Abstract
U12-dependent introns are found in small numbers in most eukaryotic genomes, but their scarcity makes accurate characterisation of their properties challenging. A computational search for U12-dependent introns was performed using the draft version of the human genome sequence. Human expressed sequences confirmed 404 U12-dependent introns within the human genome, a 6-fold increase over the total number of non-redundant U12-dependent introns previously identified in all genomes. Although most of these introns had AT-AC or GT-AG terminal dinucleotides, small numbers of introns with a surprising diversity of termini were found, suggesting that many of the non-canonical introns found in the human genome may be variants of U12-dependent introns and, thus, spliced by the minor spliceosome. Comparisons with U2-dependent introns revealed that the U12-dependent intron set lacks the ‘short intron’ peak characteristic of U2-dependent introns. Analysis of this U12-dependent intron set confirmed reports of a biased distribution of U12-dependent introns in the genome and allowed the identification of several alternative splicing events as well as a surprising number of apparent splicing errors. This new larger reference set of U12-dependent introns will serve as a resource for future studies of both the properties and evolution of the U12 spliceosome.
INTRODUCTION
Two distinct types of pre-mRNA introns, termed U2- and U12-dependent based on the spliceosome complexes that excise them during RNA processing, are found in most higher organisms (reviewed in 1). While the 99.9% of introns spliced by the major (U2-dependent) spliceosome have been extensively characterised (2,3), less is known regarding the remaining 0.1% of introns, which fall into the U12-dependent class. This minor class of introns was originally identified due to its unusual conserved donor and branch signals and highly atypical AT-AC terminal dinucleotides (4,5). More recently, analyses have found that AT-AC termini are not strictly required and identified many U12-dependent introns with GT-AG terminal dinucleotides as well as a few with other termini (6–8). Additionally, a small number of U2-dependent introns with U12-like AT-AC terminal dinucleotides have been identified, confirming that analysis of the entire splice site signal and not just the terminal dinucleotides is required for accurate classification (9).
Many genes contain both U2- and U12-dependent introns but little is known about how the two spliceosomes cooperate to identify and splice the correct introns in vivo. Distinct differences are observed, however, between the splice site signals associated with the two types of introns. U12-dependent introns exhibit strongly conserved and informative donor and branch signals, whereas U2-dependent introns exhibit only moderately informative signals at the donor and acceptor sites and a highly degenerate branch site signal. Additionally the polypyrimidine tract seen between the branch site and acceptor site of U2-dependent introns is lacking in U12-dependent introns (1).
The evolutionary history of these two classes of introns and their respective spliceosomes remains unclear. Burge et al. (7) have reported AT-AC U12-dependent introns converting to GT-AG U12-dependent introns and U12-dependent introns converting to U2-dependent introns and concluded that U12-dependent introns tend to convert to U2-dependent over evolutionary time. They also reported a biased distribution of U12-dependent introns within a variety of genomes, a result they found suggestive of a fission–fusion model of spliceosome evolution in which the U2 and U12 systems diverged in separate lineages and were later united through a merging of genetic material in a progenitor of higher eukaryotes (7).
Recent results have found a strikingly high degree of overlap between the proteins and non-coding RNAs involved in U2- and U12-dependent splicing. In addition to the U5 snRNA (10), all eight snRNP Sm proteins (11), the four proteins that constitute the splicing factor SF3b (11), and the splicing-associated protein Prp8 have been found in both the U2 and U12 spliceosomes (12). Recent evidence has indicated that splicing-associated SR proteins, long known to function in the major spliceosome, play functional roles in U12-dependent splicing as well (13). Extensive similarity in secondary structures and interactions between the set of non-coding RNAs U11, U12, U4atac and U6atac involved in the U12 spliceosome and the set U1, U2, U4 and U6 involved in the U2 spliceosome argue for homology of the two systems as well (7) as do recent results that have found the stem–loop structures of U6 and U6atac to be functionally analogous (14). Although the evolutionary implications of this high degree of overlap are not entirely clear, these findings may indicate that the U12 spliceosome evolved in the presence of the U2 spliceosome rather than in a different lineage as the fission–fusion model suggests (11).
U12-dependent introns have been identified previously through homology searches and by analysing annotated intron junctions (7). The analysis presented here represents the first large-scale search for U12-dependent introns in the recently completed human genome sequence. A greater than expected diversity in the terminal dinucleotides of U12-dependent introns was observed, giving further evidence to the idea that flexibility in these positions has played an important role in intron evolution. This analysis generated a new reference set of human U12-dependent introns 8-fold larger than the previously available set and allows a more extensive characterisation of these introns to be carried out.
MATERIALS AND METHODS
Human U12-dependent introns were identified using a two-step procedure
First, potential donor and branch site signals were identified based on statistical pattern recognition techniques. Low threshold values that detected almost all known sites while accepting a large number of false positives were used. From these signals, potential introns (donor/acceptor pairs) were generated and expressed sequence evidence was used to identify a subset of these potential introns as valid. All genomic scans used the 9 January 2001 assembly of the 7 October 2000 freeze of the human genome draft sequence (2; available from http://genome.cse.ucsc.edu/).
Candidate U12-dependent intron donor and branch sites were identified using a standard weight matrix approach (15). The weight matrix models were trained using a previously described non-redundant set of 48 U12-dependent introns from a variety of species (6). Simple pseudocounts based on genomic nucleotide frequencies (the null model) were added during the training process to avoid overfitting the model to the training set. Any sequences whose log-odds scores from the donor signal weight matrix exceeded an empirically derived bit threshold were considered potential U12-dependent intron donor sites. Potential U12-dependent acceptor signals were identified by considering all high-scoring branch signals (again using an empirically derived threshold) and including only those that had a putative acceptor site (an AC dinucleotide, for instance) within a certain distance range from the putative branch site. The traditional consensus branch site for U12-dependent introns is TTCCTTAA, although our search pattern extended slightly beyond this consensus and none of the bases were strictly required in our analysis. All potential donors and acceptors that met the above criteria and were within a certain distance of each other were considered to define a potential U12-dependent intron. For each of these cases, 64 bp of potential exon sequence, 32 bp from before and 32 bp from after the hypothetical intron were extracted and saved for later analysis.
The analysis described above involved five parameters: a donor site score threshold (nine bits), a branch site score threshold (six bits), both a minimum and a maximum branch site to acceptor site distance (8 bp, 21 bp) and a maximum intron size (20 kb). The first four of these were selected to be as inclusive as possible (based on the training data) while still minimising time required for computation, while the final parameter, maximum intron size, had to be limited to relatively small values to render the analysis computationally tractable. The analysis did, therefore, overlook some longer U12-dependent introns (see Discussion). After confirmation of introns, the distributions of donor scores, branch scores and the branch to acceptor distance were plotted and showed approximately normal distributions with the thresholds well separated from the peaks (see Fig. 2 and data not shown), suggesting that our empirical thresholds did not eliminate a large number of valid results. Parameter values for the GT-AG and AT-AC scans are provided above; parameter values for all scans are provided in the legend to Table 1.
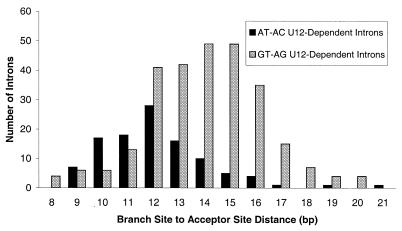
Figure 2.
Branch site to acceptor site distance for U12-dependent introns. The distance between the branch site and the acceptor site is plotted for 108 AT-AC U12-dependent introns (black bars) and 275 GT-AG U12-dependent introns (grey bars).
Table 1. Diverse terminal dinucleotides on U12-dependent introns in the human genome.
| Intron termini |
Reported in Burge et al. (7) |
Total found |
Putative splicing errors |
Total confirmed |
| GT-AG | 34 | 279 | 4 | 275 |
| AT-AC | 12 | 109 | 1 | 108 |
| AT-AG | 1 | 8 | 1 | 7 |
| GT-AT | 0 | 5 | 1 | 4 |
| AT-AT | 0 | 4 | 0 | 4 |
| GT-GG | 0 | 7 | 4 | 3 |
| AT-AA | 1 | 5 | 3 | 2 |
| GT-AA | 0 | 1 | 0 | 1 |
| GT-CA | 0 | 1 | 1 | 0 |
| GC-AG | 1 | 0 | 0 | 0 |
| Totals | 49 | 419 | 15 | 404 |
The total number of U12-dependent introns previously reported in Burge et al. (7), the original total found in this analysis, the total discarded as likely splicing errors and the final confirmed total are shown for a variety of possible intron termini. Genomic scans were also performed for introns with GT-AC, GC-AC, GG-AC, GG-AG, AT-GG and AT-CA termini. No U12-dependent introns were identified in these scans. The AT-AC and GT-AG scans considered target introns with donor site scores greater than nine bits, branch site scores greater than six bits and lengths of <20 kb. All other scans examined introns of up to 2 kb in length. The GT-AC and AT-AG scans used 11 and nine bits as donor and branch scores thresholds, the AT–AA scan used 10 and eight bits and all remaining scans used 10 and seven bits. All scans used a branch site to acceptor site range of 8–21 bp.
Expressed sequence data were used to confirm a small portion of the large set of potential U12-dependent introns as true introns. For this purpose a specialised human expressed sequence database was developed which contained 54 484 human RNA sequences from EMBL release 65 (16) and 3 268 161 human ESTs from dbEST downloaded from the NCBI on 28 February 2001 (17; available from ftp://ncbi.nlm.nih.gov/genbank/).
High-speed SSAHA similarity searches were performed looking for matches between each potential U12-dependent intron and a repeat-masked version of the database described above (18). Repeat masking was performed using DUST (R.L.Tatusov and D.J.Lipman, unpublished). SSAHA (version 1.1) was used with the following options: wordlength, 13; minprint, 39; maxstore, 50000; reportmode, replaceC. The results of this search were parsed to include only those expressed sequence matches that extended at least 15 bp on both sides of the hypothetical splice junction. Two potential introns were considered duplicate if they showed identical sequences along the full 64 bp of potential exon regions. Although such a situation could potentially result from gene duplications and represent a valid intron, redundancy in the draft sequence assembly presents an equally plausible explanation. Accordingly only one copy of each potentially duplicate intron was saved for further analysis. Introns supported by a variety of SSAHA matches extending from at least position 3 to position 61 were considered verified at this point. As SSAHA functions in a phased manner and does not necessarily report the full length of the sequence match, introns which showed support but did not meet this stringent SSAHA criterion were analysed using BLAST (19; version 2.0.6, installed locally). Introns supported by a perfect BLAST match over all 64 bp were considered as verified. The remaining set of candidate introns, which showed some support but met neither the SSAHA nor the BLAST criteria were examined and classified manually.
Scans were performed for the standard U12-dependent introns with AT-AC and GT-AG terminal dinucleotides as well as a variety of non-standard introns (Table 1). Non-standard donor signals were identified using modified training sets, which had, for instance, each GT dinucleotide at the donor position replaced with a GC dinucleotide. Non-standard acceptors were identified by using the original branch site training set but scanning the downstream region after high-scoring branch sites for the non-standard dinucleotide of interest.
Non-standard splice junctions were checked for possible ambiguities in the form of cases where a single expressed sequence could support a variety of splice junctions, as previously described (20). No such cases were found.
Distributions of U12-dependent introns in the genome were modelled using binomial distributions as previously described (7).
RESULTS
Characteristics of human U12-dependent introns
Scans of recent human genome draft sequence were performed to identify both typical AT-AC and GT-AG U12-dependent introns and atypical U12-dependent introns with a variety of other splice junctions (Table 1). The searches for AT-AC and GT-AG introns examined all candidate introns up to 20 kb in length while the other searches only examined potential introns of up to 2 kb in length. Accordingly atypical introns are likely to be somewhat under-represented in our results. Unlike the only previous large-scale U12-dependent intron search, these scans analysed unannotated genome sequence data and were neither biased nor aided by previous annotation (7).
The search for AT-AC and GT-AG introns examined ∼20 million candidate introns found by pairing high-scoring U12-dependent donor and branch site splice signals. Of these candidates, 388 were confirmed by expressed sequence data using the stringent criteria described above. Five out of these 388 were classified as likely splicing errors and removed from further analysis. The 383 AT-AC and GT-AG human U12-dependent introns reported here represent an increase of 337 (>8-fold) over the introns reported in the only similar study (7).
In total, scans for U12-dependent introns with 16 different combinations of terminal dinucleotides were performed (Table 1). A total of 419 introns, including the 388 AT-AC and GT-AG introns discussed above, met the confirmation criteria. Of the additional 31 introns, 10 were classified as likely splicing errors, leaving a total of 21 non-AT-AC or GT-AG human U12-dependent introns, distributed among six classes, including the previously documented AT-AG and AT-AA (7) as well as several previously undocumented classes. Examination of the donor and acceptor signals of the atypical U12-dependent introns reveals almost perfect conservation of both the donor and branch sites with the U12-dependent intron consensus sequences. Detailed information, including intron/exon junction sequences, for all 404 confirmed introns is available as Supplementary Material in the online version of the paper or from http://www.sanger.ac.uk/Users/rd/U12/.
Despite searches for introns starting with GC or GG, all confirmed introns showed standard AT or GT dinucleotides at the donor position, suggesting that these bases may be almost universally required for successful splicing. One GC-AG U12-dependent intron, which was missed during our analysis due to its atypical and low-scoring donor site, has been reported previously indicating that an AT or GT dinucleotide is not an absolute requirement (7). In contrast, a variety of terminal dinucleotides (including AG, AC, AT, AA and GG) were observed at the acceptor position. The diversity of terminal dinucleotides observed at the acceptor site of human U12-dependent introns confirmed recent experimental work, which indicated that a variety of dinucleotides can serve as functional U12-dependent acceptor sites in vitro (21). This flexibility fits well with the idea that the branch site serves as the primary recognition point for the 3′ end of U12-dependent introns and suggests that the mechanism of 3′ site identification may be only loosely constrained.
282 confirmed GT donor sites were also scored as U2-dependent donor sites, using a custom U2 splice predictor based on first order weight matrices (A.Levine, unpublished; 22). The vast majority of these sites scored poorly as U2 sites. Only seven out of 282 (2.5%) received a log-odds score greater than five bits and even these scores were generally well below the mean score (mean 8.66, SD 2.31) for a set of 3620 true sites scored with the U2 model.
Estimating the frequency of U12-dependent introns within the genome is a difficult problem and due to the lack of comparable data for U2-dependent introns our results do not lead to an easy solution. However, comparing the small sample of 11 U12-dependent introns we identified on chromosome 22 with the 3199 U2-dependent introns identified in a similar search for U2-dependent introns on chromosome 22 suggests that as many as 0.34% of human introns are spliced by the U12 spliceosome. This number is larger than earlier estimates that suggested roughly 0.15% of human introns were likely to be U12-dependent (7), but, due to the small sample size, must be taken as only a rough estimate.
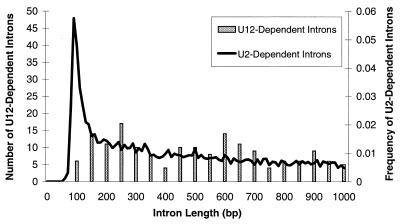
Access to this large set of confirmed U12-dependent introns allowed us to analyse several characteristics of this rare class of introns. Figure 1 compares the length distribution of the 168 confirmed U12-dependent introns with 11 402 U2-dependent introns from version 1.0 of Ensembl (2). U2-dependent introns have a two-component distribution, with a peak at ∼90 bp and an exponential-like component for longer lengths. U12-dependent introns seem to be lacking the short component of the U2-dependent intron length distribution. In contrast, U12-dependent introns show a gradual peak between 200 and 250 bp, then a slow decay. The distributions are similar for larger introns between 1 and 20 kb (U2: mean 4130 bp, SD 3720 bp; U12: mean 3600 bp, SD 3300 bp and data not shown), showing that the exponential components are similar.
Figure 1.
Length of U12- and U2-dependent introns. The length of 168 U12-dependent introns and 11 402 RefSeq-confirmed U2-dependent introns <1 kb in length are plotted. Grey bars represent the counts of U12-dependent introns grouped into 50-bp wide bins, while the black line represents the frequency of U2-dependent introns grouped into 10-bp wide bins.
The distribution of the distance between the branch site and the acceptor site for both AT-AC and GT-AG U12-dependent introns is illustrated in Figure 2. These results confirm earlier findings that this distance is much more sharply restricted in U12-dependent introns than it is for U2-dependent introns and verify suggestions (21) that AT-AC and GT-AG introns show different distributions for this distance (chi-squared test, P < 0.001). No functional relevance for this difference has been identified.
Table 2 compares the phase of 284 of the U12-dependent introns found in this study with 11 117 predominately U2-dependent introns previously analysed (23). The two distributions differ significantly (chi-squared test: P < 0.001) with the most striking difference being the bias against phase 0 introns in the U12-dependent intron data, compared with the bias toward phase 0 introns in the U2-dependent intron data. These results generally agree with previously analysed intron phase data from a smaller dataset (7).
Table 2. Phase of U2- and U12-dependent introns.
| U2-dependent introns | U12-dependent introns | |||
| |
Number |
Percentage |
Number |
Percentage |
| Phase 0 | 5263 | 47.4 | 59 | 20.8 |
| Phase 1 | 3372 | 30.3 | 118 | 41.5 |
| Phase 2 | 2482 | 22.3 | 107 | 37.7 |
| Total | 11 117 | 100 | 284 | 100 |
The number and percentage of U2- and U12-dependent introns that fall into the three possible phase classes are shown. Introns are phase 0 when the intron occurs between codons, phase 1 when the intron is found 1 base into the codon and phase 2 when the intron is found 2 bases into the codon. U2-dependent intron data is from Long et al (23).
We built frequency tables for the donor, branch and acceptor site of both AT-AC and GT-AG U12-dependent introns from our large set of these introns and provide these as Supplementary Material. Although some slight differences are evident, the consensus sequences for these sites largely agree with those derived previously (1).
Analysis of the region between the branch site and the acceptor site reveals a slight pyrimidine bias in this region. Sixty-six percent of a sample of 2191 nt from between the branch and acceptor consensus sequences were pyrimidines, while only 54% of a control set of 3060 nt from upstream of the branch site consensus sequence were pyrimidines. Although extracting a comparable set of data for U2-dependent introns is difficult, pyrimidines make up nearly 80% of the nucleotides in the 9 bp upstream of the acceptor site consensus (CAG), suggesting that the pyrimidine bias at U12-dependent introns is not as strong as it is at U2-dependent introns.
High error rates at the acceptor site in U12-dependent splicing
A surprisingly high number of introns were identified which met all confirmation criteria, yet seemed unlikely to represent real introns. In general, these introns shared donor sites with other confirmed introns yet differed slightly (1–6 bp) in acceptor site positions. In most cases one member of these pairs of introns had typical terminal dinucleotides and was strongly supported by a large number of expressed sequences while the second exhibited atypical dinucleotides and was weakly supported. In many cases the second intron led to the subsequent exon being out of frame and thus is unlikely to represent a true alternatively spliced variant of the gene. Fifteen introns exhibited these criteria and were classified as likely splicing errors (Table 3). Although a few of these so-called splicing errors may represent errors in EST sequencing, most seem likely to represent mistakes made by the U12-spliceosome.
Table 3. Putative 3′ splicing errors in U12-dependent splicing.
| Confirmed intron | Putative splicing error | 3′ Difference | |||
| ID |
Termini |
Evidence |
Termini |
Evidence |
|
| 2 | GT-AG | 8 | GT–GG | 1 | –4 |
| 14 | GT-AG | 94 | GT–CA | 1 | –1 |
| 45 | GT-AG | 7 | GT–AG | 1 | –3 |
| 92 | GT-AT | 2 | GT–GG | 1 | +3 |
| 97 | AT-AC | 7 | AT–AC | 1 | +4 |
| 122 | GT-AG | 60 | GT–AG | 1 | +2 |
| 124 | GT-AG | 266 | GT–GG | 7 | +1 |
| 127 | GT-AG | 12 | GT–AT | 1 | –2 |
| 145 | AT-AC | 12 | AT–AA | 1 | +5 |
| 216 | AT-AC | 16 | AT–AA | 1 | –3 |
| 226 | AT-AC | 3 | AT–AA | 1 | +6 |
| 236 | GT-AG | 15 | GT–GG | 1 | –3 |
| 251 | GT-AG | 7 | GT–AG | 1 | –4 |
| 290 | AT-AC | 13 | AT–AG | 1 | +2 |
| 393 | GT-AG | 24 | GT–AG | 1 | +2 |
The total evidence (number of expressed sequences) supporting a confirmed intron and an associated putative splicing error are shown. Each confirmed intron and associated splicing error share the same donor position but differ by a small number of bases (3′ difference) in the acceptor position. The ID column corresponds to the ID field on the complete intron list provided as Supplementary Material.
In total, 21 ESTs were observed confirming likely splicing errors and 5864 ESTs were observed confirming accepted introns. These numbers suggest that splicing mistakes at the 3′ end of U12-dependent introns occur at a rate of approximately one error in every 280 splices. This value likely underestimates the true error rate in U12 acceptor site selection as our analysis considered only a small subset of possible terminal dinucleotides. Similar genomic scans with other pairs of bases at the acceptor position could potentially uncover even more evidence of errors during U12-dependent splicing.
Alternative splicing of U12 introns
The approach to intron identification used for these analyses allowed us to identify alternative splicing situations in which one splice site was used in two or more confirmed introns. Among the 404 U12-dependent introns, 13 cases of such alternative splicing events were observed (Table 4). Eleven cases were identified where the same donor signal was used with a different acceptor signal and two cases were found in which different donor signals were paired with the same acceptor site. Interestingly, three of these alternative splicing events involved introns with different pairs of terminal dinucleotides. For instance 14 expressed sequences supported an AT-AT intron of length 620 bp in a hypothetical human protein (DDBJ/EMBL/GenBank accession no. NM_024549) while two expressed sequences support an AT-AC intron with the same donor site but a different acceptor site 3344 bp downstream of the donor site.
Table 4. Alternatively spliced U12-dependent introns.
| Gene |
ID |
Termini |
Length |
Evidence |
Accession no. |
| Porphobilinogen deaminase (PBG-D) | 3 | GT-AG | 1145 | 26 | X04217 |
| 4 | GT-AG | 1593 | 2 | R06263 | |
| Quinone oxidoreductase homolog-1 | 343 | AT-AC | 4501 | 3 | AA370151 |
| 342 | AT-AC | 4522 | 11 | AF029689 | |
| Von Hippel–Lindau binding protein (VBP-1) | 132 | GT-AG | 2403 | 35 | U96759 |
| 133 | GT-AG | 3187 | 1 | BF667071 | |
| Calcium channel, alpha 2/delta subunit 2 (CACNA2D2) | 247 | GT-AG | 103 | 2 | AJ251367 |
| 248 | GT-AT | 97 | 4 | AF042972 | |
| Unknown | 105 | GT-AG | 2951 | 2 | AV725561 |
| 106 | GT-AG | 5038 | 1 | AI917412 | |
| Unknown | 304 | GT-AG | 13385 | 1 | BF373273 |
| 303 | GT-AG | 13423 | 2 | BE887649 | |
| Unknown | 158 | AT-AC | 3344 | 2 | AK024780 |
| 157 | AT-AT | 620 | 14 | BE275895 | |
| Unknown | 287 | GT-AG | 1471 | 39 | AK001916 |
| 288 | GT-AG | 2747 | 1 | BE263460 | |
| Unknown | 67 | GT-AG | 605 | 17 | T50022 |
| 68 | GT-AG | 2677 | 1 | AL523899 | |
| Cullin 4a (CUL4A) | 257 | AT-AC | 8926 | 21 | AF077188 |
| 258 | AT-AC | 277 | 1 | AL560997 | |
| Unknown | 386 | GT-AG | 12503 | 2 | AK000443 |
| 387 | GT-AG | 14540 | 4 | AK022732 | |
| JNK1 protein kinase | 367 | GT-AG | 1727 | 2 | L26318 |
| 368 | GT-AG | 1301 | 3 | L35004 | |
| Unknown | 106 | GT-AG | 5038 | 1 | AI917412 |
| 107 | AT-AG | 1984 | 5 | AI023856 |
Thirteen examples of alternatively spliced U12-dependent introns are shown. For each splicing variant, the ID matching the Supplementary intron table, the intron terminal dinucleotides, the intron length, the total evidence supporting the intron and an accession number of a confirming expressed sequence are presented.
These results suggest that, at a minimum, 13 out of 404 or ~3.2% of human U12-dependent introns show truncation/extension type alternative splicing events. A bias (11 out of 13) towards alterations at the acceptor site was also observed, although the numbers are too small to draw any strong conclusions in this regard. A similar analysis of ∼3200 expressed-sequence-confirmed U2-dependent introns (of length <20 kb) on human chromosome 22 found truncation/extension alternative splicing events to occur at ~14% of introns and only negligible differences between the frequency of events involving donor and acceptor sites (data not shown).
Non-random distribution of U12-dependent introns in the genome
The distribution of U12-dependent introns within the human genome has important implications for understanding the evolutionary history of the major and minor spliceosomes. Among the 404 U12-dependent introns identified in this analysis, there were 16 cases where two or more U12-dependent introns were confirmed by the same expressed sequence, indicating that the two introns occurred within a single gene (Table 5). One of these cases (Homo sapiens NHE-6, DDBJ/EMBL/GenBank accession no. AF030409) had three U12-dependent introns (one AT-AC, two GT-AG) supported by a single expressed sequence.
Table 5. Genes with multiple U12-dependent introns.
| Gene |
U12-dependent introns |
Accession no. |
| Smg GDS-associated protein (SMAP) | GT-AG (84) | U59919 |
| AT-AC (85) | ||
| Transcription elongation factor TFIIS.h | AT-AC (239) | AJ223473 |
| GT-AG (240) | ||
| Inositol polyphosphate 5-phosphatase (5ptase) | GT-AG (321) | M74161 |
| AT-AG (322) | ||
| WDR10p-L (WDR10) | GT-AG (235) | AF244931 |
| GT-AG (236) | ||
| Diaphanous 1 (HDIA1) | AT-AC (81) | AF051782 |
| AT-AC (82) | ||
| Erythroid K:Cl cotransporter (KCC1) | GT-AG (243) | AF047338 |
| GT-AG (244) | ||
| Hypothetical transmembrane protein SBBI53 | GT-AG (381) | AF242523 |
| |
GT-AG (382) |
|
| Spermidine aminopropyltransferase |
AT-AC (312) |
AD001528 |
| |
GT-AG (313) |
|
| Dihydropyridine-sensitive L-type calcium channel alpha-1 subunit CACNL1A3 (CACNA1S) |
GT-AG (9) |
L33798 |
| |
GT-AG (10) |
|
| Hypothetical protein FLJ22028 |
GT-AG (105) |
AV725561 |
| |
AT-AG (107) |
|
| Autoantigen |
GT-AG (245) |
L26339 |
| |
GT-AG (246) |
|
| KIAA0136 gene |
AT-AC (344) |
D50926 |
| |
GT-AG (345) |
|
| Histidase |
GT-AG (98) |
D16626 |
| |
GT-AG (99) |
|
| ERCC5 excision repair protein (XPG) |
GT-AG (212) |
L20046 |
| |
AT-AT (213) |
|
| KIAA1176 protein |
GT-AG (188) |
AB033002 |
| |
GT-AG (189) |
|
| Sodium-hydrogen exchanger 6 (NHE-6) |
AT-AC (401) |
AF030409 |
| |
GT-AG (302) |
|
| GT-AG (303) |
Sixteen genes with at least two U12-dependent introns are shown. For each U12-dependent intron in the specified gene, the terminal dinucleotides and ID (matching the complete intron list provided as Supplementary Material) are provided. The accession number of a confirming expressed sequence is provided for each gene.
If we assume that U12-dependent introns are randomly distributed throughout the genome, the probability of identifying 16 or more genes with multiple U12-dependent introns among 388 genes with at least one U12-dependent intron is P < 0.009. This strongly confirms earlier reports that suggested U12-dependent introns were distributed non-randomly within genomes (7). It is worth noting that the strict requirement for multiple introns to be supported by a single expressed sequence almost certainly leads to an underestimate of the true number of genes with multiple U12-dependent introns and, thus, an overestimate of the likelihood of this distribution occurring by chance. This underestimation occurs due to the short length of most ESTs and the correspondingly small chance that a single EST would support multiple introns. Furthermore, in this analysis duplicate U12-dependent introns, which arose from gene duplications during evolution, are counted as distinct introns. If each group of duplicate introns was counted as a single intron, the likelihood of seeing this distribution arising randomly would be reduced.
DISCUSSION
The analysis presented here greatly increases both the number of U12-dependent introns identified and the diversity of these introns. The observation that a significant number of U12-dependent introns exhibit atypical terminal dinucleotides suggests that a good number of the so-called non-canonical introns identified in a variety of genomes (24) may represent variants of U12-dependent introns. Furthermore, due to the different parameters used in the searches for typical and atypical U12-dependent introns, the results presented here most likely reflect an under-representation of atypical U12-dependent introns. For instance, only 76% of AT-AC and GT-AG U12-dependent introns have lengths <2 kb. If this ratio holds for atypical U12-dependent introns as well, the 21 examples reported here should increase to roughly 27. Furthermore, scans for introns with pairs of terminal dinucleotides not considered in this study may identify additional atypical U12-dependent introns as well.
The 404 U12-dependent introns identified here represent a lower bound on the genome’s full complement of these introns for a variety of reasons. First, as noted previously, the arbitrary limit of 20 kb as the maximum intron length for AT-AC and GT-AG U12-dependent introns almost certainly excluded a significant number of true introns from our analysis. For comparison, ~5% of Ensembl U2-dependent introns confirmed by RefSeq entries are >20 kb in length (2). In addition, the threshold values used for donor and branch site scores, while chosen to be inclusive, likely excluded a small number of valid introns from the analysis.
Furthermore, the incomplete nature of the EST and mRNA sets used to confirm introns means that some number of true introns, which were identified as potential introns in the first stage of this analysis, failed to meet the confirmation criteria and were not included in the final counts. EST datasets in particular are biased towards the 5′ and 3′ ends of genes and are less likely to provide evidence for introns near the middle of larger genes.
A large majority of the human U12-dependent introns reported previously were identified in our large-scale genomic analysis. However, a few appear to have been missed. In addition to the GC-AG U12-dependent intron discussed above, intron 5 of FHIT (human fragile histidine triad gene) and intron 16 of HPS (human Hermansky–Pudlak syndrome gene), previously noted to be U12-dependent introns (7), were both missed by our analysis. Careful examination of these particular introns reveals that the FHIT intron was missed due to its exceptionally long length while the HPS intron was missed to due to its atypical and low-scoring donor and branch sites.
The large set of U12-dependent introns presented here should prove helpful for future studies regarding the evolution of the two-spliceosome system. In particular comparisons with the nearly complete mouse genome should prove useful in analysing the frequency of subtype switching between AT-AC and GT-AG U12-dependent introns, as well as intron conversion and loss.
The differences observed between the length distribution of U12- and U2-dependent introns raise interesting questions about the two splicing mechanisms. In particular the accurate pairing of donor and acceptor sites is thought to occur by two different models in higher eukaryotes, an intron definition model, which functions in the excision of small introns (25), and an exon definition model, which functions in the excision of larger introns (26). U12-dependent introns have been shown to participate to some degree in exon definition interactions (27) and one possible explanation for the relative dearth of short U12-dependent introns may be that they are recognised exclusively in an exon-dependent fashion, eliminating any selective benefit potentially associated with the short length of many U2-dependent introns.
A number of the U12-dependent introns found in the human genome occur within larger gene families, suggesting that the intron arose originally in a single ancestral gene and was duplicated along with the rest of the gene as the families grew. The presence of U12-dependent introns in some gene families, including the calcium and sodium voltage-gated cation channels (8), the matrilin family (28), the protein kinase superfamily (7) and the E2F transcription factor family, has been well studied. Our results found conservation of U12-dependent introns in the phospholipase C family, the transportin family, the diaphanous family and the cAMP-binding guanine nucleotide exchange factor family (see Supplementary Material) in addition to these previously identified gene families. Additionally, U12-dependent intron containing genes seem to be over-represented in the ras-raf signal transduction pathway, although further work is required to determine the significance of this observation.
The observation of alternative splicing of U12-dependent introns poses interesting evolutionary questions as well. If U12-dependent introns convert to U2-dependent over evolutionary time by accumulation of mutations at the splicing junctions as previously postulated (7,9), how would this work for alternatively spliced introns? In the case of an intron truncation event where two different acceptors could pair with a single donor, the intron conversion process might necessitate either the seemingly unlikely simultaneous conversion of multiple intron junctions or the loss of one of the splicing alternatives. This scenario suggests that alternatively spliced U12-dependent introns would be preferentially preserved, but is in conflict with the observation that alternative splicing is rarer at U12-dependent introns. A possible explanation may be that the U12 spliceosome is less amenable to the complex regulation patterns that alternative splicing requires and that alternative splicing, therefore, arises less frequently at U12-dependent introns. Identification of additional examples of U12-dependent alternative splicing and comparative analysis of gene structures may present the most direct way towards an understanding of these phenomena.
Although little is known about error rates of U2-dependent intron splicing, the calculation of a preliminary error rate for U12-dependent splicing presents some interesting possibilities. In particular, if errors occur with a significantly higher frequency at U12-dependent introns than at U2-dependent introns, this may point to a reason that U12-dependent introns seem to be selected against during evolution and even are found to be lacking entirely from some eukaryotes, such as Caenorhabditis elegans.
In addition to the observations made here, we hope the set of U12-dependent introns generated by this analysis will provide a useful resource for future examinations of the minor spliceosome and its evolution.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
ACKNOWLEGDEMENTS
We thank M. Clamp for providing the Ensembl intron length data and the two anonymous reviewers for many helpful suggestions. A.L. is a Winston Churchill Scholar. R.D. is supported by the Wellcome Trust.
References
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T. and Atkins,J.F. (eds), The RNA World II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 2.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M.Q. (1998) Statistical features of human exons and their flanking regions. Hum. Mol. Genet., 7, 919–932. [DOI] [PubMed] [Google Scholar]
- 4.Jackson I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall S.L. and Padgett,R.A. (1994) Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol., 239, 357–365. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P.A. and Burge,C.B. (1997) Classification of introns: U2-type or U12-type. Cell, 91, 875–879. [DOI] [PubMed] [Google Scholar]
- 7.Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]
- 8.Wu Q. and Krainer,A.R. (1999) AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol., 19, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich R.C., Incorvaia,R. and Padgett,R.A. (1997) Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell, 1, 151–160. [DOI] [PubMed] [Google Scholar]
- 10.Tarn W.Y. and Steitz,J.A. (1996) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science, 27, 1824–1832. [DOI] [PubMed] [Google Scholar]
- 11.Will C.L., Schneider,C., Reed,R. and Lührmann,R. (1999) Identification of both shared and distinct proteins in the major and minor spliceosomes. Science, 284, 2003–2005. [DOI] [PubMed] [Google Scholar]
- 12.Luo H.R., Moreau,G.A, Levin,N. and Moore,M.J. (1999) The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA, 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastings M.L. and Krainer,A.R. (2001) Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA, 7, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla G.C. and Padgett,R.A. (2001) The intramolecular stem–loop structure of U6 snRNA can functionally replace the U6atac snRNA stem–loop. RNA, 7, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staden R. (1984) Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res., 12, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker W., van den Broek,A., Camon,E., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL nucleotide sequence database. Nucleic Acids Res., 28, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST-database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 18.Ning Z., Cox,A.J. and Mullikin,J.C. (2001) SSAHA: a fast search method for large DNA databases. Genome Res., in press [DOI] [PMC free article] [PubMed]
- 19.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietrich R.C., Peris,M.J., Seyboldt,A.S. and Padgett,R.A. (2001) Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol., 21, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M.Q. and Marr,T.G. (1993) A weight array method for splicing signal analysis. Comput. Appl. Biosci., 9, 499–509. [DOI] [PubMed] [Google Scholar]
- 23.Long M., Rosenberg,C. and Gilbert,W. (1995) Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl Acad. Sci. USA, 92, 12495–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res., 29, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talerico M. and Berget,S.M. (1994) Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol., 14, 3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q. and Krainer,A.R. (1996) U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science, 274, 1005–1008. [DOI] [PubMed] [Google Scholar]
- 28.Muratoglu S., Krysan,K., Balázs,M., Sheng,H., Zákány,R., Módis,L., Kiss,I. and Deák,F. (2000) Primary structure of human matrilin-2, chromosome location of the MATN2 gene and conservation of an AT-AC intron in matrilin genes. Cytogenet. Cell Genet., 90, 323–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.