatocytes and hepatoma cells: can the exchange from C24:0 to C16:0 affect signal proteins and vitamin D receptor? Mol Biol Cell. 2015;26:2418–2425. doi: 10.1091/mbc.E15-04-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeen R, Wu G. GM1 in the nuclear envelope regulates nuclear calcium through association with a nuclear sodium-calcium exchanger. J Neurochem. 2007;103(Suppl 1):126–134. doi: 10.1111/j.1471-4159.2007.04722.x. [DOI] [PubMed] [Google Scholar]
- Ledeen RW, Wu G. Sphingolipids of the nucleus and their role in nuclear signaling. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2006;1761:588–598. doi: 10.1016/j.bbalip.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Ledeen RW, Wu G. Thematic Review Series: Sphingolipids. Nuclear sphingolipids: metabolism and signaling. The Journal of Lipid Research. 2008;49:1176–1186. doi: 10.1194/jlr.R800009-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeen R, Wu G. New findings on nuclear gangliosides: overview on metabolism and function. J Neurochem. 2011;116:714–720. doi: 10.1111/j.1471-4159.2010.07115.x. [DOI] [PubMed] [Google Scholar]
- Lépine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death & Differentiation. 2011;18:350–361. doi: 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Toman RE, Goparaju SK, Maceyka M, Nava VE, Sankala H, Payne SG, Bektas M, Ishii I, Chun J, Milstien S, Spiegel S. Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. J Biol Chem. 2003;278:40330–40336. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Su Y, Wang X. Phosphatidic acid-mediated signaling. Adv Exp Med Biol. 2013;991:159–176. doi: 10.1007/978-94-007-6331-9_9. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science (New York, NY) 1997;277(5323):232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Lucki NC, Sewer MB. Nuclear sphingolipid metabolism. Annu Rev Physiol. 2012;74:131–151. doi: 10.1146/annurev-physiol-020911-153321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Bartoccini E, Cascianelli G, Voccoli V, Baviglia MG, Magni MV, Garcia Gil M, Albi E. Effect of 1α,25-dihydroxyvitamin D3 in embryonic hippocampal cells. Hippocampus. 2010;20:696–705. doi: 10.1002/hipo.20670. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Tabellini G, Bortul R, Manzoli L, Bareggi R, Baldini G, Grill V, Zweyer M, Narducci P, Cocco L. Enhanced Nuclear Diacylglycerol Kinase Activity in Response to a Mitogenic Stimulation of Quiescent Swiss 3T3 Cells with Insulin-like Growth Factor I. Cancer Res. 2000;60:815–821. doi: 10.1016/0092-8674(93)80041-C. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Bortul R, Tabellini G, Bareggi R, Manzoli L, Narducci P, Cocco L. Diacylglycerol kinases in nuclear lipid-dependent signal transduction pathways. Cell Mol Life Sci. 2002;59:1129–1137. doi: 10.1007/s00018-002-8492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli AM, Gilmour RS, Neri LM, Manzoli L, Corps AN, Cocco L. Mitogen-stimulated events in nuclei of Swiss 3T3 cells Evidence for a direct link between changes of inositol lipids, protein kinase C requirement and the onset of DNA synthesis. FEBS Lett. 1991;283:243–246. doi: 10.1016/0014-5793(91)80598-W. [DOI] [PubMed] [Google Scholar]
- Matsushima S, Kuroda J, Ago T, Zhai P, Park JY, Xie L-H, Tian Bin, Sadoshima J. Increased Oxidative Stress in the Nucleus Caused by Nox4 Mediates Oxidation of HDAC4 and Cardiac Hypertrophy. Circulation Research. 2012;112:CIRCRESAHA.112.279760–663. doi: 10.1161/CIRCRESAHA.112.279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyas GR, Morré DJ. Subcellular distribution and biosynthesis of rat liver gangliosides. Biochimica et Biophysica Acta (BBA)-Lipids andLipid Metabolism. 1987;921:599–614. doi: 10.1016/0005-2760(87)90089-0. [DOI] [PubMed] [Google Scholar]
- Moruno-Manchon JF, Uzor NE, Blasco-Conesa MP, Mannuru S, Putluri N, Furr-Stimming EE, Tsvetkov AS. Inhibiting sphingosine kinase 2 mitigates mutant Huntingtin-induced neurodegeneration in neuron models of Huntington disease. Human Molcular genetics. 2017;26:1305–1317. doi: 10.1093/hmg/ddx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre TM, Pontsler AV, Silva AR, St Hilaire A, Xu Y, Hinshaw JC, Zimmerman GA, Hama K, Aoki J, Arai H, Prestwich GD. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc Natl Acad Sci USA. 2003;100:131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster CR, Lu C-Q, Choy PC. The existence of a soluble plasmalogenase in guinea pig tissues. Lipids. 1992;27:945–949. doi: 10.1007/BF02535569. [DOI] [PubMed] [Google Scholar]
- Melendez AJ, Carlos-Dias E, Gosink M, Allen JM, Takacs L. Human sphingosine kinase: molecular cloning, functional characterization and tissue distribution. Gene. 2000;251:19–26. doi: 10.1016/s0378-1119(00)00205-5. [DOI] [PubMed] [Google Scholar]
- Micheli M, Albi E, Leray C, Magni MV. Nuclear sphingomyelin protects RNA from RNase action. FEBS Lett. 1998;431:443–447. doi: 10.1016/s0014-5793(98)00810-2. [DOI] [PubMed] [Google Scholar]
- Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan V, Dudek SM, Jacobson JR, Moreno-Vinasco L, Huang LS, Abassi T, Mathew B, Zhao Y, Wang L, Bittman R, Weichselbaum R, Berdyshev E, Garcia JGN. Sphingosine-1–Phosphate, FTY720, and Sphingosine-1–Phosphate Receptors in the Pathobiology of Acute Lung Injury. Am J Respir Cell Mol Biol. 2013 doi: 10.1165/rcmb.2012-0411TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitcheva T, Peeva D. Phospholipid composition, phospholipase A2 and sphingomyelinase activities in rat liver nuclear membrane and matrix. Int J Biochem Cell Biol. 1995;27:995–1001. doi: 10.1016/1357-2725(95)00087-6. [DOI] [PubMed] [Google Scholar]
- Neubauer HA, Pitson SM. Roles, regulation and inhibitors of sphingosine kinase 2. FEBS J. 2013;280:5317–5336. doi: 10.1111/febs.12314. [DOI] [PubMed] [Google Scholar]
- Newton J, Hait NC, Maceyka M, Colaco A, Maczis M, Wassif CA, et al. FTY720/fingolimod increases NPC1 and NPC2 expression and reduces cholesterol and sphingolipid accumulation in Niemann-Pick type C mutant fibroblasts. The FASEB Journal. 2017;31(4):1719–1730. doi: 10.1096/fj.201601041R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Tran D-H, Hait NC, Sperber H, Qi J, Fischer K, Ieronimakis N, Pantoja M, Hays A, Allegood J, Reyes M, Spiegel S, Ruohola-Baker H. Molecular mechanism of sphingosine-1-phosphate action in Duchenne muscular dystrophy. Disease Models & Mechanisms. 2014;7:41–54. doi: 10.1242/dmm.013631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohotski J, Edwards J, Elsberger B, Watson C, Orange C, Mallon E, Pyne S, Pyne NJ. Identification of novel functional and spatial associations between sphingosine kinase 1, sphingosine 1-phosphate receptors and other signaling proteins that affect prognostic outcome in estrogen receptor-positive breast cancer. Int J Cancer. 2013;132:605–616. doi: 10.1002/ijc.27692. [DOI] [PubMed] [Google Scholar]
- Olivera A, Allende ML, Proia RL. Shaping the landscape: Metabolic regulation of S1P gradients. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2013;1831:193–202. doi: 10.1016/j.bbalip.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneer Selvam S, De Palma RM, Oaks JJ, Oleinik N, Peterson YK, Stahelin RV, Skordalakes E, Ponnusamy S, Garrett-Mayer E, Smith CD, Ogretmen B. Binding of the sphingolipid S1P to hTERT stabilizes telomerase at the nuclear periphery by allosterically mimicking protein phosphorylation. Science Signaling. 2015;8:ra58–ra58. doi: 10.1126/scisignal.aaa4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payrastre B, Nievers M, Boonstra J, Breton M, Verkleij AJ, Van Bergen en Henegouwen PM. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J Biol Chem. 1992;267:5078–5084. [PubMed] [Google Scholar]
- Pewzner-Jung Y, Tabazavareh ST. 38 Sphingoid long chain bases prevent lung infection by Pseudomonas aeruginosa. Journal of Cystic …. 2015 doi: 10.15252/emmm.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson SM, Moretti PAB, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nature Reviews Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- Pyne S, Lee SC, Long J, Pyne NJ. Role of sphingosine kinases and lipid phosphate phosphatases in regulating spatial sphingosine 1-phosphate signalling in health and disease. Cellular Signalling. 2009;21:14–21. doi: 10.1016/j.cellsig.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Raben DM, Baldassare JJ. Nuclear envelope signaling-role of phospholipid metabolism - ProQuest. … journal of histochemistry: EJH. 2000 [PubMed] [Google Scholar]
- Rossi G, Viola Magni M, Albi E. Signal transducer and activator of transcription 3 and sphingomyelin metabolism in intranuclear complex during cell proliferation. Archives of Biochemistry and Biophysics. 2007;464:138–143. doi: 10.1016/j.abb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Rovina P, Schanzer A, Graf C, Mechtcheriakova D, Jaritz M, Bornancin F. Subcellular localization of ceramide kinase and ceramide kinase-like protein requires interplay of their Pleckstrin Homology domain-containing N-terminal regions together with C-terminal domains. Biochimica Et Biophysica Acta. 2009;1791(10):1023–1030. doi: 10.1016/j.bbalip.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Saba JD, Hla T. Point-counterpoint of sphingosine 1-phosphate metabolism. Circulation Research. 2004;94:724–734. doi: 10.1161/01.RES.0000122383.60368.24. [DOI] [PubMed] [Google Scholar]
- Scassellati C, Albi E, Cmarko D, Tiberi C, Cmarkova J, Bouchet-Marquis C, Verschure PJ, Driel RV, Magni MV, Fakan S. Intranuclear sphingomyelin is associated with transcriptionally active chromatin and plays a role in nuclear integrity. Biology of the Cell. 2010;102:361–375. doi: 10.1042/BC20090139. [DOI] [PubMed] [Google Scholar]
- Schroeder F, Petrescu AD, Huang H, Atshaves BP, McIntosh AL, Martin GG, Hostetler HA, Vespa A, Landrock D, Landrock KK, Payne HR, Kier AB. Role of Fatty Acid Binding Proteins and Long Chain Fatty Acids in Modulating Nuclear Receptors and Gene Transcription. Lipids. 2008;43:1–17. doi: 10.1007/s11745-007-3111-z. [DOI] [PubMed] [Google Scholar]
- Seeds AM, York JD. Inositol polyphosphate kinases: regulators of nuclear function. Biochem Soc Symp. 2007;74:183–197. doi: 10.1042/BSS0740183. [DOI] [PubMed] [Google Scholar]
- Shiraishi T, Imai S, Uda Y. The Presence of Ceramidase Activity in Liver Nuclear Membrane. Biological and Pharmaceutical Bulletin. 2003;26:775–779. doi: 10.1248/bpb.26.775. [DOI] [PubMed] [Google Scholar]
- Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, et al. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500(7463):463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siow DL, Anderson CD, Berdyshev EV, Skobeleva A, Natarajan V, Piston SM, Wattenberg W. Sphingosine kinase localization in the control of sphingolipid metabolism. Adv Enzyme Regul. 2011;51:229–244. doi: 10.1016/j.advenzreg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013;52:424–437. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Sonoda H, Aoki J, Hiramatsu T, Ishida M, Bandoh K, Nagai Y, Taguchi R, Inoue K, Arai H. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J Biol Chem. 2002;277:34254–34263. doi: 10.1074/jbc.M201659200. [DOI] [PubMed] [Google Scholar]
- Sugiura M, Kono K, Liu H, Shimizugawa T, Minekura H, Spiegel S, Kohama T. Ceramide Kinase, a Novel Lipid Kinase MOLECULAR CLONING AND FUNCTIONAL CHARACTERIZATION. J Biol Chem. 2002;277:23294–23300. doi: 10.1074/jbc.M201535200. [DOI] [PubMed] [Google Scholar]
- Tabellini G, Bortul R, Santi S, Riccio M, Baldini G, Cappellini A, Billi AM, Berezney R, Ruggeri A, Cocco L, Martelli AM. Diacylglycerol kinase-θ is localized in the speckle domains of the nucleus. Experimental Cell Research. 2003;287:143–154. doi: 10.1016/S0014-4827(03)00115-0. [DOI] [PubMed] [Google Scholar]
- Tamiya-Koizumi K, Umekawa H, Yoshida S, Kojima K. Existence of Mg2+-dependent, neutral sphingomyelinase in nuclei of rat ascites hepatoma cells. J Biochem. 1989;106:593–598. doi: 10.1093/oxfordjournals.jbchem.a122901. [DOI] [PubMed] [Google Scholar]
- Topham MK, Prescott SM. Mammalian Diacylglycerol Kinases, a Family of Lipid Kinases with Signaling Functions. J Biol Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SGE. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2012;2:462–469. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane K, Tamiya-Koizumi K, Nagino M, Nimura Y, Yoshida S. A possible role of nuclear ceramide and sphingosine in hepatocyte apoptosis in rat liver. Journal of Hepatology. 1999;31:8–17. doi: 10.1016/S0168-8278(99)80158-5. [DOI] [PubMed] [Google Scholar]
- Tsukahara T, Haniu H, Matsuda Y. Effect of alkyl glycerophosphate on the activation of peroxisome proliferator-activated receptor gamma and glucose uptake in C2C12 cells. Biochem Biophys Res Commun. 2013;433:281–285. doi: 10.1016/j.bbrc.2013.02.101. [DOI] [PubMed] [Google Scholar]
- Tu-Sekine B, Goldschmidt HL, Raben DM. DGK-θ: Structure, Enzymology, and Physiological Roles. Frontiers in Cell and Developmental Biology. 2016;4:101. doi: 10.3389/fcell.2016.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs AN, Dammer E, Sewer MB. Sphingosine Regulates the Transcription of CYP17 by Binding to Steroidogenic Factor-1. Endocrinology. 2006;147:5249–5258. doi: 10.1210/en.2006-0355. [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Gijsbers S, Mannaerts GP, Vermeesch JR, Brys V. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q2211DNA sequence was deposited in the EMBL database (AJ011304) Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2000;1487:128–134. doi: 10.1016/S1388-1981(00)00079-2. [DOI] [PubMed] [Google Scholar]
- Vanier MT. Niemann-Pick diseases. Handbook of Clinical Neurology. 2013;113:1717–1721. doi: 10.1016/B978-0-444-59565-2.00041-1. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Lee Y-M, Michaud J, Thangada S, Ai Y, Bonkovsky HL, Parikh NS, Habrukowich C, Hla T. Vascular Endothelium As a Contributor of Plasma Sphingosine 1-Phosphate. Circulation Research. 2008;102:669–676. doi: 10.1161/CIRCRESAHA.107.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Walter W, Le Guezennec X, Kim J, Edayathumangalam RS, Lasonder E, Luger K, Roeder RG, Logie C, Berger SL, Stunnenberg HG. A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol Cell Biol. 2006;26:5226–5236. doi: 10.1128/MCB.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97:259–265. doi: 10.1016/j.yexmp.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu G, Miyagi T, Lu Z-H, Ledeen RW. Sialidase occurs in both membranes of the nuclear envelope and hydrolyzes endogenous GD1a. J Neurochem. 2009;111:547–554. doi: 10.1111/j.1471-4159.2009.06339.x. [DOI] [PubMed] [Google Scholar]
- Waters CM, Saatian B, Moughal NA, Zhao Y, Tigyi G, Natarajan V, Pyne S, Pyne NJ. Integrin signalling regulates the nuclear localization and function of the lysophosphatidic acid receptor-1 (LPA1) in mammalian cells. Biochem J. 2006;398:55–62. doi: 10.1042/BJ20060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kitano T, Kondo T, Yabu T, Taguchi Y, Tashima M, et al. Increase of nuclear ceramide through caspase-3-dependent regulation of the “sphingomyelin cycle” in Fas-induced apoptosis. Cancer Research. 2004;64(3):1000–1007. doi: 10.1158/0008-5472.can-03-1383. [DOI] [PubMed] [Google Scholar]
- Wu G, Xie X, Lu Z-H, Ledeen RW. Sodium-calcium exchanger complexed with GM1 ganglioside in nuclear membrane transfers calcium from nucleoplasm to endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:10829–10834. doi: 10.1073/pnas.0903408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lu Z-H, Wang J, Wang Y, Xie X, Meyenhofer MF, Ledeen RW. Enhanced susceptibility to kainate-induced seizures, neuronal apoptosis, and death in mice lacking gangliotetraose gangliosides: protection with LIGA 20, a membrane-permeant analog of GM1. J Neurosci. 2005;25:11014–11022. doi: 10.1523/JNEUROSCI.3635-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wu G, Lu Z-H, Ledeen RW. Potentiation of a sodium-calcium exchanger in the nuclear envelope by nuclear GM1 ganglioside. J Neurochem. 2002;81:1185–1195. doi: 10.1046/j.1471-4159.2002.00917.x. [DOI] [PubMed] [Google Scholar]
- Yamaji T, Kumagai K, Tomishige N, Hanada K. Two sphingolipid transfer proteins, CERT and FAPP2: Their roles in sphingolipid metabolism. IUBMB Life. 2008;60:511–518. doi: 10.1002/iub.83. [DOI] [PubMed] [Google Scholar]
- Yatomi Y, Yamamura S, Hisano N, Nakahara K, Igarashi Y, Ozaki Y. Sphingosine 1-Phosphate Breakdown in Platelets. J Biochem. 2004;136:495–502. doi: 10.1093/jb/mvh143. [DOI] [PubMed] [Google Scholar]
- Zhao Y, I, Gorshkova A, Berdyshev E, He D, Fu P, Ma W, Su Y, Usatyuk PV, Pendyala S, Oskouian B, Saba JD, Garcia JGN, Natarajan V. Protection of LPS-induced murine acute lung injury by sphingosine-1-phosphate lyase suppression. Am J Respir Cell Mol Biol. 2011;45:426–435. doi: 10.1165/rcmb.2010-0422OC. [DOI] [PMC free article] [PubMed] [Google Scholar]