Abstract
Of familial amyotrophic lateral sclerosis (fALS) cases, 20% are caused by mutations in the gene encoding human cytosolic Cu/Zn superoxide dismutase (hSOD1). Efficient translation of the therapeutic potential of RNAi for the treatment of SOD1-ALS patients requires the development of vectors that are free of significant off-target effects and with reliable biomarkers to discern sufficient target engagement and correct dosing. Using adeno-associated virus serotype 9 to deliver RNAi against hSOD1 in the SOD1G93A mouse model, we found that intrathecal injection of the therapeutic vector via the cisterna magna delayed onset of disease, decreased motor neuron death at end stage by up to 88%, and prolonged the median survival of SOD1G93A mice by up to 42%. To our knowledge, this is the first report to demonstrate no significant off-target effects linked to hSOD1 silencing, providing further confidence in the specificity of this approach. We also report the measurement of cerebrospinal fluid (CSF) hSOD1 protein levels as a biomarker of effective dosing and efficacy of hSOD1 knockdown. Together, these data provide further confidence in the safety of the clinical therapeutic vector. The CSF biomarker will be a useful measure of biological activity for translation into human clinical trials.
Keywords: ALS, AAV9, shRNA, SOD1, clinical vector, off-target effects, cisterna magna, neurodegeneration, motor neuron, biomarker
Development of off-target effect-free AAV9-shRNA-mediated SOD1 silencing in an ALS mouse model, with reliable biomarkers to discern sufficient target engagement and correct dosing.
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating progressive and fatal neurodegenerative disorder. Most commonly presenting as a late-onset adult disorder, ALS typically results in death from respiratory failure approximately 3–5 years from disease onset. ALS is primarily characterized by the degeneration of upper and lower motor neurons, and, as a result, patients experience widespread and debilitating muscle atrophy and weakness. In addition to impaired movement, muscle atrophy leads to further problems, including difficulty swallowing, talking, and breathing. Of ALS cases, 90% are sporadic (sALS), with the remaining 10% having an identifiable hereditary component (familial ALS [fALS]).
The first dominant gain-of-function mutations found to cause fALS were discovered in 1993 in the gene encoding the antioxidant protein superoxide dismutase 1 (SOD1).1 Since then, many pathogenic mutations have been discovered throughout the SOD1 gene, and the genetic etiology of sporadic and fALS has expanded dramatically to implicate more than 30 different genes.2, 3 More than two decades of research has demonstrated that the process of motor neuron injury in the presence of SOD1 mutations is complex, involving multiple pathophysiological processes.4 This complexity produces difficulties for neuroprotective therapy development. Therefore, the upstream strategy of lowering the expression of the mutant SOD1 protein using gene therapy is compelling, particularly as knockout mouse models show that this can be done relatively safely.5
Other than palliative care, the available treatments for ALS only marginally influence survival (riluzole) or disability progression (edaravone).6, 7 Gene therapy shows great promise for the treatment of SOD1-linked fALS. Specifically, lentiviral delivery of a short hairpin RNA (shRNA) targeting human SOD1 (hSOD1) mRNA can delay disease progression and extend survival in mice carrying the SOD1 G93A mutation.8, 9 This approach was further confirmed by Foust et al.,10 who exchanged the lentiviral vector for a self-complementary recombinant adeno-associated virus serotype 9 (scAAV9) vector10 more suitable for use in human clinical trials.11
In the present proof-of-concept study, we evaluated the therapeutic efficacy of scAAV9-shRNA-mediated SOD1 silencing delivered intrathecally via the cisterna magna in the SOD1G93A mouse model at post-natal day 1 (P1) or 40 (P40) using a vector configuration ready to be translated into a human clinical trial for SOD1-linked fALS. The findings reported here are timely and are of great relevance for clinical translation as follows: (1) evaluation of the final clinical configuration of the therapeutic vector led to remarkable efficacy in the SOD1G93A mouse model; (2) to our knowledge, this is the first report to demonstrate no significant off-target effects linked to scAAV9-mediated SOD1 silencing, providing further confidence in the specificity and safety of the clinical therapeutic vector we have developed; and (3) we report the measurement of cerebrospinal fluid (CSF) hSOD1 protein level as a biomarker of effective dosing and efficacy of SOD1 knockdown. The same approach will be used as a readout for the biological activity of our therapeutic vector when administered in fALS patients.
Results
Validation of the Clinical Therapeutic Vector for scAAV9-Mediated hSOD1 Silencing
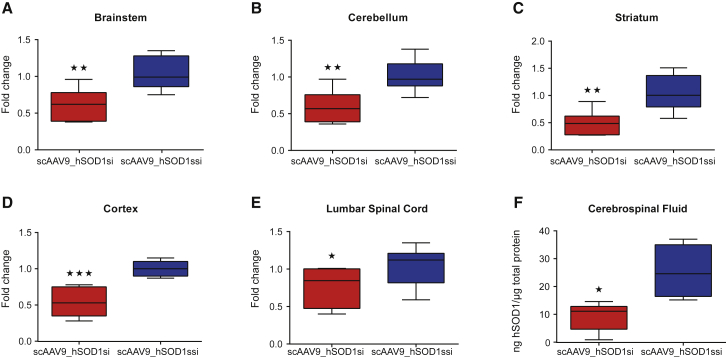
Clinical configuration of the therapeutic vector was designed by the group of Brian Kaspar at Nationwide Childrens Hospital, Columbus, OH, USA. The scAAV backbone contains an H1 promoter to drive expression of the shRNA targeted against the hSOD1 gene. We first ensured that the therapeutic vector was functional and able to mediate hSOD1 depletion. Given that our previous studies revealed that cisterna magna is an attractive route of delivery for gene transfer to the CNS,12 we set out to assess the efficiency of scAAV9_hSOD1si for SOD1 depletion in SOD1G93A mice, a widely used mouse model of ALS. P1 mice (n = 9) received scAAV9_hSOD1si (5 × 1010 vector genomes [vg]/g) via the cisterna magna. A second group of animals (n = 10) was treated with scrambled control vector scAAV9_hSOD1ssi (5 × 1010 vg/g). CNS tissue and CSF samples were collected from these mice 4 weeks post-vector delivery and used to measure hSOD1 mRNA and protein levels. scAAV9_hSOD1si-injected animals displayed lower levels of hSOD1 compared with controls in all the analyzed CNS regions. Specifically, we found a reduction in hSOD1 mRNA of 38% (Figure 1A), 43% (Figure 1B), 56% (Figure 1C), 45% (Figure 1D), and 26% (Figure 1E) in brainstem, cerebellum, striatum, cortex, and lumbar spinal cord, respectively.
Figure 1.
Cisterna Magna Delivery of scAAV9_hSOD1si in SOD1G93A Mice Efficiently Reduces hSOD1 mRNA and Protein in Brain, Spinal Cord, and CSF
(A–F) qRT-PCR analysis shows depletion of hSOD1 mRNA in brainstem (A), cerebellum (B), striatum (C), cortex (D), and lumbar spinal cord (E) 4 weeks post-cisterna magna delivery of scAAV9_hSOD1si in post-natal day 1 (P1) SOD1G93A mice. ELISA analysis shows a significant decrease in hSOD1 protein in CSF from the same group of mice (F). Data are presented as box and whiskers plot showing median, 25th and 75th percentiles, and maximum and minimum values. Data were analyzed by Student’s one-tailed t test (unpaired) (A–E, n ≥ 6; F, n ≥ 4). *p < 0.05, **p < 0.01, and ***p < 0.001.
One of the major bottlenecks in therapy development for neurodegeneration and for ALS in particular is the lack of reliable biomarkers for easy assessment of the therapeutic readouts. We therefore set out to evaluate the impact of our gene-based therapeutics at lowering the hSOD1 protein levels in the CSF of treated mice. Interestingly, analysis of CSF revealed the depletion in hSOD1 protein by 63% in scAAV_hSOD1si-treated animals when compared to controls (Figure 1F; p = 0.0142). Together, these data suggest that the therapeutic vector is functional and that CSF SOD1 protein levels can be used as a reliable biomarker to discern sufficient target engagement and correct dosing in future clinical trials in SOD1-associated ALS patients.
Single scAAV9_hSOD1si Administration at Disease Pre-onset (P1) Improves Motor Performance and Neurological Deficits and Extends Survival in SOD1G93A Mice
Widespread delivery of therapeutic genes to the CNS is a formidable challenge in ALS therapy for many reasons. First, delivery of therapeutic genes into motor neurons is restricted by the blood-brain barrier (BBB). Second, due to the severe nature of SOD1-associated ALS, SOD1 silencing would be desirable at early stages of the disease. We therefore intentionally chose the cisterna magna as the route of delivery, as we and others have reported that CSF delivery of scAAV9 produced very efficient, robust, and widespread gene transfer to the CNS in mice.12, 13, 14 To evaluate the efficacy of hSOD1 silencing on the motor phenotype in the SOD1G93A mouse model, scAAV9_hSOD1si (n = 9) or scAAV9_hSODssi (n = 10) vectors were administered intrathecally via the cisterna magna of mice at P1.
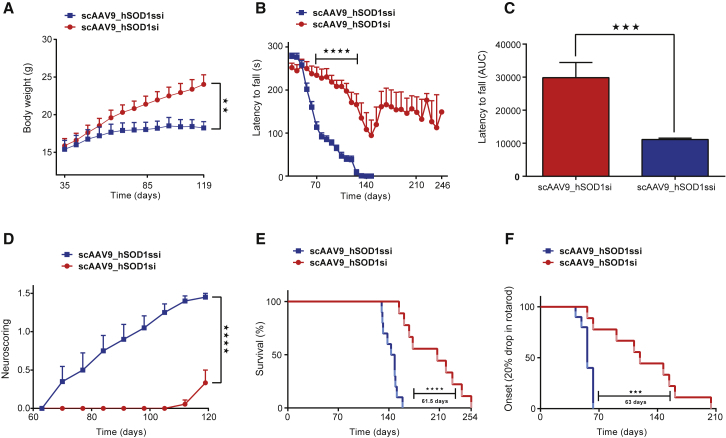
Daily inspection of the phenotype of the animals revealed that scAAV9_hSOD1si-injected mice displayed continuous body weight gain (Figure 2A), while scAAV9_hSOD1ssi-treated animals showed signs of reduced weight gain starting from P50 (Figure 2A).
Figure 2.
Cisterna Magna Delivery of scAAV9_hSOD1si at Post-natal Day 1 Improves Motor Performance and Survival in SOD1G93A Mice
(A–F) Graphs showing body weight (A), rotarod performance measured as time to fall in seconds against age in days (B), area under the curve (AUC) of rotarod performance (C), neuroscoring (D), survival analysis (E), and onset of symptoms (F) in SOD1G93A mice injected with 2.5e+10 vg/g scAAV9_hSOD1si (n = 9) or scAAV9_SOD1ssi (n = 10) via the cisterna magna at P1. Student’s t test of AUC data showed improved rotarod performance (C) in scAAV9_hSOD1si-treated mice, when compared with scAAV9_hSOD1ssi. In all graphs data are presented as the mean ± SEM. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To objectively quantify motor performance of treated SOD1G93A mice, we used the rotarod (Figures 2B and 2C) and neuroscoring (Figure 2D). Rotarod tasks detected first motor deficits around 50–60 days of age in scAAV9_hSOD1ssi control mice, which became rapidly more severe and very pronounced by 70 days of age (Figure 2B). In contrast, the scAAV9_hSOD1si-treated animals still performed normally at 110–120 days of age. Some therapeutically treated mice started to display motor performance deficits at around 140 days (Figure 2B). Neuroscoring analysis was performed to assess the splay defects in the two groups of mice. Control-injected mice displayed an increase in hind limb splay defects when compared with scAAV9_hSOD1si-treated mice (Figure 2D; p < 0.0001). Taken together, pre-onset delivery of scAAV9-mediated SOD1 silencing led to a remarkable motor improvement in the SOD1G93A mouse model of ALS. Notably, scAAV9_hSOD1si-treated mice lacked obvious paralysis and continued to remain ambulant even at end stage.
The mean onset of clinical disease (defined by 20% drop in motor performance as assessed by rotarod task) was delayed by 67 days in scAAV9_hSOD1si-treated mice (n = 9) as compared to control animals (n = 10) (123 ± 16.1 versus 56.7 ± 2.2; p < 0.001) (Figure 2F). Interestingly, lifespan analysis in scAAV9_hSOD1si-treated SOD1G93A mice revealed a median prolongation of the life expectancy by as much as 42% compared to control animals (207 ± 12.4 days versus 145.5 ± 3.1 days; p < 0.0001) (Figure 2E).
SOD1 Depletion Improves Motor Neuron Survival and Reduces Gliosis in SOD1G93A Mice
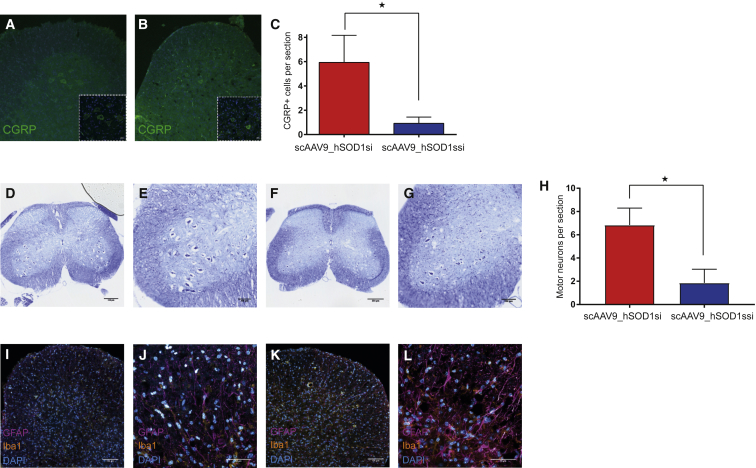
Since early anatomical changes in the SOD1G93A models of ALS are seen in lower motor neurons,15 we quantified calcitonin gene-related peptide (CGRP)-positive lumbar motor neurons at end stage.16 Cell counts revealed that the number of CGRP-labeled motor neurons in lumbar spinal cord sections was significantly higher in scAAV9_hSOD1si-treated mice (n = 5; p = 0.047) (Figure 3A) compared to scAAV9_hSOD1ssi (n = 5) (Figure 3B), which was consistent with an improvement in motor neuron survival of 87.8% (Figure 3C). Motor neurons were also quantified by Nissl staining in lumbar spinal cord sections (Figure 3H), and they showed a significantly higher number of motor neurons per section in scAAV9_hSOD1si-treated mice (n = 3; p = 0.028) (Figures 3D and 3E) compared to scAAV9_hSOD1ssi (n = 3) (Figures 3F and 3G).
Figure 3.
Cisterna Magna Delivery of scAAV9_hSOD1si in SOD1G93A Mice at Post-natal Day 1 Efficiently Improves Lumbar Motor Neuron Survival at End Stage and Reduces Gliosis
(A and B) Representative images of motor neurons in lumbar spinal cord ventral horn from mice treated at P1 with scAAV9_hSOD1si (A, main 20×, inset 63×; scale bar, 100 μm) or scAAV9_hSOD1ssi (B, main 20×, inset 63×; scale bar, 100 μm), stained for CGRP (green), and quantified in (C) (n = 20 sections/mouse, 5 mice per group). (C–H) Representative images of Nissl-stained lumbar spinal cord from mouse treated at P1 with scAAV9_hSOD1si (D, scale bar, 100 μm; E, scale bar, 250 μm) or scAAV9_hSOD1ssi (F, scale bar, 100 μm; G, scale bar, 250 μm) and quantified in (H) (n = 10 sections/mouse, 3 mice per treatment group). (I–L) Representative images of lumbar cord ventral horn from mouse treated at P1 with scAAV9_hSOD1si (I,scale bar, 50 μm; J, scale bar, 100 μm) or scAAV9_hSOD1ssi (K, scale bar, 50 μm; L, scale bar, 100 μm) and stained for GFAP (magenta), Iba1 (yellow), and DAPI (cyan). *p < 0.05.
Previous histopathological characterization of the spinal cords of SOD1G93A mice revealed increased expression of glial fibrillary acidic protein (GFAP) and ionized calcium-binding adaptor molecule 1 (Iba1), markers of astrocytes and microglia, respectively, indicative of elevated reactive gliosis and inflammation.17 Spinal cord sections from end-stage mice treated with scAAV9_hSOD1si showed a reduction in both GFAP and Iba1 staining (Figures 3I and 3J) compared to control mice (Figures 3K and 3L).
Single scAAV9_hSOD1si Administration at Disease Onset (P40) Improves Motor Performance and Neurological Deficits, and It Extends Survival in SOD1G93A Mice
As treatment of ALS patients is currently initiated only after disease diagnosis, testing the therapeutic potential of SOD1 silencing at the onset of clinical symptoms in SOD1G93A mice is mandatory. Cisterna magna delivery of scAAV9_hSOD1si (8 × 109 vg/g) was subsequently applied in SOD1G93A mice at P40. Gene transfer to the CSF through the cisterna magna can be easily translated to human clinical trials using intrathecal delivery via lumbar puncture. Using the intrathecal route of delivery provides advantages for translational applications, since a smaller quantity of the vector is required compared to systemic administration of AAV and allows for a reduced number of viral particles reaching peripheral organs, such as the liver.18, 19
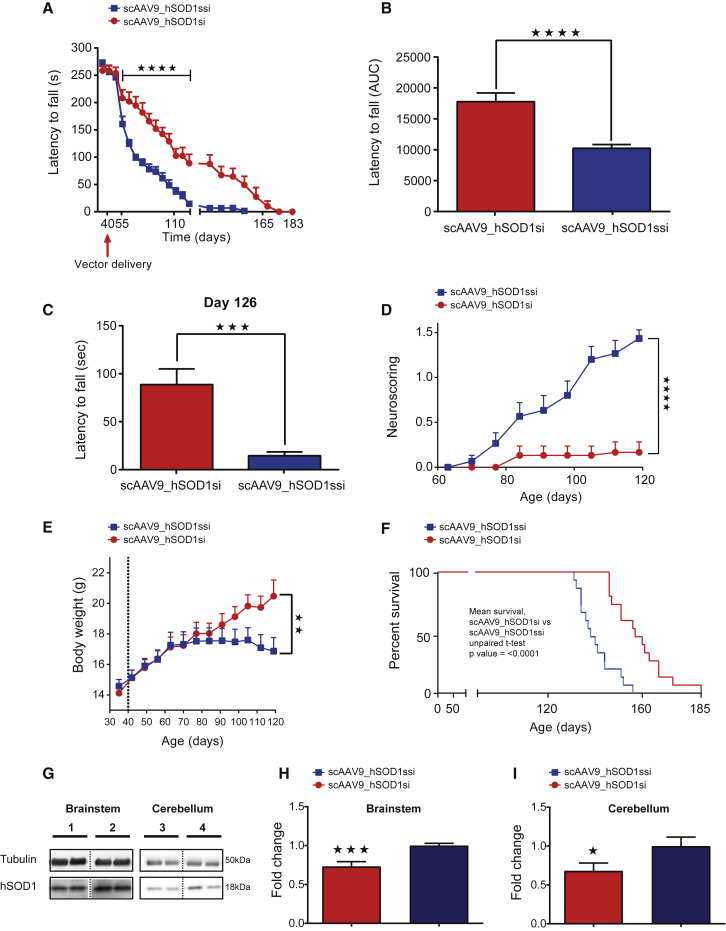
Consistent with the P1 data, assessment of neuromuscular function using the rotarod task showed a significant improvement up to 133 days of age in scAAV9_hSOD1si-treated mice when compared with scAAV9_hSOD1ssi (p < 0.001) (Figure 4A). The area under the curve (AUC) for rotarod data showed overall improved motor performance (Figure 4B) in scAAV9_hSOD1si-treated mice when compared with scAAV9_hSOD1ssi (n = 15; p < 0.0001), and scAAV_hSOD1si-treated mice had a significantly longer latency to fall (measured in seconds) at 126 days old compared to scramble-treated controls (Figure 4C). Neuroscoring analysis revealed improvement in hind limb splay defects following SOD1 depletion when compared with control mice (Figure 4D; n = 15; p < 0.0001). Body weight was significantly decreased in scAAV9_hSOD1ssi-treated mice at day 120 compared to scAAV9_hSOD1si-treated mice (n = 15; p < 0.01), as assessed by two-way ANOVA with Sidak’s post hoc test (Figure 4E). scAAV9_hSOD1ssi-treated SOD1G93A mice survived to a median age of 138 ± 1.9 days of age (n = 15) (Figure 4F). In contrast, SOD1G93A mice treated with scAAV9_hSOD1si showed a statistically significant extension in lifespan to a median of 157 ± 2.9 days of age (n = 15; p < 0.0001) (Figure 4F).
Figure 4.
Cisterna Magna Delivery of scAAV9_hSOD1si at Post-natal Day 40 Improves Motor Performance and Extends Life Expectancy in SOD1G93A Mice
(A–F) Graphs showing rotarod performance measured as time to fall in seconds against age in days (A) and area under the curve (AUC) of rotarod performance (B), histogram showing the effect of scAAV9_hSOD1si on rotarod performance at day 126 (C), neuroscoring (D), body weight (E), and survival analysis (F). (G–I) Representative western blot (G) of SOD1 knockdown in brainstem and cerebellum of P40-treated mice, where 1 and 3 are scAAV9_hSOD1si treated and 2 and 4 are scAAV9_hSOD1ssi treated, and quantification of hSOD1 protein expression in brainstem (H) and cerebellum (I) at end stage in SOD1G93A mice injected with clinical vector scAAV9_hSOD1si at P40. Western blot data were analyzed by Student’s one-tailed t test (n ≥ 9 per group), *p < 0.05 and ***p < 0.001. Student’s t test of AUC data showed improved rotarod performance (B and C) and neuroscoring (D) in scAAV9_hSOD1si-treated mice compared with scAAV9_hSOD1ssi (n = 15 per group). Student’s t test showed improved mean survival (F) in scAAV9-hSOD1si-treated mice compared with scAAV9_hSOD1ssi (n = 15 per group). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We used western blotting to determine hSOD1 protein levels in brainstem and cerebellum from scAAV9_hSOD1si and -ssi virus-treated mice at end stage following injection at P40 via cisterna magna (Figure 4G). We report a reduction in hSOD1 protein of 27% and 32% in brainstem (Figure 4H) and cerebellum (Figure 4I), respectively.
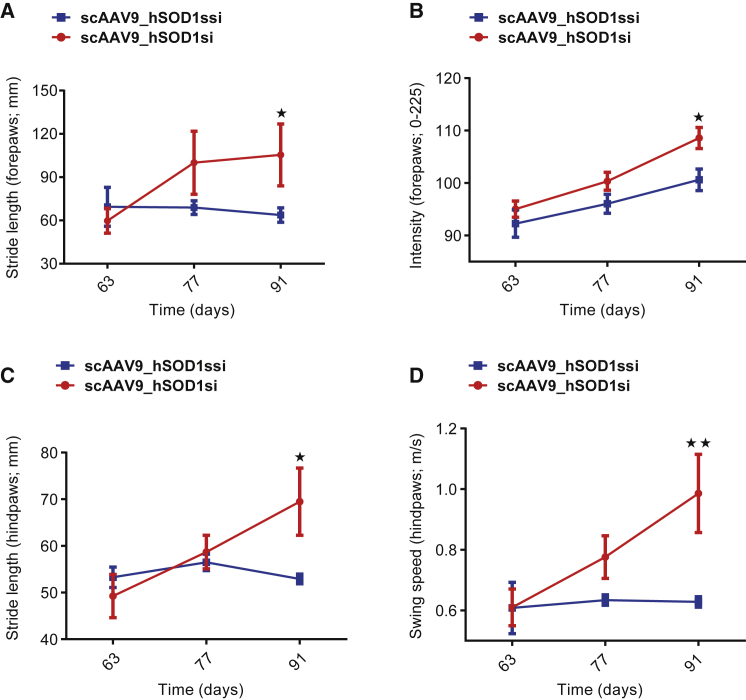
A two-way ANOVA of individual gait parameters over time showed a significant improvement in forepaw and hind paw stride length (both p < 0.05; Figures 5A and 5C), forepaw intensity (p < 0.05; Figure 5B), and swing speed (p < 0.01; Figure 5D) at 91 days in SOD1G93A transgenic mice treated with scAAV9_hSOD1si compared with scAAV9_hSOD1ssi, indicating an improvement in motor performance and reduced motor deficits.
Figure 5.
Cisterna Magna Delivery of scAAV9_hSOD1si at P40 Improves Gait Parameters in SOD1G93A Mice
(A–D) Analysis of individual gait parameters over time in SOD1G93A transgenic mice injected with scAAV9_hSOD1si (n = 5) or scAAV9_hSOD1ssi (n = 5) in the cisterna magna at post-natal day 40 (P40). Selected gait parameters were analyzed using a two-way ANOVA with Sidak’s post hoc test. Forepaw stride length in millimeters (A), forepaw pixel intensity (B), hind paw stride length in millimeters (C), and hind paw swing speed in meters per second (D) for SOD1G93A mice treated with scAAV9_hSOD1si or scAAV9_hSOD1ssi are shown. Data are presented as mean ± SEM. Differences were detectable at day 91. *p < 0.05 and **p < 0.01.
Design and Validation of Off-Target Effect Constructs In Vitro
Efficient clinical translation of an RNAi-mediated treatment for SOD1-linked fALS requires the development of RNAi reagents that are free of significant off-target effects. To test this, several shRNA constructs in addition to the therapeutic construct (scAAV9_hSOD1si) were generated and together screened for their ability to silence endogenous hSOD1 mRNA by scAAV9 viral transduction in three different cell types: (1) HEK293T cells (Figure 6A); (2) an isogenic Tet-inducible Flp-FRT HEK293 cell line expressing either wild-type (WT) or G93A hSOD1 transgenes (Figure 6B); and (3) an astrocyte cell model of ALS (induced astrocytes [iAstrocytes]) derived from direct conversion of fibroblasts from ALS patients and healthy controls into induced neuronal progenitor cells,20 which are then differentiated into astrocytes (Figure 6C). The isogenic cell line was chosen as it was considered important to have an in vitro model that was representative of both healthy and disease phenotypes but that also avoided the inherent genetic variation of other cell models, such as cells derived from patients. The iAstrocyte cell lines were used due to a number of translationally relevant attributes: (1) the availability of lines derived from healthy controls and sporadic and SOD1-fALS patients, representing the natural genetic variation found in the human population; (2) the relevance of including a glial model of ALS in this study due to the increasing volume of data pointing to a significant role for astrocyte-mediated toxicity in the pathobiology of ALS;21, 22, 23, 24 and (3) the fact that astrocytes are a likely in vivo target of the scAAV9 viral vector used in this study.
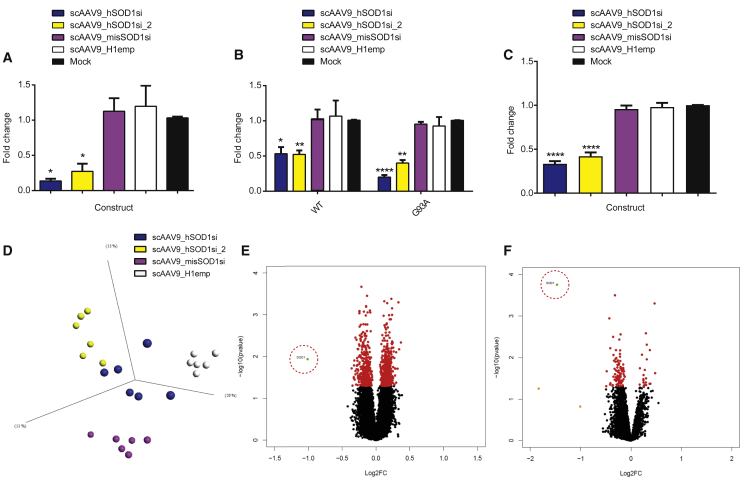
Figure 6.
In Vitro Analysis of Off-Target Gene Regulation
(A–F) Graphs showing hSOD1 mRNA level fold change as determined by qRT-PCR in HEK293T cells transduced with 50,000 vg/cell scAAV9 viral vectors expressing off-target constructs, harvested 120 hr post-transduction (n = 3) (A); isogenic Tet-inducible Flp-FRT HEK293 cells expressing either WT or G93A hSOD1 transduced with 50,000 vg/cell scAAV9 viral vectors expressing off-target constructs, harvested 120 hr post-transduction (n = 3) (B); iAstrocyte cells transduced with 250,000 vg/cell scAAV9 viral vectors expressing off-target constructs, harvested 120 hr post-transduction (n = 10) (C); PCA plot of virally transduced isogenic Tet-inducible Flp-FRT HEK293 cell microarray data colored by treatment (scAAV9_hSOD1si, blue; scAAV9_hSOD1si_2, yellow; scAAV9_misSOD1si, pink; and scAAV9_H1emp, white) (D); volcano plots showing differentially expressed genes in scAAV9_hSOD1si transduced isogenic Tet-inducible Flp-FRT HEK293 cells (E) and iAstrocytes (F) compared to scAAV9_H1emp transduced cells (Probeset color key: green, p < 0.05, fold change > 1.5; orange, p > 0.05, fold change > 1.5; red, p < 0.05, fold change < 1.5). SOD1 probeset is highlighted by red dashed circles. Data are presented as the mean ± SEM. Data were analyzed by one-way ANOVA (A and C) or two-way ANOVA (B) followed by post hoc Dunnett’s multiple comparisons test with respect to mock. *p < 0.05, **p < 0.01, and ****p < 0.0001.
Of the additional RNAi constructs cloned into scAAV9 for the purposes of investigating off-target effects, scAAV9_hSOD1si_2 was designed to act as a positive control for on-target gene regulation, as it also targets hSOD1 mRNA, albeit at a different region of the gene targeted by scAAV9_hSOD1si. The shRNA sequence used in scAAV9_hSOD1si_2 has previously shown strong therapeutic efficacy when delivered by lentivirus vector in the G93A mouse model.8 scAAV9_misSOD1si contains the same shRNA sequence as scAAV9_hSOD1si but differs by a central mismatch created by changing a 3-bp central portion of the therapeutic shRNA to its complement base pairs, resulting in a non-SOD1-targeting shRNA that contains the same seed region sequence as scAAV9_hSOD1si. The final control construct consists of an H1 promoter-only control that does not express an shRNA (scAAV9_H1emp). Both hSOD1 mRNA-targeting shRNA constructs (therapeutic construct scAAV9_hSOD1si and on-target control construct scAAV9_hSOD1si_2) were found to significantly and effectively reduce levels of hSOD1 mRNA by ≥49% relative to mock, in all cell lines (Figures 6A–6C). Neither the seed control (scAAV9_misSOD1si) nor the promoter-only control (scAAV9_H1emp) construct was observed to reduce hSOD1 mRNA significantly, relative to mock.
Gene Expression Analysis of RNAi-Mediated Knockdown of hSOD1 In Vitro Shows No Off-Target Effects
Off-Target Effects Study in Isogenic tet-Inducible Flp-FRT HEK293 Cells
A principal-component analysis (PCA) plot of the gene expression data, analyzed by two-way ANOVA multigroup comparison (p < 0.05), showed clear separation of the four different treatments into discrete groups in both inducible HEK293 cells (Figure 6D) and iAstrocytes (Figure S1). A two-group comparison (t test) in Qlucore Omics Explorer was used to investigate differential gene expression between each shRNA and the negative control (scAAV9_H1emp). Gene expression differences that displayed a p value < 0.05 were considered significant. Filtering genes by p value (p < 0.05) and fold change (≥1.5) resulted in a list of 1 differentially expressed gene for scAAV9_hSOD1si, 0 genes for scAAV9_misSOD1si, and 2 genes for scAAV9_hSOD1si_2. Only 1 differentially expressed gene was shared between the knockdown constructs scAAV9_hSOD1si and scAAV9_hSOD1si_2: SOD1, with a fold change of −2.01 and −1.53, respectively.
A volcano plot (x axis, log2 fold change; y axis, −log10 p value) for differentially expressed genes in scAAV9_hSOD1si-transduced cells compared with scAAV9_H1emp-transduced cells shows only SOD1 passing the threshold values of p < 0.05 and fold change ≥1.5 (Figure 6E). Despite sharing a seed region, scAAV9_hSOD1si and scAAV9_misSOD1si shared no common downregulated genes compared to scAAV9_H1emp. If strong off-target effects mediated by the shRNA seed region were present, it would be expected that the two shRNA constructs sharing this region, scAAV9_hSOD1si and scAAV9_misSOD1si, would cluster together and downregulate similar genes, which was not the case. The observation that scAAV9_misSOD1si did not downregulate any genes greater than 1.5-fold indicates that the seed region itself does not have any affinity for any human transcript.
Off-Target Effects Study in iAstrocyte Cell Lines
To assess the off-target effect of the therapeutic vector in a more variable and rich genetic context, gene expression data from fibroblast-derived iAstrocyte lines transduced with scAAV9_hSOD1si, scAAV9_misSOD1, and scAAV9_H1emp were similarly analyzed for differential gene expression. A two-group comparison (t test) in Qlucore Omics Explorer was used to investigate differential gene expression between each shRNA and the negative control (scAAV9_H1emp). scAAV9_H1emp was also compared to untransduced cells. Filtering genes by p value (p < 0.05) and fold change (≥1.5) resulted in a list of 1 differentially expressed gene for scAAV9_hSOD1si and 0 genes for scAAV9_misSOD1si. One gene was found to be differentially expressed between samples transduced with scAAV9_H1emp and untransduced samples: MIF4G Domain-Containing protein (MIF4GD), a protein that facilitates the translation of histone mRNA (fold change −1.66 relative to untransduced). SOD1 was the only differentially expressed gene seen in scAAV9_hSOD1si-transduced cells (fold change, −2.78 relative to scAAV9_H1emp), recapitulating the results seen in the inducible HEK cell lines and further supporting the observation that no genes other than the target gene were downregulated in vitro by the therapeutic construct. A volcano plot for differentially expressed genes in scAAV9_hSOD1si-transduced cells compared with scAAV9_H1emp-transduced cells shows SOD1 as the only named gene passing the threshold values of p < 0.05 and fold change ≥1.5 (Figure 6F).
Discussion
In summary, this study revealed that AAV9-mediated CSF delivery of functional shRNA molecules to the CNS causes sustained long-term rescue of a dominantly inherited form of ALS in a transgenic mouse model. We consider that our findings are timely and are of great relevance for clinical translation for the following reasons: (1) evaluation of the final clinical configuration of the therapeutic vector led to remarkable efficacy in a mouse model of SOD1-associated ALS; (2) gene transfer to the CSF through the cisterna magna can be easily translated to human clinical trials using intrathecal injection as the route of delivery; (3) to our knowledge, this is the first report to demonstrate that no significant off-target effects are linked to AAV9-mediated SOD1 silencing using this construct, providing further confidence in the specificity and safety of the clinical therapeutic vector used in this study; and (4) we report the measurement of CSF SOD1 protein levels as a biomarker, supporting future effective dosing and efficacy of SOD1 knockdown in human subjects.
Previous gene therapy approaches in models of SOD1-linked ALS have involved delivery of shRNA molecules to susceptible motor neuron populations using retrograde transport of lentiviral vectors following intramuscular (IM) delivery.8 Similarly, recent reports also revealed remarkable proof of concept achieved by systemic delivery (intravenous [i.v.]) of AAV9-mediated SOD1 silencing in SOD1G93A transgenic mice.10, 25 Advancing IM and i.v. approaches to the clinic, however, is challenged by the high cost of manufacturing and the large amounts of viral vector required for such modes of vector delivery.
Cisterna magna delivery allows widespread gene transfer to the CNS as previously reported in one of our studies.12 However, this mode of delivery is not specific to the brain, as several reports suggest that AAV viral particles can be detected in the periphery following CSF administration.26 The key advantages of CSF (e.g., cisterna magna) gene therapy administration are as follows: (1) a reduced amount of viral vector required for this mode of delivery when compared to systemic (i.v.) administration; and (2) the probability of reduced amounts of viral particles reaching peripheral organs, such as the liver, minimizing the risk of immune response and other potential side effects.
The considerable improvement in the life expectancy of the SOD1G93A mice observed in this report is probably a consequence of the more direct approach used in this study for targeting expression of the genetic factor causing the disease. Furthermore, the CSF mode of vector delivery used in this study and the remarkable efficacy achieved illustrate the realistic potential for advancing AAV9-based RNAi approaches to human clinical trials in SOD1 fALS. Indeed, the AAV9 vector system has already been used in spinal muscular atrophy (SMA), Batten disease, and giant axonal neuropathy clinical trials,27 which provide evidence of the safety profile of this vector system and, hence, the true potential for scaling up our small interfering RNA (siRNA) approach for human gene therapy trials in SOD1-related ALS.
In contrast to previous studies, the data reported here have been generated using a therapeutic construct that requires no further alteration before entering the clinic. The construct contains no exogenous reporter genes (e.g., GFP) and is composed of only an H1 promoter, the therapeutic shRNA, and a sophisticated stuffer sequence designed by the Kaspar laboratory. Despite the widespread use of GFP or similar reporter genes in gene therapy vectors, we have found that the removal of the GFP reporter gene and its replacement with a stuffer sequence has had no deleterious effect on the ability of the vector construct to reduce SOD1 mRNA and protein levels in vitro or extend survival in the mouse model in vivo. In fact, the percentage increase in median survival in P1-treated animals observed in this study (42%) compares favorably to that observed by Foust et al.,10 who used the CBA-GFP-containing vector and observed a percentage increase in median survival of 39% after treatment at P1. Foust et al.,10 utilized i.v. delivery of AAV9, resulting in a considerably higher dose of 3.6 × 1011 vg/g compared to the dose of 5 × 1010 vg/g we used for cisterna delivery. Our approach, i.e., intrathecal delivery has 2 major advantages: (1) similar efficacy is achieved compared to i.v. delivery while administering a lower dose of therapeutic vector, therefore yielding a potential reduction in good manufacturing practice (GMP) manufacturing cost; (2) the amount of viral particles reaching peripheral organs, such as the liver, is likely to be reduced when compared to i.v. delivery, therefore reducing the risk of side effects. Consistent with previous studies in mice and non-human primates,26 intrathecal delivery allows reducing the viral dose by 10 times and achieving comparable results.
The lack of paralysis and continued ability of scAAV9_hSOD1si-treated animals to remain ambulant even at end stage is consistent with our observation of increased motor neuron count. Expression of mutant SOD1 in non-neuronal cells, such as glia, has also been demonstrated to have an influence on the progression of motor neuron degeneration in in vivo28, 29, 30, 31 and in vitro21, 32 ALS models. Since the CSF mode of delivery of AAV9 described here targets both motor neuron and glial populations, the prevention of the deleterious effects of mutant SOD1 expression in non-neuronal cell types may also have contributed to the delayed onset of disease symptoms in these animals. Consistent with this, we observed reduced gliosis in the spinal cord of therapeutically treated mice compared to controls.
The reason behind the lack of a complete rescue is unclear. A group of AAV9_hSOD1si-treated SOD1 mice showed ALS-like symptoms with modest extension in survival and was humanely euthanized. A proportion of mice did not die as a consequence of motor impairment, since the motor function and behavior of these animals remained significantly higher compared to untreated and scramble vector-treated mice. In particular, this second group of mice suddenly looked moribund and died overnight or were humanely culled. A gene therapy study of virally mediated hSOD1 mRNA reduction by artificial microRNA in the same mouse model reported a similar preservation of limb function in treated mice, with euthanasia initiated due to severe weight loss and excessive curvature of the spine (kyphosis), which the authors postulate was a symptom of weakening of the intercostal muscles and diaphragm.33 As respiratory failure is the most common cause of death in ALS patients, this may point to a similar involvement of diaphragm weakness in the death of the treated mice in our study and of those in Stoica et al.,33 with potential future implications for targeting of this structure by therapies.
Complete rescue of the model is also likely hindered by the extremely high burden of mutant SOD1 protein in the SOD1-G93A line—many times that of a human patient—resulting from the multiple transgene copies present in the line. Insufficient transduction of CNS neurons/glia or even other peripheral organs could be behind early death or the lack of a complete rescue. Indeed we observed variability in viral particle biodistribution, suggesting technical variability during cisterna magna administration of the AAV9 virus.
Our efforts went beyond establishing in vivo proof-of-concept efficacy of the therapeutic strategy. Indeed, to our knowledge this is the first report to demonstrate that AAV9-mediated SOD1 silencing can be achieved without significant off-target effects, providing further confidence in the specificity and safety of the clinical therapeutic vector developed by the Kaspar group. Although exogenous shRNA molecules are designed to be fully complementary to their target transcript, and efforts are made in silico to search for sequences that do not have any complementarity with other genes, off-target effects can occur whereby shRNA molecules act like endogenous microRNA (miRNA) molecules. miRNA molecules in vivo often bind to the 3′ UTRs of multiple mRNA transcripts, exhibiting only partial sequence complementarity and regulating gene expression by translational inhibition.34, 35 Acting in this way, even an siRNA or shRNA designed to be highly specific to a target transcript may downregulate the expression of multiple gene products.36, 37, 38
Off-target miRNA-like silencing in shRNA molecules is mediated by nucleotides 2–8 in the anti-sense strand of an shRNA molecule, analogous to the 7-nt seed region that mediates gene repression in endogenous miRNAs.39, 40 In light of this, several control constructs were cloned into the scAAV9 plasmid backbone to allow detection of seed region-mediated off-target effects (Table S1). Previous studies into off-target effects mediated by siRNAs have shown widespread off-target gene regulation.37, 39, 41, 42 However, others have shown reduced off-target effects when using virally delivered shRNA constructs compared with transfected siRNA constructs, albeit using lentivirus rather than AAV.43, 44 Reports investigating shRNA-mediated knockdown of target genes have shown few differentially expressed genes in addition to the target gene, in accordance with the observations in this study.44, 45
The lack of observable off-target effects in vitro using our therapeutic vector is very encouraging, and it will hopefully be supported by the regulatory safety studies required for advancing this strategy to human clinical trials. Furthermore, we report the measurement of CSF SOD1 protein levels as a biomarker of effective dosing and efficacy of SOD1 knockdown. This approach is likely to prove useful in human clinical trials to assess the biological activity of the therapeutic vector. Together, our data highlight the considerable therapeutic potential of RNAi for treating a dominantly inherited neurological disease, and they provide optimism for the future translation of AAV9-RNAi from a research tool to a therapeutic agent.
Materials and Methods
Ethics Statement
All animal in vivo experiments were approved by the University of Sheffield Ethical Review Sub-Committee, the UK Animal Procedures Committee (London, UK), and they were performed according to the Animal (Scientific Procedures) Act 1986, under the Project License 40/3739. SOD1G93A mice were maintained in a controlled facility in a 12-hr dark/12-hr light photocycle (on at 7 a.m./off at 7 p.m.) with free access to food and water. The Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines have been followed in reporting this study.46 We employed a double-blind randomization process in which experimental groups were blinded to the individuals conducting viral delivery and data analyses (e.g., motor neuron counts).
AAV Construction and Preparation
The original pAV2 vector backbone was published in Foust et al.10 Briefly, within the vector inverted terminal repeats (ITRs), an H1 promoter drives expression of the shRNA targeted against the hSOD1 gene. A stuffer sequence has been added to optimize the packaging efficiency of the vector backbone kindly provided by Brian Kaspar. This plasmid has an 11-bp deletion in the downstream ITR, removing a target site that would otherwise be cleaved by the AAV endonuclease during capsid assembly and packaging. The consequence of this deletion is the formation of palindromic repeats of the viral genome, able to fold into a double-stranded DNA (dsDNA) hairpin structure referred to as a self-complementary genome. A separate vector used as a scrambled shRNA virus control (scAAV9_hSOD1ssi) contained a non-targeting shRNA under the control of the H1 promoter, as well as a cytomegalovirus (CMV)-GFP reporter gene construct.
Cisterna Magna Delivery of scAAV9_hSOD1si or scAAV9_hSOD1ssi in SOD1G93A Mice
scAAV9-mediated gene delivery was applied via the cisterna magna at two different time points: pre-onset (P1) and onset (P40) of symptoms. We chose onset as P40 as functional and structural changes in motor units are reported as starting from this time point in SOD1G93A mice.47, 48 In an initial study, 30 SOD1G93A transgenic mice were divided into two groups to be treated at P40 with either scAAV9_hSOD1si or scrambled control virus scAAV9_hSOD1ssi (12 μL total injection volume, 2 μL iodixanol and 10 μL virus; virus concentration, 1.2 × 1013 vg/mL; total dose, 5 × 1010 vg/g). In a second study, 19 SOD1G93A transgenic mice were divided into 2 groups to be treated at P1 with either scAAV9_hSOD1si (n = 9) or scrambled control virus scAAV9_hSOD1ssi (n = 10) (total dose, 8 × 109 vg/g). A cohort of 10 mice per group (scAAV9_hSOD1si and scAAV9_hSOD1ssi) was injected at P1 and sacrificed 4 weeks post-injection to assess viral biodistribution and hSOD1 mRNA and protein levels (in striatum, brainstem, cerebellum, cortex, lumbar spinal cord, and CSF). The remaining mice underwent behavioral testing to assess effects on disease progression, achieved by the use of weekly rotarod analysis after 3 consecutive days of training.
In all studies, the onset and progression of disease were estimated by neurological scoring three times per week from 60 days of age. Body weight was recorded weekly. Mice were scored for tremor, hind limb splay, and overall neurological deficit using a previously reported scoring system.49 Disease onset was defined as the point at which both enhanced tremor and defective hind limb splay were first observed. All mice continued to end-stage disease and the time to reach this stage was recorded. The humane endpoint is defined as the inability to right within 10 s of being placed onto the back or weight loss of more than 30% over 72 hr. Mice were distress scored daily from 120 days onward. All animals were perfused under terminal anesthesia, and CNS tissue was collected to assess viral biodistribution and SOD1 mRNA and protein levels. Injected animals displaying low levels of viral biodistribution, as measured by qPCR, were excluded from subsequent analysis.
Genotyping and Copy Number Analysis
Genotyping and copy number analysis of SOD1G93A mice was performed based on the protocol described in Mead et al.49 Any changes made are detailed below.
Mouse genotyping was performed on genomic DNA extracted from tail tissue using the Maxwell 16 system DNA purification kit (Promega, WI, USA). Genotyping PCRs were performed on genomic DNA in a 25-mL volume with ABgene ReddyMix (ABgene, Epsom, UK), 150 nM each of hSOD1 primers (forward, 59-CATCAGCCCTAATCCATCTGA-39; reverse, 5′-CGCGACTAACAATCAAAGTGA-3′) and control interleukin-2 receptor (IL-2R) primers (forward, 5′-CTAGGCCACAGAATTGAAAGATCT-3′; reverse, 5′-GTAGGTGGAAATTCTAGCATCATC-3′). Following PCR and agarose gel electrophoresis (3% agarose gel), IL-2R PCR products were visualized at 324 bp and hSOD1, if present, at 236 bp. Mice positive for transgene expression were then subjected to copy number analysis by qPCR using 30 ng cDNA, 2× SYBR Green PCR Master Mix (Agilent Technologies, CA, USA), and 150 nM primers used for genotyping, in a total volume of 20 mL. Following an initial denaturation at 95°C for 10 min, DNA was amplified by 40 cycles of 95°C for 15 s and 60°C for 1 min, on an MX3000P Real-Time PCR System (Stratagene). hSOD1/IL-2R ΔCt values from samples were compared to a reference DNA sample from an SOD1G93A control mouse displaying a phenotype consistent with one carrying the full 25 copies of the transgene.
CSF Collection and ELISA for hSOD1 Quantification
SOD1G93A mice were placed on a stereotaxic frame under general anesthesia by isoflurane inhalation. The head was secured with ear adaptors and the skin of the neck was shaved. A sagittal incision of the skin was made inferior to the occiput. The subcutaneous tissue and muscles (m. biventer cervicis and m. rectus capitis dorsalis major) were separated by blunt dissection with forceps. A pair of microretractors was used to hold the muscles apart. A capillary tube was inserted into the cisterna magna through the dura mater as previously described.50 Following a change in resistance to the capillary tube insertion, the CSF flowed into the tube and was transferred into a 0.5-mL Eppendorf tube containing 20 μL 0.1 M PBS. The CSF sample was spun for 4 min to pellet any contaminating cells, transferred to a fresh tube, snap frozen in liquid nitrogen, and stored at −80°C. hSOD1 was measured in CSF from SOD1G93A mice using the Human Cu/ZnSOD Platinum ELISA kit (BMS222, eBioscience, San Diego, CA, USA), according to the manufacturer’s instructions. Briefly, 1–4 μL blood-free CSF samples were collected and added to 20 μL sterile 0.1 M PBS and stored at −80°C. All samples were quantified for total protein concentration using a Bradford assay to normalize loading to total protein. The quantification for hSOD1 protein was performed in duplicate using 0.2 μg total protein per assay well, and the resulting average value was expressed as ng hSOD1/μg CSF total protein.
qRT-PCR for hSOD1 mRNA Knockdown Analysis
qRT-PCR was carried out using 2 μL total RNA diluted to a concentration of 10 ng/μL in nuclease-free water, 5 μL 2× QuantiFast SYBR Green RT-PCR Master Mix (QIAGEN), hSOD1 (forward, 5′-TCACTTTAATCCTCTATCCAGAAAACAC-3′; reverse, 5′-ACCATCTTTGTCAGCAGTCACATT-3′; 1 μM) and 18S (forward, 5′-GTAACCCGTTGAACCCCAT-3′; reverse, 5′-CCATCCAATCGGTAGTAGCG-3′; 1 μM) primers, 0.1 μL QuantiFast RT mix, and H2O to a final volume of 10 μL. Following an RT step at 50°C for 10 min and 5 min at 95°C, cDNA was amplified by 39 cycles of 95°C for 10 s and a combined annealing and extension step at 60°C for 10 s. This was followed by one cycle at 65°C for 31 s, before analysis by melt curve step. All qRT-PCR was performed on a Bio-Rad C1000 Touch Thermal Cycler. Bio-Rad CFX Manager software was used to analyze signal intensity, and relative gene expression values were determined using the ΔΔCt method, with 18S rRNA used as a reference gene.
Rotarod
Ugo Basile 7650 accelerating rotarod (set to accelerate from 3 to 37 rpm over 300 s) was used to measure motor function from day 35 onward. Rotarod training was performed over 3 consecutive days, with two trials per day. Subsequently, this test was performed at weekly intervals in the late morning. For each evaluation, the mice were tested twice, with a rest period between runs. The best performance, measured as latency to fall in seconds, was used for analysis. The minimum threshold for recording rotarod activity was 3 s.
Gait Analysis
The catwalk gait analysis system version 7.1 was used to assess gait parameters in five SOD1G93A mice per group. Mice were tested at 9, 11, and 13 weeks of age. They were placed on the catwalk apparatus in complete darkness, and the recording of gait patterns was performed in a separate room. Six runs were recorded for each mouse and two selected for analysis. The runs to be analyzed were selected based on the absence of behavioral anomalies, such as sniffing, exploration, and rearing, and where mouse locomotion was steady without noticeable accelerations or decelerations. Processing of gait data was performed with the Noldus software. Limbs were assigned manually and gait parameters were calculated automatically.
Protein Extraction and Western Blotting for hSOD1 Protein Knockdown Analysis
Spinal cords and dissected brain regions were harvested from end-stage P40 SOD1G93A mice treated with scAAV9_hSOD1si or scrambled control scAAV9_hSOD1ssi and snap frozen in liquid nitrogen. Tissue was homogenized using a dounce homogenizer in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCL [pH 7.4], 1% v/v NP-40, 0.5% w/v sodium deoxycholate, 0.1% v/v SDS, 150 mM NaCl, and 2 mM EDTA) containing 1× protease inhibitor cocktail (Sigma-Aldrich). Lysate protein concentrations were determined using the bicinchoninic assay (BCA; Thermo Scientific Pierce); 10 μg protein lysate was denatured by heating to 100°C for 5 min in the presence of 4× loading buffer (10 mL buffer contained 240 mM Tris-HCL [pH 6.8], 8% w/v SDS, 40% glycerol, 0.01% bromophenol blue, and 10% β-mercaptoethanol). Lysates were then loaded onto 4%–20% gradient mini-PROTEAN TGX precast polyacrylamide gels (Bio-Rad). Gels were run at 160 V in running buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS [pH 8.3]) until the dye front reached the bottom of the gel (∼45 min).
Separated proteins were transferred by semi-dry electrophoresis to a polyvinylidene fluoride (PVDF) membrane using the Invitrogen iBlot2 transfer system, as per standard manufacturer’s protocol. Membranes were blocked for 1 hr in 5% “milk/TBS-T”. Primary antibodies mouse anti-α-tubulin (1:5,000) and rabbit anti-hSOD1 (1:1,000) were diluted in 5% “milk/TBS-T” and incubated with the membrane overnight at 4°C. After primary antibody incubation, the membrane was washed for 3 × 15 min each in TBS-T buffer. Secondary antibodies anti-mouse horseradish peroxidase (HRP) (1:3,000) and anti-rabbit HRP (1:3,000) were diluted in 5% “milk/TBS-T” and incubated with the membrane for 2 hr at room temperature. After secondary antibody incubation, the membrane was washed for 3 × 15 min each in TBS-T buffer. Protein bands were visualized using EZ-ECL reagent (Geneflow) and the G-Box imaging system (Syngene). Densitometric analysis of protein bands was carried out using ImageJ software.
Immunofluorescence
All mice were deeply anaesthetized with pentobarbital and perfused transcardially with 100 mL room temperature 0.1 M PBS followed by 100 mL ice-cold 4% paraformaldehyde (PFA). Lumbar spinal cord was removed and post-fixed overnight in 4% PFA followed by 30% sucrose in 0.1 M PBS until sunk. Immunohistochemistry was performed according to Lukashchuk and colleagues.12 In brief, the tissues were embedded in optimal cutting temperature (OCT), and 25-μm-thick spinal cord sections were cut on a cryostat (Leica CM3050S) at −22°C free floating. Spinal cord sections were washed twice with PBS, permeablized in PBS/0.3% Triton X-100, blocked with 10% normal goat serum (NGS) in PBS/0.3% Triton X-100 solution for up to 3 hr, and incubated with primary antibody anti-CGRP (1:500; Abcam), GFAP (1:1,000, Sigma), or Iba1 (1:1,000, Abcam) diluted in 2% NGS in 0.1 M PBS/0.15% Triton X-100 overnight. The following day, sections were washed in 0.1 M PBS four times for up to 2 hr and incubated with secondary antibodies Alexa Fluor anti-mouse 568 or Alexa Fluor anti-mouse 488 (Life Technologies, CA, USA) diluted in 5% NGS/PBS for 1 hr. For visualization of nuclei, 1:2,500 Hoechst stain (Sigma, MI, USA) diluted in 0.1 M PBS was used. Images were captured using a Leica confocal microscope and were processed using LAS AF Lite software and ImageJ.
Nissl Staining and Quantification of Motor Neurons
Lumbar spinal cord was fixed overnight in 4% PFA and cryoprotected in 30% sucrose in 0.1 M PBS. OCT-embedded lumbar cord was cut by cryostat (Leica CM3050S) into 20-μm-thick sections and mounted on charged glass slides. Sections were fixed in cold acetone for 10 min, then stained with 1% w/v aqueous cresyl violet acetate solution for 2 min. Sections were rinsed with water before being destained with 0.25% acetic acid in ethanol. Sections were dehydrated by passing through an ethanol gradient and cleared by brief immersion in xylene before coverslipping. Slides were visualized using a Hamamatsu NanoZoomer and NDP.view2 viewing software. Motor neurons within the ventral horn of the lumbar cord were identified by their morphology and size. Motor neurons in every fifth section were counted, one ventral horn region per section, with a total of 10 sections per mouse counted (n = 3 mice per treatment group).
Cell Culture
HEK293T cells were cultured at 37°C and 5% CO2 in growth media consisting of DMEM (Sigma) supplemented with 10% v/v fetal bovine serum (FBS, Sigma, MI, US) and 1% v/v penicillin/streptomycin (Lonza, Basel, Switzerland).
Isogenic Tet-inducible Flp-FRT HEK293 cells were created using the Flp-In T-REx system (Invitrogen). Cells were cultured in the same conditions as HEK293T cells detailed above, with the addition of 15 μg/mL blasticidin and 100 μg/mL hygromycin and the use of tetracycline-free FBS (Biosera). Isogenic Flp-FRT cells contained either a WT or G93A transgene under the control of a tetracycline-inducible CMV promoter, allowing regulated overexpression of the transgene. Expression of the recombinant gene of interest was induced by the addition to the culture media of 1 μg/mL tetracycline during seeding of the cells into cell culture plates.
The iAstrocytes derived from human fibroblasts were generated by the process detailed in Meyer et al.20 Briefly, fibroblasts were transduced with retroviral vectors expressing four reprogramming factors (Sox2, KLF4, Oct3/4, and c-Myc). After transduction, cells were cultured in growth media containing FGF2, EGF, and heparin to promote the conversion of fibroblasts to induced neuronal progenitor cells (iNPCs). iNPCs were further differentiated to iAstrocytes by plating onto a fibronectin-coated culture dish and changing the growth media to DMEM (Sigma) supplemented with 10% FBS (Sigma), 1% v/v penicillin/streptomycin (Lonza), and 0.3% N2 (Life Technologies). iAstrocytes used in transduction studies were cultured at 37°C and 5% CO2 in the growth media used for differentiation.
Plasmid and Viral Construct Production for Off-Target Effects Analysis
Off-target analysis constructs (scAAV9_hSOD1si_2, scAAV9_misSOD1si, and scAAV9_H1emp) were cloned into the scAAV9_hSOD1si plasmid backbone using the oligonucleotide sequences listed in Table S1. scAAV9 viral vectors containing the off-target constructs were produced by the Children’s Hospital of Philadelphia (CHOP) and the viral titer was determined by qPCR.
Validation of Off-Target Constructs In Vitro
HEK293T and isogenic Tet-inducible Flp-FRT HEK293 cells were transduced 24 hr after plating with 50,000 vg/cell mixed with normal growth media. iAstrocyte cell lines were transduced 24 hr after plating with 250,000 vg/cell mixed with normal growth media. All cell lines were incubated with virus for 5 days before being harvested for analysis of hSOD1 knockdown by western blot and qRT-PCR.
Microarray Analysis of Off-Target Effects
Total RNA, extracted from Tet-inducible Flp-FRT HEK293 and iAstrocyte cells transduced with scAAV9 vectors expressing the off-target constructs (HEK293 cells, 50,000 vg/cell, collected 5 days post-transduction; iAstrocyte cells, 250,000 vg/cell, collected 5 days post-transduction), was analyzed by whole-genome microarray (HEK293 cells, 3′ IVT Express kit using GeneChip Human Genome U133 Plus 2.0 Arrays [Thermo Fisher Scientific]; iAstrocytes, GeneChip WT Plus reagent kit using Clariom S Human whole transcript arrays [Thermo Fisher Scientific]) to investigate gene expression changes elicited by treatment with the viral vectors. RNA quality was measured as the RNA integrity number (RIN) using the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) and RNA 6000 Nano LabChip kit (Agilent Technologies, CA, USA). Array data CEL files were normalized and quality control checked in Affymetrix Expression Console software. Differential gene expression between treated and control arrays was determined using Robust Multi-array (RMA)-normalized data analyzed in Qlucore Omics Explorer (Qlucore AB, Lund, Sweden).
Statistical Analyses
Data were analyzed using GraphPad Prism 6 software (GraphPad, San Diego, CA, USA). Behavioral studies were statistically analyzed using a two-way ANOVA with repeated measures followed by Sidak’s post hoc test (for rotarod and neuroscoring) and unpaired Student’s t test for AUC data of rotarod, neuroscoring, and survival. For in vivo knockdown of hSOD1 mRNA and protein determined by qRT-PCR and western blot analysis, respectively, comparisons were achieved by unpaired one-tailed Student’s t test. For immunohistochemical studies, data were analyzed using unpaired Student’s t test. For in vitro analysis of knockdown by off-target constructs in HEK293 cells and iAstrocyte cell lines, one-way ANOVA followed by Dunnett’s post hoc test was used. For in vitro analysis of knockdown by off-target constructs in isogenic Tet-inducible Flp-FRT HEK293 cell lines, two-way ANOVA followed by Dunnett’s post hoc test was used. Differential gene expression analysis from microarray data for each shRNA construct was determined by Student’s t test between that construct and the scAAV9_H1emp promoter-only control construct.
Author Contributions
T.I. and J.M.S. carried out most of the experiments. S.L., K.M., and B.K.K. designed the AAV-shRNA backbone. I.R.P.C. and K.E.L. assisted with mouse work. A.H. measured SOD1 levels in CSF. P.R.H. and G.M.H. assisted with off-target effect studies. J.M.S. and M.M. analyzed microarray data. M.A.M. and L.F. provided iAstrocytes cultures. T.I., J.M.S., L.F., P.J.S., and M.A. interpreted data. T.I., J.M.S., P.J.S., and M.A. wrote the manuscript. All authors proofread the paper. P.J.S., L.F., and M.A. conceived the study. P.J.S. and M.A. jointly co-led the project.
Conflicts of Interest
The authors declare that no conflicts of interest exist.
Acknowledgments
The pAAV2/9 plasmid was a gift from J. Wilson. M.A. is supported by a European Research Council grant (ERC Advanced Award 294745) and a MRC DPFS Award (129016). P.J.S. is an NIHR Senior Investigator (NF-SI-0617-10077) and Director of the NIHR Sheffield Biomedical Research Centre for Translational Neuroscience (IS-BRC-1215-20017) and is supported by the MND Association. L.F. is supported by an Academy of Medical Sciences (AMS)/Wellcome Trust Springboard Award and the Thierry Latran Fondation (TLF). J.M.S. is supported by a University of Sheffield PhD studentship and the Jeff Wadsworth Battelle Scholarship. K.M. is supported by a Young Investigator Development grant from the Muscular Dystrophy Association and the ALS Association.
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.04.015.
Contributor Information
Pamela J. Shaw, Email: pamela.shaw@sheffield.ac.uk.
Mimoun Azzouz, Email: m.azzouz@sheffield.ac.uk.
Supplemental Information
References
- 1.Rosen D.R. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 2.Bennion Callister J., Pickering-Brown S.M. Pathogenesis/genetics of frontotemporal dementia and how it relates to ALS. Exp. Neurol. 2014;262(Pt B):84–90. doi: 10.1016/j.expneurol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton A.E., Chiò A., Traynor B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 5.Nardo G., Trolese M.C., Tortarolo M., Vallarola A., Freschi M., Pasetto L., Bonetto V., Bendotti C. New Insights on the Mechanisms of Disease Course Variability in ALS from Mutant SOD1 Mouse Models. Brain Pathol. 2016;26:237–247. doi: 10.1111/bpa.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe K., Itoyama Y., Sobue G., Tsuji S., Aoki M., Doyu M., Hamada C., Kondo K., Yoneoka T., Akimoto M., Yoshino H., Edaravone ALS Study Group Confirmatory double-blind, parallel-group, placebo-controlled study of efficacy and safety of edaravone (MCI-186) in amyotrophic lateral sclerosis patients. Amyotroph. Lateral Scler. Frontotemporal Degener. 2014;15:610–617. doi: 10.3109/21678421.2014.959024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensimon G., Lacomblez L., Meininger V., ALS/Riluzole Study Group A controlled trial of riluzole in amyotrophic lateral sclerosis. N. Engl. J. Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 8.Ralph G.S., Radcliffe P.A., Day D.M., Carthy J.M., Leroux M.A., Lee D.C., Wong L.F., Bilsland L.G., Greensmith L., Kingsman S.M. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat. Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 9.Raoul C., Abbas-Terki T., Bensadoun J.C., Guillot S., Haase G., Szulc J., Henderson C.E., Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 10.Foust K.D., Salazar D.L., Likhite S., Ferraiuolo L., Ditsworth D., Ilieva H., Meyer K., Schmelzer L., Braun L., Cleveland D.W., Kaspar B.K. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 2013;21:2148–2159. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 12.Lukashchuk V., Lewis K.E., Coldicott I., Grierson A.J., Azzouz M. AAV9-mediated central nervous system-targeted gene delivery via cisterna magna route in mice. Mol. Ther. Methods Clin. Dev. 2016;3:15055. doi: 10.1038/mtm.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bey K., Ciron C., Dubreil L., Deniaud J., Ledevin M., Cristini J., Blouin V., Aubourg P., Colle M.A. Efficient CNS targeting in adult mice by intrathecal infusion of single-stranded AAV9-GFP for gene therapy of neurological disorders. Gene Ther. 2017;24:325–332. doi: 10.1038/gt.2017.18. [DOI] [PubMed] [Google Scholar]
- 14.Haurigot V., Marcó S., Ribera A., Garcia M., Ruzo A., Villacampa P., Ayuso E., Añor S., Andaluz A., Pineda M. Whole body correction of mucopolysaccharidosis IIIA by intracerebrospinal fluid gene therapy. J. Clin. Invest. 2013:66778. doi: 10.1172/JCI66778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci. 2014;34:15587–15600. doi: 10.1523/JNEUROSCI.2037-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azzouz M., Le T., Ralph G.S., Walmsley L., Monani U.R., Lee D.C., Wilkes F., Mitrophanous K.A., Kingsman S.M., Burghes A.H., Mazarakis N.D. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acevedo-Arozena A., Kalmar B., Essa S., Ricketts T., Joyce P., Kent R., Rowe C., Parker A., Gray A., Hafezparast M. A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis. Model. Mech. 2011;4:686–700. doi: 10.1242/dmm.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray S.J., Nagabhushan Kalburgi S., McCown T.J., Jude Samulski R. Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther. 2013;20:450–459. doi: 10.1038/gt.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinderer C., Bell P., Vite C.H., Louboutin J.P., Grant R., Bote E., Yu H., Pukenas B., Hurst R., Wilson J.M. Widespread gene transfer in the central nervous system of cynomolgus macaques following delivery of AAV9 into the cisterna magna. Mol. Ther. Methods Clin. Dev. 2014;1:14051. doi: 10.1038/mtm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer K., Ferraiuolo L., Miranda C.J., Likhite S., McElroy S., Renusch S., Ditsworth D., Lagier-Tourenne C., Smith R.A., Ravits J. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc. Natl. Acad. Sci. USA. 2014;111:829–832. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haidet-Phillips A.M., Hester M.E., Miranda C.J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M.J., Foust K.D. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlan B.A., Pehar M., Sharma D.R., Beeson G., Beeson C.C., Vargas M.R. Enhancing NAD+ Salvage Pathway Reverts the Toxicity of Primary Astrocytes Expressing Amyotrophic Lateral Sclerosis-linked Mutant Superoxide Dismutase 1 (SOD1) J. Biol. Chem. 2016;291:10836–10846. doi: 10.1074/jbc.M115.698779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pehar M., Harlan B.A., Killoy K.M., Vargas M.R. Role and Therapeutic Potential of Astrocytes in Amyotrophic Lateral Sclerosis. Curr. Pharm. Des. 2017;23:5010–5021. doi: 10.2174/1381612823666170622095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song S., Miranda C.J., Braun L., Meyer K., Frakes A.E., Ferraiuolo L., Likhite S., Bevan A.K., Foust K.D., McConnell M.J. Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat. Med. 2016;22:397–403. doi: 10.1038/nm.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biferi M.G., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Bos C., Voit T., Ferry A., Barkats M. A New AAV10-U7-Mediated Gene Therapy Prolongs Survival and Restores Function in an ALS Mouse Model. Mol. Ther. 2017;25:2038–2052. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer K., Ferraiuolo L., Schmelzer L., Braun L., McGovern V., Likhite S., Michels O., Govoni A., Fitzgerald J., Morales P. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol. Ther. 2015;23:477–487. doi: 10.1038/mt.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N. Adeno-Associated Virus-Based Gene Therapy for CNS Diseases. Hum. Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boillée S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 29.Wang L., Gutmann D.H., Roos R.P. Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum. Mol. Genet. 2011;20:286–293. doi: 10.1093/hmg/ddq463. [DOI] [PubMed] [Google Scholar]
- 30.Wang L., Sharma K., Grisotti G., Roos R.P. The effect of mutant SOD1 dismutase activity on non-cell autonomous degeneration in familial amyotrophic lateral sclerosis. Neurobiol. Dis. 2009;35:234–240. doi: 10.1016/j.nbd.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferraiuolo L., Meyer K., Sherwood T.W., Vick J., Likhite S., Frakes A., Miranda C.J., Braun L., Heath P.R., Pineda R. Oligodendrocytes contribute to motor neuron death in ALS via SOD1-dependent mechanism. Proc. Natl. Acad. Sci. USA. 2016;113:E6496–E6505. doi: 10.1073/pnas.1607496113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoica L., Todeasa S.H., Cabrera G.T., Salameh J.S., ElMallah M.K., Mueller C., Brown R.H., Jr., Sena-Esteves M. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann. Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 35.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 37.Lin X., Ruan X., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C., Hughes C.M., Shanmugam K.S., Bhattacharjee A., Meyerson M., Collins F.S. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 41.Di Rocco G., Verdina A., Gatti V., Virdia I., Toietta G., Todaro M., Stassi G., Soddu S. Apoptosis induced by a HIPK2 full-length-specific siRNA is due to off-target effects rather than prevalence of HIPK2-Δe8 isoform. Oncotarget. 2016;7:1675–1686. doi: 10.18632/oncotarget.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klinghoffer R.A., Magnus J., Schelter J., Mehaffey M., Coleman C., Cleary M.A. Reduced seed region-based off-target activity with lentivirus-mediated RNAi. RNA. 2010;16:879–884. doi: 10.1261/rna.1977810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao D.D., Senzer N., Cleary M.A., Nemunaitis J. Comparative assessment of siRNA and shRNA off target effects: what is slowing clinical development. Cancer Gene Ther. 2009;16:807–809. doi: 10.1038/cgt.2009.53. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frey D., Schneider C., Xu L., Borg J., Spooren W., Caroni P. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J. Neurosci. 2000;20:2534–2542. doi: 10.1523/JNEUROSCI.20-07-02534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegedus J., Putman C.T., Gordon T. Time course of preferential motor unit loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2007;28:154–164. doi: 10.1016/j.nbd.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Mead R.J., Bennett E.J., Kennerley A.J., Sharp P., Sunyach C., Kasher P., Berwick J., Pettmann B., Battaglia G., Azzouz M. Optimised and rapid pre-clinical screening in the SOD1(G93A) transgenic mouse model of amyotrophic lateral sclerosis (ALS) PLoS ONE. 2011;6:e23244. doi: 10.1371/journal.pone.0023244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L., Duff K. A technique for serial collection of cerebrospinal fluid from the cisterna magna in mouse. J. Vis. Exp. 2008;(21):960. doi: 10.3791/960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.