Abstract
Huntington’s disease (HD) is associated with the misfolding and aggregation of mutant huntingtin harboring an elongated polyglutamine stretch at its N terminus. A distinguishing pathological hallmark of HD is mitochondrial dysfunction. Any strategy that can restore the integrity of the mitochondrial environment should have beneficial consequences for the disease. Specific RNA aptamers were selected that were able to inhibit aggregation of elongated polyglutamine stretch containing mutant huntingtin fragment (103Q-htt). They were successful in reducing the calcium overload, which leads to mitochondrial membrane depolarization in case of HD. In one case, the level of Ca2+ was restored to the level of cells not expressing 103Q-htt, suggesting complete recovery. The presence of aptamers was able to increase mitochondrial mass in cells expressing 103Q-htt, along with rescuing loss of mitochondrial genome. The oxidative damage to the proteome was prevented, which led to increased viability of cells, as monitored by flow cytometry. Thus, the presence of aptamers was able to inhibit aggregation of mutant huntingtin fragment and restore mitochondrial dysfunction in the HD cell model, confirming the advantage of the strategy in a disease-relevant parameter.
Keywords: calcium homeostasis, intramers, membrane depolarization, mitochondrial mass, oxidative damage, reactive oxygen species
Introduction
Huntington’s disease (HD) is an autosomal, dominantly inherited neurodegenerative disorder characterized by progressive chorea, psychiatric changes, and intellectual decline.1 In HD, CAG repeat expansion occurs in the first exon of the gene IT15, which encodes a large (∼350 kDa) protein called huntingtin.2 Misfolding and aggregation of mutant huntingtin with expanded N-terminal polyglutamine (polyQ) tract has been identified as a root cause of the disease,3, 4 although loss of endogenous huntingtin function5 and RNA-based toxicity6 may also play a role. Inhibition of aggregation of mutant huntingtin is thus a validated therapeutic approach. A number of studies indicate that mitochondrial dysfunction is central to HD pathogenesis.7, 8, 9, 10, 11, 12, 13, 14 Despite normal food intake, HD patients exhibit significant weight loss.15 A study of the affected brain regions of HD patients showed decreased glucose metabolism and increased level of lactate, indicating bioenergetic defect.16 The role of mitochondrial dysfunction in HD was elucidated in a study where 3-nitropropionic acid (3-NP), a selective inhibitor of succinate dehydrogenase, was able to recapitulate the loss of medium spiny neurons in the substantia nigra and led to HD-like symptoms in several vertebrate models.17, 18 Reduction of the activities of complexes II, III, and IV has been observed in the caudate and putamen of HD patients.7 These redox enzymes are involved in the transfer of electrons from one substrate to other, as well as in an extensive network of antioxidant defenses. Mitochondrial insults can cause an imbalance between reactive oxygen species (ROS) production and its removal, resulting in net ROS production.19, 20
HD patients exhibit energy deficit, Ca2+ overload, and enhanced oxidative stress.21, 22 Mutant huntingtin interacts with and destabilizes the outer mitochondrial membrane.23 It increases the sensitivity of the mitochondrial permeability transition pore (mPTP) to Ca2+ or other apoptotic stimuli.24, 25 Mitochondria regulate calcium homeostasis by providing high capacity Ca2+ buffering system.26, 27 In HD, the damaged mitochondria are unable to cope up with Ca2+ overload, which may open up mPTP, a high-conductance pathway, leading to neuronal death.23 The enhanced ROS production increases the vulnerability of the mPTP in the presence of Ca2+ and further worsens the mitochondrial functions.28
Mutant huntingtin is reported to disrupt transcriptional regulation by interacting with transcription factors.29, 30 Reduced expression of PGC-1α, a nuclear co-activator that plays a major role in mitochondrial biogenesis, has been reported in HD models.31, 32 The level of the nuclear respiratory factors, NRF-1 and NRF-2, which are present downstream of PGC-1α, are also affected in HD.33 The repression of these transcription factors impairs mitochondrial biogenesis in HD.32, 33
Various approaches have been tried to treat HD. The use of nucleic acid aptamers is one such approach that has shown preliminary promise as a therapeutic strategy.34 Aptamers are relatively short single-stranded oligonucleotides (DNA or RNA) that assume specific, stable conformations and bind tightly and specifically to protein targets by shape complementarity.35, 36, 37 They can act as a stabilizer of the target protein in two ways: (1) by sterically hindering the unfolding of target protein monomer and (2) by inhibiting protein-protein interaction via electrostatic repulsion.35, 38 Thus, RNA aptamers, which are non-immunogenic and non-toxic, can be used to inhibit protein aggregation inside cells.
Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in various cell and animal models and recapitulate HD phenotype.39, 40, 41, 42, 43 The ease of genetic manipulation makes yeast (Saccharomyces cerevisiae) a promising model for studying human diseases and for carrying out high-throughput screenings.41, 42, 44, 45, 46 N-terminal fragments of mutant huntingtin protein (encoded by exon 1 of huntingtin gene with expanded CAG repeats) have been expressed in yeast, where these were found to induce polyglutamine length-dependent toxicity.39, 41, 42 We have earlier reported the selection of RNA aptamers against N-terminal fragment of huntingtin with expanded glutamine repeat (51Q). The selected aptamers were able to reduce the aggregation of mutant huntingtin (both 51Q and 103Q) and helped to restore defect in endocytosis associated with HD.34 In the present study, we have evaluated the effect of the presence of RNA aptamers on mitochondrial dysfunction, which is a hallmark of HD.7, 8, 9, 10, 11, 12, 13, 14, 21, 22, 23 The aptamers were able to reduce oxidative stress and associated damage to the proteome in a yeast model of HD. Aptamers facilitated the restoration of calcium homeostasis and mitochondrial membrane potential and improved cell viability.
Results
Expression of “Intramers” in Yeast Cells
RNA aptamers were selected that specifically recognized N-terminal region of huntingtin containing elongated polyQ tract.34 These aptamers were able to reduce aggregation of N-terminal mutant huntingtin in vitro and in a yeast HD model, with concomitant decrease in oxidative stress. A combination of such aptamers that bind to different regions of the N-terminal stretch showed greater reduction in aggregation of the protein in vitro.34 We wanted to investigate whether a similar result could be replicated in a cellular context and also to elucidate the mechanism by which the intramers show their ameliorative effect in the yeast HD model. If they aim for a disease-relevant parameter, they would provide a promising route to target HD pathogenesis.
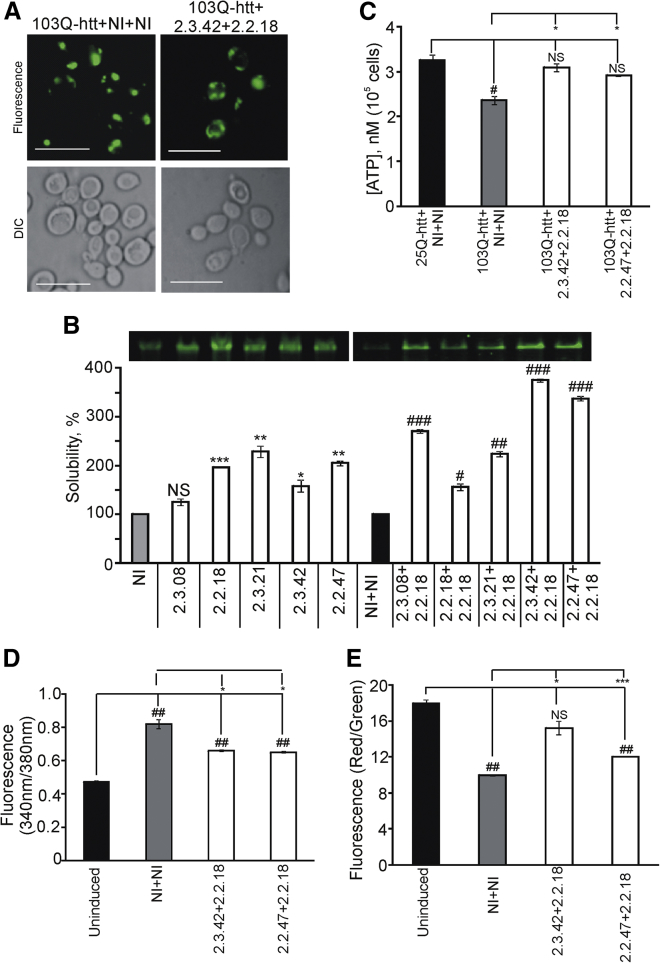
The ability of the selected RNA aptamers to function as inhibitors of protein aggregation inside the cell (intramers) was determined by expressing a pair of aptamers or non-inhibitors along with 103Q-htt in Saccharomyces cerevisiae BY4742 cells. A non-inhibitor is a sequence from the same library as aptamers that bound to N-terminal mutant huntingtin with similar affinity as aptamers but was unable to inhibit the aggregation of the latter.34 The expression of 103Q-htt was seen under a fluorescence microscope (Figure 1A). N-terminal mutant huntingtin (103Q-htt) was seen to exist in the aggregated form in the presence of a pair of non-inhibitors, whereas the presence of mHtt2.3.42+mHtt2.2.18 aptamer pair led to increase in the soluble form of the protein, seen as more diffused fluorescence. As there is no difference between the non-inhibitor (RNA sequences that bind to but do not inhibit aggregation of 103Q-htt) and aptamer (RNA sequences that bind to and inhibit aggregation of 103Q-htt) sequences except for the order of nucleic acid bases, the results reported here arise from the effect of aptamers on aggregation of 103Q-htt and are not a result of the effect of RNA sequences on the parameters measured. The increased solubilization was quantified by native PAGE analysis. The coexpression of aptamers had no effect on the expression level of 25Q-htt, a benign polyglutamine length (data not shown). Cell lysates showed higher solubilization of 103Q-htt in the presence of single aptamers or pairs of aptamers compared to the corresponding non-inhibitor controls (Figure 1B). The combinations of aptamers were chosen based on their mutually exclusive binding sites on the target, i.e., the elongated polyQ stretch.34 It is clear that pairs of aptamers in which each partner binds to different regions on the target show a stronger inhibitory effect than single aptamers. The presence of mHtt2.3.42+mHtt2.2.18 and mHtt2.2.47+mHtt2.2.18 showed maximal solubilization, i.e., ∼3.7- and ∼3.4-fold increase in intensities of bands for the soluble protein, respectively, as compared to the non-inhibitor control (Figure 1B), and these pairs were used for further studies. In case of mHtt2.2.18+mHtt2.2.18, both aptamers were the same and bound to the same region of the protein. This could account for their minimal activity among all tested aptamer pairs. It may be noted that, in case of all parameters measured, results are reported against the non-inhibitor control (NI+NI). The non-inhibitor control is also an aptamer in that it shows comparable binding affinity for the elongated polyQ stretch but is unable to inhibit protein aggregation.34
Figure 1.
Effect of Intramers on Properties of Yeast Cells Expressing 103Q-htt-EGFP
(A) Yeast cells were cotransformed with constructs for expression of 103Q-htt-EGFP and aptamers or non-inhibitors as described in the Materials and Methods section. Aggregation pattern was monitored by fluorescence microscopy. Cells expressing 103Q-htt-EGFP with a pair of aptamers or non-inhibitors (NI+NI) were analyzed by fluorescence microscopy. Representative result with one pair of RNA aptamers (mHtt2.2.18 and mHtt2.3.42) is shown. The scale bars represent 10 μm. (B) Native PAGE analysis of 103Q-htt-EGFP expressed in the presence of single aptamers or pairs of RNA aptamers along with corresponding non-inhibitor control in yeast cells. The gel was scanned with an image scanner (Typhoon Trio; GE Healthcare). The intensities of the bands for 103Q-htt-EGFP on native PAGE were quantified by densitometry (Image Quant TL; GE Healthcare). An arbitrary value of 100% was assigned to the band intensity of 103Q-htt-EGFP coexpressed with a single aptamer or a pair of non-inhibitors. Solubility in the presence of a single aptamer is compared with a single non-inhibitor sequence (NI, gray bar) whereas solubility in the presence of a pair of aptamers is compared with a pair of non-inhibitors (NI+NI, black bar). Values shown are mean ± SEM of three independent experiments. ***p < 0.001; **p < 0.01; *p < 0.05 against the intensity of the band in cells coexpressing 103Q-htt-EGFP with a non-inhibitor (gray bar). ###p < 0.001; ##p < 0.01; #p < 0.05 against the intensity of the band in cells coexpressing 103Q-htt-EGFP with a pair of non-inhibitors (black bar). (C) Intracellular ATP content in cells coexpressing 103Q-htt-EGFP in the presence of a pair of intramers or non-inhibitors was measured using BacTiter-Glo Microbial Cell Viability assay. The contents of ATP were calculated from a standard curve, which was plotted using different concentrations of ATP in culture medium. Values shown are mean ± SEM of two independent experiments. *p < 0.05 against cells coexpressing 103Q-htt with a pair of non-inhibitors (gray bar); #p < 0.05 against cells coexpressing 25Q-htt with a pair of non-inhibitors (black bar); NS, nonsignificant. (D) Intracellular calcium amount in cells coexpressing 103Q-htt in the presence of a pair of intramers or non-inhibitors was measured using Fura-2 AM dye. Values shown are mean ± SEM of three independent experiments. *p < 0.05 against cells coexpressing 103Q-htt with a pair of non-inhibitors (gray bar); ##p < 0.01 against uninduced cells harboring the empty vector (but not expressing 103Q-htt) with a pair of non-inhibitors (black bar). (E) Bar chart representing the fluorescence emission of the JC-1 dye expressed as a ratio of multimer (590 nm) to monomer (530 nm), for cells expressing 103Q-htt with a pair of intramers or non-inhibitors, is shown. Values shown are mean ± SEM of three independent experiments. ***p < 0.001, *p < 0.05 against cells coexpressing 103Q-htt and a pair of non-inhibitors (gray bar); ##p < 0.01 against uninduced cells harboring the empty vector (not expressing 103Q-htt) with a pair of non-inhibitors (black bar), N.S. not significant.
Measurement of Cellular Metabolic Activity
The measurement of cellular ATP content was carried out to analyze the effect of aptamers against 103Q-htt-mediated mitochondrial dysfunction. Cells expressing 103Q-htt showed significant reduction in the amount of ATP produced as compared to the cells expressing N-terminal huntingtin with normal polyQ stretch (25Q; Figure 1C). mHtt2.2.18, in combination with either mHtt2.3.42 or mHtt2.2.47, showed significant increase in ATP count as compared to cells expressing 103Q-htt along with a pair of non-inhibitors (Figure 1C). The level of ATP in these cells expressing aptamers was comparable to the ATP count of cells expressing 25Q-htt with a pair of non-inhibitors. As ATP is an indicator of metabolically active cells, this hints at the protective effect of aptamers against 103Q-htt-induced respiratory chain defects.
Measurement of Intracellular Calcium and Mitochondrial Membrane Potential
YAC128 HD transgenic mouse (harboring 128Q-htt) showed significantly higher level of mitochondrial matrix Ca2+ level in fibroblasts, which was associated with mtDNA damage.12 Knockdown of InsP3R1 (inositol 1,4,5-triphosphate receptor1), which has been linked to aberrant calcium signaling in HD, led to significant reduction in Ca2+ level in YAC128 fibroblasts, suggesting a link between HD and Ca2+ overload. Intracellular calcium was measured using the ratiometric calcium indicator Fura-2 AM.47, 48 Upon calcium binding, the fluorescence excitation maximum of the indicator undergoes a blue shift from 363 nm (Ca2+ free) to 335 nm (Ca2+ saturated). In the presence of Ca2+, maximum Fura-2 AM fluorescence intensity (at 510 nm) is observed at an excitation wavelength of 340 nm and at 380 nm for Ca2+-free condition.47, 48 Thus, the concentration of free intracellular Ca2+ is proportional to the ratio of fluorescence intensities when excited at 340/380 nm. The ratio was found to be the lowest for uninduced cells expressing a pair of aptamers (Figure 1D), indicating low intracellular calcium in cells that do not express 103Q-htt. The intracellular calcium level of the cells expressing 103Q-htt showed ∼45% increase as compared to uninduced cells, matching the increased level of Ca2+ in HD models reported elsewhere. Significant decrease in 340/380 nm ratio was observed in case of cells expressing 103Q-htt in the presence of a pair of aptamers compared to the non-inhibitor control expressing 103Q-htt (Figure 1D). The presence of the pairs of aptamers showed ∼20% reduction in calcium overload as compared to cells expressing 103Q-htt in the presence of a pair of non-inhibitors. This study indicated that the selected aptamers were able to rescue the cells from calcium toxicity resulting from aggregation of 103Q-htt.
Uptake of Ca2+ ions by mitochondria is reported to depolarize the mitochondrial membrane.49 The change in the mitochondrial membrane potential was measured using JC-1. This cationic dye accumulates in energized mitochondria. At low concentrations (due to low mitochondrial membrane potential), JC-1 predominantly exists as a monomer that yields green fluorescence with emission maxima at 530 ± 15 nm. At high concentrations (due to high mitochondrial membrane potential), the dye forms aggregate yielding a red- to orange-colored emission (590 ± 17.5 nm). Thus, reduction in the red aggregated fluorescence count is suggestive of depolarization, whereas an increase indicates hyperpolarization.50, 51 The relative change in mitochondrial membrane potential of the cells expressing 103Q-htt in the presence of a pair of aptamers or non-inhibitors was expressed as a change in the ratio of fluorescence intensities at 590/525 nm (the respective emission maxima of red and green fluorescence). The ratio was found to be the highest for uninduced cells expressing a pair of aptamers, indicating low membrane polarization (Figure 1E). When the cells expressed 103Q-htt in the presence of a pair of non-inhibitors, the ratio was the lowest (1.8-fold less than uninduced cells), suggesting significant polarization of the mitochondrial membrane during aggregation of 103Q-htt (Figure 1E), and correlates well with the high calcium load in these cells (Figure 1D). Significant increase in the ratio was observed in cells expressing 103Q-htt in the presence of a pair of aptamers as compared to the non-inhibitor control expressing 103Q-htt, which matched with the lower level of Ca2+ in these cells. In fact, in case of mHtt2.3.42+mHtt2.2.18 pair, the ratio was similar to that of uninduced cells (Figure 1E), which may suggest restoration of normal Ca2+ level due to solubilization of 103Q-htt. This study indicated that the presence of aptamers inhibited aggregation of 103Q-htt, which reduced Ca2+ level in the cells and also helped to maintain low mitochondrial membrane polarization.
Analysis of Mitochondrial Mass
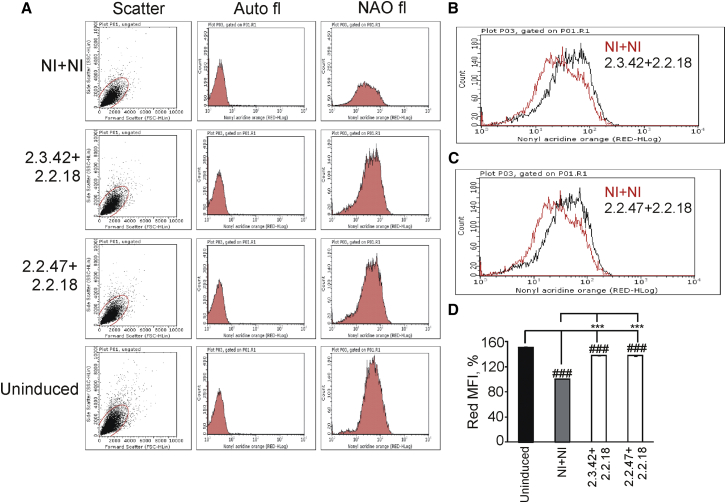
The expression of mutant huntingtin in cells leads to mitochondrial dysfunction and decrease in mitochondrial mass.7, 8, 9, 10, 11, 12, 13, 14 The effect of aptamers on mitochondrial mass was determined using 10-N-nonyl acridine orange (NAO), which binds with cardiolipin of mitochondrial membrane, thereby giving an estimate of total mitochondrial mass present inside the cell.52, 53 NAO has been used to analyze mitochondrial mass by flow cytometry.54
The cells expressing 103Q-htt in the presence of a pair of aptamers or non-inhibitors were stained with NAO and analyzed by flow cytometry for quantitative measurement of red fluorescence of the dye, which reflects mitochondrial mass of the cell population. In the absence of NAO, the cells did not exhibit any fluorescence, whereas in the presence of the dye, strong red fluorescence signal was observed in each cell population (Figure 2A). This confirmed that the red fluorescence was observed only due to the presence of the probe. As expected, the strongest signal was seen for the uninduced cells expressing a pair of aptamers, indicating the highest mitochondrial mass. In overlaid histograms of NAO fluorescence, right shift in red fluorescence was observed in cells expressing 103Q-htt in the presence of a pair of aptamers compared to non-inhibitors (Figures 2B and 2C), indicating higher mitochondrial mass in cells where aggregation of 103Q-htt was inhibited. The value of red MFI (mean fluorescence intensity) is a determinant of mitochondrial mass of each cell population. Around 33% decrease in mitochondrial mass was observed in cells expressing 103Q-htt in the presence of a pair of non-inhibitors as compared to uninduced cells (Figure 2D). The mitochondrial mass increased significantly in case of cells expressing 103Q-htt in the presence of a pair of aptamers and where the protein was partially solubilized, as compared to the non-inhibitor control. Thus, the presence of specific aptamers was able to exert a protective effect against 103Q-htt-mediated mitochondrial depletion.
Figure 2.
Determination of Mitochondrial Mass
(A) Representative scatterplots showing positioning of gates after voltage calibration. Auto-fluorescence and fluorescence of NAO for mitochondrial mass determination in cells expressing 103Q-htt in the presence of a pair of aptamers or non-inhibitors are shown. (B) Overlaid histograms of NAO fluorescence in cells expressing 103Q-htt with a pair of non-inhibitors (NI+NI) and a pair of aptamers (mHtt2.2.18+mHtt2.3.42). (C) Overlaid histograms of NAO fluorescence in cells expressing 103Q-htt with a pair of non-inhibitors (NI+NI) and a pair of aptamers (mHtt2.2.18+mHtt2.2.47). (D) The mean fluorescence intensities (MFIs) of cells were analyzed by flow cytometry after staining them with NAO. MFI was calculated to highlight the difference in the overall mitochondrial mass. 10,000 events were counted for each cell population. Values shown are mean ± SEM of three independent experiments. 100% value was assigned to the MFI of cells expressing 103Q-htt with a pair of non-inhibitors (gray bar). ***p < 0.001 against cells coexpressing 103Q-htt with a pair of non-inhibitors (gray bar) and ###p < 0.001 against uninduced cells harboring the empty vector (not expressing 103Q-htt) with a pair of non-inhibitors (black bar).
Analysis of mtDNA Loss
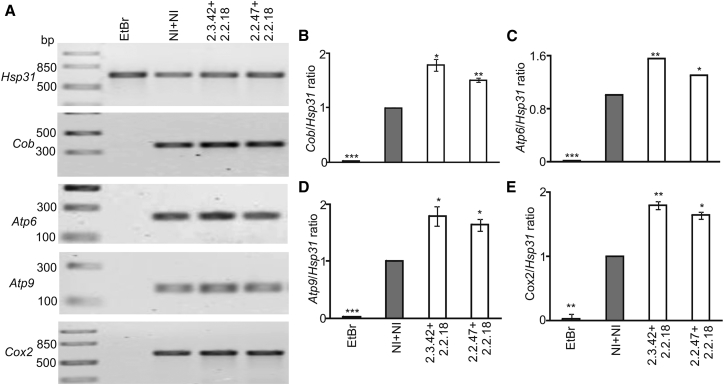
Mutant-huntingtin-mediated oxidative stress leads to loss of mtDNA.14, 28 mtDNA-encoded reporter genes Cob, Atp6, Atp9, and Cox2 were amplified along with a nuclear gene Hsp31 as the reference.54 As Cob, Atp6, Atp9, and Cox2 are located only in the mtDNA, the amount of amplified product can be directly correlated with initial copies of these genes present in the isolated genomic DNA and is indicative of mtDNA abundance.54 Yeast cells treated with EtBr were used as the negative control, as the dye is reported to inhibit the incorporation of thymidine into mtDNA, resulting in decrease in the mtDNA copies in cells exposed to EtBr.55
The cells treated with EtBr showed the least ratio (Figure 3), as the replication of mtDNA was compromised in the presence of the intercalator, validating the experimental design. The ratio of mitochondrial to nuclear marker indicates the copies of mtDNA present per nuclei. This ratio was found to be 1.8-, 1.5-, 1.8-, and 1.8-fold higher for Cob (Figure 3B), Atp6 (Figure 3C), Atp9 (Figure 3D), and Cox2 (Figure 3E) in cells expressing 103Q-htt in the presence of mHtt2.2.18+mHtt2.3.42 as compared to cells expressing 103Q-htt in the presence of a pair of non-inhibitors. Similar increases were seen in case of the second aptamer pair too (Figures 3A–3E). The results confirm the protective effect of aptamers against 103Q-htt-induced mtDNA loss.
Figure 3.
Estimation of mtDNA Abundance
(A) PCR amplification of the Cob, Atp6, Atp9, and Cox2 genes was used to assess mtDNA loss using genomic DNA isolated from cells expressing 103Q-htt with intramers or non-inhibitors. Hsp31 served as the control for nuclear DNA. Yeast cells treated with EtBr (black bars) to induce mtDNA loss were used as a negative control. (B–E) Densitometric analysis for band intensities was carried out, and the ratio of mitochondrial to nuclear marker was plotted for (B) Cob, (C) Atp6, (D) Atp9, and (E) Cox2. The ratio of intensities of mitochondrial to nuclear markers in cells expressing 103Q-htt in the presence of a pair of non-inhibitors (gray bars) was assigned an arbitrary value of 1 in each case. Values shown are mean ± SEM of three independent experiments. ***p < 0.001, **p < 0.01, and *p < 0.05 against cells coexpressing 103Q-htt with a pair of non-inhibitors in each case.
Measurement of Proteasomal Damage
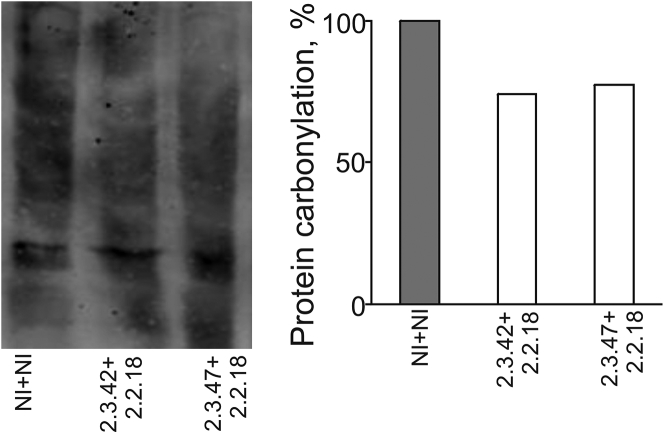
Oxidative stress is significantly elevated upon aggregation of mutant huntingtin in cells56 and in the brains of HD patients.57 Expression of selected aptamers in cells expressing 103Q-htt led to reduction in intracellular oxidative stress.34 Based on this observation, we attempted to evaluate the effect of aptamers on oxidative damage to the proteome. The higher accumulation of ROS in cells has been reported to increase the formation of carbonyl groups in amino acid residues, and this serves as a marker for proteome damage due to oxidative stress.58, 59
The protein bands in the lysates of cells expressing 103Q-htt with a pair of aptamers or non-inhibitors were transferred to a polyvinylidene fluoride (PVDF) membrane and probed with anti-DNP (dinitrophenol) antibody (Figure 4). Densitometric analysis of the bands of 2,4-dinitrophenylhydrazine hydrochloric acid (DNPH)-derivatized carbonylated proteins showed decreased intensity in cells coexpressing 103Q-htt in the presence of a pair of aptamers as compared to the non-inhibitor control (Figure 4). The presence of a pair of aptamers showed ∼25% reduction in protein carbonylation as compared to the non-inhibitor control. Thus, aptamers were able to reduce oxidative-stress-mediated proteome damage in cells expressing mutant huntingtin.
Figure 4.
Determination of Proteasomal Damage
Estimation of proteasomal damage due to oxidative stress was carried out by immunoblotting of lysates of induced cells expressing 103Q-htt with DNP (dinitrophenol) antibody as the primary antibody. Densitometric analysis of the intensities of bands (complete lanes) for damaged proteins was carried out using Image QuantTL (GE Healthcare). The intensity of the lane for cells expressing 103Q-htt in the presence of a pair of non-inhibitors was assigned an arbitrary value of 100%.
Measurement of Cell Viability
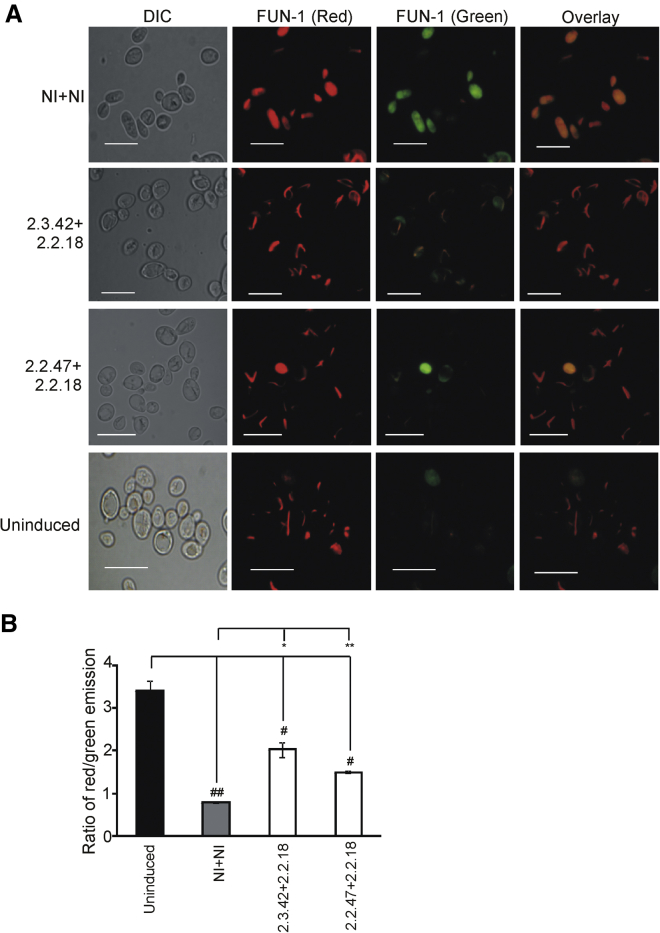
HD is characterized by neuronal cell death in striatum and caudate nucleus due to misfolding and aggregation of mutant huntingtin. The use of a single aptamers had shown a modest increased in cell survival.34 The above results show that reduction in aggregation of mutant huntingtin in the presence of a pair of aptamers was able to increase ATP content and enhance metabolic activity of the cell, reduce Ca2+ overload and concomitant membrane polarization, increase mitochondrial mass, and rescue mtDNA loss as compared to cells expressing 103Q-htt in the presence of a pair of non-inhibitors where the protein existed in the aggregated form. The effect of aptamers on the viability of cells was assessed using FUN-1 dye. This membrane-permeant dye is a two-color fluorescent viability probe that passively diffuses into yeast cells and initially stains the cytoplasm with a diffusely distributed green fluorescence. Subsequent bioprocessing of the dye by live cells results in the appearance of cylindrical intravacuolar structures (CIVSs) with compact form that exhibit a striking red fluorescence, accompanied by reduction in the cytoplasmic green fluorescence.24, 60
Uninduced cells expressing a pair of aptamers showed clearly defined CIVSs with negligible green fluorescence when analyzed by fluorescence microscopy (Figure 5A), indicating the highest viability among all the cells studied. In the cells expressing 103Q-htt with a pair of non-inhibitors, a higher number of cells with green fluorescence were observed, suggesting the presence of a larger population of non-viable cells. Diffused red fluorescence, lacking CIVSs, was observed, indicating low metabolic activity of these cells (Figure 5A). In case of cells expressing 103Q-htt in the presence of a pair of aptamers, more number of cells with distinct CIVS formation and reduced green fluorescence was seen (Figure 5A).
Figure 5.
Determination of Cell Viability
(A) Viability of cells was analyzed by staining of the cells with FUN-1. Fluorescence microscopic images of yeast cells expressing 103Q-htt with a pair of aptamers or non-inhibitors, stained with FUN-1. The scale bars represent 10 μm. (B) Bar chart representing the fluorescence emission of the FUN-1 dye in terms of ratio of red fluorescence (575 nm) to green fluorescence (535 nm), for cells expressing 103Q-htt with a pair of intramers or non-inhibitors. Values shown are mean ± SEM of three independent experiments. **p < 0.01 and *p < 0.05 against cells coexpressing 103Q-htt and a pair of non-inhibitors (gray bar); ##p < 0.01 and #p < 0.05 against uninduced cells harboring the empty vector (not expressing 103Q-htt) with a pair of non-inhibitors (black bar).
As the conversion of FUN-1 from diffused green fluorescence to red fluorescence of CIVSs requires ATP, the ratio of red (λem 575 nm) and green (λem 535 nm) fluorescence intensities is taken as a measure of metabolic activity of cells.61 The ratio was found to be the highest for uninduced cells expressing a pair of aptamers (>4-fold; Figure 5B), indicating maximum metabolic activity for cells that do not express N-terminal mutant huntingtin. Significant increase in 575/535 nm ratio was observed for cells expressing 103Q-htt in the presence of a pair of aptamers compared to those cells expressing 103Q-htt and non-inhibitors (Figure 5B). The pair of mHtt2.3.42+mHtt2.2.18 showed marginally higher metabolic activity than the mHtt2.2.47+mHtt2.2.18 pair, i.e., ∼2.5-fold and ∼2-fold, respectively. These results are also in accordance with the ATP content of the cells (Figure 1C).
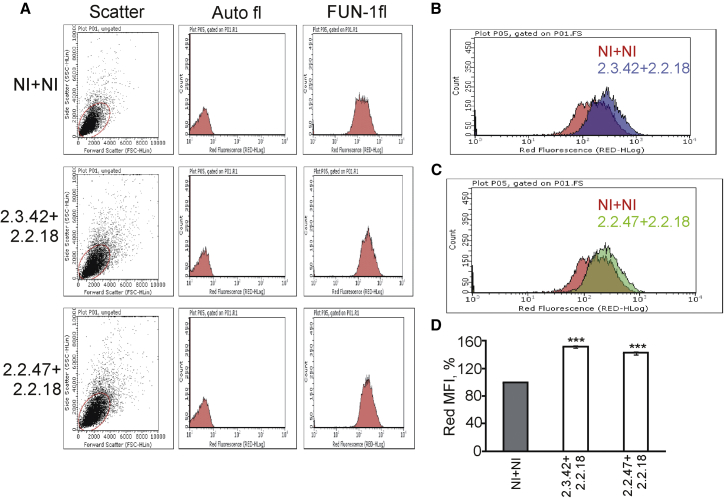
The stained cells were further analyzed by flow cytometry for quantitative measurement of red fluorescence. In the absence of FUN-1, the cells did not show any red fluorescence, whereas in the presence of the dye, strong red fluorescence signal was observed in each cell population (Figure 6A), suggesting that the red fluorescence observed in each case was only due to FUN-1. In overlaid histograms of FUN-1 fluorescence, right shift in red fluorescence was observed in cells expressing 103Q-htt in the presence of mHtt2.3.42+mHtt2.2.18 (Figure 6B) or mHtt2.2.47+mHtt2.2.18 (Figure 6C) as compared to a pair of non-inhibitors, indicating higher viability of the former. The value of red MFI is a determinant of viability of each cell population. Around 1.5-fold increase in red MFI was observed for cells expressing 103Q-htt in the presence of pairs of aptamers compared to the non-inhibitor control (Figure 6D). Thus, the aptamers were able to rescue the cells from metabolic damage caused by aggregation of 103Q-htt. This observation is also in accordance with earlier studies, which have reported higher survival of cells showing reduced aggregation of mutant huntingtin.34, 62
Figure 6.
Determination of Cell Viability by Flow Cytometry
(A) Representative scatterplots showing positioning of gates after voltage calibration. Auto-fluorescence and FUN-1 fluorescence for determination of viability of cells expressing 103Q-htt in presence of a pair of aptamers or non-inhibitors are shown. (B) Overlaid histograms of FUN-1 fluorescence in cells expressing 103Q-htt with a pair of non-inhibitors (NI+NI) and a pair of aptamers (mHtt2.2.18+mHtt2.3.42). (C) Overlaid histograms of FUN-1 fluorescence in cells expressing 103Q-htt with a pair of non-inhibitors (NI+NI) and a pair of aptamers (mHtt2.2.18+mHtt2.2.47). (D) The MFIs of cells were analyzed by flow cytometry after staining them with FUN-1. MFI was calculated to highlight the difference in cell viability. 10,000 events were counted for each cell population. Values shown are mean ± SEM of three independent experiments. 100% value was assigned to the MFIs of cells expressing 103Q-htt with a pair of non-inhibitors (gray bar). ***p < 0.001 against the cells coexpressing 103Q-htt with a pair of non-inhibitors.
Discussion
The N-terminal fragment of Drosophila huntingtin associates with mammalian vesicles and colocalizes with brain-derived neurotrophic factor (BDNF) vesicles in cortical neurons,63 in the same manner as the interaction of the N-terminal fragment of mammalian huntingtin with components of the molecular motor complex.64 The N-terminal fragment of normal huntingtin is sufficient to carry out this interaction, and elongation of polyQ stretch in this segment of mutant huntingtin is enough to observe the same kind of disruption in endocytosis as is seen following expression of full-length mutant huntingtin in mammalian cells.65 Mutations of genes involved in endocytosis in yeast lead to aggravation of toxicity due to aggregated polyQ.42 A similar defect in endocytosis has been seen in cultured mammalian cells (HEK293) expressing 103Q (same construct as used here, under the constitutive cytomegalovirus [CMV] promoter), using Texas red-tagged transferrin, which is internalized through receptor-mediated endocytosis.42 The use of aptamers was able to rescue endocytotic defect seen due to aggregation of 103Q-htt.34 Thus, the yeast model used here is a validated approach to study disease-specific parameters in HD.42, 44
Mitochondrial dysfunction is evident in HD patients, which includes defects in the respiratory chain and reduced ATP production. The post mortem samples of the caudate and putamen in HD patients showed significant reduction in the activity of complex II and III and milder reduction of complex IV.7, 28 Deficient import of TIM23 (translocase of the inner membrane) mitochondrial protein complex in HD has been identified as a factor in mutant-huntingtin-induced cell death.14 A genome-wide analysis study has shown that mitochondrial genes, when deleted, exacerbate polyglutamine-induced toxicity.66 Mitochondrial damage in HD is also suggested by the direct interaction of mutant huntingtin with brain mitochondria in the HdhQ150 knockin mouse model, which has been linked to disease progression.67 Impairment of respiration following aggregation of mutant huntingtin occurred as a result of loss of activities of complexes II and III.9 Conversely, overexpression of Hap4, the catalytic subunit of the Hap2, 3, 4, and 5 complex involved in increasing the respiratory capacity of the cell, resulted in induction of mitochondrial biogenesis and amelioration of polyQ-induced toxicity in yeast cells.10 Pharmacological intervention with bezafibrate, a pan-PPAR agonist, led to increased levels of PPARs and PGC-1α, with increased mitochondrial biogenesis and enhanced survival of transgenic R6/2 HD mice.68 Thus, a strategy like the use of RNA aptamers described here, which reduces aggregation of 103Q-htt and is able to attenuate damage to the mitochondria, is a promising strategy for further development.
Mitochondria are key regulators of calcium homeostasis, which protect the cells from the damage by calcium overload. Mitochondria scavenge excess Ca2+ from the cytosol26 or local Ca2+-rich microdomains27 and release it back during a fall in concentration. In HD, expression of mutant huntingtin reduces Ca2+ handling capacity of mitochondria and opens up mPTP, which is potentially lethal for the cell. Thus, deficits in mitochondrial Ca2+ handling likely contribute to neurodegeneration.23, 24 We found that the selected aptamers were able to reduce the elevated Ca2+ levels resulting from aggregation of 103Q-htt. These novel inhibitors of 103Q-htt aggregation exhibit the potential to alleviate various mitochondrial dysfunctions, which is a hallmark of HD. The RNA aptamers were able to restore Ca2+ homeostasis and prevented hyperpolarization of mitochondrial membrane resulting from aggregation of 103Q-htt. The mitochondrial mass and mtDNA copies were also increased. ATP production was restored, which helped the cells to sustain metabolic activity. Thus, this work reiterates the crucial role of mitochondrial damage to progression of HD and describes the beneficial role that RNA aptamers may play in ameliorating this disease-relevant characteristic.
Materials and Methods
Materials
Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody and DNPH solution were purchased from Sigma-Aldrich Chemicals, Bengaluru, India. BacTiter-Glo Microbial cell viability assay kit, Taq DNA polymerase and Wizard SV Gel, and PCR Clean-Up System were purchased from Promega, Madison, USA. PVDF membrane and syringe filter (0.2 μm) were purchased from Advanced Microdevices, Ambala, India. FUN-1 (2-chloro-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium iodide), JC-1 (tetraethylbenzimidazolylcarbocyanine iodide), Fura-2 AM (2-[6-(bis[2-([acetyloxy]methoxy)-2-oxoethyl]amino)-5-(2-[2-(bis[2-([acetyloxy]methoxy)-2-oxoethyl]amino)-5-methylphenoxy]ethoxy)-2-benzofuranyl]-5-oxazolecarboxylic acid [acetyloxy]methyl ester), 10-N-nonyl acridine orange (products of Molecular Probes), and rabbit anti-2,4-DNP antibody were purchased from Invitrogen Bio Services India, Bengaluru, India. pRSII423 was a gift from Prof. Steven Haase (Addgene plasmid no. 35464). All other reagents and chemicals used were of analytical grade or higher.
Methods
Expression of Aptamers in Yeast Cells (Intramers)
The selection of aptamers against mutant huntingtin containing 51 glutamine stretch was carried out by an in vitro iterative selection process and has been described previously.34 For expression of RNA aptamers in Saccharomyces cerevisiae cells, the aptamer sequences were cloned in the plasmid pWHE601 (Ura3 selection marker) and pRSII423 (His3 selection marker). Details of pWHE601 aptamer expression vector have been described earlier.34, 69 For simultaneous expression of a second RNA aptamer in yeast cells, pRSII423 with His3 selection marker was used. For construction of pRSII423-Adh1-mHtt2.2.18, the parent plasmid (pRSII423) was double digested with EcoRI and BamHI. The mHtt2.2.18 sequence along with Adh1 promoter and terminator was amplified from pWHE601-mHtt2.2.18. Respective restriction sites were inserted in Adh1-mHtt2.2.18 by PCR mutagenesis in the cDNA sequences using forward primer (5′-GATGATGAATTCTGGAAAAACGCCAGCAACGCG-3′) and reverse primer (5′-GATGATGGATCCAGTCTCATCCTTCAATGCTATCAT-3′). Bold sequences in forward and reverse primers denote restriction sites for EcoRI and BamHI, respectively, and underlined sequences anneal with the 5′ Adh1 promoter and 3′ Adh1 terminator sequences, respectively. The insertion of sites at the correct position was confirmed by specific amplification of the inserted sequence, followed by agarose gel electrophoresis. In both plasmid constructs, RNA aptamers were expressed under the constitutive Adh1 promoter.
Expression of Mutant Huntingtin Protein in Saccharomyces cerevisiae Cells
Saccharomyces cerevisiae BY4742 cells (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [RNQ1+]), transformed with pRS315-103Q-htt-EGFP (expressing 103Q-htt of exon1 fused to EGFP under the control of Cup1 promoter and Leu2 selection marker), were sequentially transformed with individual pWHE601-aptamer (expressing RNA aptamer under the control of Adh1 promoter and Ura3 selection marker) or pWHE601-aptamer and pRSII413-aptamer (expressing RNA aptamer under the control of Adh1 promoter and His3 selection marker). Cells were grown in SC-Leu-Ura (2% w/v dextrose) or SC-Leu-Ura-His (2% w/v dextrose) media at 30°C until optical density at 600 nm (OD600) 0.8–1.0 (mid-logarithmic phase). The cells were washed and resuspended in the corresponding media. Expression of 103Q-htt was induced with 500 μM CuSO4 for 10 hr. The aggregation profile of 103Q-htt-EGFP was monitored under a fluorescence microscope (model E600; Nikon, Japan) using 100× objective lens. Conditions for cell lysis and native PAGE analysis have been described earlier.34
Measurement of Cellular Metabolic Activity
ATP production was monitored using a luminometer (1450 LSC and luminescence counter; PerkinElmer) in the yeast cells expressing 103Q-htt in the presence of a pair of aptamers or non-inhibitors by a standardized BacTiter-Glo Microbial cell viability assay kit following the protocol described by the manufacturer. The luminescence produced is directly proportional to the amount of ATP present.
Measurement of Intracellular Calcium
Intracellular calcium was measured using the ratiometric calcium indicator Fura-2 AM.47, 48 Cells (1 × 107) were incubated with 10 μM Fura-2 AM (stock solution 1 mM; in DMSO) in dark for 30 min at 37°C. After washing with Krebs/HEPES buffer, incubation was carried out further for 15 min at room temperature in dark for de-esterification of Fura-2 AM. The excitation spectra at 380 nm (calcium free) and 340 nm (calcium complex) were monitored with fixed emission at 510 nm using a spectrofluorimeter (RF5301; Shimadzu). The relative amount of free intracellular calcium was expressed as a change in the ratio of intensities following excitation at 340 nm to 380 nm.
Measurement of Mitochondrial Membrane Potential
Mitochondrial membrane potential was measured using JC-1.50, 51 Cells (1 × 107) were incubated with 5 μM of JC-1 (stock solution 1 mM; in DMSO) for 30 min at 30°C in dark. Cells were excited at 490 nm, and emission scan was recorded between 500 and 620 nm. The ratios of the peaks at 590 nm (aggregated form of JC-1) and 530 nm (monomeric form) were used as an indicator of membrane polarization.
Analysis of Mitochondrial Mass
NAO was used to measure mitochondrial mass inside the cells.52, 54 Cells (1 × 107) were incubated with 7.5 nM nonyl acridine orange (stock solution 10 μM; in DMSO) in dark at 30°C, 200 rpm for 30 min, washed twice with PBS, and then analyzed by flow cytometry (Guava flow cytometer, Merck Millipore, USA) using the standard excitation and emission parameters for the dye (489 nm and 525 nm, respectively). The values were expressed as MFIs of the cell population.
Analysis of mtDNA Loss
Cells expressing 103Q-htt with a pair of aptamers were harvested, and genomic DNA was isolated.70 A PCR-based method was used to determine mtDNA loss.71 Primers were designed for the following mitochondrial markers: Cytochrome B (Cob): forward primer: 5′-TGATTCACCACAACCATCATCA-3′, reverse primer: 5′-GTCTCACTCTGTCCATAAACACA-3′; ATP synthase subunit a (Atp6): forward primer: 5′-ATATGCTTAAAGGACAAATTGGAGGTA-3′, reverse primer: 5′-GCAGGTACGAATAATGAGAAGAATACT-3′; ATP synthase subunit c (Atp9): forward primer: 5′-TATATTGGAGCAGGTATCTCAACAATT-3′, reverse primer: 5′-CAGAATAAACCTGTAGCTTCTGATAA-3′; Cytochrome c oxidase subunit 2 (Cox2): forward primer: 5′-TTCAGGATTCAGCAACACCAAAT-3′, reverse primer: 5′-GATACTGCTTCGATCTTAATTGG-3′; and the nuclear marker heat shock protein (Hsp31): forward primer: 5′-CGCTCTTACCTCATATAACGATG-3′, reverse primer: 5′-TTTAAAGCGTCGATGGATCTTAC-3′. These mitochondrial markers along with the nuclear marker were amplified from the yeast genomic DNA to determine their relative abundance. Cells treated with ethidium bromide (EtBr) (exposed to 20 ng/mL for multiple generations) were used as a negative control as EtBr is reported to induce the loss of mtDNA.72 Amplified PCR products were run on agarose gel, the gels were scanned, and the intensities of bands were quantified by densitometry using ImageQuant software (GE Healthcare Life Sciences, Uppsala, Sweden).
Analysis of Proteome Damage
The cells expressing 103Q-htt along with a pair of aptamers or non-inhibitors were lysed by glass beads method.73 The lysate was centrifuged at 800 g for 10 min at 4°C. Proteins in the supernatant were separated by 12% SDS-PAGE. After transferring the proteins on to PVDF membrane, the damaged proteins were derivatized using DNPH, and the level of derivatization was measured using anti-DNP antibody as the primary antibody and FITC-conjugated anti-rabbit antibody as the secondary antibody.59
Measurement of Cell Viability
The viability of cells was measured using FUN-1 dye. Cells (1 × 107) were resuspended in GH solution (2% w/v glucose in 10 mM Na-HEPES [pH 7.2]) and incubated with 25 μM FUN-1 (stock solution 10 mM; in DMSO) at 30°C, 200 rpm for 14 hr. Cells were mounted on glass slides and viewed under a fluorescence microscope (Nikon Eclipse E600; Nikon, Japan) for red and green fluorescence. The cells were also analyzed by flow cytometry (Guava flow cytometer; Merck Millipore, USA) using Guava Incyte software (Burlingame, USA) using the standard excitation and emission parameters for the dye (450/550 nm). Values were expressed as MFI of the cell population.
The metabolic activity of cells was measured using FUN-1. The cells were resuspended in GH solution (2% w/v glucose in 10 mM Na-HEPES [pH 7.2]) and incubated with 0.5 μM FUN-1 (stock solution 10 mM; in DMSO) in dark at 30°C, 200 rpm for 14 hr.74 Fluorescence intensity was measured at 500–650 nm following excitation of the samples at 488 nm using a spectrofluorimeter (RF5301; Shimadzu).
Statistical Analysis
All data were expressed as mean ± SEM of three independent experiments. For comparison between two groups, data were analyzed by Student’s t test. For comparison between more than two groups, data were analyzed by the ANOVA along with post hoc Tukey test. Statistically significant difference was considered when the p value was less than 0.05.
Author Contributions
I.R. conceived the study. K.A.P. carried out the majority of the experiments. The aptamers used in this work were selected by R.K.C. K.A.P. and I.R. analyzed the data and wrote the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors are thankful to Prof. B. Suess, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, for the gift of RNA expression vector pWHE601. For construction of pRS315-25Q/103Q-htt-EGFP, the original vector pRS315-RNQ1-RFP was received from Prof. Douglas Cyr, University of North Carolina, USA. The constructs pRS315-htt-25Q/103Q-EGFP were made by Ankan Kumar Bhadra in the lab. This work was partially supported by Department of Biotechnology (BT/PR12306/BRB/10/1346/2014). K.A.P. and R.K.C. acknowledge the award of senior research fellowships by Department of Biotechnology and Indian Council of Medical Research, respectively.
References
- 1.MacDonald M.E., Gines S., Gusella J.F., Wheeler V.C. Huntington’s disease. Neuromolecular Med. 2003;4:7–20. doi: 10.1385/NMM:4:1-2:7. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 4.Labbadia J., Morimoto R.I. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci. 2013;38:378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo E., Rigamonti D., Goffredo D., Zuccato C., Squitieri F., Sipione S. Loss of normal huntingtin function: new developments in Huntington’s disease research. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 6.Li L.-B., Bonini N.M. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. Trends Neurosci. 2010;33:292–298. doi: 10.1016/j.tins.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 8.Tabrizi S.J., Workman J., Hart P.E., Mangiarini L., Mahal A., Bates G., Cooper J.M., Schapira A.H. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann. Neurol. 2000;47:80–86. doi: 10.1002/1531-8249(200001)47:1<80::aid-ana13>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Solans A., Zambrano A., Rodríguez M., Barrientos A. Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum. Mol. Genet. 2006;15:3063–3081. doi: 10.1093/hmg/ddl248. [DOI] [PubMed] [Google Scholar]
- 10.Ocampo A., Zambrano A., Barrientos A. Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J. 2010;24:1431–1441. doi: 10.1096/fj.09-148601. [DOI] [PubMed] [Google Scholar]
- 11.Ayala-Peña S. Role of oxidative DNA damage in mitochondrial dysfunction and Huntington’s disease pathogenesis. Free Radic. Biol. Med. 2013;62:102–110. doi: 10.1016/j.freeradbiomed.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J.Q., Chen Q., Wang X., Wang Q.C., Wang Y., Cheng H.P., Guo C., Sun Q., Chen Q., Tang T.S. Dysregulation of mitochondrial calcium signaling and superoxide flashes cause mitochondrial genomic DNA damage in Huntington disease. J. Biol. Chem. 2013;288:3070–3084. doi: 10.1074/jbc.M112.407726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schapira A.H., Olanow C.W., Greenamyre J.T., Bezard E. Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: future therapeutic perspectives. Lancet. 2014;384:545–555. doi: 10.1016/S0140-6736(14)61010-2. [DOI] [PubMed] [Google Scholar]
- 14.Yano H., Baranov S.V., Baranova O.V., Kim J., Pan Y., Yablonska S., Carlisle D.L., Ferrante R.J., Kim A.H., Friedlander R.M. Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci. 2014;17:822–831. doi: 10.1038/nn.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djoussé L., Knowlton B., Cupples L.A., Marder K., Shoulson I., Myers R.H. Weight loss in early stage of Huntington’s disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins B.G., Andreassen O.A., Dedeoglu A., Leavitt B., Hayden M., Borchelt D., Ross C.A., Ferrante R.J., Beal M.F. Effects of CAG repeat length, HTT protein length and protein context on cerebral metabolism measured using magnetic resonance spectroscopy in transgenic mouse models of Huntington’s disease. J. Neurochem. 2005;95:553–562. doi: 10.1111/j.1471-4159.2005.03411.x. [DOI] [PubMed] [Google Scholar]
- 17.Brouillet E., Jacquard C., Bizat N., Blum D. 3-nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington’s disease. J. Neurochem. 2005;95:1521–1540. doi: 10.1111/j.1471-4159.2005.03515.x. [DOI] [PubMed] [Google Scholar]
- 18.Túnez I., Drucker-Colín R., Jimena I., Medina F.J., Muñoz Mdel.C., Peña J., Montilla P. Transcranial magnetic stimulation attenuates cell loss and oxidative damage in the striatum induced in the 3-nitropropionic model of Huntington’s disease. J. Neurochem. 2006;97:619–630. doi: 10.1111/j.1471-4159.2006.03724.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 20.Chiurchiù V., Orlacchio A., Maccarrone M. Is modulation of oxidative stress an answer? The state of the art of redox therapeutic actions in neurodegenerative diseases. Oxid. Med. Cell. Longev. 2016;2016:7909380. doi: 10.1155/2016/7909380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milakovic T., Johnson G.V. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J. Biol. Chem. 2005;280:30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 22.Milakovic T., Quintanilla R.A., Johnson G.V. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J. Biol. Chem. 2006;281:34785–34795. doi: 10.1074/jbc.M603845200. [DOI] [PubMed] [Google Scholar]
- 23.Panov A.V., Gutekunst C.-A., Leavitt B.R., Hayden M.R., Burke J.R., Strittmatter W.J., Greenamyre J.T. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira J.M., Chen S., Almeida S., Riley R., Gonçalves J., Oliveira C.R., Hayden M.R., Nicholls D.G., Ellerby L.M., Rego A.C. Mitochondrial-dependent Ca2+ handling in Huntington’s disease striatal cells: effect of histone deacetylase inhibitors. J. Neurosci. 2006;26:11174–11186. doi: 10.1523/JNEUROSCI.3004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira J.M., Jekabsons M.B., Chen S., Lin A., Rego A.C., Gonçalves J., Ellerby L.M., Nicholls D.G. Mitochondrial dysfunction in Huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J. Neurochem. 2007;101:241–249. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls D.G. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–317. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Fonteriz R., Matesanz-Isabel J., Arias-Del-Val J., Alvarez-Illera P., Montero M., Alvarez J. Modulation of calcium entry by mitochondria. Adv. Exp. Med. Biol. 2016;898:405–421. doi: 10.1007/978-3-319-26974-0_17. [DOI] [PubMed] [Google Scholar]
- 28.Browne S.E., Beal M.F. Oxidative damage in Huntington’s disease pathogenesis. Antioxid. Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 29.Cha J.-H.J. Transcriptional dysregulation in Huntington’s disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 30.Zucker B., Luthi-Carter R., Kama J.A., Dunah A.W., Stern E.A., Fox J.H., Standaert D.G., Young A.B., Augood S.J. Transcriptional dysregulation in striatal projection- and interneurons in a mouse model of Huntington’s disease: neuronal selectivity and potential neuroprotective role of HAP1. Hum. Mol. Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]
- 31.Chaturvedi R.K., Calingasan N.Y., Yang L., Hennessey T., Johri A., Beal M.F. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington’s disease following chronic energy deprivation. Hum. Mol. Genet. 2010;19:3190–3205. doi: 10.1093/hmg/ddq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Taherzadeh-Fard E., Saft C., Akkad D.A., Wieczorek S., Haghikia A., Chan A., Epplen J.T., Arning L. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol. Neurodegener. 2011;6:32. doi: 10.1186/1750-1326-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhary R.K., Patel K.A., Patel M.K., Joshi R.H., Roy I. Inhibition of aggregation of mutant huntingtin by nucleic acid aptamers in vitro and in a yeast model of Huntington’s disease. Mol. Ther. 2015;23:1912–1926. doi: 10.1038/mt.2015.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 36.Kaur G., Roy I. Therapeutic applications of aptamers. Expert Opin. Investig. Drugs. 2008;17:43–60. doi: 10.1517/13543784.17.1.43. [DOI] [PubMed] [Google Scholar]
- 37.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel K.A., Sethi R., Dhara A.R., Roy I. Challenges with osmolytes as inhibitors of protein aggregation: Can nucleic acid aptamers provide an answer? Int. J. Biol. Macromol. 2017;100:75–88. doi: 10.1016/j.ijbiomac.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 40.Cooper J.K., Schilling G., Peters M.F., Herring W.J., Sharp A.H., Kaminsky Z., Masone J., Khan F.A., Delanoy M., Borchelt D.R. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 41.Krobitsch S., Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meriin A.B., Zhang X., Miliaras N.B., Kazantsev A., Chernoff Y.O., McCaffery J.M., Wendland B., Sherman M.Y. Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol. Cell. Biol. 2003;23:7554–7565. doi: 10.1128/MCB.23.21.7554-7565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss K.R., Kimura Y., Lee W.C., Littleton J.T. Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington’s disease model. Genetics. 2012;190:581–600. doi: 10.1534/genetics.111.133710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Outeiro T.F., Giorgini F. Yeast as a drug discovery platform in Huntington’s and Parkinson’s diseases. Biotechnol. J. 2006;1:258–269. doi: 10.1002/biot.200500043. [DOI] [PubMed] [Google Scholar]
- 45.Smith M.G., Snyder M. Yeast as a model for human disease. Curr. Protoc. Hum. Genet. 2006;Chapter 15 doi: 10.1002/0471142905.hg1506s48. Unit 15.6. [DOI] [PubMed] [Google Scholar]
- 46.Mason R.P., Giorgini F. Modeling Huntington disease in yeast: perspectives and future directions. Prion. 2011;5:269–276. doi: 10.4161/pri.5.4.18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grynkiewicz G., Poenie M., Tsien R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 48.Roe M.W., Lemasters J.J., Herman B. Assessment of Fura-2 for measurements of cytosolic free calcium. Cell Calcium. 1990;11:63–73. doi: 10.1016/0143-4160(90)90060-8. [DOI] [PubMed] [Google Scholar]
- 49.Chalmers S., McCarron J.G. The mitochondrial membrane potential and Ca2+ oscillations in smooth muscle. J. Cell Sci. 2008;121:75–85. doi: 10.1242/jcs.014522. [DOI] [PubMed] [Google Scholar]
- 50.Reers M., Smiley S.T., Mottola-Hartshorn C., Chen A., Lin M., Chen L.B. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol. 1995;260:406–417. doi: 10.1016/0076-6879(95)60154-6. [DOI] [PubMed] [Google Scholar]
- 51.Qian Y., Kachroo A.H., Yellman C.M., Marcotte E.M., Johnson K.A. Yeast cells expressing the human mitochondrial DNA polymerase reveal correlations between polymerase fidelity and human disease progression. J. Biol. Chem. 2014;289:5970–5985. doi: 10.1074/jbc.M113.526418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallet P.F., Maftah A., Petit J.M., Denis-Gay M., Julien R. Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur. J. Biochem. 1995;228:113–119. doi: 10.1111/j.1432-1033.1995.tb20238.x. [DOI] [PubMed] [Google Scholar]
- 53.Jacobson J., Duchen M.R., Heales S.J. Intracellular distribution of the fluorescent dye nonyl acridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass. J. Neurochem. 2002;82:224–233. doi: 10.1046/j.1471-4159.2002.00945.x. [DOI] [PubMed] [Google Scholar]
- 54.Goswami A.V., Samaddar M., Sinha D., Purushotham J., D’Silva P. Enhanced J-protein interaction and compromised protein stability of mtHsp70 variants lead to mitochondrial dysfunction in Parkinson’s disease. Hum. Mol. Genet. 2012;21:3317–3332. doi: 10.1093/hmg/dds162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desjardins P., Frost E., Morais R. Ethidium bromide-induced loss of mitochondrial DNA from primary chicken embryo fibroblasts. Mol. Cell. Biol. 1985;5:1163–1169. doi: 10.1128/mcb.5.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hands S., Sajjad M.U., Newton M.J., Wyttenbach A. In vitro and in vivo aggregation of a fragment of huntingtin protein directly causes free radical production. J. Biol. Chem. 2011;286:44512–44520. doi: 10.1074/jbc.M111.307587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorolla M.A., Reverter-Branchat G., Tamarit J., Ferrer I., Ros J., Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic. Biol. Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 58.Madian A.G., Regnier F.E. Proteomic identification of carbonylated proteins and their oxidation sites. J. Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh K., Saleh A.A., Bhadra A.K., Roy I. Hsp104 as a key modulator of prion-mediated oxidative stress in Saccharomyces cerevisiae. Biochem. J. 2013;454:217–225. doi: 10.1042/BJ20121806. [DOI] [PubMed] [Google Scholar]
- 60.Millard P.J., Roth B.L., Thi H.P., Yue S.T., Haugland R.P. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 1997;63:2897–2905. doi: 10.1128/aem.63.7.2897-2905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwolek-Mirek M., Zadrag-Tecza R. Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Res. 2014;14:1068–1079. doi: 10.1111/1567-1364.12202. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N.R., Doi H., Kurosawa M., Nekooki M., Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 63.Zala D., Hinckelmann M.-V., Saudou F. Huntingtin’s function in axonal transport is conserved in Drosophila melanogaster. PLoS ONE. 2013;8:e60162. doi: 10.1371/journal.pone.0060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pardo R., Molina-Calavita M., Poizat G., Keryer G., Humbert S., Saudou F. pARIS-htt: an optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol. Brain. 2010;3:17. doi: 10.1186/1756-6606-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papsdorf K., Kaiser C.J., Drazic A., Grötzinger S.W., Haeßner C., Eisenreich W., Richter K. Polyglutamine toxicity in yeast induces metabolic alterations and mitochondrial defects. BMC Genomics. 2015;16:662. doi: 10.1186/s12864-015-1831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orr A.L., Li S., Wang C.-E., Li H., Wang J., Rong J., Xu X., Mastroberardino P.G., Greenamyre J.T., Li X.-J. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johri A., Calingasan N.Y., Hennessey T.M., Sharma A., Yang L., Wille E., Chandra A., Beal M.F. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2012;21:1124–1137. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suess B., Hanson S., Berens C., Fink B., Schroeder R., Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular Cloning, a Laboratory Manual. [Google Scholar]
- 71.Blank H.M., Li C., Mueller J.E., Bogomolnaya L.M., Bryk M., Polymenis M. An increase in mitochondrial DNA promotes nuclear DNA replication in yeast. PLoS Genet. 2008;4:e1000047. doi: 10.1371/journal.pgen.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goldring E.S., Grossman L.I., Marmur J. Petite mutation in yeast. II. Isolation of mutants containing mitochondrial deoxyribonucleic acid of reduced size. J. Bacteriol. 1971;107:377–381. doi: 10.1128/jb.107.1.377-381.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Einhauer A., Schuster M., Wasserbauer E., Jungbauer A. Expression and purification of homogenous proteins in Saccharomyces cerevisiae based on ubiquitin-FLAG fusion. Protein Expr. Purif. 2002;24:497–504. doi: 10.1006/prep.2001.1595. [DOI] [PubMed] [Google Scholar]
- 74.Eggleston M.D., Marshall P.A. Saccharomyces cerevisiae samples stained with FUN-1 dye can be stored at -20 degrees C for later observation. J. Microsc. 2007;225:100–103. doi: 10.1111/j.1365-2818.2007.01720.x. [DOI] [PubMed] [Google Scholar]