Abstract
Coding and non-coding mutations in DNA contribute significantly to phenotypic variability during evolution. However, less is known about the role of epigenetics in this process. Although previous studies have identified eye development genes associated with the loss of eyes phenotype in the Pachón blind cave morph of the Mexican tetra Astyanax mexicanus, no inactivating mutations have been found in any of these genes. Here we show that excess DNA methylation-based epigenetic silencing promotes eye degeneration in blind cave Astyanax mexicanus. By performing parallel analyses in Astyanax mexicanus cave and surface morphs and in the zebrafish Danio rerio, we have discovered that DNA methylation mediates eye-specific gene repression and globally regulates early eye development. The most significantly hypermethylated and down-regulated genes in the cave morph are also linked to human eye disorders, suggesting the function of these genes is conserved across the vertebrates. Our results show that changes in DNA methylation-based gene repression can serve as an important molecular mechanism generating phenotypic diversity during development and evolution.
Introduction
Subterranean animals offer an excellent opportunity to study morphological, molecular and physiological changes that allow organisms to adapt to unique environments. Loss of eyes is one of the most common morphological features of cave-adapted animals, including many fish species. Blind cave fish (CF) morphs of Astyanax mexicanus evolved from surface fish (SF) during a few million years of isolation in dark Mexican caves1, with recent studies suggesting that regression of eyes evolved as part of a strategy to conserve energy in fish adapted to dark and nutrient deficient caves2. Although a number of studies have examined molecular mechanisms underlying eye loss in Pachón cave-derived Astyanax mexicanus CF, recent sequencing of the Pachón CF genome and other studies revealed no inactivating null mutations in essential eye development genes3-7. In contrast, genome sequencing of another subterranean animal, the naked mole rat Heterocephalus glaber, showed combined functional loss of more than a dozen key eye genes due to inactivating mutations8. The absence of obvious null mutations in essential eye genes despite dramatic effects on eye development suggests the possibility that Pachón cavefish might harbor cis-regulatory mutations in causing altered expression of eye genes and epigenetic regulators, and most likely the epigenetic regulation indirectly affecting the expression of large numbers of target eye genes at the same time. DNA methylation is one such epigenetic mechanism which is known to be able to simultaneously alter the expression of large cohorts of genes9. To test the possibility that altered DNA methylation plays a role in Pachón cave fish eye loss, we used CF and SF morphs of Astyanax mexicanus as well as wild type and DNA methylation and demethylation-deficient zebrafish Danio rerio to examine whether DNA methylation regulates eye formation, and whether eye loss in Pachón cave fish evolved at least in part through hypermethylation of key eye genes.
Results and Discussion
At 36 hpf Astyanax mexicanus CF and SF embryos are superficially indistinguishable with properly formed lenses and optic cups in both morphs (Fig. 1a,b). By five days of development, however, degeneration of eye tissue is clearly evident (Fig. 1c,d), and by adulthood CF eyes are completely absent (Fig. 1e,f)10. Eye regression is preceded by decreased expression of a number of different eye-specific genes, including the crystallins crybb1, crybb1c, and cryaa (Fig. 1g). The expression of large sets of genes can be repressed by DNA methylation based epigenetic silencing, as perhaps most famously shown for X chromosome inactivation11. New epigenetic DNA methyl “marks” are added by specific enzymes known as de novo DNA methyltransferases (DNMTs)12, and we recently showed that one of these enzymes, dnmt3bb.1, is expressed in zebrafish hematopoietic stem and progenitor cells (HSPC) where its loss leads to failure to maintain HSPCs13. We found that dnmt3bb.1 is also expressed in the ciliary marginal zone (CMZ) of the developing eye, a specialized stem cell-containing tissue surrounding the lens that is responsible for generating neurons and other eye cell types14-16 (Fig. 1h). In addition to their hematopoietic defects dnmt3bb.1y258 null mutant larvae and adults have enlarged eyes compared to their wild type (WT) siblings (Fig. 1i-k) with retinal hyperplasia (Supplementary Fig. 1), and increased expression of a number of different eye genes, including opn1lw1, gnb3a, and crx (Fig. 1l). Interestingly, the closely related Astyanax mexicanus dnmt3bb.1 gene, which also maps to an eye size QTL6, shows 1.5-fold increased expression in Pachón CF compared to SF (Fig. 1m and Supplementary Fig. 2). The inverse correlation between eye size and dnmt3bb.1 expression in Danio rerio and Astyanax mexicanus led us to hypothesize that excessive Dnmt3bb.1-dependent methylation may globally repress expression of eye genes in CF.
Figure 1. Eye phenotypes in Astyanax mexicanus surface and cave fish, and in zebrafish Danio rerio wild type and dnmt3bb.1y258 mutant animals.
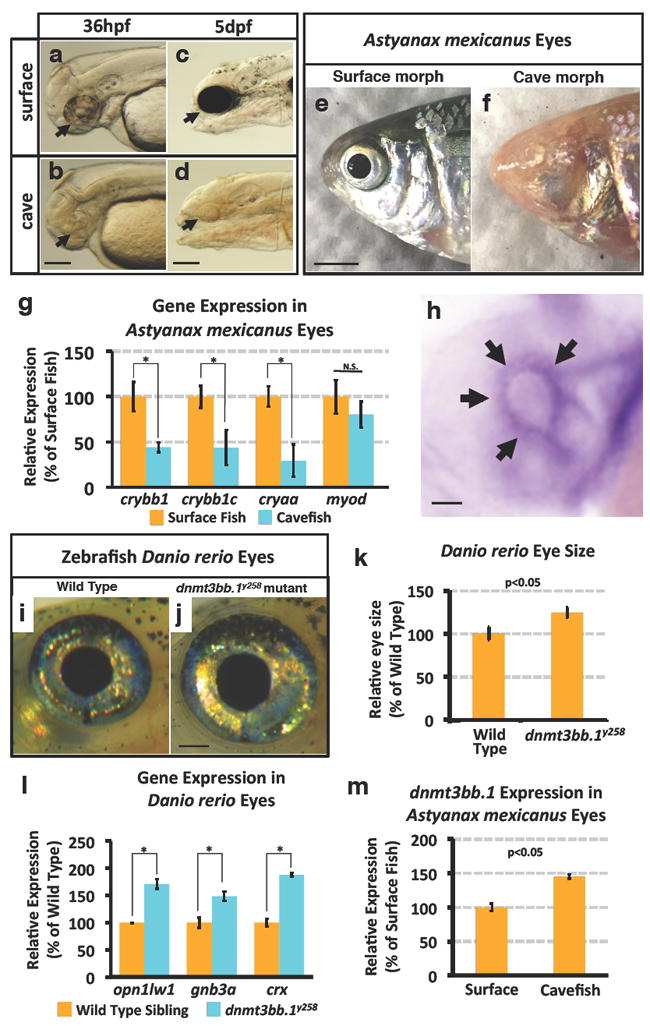
a-d, Transmitted light photomicrographs of the heads of 36 hpf (a,b) and 5 dpf (c,d) surface (a,c) and cavefish (b,d) morphs of A. mexicanus. Black arrow mark the developing eyes in CF and SF. e,f, Photographic images of the heads of adult surface (e, with eyes) and cave (f, eyeless) morphs of A. mexicanus. g, Quantitative RT-PCR analysis of the percent relative expression of crybb1, crybb1c, cryaa, and myod in isolated heads from 54 hpf surface (orange columns) and cave (blue columns) morphs of A. mexicanus, normalized to surface fish levels. Histograms show mean, error bars represent s.e.m of three biological replicates; two-tailed t-test, *p<0.005. h, Whole mount in situ hybridization of a 36 hpf zebrafish eye probed for dnmt3bb.1, showing expression in the ciliary marginal zone (CMZ, arrows). i,j, Photographic images of the eyes of adult wild type sibling (i) and dnmt3bb.1y258 mutant (j) zebrafish. k, Quantitation of eye size in three week old wild type sibling and dnmt3bb.1y258 mutant animals. Histograms show mean, error bars represent s.d. in the relative eye size (n =18) two-tailed t-test, *p<0.05. l, Quantitative RT-PCR analysis of the percent relative expression of opn1lw1, gnb3a, and crx in adult wild type sibling (orange columns) and dnmt3bb.1y258 mutant (blue columns) zebrafish eyes, normalized to wild type sibling levels. Histograms show mean, error bars represent s.e.m of three biological replicates; two-tailed t-test, *p<0.005. m, Quantitative RT-PCR analysis of the percent relative expression of dnmt3bb.1 in surface and cave morphs of A. mexicanus, normalized to surface fish levels. All images are lateral views, rostral to the left. Histograms show mean, error bars represent s.e.m of three biological replicates, p<0.05 using t-test. Scale bars 100μM in b for a,b, 250μM in d for c,d, 3mm in e for e,h, 50μM for h and 1mm in j for i,j.
To more comprehensively examine whether increased DNA methylation correlates with reduced eye gene expression in CF versus SF, we performed combined RNAseq and whole genome bisulfite sequencing on RNA and DNA simultaneously co-isolated from 54 hpf CF or SF eyes, during a critical period for eye development in Pachón CF10 (Fig. 2a). RNAseq analysis confirmed increased expression of dnmt3bb.1 in CF eyes (Fig. 2b,c). The RNAseq data also revealed that a large number of different eye development genes show reduced expression in CF eyes (Fig. 2c and Supplementary data 1). As in the naked mole rat, visual perception (GO:0007601) and electrophysiology of eye are the top down-regulated biological processes found using Gene Ontology (GO) (Fig. 2d) and Ingenuity Pathway Analysis (Supplementary Fig. 3), respectively. Ingenuity pathway analysis also predicted that the phototransduction pathway is the most affected signaling pathway in cavefish eyes compared to surface eyes (Supplementary Fig. 3).
Figure 2. Gene expression changes in cave versus surface fish morphs of Astyanax mexicanus.
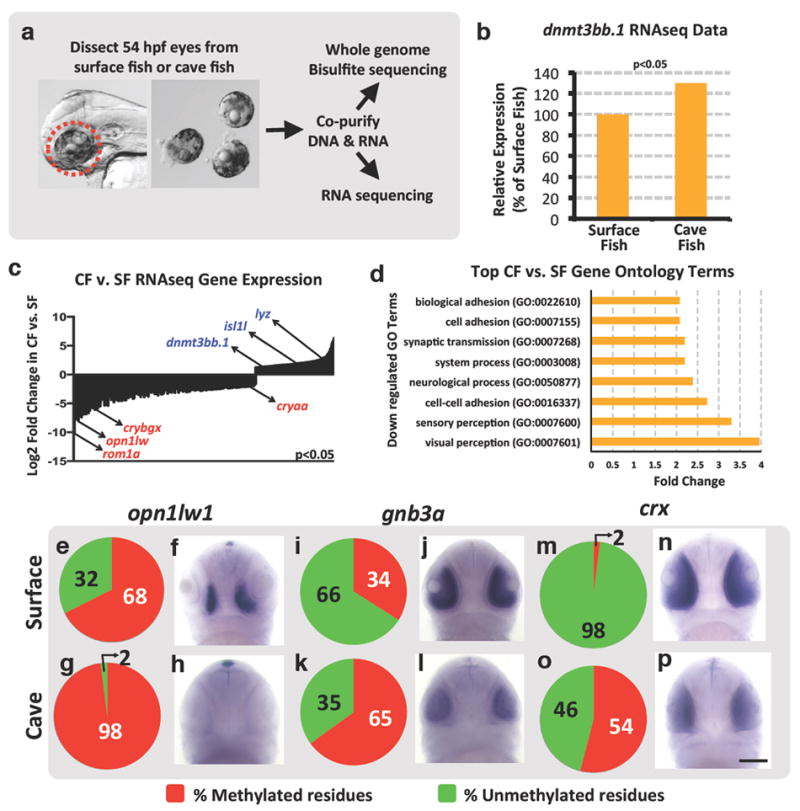
a, Diagram showing the workflow for obtaining larval eyes from A. mexicanus and for co-isolating eye DNA and RNA for whole genome assessment of DNA methylation and gene expression, respectively. b, Percent relative expression of dnmt3bb.1 in the eyes of A. mexicanus cave vs. surface fish morphs by comparison of their respective RNAseq data sets (2-replicates), normalized to surface fish levels, FPKM p<0.05. c, Log2 fold differential expression (p<0.05) of genes in A. mexicanus cave vs. surface fish morph RNAseq data sets, with down-regulated (red) or up-regulated (blue) expression of selected genes in CF noted. d, Listing of the Gene Ontology (GO) terms showing the greatest down-regulation in cave fish compared to surface fish. e-p, Assessment of opn1lw1(e-h), gnb3a (i-l), and crx (m-p) promoter DNA methylation (e,g,i,k,m,o) and gene expression (f,h,j,l,n,p) in 54 hpf surface (e,f,i,j,m,n) or cave (g,h,k,l,o,p) morphs of A. mexicanus. Panels show pie chart graphical representation of the percentage methylation of the promoter CpG (e,g,i,k,m,o), and representative whole mount in situ hybridization of the larval heads (f,h,j,l,n,p) using the probes noted (ventral views, rostral up, minimum 20-25 embryos were analyzed by in situ hybridization). Scale bar 100μM in p for f,h,j,l,n, and p.
Previous studies have shown that promoter methylation is highly correlated with gene repression17. One hundred and twenty-eight genes show substantially increased methylation within the 2 Kb of genomic DNA upstream from the transcriptional start site in CF versus SF (≥ 15% increase, p ≤ 0.05) and decreased expression in CF eyes compared to SF eyes (fold decrease ≤ 1.5, p ≤ 0.05) (Supplementary data 2). These include 39 and 26 genes annotated as having eye expression in mice and humans respectively (Supplementary data 3). Interestingly, nineteen of these genes have been previous linked to human eye disorders (Table 1). These include opn1lw1, an opsin associated with cone-rod dystrophy and colorblindness18,19, gnb3a, defective in autosomal recessive congenital stationary night blindness20,21, and crx, a photoreceptor-specific transcription factor whose loss leads to blindness in humans22,23. Targeted bisulfite sequencing of DNA amplified from the opn1lw1, gnb3a, and crx promoter regions from 54 hpf SF or CF eyes confirms increased methylation of CpGs in the 5’ upstream sequences of each of these three genes (Fig. 2e,g,i,k,m,o, Supplementary Fig. 4). Whole mount in situ hybridization using opn1lw1, gnb3a, and crx probes also verifies strongly decreased expression of the three genes in CF versus SF eyes (Fig. 2f,h,j,l,n,p). Together, these data show that expression of key human eye disease-associated eye development genes is reduced in Pachón CF compared to their SF relatives, and that many of these eye genes also display increased promoter methylation. Interestingly, the crx transcription factor is itself necessary for proper expression of a large number of additional eye genes23, many of which are also strongly reduced in CF eyes, even though most of these genes are not themselves methylated (Supplementary Fig. 5).
Table 1. Cavefish genes with significant promoter hypermethylation and reduced expression are associated with human eye disorders.
Nineteen genes with both substantially increased methylation within the 2 Kb of genomic DNA upstream from the transcriptional start site (≥ 15% increase, p ≤ 0.05) and substantially decreased gene expression (fold decrease ≤ 1.5, p ≤ 0.05) in CF eyes compared to SF eyes that have also been linked to human eye disorders.
| Eye Gene | Log2 Fold Change in Gene Expression | Change in Promoter Methylation (%) | Associated Human Eye Disease |
|---|---|---|---|
| STX3 | -2.6545922 | 15.0532125 | Microvillus inclusion disease |
| IMPG1 | -2.0676398 | 15.7283333 | Macular dystrophy |
| GNB3 | -2.1535092 | 15.7549858 | Congenital nigh blindness |
| RS1 | -4.8162527 | 16.2271036 | Retinoschisis |
| ELOVL4 | -2.7387552 | 16.9231332 | Stargard disease |
| TRPM1 | -1.5194777 | 17.1153846 | Congenital night blindness |
| KCNV2 | -2.6133734 | 19.2768429 | Retinal cone dystrophy |
| HEPACAM | -1.292728 | 19.2838136 | Macrocephaly |
| CRX | -1.3492741 | 20.3620072 | Retinitis pigmentosa |
| PDE6H | -2.4364817 | 20.8408953 | Retinal cone dystrophy |
| PRPH2 | -4.6162474 | 21.9236441 | Retinitis pigmentosa |
| TMEM98 | -1.3082467 | 23.6850216 | Nanophthalmia |
| GRK1 | -4.3915902 | 24.7866469 | Oguchi disease |
| MYO7A | -1.342215 | 31.2060616 | Usher syndrome |
| ATP6V0A1 | -1.4653532 | 32.7039627 | Macroautophagy |
| TDRD7 | -1.8275097 | 34.9881388 | Cataract |
| OPN1MW | -7.2622181 | 36.788428 | Cone rod dystrophy |
| CRYGB | -4.1315449 | 39.2857143 | Polar cataract |
| OPN1LW | -4.1525329 | 47.985771 | Cone rod dystrophy |
Increased expression of eye genes such as opn1lw1, gnb3a, and crx (Fig. 1l) and increased eye size (Fig. 1m) in dnmt3bb.1 mutant zebrafish is consistent with the hypothesis that DNA methylation represses eye gene expression to restrain or limit eye development. To experimentally test this idea, we examined whether increasing DNA methylation levels results in decreased eye gene expression and reduced eye development, using previously described zebrafish Ten-eleven translocation methylcytosine dioxygenase (TET) mutants that display global DNA hypermethylation24. Unlike DNMTs, which add methyl groups to cytosines to generate 5-methylcytosine, TET proteins oxidize methylcytosine to hydroxyl-methylcytosine, promoting DNA demethylation25,26. Like mammals, zebrafish have three TET enzymes with partially redundant functions, Tet1, Tet2, and Tet3. Tet2 and Tet3 are the main TET proteins responsible for converting methylcytosine into hydroxymethylcytosine; zebrafish tet1-/-, tet2-/-, tet3-/- triple mutants do not show additional phenotypes compared to tet2-/-, tet3-/- double mutants24. We found that tet2-/-, tet3-/- double mutants have smaller eyes than their wild type siblings (Fig. 3a-c). Targeted bisulfite sequencing and qRT-PCR on nucleic acids from whole eyes dissected from 48 hpf tet2-/-, tet3-/- double mutants and their wild type siblings also revealed increased crx and gnb3a promoter methylation (Fig. 3d-g, Supplementary Fig. 6) and reduced crx and gnb3a gene expression (Fig. 3h) in the double mutants. Like dnmt3bb.1 mutants, tet2-/-, tet3-/- double mutants also show hematopoietic defects. In addition, after 48 hpf tet2-/-, tet3-/- double mutants begin to deteriorate with defects in brain development and do not survive beyond 5 dpf. These additional phenotypes may arise as a result of global DNA hypermethylation in these mutants.
Figure 3. Eye phenotype and associated gene expression changes in wild type and DNA methylation-deficient Danio rerio.
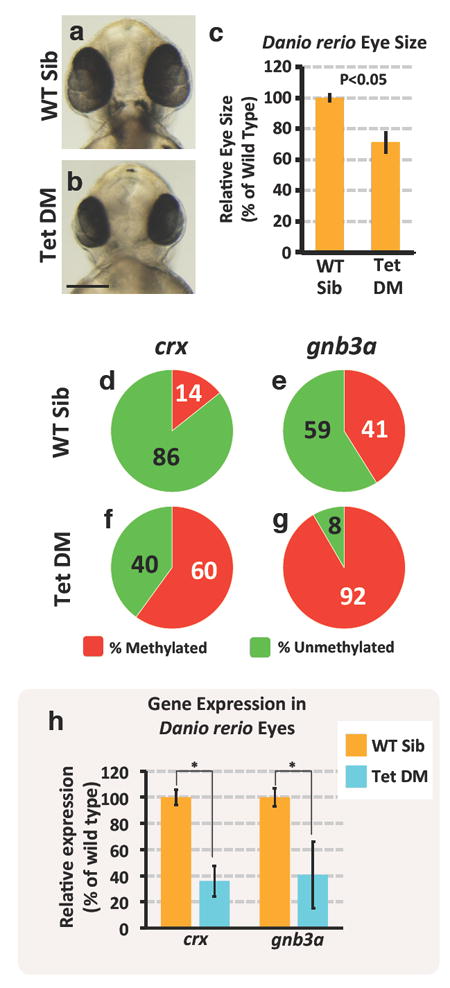
a-b, Transmitted light photomicrographs of the heads of 48 hpf wild type sibling (a) and tet2-/-, tet3-/- double mutant (b) embryos. c, Quantitation of eye size in 48 hpf wild type sibling and tet2-/-, tet3-/- double mutant embryos. Histograms show mean, error bars represent s.d. in the relative eye size (n =10), two-tailed t-test, *p<0.05. d-g, Assessment of crx (d,f) and gnb3a (e,g), promoter DNA methylation in isolated eyes from 48 hpf wild type sibling (d,e) and tet2-/-, tet3-/- double mutant (f,g) animals. h, Quantitative RT-PCR analysis of the percent relative expression of crx and gnb3a in isolated eyes from 48 hpf wild type sibling (orange columns) and tet2-/-, tet3-/- double mutant (blue columns) animals. Histograms show mean, error bars represent s.e.m of three biological replicates; two-tailed t-test, *p<0.005. Scale bar 100μM in b for a and b.
To further test whether excess DNA methylation contributes to failure to maintain eye development in Pachón CF, we examined whether eye loss could be “rescued” by pharmacological inhibition of DNA methylation. 5-Azacytidine (Aza) is a well-characterized inhibitor of DNA methylation27 approved for treatment of aberrant DNA methylation in myelodysplastic syndrome patients28. Since systemic administration of Aza to zebrafish embryos leads to severe pleiotropic defects and early lethality29, we carried out single injections of either Aza or control DMSO carrier into the vitreous chamber of the left eye of 42-48 hpf CF embryos, and then scored phenotypes in both injected left and control right eyes at 5 days post fertilization (Fig. 4a). A single early injection of Aza into the left eye resulted in larger eyes than either the control uninjected right eyes in the same animals or the DMSO-injected left eyes of other animals (Fig. 4b). Histological analysis confirmed that Aza-injected 5 dpf CF eyes are significantly larger than uninjected or DMSO injected controls, and that they possess a more organized structure including morphologically well defined lens and retinal layers (Fig. 4c-e). Targeted promoter bisulfite sequencing of gnb3a from Aza-injected cavefish eyes revealed reduced DNA methylation at this locus (Supplementary Fig. 7A). Increased expression of gnb3a and crx was also noted in cavefish eyes injected with either Aza or the more “DNA specific” drug 5-aza-2-deoxycytidine (Aza-D) (Supplementary Fig. 7B), suggesting that either inhibiting the excess methylation of eye genes in cavefish results in increased expression of these genes and at least a partial “rescue” of eye development.
Figure 4. Partial rescue of cavefish eyes by AZA-mediated inhibition of eye DNA methylation.
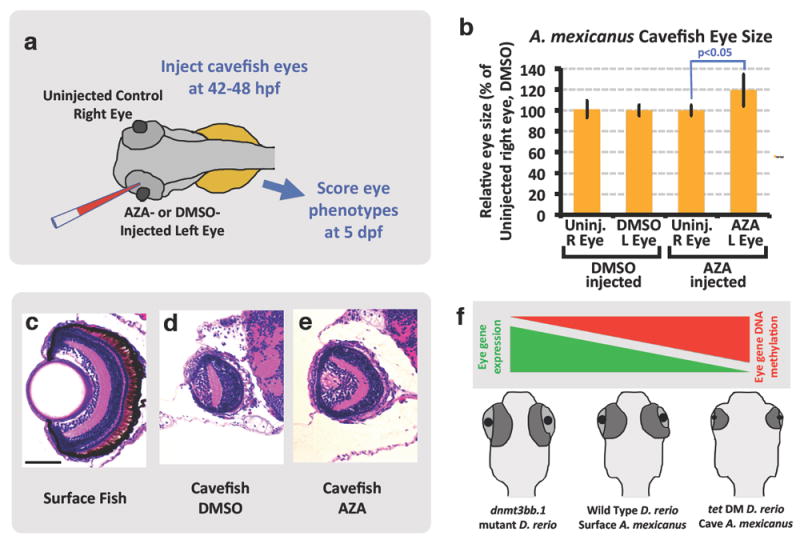
a, Schematic diagram showing the experimental procedure for injection of DMSO or AZA into 42-48 hpf cavefish embryo eyes. b, Quantitation of eye size in 5 dpf cavefish embryos injected in the left eye with DMSO or AZA. Histograms show mean, error bars represent s.d. in the relative eye size (18 DMSO and 12 Aza injected embryos) two-tailed t-test, *p<0.05. c-e, Histological analysis of H&E stained 5 dpf surface fish eye (c), DMSO injected cavefish eye (d) and AZA injected cavefish eye (e) (four to eight embryos per condition were sectioned and analyzed by H&E staining). f, Model depicting the role of DNA methylation in teleost eye development and degeneration. Hypermethylation and down-regulation of eye gene expression in cavefish and zebrafish tet2/3 double mutants leads to eye degeneration. Scale bar 100μM in a for a-c.
Together, our results suggest that DNA methylation plays a critical role in teleost eye development, and that increased DNA methylation-based eye gene repression is a major molecular mechanism underlying CF eye degeneration (Fig. 4f). Many of the key eye genes down-regulated in cave fish are conserved in humans and linked to eye disease and/or blindness, suggesting potential conserved function for these genes across evolution. Although a central role for DNA methylation in development and disease has been well-documented30,31, our results suggest that small genetic changes that alter epigenetic regulation can play an important role in rapid adaptive evolution by triggering dramatic changes in the expression of large sets of genes. There is precedent for epigenetic changes resulting in dramatically altered developmental fates; for example, nutrition-driven changes in DNA methylation regulate the switch between queen and worker caste specification in honey bees (Apis mellifera)32. Our findings indicate that eye loss in Pachón cavefish occurs via distinct molecular mechanisms compared to naked mole rats, where inactivating mutations are found in multiple key eye genes. This could reflect differences in their evolutionary timescales. Cavefish evolved over the past one to five million years1, while naked mole rats evolved seventy-three million years ago8, allowing sufficient time to fix and select acquired mutations in genes essential for eye development. It remains to be seen whether altered epigenetic regulation has been used to generate phenotypic variability in other rapidly evolved animals.
Methods
Fish stocks and embryos
Zebrafish lines used in this study include dnmt3bb.1y258 (Ref. 13) and tet2mk17, tet3mk18 (Ref. 24) mutants and the EK wild type line. Surface and Pachon cave populations of Astyanax mexicanus are used in this study. Fish were spawned naturally and embryos were raised and staged as described previously33,34. All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocols (12-039 and 18-016) of the National institute of child health and human development.
Genomic DNA isolation, bisulfite conversion, and sequencing
Surface and cavefish embryos were raised to described stages. Dechorionated embryos were transferred into 1X PBS without calcium and magnesium. Eyes were surgically removed using a pair of fine tip tungsten needles. Total cellular RNA and DNA was isolated from harvested eyes using ZR Duet DNA/RNA miniprep kit (Zymo Research). For whole genome bisulfite sequencing, 200 ng of purified genomic DNA was bisulfite converted using EZ DNA methylation-Lightning kit (Zymo research). Next Gen sequencing libraries were generated from the bisulfite converted DNA using TruSeq DNA methylation kit and TruSeq DNA methylation Index PCR primers (Illumina). Sequencing libraries of two biological replicates from surface and cavefish were run separately on two FlowCells of an Illumina HiSeq2500 sequencer operated in the RapidRun mode with V2 chemistry to yield about 400 million read pair reads (2 × 100bp) for each sample. Raw single-end sequence data was trimmed for quality and adapter sequence using Trimmomatic software, trimming leading or trailing bases at quality < 5 as well as using a sliding window requiring a 4 bp average of quality > 15. Reads trimmed below 50 bp were discarded. Following trimming, reads were aligned using Bismark software against a bisulfite converted Pachon cavefish genome using non-directional alignment. Using defined gene coordinates, gene body and promoter -2kb of TSS regions were assigned and combined conversions and non-conversions were summed to provide an overall bisulfite conversion rate per region. The ratio of conversions was then tested for change between conditions using a two-proportion z-test testing the hypothesis that the proportion of bisulfite conversion had altered between conditions. For targeted bisulfite analysis, bisulfite converted DNA was PCR amplified using primers designed by MethPrimer35 (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). One Taq Hotstart 2X master mix in standard buffer DNA polymerase (NEB) was used to amplify bisulfite converted DNA. PCR amplicons were purified and cloned using pCRII-TOPO TA cloning kit (Thermofisher). Miniprep plasmid DNA was sequenced using T7 or SP6 sequencing primers. Sequencing results were analyzed using QUMA36 (http://quma.cdb.riken.jp/).
RNA isolation and sequencing
Total cellular RNA and DNA was isolated from the harvested eyes using ZR Duet DNA/RNA miniprep kit (Zymo Research). 300-900ng poly-A enriched RNA was converted to indexed sequencing libraries using the TruSeq Standard mRNA library prep kit (Illumina). Libraries for two biological replicates of surface and cavefish were combined and run on one FlowCell of an Illumina HiSeq 2500 sequencer in RapidRun mode with V2 chemistry. 100 million read pairs (2 × 100) per sample were sequenced. Paired-end reads were trimmed using trimmomatic and aligned to Pachon cavefish genome using RNA-STAR, quantitation performed with subread feature Counts and differential expression analysis performed with count data via DESeq2. Human, mouse and zebrafish homologs of cavefish genes were identified using Ensembl BioMart. Ingenuity pathway and Panther GO term enrichment analyses were carried out on RNA sequencing data to identify major signaling pathways and networks affected based on differentially expressed genes.
RNA isolation, cDNA synthesis and qRT-PCR
Total cellular RNA was isolated from harvested eyes and other tissues as mentioned in the text using ZR Duet DNA/RNA miniprep kit (Zymo Research). Equal amounts of RNA were converted into cDNA using the ThermoScript RT-PCR system (Invitrogen). Resulting cDNA was used in qPCR using SsoAdvanced™ Universal SYBR® Green Supermix (Biorad) on a CFX96 Real Time system. A minimum of three biological replicates were performed. Primers used in this study are listed in Supplementary Data 4.
Riboprobe synthesis, In situ hybridizations and histology
Antisense riboprobes were generated using Roche DIG and FITC labeling mix. Portions of the coding regions of genes were PCR amplified using One Taq Hotstart 2X master mix in standard buffer DNA polymerase (NEB) and cloned into pCRII-TOPO TA vector (Thermofisher). Sequence verified clones were used to generate antisense riboprobes using appropriate enzymes. The zebrafish dnmt3bb.1 probe was generated as described previously13. Whole mount in situ hybridization was carried out as described previously with a few modifications. To reduce non-specific hybridization and enhance signal to noise ratio we used 5% dextran sulfate (Sigma) in the hybridization buffer and pre-adsorbed anti-DIG and anti-FITC antibodies to whole cavefish powder. For histology, embryos and tissue samples were fixed using 4% para-formaldehyde overnight at 4°C and subsequently passed through ascending grades of alcohol followed by paraffin embedding. Sections were stained using hematoxylin and eosin (H&E).
Microinjection
Microinjection of 5-Azacytidine or 5-Aza-2’-deoxycytidine (Sigma) into Pachon cavefish embryonic eyes was carried out at 42-48 hpf embryos. Embryos were mounted laterally in low melting point agarose. Injection needles were pulled from filament-containing glass capillaries (World Precision Instruments Cat. No. TW100F-4) using a needle puller (Sutter Instruments). Needles were back loaded and a single 1-2 nl bolus of either 100μM 5-Azacytidine or 5-Aza-2’-deoxycytidine in 5% DMSO or 5% DMSO carrier alone was delivered into the vitreous of the left eye using a Pneumatic Picopump (World Precision Instruments). Embryos were removed from the agarose, allowed to continue to develop until 5 dpf, and then scored for eye size and/or used for histological analysis of the eyes. Injected embryos with axis or brain deformities were discarded from the analysis.
Eye size quantifications
For eye size measurements, embryos were imaged at same magnifications for mutants, treated and controls. All eye size measurements were performed by normalizing the area of the eyes to the head. Student’s t-test (paired) was performed on the normalized eye sizes.
Supplementary Material
Acknowledgments
We thank members of the Weinstein and Jeffery lab for their support, help, and suggestions. We thank NICHD’s Molecular Genomics Laboratory for bisulfite and RNA sequencing assistance. We also thank members of the zebrafish and cavefish community for sharing reagents and protocols. We thank Dr. Karuna Sampath for her comments on the manuscript. We thank Dr. Suzanne McGaugh for her suggestions on cavefish sequence alignments and Dr. Mary Goll for providing the zebrafish tet2,3 double mutant line. Work in the Weinstein and Jeffery labs is supported by the intramural program of the NICHD and by R01EY024941, respectively.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions
A.V.G. and B.M.W. designed the study with inputs from K.A.T. and W.J. A.V.G. and K.A.T. performed the experiments with help from L.M., D.C. and A.D. J.I. analyzed the sequencing data. D.C., A.D., S.S. provided fish husbandry support. A.V.G. and B.M.W. wrote the manuscript with input from all the authors.
The authors declare no competing financial interests.
Data Availability Sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE109006.
References
- 1.Gross JB, Meyer B, Perkins M. The rise of Astyanax cavefish. Dev Dyn. 2015 doi: 10.1002/dvdy.24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran D, Softley R, Warrant EJ. The energetic cost of vision and the evolution of eyeless Mexican cavefish. Sci Adv. 2015;1:e1500363. doi: 10.1126/sciadv.1500363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinaux H, et al. Lens defects in Astyanax mexicanus Cavefish: evolution of crystallins and a role for alphaA-crystallin. Dev Neurobiol. 2015;75:505–521. doi: 10.1002/dneu.22239. [DOI] [PubMed] [Google Scholar]
- 4.Casane D, Retaux S. Evolutionary Genetics of the Cavefish Astyanax mexicanus. Adv Genet. 2016;95:117–159. doi: 10.1016/bs.adgen.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Parkhurst A, Jeffery WR. The role of a lens survival pathway including sox2 and alphaA-crystallin in the evolution of cavefish eye degeneration. Evodevo. 2014;5:28. doi: 10.1186/2041-9139-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGaugh SE, et al. The cavefish genome reveals candidate genes for eye loss. Nat Commun. 2014;5:5307. doi: 10.1038/ncomms6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinaux H, et al. De novo sequencing of Astyanax mexicanus surface fish and Pachon cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PLoS One. 2013;8:e53553. doi: 10.1371/journal.pone.0053553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nature reviews Genetics. 2016;17:551–565. doi: 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina: Evidence from the blind cavefish Astyanax. Dev Biol. 2007;311:512–523. doi: 10.1016/j.ydbio.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr Med Chem. 2010;17:4052–4071. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gore AV, et al. Epigenetic regulation of hematopoiesis by DNA methylation. Elife. 2016;5:e11813. doi: 10.7554/eLife.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seritrakul P, Gross JM. Expression of the de novo DNA methyltransferases (dnmt3 - dnmt8) during zebrafish lens development. Dev Dyn. 2014;243:350–356. doi: 10.1002/dvdy.24077. [DOI] [PubMed] [Google Scholar]
- 15.Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Y, et al. The ciliary marginal zone of the zebrafish retina: clonal and time-lapse analysis of a continuously growing tissue. Development. 2016;143:1099–1107. doi: 10.1242/dev.133314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014;30:75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Ayyagari R, et al. Bilateral macular atrophy in blue cone monochromacy (BCM) with loss of the locus control region (LCR) and part of the red pigment gene. Mol Vis. 1999;5:13. [PubMed] [Google Scholar]
- 19.Winderickx J, et al. Defective colour vision associated with a missense mutation in the human green visual pigment gene. Nat Genet. 1992;1:251–256. doi: 10.1038/ng0792-251. [DOI] [PubMed] [Google Scholar]
- 20.Arno G, et al. Recessive Retinopathy Consequent on Mutant G-Protein beta Subunit 3 (GNB3) JAMA Ophthalmol. 2016;134:924–927. doi: 10.1001/jamaophthalmol.2016.1543. [DOI] [PubMed] [Google Scholar]
- 21.Vincent A, et al. Biallelic Mutations in GNB3 Cause a Unique Form of Autosomal-Recessive Congenital Stationary Night Blindness. Am J Hum Genet. 2016;98:1011–1019. doi: 10.1016/j.ajhg.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaroop A, et al. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- 23.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, et al. Overlapping Requirements for Tet2 and Tet3 in Normal Development and Hematopoietic Stem Cell Emergence. Cell Rep. 2015;12:1133–1143. doi: 10.1016/j.celrep.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 28.Raj K, Mufti GJ. Azacytidine (Vidaza(R)) in the treatment of myelodysplastic syndromes. Ther Clin Risk Manag. 2006;2:377–388. doi: 10.2147/tcrm.2006.2.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- 30.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nature reviews Genetics. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 32.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 33.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 34.Elipot Y, Legendre L, Pere S, Sohm F, Retaux S. Astyanax transgenesis and husbandry: how cavefish enters the laboratory. Zebrafish. 2014;11:291–299. doi: 10.1089/zeb.2014.1005. [DOI] [PubMed] [Google Scholar]
- 35.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 36.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


