Abstract
Inflammation is a complex biological host reaction to tissue damage, infection and trauma. Extensive study of the inflammatory response has led to the identification of several protein kinases that are essential for signaling and could be potential therapeutic targets. The RSK family of kinases has multiple cellular functions. In our study, we found that RSK2 is a mediator for inflammation signaling and interacts with TRAF6. In vitro kinase assay results indicated that RSK2 strongly phosphorylates TRAF6 at serines 46, 47 and 48. Ectopic over-expression of TRAF6 or knocking down RSK2 expression confirmed that RSK2 is a positive regulator of TRAF6 K63 ubiquitination. TRAF6 is also required for RSK2 ubiquitination. TRAF6 bridges the TNF receptor superfamily and intracellular signaling for the induction of pro-inflammatory cytokines. We developed a colon inflammation model using RSK2 wild type (WT) and knockout (KO) mice. As expected, F4/80 and CD3 infiltration were significantly upregulated in WT mice compared to RSK2 KO mice. Furthermore, inflammation signaling, including Ikkα/β, p38 and JNKs, was dramatically up-regulated in WT mice. Colon tissue immunoprecipitation results further confirmed that TRAF6 K63 ubiquitination was lower in RSK2 KO mice. Overall, these results indicate that phosphorylation of TRAF6 (S46, 47, 48) by RSK2 is required for TRAF6 K63 ubiquitination and inflammation signaling.
Keywords: RSK2, TRAF6, ubiquitination, inflammation
Introduction
The inflammatory response includes the formation of leukocyte infiltration and cytokine network (1). To understand the role of inflammation, understanding how it contributes to physiological and pathological processes is important. Many types of cancer can arise from inflammation (2, 3). Notably, inflammation has many similarities as well as differences with tumor formation. Colitis-associated cancers (CAC), such as chronic inflammation and injury-dysplasia carcinomas (4), comprise crucial signaling processes, including adaptive immunity and innate immunity.
The serine/threonine kinase RSK2 has 2 catalytic domains (5). When activated by growth factors, neurotransmitters, serum or phorbol esters, RSK proteins can phosphorylate substrate proteins (6–8), which are involved in various cellular events such as gene expression, differentiation and transformation (9). RSKs are downstream of the Ras-extracellular signal-regulated kinases (ERKs)/mitogen-activated protein kinases (MAPKs) signaling cascade (10). MAPKs control many signaling pathways and have been studied in diseases with chronic inflammation, such as inflammatory bowel disease (IBD) (11). Although the RSK family of proteins has been extensively studied, little is known regarding their roles in the immune system. Tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6) belongs to a group of six closely related TRAF proteins (12), which have emerged as major signal transducers that link the TNF receptor (TNFR) superfamily to intracellular signaling events. TRAF6 also mediates signaling downstream of the interleukin-1 (IL-1) receptor/Toll-like receptor (IL-1R/TLR) superfamily (12, 13). Through pathogen-associated molecular patterns, TLRs play an important role in detecting invading pathogens (14). MyD88 recruitment leads to the interaction and phosphorylation of IL-1R–associated kinases and the subsequent activation of TRAF6 and TGF-β-activated kinase 1 (TAK1)(14, 15). The TAK1 kinase complex activation occurs through the TRAF6-catalyzed unique Lys63 (K63)-linked poly-Ub chain, not the conventional Lys48 (K48)-linked poly-Ub chains, which mediate protein degradation. The in vivo knockdown of TRAF6 thus blocked NF-κB activation by IL-1β and lipopolysaccharide (LPS) (16, 17). TAK1 activates IKK for NF-κB signaling and promotes the induction of pro-inflammatory cytokines. MKK6 and MKK7 are also phosphorylated by TAK1, which leads to the activation of the JNKs and p38 MAP kinase pathways, respectively (18). Importantly, TRAF6 E3 ligase ubiquitinates itself (19). The auto-ubiquitination of TRAF6 occurs through K63 linkages, which require an intact RING domain and the E2 enzyme UBc13/Uev. The regulation of K63-linked TRAF6 auto-ubiquitination is a pivotal upstream mediator for IKK activation and other TRAF6-mediated signaling processes (20). TRAF6 also mediates LPS-induced Src family kinase (SFK) activation through TLR4 signaling (21) However, other post-translational modification-related K63-linked TRAF6 auto-ubiquitination activities remain unclear.
In this study, we demonstrated that RSK2 is required for the inflammation signaling pathway in that RSK2 binds and phosphorylates TRAF6 on serines 46, 47 and 48. These novel phosphorylation sites mediate it’s K63 ubiquitination and spontaneous downstream inflammation signaling. Moreover, these results were further confirmed in an RSK2 knockout (KO) AOM/DSS mouse model, in which both TRAF6 K63 ubiquitination and inflammation signaling were down-regulated compared with Balb/c WT mice. Overall, our findings suggest that TRAF6 is a novel molecule phosphorylated by RSK2, and that TRAF6 phosphorylation is essential for inflammation signaling through TRAF6 K63 ubiquitination.
Results
RSK2 is required for the IL1β– and LPS-mediated inflammation pathway
The MAPK signaling pathway is involved in multiple cellular processes, such as proliferation and cell cycle regulation, and also mediates the inflammation signaling pathway. In vitro and in vivo studies indicate that, ERK1/2 are implicated in inflammation (11), We propose that RSK, as a kinase downstream of ERKs may also be involved in inflammation signaling. To determine if RSK plays a role in inflammation through LPS- or IL-1β–mediated signaling, knockdown of RSK2 was conducted in RAW cells (Fig. 1A). Both sh-mock- and sh-RSK2-transfected RAW cells were stimulated with LPS. Results showed that phosphorylation and activation of Ikkα/β, IKBα, JNKs and p38 were increased at 15 min after LPS treatment (Fig. 1A). Also, RSK2 knockdown by sh-RSK2 suppressed LPS-induced phosphorylation of Ikkα/β, IKBα, JNKs and p38, and also decreased the total RSK2 protein levels. (Fig. 1A). To further confirm a role of RSK2 in inflammation signaling, RSK2+/+ and RSK2 −/− MEFs were incubated with IL-1β. RSK2 deficiency was confirmed by Western blot. Also, phosphorylation and activation of Ikkα/β, IKBα, JNKs and p38 were strongly induced in RSK2-expressing cells compared with RSK2-deficient murine embryonic fibroblasts (MEFs; Fig. 1B). Additionally, the levels of TNFα mRNA induced by IL-1β were significantly higher in RSK2+/+ MEFs, as compared with RSK2 −/− MEFs (Fig. 1C). Overall, these results provide strong evidence showing that RSK2 acts as a pivotal regulator of inflammation in the IL1β– and LPS-mediated pathways.
Figure 1. The RSK2-mediated inflammation pathway.
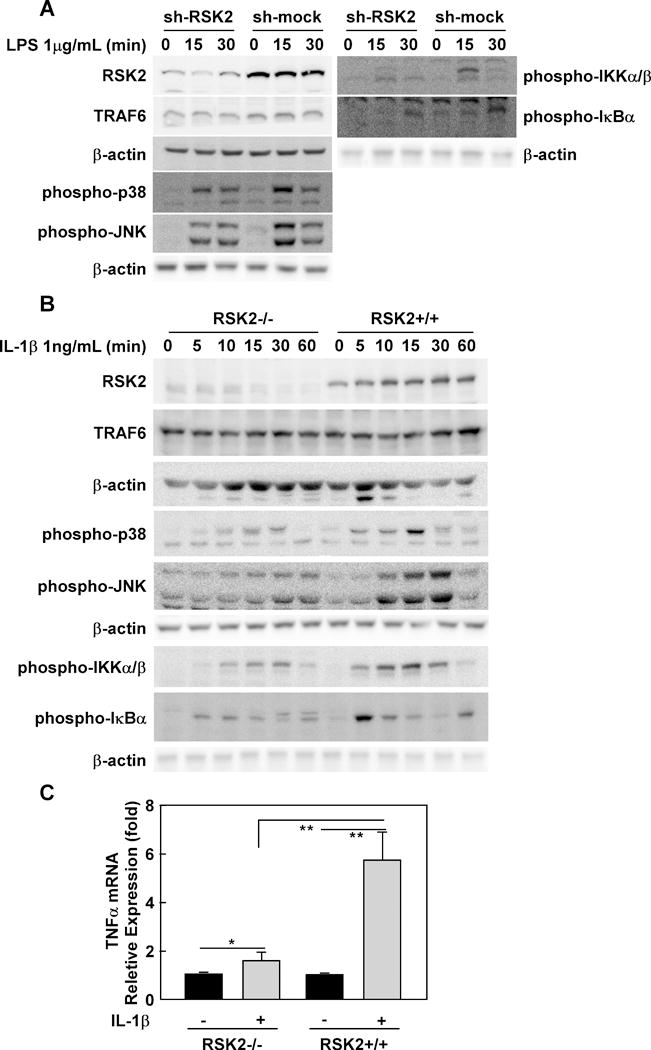
(A) RSK2 knockdown suppresses LPS-induced inflammation signaling. RAW cells were infected with an sh-mock or sh-RSK2 viral vector and each stable cell line was established. RSK2 knockdown was confirmed by Western blot (panel 7 from top). Sh-RSK2 and sh-mock cells were cultured and stimulated with LPS (1 μg/mL). Phosphorylated Ikkα/β, IKBα, JNKs, p38 and total TRAF6 proteins were visualized by Western blotting. (B) RSK2 deficiency blocks IL-1β–induced inflammation signaling. RSK2+/+ and RSK2−/− murine embryonic fibroblasts (MEFs) were cultured to 90–95% confluence, and subsequently starved in 0.1% FBS-DMEM for 24 h. The cells were stimulated with IL-1β (1 ng/mL) and harvested at the indicated time point. Western blotting was conducted using whole cell lysates from RSK2+/+ and RSK2−/− MEFs as indicated. The levels of phosphorylated Ikkα/β, IKBα, JNKs, p38 and total RSK2 proteins were visualized by Western blot. Each assay was performed 3 times and representative blots of similar results are shown. (C) RSK2 deficiency inhibits the induction of TNFα expression by IL-1β. Graph data are shown as means ± S.D. of values obtained from triplicate experiments and significant differences were evaluated using the Student’s t- test (*, p < 0.05; **, p < 0.01).
RSK2 interacts with TRAF6
Next, we examined whether the RSK2 protein could bind to TRAF6 directly. After co-expression of both proteins into 293 cells, an immunoprecipitation (IP) experiment was conducted. We confirmed the binding between TRAF6 and RSK2 in both pulled down Flag-tagged TRAF6 (Fig. 2A, lane 3) and Xpress-tagged RSK2 (Fig. 2B, lane 3). To further verify this interaction, we performed an endogenous binding assay using RAW cells. The results showed that LPS stimulation significantly enhanced RSK2 and TRAF6 binding (Fig. 2B). We also conducted identical experiments to examine the interaction between Flag-tagged full length (FL) TRAF6 and truncated Xpress-RSK2 mutants (Fig. 2C). The vectors, including a full-length RSK2 (RSK2-FL) and a constructed CTD RSK2 (RSK2-D1), NTD-deleted RSK2 (RSK2-D2) and a CTD-deleted RSK2 (RSK2-D3; Fig. 3A) (22) were co-expressed in 293 cells. The binding of RSK2 to TRAF6 FL was diminished in the RSK2-D1 mutant, which does not include the linker region (Fig. 2C, lane 3). Additionally, both NTD- and CTD-deleted RSK2 proteins could interact with TRAF6 FL (Fig. 2C, lanes 4 and 5). These data indicated that the linker region might be important in the RSK2 interaction with TRAF6. To determine the region of TRAF6 that mediates binding with RSK2, truncated mutants of TRAF6 (23) were individually transfected into 293 cells with pcDNA4-His/Xpress-RSK2, and expression was confirmed (Fig. 2D, lower panel). IP results using a Flag-HRP antibody showed that the deletion mutant TRAF6 D2 co-precipitated with RSK2 (Fig. 2D, upper panel, middle lane). These results suggested that the middle portion including the ZnF domain and the coil-coil domain of TRAF6 are important for the interaction with RSK2. Based on the overall results, we conclude that TRAF6 is a novel binding partner of RSK2.
Figure 2. RSK2 binds with TRAF6.
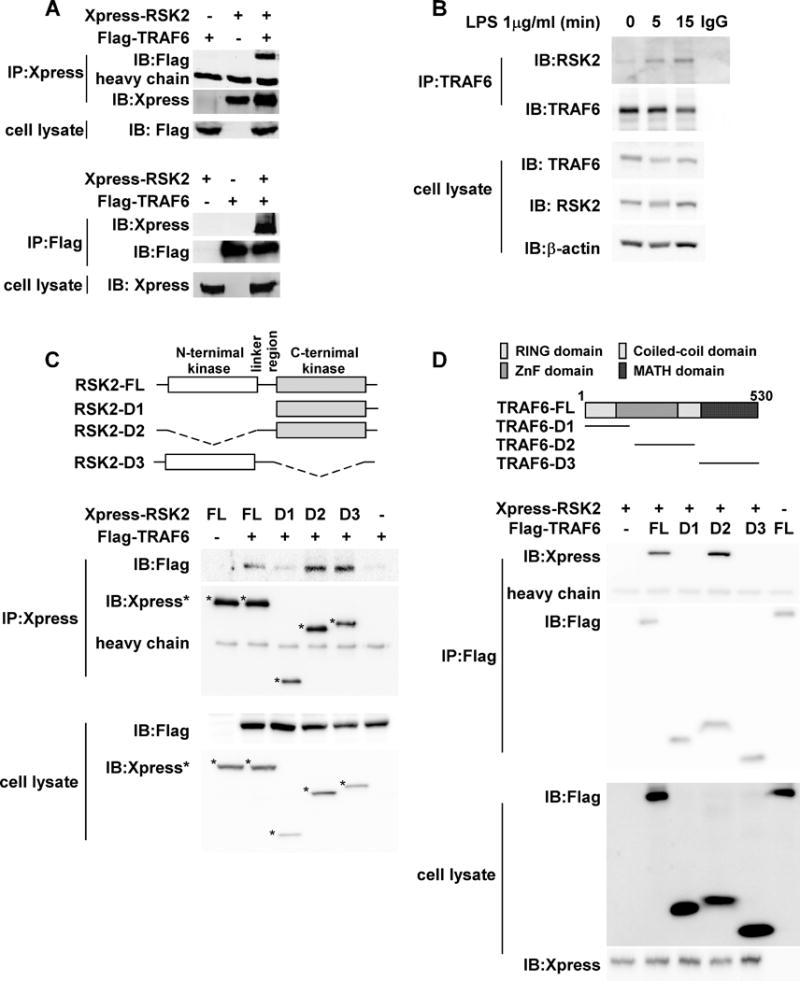
(A) TRAF6 and RSK2 bind with each other. To confirm the binding of TRAF6 and RSK2, the Flag-TRAF6 construct was co-transfected with Xpress-RSK2 into 293 cells. After culturing for 36 h, cells were disrupted with NP-40 lysis buffer and immunoprecipitated (IP) with an Xpress (upper panels) or Flag (lower panels) monoclonal antibody, respectively. Co-immunoprecipitated Flag-TRAF6 or Xpress-RSK2 was detected using a specific antibody. (B) LPS stimulation enhances RSK2 binding with TRAF6. RAW cells were stimulated with 1 μg/mL LPS and harvested at the indicated time point. The proteins were extracted with NP40 cell lysis buffer and immunoprecipitated with a TRAF6 antibody and co-immunoprecipitated RSK2 was detected with a specific antibody. Total TRAF6 was detected as an IP control. (C) Identification of the domain of RSK2 that binds with TRAF6. Structure and schematic diagrams of RSK2 deletion mutant constructs (upper panel). To identify the domain of RSK2 to which TRAF6 binds, three Xpress-RSK2 deletion constructs were individually co-transfected with Flag-TRAF6 into 293 cells. After culturing for 48 h, cells were disrupted with NP-40 cell lysis buffer and immunoprecipitated with an Xpress monoclonal antibody. Co-immunoprecipitated Flag-TRAF6 was detected by Western blot. Flag-TRAF6 or Xpress-RSK2 was detected in the whole cell lysate. (D) Identification of the domain of TRAF6 that binds RSK2. Structure and schematic diagrams of TRAF6 deletion mutant constructs (upper panel). To identify the domain of TRAF6 to which RSK2 binds, three Flag-TRAF6 deletion constructs were individually co-transfected with Xpress-RSK2 into 293 cells. After culturing for 48 h, cells were disrupted with NP-40 cell lysis buffer and immunoprecipitated with a Flag monoclonal antibody. Co-immuno- precipitated Xpress-RSK2 was detected by Western blot. Flag-TRAF6 or Xpress-RSK2 was detected in the whole cell lysate. Each assay was performed 3 times and representative blots of similar results are shown.
Figure 3. RSK2 phosphorylates TRAF6 at serines 46, 47 and 48.
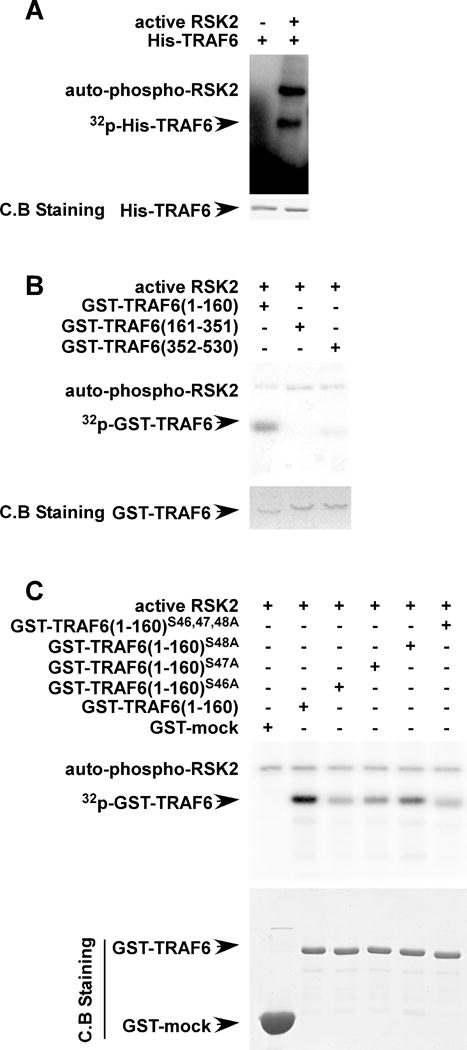
(A) RSK2 phosphorylates TRAF6. To confirm the phosphorylation of TRAF6 by active RSK2, a GST-TRAF6 fusion protein was purified and directly subjected to an in vitro kinase assay with [γ-32P] ATP and active RSK2, and the results visualized by autoradiography. (B) Identification of the TRAF6 domain that is phosphorylated by RSK2. Each GST-truncated deletion mutant GST-TRAF6 (1–160), GST-TRAF6 (161–351) or GST-TRAF6 (352–530) was constructed as shown and the GST-fusion proteins were purified. The proteins were directly subjected to an in vitro kinase assay with [γ-32P] ATP and active RSK2. The phosphorylated GST-TRAF6 proteins were visualized by autoradiography. Purified GST fusion proteins were stained with Coomassie blue (C.B.). (C) RSK2 phosphorylates TRAF6 at serines 46, 47 and 48. GST-proteins, including wild type GST-TRAF6 (1–160), mutant GST-TRAF6 (1–160; S46, 47, 48A), mutant GST-TRAF6 (1–160; S46A), GST-TRAF6 (1–160; S47A) and mutant GST-TRAF6 (1–160; S48A) were purified and directly subjected to an in vitro kinase assay with [γ-32P] ATP and active RSK2 and results were visualized by autoradiography.
RSK2 phosphorylates TRAF6
The serine/threonine kinase RSK2 phosphorylates multiple substrates and is involved in several cellular functions, however its role in the immune system is unclear. As previously reported, the post-translational modification of TRAF6 is important for its function (21, 24). Results of an in vitro kinase assay showed that the full-length mouse TRAF6 was strongly phosphorylated by RSK2 (Fig. 3A). To determine the regions of TRAF6 phosphorylated by RSK2, we constructed a series of GST-TRAF6 deletion fragments (Fig. 3B). Proteins were expressed and purified in E.Coli and then were used for an in vitro phosphorylation assay. The results showed that RSK2 phosphorylated TRAF6 in the region spanning amino acids 1 to 160 (Fig. 3B, first lane), which includes a ring finger domain. We then performed a LTQ Orbitrap hybrid mass spectrometer analysis (Supplemental Table) and identified the residues in TRAF6 that are phosphorylated by RSK2. We found that that RSK2 phosphorylated serines 46, 47 and 48 on TRAF6 and these results were further confirmed by an in vitro kinase assay. Full-length mouse TRAF6 was strongly phosphorylated by RSK2 (Fig. 3C, second lane); however, mutation of TRAF6 serine 46, 47 and 48 to alanine (TRAF6 S46A/S47A/S48A) strongly suppressed the phosphorylation by RSK2 (Fig. 3C, sixth lane). These data further confirmed that RSK2 acts as upstream kinase for the phosphorylation of TRAF6.
RSK2 is required for TRAF6 K63 ubiquitination
The post-translational modification of proteins by ubiquitin regulates multiple biological functions (25). Ubiquitination usually serves as a signal for protein degradation, such as ubiquitination through K48; however, ubiquitination through K63 plays an important role in signaling activation (25). Post-translational modifications of TRAF6 include phosphorylation and ubiquitination (15, 19, 24). For identification of the lysine residues on ubiquitin that are involved in forming the poly-ubiquitin chains on TRAF6, wild type ubiquitin, Lys48 ubiquitin or Lys63 ubiquitin along with RSK2 or sh-RSK2 were transfected into 293 cells. Data showed that ubiquitination occurred through wild type and K63, but not through K48 (Fig. 4A, lanes 2, 4 and 6). Knock down of RSK2 expression was confirmed by anti-RSK2 (Fig. 4A, lower panel) and ubiquitination of TRAF6 was attenuated in both wild type ubiquitin and Lys63 ubiquitin co-expression. These data provide evidence showing that an RSK2 interaction with TRAF6 is required for TRAF6 K63 ubiquitination. We further tested this hypothesis in RSK2+/+ and RSK2−/− MEFs, and TRAF6 K63 ubiquitination was strongly detected in RSK2-expressing MEFs, but, as expected, not in RSK2-null MEFs, (Fig. 4B). Overall, these data confirmed that RSK2 mediates TRAF6 K63-linked ubiquitination.
Figure 4. RSK2 is required for TRAF6 K63 ubiquitination.
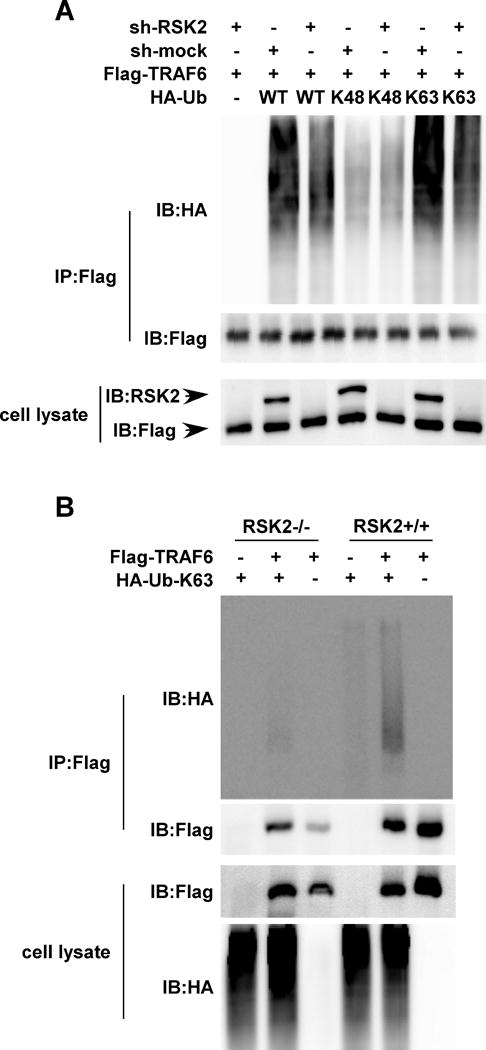
(A) RSK2 knockdown abrogates TRAF6 ubiquitination. Flag-TRAF6 was co-transfected with or without sh-RKS2 and the HA-Ub-WT, HA-Ub-K48, or HA-Ub-K63 plasmid, respectively. Whole cell lysates were subjected to immunoprecipitation (IP) with anti-Flag followed by Western blot analysis with anti-HA (upper panel) and anti-Flag (middle panel). RSK2 and Flag-TRAF6 proteins were detected by Western blot (lower panel). (B) RSK2 deficiency blocks TRAF6 K63 ubiquitination. Flag-TRAF6 was co-transfected with or without the HA-Ub (K63) plasmid into RSK2+/+ or RSK2−/− MEFs as indicated. Whole cell lysates were subjected to IP with anti-Flag followed by Western blot analysis with anti-HA (upper panel) and anti-Flag (middle panel). TRAF6 proteins were detected using a Flag antibody (lower panel). Each assay was performed 3 times and representative blots of similar results are shown.
TRAF6 phosphorylation induces K63 ubiquitination-mediated inflammation
Previous studies reported that TRAF6 K63 ubiquitination is critical for inflammation. Based on our study results, we expected that the RSK2-mediated phosphorylation of TRAF6 would regulate its K63 ubiquitination. Our data revealed that wild type TRAF6 ubiquitination was detected with co-transfected Lys63-linked ubiquitin, whereas a TRAF6 serine 46 alanine mutation (TRAF6 S46A), serine 47 alanine mutation (TRAF6 S47A) or serine 48 alanine mutation (TRAF6 S48A) attenuated TRAF6 K63 ubiquitination. Furthermore, a triple mutation of TRAF6 (S46A/S47A/S48A) almost totally abolished TRAF6 K63 ubiquitination. These data indicated that the TRAF6 K63 ubiquitination requires RSK2-mediated phosphorylation (Fig. 5A). TRAF6 K63 ubiquitination is important for IL-1β– and LPS-mediated inflammation signaling. To examine TRAF6 K63 ubiquitination mediates IKK, JNKs and p38 inflammation signaling. We overexpressed wild type TRAF6 or TRAF6 (S46, 47, 48A) in RAW cells and stimulated cells with LPS and found that phosphorylation and activation of Ikkα/β, IKBα, JNK and p38 were strongly induced in wild type TRAF6 cell compared with mutant TRAF6 (S46, 47, 48A) cells (Fig. 5B, lanes 2 and 3). Inflammation signaling was strongly blocked by the TRAF6 C70A mutant (Supplemental Fig. 1A). To confirm that RSK2-mediated phosphorylation of TRAF6 is required for this signaling, RAW cells transfected with the wild type TRAF6 or mutant (S46A/S47A/S48A) plasmid were stimulated with LPS. Cell lysates were pulled down with Flag-tagged TRAF6 and TRAF6 ubiquitination was detected with a ubiquitination antibody. Our data showed that with wild type TRAF6 overexpression, ubiquitination was detected and substantially increased by LPS (Fig. 5C, lanes 2 and 4). However, with mutant TRAF6 (S46A/S47A/S48A) overexpression, LPS stimulation could not induce ubiquitination of TRAF6 (Fig. 5C, lanes 3 and 5). We observed similar results in RSK2+/+ MEFs with IL-1β stimulation (Fig. 5D). These results demonstrate that TRAF6 K63 ubiquitination might be regulated by an RSK2-mediated phosphorylation-dependent mechanism and phosphorylation of TRAF6 at Ser46, 47 and 48 enhances its ubiquitin-mediated inflammation signaling.
Figure 5. TRAF6 serine 46,47 and 48 phosphorylation mediated inflammation pathway.
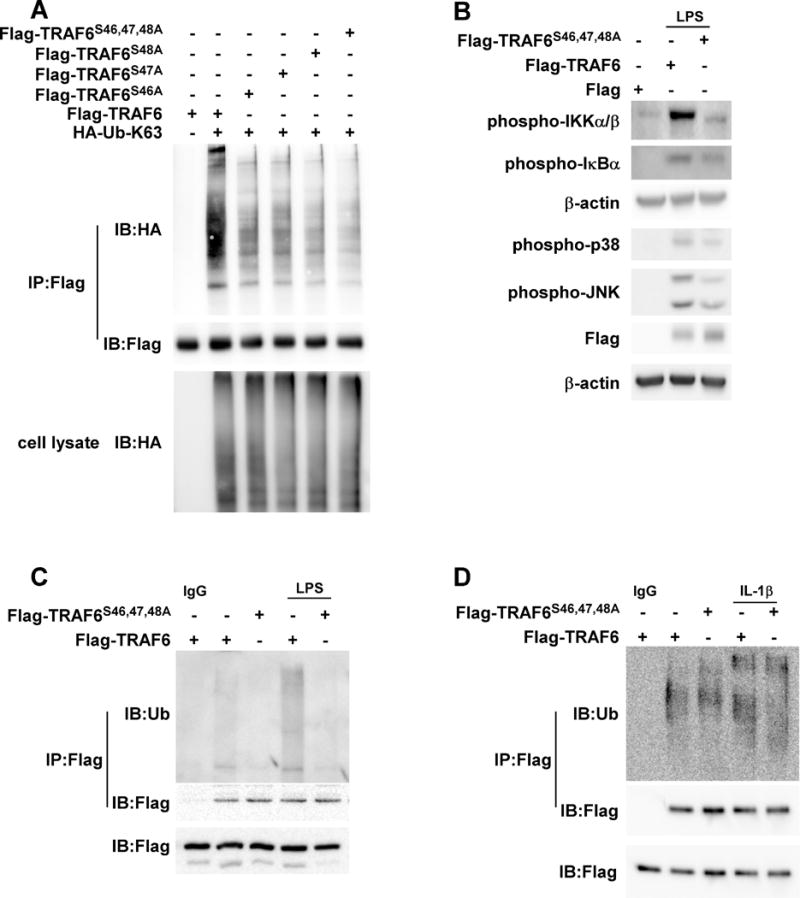
(A) Serines 46, 47 and 48 phosphorylation mediates TRAF6 K63 ubiquitination. Mutant Flag-TRAF6 (S46, 47, 48A), mutant Flag-TRAF6 (S48A), Flag-TRAF6 (S47A), mutant Flag-TRAF6 (S46A) or Flag-TRAF6 was co-transfected with HA-Ub-K63 as indicated. After imunoprecipitation (IP) with anti-Flag, HA-Ub or Flag-TRAF6 was detected by Western blot analysis. (B) Mutant Flag-TRAF6 (S46, 47, 48A) blocks the LPS-induced inflammation pathway. Mutant Flag-TRAF6 (S46, 47, 48A) or Flag-TRAF6 was individually overexpressed in RAW cells. After LPS induction, phosphorylated Ikkα/β, IKBα, JNKs and p38 protein levels were visualized by Western blot using specific antibodies. Equal protein loading was confirmed using a β-actin antibody on the same membrane. (C) Mutant Flag-TRAF6 (S46, 47, 48A) blocks LPS-induced TRAF6 K63 ubiquitination. Mutant Flag-TRAF6 (S46, 47, 48A) or Flag-TRAF6 was overexpressed in RAW cells as indicated. After LPS induction, whole cell lysates were subjected to IP with anti-Flag followed by Western blot analysis with anti-Ub or anti-Flag as indicated. (D) Mutant Flag-TRAF6 (S46, 47, 48A) suppresses IL-1β-induced TRAF6 K63 ubiquitination. Mutant Flag-TRAF6 (S46, 47, 48A) or Flag-TRAF6 was overexpressed in MEFs as indicated. After IL-1β induction, whole cell lysates were subjected to IP with anti-Flag followed by Western blot analysis with anti-Ub or anti-Flag as indicated. Each assay was performed 3 times and representative blots of similar results are shown.
TRAF6 acts as an RSK2 E3 ligase
Previous findings suggest that TRAF6 binds to the E2 enzyme, causing it to poly-ubiquitinate itself and other downstream molecules (19). To determine whether TRAF6 regulates RSK2 activity, RSK2 with or without TRAF6 was transfected into 293 cells along with the HA-Ub plasmid. Results indicated that RSK2 ubiquitination levels were higher in cells expressing TRAF6 (Fig. 6A). When the key Cys residue (C70) in the TRAF6 RING domain was mutated to Ala (C70A), the RING-dependent Ub ligase activity was abolished (26). Co- transfection of cells with RSK2 and wild type TRAF6 or TRAF6 (C70A) significantly reduced ubiquitinated RSK2 in RING-dependent Ub ligase activity-deficient TRAF6 (Fig. 6B). Lys48-linked polyubiquitin is mostly known to target proteins for degradation, whereas Lys63-linked ubiquitin chains have been linked to numerous cellular events such as the regulating protein kinase activity and act as protein adaptors (27–30). To identify the lysine residue involved in forming the poly-ubiquitin chains on RSK2, HA-Ub WT, K48 or K63 ubiquitin along with RSK2 and TRAF6 were co-expressed in 293 cells. To prevent RSK2 degradation through K48-linked ubiquitin, cells were treated with the proteasome inhibitor MG132. We found that RSK2 was more heavily coupled to Lys63 ubiquitin as compared with Lys 48 (Fig. 6C, lanes 2 and 3). In addition, Lys63 ubiquitin, but not Lys48 ubiquitin (Fig. 6C, lanes 2 and 5), was more diminished in RING-dependent Ub ligase activity deficient-TRAF6 (TRAF6 (C70A) cells, as compared to wild type TRAF6 (Fig. 6C, lanes 3 and 6). Overall, these data indicate that TRAF6 catalyzes Lys63-ubiquitination of RSK2.
Figure 6. TRAF6 is required for RSK2 ubiquitination.
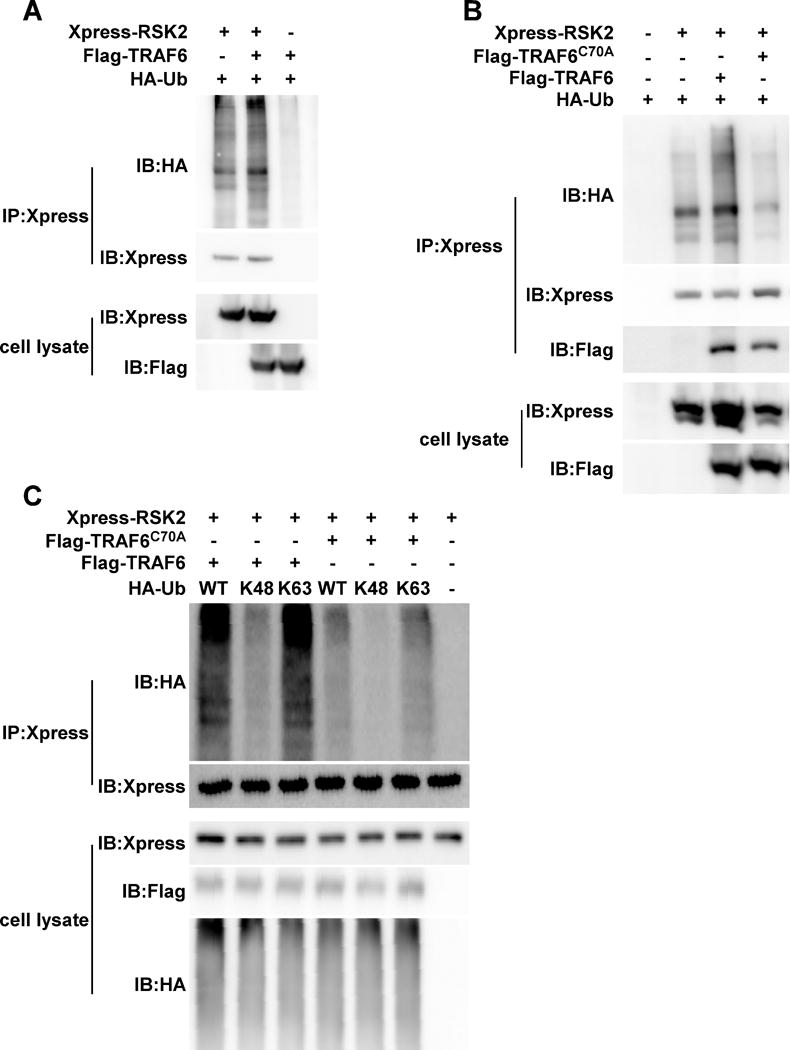
(A) TRAF6 mediates RSK2 ubiquitination. Xpress-RSK2, Flag-TRAF6 and HA-Ub were co-transfected as indicated. Whole cell lysates were subjected to immunoprecipitaton (IP) with anti-Xpress followed by Western blot analysis with anti-HA (upper panel) and anti-Xpress (middle panel). (B) TRAF6 (C70A) abrogates RSK2 ubiquitination. Xpress-RSK2 and HA-Ub were co-transfected with Flag-TRAF6 or Flag- TRAF6 (C70A), respectively. Whole cell lysates were subjected to IP with anti-Xpress followed by Western blot analysis with anti-HA (upper panel) and anti-Xpress (panel 2 from top). (C) TRAF6 (C70A) abrogates RSK2 K63 ubiquitination. Xpress-RSK2 was co-transfected with Flag-TRAF6 or Flag- TRAF6 (C70A), respectively, and then overexpressed with a HA-Ub-WT, HA-Ub-K48, or HA-Ub-K63 plasmid as indicated. Whole cell lysates were subjected to IP with anti-Xpress followed by Western blot analysis with anti-HA (upper panel) and anti-Xpress (panel 2). Xpress-RSK2, Flag-TRAF6 and HA-Ub proteins were detected by anti-Xpress, anti-Flag and anti-HA. Each assay was performed 3 times and representative blots of similar results are shown.
AOM/DSS induces colon epithelia inflammation more in RSK2 WT compared to KO mice
The murine colon carcinogenesis model, induced by azoxymethane (AOM)/dextran sodium sulfate (DSS), is an important animal model for examining colon inflammation. After several cycles of DSS administration, inflammation is present in the mucosa of treated mice. The lamina propria contains a dense infiltrate of immune cells, including macrophages, dendritic cells, and T-cells (31). The F4/80 glycoprotein is a specific macrophage cell-surface marker (32). CD3 works as a T-cell co-receptor to help activate the cytotoxic T-cell (33). Immunofluorescence staining for immune cell subsets revealed that RSK2 wild-type mice, compared with RSK2 deficient mice, exhibited a marked multi-lineage increase in infiltrates, include CD3+ T cells as well as F4/80 expressing macrophages (Fig. 7A). To determine whether RSK2 is active in the AOM/DSS mice model, wild type mouse colon tissue was subjected to an IP assay. The results showed that RSK2 and its activation, were required for colon colitis formation in the AOM/DSS treatment groups (Supplemental Fig. 1B and C). To further confirm and extend these observations, we performed immunohistochemistry on WT and RSK2 KO mouse epithelial colon tissue to determine the phosphorylation levels of JNKs, p38, IKKα/β, NFκB p65 and RSK (Fig. 7B). Results indicated increased inflammation signaling. Also, COX2, a molecular marker induced by inflammatory stimuli (34), was higher in the WT tissue sample. To explore the relationship of TRAF6 K63 ubiquitination with these inflammation changes, tissue samples from WT and RSK2 KO mice were disrupted and lysates were pulled down with a TRAF6 antibody and Lys63 ubiquitin was detected. Data showed that ubiquitination occurred in both wild type and RSK2 KO mice especially after AOM/DSS treatment, and further analysis showed that TRAF6 ubiquitination was stronger in the five randomly picked wild type mice compared with RSK2 KO mice (Fig. 7C, panels 1 and 5). Along with TRAF6 K63 ubiquitination, RSK2-deficient mice showed less phosphorylation and activation of Ikkα/β and IKBα compared with wild type mice (Fig. 7C, panels 3 and 4). Furthermore, TNFα mRNA levels were significantly higher in the colon tissue of WT mice, as compared to RSK2 KO mice, after AOM/DSS treatment (Fig. 7D).
Figure 7. Less inflammation signaling is observed in RSK2 KO mice compared with wild type controls.
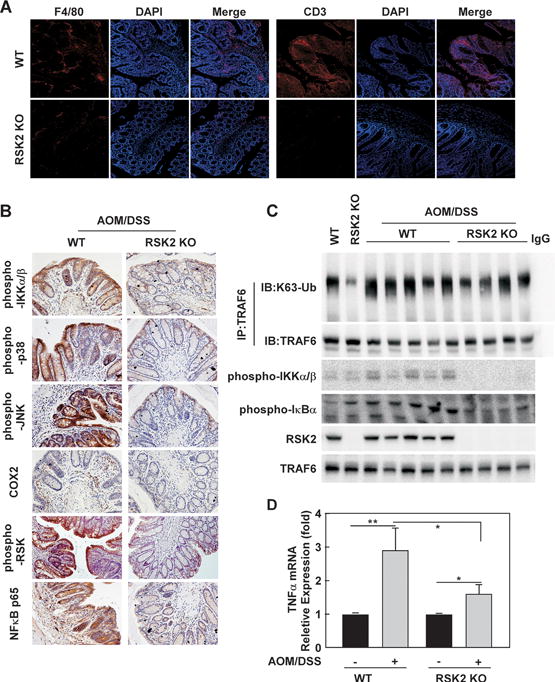
(A) F4/80 and CD3 are down-regulated in RSK2 KO mice. RSK2 KO and wild type control colon tissues were fixed and subjected to immunofluorescence analysis. Sections were stained as described in Methods using anti-F4/80 and anti-CD3 followed by Alexa568-conjugated goat anti-mouse IgG2A. Nuclei were counterstained with DAPI. Representative images from each group (n = 5) are shown. (B) RSK2 deficiency reduces inflammation signaling. RSK2 KO and wild type control colon tissues were fixed and subjected to immunohistochemical analysis. Expression of phosphorylated Ikkα/β, p38, JNK, RSK, COX2 and NFκB p65 was visualized by microscopy. Representative images from each group (n = 5) are shown. (C) TRAF6 K63 Ub is lower in RSK2 KO mice. RSK2 KO and wild type control colon tissues were disrupted and subjected to immunoprecipitation with anti-TRAF6 followed by Western blot analysis with anti-K63-Ub (upper panel) and anti-TRAF6 (panel 2). Phosphorylated Ikkα/β and IKBα proteins levels were visualized by Western blot using specific antibodies. RSK2 protein level was detected to confirm RSK2 deficiency. Each assay was performed 3 times and representative blots of similar results are shown. (D) Induction of TNFα mRNA expression by AOM/DSS treatment in WT and RSK2 KO mice. Graph data are shown as means ± S.D. of values (n=10). The asterisks indicate a significant difference (*, p < 0.05; **, p < 0.01).
Discussion
Ubiquitination, is a posttranslational modification that can modulate protein function and is involved in nearly all aspects of cell biology (35). Ubiquitination not only targets proteins for degradation, but can change the conformation and activity of the target proteins: Lys48-linked chains generally result in protein degradation by the proteasome; Lys63-linked chains play non-degradative roles in inflammation signaling and NF-κB activation (36). Ubiquitination is essential for inflammation signaling. Pathogen-recognition receptors (PRRs) including TLRs defend us from invading pathogens by recognizing pathogen-associated molecular patterns (37). These receptors can initiate MAPK signaling activation and trigger the expression of pro-inflammatory cytokines and chemokines (38). RSKs are a family of Ser/Thr kinases that play important roles in regulating cell viability and metabolism and are downstream of the Ras-MAPK signaling pathway. The activation and phosphorylation of RSK at the plasma membrane is followed by its translocation and accumulation in the nucleus (39). The recruitment of RSK to membranes results in its activation, suggesting that RSK activation requires membrane-associated activating stimuli (10). Although membrane recruitment of RSK2 is a critical step for RSK2 activation and phosphorylation, how RSK2 is recruited to the plasma membrane is unclear. Akt K63 poly-ubiquitination has been previously reported to affect its recruitment to the membrane, required for its activation.(40). Ubiquitin can affect protein trafficking (25, 41), therefore we determined whether we could detect the ubiquitination of RSK2 in cells. We found that TRAF6 can ubiquitinate RSK2 (Fig. 6A) and mutant TRAF6 (C70A) abrogates TRAF6 E3 ligase activity and blocks RSK2 ubiquitination (Fig. 6B). Furthermore, we showed that RSK2 was coupled with K63-ubiqitin-mediated protein activation, but not K48-ubiqitin mediated protein degradation (Fig. 6C). We also discovered that RSK2 ubiquitination is required for its kinase activity and function (Fig. 7B and Supplemental Fig. 1C).
The AGC kinases are Ser/Thr protein kinases named after the protein kinase A, C, and G families (PKA, PKC, PKG) (42). AGC kinases contain several conserved sequences. One of these sequences is called the hydrophobic motif and upon stimulation, it is phosphorylated and translocated to the membrane where it becomes activated (43). The phosphorylated hydrophobic motif of RSK serves as a docking site for PDK1 (10). Here, we found that the RSK2 linker region binds with TRAF6 (Fig. 2C) and mediates TRAF6 phosphorylation (Fig. 3) and K63 ubiquitination (Fig. 5). We also discovered that TRAF6-mediated ubiquitination of RSK2 is required for its activity (Fig. 7B and Supplemental Fig. 1C); however, the detailed mechanisms regarding the ubiquitin-mediated regulation of the RSK2 kinase remain unclear and could be an interesting direction for future work.
Crosstalk between different types of post-translational modifications, is an emerging theme in eukaryotic biology (44). Many parallels exist between phosphorylation and ubiquitination. Both modifications are used during several cellular processes. The multiple connections between phosphorylation and ubiquitination act either positively or negatively. As previously reported, TRAF6 is phosphorylated by MST4 at Thr463 and Thr486 impairing its homo-oligomeric association, which then impedes TRAF6 K63-ubiqitin and NFκB signaling (24). In addition, LPS increases TRAF6 poly-ubiquitination association with SFKs and SFK increases tyrosine phosphorylation of TRAF6. TRAF6 acts as a crucial signaling mediator, coupling the LPS interaction with TLR4 to the activation of SFK and the loss of the integrity of the endothelial barrier (21). Our study revealed that TRAF6 is phosphorylated by RSK2 and mediates LPS- induced inflammation signaling (Fig. 5B) and is required for its K63 ubiquitination (Fig. 5C). The phosphorylation of Ser/Thr/Tyr residues can change the properties of the ubiquitin molecule by affecting charge and surface properties. This is important as most ubiquitin surfaces are functional for ubiquitin binding and machinery (35). Overall, our results showed that RSK2 interacts with and phosphorylates TRAF6 at serines 46,47 and 48. TRAF6 phosphorylation accelerates TRAF6 K63 ubiquitination and mediates RSK2 ubiquitination and activation. This feedback loop was further confirmed in vivo using a colon inflammation model. Compared with wild type mice, RSK2 deficient mice showed a low level of TRAF6 K63 ubiquitination. Overall, these results revealed that TRAF6 phosphorylation and ubiquitination act as positive regulators in the inflammation pathway.
Materials and Methods
Reagents and plasmids
Fetal bovine serum (FBS) and Dulbecco’s modified Eagle’s medium (DMEM) were from Life Technologies, Inc. (Grand Island, NY). Antibodies against phosphor-RSK2 (#11989), phosphor-Ikkα/β (#2697), phosphor-IKBα (#2859), phosphor-JNKs (#4668), phosphor-p38 (#4511) and COX2 (#12282) were from Cell Signaling Technology, Inc. (Beverly, MA). Antibodies against TRAF6 (sc-8409), Ub (sc-166553), and NFκB p65 (sc-8008) were from Santa Cruz Biotechnology, Inc. β-actin antibody was from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Anti-Xpress (R91025) was from Thermo Fisher Scientific Corporation (Carlsbad, CA). Active RSK2 and anti-Flag (F3165) were from Upstate Biotechnology, Inc. (Charlottesville, VA). The pRK5-HA-Ub-Wt, pRK5-HA-Ub-K48 and pRK5-HA-Ub-K63 plasmids were purchased from Addgene (Cambridge, MA). The pCDNA3-Flag-TRAF6 plasmid was kindly provided by Dr. Peter ten Dijke (23), and the pCDNA4-Xpress-RSK2 plasmid was described previously (7).
Cell culture
Mouse macrophage RAW cells (RAW264.7) and 293 cells were purchased from the American Type Culture Collection (ATCC; Rockville, MD) and cultured following ATCC instructions. Mouse embryonic fibroblasts (MEFs) from wild type and RSK2 KO mice were prepared following the approved University of Minnesota IACUC protocol. All cell lines were recently authenticated and cytogenetically tested before being frozen. Cells were tested to confirm that there was no mycoplasma contamination. Thawed cells were maintained in culture for a maximum of 8 weeks. Cells were split at 80–90% confluence and media were changed every 3 days.
Ubiquitination assays
In vivo Ubiquitination analysis was performed by immunoprecipitation assays (45, 46). Before harvesting, cells were treated with MG132 for 4 h. Cells were disrupted in lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM sodium-deoxycholate and 2% SDS) supplemented with protease inhibitors and 50 mM sodium fluoride. Lysates were sonicated, diluted by 9X volume of dilution buffer and centrifuged at 20,000 ×g at 4°C for 30 min. Supernatant fractions were incubated with an Xpress or HA antibody with protein A/G-Sepharose beads (GE healthcare) overnight at 4°C. After washing three times with washing buffer, bound proteins were recovered by boiling in 2X SDS loading buffer and separated by SDS/PAGE.
Western blotting and immunoprecipitation
Whole cell extracts were prepared by disrupting cells in NP40 lysis buffer. For the IP assay, cell lysates were incubated with an Xpress or Flag antibody with protein A/G-Sepharose beads overnight at 4°C. After washing three times with NP40 lysis buffer, proteins were separated by SDS/PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Amersham, Pittsburgh, PA). After being blocked and hybridized with specific primary antibodies at 4°C overnight, proteins were visualized using an enhanced chemiluminescence reagent and the ImageQuant LAS4000 system (GE Healthcare, Piscataway, NJ).
In vitro kinase assay
Truncated TRAF6, amino acids 1–160, 161–251 and 252–530, was amplified from the full-length mouse TRAF6 cDNA and then sub-cloned into the pGEX-5XC vector. Site-directed mutagenesis (Ser46, 47, 48 single or triple mutated to alanine) was conducted with the Site-Directed Mutagenesis Kit (Stratagene, LA Jolla, CA). GST-TRAF6, point mutant TRAF6, and truncated TRAF6 were utilized for performing an in vitro kinase assay in the presence of active RSK2. The reaction was conducted at 30°C for 20 min in a mixture containing 50 μM unlabeled ATP and 10 μCi of [γ-32P] ATP. After being boiled, proteins were resolved by SDS/PAGE, and then subjected to autoradiography or Coomassie blue staining.
AOM-initiated and DSS-promoted colon carcinogenesis
At 8 to 10 weeks of age, Balb/c wild-type (WT) and RSK2 knockout (KO) mice (20 mice per group, the male: female ratio was 1:1), were injected once subcutaneously with azoxymethane (AOM; Sigma; A5486; 10 mg/kg bodyweight), followed by exposure to three cycles of 2% DSS (MPBiomedicals, 160110) in drinking water (1 cycle: 5 days of DSS, 16 days of fresh water). All animal care and experimental procedures were performed under the guidelines of the University of Minnesota Institutional Animal Care and Use Committee. To meet the criteria for achieving statistical significance or the observation of real differences, 20 animals per group are typically recruited, and treated without investigator blinding. Mice were randomly grouped by body weight and age. After euthanasia, the colon tissue was flushed with PBS. Colon lesions were measured and snap-frozen for further analysis. The fixed colon tissue was processed by the Swiss-roll technique and embedded in paraffin for histopathological examination, in a subset of animals (47).
Immunofluorescence and immunohistochemistry staining
Mouse tissues were embedded in paraffin and subjected to immunofluorescence or immunohistochemistry. For the immunofluorescence assay, tissues were de-paraffinized and hydrated and then hybridized with the F4/80 and CD3 antibodies (red fluorescence). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; blue fluorescence). For the immunohistochemistry assay, tissues were hybridized with phosphor-IKKα/β, p38, JNKs, RSK, COX2 and NFκB p56 antibody and biotinylated secondary antibody. An ABC kit (Vector Laboratories, Inc., Burlingame, CA) was used to detect protein targets following the manufacturer’s instructions. After developing with 3,3′-diaminobenzidine, the sections were counterstained with hematoxylin and observed by microscope (200X) and Image-Pro Plus software (v. 6.1; Media Cybernetics, Inc., Bethesda, MD).
Statistical analysis
All the statistical analysis was conducted using GraphPad Prism 5.0 software. Significant differences comparing two groups were calculated using the Student’s t-test. One-way ANOVA was used to compare data between 3 or more groups. Values are expressed as mean values ± S.D. and a p value < 0.05 was considered significant. All data shown represents results from at least 3 independent experiments, conducted in triplicate.
Supplementary Material
Acknowledgments
We thank Simin Zhao and Todd Schuster for supporting experiments, Nicki Brickman and Dr. Tia Rai for assistance in submitting our manuscript (The Hormel Institute, University of Minnesota). This work was funded by The Hormel Foundation, National Institutes of Health grants CA166011, CA187027, CA196639 and NSF of Henan Province China No. 162300410337.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Author contributions
K.Y., S.L., and Z.D. study conception and research design; K.Y., S.L., D.L., H.Y., C.P., J.R., T.L., H.C., G.J., Z.Z., Y.H., and W.M performed research; K.Y., S.L., and D.L. analysis and interpretation of the data; K.Y., A.M.B., and Z.D. wrote the paper.
Supplemental Information accompanies the paper on the Oncogene website.
References
- 1.Zhang JM, An J. Cytokines, inflammation, and pain. International anesthesiology clinics. 2007;45(2):27–37. doi: 10.1097/AIA.0b013e318034194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7(3):211–7. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–14 e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 5.Malakhova M, Tereshko V, Lee SY, Yao K, Cho YY, Bode A, et al. Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nature structural & molecular biology. 2008;15(1):112–3. doi: 10.1038/nsmb1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K, Cho YY, Yao K, Nadas J, Kim DJ, Cho EJ, et al. Eriodictyol inhibits RSK2-ATF1 signaling and suppresses EGF-induced neoplastic cell transformation. J Biol Chem. 2011;286(3):2057–66. doi: 10.1074/jbc.M110.147306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YY, Yao K, Bode AM, Bergen HR, 3rd, Madden BJ, Oh SM, et al. RSK2 mediates muscle cell differentiation through regulation of NFAT3. J Biol Chem. 2007;282(11):8380–92. doi: 10.1074/jbc.M611322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu F, Zykova TA, Peng C, Zhang J, Cho YY, Zheng D, et al. Phosphorylation of H2AX at Ser139 and a new phosphorylation site Ser16 by RSK2 decreases H2AX ubiquitination and inhibits cell transformation. Cancer Res. 2011;71(2):393–403. doi: 10.1158/0008-5472.CAN-10-2012. [DOI] [PubMed] [Google Scholar]
- 9.Cho YY, Yao K, Kim HG, Kang BS, Zheng D, Bode AM, et al. Ribosomal S6 kinase 2 is a key regulator in tumor promoter induced cell transformation. Cancer Res. 2007;67(17):8104–12. doi: 10.1158/0008-5472.CAN-06-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9(10):747–58. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 11.Broom OJ, Widjaya B, Troelsen J, Olsen J, Nielsen OH. Mitogen activated protein kinases: a role in inflammatory bowel disease? Clin Exp Immunol. 2009;158(3):272–80. doi: 10.1111/j.1365-2249.2009.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25(11):1096–105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 13.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, Wang L, Zhang M, Yuan C, Gao C. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J Immunol. 2012;188(6):2567–74. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 16.Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26(22):3214–26. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunological reviews. 2012;246(1):95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang G, Shi LZ, Chi H. Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine. 2009;48(3):161–9. doi: 10.1016/j.cyto.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282(6):4102–12. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2(3):a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu A, Gong P, Hyun SW, Wang KZ, Cates EA, Perkins D, et al. TRAF6 protein couples Toll-like receptor 4 signaling to Src family kinase activation and opening of paracellular pathway in human lung microvascular endothelia. J Biol Chem. 2012;287(20):16132–45. doi: 10.1074/jbc.M111.310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho YY, Yao K, Pugliese A, Malakhova ML, Bode AM, Dong Z. A regulatory mechanism for RSK2 NH(2)-terminal kinase activity. Cancer Res. 2009;69(10):4398–406. doi: 10.1158/0008-5472.CAN-08-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang J, Zhang L, van Dam H, ten Dijke P. UBE2O negatively regulates TRAF6-mediated NF-kappaB activation by inhibiting TRAF6 polyubiquitination. Cell Res. 2013;23(3):366–77. doi: 10.1038/cr.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao S, Zhang Z, Li C, Huang M, Shi Z, Wang Y, et al. The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat Immunol. 2015;16(3):246–57. doi: 10.1038/ni.3097. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–5. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 26.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nature structural & molecular biology. 2009;16(6):658–66. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–8. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bekes M, Salvesen GS. The CULt of caspase-8 ubiquitination. Cell. 2009;137(4):604–6. doi: 10.1016/j.cell.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 29.Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, et al. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40(1):75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell. 2010;40(1):63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 31.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med. 2005;201(10):1615–25. doi: 10.1084/jem.20042307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Becker D, Vass T, White J, Marrack P, Kappler JW. A conserved CXXC motif in CD3epsilon is critical for T cell development and TCR signaling. PLoS biology. 2009;7(12):e1000253. doi: 10.1371/journal.pbio.1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31(5):986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13(8):508–23. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 37.Zinngrebe J, Montinaro A, Peltzer N, Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014;15(1):28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corn JE, Vucic D. Ubiquitin in inflammation: the right linkage makes all the difference. Nature structural & molecular biology. 2014;21(4):297–300. doi: 10.1038/nsmb.2808. [DOI] [PubMed] [Google Scholar]
- 39.Richards SA, Dreisbach VC, Murphy LO, Blenis J. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol Cell Biol. 2001;21(21):7470–80. doi: 10.1128/MCB.21.21.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang WL, Wu CY, Wu J, Lin HK. Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 2010;9(3):487–97. doi: 10.4161/cc.9.3.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 42.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 43.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370(Pt 2):361–71. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730–8. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 45.Zhou F, Zhang X, van Dam H, Ten Dijke P, Huang H, Zhang L. Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J Biol Chem. 2012;287(14):11002–10. doi: 10.1074/jbc.M111.328187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choo YS, Zhang Z. Detection of protein ubiquitination. Journal of visualized experiments: JoVE. 2009;(30) doi: 10.3791/1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson LA, Callaway ES, Kim E, Weeks BR, Fan YY, Allred CD, et al. Targeted Deletion of p53 in Lgr5-Expressing Intestinal Stem Cells Promotes Colon Tumorigenesis in a Preclinical Model of Colitis-Associated Cancer. Cancer Res. 2015;75(24):5392–7. doi: 10.1158/0008-5472.CAN-15-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


