Abstract
Specificity protein (Sp1) plays an important role in invasion-metastasis cascade. Sp1 regulation on protein coding genes has been extensively investigated; however, little is known about its regulation on protein non-coding genes. In this study, miR-3178 is reported as a novel target of Sp1 in multiple cancer cell models. Sp1 functions as its transcriptional suppressor as evidenced by luciferase reporter and chromatin immunoprecipitation (ChIP) assays. In line with the pro-metastatic role of Sp1, miR-3178 exerts anti-metastasis function. Overexpression of miR-3178 inhibits both migration and invasion of highly metastatic prostate, lung, and breast cancer cells whereas antagonizing miR-3178 promotes those events in their lowly metastatic counterparts. The in vivo study demonstrates that miR-3178 suppresses the tail vein inoculated prostate cancer cells to form colonies in lung, lymph node, and liver of BALB/c nude mice. miR-3178 directly targets the 3′ UTR of TRIOBP-1 and TRIOBP-5, two isoforms of TRIOBP expressed in prostate, lung, and breast cancer cells. Overexpression of TRIOBP-1 could rescue miR-3178 inhibition on cell migration and invasion. Collectively, our findings reveal the regulatory axis of Sp1/miR-3178/TRIOBP in metastasis cascade. Our results suggest miR-3178 as a promising application to suppress metastasis in Sp1-overexpressed cancers.
Keywords: Sp1, miR-3178, TRIOBP, metastasis, cancer
Introduction
Metastasis is one of the most important hallmarks of cancer and accounts for ∼90% of cancer-associated death.1 The dissemination of cancer cells from the primary site and forming new colonies in the secondary site involves a multi-step process. The transcriptional factor Sp1 plays an important role in each of the crucial events of metastasis: adhesion; invasion; migration; and angiogenesis. The cell surface adhesive molecule cadherin,2 epithelial-mesenchymal transition (EMT) drivers such as ZEB13 and Snail,4 and angiogenesis regulators EGFR5 and VEGFR36 are all recognized as Sp1 downstream targets.
Sp1 is ubiquitously expressed and frequently dysregulated in various types of cancers.7, 8, 9 Sp1 can recognize and bind to the consensus GC-box 5′-(G/T)GGGCG G(G/A)(G/A)(G/T)-3′ of target genes.10 About 12,000 Sp1 sites are estimated to exist in the human genome.11 Sp1 regulation on protein coding genes has been extensively investigated; however, little is known about its regulation on protein non-coding genes. As protein non-coding genes account for ∼97% of the genome,12 it is necessary to unveil Sp1 regulation on protein non-coding genes.
Recently, microRNAs (miRNAs) were implicated in cancer metastasis and emerged as “metastamir” or metastasis suppressor.13, 14 miRNAs are small non-coding RNAs with ∼20 nt in length that can post-transcriptionally regulate the expressions of multiple target genes. Binding of miRNAs to the 3′ UTR of their target mRNAs results in either translation suppression or mRNA degradation.15,16 In the present study, miR-3178 was reported as a novel target of Sp1. miR-3178 could suppress cancer cell migration and invasion by targeting Trio and F-actin binding protein (TRIOBP) in prostate, lung, and breast cancer models. Our findings reveal the regulatory axis of Sp1/miR-3178/TRIOBP in cancer metastasis.
Results
Sp1 Is a Transcriptional Suppressor of miR-3178
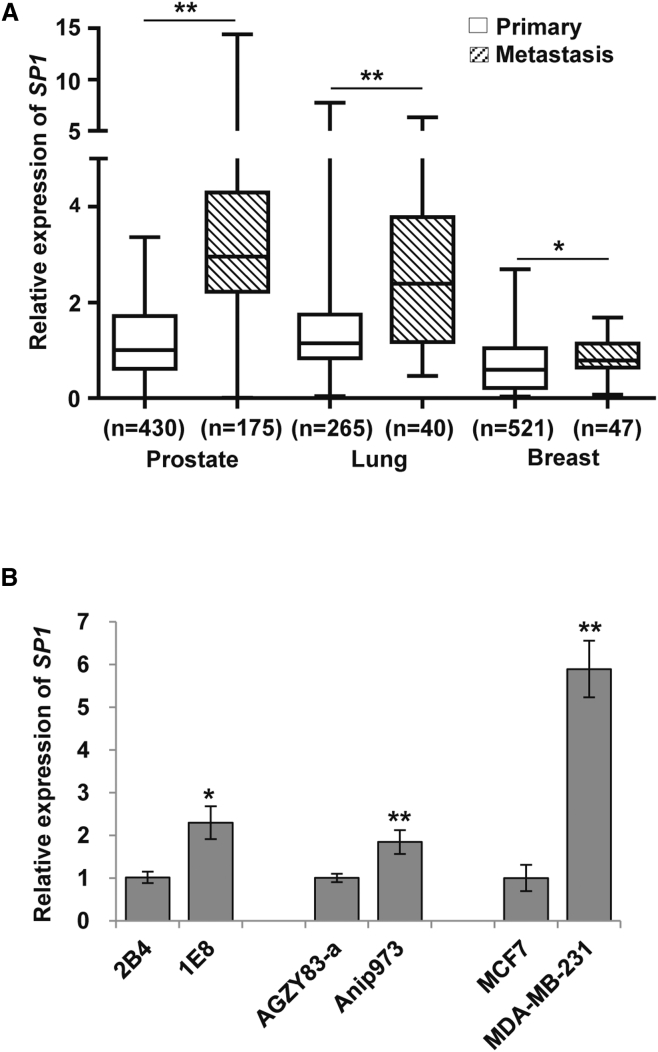
As Sp1 regulates a broad spectrum of genes associated with invasion-metastasis cascade,4, 17 we were interested to know the SP1 expression pattern in metastatic against primary tumors. Oncomine18 database was searched and SP1 expression was analyzed in cancer patients with prostate, lung, and breast cancers. Different expressions of SP1 between metastatic and primary tumors were compared, and significant upregulation of SP1 was observed in metastatic prostate (1.2 versus 3.6), lung (1.4 versus 2.7), and breast (0.7 versus 0.9) cancers (Figure 1A). Similar results were observed in prostate, lung, and breast cancer cell lines with different metastatic potentials. PC-3M-1E8 and PC-3M-2B4 are highly and lowly metastatic sublines selected from human prostate cancer PC-3M cells, respectively.19 Highly metastatic Anip973 is developed from lung adenocarcinoma AGZY83-a with lowly metastatic ability.20 MDA-MB-231 and MCF-7 are two breast cancer cell lines with different metastatic potentials.21, 22 The expressions of SP1 in highly metastatic 1E8, Anip973, and MDA-MB-231 cells were significantly higher compared with their lowly metastatic counterparts (Figure 1B).
Figure 1.
Upregulation of SP1 in Metastatic Cancers
(A) Oncomine data show SP1 mRNA overexpression in metastatic versus primary prostate, lung, and breast tumor tissues. (B) SP1 expression in prostate (PC-3M-1E8 versus PC-3M-2B4), lung (Anip973 versus AGZY83-a), and breast (MDA-MB-231versus MCF-7) cancer cell lines with highly or lowly metastatic potentials is shown. Experiments were repeated three times, and results were shown as mean ± SD. *p < 0.05 and **p < 0.01.
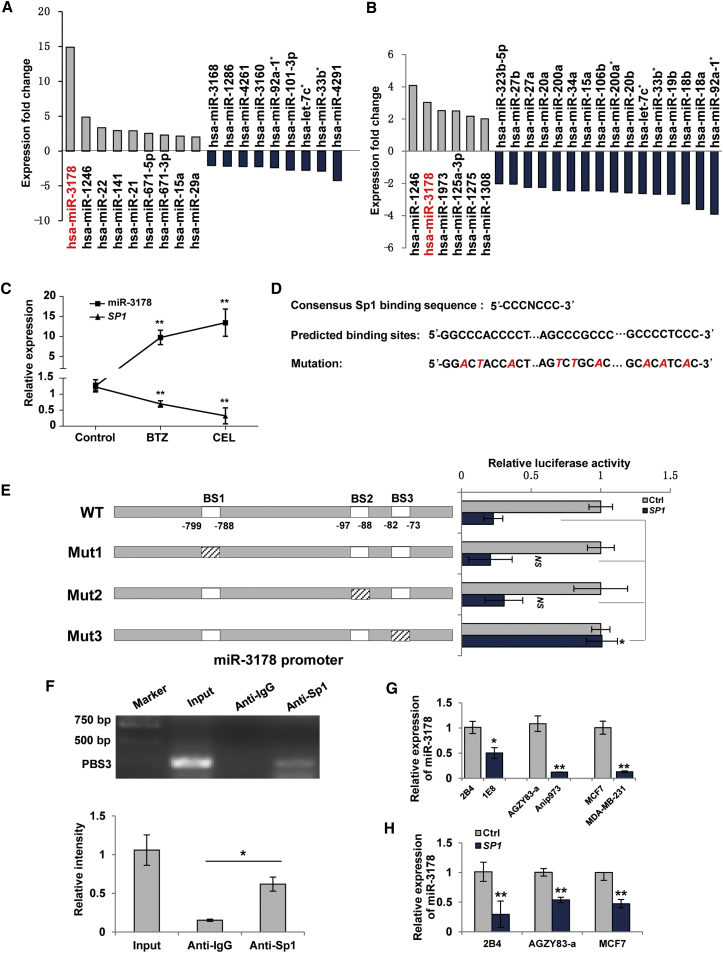
We treated prostate cancer cells with authentic proteasome inhibitor bortezomib or celastrol with proteasome inhibitory activity and performed miRNA profiling assay. miR-3178 was scored top 1 or 2 in upregulated miRNAs after both treatments (Figures 2A and 2B). Treatments with bortezomib or celastrol led to decreased expression of SP1.23, 24 Our qRT-PCR results confirmed that SP1 was downregulated in prostate cancer cells post-bortezomib or celastrol treatment, whereas miR-3178 was upregulated (Figure 2C), suggesting a negative relationship. JASPAR and PROMO were used to search for miR-3178 transcription factors, among which Sp1 was scored top 5 (data not shown). Three possible Sp1 binding sites (BSs) were predicted across a 1.5 kb sequence upstream of miR-3178 (Figure 2D). Luciferase reporter constructs containing wild-type (WT) miR-3178 promoter sequence or that with mutations at the three predicted BSs (Mut1, Mut2, and Mut3) were generated. Sp1 significantly suppressed the luciferase activity with WT miR-3178 promoter. Mutation in the first two BSs (Mut1 and Mut2) failed to rescue miR-3178 luciferase activity, whereas Mut3 could prevent the loss of luciferase activity of miR-3178 (Figure 2E), indicating that Sp1 binds to the BS3 to suppress miR-3178 transcription. Separately, chromatin immunoprecipitation (ChIP) assay was also performed to determine the specific binding of Sp1 to BS3. PC-3M-1E8 cells were fixed by 1% formaldehyde and harvested. Nuclear proteins were isolated and immunoprecipitated by Sp1 or immunoglobulin G (IgG) antibody and then DNA was extracted and amplified using PCR. DNA fragments containing BS3 were specifically amplified (62% of input), further confirming that Sp1 could bind to BS3 (Figure 2F).
Figure 2.
Sp1 Negatively Regulates miR-3178 by Binding to Its Promoter Region
(A and B) miRNA profiling analysis. LNCaP cells were treated with 100 nM bortezomib (BTZ) (A) or 2.5 μM celastrol (CEL) (B) for 12 hr. miRNAs with over 2-fold changes against control were shown. (C) Expressions of SP1 and miR-3178 post-treatments as (A) and (B) in LNCaP cells are shown. (D) Consensus Sp1 sites and predicted binding sites (BSs) of Sp1 in miR-3178 promoter are shown. Mutations of each BS were indicated by italic red cases. (E) Luciferase reporter assay is shown. Luciferase reporter constructs were generated as schematic depiction and transfected into 1E8 cells in the presence of SP1 or control (Ctrl) plasmid. Wild-type (WT) and mutant Sp1 sites (Mut1–3) were indicated by blank and italic dash, respectively. NS, non-significant difference. (F) ChIP assay is shown. DNA was immunoprecipitated with anti-IgG or anti-Sp1 antibody and amplified by PCR using primer specific for BS3. Input chromatin before immunoprecipitation was used as control. Experiments were repeated three times, and results were shown as mean ± SD in the lower panel. (G) Expressions of miR-3178 in prostate, lung, and breast cancer cell lines are shown. (H) Expression of miR-3178 in lowly metastatic cancer cells after ectopic expression of SP1 is shown. Values were shown as mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01.
Furthermore, endogenous expression levels of SP1 were found negatively correlated with miR-3178, showing SP1 highly expressed cells had low level of miR-3178 in prostate, lung, and breast cancer cells (Figure 2G versus Figure 1B). Ectopic expression of SP1 in lowly metastatic 2B4, AGZY83-a, and MCF7 cells decreased miR-3178 expression by 71%, 46%, and 53%, respectively (Figure 2H). These results indicate that Sp1 negatively regulates miR-3178 by binding to its promoter region in prostate, breast, and lung cancer cells.
miR-3178 Inhibits the Migration and Invasion Abilities of Highly Metastatic Cancer Cells
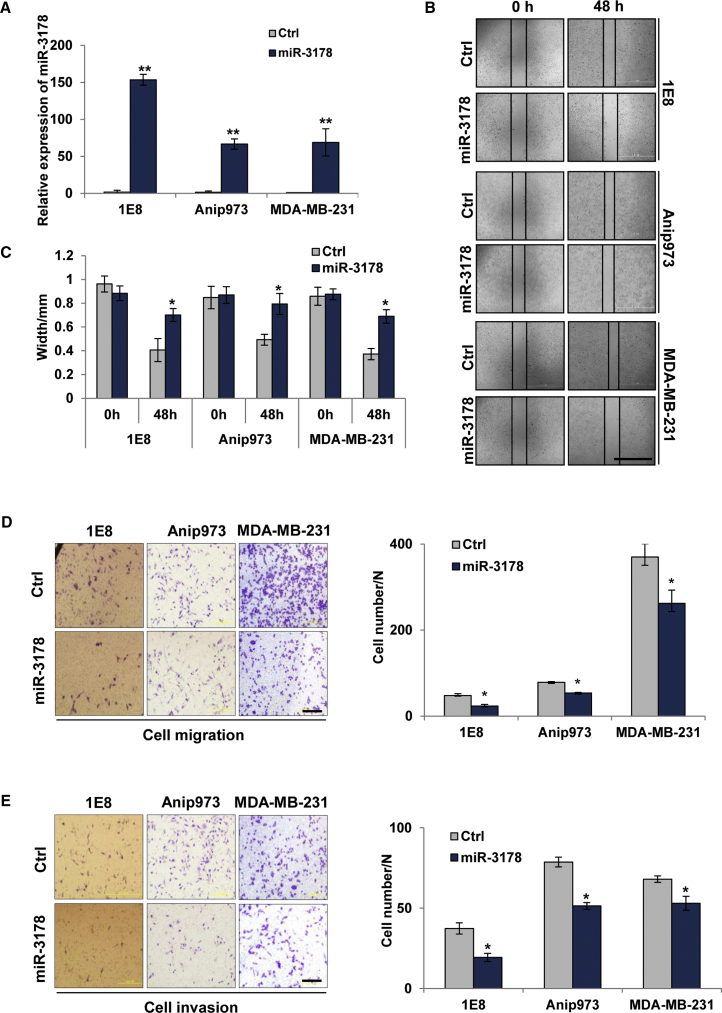
Considering the pro-metastatic function of Sp1 and its negative regulation on miR-3178, we sought to determine whether miR-3178 was a metastasis suppressor. We enhanced miR-3178 expression in highly metastatic cancer cells (1E8, Anip973, and MDA-MB-231; Figure 3A). Indeed, miR-3178 led to significant suppression on cell mobility, as shown in wound healing assay (Figure 3B), and ∼30% inhibition was observed in the three cell models (Figure 3C). In addition, miR-3178 decreased migration and invasion of highly metastatic 1E8, Anip973, and MDA-MB-231 cells compared with control (Figures 3D and 3E), showing ∼52%, 32%, and 29% inhibition on migration (Figure 3D) and 49%, 35%, and 22% inhibition on invasion in prostate, lung, and breast cancer cells, respectively (Figure 3E).
Figure 3.
miR-3178 Inhibits the Migration and Invasion Abilities of Highly Metastatic Cancer Cells
Highly metastatic cancer cells were transfected with 10 nM of miR-3178 mimics and control (Ctrl) for 24 hr. (A) miR-3178 expression after transfection is shown. (B and C) Wound healing assay (B) is shown and analyzed (C). The scale bar represents 2 mm. (D and E) Cell migration (D) and invasion (E) are shown by Transwell assay. The scale bars represent 200 μm. All experiments were repeated three times, and results were shown as mean ± SD. *p < 0.05 and **p < 0.01.
miR-3178 Inhibition Enhances the Migration and Invasion Abilities of Lowly Metastatic Cancer Cells
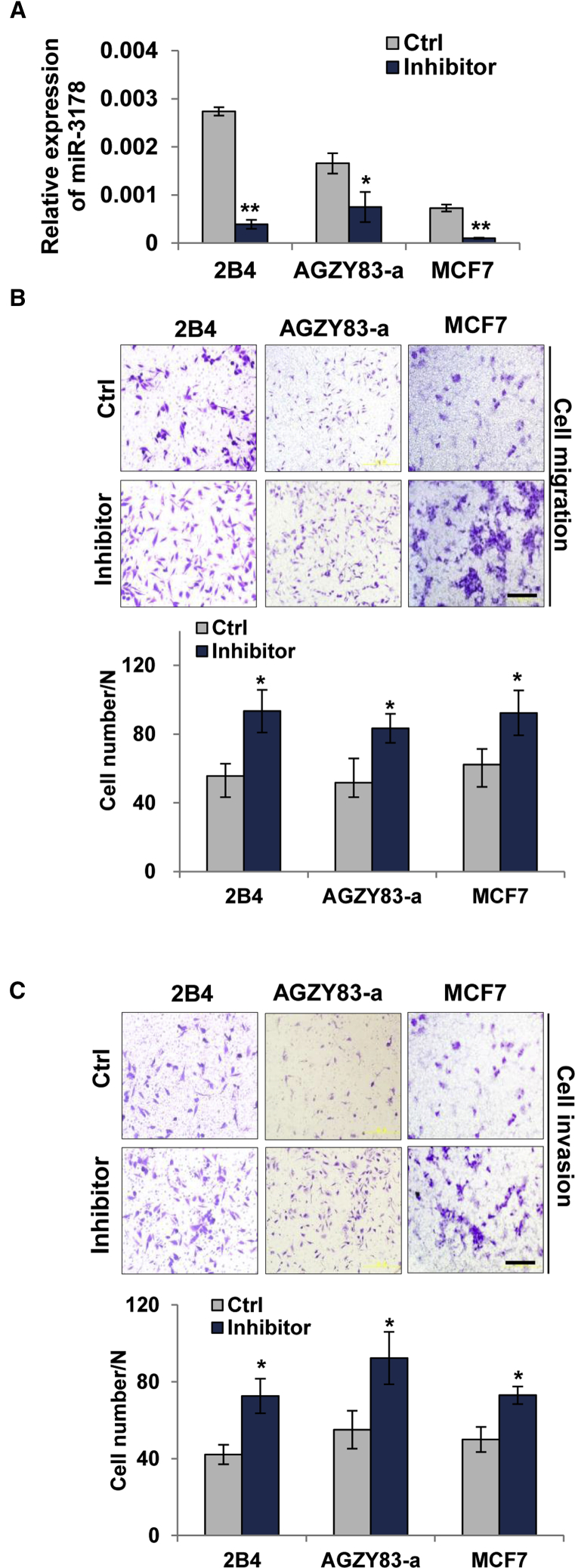
In lowly metastatic 2B4, AGZY83-a, and MCF7 cancer cells, miR-3178 inhibitor (20 nM) was transfected to reduce miR-3178 expression (Figure 4A) and then the migration and invasion abilities were evaluated. MCF7 cells that migrated through the Transwell chamber were increased by ∼46% after the miR-3178 inhibitor was transfected (Figure 4B). An even greater increase of migration (∼68%) was observed in AGZY83-a cells after miR-3178 inhibition (Figure 4B). In addition, the invasion abilities of MCF7 and AGZY83-a cells were also enhanced dramatically (by a 48% and 60% increase, respectively) after miR-3178 inhibitor transfection (Figure 4C). Neither migration nor invasion was significantly changed in 2B4 cells after transfection with the same amount of miR-3178 inhibitor (data not shown). As the endogenous level of miR-3178 in 2B4 cells was higher compared with the other two lowly metastatic cancer cells AGZY83-a and MCF7 (Figure 4A), a higher concentration of inhibitor (40 nM) was transfected into 2B4 cells to determine its effects on cell invasion and migration. The expression of miR-3178 was decreased by 85% (Figure 4A), whereas the migration and invasion abilities of 2B4 cells were increased by 66% and 71%, respectively (Figures 4B and 4C). Therefore, a high concentration of miR-3178 inhibitor was sufficient to compete with endogenous miR-3178, causing enhanced invasion and migration in 2B4 cells. The above results demonstrate that miR-3178 inhibition results in increased migration and invasion abilities in lowly metastatic cancer cells.
Figure 4.
Antagonizing miR-3178 Promotes the Migration and Invasion Abilities of Lowly Metastatic Cancer Cells
Lowly metastatic cancer cells were transfected with 20 nM (AGZY83-a and MCF-7) or 40 nM (2B4) of miR-3178 inhibitor (inhibitor) or control (Ctrl) for 24 hr. (A) miR-3178 expression after miR-3178 inhibitor transfection is shown. (B and C) Cell migration (B) and invasion (C) are shown by Transwell assay. Scale bars represent 200 μm. All experiments were repeated three times, and results were shown as mean ± SD. *p < 0.05 and **p < 0.01.
TRIOBP-5 and TRIOBP-1 Are Targets of miR-3178
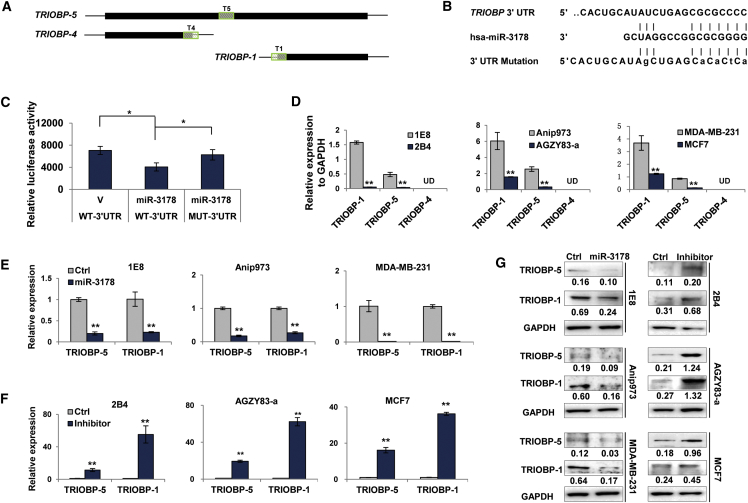
TRIOBP is mainly expressed in 3 isoforms, TRIBOP-5, TRIBOP-4, and TRIBOP-1 (Figure 5A).25 Database (TargetScan, miRanda, and miRbase) analysis identified TRIBOP-5 and TRIBOP-1 as the putative targets of miR-3178 because they share the same 3′ UTR sequences (Figure 5B). To confirm that miR-3178 could interact with their 3′ UTR, luciferase reporter constructs containing TRIOBP 3′ UTR WT (WT-3′ UTR) or mutant predicted binding site (MUT-3′ UTR) (Figure 5C) were generated and co-transfected with miR-3178 or control oligos. Luciferase activity of WT-3′ UTR decreased by 46% with miR-3178 transfection compared with control; however, it remained unchanged in the presence of miR-3178 when the putative binding site was mutated (Figure 5D), demonstrating that miR-3178 acted at the 3′ UTR putative site of TRIOBP-5 and TRIBOP-1.
Figure 5.
miR-3178 Inhibits TRIOBP
(A) Schematic mRNA structure of TRIOBP. T5, T4, and T1 represent amplification region by primers specific for TRIOBP-5, TRIOBP-4, and TRIOBP-1, respectively. Black box, open reading frame; lines, 5′ and 3′ UTR. (B and C) Luciferase reporter assay is shown. Luciferase reporter constructs containing TRIOBP 3′ UTR wild-type (WT-3′ UTR) or mutant predicted binding site (MUT-3′ UTR) were generated (B) and co-transfected with pcDNA3.1-miR-3178 (miR-3178) or empty vector (V) for 24 hr and then luciferase activity was determined (C). (D) Expression of TRIOBP in prostate, lung, and breast cancer cells is shown. UD, undetected. (E–G) miR-3178 inhibits TRIOBP mRNA and protein expression. Highly metastatic cancer cells (1E8, MDA-MB-231, and Anip973) were transfected with miR-3178 mimics (miR-3178) whereas lowly metastatic cancer cells (2B4, AGZY83-a, and MCF7) were transfected with miR-3178 inhibitor (inhibitor) for 24 hr, followed by qRT-PCR (E and F) and western blotting (G). Ctrl, control. All experiments were repeated three times, and results were shown as mean ± SD. *p < 0.05 and **p < 0.01.
qRT-PCR using isoform-specific primers (Figure 5A) confirmed the regulation of miR-3178 on TRIOBP-5 and TRIOBP-1 in all cell models. TRIOBP-1 was the major isoform expressed in these cells whereas TRIOBP-5 was expressed at much lower level (Figure 5D) and TRIOBP-4 was not detected (Figure 5D). Furthermore, expressions of TRIOBP-5 and TRIOBP-1 were negatively correlated with endogenous level of miR-3178. Highly metastatic cancer cells (1E8, MDA-MB-231, and Anip973) with low level of miR-3178 showed higher expression levels of TRIOBP-5 and TRIOBP-1 than that in their counterparts with high level of miR-3178 (2B4, MCF7, and AGZY83-a; Figure 5D). Transfection of miR-3178 into 1E8, MDA-MB-231, and Anip973 cells led to significant decreases on both isoforms compared with control (Figure 5E) whereas antagonizing miR-3178 caused dramatic increases of TRIOBP-5 and TRIOBP-1 in low metastatic cells (2B4, MCF7, and AGZY83-a; Figure 5F). Similar to TRIOBP mRNA alterations, influence of miR-3178 on TRIOBP was also reflected at protein levels (Figure 5G). All these results demonstrate that miR-3178 targets TRIOBP-1 and TRIOBP-5 in prostate and lung as well as breast cancer cells.
miR-3178 Inhibits Prostate Cancer Metastasis In Vivo
Whether miR-3178 could suppress metastasis in vivo was determined next. The red fluorescent protein dsRed was introduced into 1E8 cells to trace cancer metastasis. 1E8-dsRed cells with miR-3178 stable expression or control vector were injected into BALB/c nude mice through the tail vein, and metastasis of cancer cells was monitored through dsRed fluorescence (Figure 6A). The major sites that cancer cells colonized were lung and axillary nodes, as shown by high fluorescence intensity (Figures 6B and 6C). Weak dsRed signal was detected in liver (Figure 6D). Compared with control, overexpression of miR-3178 led to significant decrease in fluorescence intensity, showing decrease of 45% in mice body, 27% in lung, and 32% in lymph nodes (Figure 6E). Although the fluorescent signals from liver were too weak to calculate, difference could be observed between control and miR-3178 groups (Figure 6D).
Figure 6.
miR-3178 Inhibits Cancer Cell Metastasis In Vivo
(A–E) Metastasis detection by fluorescence signals. 1E8 cells stably expressing miR-3178 and dsRed fluorescent protein were injected into BALB/c nude mice (n = 9) through tail vein (2 × 106 per inoculation). Mice were sacrificed after 7 weeks, and fluorescence signals of whole body (A) and specific organs (B, lung; C, lymph node; D, liver) were detected and analyzed (E). (F) H&E staining is shown. Arrowheads indicate cancer metastasis foci. The scale bar represents 4×, 2 mm; 10×, 500 μm; and 40×, 200 μm. (G) 1E8 cells were co-transfected with miR-3178 and pCNDA3.1-TRIOBP-1 (TRIOBP-1) or control vector (Ctrl) for 24 hr, followed by western blotting and Transwell assays. *p < 0.05 and **p < 0.01.
H&E staining revealed extensive metastasis of 1E8 cells to lung, lymph node, and liver in control group, whereas it was significantly suppressed in miR-3178 group (Figure 6F). Due to occupancy of prostate cancer cells, the numbers of alveolar cavities were decreased in the control group compared with the miR-3178 overexpressed group (Figure 6F). Clear boundaries between tumor and non-tumorous regions could be observed in lymph nodes and liver tissue. Prostate cancer cells appeared as crowded and clustered sub-populations with high nuclear/cytoplasmic ratio, enlarged and/or multiple nuclear phase, and tight contact between cells, showing typical characteristics of cancer cells (Figure 6F). These metastasis colonies formed by 1E8 cells in lymph node and liver were decreased by miR-3178 compared with control (Figure 6F).
Whether miR-3178 exerted anti-metastasis function through its target TRIOBP was determined. TRIOBP-1 has been reported to regulate cytoskeleton organization.26 Protein structure analysis indicates that the central coiled-coil domain of TRIOBP-1 is responsible for actin binding.27, 28 Then, we cloned TRIOBP-1, as it was the major isoform in prostate cancer cells. miR-3178 was co-transfected with or without TRIOBP-1 into 1E8 cells. Again, miR-3178 caused significant decrease in TRIOBP-1 protein level (Figure 6G) and dramatic reduction on cell migration and invasion (56 ± 4 versus 28 ± 2 and 26 ± 6 versus 9 ± 2, respectively). However, co-transfection of TRIOBP-1 could reverse the effect of miR-3178, showing that both migration and invasion were increased to comparable levels to control (Figure 6G). These results demonstrate that TRIOBP-1 protein serves as the downstream target of miR-3178, and miR-3178 exerts its anti-metastasis function at least partially through inhibiting TRIOBP-1 expression.
Discussion
Sp1 is ubiquitously expressed and dysregulated in various cancers. About 12,000 Sp1 sites are estimated existing in human genome; among them, Sp1 regulation on protein coding genes has been extensively investigated,4,29,30 whereas its regulation on non-protein coding genes was less studied. Recently, few studies explored Sp1 regulation on miRNAs. For example, Sp1 activated miR-19a, which could decrease RHOB and promote pancreatic cancer progression,31 and Sp1 could also increase miR-182 expression and enhance invasive and migratory abilities of lung cancer cell lines;32 besides, Sp1 could transactivate miR-520d-5p by binding to its upstream promoter region.33 In addition, Sp1 and miR-22 were found negatively regulating each other in colorectal cancer.34 In the present study, miR-3178 was reported as a novel target of Sp1. Luciferase activity assay showed that Sp1 could suppress the promoter activity of miR-3178. The promoter of miR-3178 harbored three predicted Sp1 binding sites, among which the third binding site (BS3) was responsible for Sp1-mediated transcription suppression. Mutation of BS3 could rescue Sp1 suppression on miR-3178 transcription, and ChIP assay confirmed the specific binding of Sp1 to BS3 (Figures 2E and 2F). In addition, endogenous expression level of SP1 was also negatively correlated with miR-3178 and overexpression of SP1 resulted in downregulation of miR-3178 in prostate, lung, and breast cancer cells. We noticed a 2-fold higher expression of SP1, whereas almost 10-fold lower expression of miR-3178 was detected in Anip973 than that in AGZY83-a cells (Figure 1B), suggesting that miR-3178 might also be regulated by other factors in addition to Sp1 in these two lung cancer cell lines. Snail, which is upregulated in Anip973 cells compared with AGZY83-a,35 was also predicted to regulate miR-3178. In addition, KLF4, MEF2C, GLI2, and Sharp1 (also named BHLHE41 or DEC2), along with Sp1, were listed as the top 5 predicted transcriptional factors of miR-3178. GLI2 can promote migration and invasion of osteosarcoma cells.36 Overexpression of KLF4 leads to reduction on migration and invasion of cancer cells.37 As a metastasis suppressor,38 Sharp1 competes with Sp1 for the binding to Twist1 promoter region and suppresses Twist1 transcription acitivity.39 These metastasis regulators might also contribute to miR-3178 regulation in highly metastatic Anip973 cells.
Downregulation of miR-3178 was detected in patients with positive lymphatic metastasis of the primary gastric tumor,40 suggesting that it might contribute to suppression of gastric cancer to lymphatic metastasis. Subsequent observation linked this miRNA to suppression of invasion and angiogenesis in hepatocellular carcinoma in vitro.41 In contrast to these observations, miR-3178 was found upregulated specifically in liver metastasis of colorectal cancer tissues rather than in primary tumors.42 Our results clearly demonstrate that miR-3178 is a metastasis suppressor in the tested cancer cell models. First, miR-3178 expression was significantly decreased in cancer cell lines with highly metastatic capacity compared with their lowly metastatic counterparts. Second, using gain- and loss-of-function approaches, we showed that upregulation of miR-3178 suppressed whereas downregulation of miR-3178 promoted both migration and invasion abilities of cancer cells. Most importantly, in vivo study confirmed the anti-metastasis function of miR-3178, showing that miR-3178 decreased the metastasis of prostate cancer cells to lymph nodes, lung, as well as liver. Moreover, according to data from Oncomine database, metastatic prostate, lung, or breast cancer expressed higher levels of SP1 compared with the primary tumors they were derived from. This observation is in accordance with the anti-metastasis function of miR-3178, as Sp1 was shown to be a transcription suppressor of miR-3178. The discrepancy between our finding and recent observation showing that miR-3178 was upregulated in liver metastatic colorectal tissues versus non-tumoral tissues might be caused by different cancer types or the highly heterogeneous property within certain types of cancer. Therefore, the expression levels of miR-3178 in metastatic versus primary prostate, lung, or breast cancer samples need to be determined in the future. This is the limitation of our current study.
MicroRNAs mediate intracellular effects by targeting multiple protein coding genes through binding to the 3′ UTR regions. In this study, TRIOBP was found to be the downstream target for miR-3178 to mediate anti-metastasis function. TRIOBP is an actin-bundling protein that is classified into three alternative splicing isoforms, the long isoform TRIOBP-5 and two shorter isoforms, TRIOBP-4 and TRIOBP-1 (also named Tara). TRIOBP-1 is ubiquitously expressed, whereas TRIOBP-4 and TRIOBP-5 are predominantly expressed in the inner ear and the retina of normal adults.27 In pancreatic cancer cells, TRIOBP-4 and TRIOBP-5 were reported upregulated and knockdown of TRIOBP-4 and TRIOBP-5 inhibited the filopodial formation and motility of pancreatic cancer cells.43 TRIOBP-4 was also detected in other human cancer cell lines, such as HeLa.44 However, it was not detected in the prostate, lung, and breast cancer cell lines tested in this study. TRIOBP-5 was marginally detected, whereas TRIOBP-1 was the major isoform expressed in those cells (Figures 5D and 5G). Luciferase reporter assay indicated that miR-3178 could bind to the 3′ UTR of TRIOBP-1, which is shared by TRIOBP-5, causing decrease in luciferase activity. Moreover, miR-3178 could target TRIOBP-1 for mediating anti-metastasis in cancers. miR-3178 overexpression caused decrease of TRIOBP-1 in highly metastatic cancer cell lines, whereas inhibition of miR-3178 led to increase of TRIOBP-1 in lowly metastatic cancer cell lines (Figures 5E–5G). TRIOBP-1 regulates cytoskeleton organization by stabilizing F-actin, thus promoting cell spreading.27 It also decreases E-cadherin expression.45 Ectopic expression of TRIOBP-1 could rescue miR-3178 inhibition on both cell migration and invasion, demonstrating that TRIOBP-1 is the major downstream target of miR-3178, inhibition of which contributes to suppression of cancer metastasis (Figure 6G).
In summary, we reveal the role of Sp1/miR-3178/TRIOBP axis in metastasis regulation of prostate, lung, and breast cancer cells. Our results demonstrate the anti-metastasis function of miR-3178 in multiple cancer models, providing a possibility of suppressing metastasis through enhancement of miR-3178 expression in Sp1 overexpressed cancers.
Materials and Methods
Cell Culture
Human prostate cancer cell lines PC-3M-1E8 and PC-3M-2B4 were from Dr. Xiaoguang Yu (Harbin Medical University, Harbin, China). Human lung adenocarcinoma AGZY83-a and Anip973 were gifts from Dr. Li Yu (Harbin Institute of Technology, Harbin, China). Human breast cancer cell lines MDA-MB-231 and MCF7 were obtained from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air in RPMI 1640 (for prostate and lung cancer cells) or DMEM (for breast cancer cells) supplemented with 10% fetal bovine serum (Gibco-BRL, Grand Island, NY, USA).
Primers and Oligos
Primers used in all experiments were synthesized at Comate Biosciences (Jilin, China), and miR-3178 mimics and according inhibitors were purchased from Ribio Company (Guangzhou, China). Specific primers for TRIOBP-1, TRIOBP-4, and TRIOBP-5 were designed with reference.43 The primer and oligo sequences are listed in Table 1.
Table 1.
Sequences of RNA Oligos and Primers
| Oligos and Primers | Sequences |
|---|---|
| RNA Oligos | |
| miR-3178 mimic | 5′-GGGGCGCGGCCGGAUCG-3′ |
| Negative control | 5′-UUUGUACUACACAAAAGUACUG-3′ |
| qRT-PCR Primers | |
| GAPDH | F: 5′-AATCCCATCACCATCTTCCA-3′ |
| R: 5′-TGGACTCCACGACGTACTCA-3′ | |
| miR-3178 | F: 5′-GGGGCGCGGCCGGAUCG-3′ |
| R: 5′-GCTGTCAACGATACGCTACGTA-3′ | |
|
U6 |
F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| Primers for Cloning | |
| TRIOBP 3′ UTR | F: 5′-TATCTGAGTGTGCTCCTCGCACT-3′ |
| R: 5′-CGACGCGTCCTGTTCTGCCAGCCATC-3′ | |
| miR-3178 binding site mutation | F: 5′-TATCTGAGTGTGCTCCTCGCACT-3′ |
| R: 5′-AGTGCGAGGAGCACACTCAGATA-3′ | |
| miR-3178 promoter | F: 5′-CCGCTCGAGAGATCGCACCACTGCACTCCAGC-3′ |
| R: 5′-CCCAAGCTTGTAGCGCAGGCGCTGAGTCCAGG-3′ | |
| miR-3178 promoter mutation 1 | F: 5′-GGACTACCCATCCGCTTCATTTC-3′ |
| R: 5′-GAAATGAAGCGGATGGGTAGTCC-3′ | |
| miR-3178 promoter mutation 2 | F: 5′-GACAGAAGTCTGCACCCAGAAGC-3′ |
| R: 5′-GCTTCTGGGTGCAGACTTCTGTC-3′ | |
| miR-3178 promoter mutation 3 |
F: 5′-CAGAAGCACATCACACAGTAGGC-3′ |
| R: 5′-GCCTACTGTGTGATGTGCTTCTG-3′ | |
| Primers for ChIP | |
| Predicted binding site 3 | F: 5′-GCCGCGCGGGTTAACTAGA-3′ |
| R: 5′-CAGGACGCTCTCGACCGAT-3′ | |
Real-Time qPCR
Total RNA (1.0 μg) was extracted using TRIzol reagent (Thermo Fisher, Waltham, MA, USA) and used to synthesize cDNA with EasyScript First-Strand cDNA Synthesis SuperMix (Transgen, Beijing, China). qPCR was performed using SYBR Premix Ex Taq II (Takara, Otsu, Shiga, Japan).
Plasmid Construction and Cell Transfection
DNA fragment of miR-3178 stem-loop sequence was amplified by PCR from human genome DNA, cloned into pcDNA3.1 or pdsRed2-C1 vector at sites between EcoRI and XhoI (Takara, Otsu, Shiga, Japan). The fragment of TRIOBP 3′ UTR was amplified and cloned into pMIR-REPORT vector between EcoRI and BamHI (Takara, Otsu, Shiga, Japan).
Cell transfection was performed using Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s instructions. For stable transfection, pdsRed2-C1 or pdsRed2-C1-miR-3178 vector was transfected into PC-3M-1E8 cells. G418 (600 μg/mL) was used for stable colonies selection.
Microarray
LNCaP cells were treated with 100 nM of bortezomib or 2.5 μM of celastrol for 12 hr, and total RNA was extracted using TRIzol reagent following protocol described above. Microarray was performed on Affymetrix GeneChip 3000 TG System (Tianjin Biochip, Tianjin, China) using biotin as probe, and results were analyzed by Partek GS software. The raw data have been deposited in ArrayExpress: E-MTAB-6215.
Western Blotting
Whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 0.8% Triton X-100, and protease inhibitor cocktail (Roche, Mannheim, Germany). Western blotting was performed as described previously.46
Wound Healing Assay
Cells (3 × 105 per well) were seeded in 6-well plate and starved in serum-free medium for 24 hr and then the linear wound of cell monolayer was created by scratching confluent cells using a plastic pipette tip. The scratched cells were washed with PBS to remove debris, and the remaining cells were photographed after 48-hr incubation at 37°C.
Transwell Assay
Cell migration assay was performed using Transwell chambers with an 8-μm-pore-sized polycarbonate filter membrane (Corning Costar, Cambridge, MA, USA). Cells were trypsinized and resuspended in serum-free medium. Upper wells were filled with cell suspensions in serum-free medium, and lower wells were filled with medium containing 10% fetal bovine serum. After incubation for 48 hr at 37°C with 5% CO2, the lower side of filter membrane was fixed with methanol and stained with crystal violet (Sinopharm Chemical Reagent, Shanghai, China). The migrated cells were counted under microscope and quantified by ImageJ software (NIH, Bethesda, MD, USA).
Luciferase Reporter Assay
Cells were seeded into 24-well plate (1 × 105 cells per well). After attachment, 0.3 μg of pcDNA3.1-miR-3178 was co-transfected, with 0.3 μg of pMIR-WT-3′ UTR or pMIR-MUT-3′ UTR in the presence of 0.03 μg of pRLSV40, into cells for 48 hr. Luciferase activities were assessed by dual-luciferase reporter assay system (Promega, Fitchburg, WI, USA).
In Vivo Study
BALB/c nude mice aged at 4 weeks were obtained from Cavens Lab Animal Company (Jiangsu, China). All mice were housed and manipulated with humanistic care according to the protocols approved by Medical Experimental Animal Care Commission of Harbin Institute of Technology. The cells (2 × 106) were injected through the tail vein. Mice were sacrificed, and fluorescence signals were measured 7 weeks later. Lung, liver, and lymph nodes were fixed in 10% formalin for 48 hr and then embedded in paraffin. The tissues were sectioned at 7-μm thickness and then H&E staining was performed as previous description.47
ChIP Assay
ChIP assay was performed according to previous description.48 Briefly, about 1 × 107 PC-3M-1E8 cells were first fixed in 1% formaldehyde for 10 min at 37°C. Then, fixed cells were harvested and lysed with dounce homogenizer. Nuclei were collected and resuspended in hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, and 10 mM KCl). The nuclei were sonicated to an average DNA length of 200–1,000 bp. The sheared DNA solution was precleared by adding 60 μL of protein A/G Sepharose beads (MedChem Express, Monmouth Junction, NJ, USA) and shaken at 4°C for 1 hr. Precleared solution was then incubated with 2 μg of Sp1 or IgG antibody (Proteintech, Rosemont, IL, USA) at 4°C for 2 hr, followed by incubation of protein A/G Sepharose beads at 4°C for 1 hr. Then, the immune complexes were collected and cross-links were reversed by adding NaCl to a final concentration of 200 mM and incubation at 65°C for 4 hr. DNA was extracted using phenol-chloroform and analyzed by PCR. The sequences of PCR primers are listed in Table 1.
Statistical Analysis
Microsoft Excel software was used for data statistical analysis. Results were described as mean ± SD. Statistical significance between two groups was determined with Student’s t test.
Author Contributions
H.Y. designed the study and wrote the paper. H.W. performed the experiments, analyzed the data, and drafted the manuscript. K.L., Y.M., X.H., Q.Y., and Z.L. performed the experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors thank Dr. Yu Li (Harbin Institute of Technology) for providing Sp1 expression vectors and prostate and lung cancer cell lines. This work was supported by the Heilongjiang Postdoctoral Fund (LBH-Z16072), the China Postdoctoral Science Foundation (2017M610201), and the Harbin Science and Technology Program (2017RAQXJ182).
References
- 1.Cai Y., Yu X., Hu S., Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nam E.H., Lee Y., Zhao X.F., Park Y.K., Lee J.W., Kim S. ZEB2-Sp1 cooperation induces invasion by upregulating cadherin-11 and integrin α5 expression. Carcinogenesis. 2014;35:302–314. doi: 10.1093/carcin/bgt340. [DOI] [PubMed] [Google Scholar]
- 3.Kolesnikoff N., Attema J.L., Roslan S., Bert A.G., Schwarz Q.P., Gregory P.A., Goodall G.J. Specificity protein 1 (Sp1) maintains basal epithelial expression of the miR-200 family: implications for epithelial-mesenchymal transition. J. Biol. Chem. 2014;289:11194–11205. doi: 10.1074/jbc.M113.529172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian Y., Yao W., Yang T., Yang Y., Liu Y., Shen Q., Zhang J., Qi W., Wang J. aPKC-ι/P-Sp1/Snail signaling induces epithelial-mesenchymal transition and immunosuppression in cholangiocarcinoma. Hepatology. 2017;66:1165–1182. doi: 10.1002/hep.29296. [DOI] [PubMed] [Google Scholar]
- 5.Xu K., Shu H.K. EGFR activation results in enhanced cyclooxygenase-2 expression through p38 mitogen-activated protein kinase-dependent activation of the Sp1/Sp3 transcription factors in human gliomas. Cancer Res. 2007;67:6121–6129. doi: 10.1158/0008-5472.CAN-07-0141. [DOI] [PubMed] [Google Scholar]
- 6.Hertel J., Hirche C., Wissmann C., Ebert M.P., Höcker M. Transcription of the vascular endothelial growth factor receptor-3 (VEGFR3) gene is regulated by the zinc finger proteins Sp1 and Sp3 and is under epigenetic control: transcription of vascular endothelial growth factor receptor 3. Cell Oncol. (Dordr.) 2014;37:131–145. doi: 10.1007/s13402-014-0169-5. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Wang Y. Bioinformatics analysis of gene expression data for the identification of critical genes in breast invasive carcinoma. Mol. Med. Rep. 2017;16:8657–8664. doi: 10.3892/mmr.2017.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan H., Cai J., Zhang N., Wu J., Yuan J., Li J., Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int. J. Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 9.Jiang N.Y., Woda B.A., Banner B.F., Whalen G.F., Dresser K.A., Lu D. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 2008;17:1648–1652. doi: 10.1158/1055-9965.EPI-07-2791. [DOI] [PubMed] [Google Scholar]
- 10.Briggs M.R., Kadonaga J.T., Bell S.P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 11.Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 12.Olive V., Minella A.C., He L. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci. Signal. 2015;8:re2. doi: 10.1126/scisignal.2005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Hurst D.R., Edmonds M.D., Welch D.R. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 17.Yen W.H., Ke W.S., Hung J.J., Chen T.M., Chen J.S., Sun H.S. Sp1-mediated ectopic expression of T-cell lymphoma invasion and metastasis 2 in hepatocellular carcinoma. Cancer Med. 2016;5:465–477. doi: 10.1002/cam4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Zheng J., Fang W., You J., Wang J., Cui X., Wu B. [Isolation and characterization of human prostate cancer cell subclones with different metastatic potential] Zhonghua Bing Li Xue Za Zhi. 1999;28:361–364. [PubMed] [Google Scholar]
- 20.Meng X., Li Y., Zhang G. [Molecular cloning of tumor metastasis related genes from human lung adenocarcinoma cells by mRNA differential display] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 1996;14:129–133. [Google Scholar]
- 21.Rizwan A., Cheng M., Bhujwalla Z.M., Krishnamachary B., Jiang L., Glunde K. Breast cancer cell adhesome and degradome interact to drive metastasis. NPJ Breast Cancer. 2015;1:15017. doi: 10.1038/npjbcancer.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soule H.D., Vazguez J., Long A., Albert S., Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 23.Chadalapaka G., Jutooru I., Safe S. Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis. 2012;33:886–894. doi: 10.1093/carcin/bgs102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S., Liu Z., Xie Z., Pang J., Yu J., Lehmann E., Huynh L., Vukosavljevic T., Takeki M., Klisovic R.B. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111:2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitajiri S., Sakamoto T., Belyantseva I.A., Goodyear R.J., Stepanyan R., Fujiwara I., Bird J.E., Riazuddin S., Riazuddin S., Ahmed Z.M. Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell. 2010;141:786–798. doi: 10.1016/j.cell.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seipel K., O’Brien S.P., Iannotti E., Medley Q.G., Streuli M. Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J. Cell Sci. 2001;114:389–399. doi: 10.1242/jcs.114.2.389. [DOI] [PubMed] [Google Scholar]
- 27.Shahin H., Walsh T., Sobe T., Abu Sa’ed J., Abu Rayan A., Lynch E.D., Lee M.K., Avraham K.B., King M.C., Kanaan M. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am. J. Hum. Genet. 2006;78:144–152. doi: 10.1086/499495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradshaw N.J., Yerabham A.S.K., Marreiros R., Zhang T., Nagel-Steger L., Korth C. An unpredicted aggregation-critical region of the actin-polymerizing protein TRIOBP-1/Tara, determined by elucidation of its domain structure. J. Biol. Chem. 2017;292:9583–9598. doi: 10.1074/jbc.M116.767939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng M., Hu Y., Song W., Duan S., Xu Q., Ding Y., Geng J., Zhou J. MIER3 suppresses colorectal cancer progression by down-regulating Sp1, inhibiting epithelial-mesenchymal transition. Sci. Rep. 2017;7:11000. doi: 10.1038/s41598-017-11374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajpai R., Nagaraju G.P. Specificity protein 1: its role in colorectal cancer progression and metastasis. Crit. Rev. Oncol. Hematol. 2017;113:1–7. doi: 10.1016/j.critrevonc.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Tan Y., Yin H., Zhang H., Fang J., Zheng W., Li D., Li Y., Cao W., Sun C., Liang Y. Sp1-driven up-regulation of miR-19a decreases RHOB and promotes pancreatic cancer. Oncotarget. 2015;6:17391–17403. doi: 10.18632/oncotarget.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W.B., Chen P.H., Hsu T., Fu T.F., Su W.C., Liaw H., Chang W.C., Hung J.J. Sp1-mediated microRNA-182 expression regulates lung cancer progression. Oncotarget. 2014;5:740–753. doi: 10.18632/oncotarget.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan L., Yu J., Tan F., Ye G.T., Shen Z.Y., Liu H., Zhang Y., Wang J.F., Zhu X.J., Li G.X. SP1-mediated microRNA-520d-5p suppresses tumor growth and metastasis in colorectal cancer by targeting CTHRC1. Am. J. Cancer Res. 2015;5:1447–1459. [PMC free article] [PubMed] [Google Scholar]
- 34.Xia S.S., Zhang G.J., Liu Z.L., Tian H.P., He Y., Meng C.Y., Li L.F., Wang Z.W., Zhou T. MicroRNA-22 suppresses the growth, migration and invasion of colorectal cancer cells through a Sp1 negative feedback loop. Oncotarget. 2017;8:36266–36278. doi: 10.18632/oncotarget.16742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K., Li X.Y., Wang Z.M., Han Z.F., Zhao Y.H. miR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3598–3604. [PubMed] [Google Scholar]
- 36.Nagao-Kitamoto H., Nagata M., Nagano S., Kitamoto S., Ishidou Y., Yamamoto T., Nakamura S., Tsuru A., Abematsu M., Fujimoto Y. GLI2 is a novel therapeutic target for metastasis of osteosarcoma. Int. J. Cancer. 2015;136:1276–1284. doi: 10.1002/ijc.29107. [DOI] [PubMed] [Google Scholar]
- 37.Dang D.T., Chen X., Feng J., Torbenson M., Dang L.H., Yang V.W. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–3430. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montagner M., Enzo E., Forcato M., Zanconato F., Parenti A., Rampazzo E., Basso G., Leo G., Rosato A., Bicciato S. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 39.Asanoma K., Liu G., Yamane T., Miyanari Y., Takao T., Yagi H., Ohgami T., Ichinoe A., Sonoda K., Wake N., Kato K. Regulation of the mechanism of TWIST1 transcription by BHLHE40 and BHLHE41 in cancer cells. Mol. Cell. Biol. 2015;35:4096–4109. doi: 10.1128/MCB.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang B., Jing C., Wang J., Guo X., Chen Y., Xu R., Peng L., Liu J., Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin. Transl. Oncol. 2014;16:374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 41.Li W., Shen S., Wu S., Chen Z., Hu C., Yan R. Regulation of tumorigenesis and metastasis of hepatocellular carcinoma tumor endothelial cells by microRNA-3178 and underlying mechanism. Biochem. Biophys. Res. Commun. 2015;464:881–887. doi: 10.1016/j.bbrc.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 42.Sayagués J.M., Corchete L.A., Gutiérrez M.L., Sarasquete M.E., Del Mar Abad M., Bengoechea O., Fermiñán E., Anduaga M.F., Del Carmen S., Iglesias M. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget. 2016;7:72908–72922. doi: 10.18632/oncotarget.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao J., Wang S., Gunther L.K., Kitajiri S., Li C., Sakamoto T. The actin-bundling protein TRIOBP-4 and -5 promotes the motility of pancreatic cancer cells. Cancer Lett. 2015;356(2 Pt B):367–373. doi: 10.1016/j.canlet.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Bao J., Bielski E., Bachhawat A., Taha D., Gunther L.K., Thirumurugan K., Kitajiri S., Sakamoto T. R1 motif is the major actin-binding domain of TRIOBP-4. Biochemistry. 2013;52:5256–5264. doi: 10.1021/bi400585h. [DOI] [PubMed] [Google Scholar]
- 45.Yano T., Yamazaki Y., Adachi M., Okawa K., Fort P., Uji M., Tsukita S., Tsukita S. Tara up-regulates E-cadherin transcription by binding to the Trio RhoGEF and inhibiting Rac signaling. J. Cell Biol. 2011;193:319–332. doi: 10.1083/jcb.201009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y., Zhang Z., Li K., Gong L., Yang Q., Huang X., Hong C., Ding M., Yang H. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis. 2017;8:e2924. doi: 10.1038/cddis.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H., Chen D., Cui Q.C., Yuan X., Dou Q.P. Celastrol, a triterpene extracted from the Chinese “Thunder of God Vine,” is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 48.Guo J., Huang X., Wang H., Yang H. Celastrol induces autophagy by targeting AR/miR-101 in prostate cancer cells. PLoS One. 2015;10:e0140745. doi: 10.1371/journal.pone.0140745. [DOI] [PMC free article] [PubMed] [Google Scholar]