Summary
The apolipoprotein E4 (APOE4) variant is the single greatest genetic risk factor for sporadic Alzheimer’s disease (sAD). However, the cell type-specific functions of APOE4 in relation to AD pathology remain understudied. Here, we utilize CRISPR/Cas9 and induced pluripotent stem cells (iPSCs) to examine APOE4 effects on human brain cell types. Transcriptional profiling identified hundreds of differentially expressed genes in each cell type, with the most affected involving synaptic function (neurons), lipid metabolism (astrocytes) and immune response (microglia-like cells). APOE4 neurons exhibited increased synapse number and elevated Aβ42 secretion relative to isogenic APOE3 cells while APOE4 astrocytes displayed impaired Aβ uptake and cholesterol accumulation. Notably, APOE4 microglia-like cells exhibited altered morphologies, which correlated with reduced Aβ phagocytosis. Consistently, converting APOE4 to APOE3 in brain cell types from sAD iPSCs was sufficient to attenuate multiple AD-related pathologies. Our study establishes a reference for human cell type-specific changes associated with the APOE4 variant.
Introduction
Late-onset, sAD represents about 95% of all Alzheimer’s disease cases (Alzheimer’s Association, 2016). sAD etiology is likely due to complex interactions among different brain cell types leading to interconnected cellular pathologies (de Strooper and Karran, 2016). This dysfunction results in the pathological hallmarks of AD: senile plaques, neurofibrillary tangles, neurodegeneration and cognitive dysfunction (Canter et al., 2016). Critically important in the regulation of these processes is the balance between production and clearance of Aβ peptides from the brain. Aβ peptides, the main constituent of senile plaques, are produced mostly by neurons in an activity-dependent manner, and various astrocyte- and microglial-dependent mechanisms are thought to promote breakdown or clearance of Aβ from the brain (Bero et al., 2011; de Strooper and Karran, 2016). In contrast to the longstanding view of Alzheimer’s as a neuron-centric disease, recent genetic studies have identified numerous non-neuronal genes as important risk factors for sAD (Lambert et al., 2013).
The earliest identified and most significantly associated genetic risk factor for sAD is the E4 allele of the APOE which markedly increases AD risk relative to the APOE3 allele, while the APOE2 allele is considered protective (Corder et al., 1993; Lambert et al., 2013; Strittmatter et al., 1993). A single amino acid difference between APOE3 (Cys112) and APOE4 (Arg112) results in a protein conformational change that affects binding to apolipoprotein receptors, lipids and Aβ (Liu et al., 2013). Brain APOE is mainly produced by astrocytes and secreted to the extracellular space where it serves as the primary cholesterol carrier (Kim et al., 2009). Importantly, APOE is expressed by other brain cell types including neurons and microglia, where its expression can be altered under neuropathological conditions (Keren-Shaul et al., 2017; Mathys et al., 2017; Xu et al., 2006).
While the molecular etiology of AD driven by familial AD (fAD) mutations is relatively well understood, the specific impact of sAD risk factors including APOE4 remains much less clear. Studies using mouse models expressing humanized APOE isoforms, cell lines and postmortem human samples have revealed multiple AD-related pathological phenotypes impacted by APOE4, including reduced Aβ clearance, tauopathy, increased neuronal toxicity and mitochondrial dysfunction, though the cell type-specific changes remain largely unexplored (Kim et al., 2009; Liu et al., 2013; Shi et al., 2017). However, findings from animal models are often hampered by concerns about translatability to humans due to species differences while studies using human cells are limited by accessibility to the relevant cell types, and an inability to model complex disease in vitro (Calcoen et al., 2015). Recent advances in genome editing and differentiation protocols for generating 2D and 3D cultures from human iPSCs now allow for a more systematic examination of APOE4 effects on the different brain cell types in a human in vitro model. (Paquet et al., 2016; Raja et al., 2016; Ran et al., 2013; Wang et al., 2018).
To better understand how APOE4 affects major brain cell types involved in AD pathogenesis, we utilized CRISPR/Cas9 to create isogenic iPSC lines harboring homozygous APOE4 alleles from unaffected parental APOE3 cells. We found that APOE4 iPSC-derived neurons, astrocytes and microglia-like cells recapitulated phenotypes associated with AD at multiple levels. The critical role for APOE4 in AD pathogenesis is underscored by our observation that conversion of APOE4 in sAD patient-derived iPSCs to APOE3 was sufficient to alleviate most of the AD-related phenotypes observed in sAD iPSC-derived neurons, glia, and organoids.
Results
Generating isogenic homozygous APOE4 iPSCs
To enable systematic analysis of APOE4 effects on specific brain cell types (Figure 1A) we used CRISPR/Cas9 gene editing to generate APOE4 iPSCs from parental APOE3 cells derived from an unaffected subject. Successful editing resulted in a Cys112Arg substitution in the APOE protein product which was verified by Sanger sequencing of colonies derived from single putatively edited cells (Figure S1A; see Methods). Whole exome sequencing of our edited line as well as the parental iPSC line showed no unintended off target mutations (Figure S1B–D), while karyotyping analysis revealed no chromosomal abnormalities in our isogenic lines (Figure S1E). Further, both parental and genome-edited iPSCs maintained comparable expression of pluripotency makers (Figure S1F).
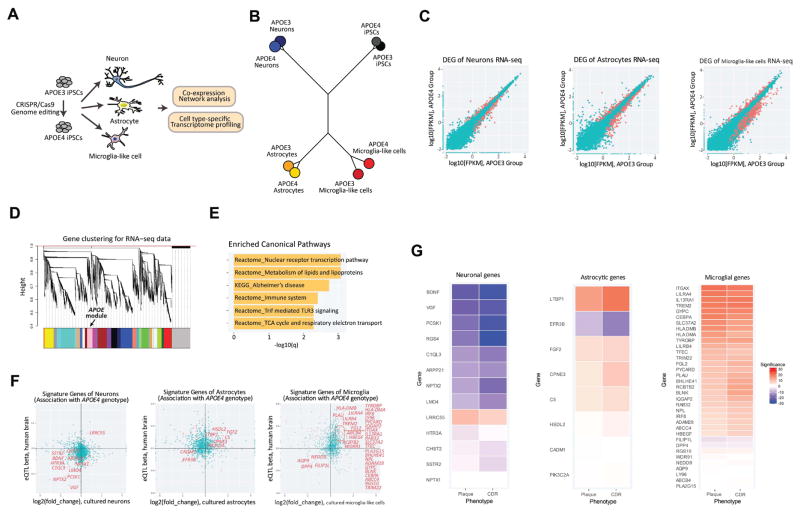
Figure 1. Altered gene expression in APOE4 neural and glial cells is correlated with severe AD clinical phenotypes.
(A) Schematics of RNA-seq with neuron, astrocytes and microglia-like cells derived from isogenic iPSCs homozygous for either APOE3 or APOE4.
(B) Phylogenetic dendrogram of RNA-seq data.
(C) Scatter plots of RNA-seq gene quantification values (log10FPKM) of APOE3 and APOE4 groups in iPSC-derived brain cell types. DEGs are colored in red.
(D) Dendrogram of RNA-seq co-expression gene network visualizing clustering of gene expression patterns in iPSC-derived cells. Color bars indicate different gene modules, with APOE module colored in pink.
(E) Top six enriched canonical pathways of APOE gene module.
(F) Expression changes of cell type signature genes in iPSC-derived corresponding APOE4 cell types were compared to APOE4 allele-associated eQTL effect sizes of those signature genes in postmortem human brain samples. The red dots in the plots showed overlap of significantly altered genes in both human brain samples and induced brain cell types. Among them, genes with concordant changes are pinpointed in the scatter plots.
(G) Expression of the concordant genes in control and MCI brains were correlated with two AD phenotypical measurements: neocortical plaque density and clinical dementia rating (CDR). Statistical p-values of Spearman’s correlation are visualized in heatmaps, with blue or red colors indicating negative or positive correlation.
Generation of isogenic APOE iPSC-derived neurons, astrocytes, and microglia-like cells
Next, we derived neurons, astrocytes, and microglia-like cells from our isogenic APOE3 and APOE4 iPSC lines. We generated neurons following the neurogenin2-mediated differentiation protocol (Zhang et al., 2013) with some modifications (Figure S2A, see Methods). To obtain a pure population, we cultured neurons supplemented with conditioned media from control human astrocytes, rather than co-culturing in direct contact with astrocytes. Although we cannot exclude potential effects on neurons by APOE proteins in the conditioned media released by APOE3 astrocytes, this approach allowed us to investigate the effect of neuronal APOE variation in human induced neurons. To generate astrocytes, we derived iPSCs into neural progenitor cells (NPCs), which were subsequently induced to differentiate into astrocytes, while microglia-like cells were generated using a recently described protocol (Chen et al., 2014; Muffat et al., 2016) (Figure S2B and S2C, see Methods).
To obtain comprehensive transcriptional profiles of the three derived cell types, we next performed RNA sequencing (RNA-seq). Pairwise correlation of the transcriptomes showed segregation of iPSC-derived neurons, astrocytes, and microglia-like cells into their corresponding cell types regardless of APOE status (Figure 1B). The transcriptome signature of each cell type clustered with the corresponding cell type derived from either human brain samples or iPSCs reported by other groups (Figure S2D, see Methods), validating the identity of our iPSC-derived neural and glial cells.
APOE3 to APOE4 switch dramatically alters transcriptomes of iPSC-derived neurons and glia
APOE variants were recently shown to affect gene expression in various cell types (Huang et al., 2017; Shi et al., 2017; Theendakara et al., 2016). We analyzed the effect of converting APOE3 to APOE4 on expression of genes in iPSCs and iPSC-derived neural and glial cells. We found many genes differentially regulated by APOE genotype in each cell type (Figure 1C), with iPSCs showing the lowest number of differentially expressed genes (DEGs) (Supplementary table 1). To identify genes whose expression is strongly correlated with APOE, we pooled all RNA-seq samples from all cell types of both genotypes and performed co-expression network analysis (see Methods). We found 857 genes having similar expression patterns to APOE (Figure 1D, Supplementary table 2). Gene ontology (GO) analysis indicated that expression of genes involved in lipid metabolism, immune responses or associated with AD was regulated synchronously with APOE (Figure 1E, Supplementary table 3). Considering the widespread effect of APOE4 on transcription, we asked whether DEGs observed in APOE4 cell types are regulated by a common set of transcription factors. Transcription motif analysis showed that, in addition to the three orphan motifs not associated with any known transcription factors, the motifs for nuclear factor 1 (NF1), activator protein 1 (AP-1) and nuclear factor kB (NF-kB) were markedly enriched in the promoters of genes in APOE module (Figure S2E). Among them, only the binding site for AP-1 was significantly enriched in DEGs affected by APOE4 in all of induced brain cell types. We also performed miRNA motif analysis and found no common motifs in DEGs across the brain cell types and APOE coregulated genes (Supplementary table 4).
We then asked whether altered gene transcription by the APOE4 variant in iPSC-derived brain cell types parallels APOE4-dependent changes in human brain samples. This analysis allows us to identify ‘gene hits’ and related pathways, potentially representing perturbed biological processes driven by the APOE4 variant. We compared our datasets with those from 880 human brain samples (normal controls and mild cognitive impairment subjects, see Methods). We selected ‘signature’ genes for each cell type, with enriched expression in neurons, astrocytes or microglia based on a previous report (Zhang et al., 2016), and performed expression quantitative trait loci (eQTLs) analysis based on APOE4 genotype. These data were then compared to our RNA-seq results from each cell type. A significant number of neuronal signature genes exhibited concordant APOE genotype-dependent changes between the human samples and our iPSC-derived neurons (p=0.042) (i.e., positively associated with APOE4 status in human, and up-regulated in APOE4 derived neurons, or vice versa). Among these coordinately regulated genes, more than half of them are known regulators of synaptic function (7 out of 12 genes, p=2.59E-10), and most were negatively associated with APOE4 (Figure 1F). For astrocyte signature genes, 8 genes were up- or down-regulated based on APOE4 genotype in both human brain and iPSC-derived astrocytes (Figure 1F). Although this number of overlapping genes was not statistically significant (p=0.23), it is interesting to note that three of those genes – PIK3C2A, CPNE3, EFR3B, are known to be involved in phospholipid metabolic process (p=5.39E-7). For microglia signature genes, there was a significant overlap between human brain and induced microglia-like cells (p=1.6E-6) (Figure 1F). In contrast to neuronal signature genes, most of the microglia signature genes whose expression was affected by APOE4 in both human brain and microglia-like cells were up-regulated. Strikingly, about half of them were associated with immune system process (p=2.22E-9), suggesting that APOE4 impacts microglial functions related to the immune response. Expression of these APOE4-associated genes in each cell type showed significant negative or positive correlation with clinically defined AD phenotypes (neocortical plaque density and clinical dementia rating) in human patients (Figure 1G). Taken together, our transcriptome profiling showed that relative to APOE3, the APOE4 variant alters the expression of several pathways implicated in the development of AD in multiple brain cell types.
Increased synapses, early endosomes and Aβ42 secretion in APOE4 neurons
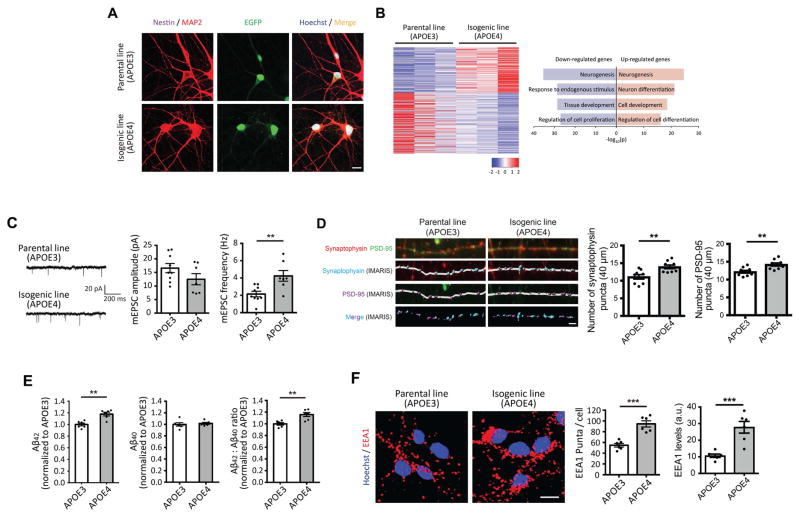
In the brain, neurons play an essential role in AD pathology as both the main producers of Aβ and the cell type most susceptible to damage. Despite this, the effect of neuronal APOE4 genotype on synaptic function and AD pathology is not clear. Immunostaining results showed that neurons derived from both APOE3 and APOE4 iPSCs strongly expressed neuronal markers such as MAP2, but not neural progenitor markers such as nestin, after 4 weeks of induction (Figure 2A). RNA-seq revealed 445 genes that were significantly differentially expressed between APOE3 and APOE4 neurons (Figure 1E and 2B). GO analysis of these DEGs revealed that cell proliferation-related transcripts were down-regulated by APOE4, whereas neuron differentiation-associated genes were up-regulated (Figure 2B). To investigate the possibility of different synaptic activity between APOE3 and APOE4-carrying neurons, we performed intracellular recording on APOE3 and APOE4 neurons. We observed increased miniature excitatory postsynaptic current (mEPSC) frequencies with indistinguishable mEPSC amplitudes in APOE4 neurons compared to APOE3 controls (Figure 2C), suggesting increased release of neurotransmitter or elevated synaptic density in APOE4 neurons. Consistent with the latter possibility, we observed an increase in number of synapses by immunostaining with the presynaptic marker, synaptophysin, and the postsynaptic marker, PSD-95, in APOE4 vs. APOE3 neurons (Figure 2D). Increased synaptic activity has been shown to correlate with increased Aβ production (Bero et al., 2011; Das et al., 2013). To determine whether APOE4 neurons exhibit altered Aβ release, we measured the levels of Aβ secreted by APOE3 and APOE4 neurons by ELISA. Aβ42 levels were 20% higher from APOE4 cultures compared to APOE3 cultures, although Aβ40 levels were indistinguishable (Figure 2E).
Figure 2. Increased number of synapses, early endosomes and Aβ42 secretion in APOE4 neurons.
(A) Immunocytochemistry with MAP2 and nestin antibodies in neurons. Scale bar = 10 μm.
(B) Heatmaps of z-score values and barplots of enriched GO terms for DEGs between APOE3 and APOE4 neurons.
(C) mEPSCs amplitude and frequency in iPSC-derived neurons. Scale bar = 20 pA and 200 ms. n=7~9 from three independent cultures.
(D) Immunocytochemistry with synaptophysin and PSD-95 antibodies in neurons. Synaptophysin and PSD-95 signals were analyzed by IMARIS. n=9 from four independent cultures. Scale bar = 2 μm.
(E) Secreted levels of Aβ42 and Aβ40 from iPSC-induced neurons were measured by ELISA and normalized to APOE3 neurons. n=6 per group.
(F) Immunocytochemistry with EEA1 antibody in neurons. Scale bar = 10 μm. n=6 from three independent cultures. **P<0.01, ***P<0.001.
In addition to elevated synaptic activity, endosomal abnormalities can elevate Aβ production by active cleavage of amyloid precursor protein (APP) via β-site APP-cleaving enzyme 1 (BACE1), generating β-CTF, a precursor of Aβ (Toh and Gleeson, 2016). Further, increased number and/or size of early endosomes has been described in the brains of AD patients (Cataldo et al., 2000). We examined early endosomes in neurons by immunostaining with an antibody against early endosome antigen 1 (EEA1). We found that the number of EEA1 puncta was higher in APOE4 neurons compared to their APOE3 counterparts. Total EEA1 levels, quantified as the product of ‘mean intensity of signal’ and ‘area covered by signal’, were also increased in APOE4 neurons (Figure 2F). These findings indicate that induced neurons harboring the APOE4 allele exhibit disruption in pathways related to synaptic formation, associated with supernumerary synapses and increased frequency of synaptic transmission. Furthermore, APOE4 neurons contain a greater number of early endosomes than APOE3 controls.
APOE4 astrocytes exhibit compromised Aβ uptake
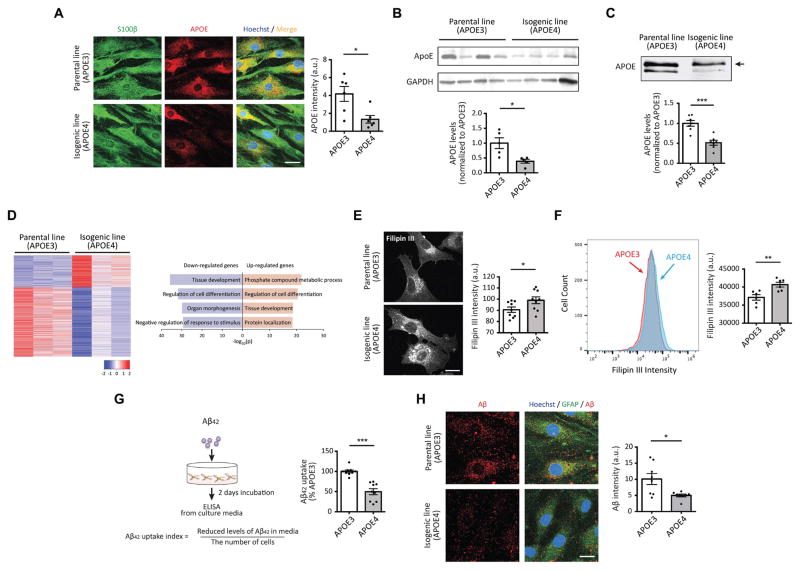
Astrocytes are the major source of secreted APOE, supplying lipids such as cholesterol to other cell types, including neurons, in the brain (Kim et al., 2009; Liu et al., 2013). Although astrocytes are known to have executive coordinating functions for neuronal activity and to play important protective roles in neurodegenerative disease (Sidoryk-Wegrzynowicz et al., 2011; Wyss-Coray et al., 2003), the effects of APOE genotype on astrocyte function remain incompletely understood. Immunostaining data showed that both APOE3 and APOE4 iPSC-derived astrocytes strongly express the astrocyte-specific marker S100β (Figure 3A). We examined APOE protein levels in these cells by immunostaining, finding significantly reduced APOE intensity in APOE4 vs. APOE3 astrocytes (Figure 3A). Immunoblotting confirmed less APOE expression in astrocytes harboring the APOE4 allele compared to APOE3 astrocytes (Figure 3B). Similarly, there was a reduction of secreted APOE in APOE4 astrocytes cultures (Figure 3C). We then compared the transcriptomes of APOE3 and APOE4 astrocytes, noting that in parallel with reduced APOE protein, APOE transcription was also reduced by the APOE4 genotype. In total, we found 1,327 DEGs (418 genes up-regulated; 909 genes down-regulated) in APOE4 compared to APOE3 astrocytes (Figure 1E and 3D). GO analysis indicated that genes related to tissue development were down-regulated whereas phosphate-containing compound metabolic process-associated genes such as CROT, LPGAT1 and PLPP3, which are known to be involved in lipid metabolism, were up-regulated in APOE4 astrocytes (Figure 3D). Our transcriptome data further showed that the expression of numerous other genes with functions in lipid transport were decreased (Figure S3A).
Figure 3. APOE4 astrocytes exhibited reduced APOE protein levels and impaired clearance of extracellular Aβ42.
(A) Immunocytochemistry with APOE antibody in astrocytes homozygous for either APOE3 or APOE4. Scale bar = 10 μm. n=6 per group.
(B, C) Immunoblotting with APOE antibody in cell pellets (C) or cultured media (D) from APOE3 or APOE4 iPSC-induced astrocytes. Bar graphs represent relative immunoreactivity of APOE in each group normalized to APOE3 genotype. n=5~7 per group. The upper bands (arrow) in panel C likely correspond to the sialiated form of APOE.
(D) Heatmaps of z-score values and barplots of enriched GO terms for DEGs between APOE3 and APOE4 astrocytes.
(E) iPSC-derived astrocytes were fixed and incubated with filipin III for 1 hr followed by imaging. The bar graph represents the intensity of filipin III in the images. Scale bar = 10 μm. n=9 images from three independent cultures.
(F) iPSC-derived astrocytes were sorted based on the intensity of filipin III. The bar graph represents the intensity of filipin III in sorted cells. n=6 per group.
(G) iPSC-derived astrocytes were incubated with Aβ42 oligomers for 2 days, then residual Aβ42 were measured by ELISA. (Right) The Aβ42 clearance index was calculated as described and normalized to APOE3 astrocytes. n=9 per group.
(H) Immunocytochemistry with Aβ and GFAP antibodies in astrocytes. (Right) Relative immunoreactivity of Aβ overlapped with GFAP was normalized to APOE3 astrocytes. Scale bar = 10 μm. n=8 images from three independent cultures. *P<0.05, **P<0.01, ***P<0.001.
Because one of the major functions of APOE is to transport lipids including cholesterol between cells, we explored whether APOE4 astrocytes are impaired in cholesterol transport. We measured the cellular levels of cholesterol in APOE3 and APOE4 astrocytes by staining with filipin III, a compound known to fluoresce upon binding to cholesterol. We observed stronger signals of fluorescent filipin III in APOE4 vs. APOE3 astrocytes (Figure 3E). We next used flow cytometry analysis to better quantify the filipin III signal in individual cells, finding higher average fluorescent intensity in APOE4 compared to APOE3 astrocytes (Figure 3F). We then examined levels of cholesterol in cultured media, including another isogenic clone of APOE3 -> APOE4 (APOE4#2) to control for possible variance among colonies. We observed increased levels of secreted cholesterol in both APOE4 and APOE4#2 astrocyte cultures compared to those in APOE3 cultures (Figure S3B), suggesting increased biosynthesis of cholesterol in APOE4 astrocytes.
Astrocytes can uptake oligomeric and fibrillar Aβ42in vitro and in vivo, which contributes to Aβ clearance and prevents the detrimental effect of Aβ accumulation seen in AD (Koistinaho et al., 2004; Wyss-Coray et al., 2003). APOE forms lipoprotein complexes with various lipids including cholesterol, and it has been shown that APOE4-containing lipoproteins have impaired ability to bind and uptake extracellular Aβ, compared to those of APOE3 (Kim et al., 2009; Liu et al., 2013). However, it has not been directly or conclusively determined whether APOE4 impacts the ability of astrocytes to uptake Aβ. To test this directly, we followed standard protocols by incubating iPSC-derived astrocytes with synthetic oligomeric Aβ42 for 2 days to allow sufficient time for Aβ clearance (Wyss-Coray et al., 2003, see Methods). To quantify Aβ42 uptake, we measured the fraction of oligomeric Aβ42 removed from the media after the two-day incubation (Figure 3G, see Methods). We confirmed that our astrocytes did not secrete any detectable amount of Aβ, ruling out endogenous Aβ as a contributing source to the media (data not shown). Also, Aβ treatment had no effect on cell viability or proliferation of APOE3 or APOE4 astrocytes (Figure S3C). APOE4 astrocytes were notably less efficient than APOE3 astrocytes in clearing Aβ42 from the media (Figure 3G). In complementary experiments to directly measure Aβ42 uptake by astrocytes, we performed immunostaining with antibodies against Aβ and the astrocyte marker GFAP. Data indicated that less Aβ42 was taken up by APOE4 astrocytes (Figure 3H). Since lysosomal activity can affect Aβ levels, we also examined lysosome-dependent degradation of Aβ42 by measuring intracellular Aβ with or without lysosomal inhibitors following Aβ42 treatment. We observed increased Aβ42 accumulation following lysosome inhibition in APOE3 astrocytes compared to APOE4 astrocytes, suggesting reduced lysosome-dependent Aβ42 degradation in APOE4 astrocytes (Figure S3D). Taken together, our results indicate that harboring the APOE4 allele rather than APOE3 negatively affects the expression of APOE, alters cholesterol metabolism, and impairs uptake of Aβ42 in human iPSC-derived astrocytes.
APOE4 microglia-like cells exhibit inflammatory gene activation and less efficient Aβ42 uptake
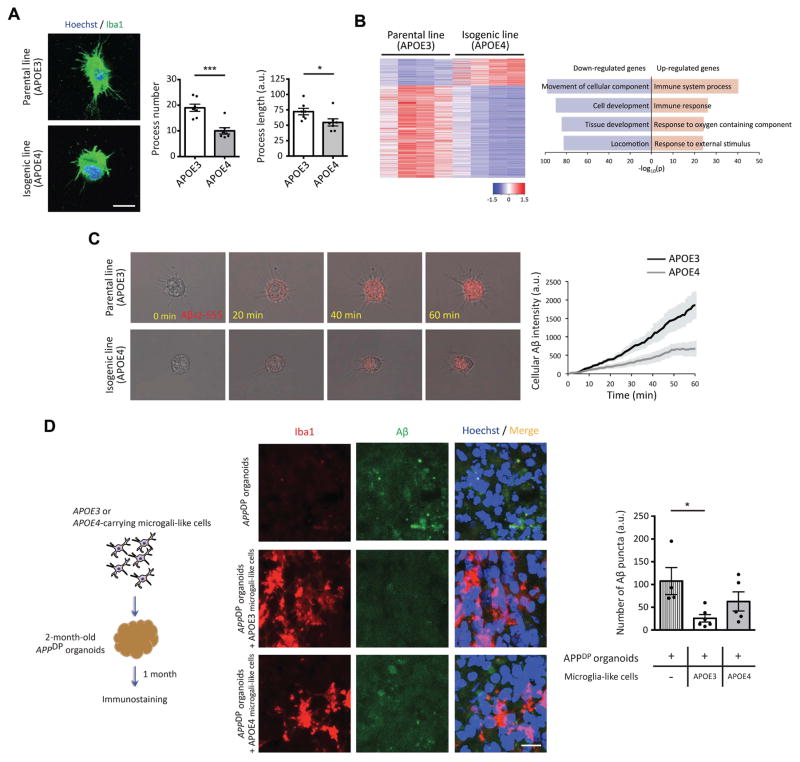
Recent transcriptome analysis from human brain showed abundant expression of APOE in microglia (Gosselin et al., 2017); however the potential effects of the APOE4 variant on microglial gene expression and function are not known. Microglia, distinguished by their elaborated morphology reflecting their phagocytic function, are important mediators of inflammation in the nervous system. Recent genetic and functional studies have highlighted critical roles for microglia in brain health and disease, and identified dysfunction of this cell type as an important driver of sAD progression (Efthymiou and Goate, 2017; Keren-Shaul et al., 2017). Our iPSC–derived microglia-like cells clearly exhibit a ramified morphology characteristic of microglia (Muffat, 2016). Noticeably, in the course of generating microglia-like cells, we observed that those harboring the APOE4 variant displayed fewer and shorter processes (Figure 4A). To gain further insights into potential effects of APOE4 on this cell type, we compared the transcriptomes of APOE3 and APOE4 microglia-like cells. We found 1,460 DEGs in APOE4 microglia-like cells compared to APOE3 cells, more than in any other cell type we examined (Figure 1E and 4B). Interestingly, many more of the DEGs were down-regulated (1,131 genes) than upregulated (329 genes) (Figure 4B, left panel). While down-regulated genes were shown to be strongly associated with cell movement and development, a third of the 329 up-regulated genes in APOE4 microglia-like cells were associated with immune responses (Figure 4B, right panel and Figure S4A). These results suggest that APOE4 likely renders microglia more prone to promote inflammation, and may also alter their phagocytic behavior. To explore the latter idea further, we examined the phagocytic activity of these cells toward Aβ. Since microglia take up Aβ much faster compared to astrocytes (Fu et al., 2012), we could monitor the uptake of fluorescently-tagged Aβ42 (Aβ42-555) by microglia-like cells in real time. During the 1hr imaging period, we found that microglia-like cells harboring the APOE4 variant took up Aβ42 much more slowly than APOE3 cells (Figure 4C, Supplementary movie 1 and 2).
Figure 4. Altered process complexity and impaired clearance of extracellular Aβ in APOE4 microglia-like cells.
(A) Immunocytochemistry with Iba1 antibody in APOE3 or APOE4 microglia-like cells. Bar graphs represent the number or length of process in microglia-like cells. Scale bar = 10 μm. n=7 images from four independent cultures.
(B) Heatmaps of z-score values and barplots of enriched GO terms for DEGs between APOE3 and APOE4 microglia-like cells.
(C) Induced microglia-like cells were treated with Aβ42-555, and its uptake was monitored under the microscope for 1 hr. (Right) The intensity of cellular Aβ42 in APOE3 and APOE4 microglia-like cells was plotted against time (min).
(D) Schematics to determine Aβ clearance in APPDP organoids by microglia-like cells. (Right) Three months APPDP organoids cultured without microglia-like cells or with either APOE3 or APOE4 microglia-like cells were fixed and subjected to immunostaining with Iba1 and Aβ antibodies. Bar graph represents the number of Aβ puncta. Scale bar = 10 μm. n=4~6 organoids per group. *P<0.05, ***P<0.001.
We recently showed that cerebral organoids generated from iPSC lines harboring fAD mutations such as APP duplication (APPDP) or PSEN1 mutation develop Aβ aggregates and hyperphosphorylated tau in two to three months (Raja et al, 2016). To investigate the ability of induced microglia-like cells to impact AD pathology in 3D organoid systems, we co-cultured APOE3 or APOE4-carrying microglia-like cells with two-month-old APPDP organoids that display Aβ aggregates. After one month of co-culture, we observed that similar numbers of microglia-like cells were incorporated into APPDP organoids regardless of APOE genotype (Figure S4B). However, unlike microglia-like cells (without Aβ treatment) in 2D-culture systems, APOE4 microglia-like cells embedded in APPDP organoids exhibited longer processes compared to APOE3 cells, without a change in process number (Figure S4C and S4D). Shorter process length of microglia in AD brains has been shown to positively correlate with microglial Aβ uptake (Iaccarino et al., 2016; Sarius and Heneka, 2017). Therefore, these data suggested that microglia-like cells with the APOE4 variant may be less able to sense and respond to extracellular Aβ. We observed that Aβ accumulation in organoids containing APOE3 microglia-like cells was significantly less than that in APPDP organoids without co-cultured microglia-like cells (Figure 4D). However, APPDP organoids co-cultured with microglia-like cells carrying the APOE4 allele exhibited more extracellular Aβ aggregates than those co-cultured with APOE3 microglia-like cells (Figure 4D, Supplementary movie 3 and 4). Our data suggest that APOE4 negatively impacts several aspects of microglial function that potentially hinder the ability of microglia to clear extracellular Aβ from AD brains, and may also influence the brain inflammatory profile.
APOE4 organoids accumulate Aβ and exhibit elevated tau phosphorylation
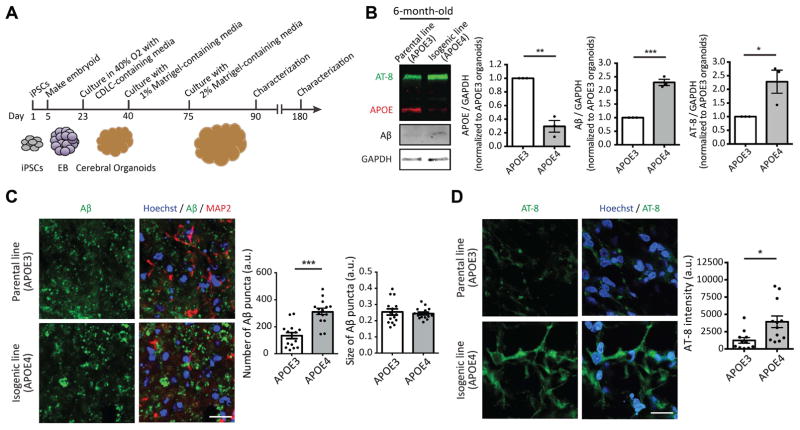
We next determined whether APOE4 organoids would develop hallmarks of AD similar to fAD organoids as described above (Figure 5A). iPSCs used to generate organoids initially differentiate into NPCs and neurons, with astrocytes being detected at later time points (Quadrato et al., 2017). Thus, organoids serve initially as a 3D neuronal model, and later as a model incorporating both neurons and astrocytes, which each exhibit APOE4-dependent defects as described above. We found that unlike fAD organoids, which developed Aβ aggregates after two months (Raja et al., 2016), APOE4 organoids did not develop detectable Aβ accumulation by immunoblot assay at this time point (Figure S5A). Further, we found that APOE protein was undetectable in three-month-old organoids, consistent with low expression of APOE in neurons, and few astrocytes at this stage of organoid development (Figure S5A). As expected, there was an increase in the number of GFAP-positive astrocytes by six months of age, in parallel with increased APOE protein levels (Figure 5B and S5B). We found that the levels of APOE protein in APOE4 organoids were significantly lower compared to APOE3 organoids (Figure 5B) with both having similar number of total astrocytes at this stage (Figure S5B). Remarkably, six-month-old APOE4 organoids exhibited increased Aβ accumulation and tau phosphorylation (p-S202/T205) compared to APOE3 organoids (Figure 5B). Immunostaining further demonstrated an increased number of Aβ aggregates and elevated p-tau levels in six-month-old APOE4 organoids compared to their APOE3 counterparts (Figure 5C and D). Taken together, these data indicate that while APOE4 organoids exhibit delayed development of AD pathology relative to fAD organoids, the APOE4 variant alone is sufficient to cause AD hallmarks in cerebral organoids.
Figure 5. Increased levels of Aβ and p-tau in APOE4 iPSC-derived cerebral organoids.
(A) Schematics for generating cerebral organoids from iPSCs.
(B) Six months organoids from APOE3 or APOE4 iPSCs were lysed and subjected to immunoblotting with APOE, Aβ, p-tau (AT-8) and GAPDH antibodies. Bar graphs represent relative immunoreactivity of APOE, Aβ and AT-8 normalized to GAPDH. n=3 per group.
(C, D) Six months organoids from APOE3 or APOE4 iPSCs were fixed and subjected to immunostaining with Aβ, MAP2 and p-tau (AT-8) antibodies. Bar graphs represent the number and size of Aβ puncta (C) or immunoreactivity of AT-8 in organoids (D). Scale bar = 20 μm. n=15 images from 5~6 organoids. *P<0.05, **P<0.01, ***P<0.001.
Converting APOE4 to APOE3 attenuates AD-related phenotypes in neurons, glia, and organoids derived from sAD iPSCs
Because APOE4 carries the highest known risk for sAD, we next tested whether converting APOE4 in iPSCs from an sAD patient to APOE3 was sufficient to ameliorate the molecular and cellular phenotypes of various iPSC-derived brain cell types we have uncovered. We used a similar 12 CRISPR/Cas9 approach as described above to create isogenic iPSCs homozygous for APOE3 from sAD iPSCs homozygous for APOE4 (Figure S6A and see Methods). Karyotyping analysis confirmed that there were no detectable chromosomal rearrangements in these isogenic lines (Figure S6B). Whole-exome sequencing identified, other than the expected APOE4 to APOE3 change, a single variant in the TRIOBP gene causing replacement of Ala with Val (Figure S6C and S6D). TRIOBP is a TRIO and F-actin binding protein, which is known to be involved in actin cytoskeleton organization (Seipel et al., 2001). While mutations of TRIOBP have been associated with autosomal recessive nonsyndromic deafness (Shahin et al., 2006), there is no known link of TRIOBP with AD or any other neurodegenerative diseases. We then generated neurons, astrocytes, microglia-like cells and organoids from these two iPSC lines.
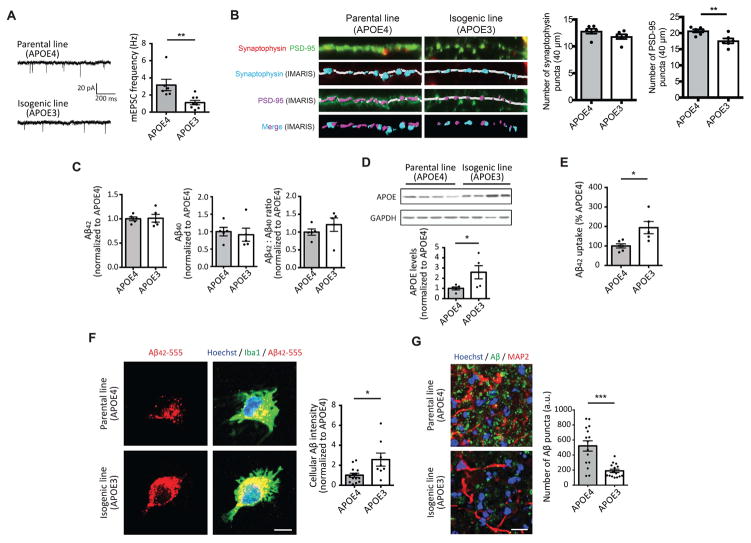
We first characterized induced neurons and found that APOE3 neurons exhibited reduced mEPSC frequencies and fewer synapses compared to APOE4 neurons (Figure 6A and B). However, we did not observe a significant difference in the levels of secreted Aβ42 or Aβ40 (Figure 6C) between the two groups. Also, while converting APOE4 to APOE3 appeared to reduce the number of early endosomes in neurons, the effect did not reach significance (P=0.06, Figure S6E). In induced astrocytes, we found higher APOE protein levels and less accumulation of intracellular cholesterol in APOE3 than APOE4 astrocytes (Figure 6D, S6F and S6G). Moreover, converting APOE4 to APOE3 enhanced the ability of astrocytes and microglia-like cells to uptake extracellular Aβ (Figure 6E and 6F). We also addressed the contribution of the APOE4 variant to Aβ accumulation in sAD cerebral organoids, and found that after six months of culture, APOE3 organoids displayed less Aβ compared to age-matched APOE4 organoids (Figure 6G). Overall, these results indicate that most of the AD-related phenotypes observed in APOE4 iPSC-derived brain cell types and organoids could be reversed by editing to APOE3, confirming the central role of APOE4 in sAD pathology.
Figure 6. Converting APOE4 to APOE3 attenuates AD-related phenotypes in sAD iPSC-derived neurons, glial and organoids.
(A) mEPSCs amplitude (Left) and frequency (Right) in iPSC-derived neurons from APOE4 or APOE3 iPSCs. Scale bar = 20 pA and 200 ms. n=6~8 from three independent cultures.
(B) Immunocytochemistry with synaptophysin and PSD-95 antibodies in neurons derived from APOE4 or APOE3 iPSCs. Synaptophysin and PSD-95 signals were analyzed by IMARIS. Scale bar = 2 μm. n=6 from three independent cultures.
(C) Secreted levels of Aβ42 and Aβ40 from iPSC-induced neurons were measured by ELISA and normalized to APOE4 neurons. n=5 per group.
(D) Relative immunoreactivity of APOE in induced APOE4 or APOE3 astrocytes was normalized to APOE4 astrocytes. n=5 per group.
(E) iPSC-derived astrocytes were incubated with Aβ42 oligomers for 2 days, then residual Aβ42 were measured by ELISA. The Aβ42 clearance index was calculated and normalized to APOE4 astrocytes. n=5~6 per group.
(F) Immunocytochemistry with Aβ and Iba1 antibodies in APOE3 or APOE4 microglia-like cells. (Right) Relative immunoreactivity of Aβ overlapped with Iba1 was normalized to that of APOE4 microglia-like cells. Scale bar = 5 μm. n=8~14 images from four independent cultures.
(G) Six months organoids from APOE4 or APOE3 iPSCs were subjected to immunostaining with Aβ and MAP2 antibodies. The bar graph represents the number of Aβ puncta in organoids. Scale bar = 20 μm. n=15 images from 5~6 organoids. *P<0.05, **P<0.01, ***P<0.001.
Discussion
Early maturation of APOE4 neurons
Our transcriptional analysis revealed roughly equal numbers of up- and down-regulated genes in iPSC-derived neurons. GO analysis showed that the expression of genes related to neuronal differentiation was up-regulated by APOE4. Consistently, the increase in mEPSC frequency and number of synapses we observed in APOE4 neurons is suggestive of enhanced neuronal differentiation/maturation. We observed similar increases in synapse numbers in PSEN1 M146I iPSC-derived neurons (data not shown), indicating that early neuronal maturation could be a common phenotype of neurons with AD-linked mutations.
Neuronal signature genes whose expression was correlated with APOE4 genotype in both human and iPSC-derived samples were mostly down-regulated and associated with synaptic functions (Figure 1F and 1G). One of the significantly down-regulated genes, BDNF, was shown to have reduced expression in human APOE4 carriers, which paralleled cognitive impairment (Liu et al., 2015; Alvarez et al., 2016). Our analysis of exome sequencing data from AD patients and unaffected controls showed a strong association between a SNP in the BDNF gene (E6K) with neocortical plaque density (Figure S2F, see Methods), although whether this SNP leads to loss-of-function remains to be clarified. Furthermore, elevating BDNF in the brains of AD rodent models also attenuated AD-related pathology (Zhang et al., 2015). Thus, BDNF is a promising candidate that could account for some of the observed APOE4 neuronal phenotypes.
Our motif analysis identified high enrichment for transcriptional activator AP-1 binding sites in promoters of genes within the APOE module from human brain, and DEGs affected by APOE4 in our induced brain cells (Figure S2E). It was recently shown that E4 was the most potent APOE isoform in eliciting AP-1 activation in neurons (Huang et al., 2017). Our analysis suggests that APOE4-dependent effects on AP-1 activation are not restricted to neurons, though further study will be required to determine whether common or distinct regulatory pathways operate in each cell type, and whether these mechanisms could be targeted for therapeutic development.
APOE4 effects on astrocytes
We found that the levels of APOE protein and mRNA were reduced in APOE4 astrocytes, suggesting that the APOE4 variant can negatively impact it’s own transcription. Consistently, studies from both humans and mouse models have reported similar reductions in APOE4 protein relative to APOE3 (Mooijaart et al., 2006; Shi et al., 2017). In contrast, a recent study of iPSC-derived astrocytes showed similar APOE levels in APOE3 vs. APOE4 cells; however these findings may be confounded by the use of non-isogenic backgrounds (Zhao et al., 2017). Interestingly, we found that the reduced APOE4 mRNA and protein levels in iPSC-derived cell types were specific to astrocytes (data not shown), indicating the effect is cell-type specific.
Our gene expression analyses indicated the APOE4 variant regulates expression of numerous lipid metabolism and transport genes (Figures 1E and 1F, 3D and S3A). One of the intriguing phenotypes we found was the accumulation of intracellular and extracellular cholesterol in APOE4 astrocyte cultures. Cholesterol and other lipids are crucial for a wide range of cellular functions in the brain, and altered metabolism or transport appear to be associated with multiple pathological phenotypes in various neurodegenerative disorders (Göritz et al., 2002; Mauch et al., 2001). Remarkably, the APOE4 variant appears to regulate both the metabolism and transport of lipids such as cholesterol, with potential effects on each of the three brain cell types. For instance, cholesterol-rich lipid rafts serve as a platform for APP processing to Aβ (Cheng et al., 2007), increased cholesterol can result in enlarged early endosomes (Marquer et al., 2014), and altered cholesterol levels can affect gene expression (Goldstein et al., 2006). Therefore, additional experiments from each of the APOE4 brain cell types have the potential to yield important insights into these aspects of APOE biology.
Importantly, we also found that APOE4 astrocytes were less efficient in uptake and clearance of Aβ compared to their APOE3 counterparts. APOE4 is known to form smaller lipoprotein complexes with reduced affinity for Aβ, as well as for APOE receptors, compared to APOE3 (Kim et al., 2009). Combined with reduced astrocyte APOE4 protein expression, these factors would be predicted to strongly impair Aβ clearance by astrocytes and other cell types. However, the role of APOE itself in Aβ clearance remains contentious; some in vivo studies have shown that APOE is required for Aβ clearance, while others have observed increased extracellular Aβ clearance and attenuation of tauopathy in the absence of APOE (Holtzman et al., 1999; Koistinaho et al., 2004; Liao et al., 2014a; Shi et al., 2017). Further work will be required to clarify this issue, as well as to determine whether the deficit in Aβ uptake by APOE4 astrocytes is due solely to differences in levels and function of secreted APOE4 vs. APOE3 protein, or if unrelated deficits in cellular function are more important.
Impaired Aβ uptake and inflammatory alterations in APOE4 microglia-like cells
In support of cell-intrinsic APOE4-dependent deficits in Aβ uptake, we found that APOE4 microglia-like cells also exhibited reduced Aβ uptake relative to their APOE3 counterparts; this despite internalizing Aβ much more quickly than astrocytes (1hr for microglia-like cells in Figure 4C compared to 2 days for astrocytes in Figure 3H). We further found that APOE4 microglia-like cells exhibited altered morphologies compared to APOE3 cells. In 2D culture (without Aβ treatment), APOE4 microglia-like cells had fewer and shorter processes; while after embedding in 3D neuronal cultures (that produce Aβ), the same cells had longer processes than their APOE3 counterparts. These observations are consistent with impairment in the ability of APOE4 microglia-like cells to effectively sense and/or respond to Aβ in their environment. Given the importance of microglia surveillance and response functions to brain health, such impairment could have considerable detrimental effect for APOE4 carriers (Salter and Beggs, 2014).
Multiple studies show a positive relationship between up-regulation of immune-related genes and APOE4 genotype (Keren-Shaul et al., 2017; Mathys et al., 2017). Our study suggests that carrying the APOE4 variant could be sufficient to convert resting microglia into immune-active. This alteration likely associates with morphological changes and may underlie the observed alterations in Aβ uptake. It will be important to determine whether redirecting these cells from a “reactive” to a “homeostatic” state could restore their Aβ uptake ability.
Among the up-regulated immune genes we observed in APOE4 microglia-like cells was IRF8, which was shown to induce transcription of many immune-related genes and transform microglia into a reactive state (Masuda et al., 2012). Thus, we asked whether the immune related transcriptional changes in APOE4 microglia-like cells could be mediated via IRF8. Using available IRF8 ChIP-Seq data from mouse bone marrow (Olsson et al., 2016) we observed that up-regulated genes indeed had higher IRF8 signals, consistent with an important role for this protein in APOE4-related transcriptional changes (Figure S4E). Interestingly, we also observed that expression of TREM2 (and its signaling adaptor TYROBP), which is crucial for microglial function and is itself a highly significant AD risk gene (Ulrich and Holtzman, 2016), was positively correlated with APOE4 genotype (Figure 1F and S4F). This is consistent with recent studies showing increased levels of soluble TREM2 in cerebrospinal fluid of AD patients (Heslegrave et al., 2016); however, the mechanisms by which TREM2 and TYROBP are regulated by APOE variants will require further investigation.
Late onset of amyloid accumulation in APOE4 organoids compared to fAD organoids
Increased levels of Aβ and p-tau in APOE4 vs. ApoE3 organoids were apparent after six months of culture, which is relatively delayed compared to two months of culture for organoids with fAD mutations (Raja et al., 2016). We observed increased number of synapses and early endosomes in APOE4 neurons, which could contribute to elevated Aβ generation; however the increase of Aβ42 secretion by APOE4 vs. APOE3 neurons is much less than the three-fold increase reported in fAD neurons vs. controls (Paquet et al., 2016). Astrocyte abundance in organoids also increases with duration of culture (Quadrato et al., 2017), consistent with our ability to detect APOE protein at 6 month of culture but not at 3 months (Figure 4B). Thus, both moderate increases of Aβ secretion by neurons and impaired Aβ clearance by astrocytes could contribute to Aβ accumulation in APOE4 organoids at six months of age.
Amelioration of AD-related phenotypes by switching APOE4 to APOE3
To compliment our studies of APOE4-dependent defects in brain cell types, we edited the APOE4 allele of iPSCs derived from a sAD patient to APOE3. We were able to reverse most of the AD-associated phenotypes we examined by this APOE4 to APOE3 conversion, underscoring the critical importance of this genetic variant. However, we did not see a reduction in Aβ42 secretion from APOE3 vs, APOE4 sAD neurons (Figure 6D). Similarly, the reduction in early endosome numbers in APOE3 sAD neurons was not statistically significant although there was a trend compared to APOE4 sAD neurons (Figure S6E). It is likely that additional factors in this sAD iPSC-derived cell line confound the AD-related pathologies we observe.
The APOE2 allele, which has been suggested to have a protective effect on the onset of AD, is also of great interest. However, compared to the APOE4 variant, our knowledge of the cellular and molecular alterations associated with APOE2 are quite limited. As such, it is not clear whether the effects of the APOE2 allele would be completely opposite to those of APOE4 on the examined cell types and organoids. Investigating the nature of protective effects of APOE2 on AD pathology using the human iPSC model systems warrants further study.
In summary, our study suggests that APOE4 impacts AD pathology mainly through impairment of astrocyte- and microglia-mediated Aβ clearance. Moreover, there are notable abnormalities associated with lipid metabolism in APOE4 astrocytes and potentially microglia. In this regard, targeting glia-mediated Aβ clearance and lipid biogenesis/metabolism could be potential targets for therapeutic intervention. Furthermore, determining whether APOE4 mutation results in loss of functions or gain of toxic functions will be critical to tailor appropriate therapeutic interventions for APOE4 carriers. Our human APOE isogenic iPSCs offer a powerful set of useful tools to address these important questions.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Aβ (D54D2) | Cell Signaling Technology | Cat#8243 |
| Rabbit monoclonal anti-APOE (clone EP1374Y) | Abcam | Cat#AB52607, RRID: AB_2258476 |
| Mouse monoclonal anti-p-tau (clone AT-8) | Thermo Fisher Scientific | Cat#MN1020, RRID: AB_223647 |
| Rat monoclonal anti-CD11b-APC | Miltenyi Biotec | Cat#130-091-241, RRID: AB_244268 |
| Mouse monoclonal anti-EEA1 | BD Transduction Laboratiories | Cat#610457, RRID: AB_397830 |
| Mouse monoclonal anti-GAPDH (clone 6C5) | Santa Cruz Biotechnology | Cat#SC-32233, RRID: AB_627679 |
| Rabbit polyclonal anti-GFAP | Millipore | Cat#AB5804, RRID: AB_2109645 |
| Mouse monoclonal anti-GLAST-PE | Mitenyi Biotec | Cat#130-098-804, RRID: AB_2660782 |
| Rabbit monoclonal anti-Iba1 | Wako Chemicals | Cat#019-19471, RRID: AB_2665520 |
| Goat polyclonal anti-Iba1 | Novus Biologicals | Cat# NB 100-1028, RRID:AB_521594 |
| Chichen polyclonal anti-MAP2 | BioLegend | Cat#822501, RRID: AB_2564858 |
| Rabbit polyclonal anti-Nestin | Millipore | Cat#AB91107, RRID: AB_91107 |
| Rabbit polyclonal anti-PSD-95 | Abcam | Cat#AB18258, RRID: AB_444362 |
| Rabbit polyclonal anti-PSD-95 | Cell Signaling Technology | Cat# 3450S, RRID:AB_2292883 |
| Mouse monoclonal anti-S100β | Sigma-Aldrich | Cat#AMAB91038, RRID: AB_2665776 |
| Goat polyclonal anti-SOX2 (clone Y-17) | Santa Cruz Biotechnology | Cat#SC-17320, RRID: AB_2286684 |
| Mouse monoclonal anti-Synaptophysin (clone SVP-38) | Sigma-Aldrich | Cat#S5768, RRID: AB_477523 |
| Mouse polyclonal anti-TRA-1-60 | Stemgent | Cat#09-0068, RRID: AB_2233143 |
| Rabbit monoclonal anti-TREM2 (clone D814C) | Cell Signaling Technology | Cat#91068, RRID: AB_2721119 |
| Rabbit monoclonal anti-IDE | Abcam | Cat#AB133561 |
| Mouse monoclonal anti-β-actin | Sigma-Aldrich | Cat# A5441, RRID:AB_476744 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Apolipoprotein E4 human | Sigma-Aldrich | Cat#A3234 |
| Apolipoprotein E3 human | Sigma-Aldrich | Cat#SRP4696 |
| Beta-Amyloid (1-42), Human | Anaspec | Cat#AS-20276 |
| Beta-Amyloid (1-42) HiLyte Fluor555-labeled, Human | Anaspec | Cat#AS-60480-01 |
| Leupeptin | Sigma-Aldrich | Cat#L2884 |
| Chloroquine diphosphate salt | Sigma-Aldrich | Cat#C6628 |
| Ammonium chloride | Sigma-Aldrich | Cat#A9434 |
| Critical Commercial Assays | ||
| Amyloid beta 40 Human ELISA Kit | Thermo Fisher Scientific | Cat#KHB3482 |
| Amyloid beta 42 Human ELISA Kit | Thermo Fisher Scientific | Cat#KHB3442 |
| Cholesterol Assay Kit (cell-based) | Abcam | Cat#AB133116 |
| Cholesterol Assay Kit - HDL and LDL/VLDL | Abcam | Cat#AB65390 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat#G7570 |
| Deposited Data | ||
| Raw data lifes for RNA-seq | This paper | NCBI GEO: GSE102956 |
| Experimental Models: Cell Lines | ||
| Human induced-pluripotent stem cell line_AG09173 | Coriell | Cat#AG09173 |
| Human induced-pluripotent stem cell line_AG10788 | Coriell | Cat#AG10788 |
| Oligonucleotides | ||
|
APOE sgRNA oligomer pair (Forward): 5′- CACCGCCTCGCCGCGGTACTGCACC-3′ |
This paper | N/A |
|
APOE sgRNA oligomer pair (Reverse): 5′- AAACGGTGCAGTACCGCGGCGAGGC-3′ |
This paper | N/A |
| ssODN for APOE3 -> APOE4: 5′- GAGGAGACGCGGGCACGGCTGTCCAAGGAGCTGC AGGCGGCGCAGGCCCGGCTGGGCGCGGACATGGAG GACGTGCGCGGCCGGCTGGTGCAGTACCGCGGCGA GGTGCAGGCCATGCTCGGCCAGAGCACCGAGGAGC TGCGGGTGCGCCTCGCCTCCCACCTGCGCAAGCTG CGTAAG-3-3′ |
This paper | N/A |
| ssODN for APOE4 -> APOE3: 5′- GAGGAGACGCGGGCACGGCTGTCCAAGGAGCTGC AGGCGGCGCAGGCCCGGCTGGGCGCGGACATGGAG GACGTGTGCGGCCGGCTGGTGCAGTACCGCGGCGA GGTGCAGGCCATGCTCGGCCAGAGCACCGAGGAGC TGCGGGTGCGCCTCGCCTCCCACCTGCGCAAGCTG CGTAAG -3′ |
This paper | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | Cat#48138 |
| FUdeltaGW-rtTA | Addgene | Cat#19780 |
| pLV-TetO-hNGN2-eGFP-Puro | Sudhof lab | |
| pRSV-Rev | Addgene | Cat#12253 |
| pCMV-VSV-G | Addgene | Cat#8545 |
| Software and Algorithms | ||
| ImageJ | NIH | http://imagej.nih.gov/ij/index.html |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| IMARIS | Bitplane | https://www.bitplane.com/imaris |
| STAR | Gingeras Lab | http://labshare.cshl.edu/shares/gingeraslab/www-data/dobin/STAR |
| CUFFLINKS | Trapnell Lab | http://cole-trapnell-lab.github.io/cufflinks/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Li-Huei Tsai (lhtsai@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human induced-pluripotent stem cells
Unaffected (Coriell #AG09173, female, Age 75) and sporadic AD (Coriell #AG10788, female, Age 87) iPSC lines have been fully characterized and the APOE genotype was confirmed by sequencing. iPSCs were cultured on irradiated mouse embryonic fibroblasts (MEFs, MTI-GlobalStem) in human ES (hES) media [DMEM/F12, HEPES media (Gibco) supplemented with 20% knockout serum replacement (KSR) (Gibco), 1X non-essential amino acids (NEAA), 1X GlutaMAX, (Life Technologies), 12 nM beta-fibroblast growth factor (FGF2, PeproTech) and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich)]. iPSCs were maintained at 37 °C and 5% CO2 in a humidified incubator.
METHOD DETAILS
Generation of isogenic iPSC lines
Preparation of the CRISPR/Cas9-ApoE sgRNA plasmid
We prepared CRISPR/Cas9-APOE sgRNA plasmid following published protocol published (Ran et al., 2013). Briefly, we designed a sgRNA sequence (5′-CCTCGCCGCGGTACTGCACC-3′) within 10 nucleotides from the target site corresponding to amino acid 112 using the CRISPR Design tool (http://crispr.mit.edu). We annealed the oligomer pairs (5′-CACCGCCTCGCCGCGGTACTGCACC-3′ and 5′-AAACGGTGCAGTACCGCGGCGAGGC-3′) and cloned them into pSpCas9-2A-GFP (PX458) plasmid (Addgene #48138). Plasmid DNA was submitted for Sanger sequencing to confirm correct APOE sgRNA sequence. We also designed single-strand oligodeoxynucleotides (ssODN) to convert APOE4 to APOE3, or APOE3 to APOE4 with a silent mutation at the protospacer adjacent motif (PAM) site to prevent recurrent Cas9 cutting in edited cells.: APOE3 -> APOE4: GAGGAGACGCGGGCACGGCTGTCCAAGGAGCTGCAGGCGGCGCAGGCCCGGCTGGGCGCGGACATGGAGGACGTGCGCGGCCGGCTGGTGCAGTACCGCGGCGAGGTGCAGGCCATGCTCGGCCAGAGCACCGAGGAGCTGCGGGTGCGCCTCGCCTCCCACCTGCGCAAGCTGCGTAAG; APOE4 -> APOE3:GAGGAGACGCGGGCACGGCTGTCCAAGGAGCTGCAGGCGGCGCAGGCCCGGCTGGGCGCGGACATGGAGGACGTGTGCGGCCGGCTGGTGCAGTACCGCGGCGAGGTGCAGGCCATGCTCGGCCAGAGCACCGAGGAGCTGCGGGTGCGCCTCGCCTCCCACCTGCGCAAGCTGCGTAAG.
Electroporation
iPSCs with 70 to 80% confluence were dissociated by treating with accutase (Thermo Fisher Scientific) and 10 μM ROCK inhibitor (Tocris) for 20 min. After spinning down iPSCs at 160 × g for 3 min, the number of cells was counted and 5 million cells were subjected to electroporation. Electroporation was performed using Nucleofector – Amaxa and Human Stem Cell Nucleofector Kit 1(Lonza) according to the manufacturer’s instructions. In brief, cells were resuspended in 100 μl of reaction buffer from the kit and 7.5 μg of CRISPR/Cas9-APOE sgRNA plasmid and 15 μg of ssODN were added to cell suspension. After electroporation with A-23 protocol, we resuspended cells with hES media with 10 μM ROCK inhibitor and seeded them onto plates containing MEFs.
Fluorescence-activated cell sorting (FACS)
Two days after electroporation, we performed FACS to isolate GFP-expressing iPSCs. Briefly, dissociated cells using accutase/ROCK inhibitor for 20 min were washed and resuspended with DPBS. Then, cells were filtered using Falcon polystyrene test tubes (#352235, Corning) and transferred to Falcon polypropylene test tubes (#352063, Corning). Sorting was performed by BD FACSAria IIU in FACS Facility at the Whitehead Institute. Sorted cells were suspended in recovery medium [hES medium with 1X Penicillin-Streptomycin (P/S) (Gemini Bio-products) and ROCK inhibitor] and plated onto 6-well plates containing MEFs (50K cells/well).
Colony inspection
Each colony was transferred to one well of 12-well plate coated with MEFs and maintained until the colony grew big enough to be transferred to another 12-well plate. After the second transfer, iPSCs in the original plate were dissociated and genomic DNA was extracted. Primers (5′-CTGGAGGAACAACTGACCCC-3′ and 5′-CTCGAACCAGCTCTTGAGGC-3′) were used to amplify DNA in APOE gene and PCR products were submitted to GENEWIZ for Sanger sequencing.
Karyotyping
In order to identify and evaluate the size, shape, and number of chromosomes in iPSCs, we performed karyotyping after colony inspection. iPSCs were cultured on hESC-qualified matrigel (Corning) coated T25 cell culture flasks in mTeSR1 media (STEMCELL Technologies) until 60% confluence, and then sent to Cell Line Genetics for karyotyping.
Differentiation of iPSCs into neurons
We generated excitatory neurons from iPSCs according to published protocol (Zhang et al., 2013). Briefly, iPSCs were prepared on matrigel coated 6-well plates at a density of 4×106 cells/well. Two hours later, lentivirus with rtTA and Ngn2-GFP expression vectors were suspended in mTeSR™1 media with 2 μM thiazovivin (Tocris) and added to the iPSCs plates. After 24 hr, the culture medium was fully changed with N2(1X)/DMEM/F12/NEAA(1X) (Thermo Fisher Scientific) medium containing 10 ng/ml BDNF (Peprotech), 10 ng/ml NT-3 (Peprotech), 0.2 μg/ml laminin (Corning), and 2 μg/ml doxycycline (Sigma-Aldrich) to induce TetO gene expression. The following day, old media was removed and replaced with Neurobasal/B27(1X)/GlutaMAX(1X) (Thermo Fisher Scientific) medium containing 10 ng/ml BDNF, 10 ng/ml NT-3, 0.2 μg/ml laminin, 2 μg/ml doxycycline and 1 μg/ml puromycin (Millipore). Two days after puromycine treatment, Ngn2-GFP expressing cells were purified by FACS and plated onto matrigel-coated plates containing human astrocyte (ScienCell, #1850) in conditioned medium containing 10 ng/ml BDNF, 10 ng/ml NT-3, 0.2 μg/ml laminin, 2 μg/ml doxycycline, and 1 μg/ml puromycin. The next day, 1 μM Ara-C (Sigma-Aldrich) was added to cultured media. Three days after Ara-C treatment, media was removed and replaced with astrocyte conditioned medium containing 10 ng/ml BDNF, 10 ng/ml NT-3, 0.2 μg/ml laminin and 0.5 μg/ml doxycycline. One half plate volume of media was replaced with new media every 4 days until the cells were ready for experimental assays.
Differentiation of iPSCs into astrocytes
iPSCs were cultured in 6-well plate coated with hESC-qualified matrigel (Corning) in mTeSR™1 media until cells had reached ~100% confluence. Then, cells were washed with DPBS and 2 ml of neural induction media [1:1 mixture of N-2 (DMEM/F-12 GlutaMAX, 1X N-2, 5 μg/ml insulin, 1 X NEAA, 100 μM 2-mercaptoethanol, and 1X P/S) and B-27 (Neurobasal medium, 1X B-27, 1X GlutaMAX, and 1X P/S media)] were added. Cells were treated with 1 μM Dorsomorphin and 10 μM SB431542 for 12 days, then passaged to new matrigel coated plates using accutase. Cells were cultured in neural maintenance media until the neural rosette structures became visible under the microscope (around days 16–24). Then, cells were passaged again with accutase and seeded at 4×106 cells/well density into matrigel coated 6-well plate. The day after passage, 20 ng/ml FGF2 (Peprotech) and 10 ng/ml BMP4 (Peprotech) were added into neural maintenance media. Media was changed every other day for 28 days and FACS were performed using GLAST antibody (Miltenyi Biotec) to purify astrocytes. After sorting, cells were seeded at 2.5×105 cells/well density into 6-well plate and cultured in astrocyte media (Sciencell) every other day for 7 days. After 7 days, we performed FACS with GLAST antibody again to increase the purity of astrocyte cultures. These cells were used for all experiments.
Generation of induced microglia-like cells
We derived microglia-like cells from iPSCs as previously described (Muffat et al., 2016). Briefly, iPSCs on MEFs-coated plates were dissociated with Collagenase IV (Thermo Fisher Scientific). iPSCs were then resuspended in MGD media [Neurobasal media supplemented with 0.5X Gem21 (Gemini Bio-products), 0.5X Neuroplex N2 (Gemini Bio-products), 0.2% Albumax I (Thermo Fisher Scientific), 5 mM sodium chloride (Sigma-Aldrich), 1X sodium pyruvate, 1X P/S, 1X GlutaMAX, 3.5 ng/ml biotin (Sigma-Aldrich), 10 μM ascorbic acid (Sigma-Aldrich) and 1.7% lactic syrup (Sigma-Aldrich)] with 10 ng/ml IL-34 (Peprotech) and 10 ng/ml M-CSF1(Peprotech)], and cultured in ultra-low attachment 6-well plates (Corning). Once the phase-bright neutralized spheroids and cystic bodies appeared, Embryoid bodies were gently triturated to shear off cells of interest, and supernatants were transferred to a single well of Primaria 6-well plate (Corning). Attached cells showed morphological characteristics of microglia and microglia precursors. Cells from 6 consecutive productions were pooled and purified by FACS using CD11b antibody (Miltenyi Biotec). Collected cells were further maintained in MGD media with 100 ng/ml IL-34 and 5 ng/ml M-CSF for all experiments.
Organoids culture
We derived organoids according to our published protocol (Raja et al., 2016). Briefly, iPSCs were maintained on MEFs until 80% confluence and dissociated using accutase. Then, cells were plated onto 0.1% gelatin (Millipore) coated 10 cm dish (Falcon) for 45 min and non-adherent iPSCs were collected and seeded at a density of 1.2×104 cells/well into V-shape 96-well plates (VWR) pre-coated with 1% pluronic acid (Sigma-Aldrich). Cells were maintained for 18 days in media consisted of Glasgow-MEM (Thermo Fisher Scientific) with 20% KSR, 1X sodium pyruvate, 1X NEAA, 0.1 mM 2-mercaptoethanol, 20 μM Rock inhibitor, 5 μM SB431532 (Tocris), and 3 μM IWRe1 (Tocris). Dorsomorphin (2 μM, Tocris) was added to media after the first three days. Media was changed every other day for 18 days and then organoids were transferred to non-adherent EZsphere dishes (Nacalai USA). At day 18, organoids were cultured in media with DMEM/F12 supplemented with 1X Chemically Defined Lipid Concentrate (Thermo Fisher Scientific) and 1X N2-supplement in an incubator (5% CO2 and 40% O2). At day 35, 5 μM heparin (Sigma-Aldrich), 10% FBS (Gemini Bio-products) and 1% matrigel (Corning) were added to media until day 70. At day 70, the concentration of matrigel was increased to 2% and 1X B27 supplement was added for the remainder of the culture period. Media in the dishes was changed every 4 to 5 days.
Bioinformatics
Genomic variant analysis for genome-edited iPSCs
Exome-seq data (76-bp paired-end) were generated at the sequencing core facility of the Broad Institute of MIT & Harvard. Our data processing pipeline was established based on “GATK Best Practices” recommended by Broad GATK team. Briefly, the raw fastq files were first mapped to human hg19 assembly using BWA mapper (version 7, mem option) (Li and Durbin, 2010); PCR duplicates were removed using MarkDuplicates function of Picard software package (http://broadinstitute.github.io/picard/). RealignerTargetCreator, IndelRealigner and BaseRecalibrator modules of GATK tools (McKenna et al., 2010) were further employed to perform local realignment and recalibration. HaplotypeCaller of GATK tools was then used to call variants in exonic regions with stand_emit_conf of 10 and stand_call_conf of 30. SNP and INDEL variants were selected using SelectVariants of GATK tools. Genomic variants from genome-edited iPSCs were compared to the variants from their parental lines to identify unique variants to genome-edited iPSCs. Further removal of variants overlapping with repeatmasker regions and variants with low DP and QUAL scores were carried out before functional annotation of exonic variants using ANNOVAR package (Wang et al., 2010). All potential unique variants identified from the above variant calling pipeline were finally examined manually by overlaying bam traces of genome-edited iPSCs with their parental lines in IGV browser (Robinson et al., 2011).
RNA-seq analysis of iPSC-derived cell lines
Extracted total RNA was subject to QC using an Advanced Analytical-fragment Analyzer before library preparation using Illumina Neoprep stranded RNA-seq library preparation kit. Libraries were pooled for sequencing using Illumina HiSeq2000 or NextSeq500 platforms at the MIT Biomicro Center. The raw fastq data were aligned to human hg19 assembly using STAR 2.4.0 RNA-seq aligner (Dobin et al., 2013). Mapped RNA-seq reads covering the edited APOE3/4 site were used to validate data genotypes. Gene raw counts were generated from the mapped data using featureCounts tool (Liao et al., 2014b). The mapped reads were also processed by Cufflinks2.2 (Trapnell et al., 2012) with hg19 reference gene annotation to estimate transcript abundances. Gene differential expression test between APOE3 and APOE4 groups of each cell type was performed using Cuffdiff module with adjusted q-value <0.05 for statistical significance. Geometric method was chosen as the library normalization method for Cuffdiff. Color-coded scatter plots were used to visualize group FPKM values for differentially expressed genes and other genes. Gene raw count matrix was processed by edgeR package (Robinson et al., 2010) to generate logCPM values (CPM – counts per million) for data clustering analysis. Phylogenetic tree was constructed based on Euclidean distance of logCPM values of group average count values and ward.D2 option of hclust tool. Correlation heatmap was generated using replicate logCPM matrix combed with data matrix of publicly available iPSC-derived neuron/microglia RNA-seq data (Busskamp et al., 2014; Muffat et al., 2016) and human fetal brain astrocyte RNA-seq data (Crowe et al., 2016). In the correlation heatmap, the correlation matrix was reordered based on average linkage hierarchical clustering with rectangular boxes to visualize sample clusters. Z-scores of replicate expression FPKM values for differentially expressed genes were visualized in heatmaps for different cell types. Gene ontology was performed using Broad GSEA tool (Subramanian et al., 2005) and TOPPGENE tool (Chen et al., 2009).
Gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) was applied to explore gene co-expression pattern in iPSC-derived brain cell types. Briefly, raw count data from 26 RNA-seq samples were processed by edgeR to generate logCPM matrix before calculating a set of soft-thresholding powers for signed network construction. A power value of 10 (SFT.R.sq>0.8, approximately scale-free) was chosen to compute adjacency matrix for a signed co-expression network. The topological overlap matrix (TOM) was then calculated, and average linkage hierarchical clustering was used to group genes based on their topological overlap. A dynamic tree-cutting method was used (minClusterSize=100 and cutHeight=0.999) to create gene co-expression modules and the results were visualized in a dendrogram.
GWAS analysis of human brain exome-seq data and eQTL analysis of human brain RNA-seq data
Human brain (normal control and mild cognitive impairment subjects) exome-seq and RNA-seq data were collected from NIH-AMP/AD consortium. Those data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine. From 341 exome-seq data, a total of 117,132 SNPs and INDELs were reported within exome-seq covered regions. Further trait association was performed using Matrix_eQTL tool (Shabalin, 2012) to associate genomic variants with mean neocortical plaque density value (race and gender as covariates). The calculated p-values were visualized in a Manhattan plot. Variants with p-value less than 1×10−5 were subject to further functional annotation with ANNOAR. Normalized gene count matrix for 880 RNA-seq data generated from postmortem brain in four different areas (BM10, BM22, BM36, BM44) were filtered to get expression matrices for genes specifically expressed in neurons, microglia or astrocytes. Cell specific gene sets were defined based on Dr. Ben Barres’ human adult brain cell-type specific RNA-seq data (Zhang et al., 2016) with a cutoff of 2-fold higher expression in a particular cell type than any other cell type. Expression matrices for cell specific gene sets were then used to examine variation of gene expression associated with APOE4 homozygous mutation by Matrix_eQTL (calculation of effect size β and statistical p-value). In parallel, expression fold change and statistical p-value between APOE3 and APOE4 groups for cell specific genes in iPSC-derived corresponding cell types were collected for comparison with human brain data. Scatterplots of fold change vs. effect size for neuronal, microglial and astrocytic genes were generated with statistically significance (p<0.05 for both data) coded by different colors. Spearman’s correlation analysis of gene expression with pathological plaque density and clinical dementia rating (CDR) was performed using customized R scripts. Heatmaps were generated using combined information of correlation rho value and statistical p-value. Spearman’s rho value and p-value were calculated using Spearman’s rank correlation test, which assesses relationships of ordinal variables.
Aggregation plot of average Irf8 ChiP-Seq intensity signals around gene transcription start sites
With One set of Irf8 ChIP-Seq data (GSE70237) produced in mouse bone marrow (so the cell types could be considered as similar to our microglia-like cells, Olsson et al., 2016), we performed ChIP-Seq analysis using MACS2 software tool (Zhang et al., 2008) to generate fold enrichment of Irf8 against input across the entire mouse genome. The up and down-regulated genes (DEGs - differentially expressed genes) in APOE4 microglia-like cells compared to APOE3 microglia-like cells were converted to the mouse orthologs, and enriched Irf8 ChIP-Seq signals around the transcription start sites of DEGs were aggregated for visualization.
Immunoblot analysis
iPSC-derived brain cell types or organoids were collected in 1.5 ml tubes and washed three times with DPBS. The samples were then lysed with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitors. Lysates were spun at 13,000 rpm for 15 min and only supernatants were transferred to new tubes. Protein concentration of samples was measured (Bio-Rad Protein Assay) and equal amount of protein was loaded for electrophoresis.
Immunostaining analysis
Neurons and glial cells
After growing on glass coverslips, cells were washed three times with DPBS and fixed with 4% paraformaldehyde in PBS for 15 min. After fixation, cells were washed with DBPS and permeabilized with blocking solution containing 0.1% Triton X-100, 10% donkey serum, 2% BSA and 1 M glycine in PBS for 1 hr at room temperature. Then cells were incubated with appropriate antibodies overnight at 4 °C. The following day, cells were incubated with fluorescently conjugated secondary antibodies (Molecular Probes) and Hoechst 33342 (Invitrogen) for 1 hr at room temperature and washed three times with DPBS before mounting on the glass slide. For the quantification of synaptophysin and PSD-95 puncta in iPSC-derived neurons, IMARIS software (Bitplane) was used. The path of process was generated based on PSD-95 signals with the filaments module and puncta were marked based on the threshold we set (For synaptophysin, surface area detail level was set to 0.5 μM, and intensity threshold with background subtraction was set to 1.0 μM/for PSD-95, surface area detail levels to 0.25 μM, and intensity threshold with background subtraction to 0.5 μM). Marked puncta then digitalized for the quantification.
Organoids
After culturing in incubator for three months or six months, organoids were washed three times with DPBS and fixed with 4% paraformaldehyde in PBS for 15 min. After fixation, organoids were dehydrated with 30% sucrose in PBS at 4°C. Organoids were then embedded with Optical cutting temperature (OCT) compound (VWR) and frozen by dry ice. The frozen block with organoids were sectioned at −20 °C using cryosection with 20 μm thickness and transferred to the glass slide. Organoid samples on the glass slide were washed with DBPS and permeabilized with blocking solution containing 0.1% Triton X-100, 10% donkey serum, 2% BSA and 1 M glycine in PBS for 1 hr at room temperature. Then samples were incubated with appropriate antibodies overnight at 4 °C. Next day, samples were incubated with fluorescently conjugated secondary antibodies and Hoechst 33342 for 1 hr at room temperature and washed three times with PBS before mounting with the glass coverslip.
Electrophysiology
Six weeks old iPSC-derived neurons were subjected for whole cell recording to measure miniature excitatory postsynaptic currents (mEPSCs). The external solution consists of 130 mM NaCl, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES and 10 mM glucose (pH=7.4). TTX (1 μM) and picrotoxin (50 μM) were added to block spontaneous firing and GABA-mediated currents. EGFP-positive neurons were held at −70 mV with recording pipettes containing 145 mM CsCl mM, 5 NaCl mM, 10 HEPES-CsOH mM, 10 EGTA mM, 4 MgATP mM and 0.3 mM Na2GTP. Recording was performed using an Axoclamp 200B amplifier with a Digidata 1440A A-D converter (Axon Instruments), and all data were analyzed by the use of pClamp 10 software (Axon Instruments).
Synaptophysin puncta quantification in iPSC-derived neurons
Dendritic branches 40 μm away from cell body were subjected for data analysis. In branches, only synaptophysin puncta within 2 μm from PSD-95 signals were quantified.
Measurement of secreted Aβ levels in iPSC-derived neurons by ELISA
iPSC-derived neurons were prepared in 96-well plates. After six weeks of differentiation, media was fully changed and kept for two days. Then, media were collected and secreted Aβ levels were measured using human Aβ42 or Aβ40 ELISA kit.
Cholesterol assay
Cholesterol levels in iPSC-derived astrocytes or astrocyte cultured media were measured using cholesterol assay kits (Abcam) following the manufacturer’s instructions. For the measurement of cholesterol levels in astrocyte, cells were plated on glass coverslips. Astrocytes were washed three times with DPBS and fixed with 4% paraformaldehyde in PBS for 15 min. Cells were treated with filipin III for 1 hr and washed with DPBS. Fluorescence from cholesterol-bound filipin III was detected by microplate reader (EnSpire, Perkin Elmer), microscope or FACS. To measure secreted cholesterol levels in iPSC-derived astrocytes, cells were prepared in 24-well plates. The day after passaging, media were fully changed. Two days later, media were collected and secreted cholesterol levels were measured using an EnSpire plate reader (Perkin Elmer).
Microscopy
All images were captured using Zeiss LSM 710 confocal microscope and Zen software, and analyzed using ImageJ (National Institutes of Health) or IMARIS software.
Aβ42 uptake assay
iPSC-induced astrocytes
To prepare oligomerized Aβ42, Aβ42 peptide (AnaSpec) was dissolved in 1% NH4OH at 1 mg/ml and sonicated. Lyophilized Aβ42 was further dissolved in water, filtered and incubated at 37 °C for 1 day before use. iPSC-derived astrocytes were seeded in 24-well plates (3×104/well) for 2 days and incubated with oligomeric Aβ42 (250 ng/ml) for additional 2 days. Aβ42 oligomer was also added to media only (without cells) to measure total levels of Aβ42 2 days after culture. The levels of Aβ42 in cultured media 2 days after treatment were measured by human Aβ42 ELISA kit (Invitrogen) following the manufacturer’s instructions. Reduced levels of Aβ42 by iPSC-derived astrocytes were calculated by subtracting remaining Aβ42 from total Aβ42. Aβ42 uptake index was obtained from dividing reduced levels of Aβ42 by the number of cells measured by CellTiter-Glo cell viability assay (Promega).
Induced microglia-like cells
For imaging Aβ42 uptake by microglia-like cells, Aβ42 labeled with HiLyte Fluor-555 peptide (AnaSpec) was used. The peptide was dissolved in 1% NH4OH at 10 mg/ml and further diluted with PBS at 1 mg/ml. Microglia-like cells were seeded at (2×104/well) for 2 days before the experiment. Microglia-like cells were treated with Aβ42-555 (1 μg/ml) for 1 hr for live imaging. Time series experiments consisted of sixty 512×512 frames with an interval of 1 min. Image stacks were exported to ImageJ, and fluorescence intensities were measured.
Lysosomal inhibitors treatment in iPSC-derived astrocytes
iPSC-derived astrocytes were prepared in 6-well plates. The day after passaging, media was fully changed and incubated with lysosomal inhibitors [leupeptin (50 μM), chloroquine (50 μM) and ammonium chloride (10 mM) for 48 hr. Cells were washed with DPBS three times and lysed with RIPA buffer containing protease and phosphatase inhibitors. Lysates were spun at 13,000 rpm for 15 min and only supernatants were transferred to new tubes.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Statistical analyses were performed using Prism 6 (GraphPad Software). ANOVAs followed by Tukey’s test, Dunnett’s test or unpaired Student’s t-tests were used. All data are represented as mean ± s.e.m.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE102956.
Supplementary Material
The uptake of fluorescently-tagged Aβ42 (Aβ42-555) by microglia-like cells was monitored for 1 hr.
The uptake of fluorescently-tagged Aβ42 (Aβ42-555) by microglia-like cells was monitored for 1 hr.
The number of down-regulated and up-regulated genes in APOE4 carrying iPSCs, iPSC-derived neurons/astrocytes/microglia-like cells compared to APOE3 cells.
List of genes which expression patterns are similar to APOE
Gene ontology analysis was performed with genes in the same modules.
miRNA motif analysis was performed with genes whose expression was affected by the APOE4 alleles.
90 days old APPDP organoids (co-cultured with APOE3-carrying microglia-like cells for 30 days) were stained for Aβ along with microglia marker Iba1 and Hoechst for nuclear staining. The images were converted into movie using the IMARIS software.
90 days old APPDP organoids (co-cultured with APOE4-carrying microglia-like cells for 30 days) were stained for Aβ along with microglia marker Iba1 and Hoechst for nuclear staining. The images were converted into movie using the IMARIS software.
Acknowledgments
The authors would like to dedicate this study to Susan L. Lindquist who provided invaluable advices and support to this work. We thank Drs. Priyanka Narayan and Jennie Z. Young for helpful comments on the manuscript, and members of Tsai lab, Lindquist lab and Neurodgeneration Consortium for fruitful advice and discussion. Dr. Thomas Südhof for kindly providing pLV-TetO-hNGN2-eGFP-Puro plasmid. This work is supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A6A3A03055359) to J.S, the National Institutes of Health (NIH) grants RF1 AG048056 and RC1 AG036106, a grant from the Glenn Foundation for Medical Research to B.A.Y., and NIH grant RF1 AG048029, the Robert A. and Renee E. Belfer Family Foundation, and Cure Alzheimer’s Fund to L.-H.T.
Footnotes
Declaration of interests
The authors declare no competing interests.
Author contributions
Conceptualization, Y.-T.L., J.S., and L.-H.T.; Methodology, Y.-T.L., J.S., F.G., H.M.F., E.G., W.K.R., J.P. and H.P.C.; Investigation, Y.-T.L., J.S., F.G., H.-L.W., J.C., R.R., O.K., F.A., Z.P., B.M, C.J.Y., S.E. and D.D.; Resource, F.G., T.K., B.A.Y and L.-H.T.; Writing-Original Draft, J.S. and H.P.C.; Writing-Review & Editing, Y.-T.L., J.S., F.G., J.P., H.P.C. and L.-H.T.; Visualization, J.S. and F.G.; Supervision, L.-H.T.; Funding Acquisition, J.S., B.A.Y. and L.-H.T.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alzheimer’s Association. Alzheimer’s disease facts and Figures. Alzheimer’s & Dementia. 2016;12(4) doi: 10.1016/j.jalz.2016.03.001. http://www.alz.org. [DOI] [PubMed] [Google Scholar]
- Alvarez XA, Alvarez I, Iglesias O, Crespo I, Figueroa J, Aleixandre M, Linares C, Granizo E, Garcia-Fantini M, Marey J, et al. Synergistic Increase of Serum BDNF in Alzheimer Patients Treated with Cerebrolysin and Donepezil: Association with Cognitive Improvement in ApoE4 Cases. Int J Neuropsychopharmacol. 2016 doi: 10.1093/ijnp/pyw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME, Lee JM, Holtzman DM. Neuronal activity regulates the regional vulnerability to amyloid-β deposition. Nat Neurosci. 2011;14:750–756. doi: 10.1038/nn.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busskamp V, Lewis NE, Guye P, Ng AHM, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol. 2014;10:760. doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcoen D, Elias L, Yu X. What does it take to produce a breakthrough drug? Nat Rev Drug Discov. 2015;14:161–162. doi: 10.1038/nrd4570. [DOI] [PubMed] [Google Scholar]
- Canter RG, Penney J, Tsai LH. The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature. 2016;539:187–196. doi: 10.1038/nature20412. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jiang P, Xue H, Peterson SE, Tran HT, McCann AE, Parast MM, Li S, Pleasure DE, Laurent LC, et al. Role of astroglia in Down’s syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat Commun. 2014;5:4430. doi: 10.1038/ncomms5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Vetrivel KS, Gong P, Meckler X, Parent A, Thinakaran G. Mechanisms of disease: new therapeutic strategies for Alzheimer’s disease--targeting APP processing in lipid rafts. Nat Clin Pract Neurol. 2007;3:374–382. doi: 10.1038/ncpneuro0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Crowe EP, Tuzer F, Gregory BD, Donahue G, Gosai SJ, Cohen J, Leung YY, Yetkin E, Nativio R, Wang LS, et al. Changes in the Transcriptome of Human Astrocytes Accompanying Oxidative Stress-Induced Senescence. Front Aging Neurosci. 2016;8:208. doi: 10.3389/fnagi.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Scott DA, Ganguly A, Koo EH, Tang Y, Roy S. Activity-Induced Convergence of APP and BACE-1 in Acidic Microdomains via an Endocytosis-Dependent Pathway. Neuron. 2013;79:447–460. doi: 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou AG, Goate AM. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol Neurodegener. 2017;12 doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Liu B, Frost JL, Hong S, Jin M, Ostaszewski B, Shankar GM, Costantino IM, Carroll MC, Mayadas TN, et al. Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Aβ by microglia. Glia. 2012;60:993–1003. doi: 10.1002/glia.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Göritz C, Mauch DH, Nägler K, Pfrieger FW. Role of glia-derived cholesterol in synaptogenesis: new revelations in the synapseglia affair. J Physiol Paris. 2002;96:257–263. doi: 10.1016/s0928-4257(02)00014-1. [DOI] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:6344. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslegrave A, Heywood W, Paterson R, Magdalinou N, Svensson J, Johansson P, Öhrfelt A, Blennow K, Hardy J, Schott J, et al. Increased cerebrospinal fluid soluble TREM2 concentration in Alzheimer’s disease. Mol Neurodegener. 2016;11:3. doi: 10.1186/s13024-016-0071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Wu S, Bhat P, Parsadanian M, Fagan AM, Chang LK, Sun Y, Paul SM. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. J Clin Invest. 1999;103:R15–R21. doi: 10.1172/JCI6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YWA, Zhou B, Wernig M, Südhof TC. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell. 2017b;168:427–441. e21. doi: 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature. 2016;540:230–235. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290. e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, Higgs R, Liu F, Malkani S, Bales KR, et al. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat Med. 2004;10:719–726. doi: 10.1038/nm1058. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Hori Y, Hudry E, Bauer AQ, Jiang H, Mahan TE, Lefton KB, Zhang TJ, Dearborn JT, Kim J, et al. Anti-ApoE antibody given after plaque onset decreases Aβ accumulation and improves brain function in a mouse model of Aβ amyloidosis. J Neurosci. 2014a;34:7281–7292. doi: 10.1523/JNEUROSCI.0646-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014b;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Jiao SS, Wang YR, Bu XL, Yao XQ, Xiang Y, Wang QH, Wang L, Deng J, Li J, et al. Associations Between ApoEε4 Carrier Status and Serum BDNF Levels--New Insights into the Molecular Mechanism of ApoEε4 Actions in Alzheimer’s Disease. Mol Neurobiol. 2015;51:1271–1277. doi: 10.1007/s12035-014-8804-8. [DOI] [PubMed] [Google Scholar]
- Marquer C, Laine J, Dauphinot L, Hanbouch L, Lemercier-Neuillet C, Pierrot N, Bossers K, Le M, Corlier F, Benstaali C, et al. Increasing membrane cholesterol of neurons in culture recapitulates Alzheimer’s disease early phenotypes. Mol Neurodegener. 2014;9:60. doi: 10.1186/1750-1326-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Tsuda M, Yoshinaga R, Tozaki-Saitoh H, Ozato K, Tamura T, Inoue K. IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype. Cell Rep. 2012;1:334–340. doi: 10.1016/j.celrep.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, Tsai LH. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21:366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch DH, Nägler K, Schumacher S, Göritz C, Müller EC, Otto A, Pfrieger FW. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooijaart SP, Berbée JFP, van Heemst D, Havekes LM, de Craen AJM, Slagboom PE, Rensen PCN, Westendorp RGJ. ApoE plasma levels and risk of cardiovascular mortality in old age. PLoS Med. 2006;3:e176. doi: 10.1371/journal.pmed.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One. 2016;11:e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, Beggs S. Sublime microglia: expanding roles for the guardians of the CNS. Cell. 2014;158:15–24. doi: 10.1016/j.cell.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Sarius H, Heneka MT. Microglia in Alzheimer’s disease. J Clin Invest. 2017;127:3240–3249. doi: 10.1172/JCI90606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, O’Brien SP, Iannotti E, Medley QG, Streuli M. Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J Cell Sci. 2001;114:389–399. doi: 10.1242/jcs.114.2.389. [DOI] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Sobe T, Abu Sa’ed J, Abu Rayan A, Lynch ED, Lee MK, Avraham KB, King MC, Kanaan M. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet. 2006;78:144–152. doi: 10.1086/499495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoryk-Wegrzynowicz M, Wegrzynowicz M, Lee E, Bowman AB, Aschner M. Role of astrocytes in brain function and disease. Toxicol Pathol. 2011;39:115–123. doi: 10.1177/0192623310385254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USa. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USa. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theendakara V, Peters-Libeu CA, Spilman P, Poksay KS, Bredesen DE, Rao RV. Direct Transcriptional Effects of Apolipoprotein E. J Neurosci. 2016;36:685–700. doi: 10.1523/JNEUROSCI.3562-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh WH, Gleeson PA. Dysregulation of intracellular trafficking and endosomal sorting in Alzheimer’s disease: controversies and unanswered questions. Biochem J. 2016;473:1977–1993. doi: 10.1042/BCJ20160147. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JD, Holtzman DM. TREM2 Function in Alzheimer’s Disease and Neurodegeneration. ACS Chem Neurosci. 2016;7:420–427. doi: 10.1021/acschemneuro.5b00313. [DOI] [PubMed] [Google Scholar]
- Wang C, Najm R, Xu Q, Jeong DE, Walker D, Balestra ME, Yoon SY, Yuan H, Li G, Miller ZA, et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small molecule structure corrector. Nat Med. 2018 doi: 10.1038/s41591-018-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med. 2003;9:453–457. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006;26:4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fang Y, Lian Y, Chen Y, Wu T, Zheng Y, Zong H, Sun L, Zhang R, Wang Z, et al. Brain-derived neurotrophic factor ameliorates learning deficits in a rat model of Alzheimer’s disease induced by aβ1-42. PLoS One. 2015;10:e0122415. doi: 10.1371/journal.pone.0122415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChiP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MSB, Li G, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Davis MD, Martens YA, Shinohara M, Graff-Radford NR, Younkin SG, Wszolek ZK, Kanekiyo T, Bu G. APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum Mol Genet. 2017;26:2690–2700. doi: 10.1093/hmg/ddx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The uptake of fluorescently-tagged Aβ42 (Aβ42-555) by microglia-like cells was monitored for 1 hr.
The uptake of fluorescently-tagged Aβ42 (Aβ42-555) by microglia-like cells was monitored for 1 hr.
The number of down-regulated and up-regulated genes in APOE4 carrying iPSCs, iPSC-derived neurons/astrocytes/microglia-like cells compared to APOE3 cells.
List of genes which expression patterns are similar to APOE
Gene ontology analysis was performed with genes in the same modules.
miRNA motif analysis was performed with genes whose expression was affected by the APOE4 alleles.
90 days old APPDP organoids (co-cultured with APOE3-carrying microglia-like cells for 30 days) were stained for Aβ along with microglia marker Iba1 and Hoechst for nuclear staining. The images were converted into movie using the IMARIS software.
90 days old APPDP organoids (co-cultured with APOE4-carrying microglia-like cells for 30 days) were stained for Aβ along with microglia marker Iba1 and Hoechst for nuclear staining. The images were converted into movie using the IMARIS software.