Abstract
Mycobacteria show peculiar aggregated outgrowth like biofilm on the surface of solid or liquid media. Biofilms harbor antibiotic resistant bacteria in a self-produced extracellular matrix that signifies the bacterial fate to sedentary existence. Despite years of research, very little is known about the mechanisms that contribute to biofilm formation. LuxS has been previously known to play a role in biofilm formation in Autoinducer-2 dependent manner. We here show the effect of LuxS product-homocysteine, on the biofilm forming ability of non-tuberculous mycobacteria, Mycobacterium smegmatis and Mycobacterium bovis BCG showing AI-2 independent phenotypic effect of LuxS. Exogenous supplementation of homocysteine in the culture media leads to aberrant cording, pellicle outgrowth, and biofilm formation. Thus, our study contributes to the better understanding of the mechanism of mycobacterial biofilm formation and sheds light on the role of LuxS product homocysteine. In addition, we highlight the contribution of activated methyl cycle in bacterial quorum sensing.
Keywords: Homocysteine, S-adenosyl methionine, Biofilm, Mycobacteria, Methylation
Introduction
Biofilms are congregation of heterogeneous population of microorganisms that are surface associated and encapsulated in a self-produced matrix [1]. This architecturally complex structure helps the bacteria to evolve, disseminate and acquire extraordinary level of resistance to most of the therapeutic agents by providing a stable permeability barrier [2, 3]. Biofilm related infections are emerging due to low susceptibility of microbial consortia to antimicrobial drugs and host immune responses [4, 5]. Microorganisms like Candida albicans, Pseudomonas aeruginosa, Staphylococcus epidermidis and Mycobacterium tuberculosis pose serious health concerns due to biofilm mediated resistance to environmental challenges [6–9]. Mycobacterium genera includes many environmental organisms which are non-pathogenic but can cause human infections in special circumstances [10]. These non-tuberculous mycobacteria (NTM) are opportunistic bacteria that can cause chronic infections or outbreaks associated with environmental sources [11]. Biofilms formed by NTMs are tolerant to anti-bacterial agents, thus posing a serious health challenge to control nosocomial infections. Therefore, a proper understanding of the molecular events involved in this transition of planktonic cells to pellicle growth can provide useful insights into drug resistance mechanism [12].
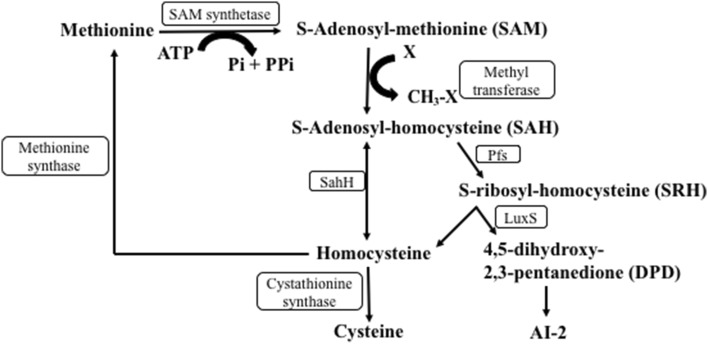
S-adenosyl methionine (SAM) is a major methyl group donor in all the methylation reactions (Fig. 1) [13]. Previous reports have identified Autoinducer-2 (AI-2), a product of LuxS, as an important molecule for interspecies communication in bacteria [14, 15]. However, this interpretation is complicated by dual role of LuxS in signaling and activated methyl cycle (AMC), a crucial metabolic pathway. AI-2 is derived from SAM through a series of enzymatic reactions [16, 17]. SAM forms the toxic intermediate S-adenosyl homocysteine (SAH) on donating methyl group. This S-adenosyl homocysteine (SAH) is further hydrolyzed to S-ribosyl homocysteine (SRH) [18]. Where LuxS cleaves SRH to form homocysteine and AI-2 precursor, S-adenosyl homocysteine hydrolase (SahH) synthesizes homocysteine directly from SAH [19]. LuxS mutant in Streptococcus sanguinis and Escherichia coli has been shown to be defective in biofilm formation. Complementation with SahH and not AI-2 restores this biofilm defect [20], indicating an unexplored role of AMC in bacterial quorum sensing.
Fig. 1.
Activated methyl cycle with homocysteine as a key intermediate. Schematic representation of activated methyl cycle. Homocysteine is synthesized either by a two-step mechanism involving Pfs and LuxS enzyme to produce AI-2 precursor and homocysteine or directly from S-adenosylhomocysteine in a one-step reaction using SahH enzyme
Homocysteine is a key intermediate for de novo synthesis of methionine in the cell which is further utilized for the production of SAM [19]. Role of SahH in maintaining the metabolic flux of SAM and SAH and its regulation through phosphorylation has been previously documented [19]. SahH mediated biofilm restoration of LuxS deficient cells prompted us to understand this concept in detail. Since SahH and LuxS both form homocysteine, we study the effect of exogenous homocysteine supplementation on the biofilm formation of M. smegmatis and M. bovis BCG. Our study explored the involvement of AMC in cording, pellicle formation and quorum sensing. Such phenotypic analysis revealed a novel link between biofilm formation and AMC.
Materials and Methods
Bacterial Growth Conditions
The mycobacterial cultures were grown as described earlier. M. smegmatis mc2 155 and M. bovis BCG cells were cultured and maintained as described before [21–23]. The cells were grown in Middlebrook 7H9 (Difco) broth supplemented with 0.5% glycerol, 0.5% Tween-80 (Sigma) and 1% ADC (bovine serum albumin [fraction V], dextrose, and catalase; Difco) for M. smegmatis and 5% for M. bovis BCG at 37 °C with shaking at 220 rpm for 2–4 days. Middlebrook 7H11 (Difco) was used as solid media containing 0.5% glycerol and 10% OADC (oleic acid, bovine serum albumin [fraction V], dextrose, and catalase) supplement. Effect of homocysteine was studied in Sauton’s minimal media (0.5 g KH2PO4, 0.5 g MgSO4∙7H2O, 2 g citric acid, 0.05 g ferric ammonium citrate, 60 mL glycerol, 4 g asparagine, 0.1 mL 1% ZnSO4 and make it to 1 L with distilled water. The pH was adjusted to 7.4 using 1 M NaOH and the media was sterilized before using).
Cording Formation in M. smegmatis
Cording formation was assessed for Mycobacterial cultures as previously described method [24]. M. smegmatis cells were grown till exponential phase in 7H9 media with 0.5% Tween-80. 5 μL of this starter culture was dropped onto a solid agar medium (Difco) with or without homocysteine (Sigma). The plate was then incubated at 37 °C for 5 days. The colonies thus appeared was imaged for the cording phenotype.
Pellicle Formation
The M. smegmatis cells were grown till exponential phase in 7H9 media supplemented with 0.5% Tween-80 and the secondary cultures were inoculated in 5 mL Middlebrook 7H9 media with no Tween at an OD600 = 0.01 as described. The cultures were kept at 37 °C for 48 h without shaking for pellicle formation as described [25].
Biofilm Formation in M. smegmatis and M. bovis
The M. smegmatis and M. bovis cells were grown until log phase in Sauton’s minimal media with 0.5% Tween-80 and secondary cultures were inoculated in Sauton’s minimal media in the absence of Tween-80 to an OD = 0.01 and aliquoted in a 96-well plate in triplicate. Homocysteine was added at different concentrations (0.1, 0.2, 0.3, 0.4 and 0.5 mM) for M. smegmatis and (0.1, 0.5 and 0.8 mM) for M. bovis. The plates were sealed and incubated at 37 °C for 5 days without shaking and biofilms were observed in control wells. Images were captured after biofilm development was seen in the control wells as described previously [26].
Results
Homocysteine Affects Cording of Mycobacteria
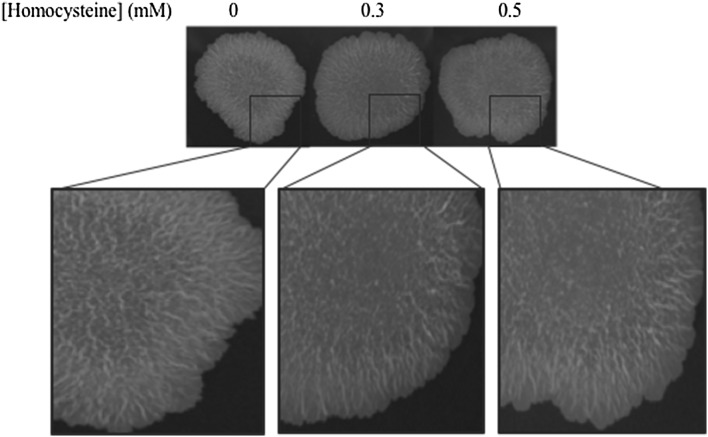
Macrophages form extracellular traps in response to extracellular aggregates of mycobacteria [27]. Such aggregates are tight bundles or serpentine cords formed by the bacteria. Previous studies reflect the involvement of gene methylation in cellular aggregation. We here study this mycobacterial surface property in the presence of homocysteine. M. smegmatis cells grown on a solid 7H11 agar with varying concentrations of homocysteine (0, 0.3 and 0.5) for 5 days showed distinct colony morphologies (Fig. 2). The bacterial growth showed cords in the center of the colony. The cord formation is altered in the center with increasing concentration of homocysteine. Thus, exogenous homocysteine is seen to modulate the molecular factors which are involved in cord formation, giving rise to colonies with altered cording phenotype.
Fig. 2.
Physiological effects of homocysteine. M. smegmatis mc2 155 cells were grown on solid agar medium containing 0.3 and 0.5 mM homocysteine. The images show modified cording phenotype at the centre of the colonies in the presence of homocysteine
Homocysteine Inhibits Biofilm Forming Ability of M. smegmatis
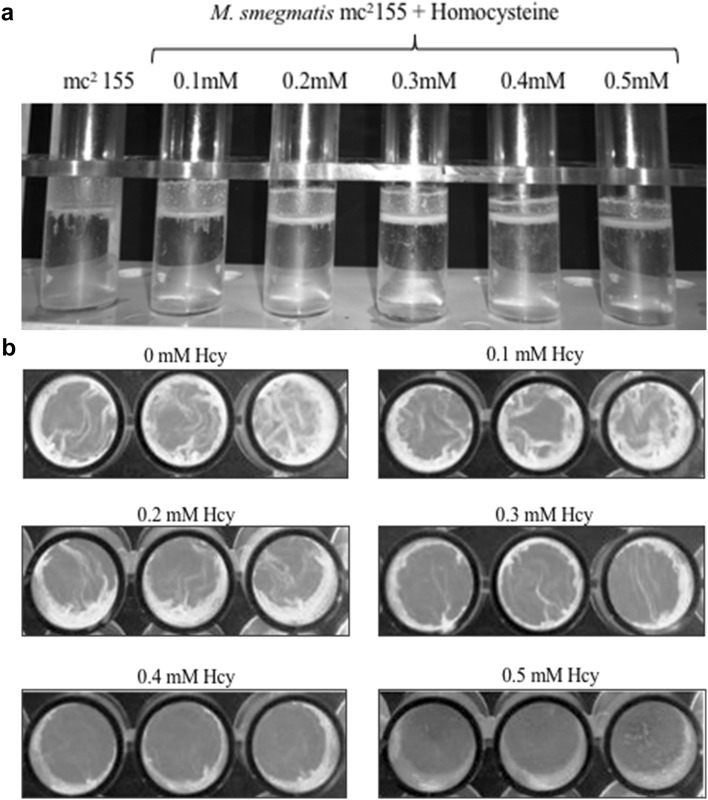
The effect of SahH complementation on regaining the biofilm forming ability of S. sanguins [28] led us to explore the role of its product-homocysteine in forming biofilm. The lipid rich mycobacterial cell wall forms a surface pellicle in liquid media when grown in the absence of detergent. M. smegmatis mc2 155 cells were grown in a static condition with increasing concentrations of homocysteine (0–0.5 mM) in the absence of detergent to analyze the pellicle formation at air–liquid interface (Fig. 3a). Significant pellicle formation occurs at the surface of broth without homocysteine. With increase in homocysteine concentration, cells become defective in pellicle formation with maximum defect observed at 0.5 mM homocysteine.
Fig. 3.
Effect of homocysteine on M. smegmatis pellicle and biofilm formation. a M. smegmatis cells were grown in liquid media with increasing homocysteine concentration in the absence of Tween-80 at static condition. The images showed decreased pellicle formation on addition of 0.4 mM homocysteine. b M. smegmatis cells were grown in minimal media under non-shaking conditions and in the absence of Tween-80 to develop biofilm. The images show decrease in biofilm as concentration of homocysteine (Hcy) is increased. Biofilm formation was completely absent after 0.4 mM homocysteine
Pellicles have been recognized as biofilm at air–liquid interface. Since, homocysteine addition exhibits aberrant pellicle phenotype, we investigated the consequences of elevated homocysteine on biofilm formation. M. smegmatis cells were grown with increasing concentrations of homocysteine (0, 0.1, 0.2, 0.3, 0.4 and 0.5 mM) in static conditions. M. smegmatis cells in a detergent-free media form biofilm at air–liquid interface. Cells grown in absence of homocysteine showed biofilm formation after 5 days (Fig. 3b). In contrast, with subsequent increase in homocysteine concentrations, bacteria were unable to form biofilm. At 0.4 mM homocysteine, biofilm forming ability of the cells was completely lost. Thus, addition of homocysteine to M. smegmatis cells significantly affect its capacity to form cords and biofilms.
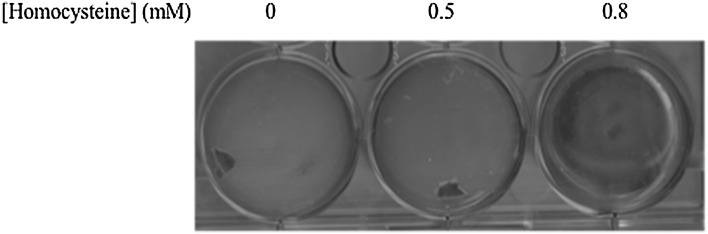
Homocysteine Affects Biofilm Formation in M. bovis BCG
M. bovis BCG is an attenuated form of M. bovis, a member of M. tuberculosis complex and is known to cause bovine tuberculosis [29]. Previous reports suggested M. bovis BCG biofilm formation and its role in disease pathogenesis [30]. Our study show the metabolite mediated abrogation of biofilm formation in M. bovis BCG. The results corroborate with our hypothesis as M. bovis BCG showed similar phenotype as that of M. smegmatis. We used a higher concentration of homocysteine (0.8 mM) with this strain. As we increased the concentration of homocysteine, the cells kept on losing the biofilm forming ability with complete loss at 0.8 mM homocysteine (Fig. 4). Thus, our results show that elevated homocysteine concentrations completely abrogated the biofilm forming tendency of mycobacteria, thus affecting its pathogenesis.
Fig. 4.
Homocysteine affects M. bovis BCG biofilm formation. M. bovis BCG was grown in the same condition as that of M. smegmatis cells. Loss in biofilm formation was observed at 0.8 mM homocysteine
Discussion
The air–liquid interface is a favorable niche for mycobacteria due to increased accessibility to oxygen [31]. Hypoxia induced surface drifting of bacteria forms a biofilm like structure known as pellicle [32]. The molecular basis of this hypha like growth remains ill defined, however, such pellicles provide a drug tolerant phenotype to the bacteria. Mycobacteria forms multicellular community by forming cord like structures [27]. We tested these surface characteristics of mycobacteria through modifying metabolite levels of activated methyl cycle in the cellular system. Thus, addition of homocysteine to the cell media confers a distinct phenotype to the colonies and surface biofilms.
Mycobacterium smegmatis colonies were allowed to form cords in the presence and absence of homocysteine. The normal cording feature in the absence of homocysteine was altered in the center of the colony at 0.3 and 0.5 mM homocysteine. This led us to further examine the pellicle formation in mycobacteria. On titrating homocysteine from 0.1 to 0.5 mM, there is a significant abrogation in the pellicle growth. Since, pellicles have recently been identified as biofilms, we tested the biofilm forming ability of M. smegmatis and M. bovis BCG in presence of homocysteine. Biofilm formation of mycobacteria was reduced when grown in minimal media with exogenous homocysteine supplementation, with a complete loss at 0.5 mM in M. smegmatis and 0.8 mM in M. bovis BCG. For M. bovis BCG, the biofilm defect was observed at a higher concentration of 0.8 mM which could be due to its slow metabolism or higher pathogenicity. Virulent bacteria showed decreased cell wall permeability leading to less effective transport of extracellular substances [33].
Biofilm formation in mycobacteria has been extensively studied with many breakthroughs to develop major genetic tools [34]. There have been major implications of biofilm formation in bacterial virulence and pathogenesis. Biofilm defective mutants of Mycobacterium avium failed to colonize and translocate through bronchial epithelial cells [35]. Also, Mycobacterium ulcerans colonizes the host and disseminate using biofilm which causes the pathological feature of Buruli ulcers [36, 37]. Thus, a proper knowledge of such underlying mechanisms of virulence is a major concern. The lipid rich mycobacterial cell wall consists of mycolic acids in the inner; and glycolipids and phospholipids on the outer surface giving a distinct surface characteristic to mycobacteria [38]. During the course of biofilm formation, mycobacteria alters the mycolic acid composition from long chain fatty acids (C70–C90) in free living planktonic state to shorter chain version (C56–C68), possibly to aggregate and develop into a mature biofilm [2]. A recent study has established the role of GroEL in the mycolic acid biosynthesis thus affecting biofilm formation [26, 39]. Mycolic acid methylation through Mma4 has a major role in M. tuberculosis virulence as mma4 mutants were susceptible to IL-12 dependent killing [40]. The methylated sugars and lipids on the mycobacterial cell wall confer hydrophobicity to the cell [41]. Such methylated sugar containing glycopeptidolipids are important for biofilm formation and colony morphology [42, 43].
Such methylation events occur in a SAM-dependent manner, which are required for chemotaxis, cell growth and development. Affecting SAM or methionine biosynthesis have been known to affect infection related processes [44, 45]. Homocysteine rules an important metabolic status where it is required for metabolite synthesis such as SAM, SAH, adenosine, methionine and cysteine and its upregulation either through SahH or its artificial supplementation in the medium directly affects cellular level of methylation. Eukaryotic SahH has been well known to regulate RNA or DNA methylation [46, 47]. SahH mediated biofilm formation in LuxS mutant and disruption of biofilm in a homocysteine supplementation medium hints the involvement of methylation in bacterial surface characteristics and thus quorum sensing.
Since biofilms are antibiotic tolerant [48] and are less susceptible to host immune responses, developing treatments against them requires a deep understanding of their physiological traits. We found that dysregulation of methionine synthesis through homocysteine is what impairs the biofilm formation in cultures with increased homocysteine. Thus, this defect in biofilm formation due to additional homocysteine shows the quorum sensing contribution of the methyl cycle.
Acknowledgements
We thank Anshika Singhal for her valuable suggestions in the experiments and the manuscript.
Abbreviations
- SAM
S-adenosyl methionine
- AI-2
Autoinducer-2
- AMC
Activated methyl cycle
- SAH
S-adenosyl homocysteine
- SahH
S-adenosyl homocysteine hydrolase
- NTM
Non-tuberculous mycobacteria
- SRH
S-ribosyl homocysteine
Author’s Contributions
RV conceived and designed the experiments. YH and YS contributed materials, reagents and analysis tools. All authors have read and approved the manuscript.
Funding
This work was supported by the Council of Scientific and Industrial Research (CSIR), India, J.C. Bose fellowship (SERB) (to Y.S.); and CSIR senior research fellowship (to R.V.).
Compliance with Ethical Standards
Competing interest
The authors declare that they have no competing interest.
Contributor Information
Yasha Hasija, Email: yashahasija@gmail.com.
Yogendra Singh, Email: ysinghdu@gmail.com.
References
- 1.Steinberg N, Kolodkin-Gal I. The matrix reloaded: how sensing the extracellular matrix synchronizes bacterial communities. J Bacteriol. 2015;197:2092–2103. doi: 10.1128/JB.02516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambrano MM, Kolter R. Mycobacterial biofilms: a greasy way to hold it together. Cell. 2005;123:762–764. doi: 10.1016/j.cell.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37:121–140. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]
- 4.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Kalia VC, Wood TK, Kumar P. Evolution of resistance to quorum-sensing inhibitors. Microb Ecol. 2014;68:13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, Alam A, Rani M, Ehtesham NZ, Hasnain SE. Biofilms: survival and defense strategy for pathogens. Int J Med Microbiol. 2017;307:481–489. doi: 10.1016/j.ijmm.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes RA, Monteiro DR, Arias LS, Fernandes GL, Delbem ACB, Barbosa DB. Virulence factors in Candida albicans and Streptococcus mutans biofilms mediated by farnesol. Indian J Microbiol. 2018;58:138–145. doi: 10.1007/s12088-018-0714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia VC. Microbes, antimicrobials and resistance: the battle goes on. Indian J Microbiol. 2014;54:1–2. doi: 10.1007/s12088-013-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maji A, Misra R, Dhakan DB, Gupta V, Mahato NK, Saxena R, Mittal P, Thukral N, Sharma E, Singh A, Virmani R. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol. 2018;20:402–419. doi: 10.1111/1462-2920.14015. [DOI] [PubMed] [Google Scholar]
- 10.Faria S, Joao I, Jordao L. General overview on nontuberculous mycobacteria, biofilms, and human infection. J Path. 2015;2015:809014. doi: 10.1155/2015/809014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa S, Bandeira M, Carvalho PA, Duarte A, Jordao L. Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int J Mycobacteriol. 2015;4:36–43. doi: 10.1016/j.ijmyco.2014.11.065. [DOI] [PubMed] [Google Scholar]
- 12.Esteban J, García-Coca M. Mycobacterium Biofilms. Front Microbiol. 2018;8:2651. doi: 10.3389/fmicb.2017.02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obeid R. The metabolic burden of methyl donor deficiency with focus on the betaine homocysteine methyltransferase pathway. Nutrients. 2013;5:3481–3495. doi: 10.3390/nu5093481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federle MJ, Bassler BL. Interspecies communication in bacteria. J Clin Investigat. 2003;112:1291–1299. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. In: Collin M, Schuch R, editors. Bacterial sensing and signaling. Basel: Karger Publishers; 2009. pp. 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winzer K, Hardie KR, Burgess N, Doherty N, Kirke D, Holden MT, Linforth R, Cornell KA, Taylor AJ, Hill PJ, Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3 (2H)-furanone. Microbiol. 2002;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- 17.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making `sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 18.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 19.Singhal A, Arora G, Sajid A, Maji A, Bhat A, Virmani R, Upadhyay S, Nandicoori VK, Sengupta S, Singh Y. Regulation of homocysteine metabolism by Mycobacterium tuberculosis S-adenosylhomocysteine hydrolase. Sci Rep. 2013;3:2264. doi: 10.1038/srep02264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singhal A, Arora G, Virmani R, Kundu P, Khanna T, Sajid A, Misra R, Joshi J, Yadav V, Samanta S, Saini N. Systematic analysis of mycobacterial acylation reveals first example of acylation-mediated regulation of enzyme activity of a bacterial phosphatase. J Biol Chem. 2015;290:26218–26234. doi: 10.1074/jbc.M115.687269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, Singh Y. Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J Bacteriol. 2011;193:5347–5358. doi: 10.1128/JB.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta M, Sajid A, Arora G, Tandon V, Singh Y. Forkhead-associated domain-containing protein Rv0019c and polyketide-associated protein PapA5, from substrates of serine/threonine protein kinase PknB to interacting proteins of Mycobacterium tuberculosis. J Biol Chem. 2009;284:34723–34734. doi: 10.1074/jbc.M109.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julián E, Roldán M, Sánchez-Chardi A, Astola O, Agustí G, Luquin M. Microscopic cords, a virulence-related characteristic of Mycobacterium tuberculosis, are also present in nonpathogenic mycobacteria. J Bacteriol. 2010;192:1751–1760. doi: 10.1128/JB.01485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambandan D, Dao DN, Weinrick BC, Vilchèze C, Gurcha SS, Ojha A, Kremer L, Besra GS, Hatfull GF, Jacobs WR. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. MBio. 2013;4:e00222-13. doi: 10.1128/mBio.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora G, Sajid A, Virmani R, Singhal A, Kumar CS, Dhasmana N, Khanna T, Maji A, Misra R, Molle V, Becher D. Ser/Thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microbiomes. 2017;3:7. doi: 10.1038/s41522-017-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalsum S, Braian C, Koeken VA, Raffetseder J, Lindroth M, van Crevel R, Lerm M. The cording phenotype of Mycobacterium tuberculosis induces the formation of extracellular traps in human macrophages. Front Cell Infect Microbiol. 2017;7:278. doi: 10.3389/fcimb.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redanz S, Standar K, Podbielski A, Kreikemeyer B. Heterologous expression of sahH reveals that biofilm formation is autoinducer-2-independent in Streptococcus sanguinis but is associated with an intact activated methionine cycle. J Biol Chem. 2012;287:36111–36122. doi: 10.1074/jbc.M112.379230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubica T, Agzamova R, Wright A, Rakishev G, Rüsch-Gerdes S, Niemann S. Mycobacterium bovis isolates with M. tuberculosis specific characteristics. Emerg Infect Dis. 2006;12:763–765. doi: 10.3201/eid1205.050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adetunji VO, Kehinde AO, Bolatito OK, Chen J. Biofilm formation by Mycobacterium bovis: influence of surface kind and temperatures of sanitizer treatments on biofilm control. Biomed Res Int. 2014;2014:210165. doi: 10.1155/2014/210165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Totani T, Nishiuchi Y, Tateishi Y, Yoshida Y, Kitanaka H, Niki M, Kaneko Y, Matsumoto S. Effects of nutritional and ambient oxygen condition on biofilm formation in Mycobacterium avium subsp. hominissuis via altered glycolipid expression. Sci Rep. 2017;7:41775. doi: 10.1038/srep41775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossignol T, Ding C, Guida A, d’Enfert C, Higgins DG, Butler G. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot Cell. 2009;8:550–559. doi: 10.1128/EC.00350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojha AK, Jacobs WR, Hatfull GF. Genetic dissection of mycobacterial biofilms. In: Parish T, Roberts DM, editors. Mycobacteria protocols. New York: Humana Press; 2015. pp. 215–226. [Google Scholar]
- 35.Yamazaki Y, Danelishvili L, Wu M, Hidaka E, Katsuyama T, Stang B, Petrofsky M, Bildfell R, Bermudez LE. The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol. 2006;8:806–814. doi: 10.1111/j.1462-5822.2005.00667.x. [DOI] [PubMed] [Google Scholar]
- 36.Marsollier L, Brodin P, Jackson M, Korduláková J, Tafelmeyer P, Carbonnelle E, Aubry J, Milon G, Legras P, Saint André JP, Leroy C. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 2007;3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora G, Sajid A, Singhal A, Joshi J, Virmani R, Gupta M, Verma N, Maji A, Misra R, Baronian G, Pandey AK. Identification of Ser/Thr kinase and forkhead associated domains in Mycobacterium ulcerans: characterization of novel association between protein kinase Q and MupFHA. PLOS Negl Trop Dis. 2014;8:e3315. doi: 10.1371/journal.pntd.0003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson JH, McConville MJ, Haites RE, Coppel RL, Billman-Jacobe H. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J Biol Chem. 2000;275:24900–24906. doi: 10.1074/jbc.M000147200. [DOI] [PubMed] [Google Scholar]
- 39.Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Dao DN, Sweeney K, Hsu T, Gurcha SS, Nascimento IP, Roshevsky D, Besra GS, Chan J, Porcelli SA, Jacobs WR., Jr Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog. 2008;4:e1000081. doi: 10.1371/journal.ppat.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson M, Brennan PJ. Polymethylated polysaccharides from Mycobacterium species revisited. J Biol Chem. 2009;284:1949–1953. doi: 10.1074/jbc.R800047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stadthagen G, Sambou T, Guerin M, Barilone N, Boudou F, Korduláková J, Charles P, Alzari PM, Lemassu A, Daffé M, Puzo G. Genetic basis for the biosynthesis of methylglucose lipopolysaccharides in Mycobacterium tuberculosis. J Biol Chem. 2007;282:27270–27276. doi: 10.1074/jbc.M702676200. [DOI] [PubMed] [Google Scholar]
- 43.Schorey JS, Sweet L. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology. 2008;18:832–841. doi: 10.1093/glycob/cwn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basavanna S, Chimalapati S, Maqbool A, Rubbo B, Yuste J, Wilson RJ, Hosie A, Ogunniyi AD, Paton JC, Thomas G, Brown JS. The effects of methionine acquisition and synthesis on Streptococcus pneumoniae growth and virulence. PLoS ONE. 2013;8:e49638. doi: 10.1371/journal.pone.0049638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brambila-Tapia AJL, Poot-Hernández AC, Perez-Rueda E, Rodríguez-Vázquez K. Identification of DNA methyltransferase genes in human pathogenic bacteria by comparative genomics. Indian J Microbiol. 2016;56:134–141. doi: 10.1007/s12088-015-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barić I, Fumić K, Glenn B, Ćuk M, Schulze A, Finkelstein JD, James SJ, Mejaški-Bošnjak V, Pažanin L, Pogribny IP, Radoš M. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc Natl Acad Sci. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mull L, Ebbs ML, Bender J. A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics. 2006;174:1161–1171. doi: 10.1534/genetics.106.063974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalia VC. In search of versatile organisms for quorum-sensing inhibitors: acyl homoserine lactones (AHL)-acylase and AHL-lactonase. FEMS Microbiol Lett. 2014;359:143. doi: 10.1111/1574-6968.12585. [DOI] [PubMed] [Google Scholar]