Abstract
Green silver nanoparticle (AgNP) biosynthesis is facilitated by the enzyme mediated reduction of Ag ions by plants, fungi and bacteria. The antimicrobial activity of green AgNPs is useful to overcome the challenge of antimicrobial resistance. Antimicrobial properties of biosynthesized AgNPs depend on multiple factors including culture conditions and the microbial source. The antimicrobial activity of AgNPs biosynthesized by Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923 and Acinetobacter baumannii (confirmed clinical isolate) were investigated in this study. Biosynthesis conditions (AgNO3 concentration, pH, incubation temperature and incubation time) were optimized to obtain the maximum AgNP yield. Presence of AgNPs was confirmed by observing a characteristic UV–Visible absorbance peak in 420–435 nm range. AgNP biosynthesis was optimal at 0.4 g/L AgNO3 concentration under alkaline conditions at 60–70 °C. The biosynthesized AgNPs showed higher stability compared to chemogenized AgNPs in the presence of electrolytes. AgNPs synthesized by P. aeruginosa were the most stable while NPs of S. aureus were the least stable. AgNPs synthesized by P. aeruginosa and S. aureus showed good antimicrobial potential against E. coli, P. aeruginosa, S. aureus, MRSA and Candida albicans. AgNPs synthesized by S. aureus had greater antimicrobial activity. The antimicrobial activity of NPs may vary depending on the size and the morphology of NPs.
Keywords: Silver nanoparticle, Antimicrobial activity, UV–Visible spectroscopy, Colony forming units, TEM
Introduction
Green synthesis of nanoparticles (NPs) using bacteria has several advantages due to its inherent ecofriendly and economical nature. Green biosynthesis offers an alternative approach which can substantially overcome the environmental challenges posed by physical and chemical methods and do not involve hazardous chemicals and intolerant reaction conditions. The method is based on the ability of environmental microorganisms to remediate metals through enzymatic reduction of metal ions [1]. During this process, metal ions can be converted into nanoparticles of varying sizes and morphologies [2].
Among metal nanoparticles such as gold [3], zinc oxide [4] and titanium dioxide [5], silver nanoparticles have a wide spectrum of applications in the field of medicine due to its inherent antimicrobial properties. Further they are used in biotechnological applications, in production of biohydrogen [6], as biocatalysts and biosensors [7]. Several groups have reported AgNP biosynthesis using different bacterial including Pseudomonas aeruginosa, Acinetobacter calcoceticus, Escherichia coli, Bacillus subtilis, Lactobacills species etc. (Table 2).
Table 2.
Biological synthesis of AgNPs by various microorganisms
| Microorganism | Particle size (nm) | Particle structure and distribution | Maximum absorption wavelength (nm) | References |
|---|---|---|---|---|
| Bacillus cereus | 2–16 | Spherical | 420 | [9] |
| P. aeruginosa strain BS-161R | 8–24 | Spherical, anisotropic | 430 | [26] |
| P. aeruginosa | 20–50 | Spherical, monodispersed | 455 | [1] |
| P. aeruginosa strain KUPSB12 | 50–85 | Spherical | 442 | [27] |
| Aspergillus flavus strain NJP08 | 10–35 | Spherical, monodispersed | 421 | [11] |
| Acinetobacter calcoaceticus strain LRVP54 | 8–12 | Crystalline, polydispersed | 410–440 | [10] |
| E. coli | 20–50 | Spherical | 400 | [28] |
| Penicillium strain | > 100 | Colloidal, polydispersed | 440 | [24] |
| P. aeruginosa ATCC 27853 | 33–300 | Spherical | 420–430 | [14] |
| Bacillus sp | 65–70 | Spherical and pseudospherical | 420 | [29] |
| A. flavus strain NCIM650 | 37.3–52.92 | nd | 420 | [30] |
| Emericella nidulans strain APF4 | 10–450 | Hexagonal, monodispersed | 450 | [31] |
| P. aeruginosa | 20–100 | Crystalline | 420–430 | [18] |
These bacterial biosynthesized AgNPs vary in the morphology, size and antimicrobial activity depending on the conditions of biosynthesis [8]. An important consideration in microbial nanosynthesis is the selection of an appropriate microorganism which can produce the targeted NPs with a particular particle shape and size range. Majority of the reported bacterial synthesized AgNPs were spherical, while other morphological forms such as triangular, hexagonal, cuboidal, rod shpaed, Quasi-spherical and nanoplates have been observed by some groups [8].
A major challenge in green nanoparticle synthesis involves the control of the morphology, size and ability for mono-dispersity in solution [9]. The intrinsic microbial properties including the cellular machinery and growth requirements of bacteria are crucial in the biosynthesis process [2, 8, 10]. Thus, optimization of conditions for nanoparticle biosynthesis is important to obtain maximum yields [7]. Biosynthesized AgNPs are coated with extracellular proteins [11] and various biomolecules [12] present in the solution which contributes to higher stability. Despite the many advantages, purification of green NPs continues to be an important deterrent in scaling up green nanoparticle production for industrial applications. Although several approaches such as use of genetically engineered microorganisms have been applied for improved nanoparticle biosynthesis [13], thus far the inability to obtain large quantities of purified NPs using biological methods is a major challenge which merits intense investigation.
The present study compared the silver nanoparticle biosynthesis and morphological and antimicrobial characteristics of AgNPs synthesized from four selected microorganisms namely, P. aeruginosa, S. aureus, A. baumannii and E. coli.
Materials and Methods
Production of Biomass
Pseudomonas aeruginosa ATCC 27853, S. aureus ATCC 25923, E. coli ATCC 25922 and A. baumannii (confirmed clinical strain) were obtained from the culture collection of the Department of Microbiology, University of Sri Jayewardenepura. The cultures were incubated at 37 °C for 72 h in nutrient broth (Himedia, India) on a shaker at 150 rpm. The culture supernatant was obtained following centrifugation at 6000 rpm for 15 min. The supernatant was subsequently used for silver NP biosynthesis.
Biosynthesis of Silver Nanoparticles
Biosynthesis of silver nanoparticles was carried out as described by Peiris et al. [14]. Briefly, the culture supernatants of selected bacteria were mixed with different concentrations of AgNO3 (Park, UK) and incubated in the dark for different time periods at varying temperatures to allow nanoparticle formation. The absorption spectrum of the solution was measured using a UV–Visible spectrophotometer (PerkinElmer Lambda 35, USA).
Optimization of Culture Parameters for AgNP Synthesis
Reaction parameters (AgNO3 concentration, pH, incubation temperature and incubation time) were optimized to obtain the maximum yield of nanoparticles. AgNO3 concentrations ranging from 0.05, 0.1, 0.2, 0.3 and 0.4 g/L were added to the culture supernatants at different pH (6–11) conditions and temperatures (0, 28, 37, 42, 60 and 70 °C) for AgNP biosynthesis. Presence of AgNPs in the solution was confirmed by the characteristic UV–Visible absorption peak at approximately 420 nm. The pH of the reaction solution was adjusted using 1 M HCl and 1 M NaOH solutions. To study the best reaction time for maximum synthesis of AgNPs, the optimal conditions of pH, AgNO3 concentration and temperature were applied to the culture supernatants at different incubation periods (24, 48, 72, 96 and 120 h).
Characterization of Nanoparticles
UV–Visible Spectrophotometry
Biosynthesized AgNP solutions were diluted 1:10 in sterile distilled water and used for characterization using UV–Visible spectrophotometer. The UV–Visible spectrum was obtained in the range of 300–800 nm using a PerkinElmer Lambda 35 UV–Visible spectrophotometer.
Transmission Electron Microscopy (TEM)
TEM imaging (JEOL JEM 2100, Japan) was performed using Drop casting method on a holey carbon coated Cu grid (Accelerating voltage 200 kV).
Fourier Transform Infrared Spectroscopy (FT-IR)
Fourier transform infrared spectra of AgNPs were recorded to provide an evidence for the interaction of functional groups involved in the reduction of AgNO3. The NP solutions were dried at room temperature and the powder form was analyzed at a spectral range 4000–400 cm−1 at a resolution of 4 cm−1 (Thermo Scientific Nicolet iS10 FT-IR spectrometer). The FT-IR analysis of cell-free extracts was also performed which served as a reference.
Stability of AgNPs
The synthesized AgNP solutions were stored at 30 °C under normal light conditions. The stability of green silver NPs under electrolytic conditions in the presence of sodium chloride (NaCl) were studied by adding aliquots of 5 M NaCl into 2 mL of AgNP solution. UV–Visible spectra were obtained in the range of 350–800 nm. Variability of the UV–visible absorbance with NaCl concentration was compared. The procedure was repeated for chemogenized AgNP solution synthesized according to Rashid et al. [15] for comparison.
Antimicrobial Activity of Nanoparticles
Agar Diffusion Assay
The antimicrobial activity of synthesized AgNPs against P. aeruginosa ATCC 27853, S. aureus ATCC 25923, E. coli ATCC 25922, Methicillin resistant S. aureus (MRSA) (clinical strain) and Candida albicans ATCC 10231 were determined using the well diffusion assay. Organisms were cultured overnight at 37 °C and suspensions of 0.5 McFarland standard were prepared for each organism. Standard inocula (100 µL) of each bacterium or Candida was lawned on Mueller–Hinton Agar (MHA, Himedia, India)/Sabourauds Dextrose Agar (SDA, Himedia, India). Wells were cut in the agar medium using a sterile 9 mm cork borer. The bottom of the wells were sealed with molten agar. Wells were loaded with green AgNP solution, negative controls (consisting of culture supernatants of each organism and sterile nutrient broth) and positive control (0.5% AgNO3 solution) and incubated at 37 °C for 24 h. All experiments were done in triplicates. Zones of inhibition (ZOI) were measured and the average of ZOI was calculated.
Plate Coating Method
Sterile 6 cm petri dishes were coated with (A) Biosynthesized silver NPs and (B) bacterial culture supernatant as the negative control. The prepared nanoparticle solutions (2 mL) were added into individual petri dishes and allowed to completely dry for 24 h inside a class II biological safety cabinet. Subsequently 2 mL of bacterial suspension (0.5 McFarland standard) was added to each petri dish and incubated for different time intervals (5, 10, 15, 30, 45, 60 min) at room temperature. Viable cell counts at different contact time points were obtained by spread plate method. The number of colony forming units (CFU) per millilitre was calculated at each incubation time. All experiments were carried out in triplicates.
Statistical Analysis
The viable counts obtained at different contact times were used to calculate the percentage reduction using the following equation.
For graph preparation, OriginPro 9.0 software was used.
Results and Discussion
Optimization of Culture Conditions for AgNP Synthesis
In this study, all the selected organisms synthesized AgNPs successfully under different optimized conditions. AgNP biosynthesis can be affected by multiple factors such as the culture conditions including pH, temperature, culture medium and the microbial species [16]. Microbial reduction of Ag ions into NPs can be carried out intracellularly or extracellularly [2, 8]. Culture supernatants contain microbial products and media components that facilitate this reaction extracellularly [8]. Proteins such as nitrate reductase stabilize NPs and reduce aggregation of particles which help to maintain the nano size [1].
Addition of AgNO3 to the four culture supernatants resulted in a colour change from pale yellow to dark brown indicating AgNP synthesis within one hour (Fig. 1). The intensity of the colour of the solution increased further during incubation. The UV–Visible spectra obtained around 420–435 nm range confirmed AgNP formation in this study. The optimum AgNO3 concentrations for all four supernatants were 0.4 g/L (Table 1). Other studies have reported varying time parameters for the occurrence of colour change resulting in AgNP biosynthesis, [17, 18]. The observed color development is also known to be associated with the shape and size of AgNPs [19, 20].
Fig. 1.
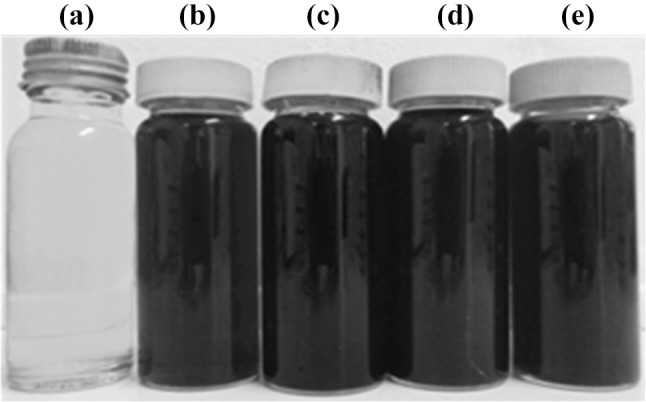
The cell free filtrates, a without AgNO3 and after addition of AgNO3 for biosynthesis of AgNPs, b P. aeruginosa, c E. coli, d A. baumannii and e S. aureus with AgNO3 after 72 h of incubation
Table 1.
Optimized culture conditions for AgNP biosynthesis
| Organism | Optimized culture conditions | |||
|---|---|---|---|---|
| AgNO3 concentration (g/L) | Culture pH | Incubation temperature (oC) | Incubation time (h) | |
| P. aeruginosa (P-AgNPs) | 0.4 | 9.0 | 70 | 96 |
| E. coli (E-AgNPs) | 0.4 | 9.0 | 60 | 72 |
| A. baumannii (A-AgNPs) | 0.4 | 9.0 | 60 | 72 |
| S. aureus (S-AgNPs) | 0.4 | 9.0 | 60 | 72 |
Alkaline pH favoured optimum AgNP biosynthesis. Optimum pH for the biosynthesis of all four AgNP samples was pH 9. The optimum incubation temperatures were 70 °C for P-AgNPs, 60 °C for E-AgNPs, A-AgNPs and S-AgNPs while maximum UV absorbance was observed at 72 h for three of the AgNPs except for P-AgNPs where the optimum incubation time was 96 h (Table 1). Van Dong et al. [19] described the effect of pH on the size and shape of the synthesized AgNPs where at high alkaline pH, spherical and rod shaped AgNPs were produced due to the fast reaction rate while triangular or polygonal shaped AgNPs resulted at low pH due to a slow reaction rate. AgNPs have several reported morphologies including triangular [21], polyhedral [21, 22], nanoprisms [19], spherical [19, 21] and rod shape [21]. These results indicate the impact of the culture conditions on the microbial metabolic activities which can be harnessed for efficient AgNP production.
The AgNO3 concentration can also affect the size and morphology of the AgNPs. At higher AgNO3 concentrations there is an increase in the rate of reduction of silver ions to AgNPs [14]. High silver salt concentrations can result in aggregated NPs with larger size [23]. In a study by Singh et al. [10] 0.7 mM AgNO3 concentration was found to be optimum for AgNP synthesis by Acinetobacter calcoaceticus. The present study reports AgNP biosynthesis by a clinical isolate of A. baumannii. To our knowledge, Silver nanoparticle biosynthesis by A. baumannii has not been reported previously. The microbial AgNP synthesis was favoured by alkaline pH while under acidic conditions, particles tend to aggregate and become unstable [24].
Morphology and Size Determination of AgNPs by Transmission Electron Microscopy
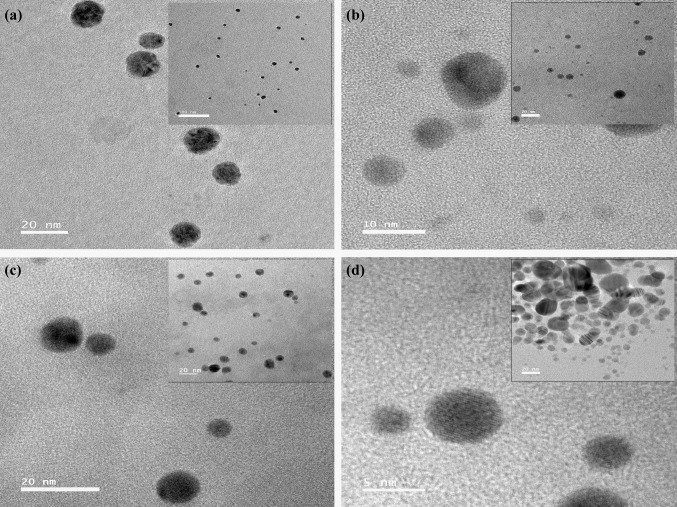
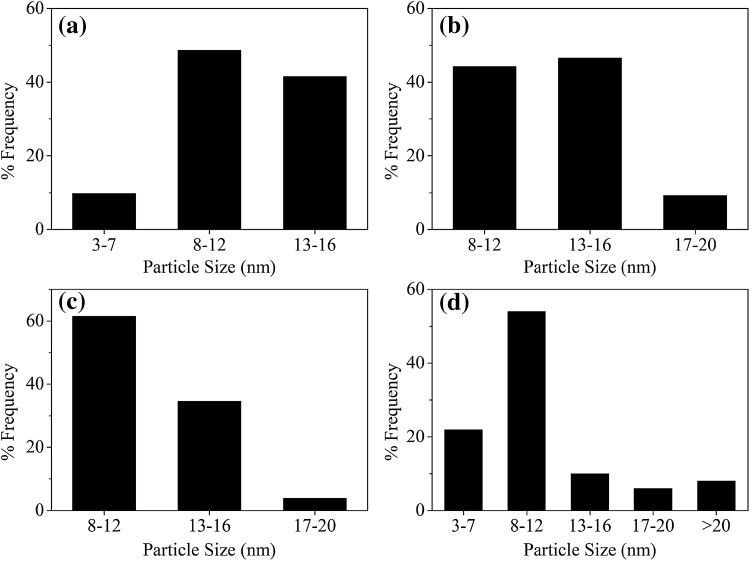
The morphology of AgNPs biosynthesized at the optimized parameters were determined using TEM. In this study, all AgNPs were found to be of spherical morphology (Fig. 2). The average sizes of the NPs were 11.14 ± 6.59 nm (S-AgNPs), 11.71 ± 2.73 nm (P-AgNPs), 12.87 ± 2.95 nm (E-AgNPs) and 12.22 ± 2.45 nm (A-AgNPs). S-AgNPs had the largest size distribution having a size range between 3.4–33.0 nm. An important finding was that all other bacterial synthesized NPs resulted in less than 20 nm size AgNPs (Fig. 3).
Fig. 2.
TEM images of AgNPs synthesized from different bacteria. a P-AgNPs, b E-AgNPs, c A-AgNPs and d S-AgNPs (scale bar given)
Fig. 3.
Particle size distribution of AgNPs synthesized from different bacteria. a P-AgNPs, b E-AgNPs, c A-AgNPs and d S-AgNPs
When considering the TEM results the smaller AgNPs were produced by S. aureus and P. aeruginosa which gave a UV–visible absorbance peak at 430 and 427 nm respectively. The highest frequency of AgNPs by S. aureus, P. aeruginosa and A. baumannii were of the size range 8–12 nm while for E. coli it was 13–16 nm. According to other studies [1, 25] the optimized conditions in this study resulted in very small AgNPs than previously reported for P. aeruginosa, S. aureus (< 8 nm) indicating stronger antimicrobial potential due to high surface area to volume ratio. Table 2 shows a comparison of AgNPs from various bacterial sources found in recent literature.
FT-IR Analysis
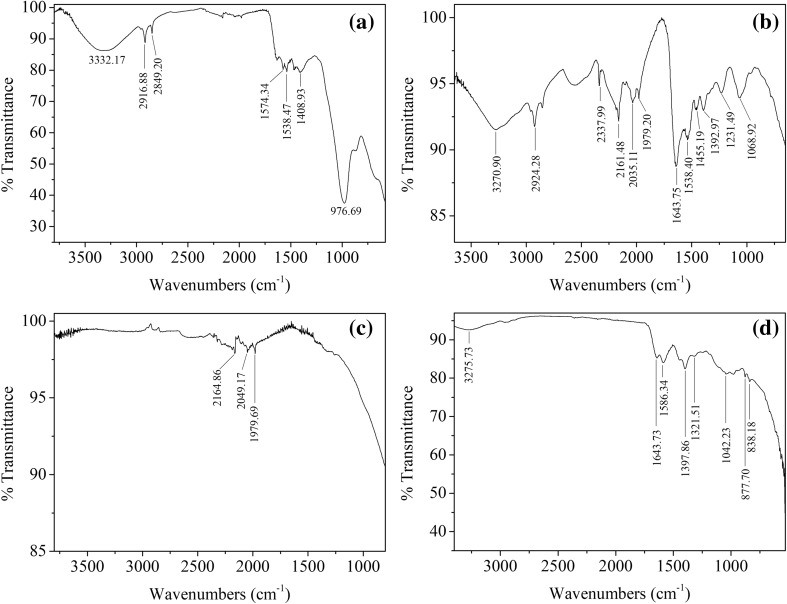
FT-IR analysis of P-AgNPs revealed four prominent bands at 1643, 1586, 1397 and 1042 cm−1. A-AgNPs had FT-IR bands at 1643.75, 2161.48 and 2924.28 cm−1 while, E-AgNPs had one prominent band at 2916.88 cm−1. FT-IR of S-AgNPs, demonstrated bands at 2164.86, 2049.17 and 1979.69 cm−1 (Fig. 4).
Fig. 4.
FT-IR spectra of AgNPs. a E-AgNPs, b A-AgNPs, c S-AgNPs and d P-AgNPs
The biosynthesized silver nanoparticles have increased stability due to the coating of the NPs by microbial and media components [26]. In the FTIR spectra (Fig. 4), 2924, 2916, 1538 and 1455 cm−1 modes are due to NH stretch vibrations present in the amide linkages of primary and secondary amines of proteins [32–34]. The peak 1397 cm−1 shows the presence of nitrocompounds from proteins or enzymes [35]. The peak at 2849 cm−1 can be assigned to the C–H symmetrical stretch vibration of alkanes [32]. According to Jeevan et al. [18] a band around 1643 cm−1 in P-AgNP and A-AgNP samples, may indicate –C=O carbonyl groups and –C=C– stretching. Percentage transmittance of 1042 cm−1 refers to –C–N– stretching vibrations. This shows the presence of proteins in the biologically synthesized AgNP solutions which support stabilization of AgNPs. Protein components of the medium can bind to AgNPs through free amino or cysteine groups and cause capping of biosynthesized AgNPs which can help to stabilize the nanoparticles in solution and prevent agglomeration [36].
Stability of Green AgNPs
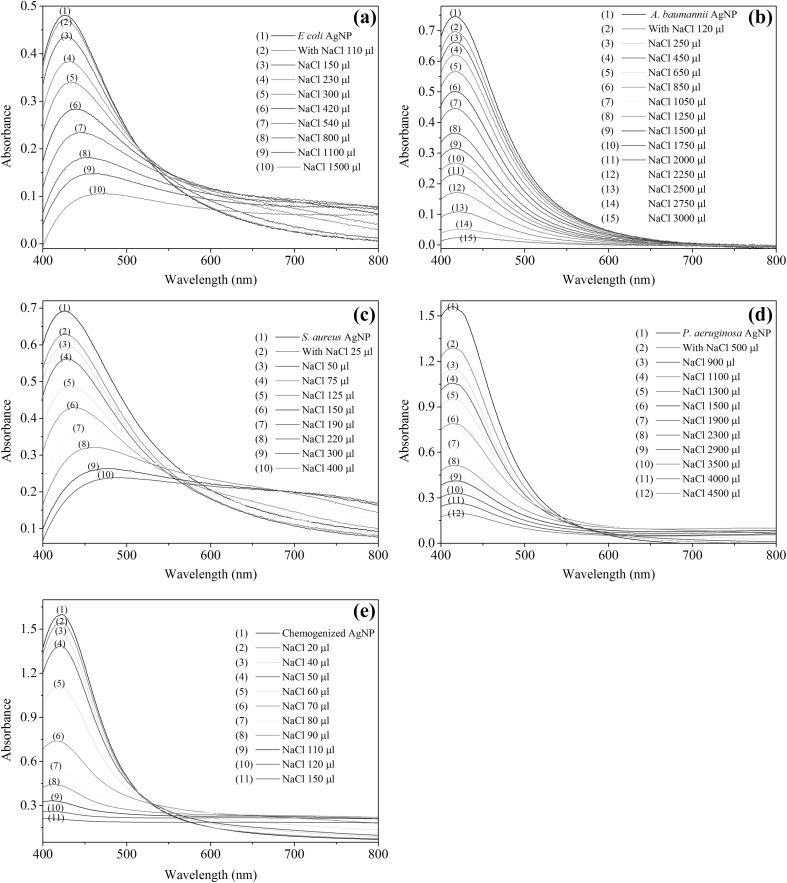
The stability of the biosynthesized AgNPs and the chemogenized AgNPs were compared in the presence of electrolytes (5 M NaCl) (Fig. 5).
Fig. 5.
Stability of a E-AgNPs, b A-AgNPs, c S-AgNPs, d P-AgNPs and e Chemogenized AgNPs in the presence of NaCl
The stability of AgNPs is considered to be inversly propotional to the rate of flocculation. The P-AgNPs and A-AgNPs remained stable even after adding more than 3000 µL of 5 M NaCl, however S-AgNPs lost their stability at much lower volume (150 µL) of 5 M NaCl while E-AgNPs were unstable after addition of 1500 µL of 5 M NaCl. Of the four NPs, P-AgNPs were the most stable followed by A-AgNPs and E-AgNPs while S-AgNPs were found to be least stable. Chemogenized AgNPs were found to be less stable compared to biosynthesized AgNPs suggesting that membrane bound proteins prevent aggregation of green AgNPs thereby promoting stability [37].
Antimicrobial Activity
Zones of inhibition (ZOI) were observed for AgNPs synthesized by all organisms tested. E-AgNPs exhibited inhibition zones against E. coli and P. aeruginosa, while S. auerus, MRSA and C. albicans did not show any ZOI. A-AgNPs had ZOI against E. coli, P. aeruginosa and C. albicans (Table 3).
Table 3.
Average zones of inhibition of AgNPs from four different bacteria against selected pathogens
| Test organism | Average ZOI (mm) obtained from various bacterial AgNPs | ||||
|---|---|---|---|---|---|
| E. coli | A. baumannii | S. aureus | P. aeruginosa | 0.5% AgNO3 solution | |
| E. coli | 10.0 | 13.3 | 14.7 | 13.0 | 16.0 |
| P. aeruginosa | 10.0 | 14.7 | 13.0 | 12.0 | 17.7 |
| S. aureus | – | – | 12.7 | 12.3 | 12.0 |
| MRSA | – | – | 12.7 | 12.7 | 12.0 |
| C. albicans | – | 10.0 | 12.7 | 14.0 | 10.0 |
In this study, well diffusion results and plate coating results suggest that E-AgNPs had the lowest antimicrobial activity while P-AgNPs and S-AgNPs had highest antimicrobial activity. In the well diffusion study, only P-AgNPs and S-AgNPs resulted in inhibition of Gram positive test organisms S. aureus and MRSA.
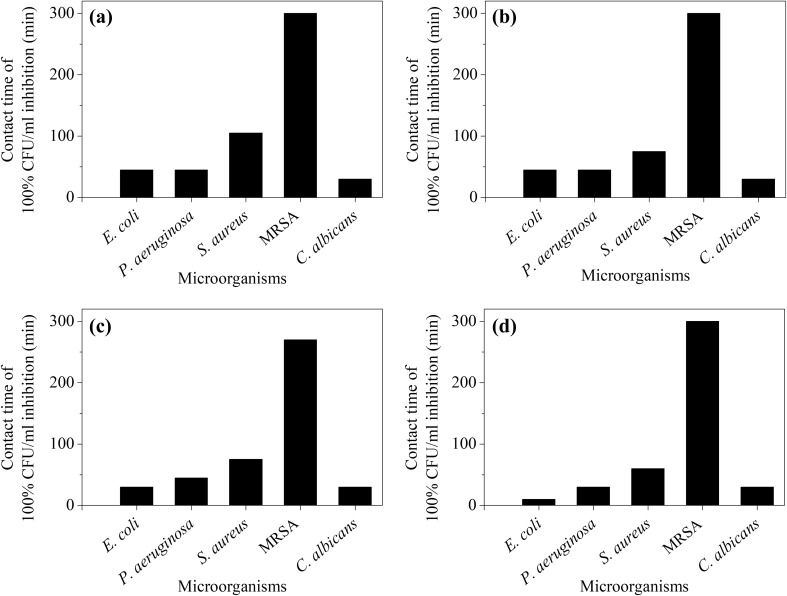
The plate coating method was performed to determine the percentage inhibition of CFU/mL at different contact times in the presence of the tested AgNPs. The results of the plate coating assay indicates that AgNPs synthesized by all four microorganisms could completely (100%) inhibit P. aeruginosa, E. coli and C. albicans when in contact for less than 60 min. P-AgNPs, A-AgNPs and S-AgNPs completely inhibited S. aureus after 75 min, while E-AgNPs completely inhibited S. aureus following 105 min of contact. However, complete inhibition of MRSA was achieved after 300 min of contact with P-AgNPs, E-AgNPs and A-AgNPs while for S-AgNPs, the contact time required was 270 min (Fig. 6).
Fig. 6.
Contact time required for 100% inhibition of selected pathogens by AgNPs. a E-AgNPs, b A-AgNPs, c S-AgNPs and d P-AgNPs in CFU/mL
Antimicrobial properties of synthesized AgNPs were higher against gram negative bacteria compared to Gram positive bacteria possibly due to the difference in cell membrane thickness [9, 28, 38]. Spherical silver NPs have increased antimicrobial activity due to high surface area to volume ratio [10]. Their larger surface area favours binding with various ligands. According to Cicek et al. [39] due to its smaller size, AgNPs can move into microbial cells through cell membranes and the antimicrobial activity tends to increase with the reduced size. Silver NPs can get attached to the cell membranes and release silver ions slowly [1]. AgNPs is suggested to cause pit formation in the cell wall of bacteria resulting in increased permeability and leakage of cellular contents through the plasma membrane. Further a study by Song et al. [40] reported a higher concentration of AgNPs were required against Gram positive bacteria compared to Gram negative bacteria. This observation which supports our findings can be explained by the difference in the bacterial cell wall, where Gram positive bacteria contain a thick cell wall containing peptidoglycan which can resist the passage of the nanoparticles compared to the thinner cell wall of Gram negative bacteria. Evidence shows release of Ag ions from AgNPs within the cell and on the cell membrane [41], where the silver ions are suggested to interact with thiol groups on proteins thereby altering the structure and functions of enzymes resulting in bacterial death [42]. Further, the negatively charged Gram positive bacterial cell wall bind with more Ag ions which results in less number of AgNPs interacting with the plasma membrane thereby protecting the plasma membrane from the reactive oxygen species generated by the AgNPs leading to increased resistance observed. [43]. Further silver NPs mediate binding with DNA, protein denaturation, and respiratory functions leading to cell death [1, 9, 38]. In conclusion, AgNPs synthesized from selected bacteria yielded less than 20 nm NPs which showed effective antimicrobial activity and was found to be a cost effective and eco-friendly method.
Acknowledgements
The authors acknowledge the financial support (Grant No. ASP/01/RE/MED/2016/42) and laboratory facilities provided by the University of Sri Jayewardenepura.
Abbreviations
- AgNPs
Silver nanoparticles
- ZOI
Zone of inhibition
- CFU
Colony forming units
Contributor Information
M. M. K. Peiris, Email: mudarapeiris@gmail.com
S. S. N. Fernando, Email: fneluka@sjp.ac.lk
P. M. Jayaweera, Email: pradeep@sjp.ac.lk
N. D. H. Arachchi, Email: nuwan.wcsg@gmail.com
T. D. C. P. Guansekara, Email: chinthika@sjp.ac.lk
References
- 1.Oza G, Pandey S, Shah R, Sharon M (2012) Extracellular fabrication of silver nanoparticles using Pseudomonas aeruginosa and its antimicrobial assay. Pelagia Res Lib Adv Appl Sci Res 3:1776–1783. https://www.researchgate.net/profile/Dr_Sunil_Pandey2/publication/230707314_Extracellular_Fabrication of Silver Nanoparticles using Pseudomonas aeruginosa and its Antimicrobial Assay/links/0912f5034 8c53f0b10000000.pdf. Accessed 27 Aug 2017
- 2.Li X, Xu H, Chen Z-S, Chen G. Biosynthesis of nanoparticles by microorganisms and their applications. J Nanomater. 2011;2011:1–16. [Google Scholar]
- 3.Otari SV, Patel SK, Jeong J-H, Lee JH, Lee J-K. A green chemistry approach for synthesizing thermostable antimicrobial peptide-coated gold nanoparticles immobilized in an alginate biohydrogel. RSC Adv. 2016;6:86808–86816. doi: 10.1039/C6RA14688K. [DOI] [Google Scholar]
- 4.Hölken I, Hoppe M, Mishra YK. Complex shaped ZnO nano-and microstructure based polymer composites: mechanically stable and environmentally friendly coatings for potential antifouling applications. Phys Chem Chem Phys. 2016;18:7114–7123. doi: 10.1039/C5CP07451G. [DOI] [PubMed] [Google Scholar]
- 5.Hunagund SM, Desai VR, Barretto DA, Pujar MS, Kadadevarmath JS, Vootla S, Sidarai AH. Photocatalysis effect of a novel green synthesis gadolinium doped titanium dioxide nanoparticles on their biological activities. J Photochem Photobiol. 2017;346:159–167. doi: 10.1016/j.jphotochem.2017.06.003. [DOI] [Google Scholar]
- 6.Patel SK, Lee J-K, Kalia VC. Nanoparticles in biological hydrogen production: an overview. Indian J Microbiol. 2017 doi: 10.1007/s12088-017-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otari S, Pawar S, Patel SK, Singh RK, Kim S-Y, Lee JH, Zhang L, Lee J-K. Canna edulis leaf extract-mediated preparation of stabilized silver nanoparticles: characterization, antimicrobial activity, and toxicity studies. J Microbiol Biotechnol. 2017;27:731–738. doi: 10.4014/jmb.1610.10019. [DOI] [PubMed] [Google Scholar]
- 8.Singh R, Shedbalkar UU, Wadhwani SA, Chopade BA. Bacteriagenic silver nanoparticles: synthesis, mechanism, and applications. Appl Microbiol Biotechnol. 2015;99:4579–4593. doi: 10.1007/s00253-015-6622-1. [DOI] [PubMed] [Google Scholar]
- 9.Gurunathan S (2014) Rapid biological synthesis of silver nanoparticles and their enhanced antibacterial effects against Escherichia fergusonii and Streptococcus mutans. Arab J Chem (in press)
- 10.Singh R, Wagh P, Bellare J. Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics. Int J Nanomed. 2013;8:4277–4290. doi: 10.2147/IJN.S48913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain N, Bhargava A, Majumdar S, Tarafdar J, Panwar J. Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus njp08: a mechanism perspective. Nanoscale. 2011;3:635–641. doi: 10.1039/C0NR00656D. [DOI] [PubMed] [Google Scholar]
- 12.Krishna IM, Reddy GB, Veerabhadram G, Madhusudhan A. Eco-friendly green synthesis of silver nanoparticles using Salmalia malabarica: synthesis, characterization, antimicrobial, and catalytic activity studies. Appl Nano Sci. 2016;6:681–689. doi: 10.1007/s13204-015-0479-6. [DOI] [Google Scholar]
- 13.Park TJ, Lee SY, Heo NS, Seo TS. In vivo synthesis of diverse metal nanoparticles by recombinant Escherichia coli. Angew Chem Int Ed. 2010;49:7019–7024. doi: 10.1002/anie.201001524. [DOI] [PubMed] [Google Scholar]
- 14.Peiris MK, Gunasekara CP, Jayaweera PM, Arachchi ND, Fernando N. Biosynthesized silver nanoparticles: are they effective antimicrobials? Mem Inst Oswaldo Cruz. 2017;112:537–543. doi: 10.1590/0074-02760170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rashid MU, Bhuiyan MKH, Quayum ME. Synthesis of silver nano particles (Ag-NPs) and their uses for quantitative analysis of vitamin c tablets. Dhaka Univ J Pharm Sci. 2013;12:29–33. doi: 10.3329/dujps.v12i1.16297. [DOI] [Google Scholar]
- 16.Gurunathan S, Kalishwaralal K, Vaidyanathan R, Venkataraman D, Pandian SRK, Muniyandi J, Hariharan N, Eom SH. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf B. 2009;74:328–335. doi: 10.1016/j.colsurfb.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 17.El-Shanshoury AE-RR, ElSilk SE, Ebeid ME. Extracellular biosynthesis of silver nanoparticles using Escherichia coli ATCC 8739, Bacillus subtilis ATCC 6633, and Streptococcus thermophilus ESh1 and their antimicrobial activities. ISRN Nanotechnol. 2011;2011:1–7. doi: 10.5402/2011/385480. [DOI] [Google Scholar]
- 18.Jeevan P, Ramya K, Rena AE (2012) Extracellular biosynthesis of silver nanoparticles by culture supernatant of Pseudomonas aeruginosa. Indian J biotechnol 11:72–76 https://pdfs.semanticscholar.org/885a/b93862e3deb07611b4325c3234a7890e3b6d.pdf. Accessed 15 June 2016
- 19.Van Dong P, Ha CH, Binh LT, Kasbohm J. Chemical synthesis and antibacterial activity of novel-shaped silver nanoparticles. Int Nano Lett. 2012;2:1–9. doi: 10.1186/2228-5326-2-1. [DOI] [Google Scholar]
- 20.Zielińska A, Skwarek E, Zaleska A, Gazda M, Hupka J. Preparation of silver nanoparticles with controlled particle size. Procedia Chem. 2009;1:1560–1566. doi: 10.1016/j.proche.2009.11.004. [DOI] [Google Scholar]
- 21.Rout A, Jena PK, Sahoo D, Bindhani BK (2014) Green synthesis of silver nanoparticles of different shapes and its antibacterial activity against Escherichia coli. Int J Curr Microbiol App Sci 3:374–383 https://www.ijcmas.com/vol-3-4/Anandini%20Rout,%20et%20al.pdf. Accessed 15 Aug 2017
- 22.Logaranjan K, Raiza AJ, Gopinath SCB, Chen Y, Pandian K. Shape- and size-controlled synthesis of silver nanoparticles using Aloe vera plant extract and their antimicrobial activity. Nanoscale Res Lett. 2016;11:1–9. doi: 10.1186/s11671-016-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobczak-Kupiec A, Malina D, Wzorek Z, Zimowska M. Influence of silver nitrate concentration on the properties of silver nanoparticles. IET Micro Nano Lett. 2011;6:656–660. doi: 10.1049/mnl.2011.0152. [DOI] [Google Scholar]
- 24.Sadowski Z, Maliszewska I, Grochowalska B, Polowczyk I, Kozlecki T (2008) Synthesis of silver nanoparticles using microorganisms. Mater Sci Poland 26:419–424 https://pdfs.semanticscholar.org/b2bc/87b05b455a7d00220465c6d231087e3fbfe4.pdf. Accessed 27 Aug 2017
- 25.Nanda A, Saravanan M. Biosynthesis of silver nanoparticles from Staphylococcus aureus and its antimicrobial activity against MRSA and MRSE. Nanomedicine. 2009;5:452–456. doi: 10.1016/j.nano.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Kumar CG, Mamidyala SK. Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloids Surf B. 2011;84:462–466. doi: 10.1016/j.colsurfb.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 27.Paul D, Sinha SN (2014) Extracellular synthesis of silver nanoparticles using Pseudomonas aeruginosa KUPSB12 and its antibacterial activity. Jordan J of Biol Sci 7:245–250 http://jjbs.hu.edu.jo/files/v7n4/Paper%20Number%202m.pdf. Accessed 01 Sept 2017
- 28.Kushwaha A, Singh VK, Bhartariya J, Singh P, Yasmeen K (2015) Isolation and identification of E. coli bacteria for the synthesis of silver nanoparticles: characterization of the particles and study of antibacterial activity. Eur J Exp Biol 5:65–70 http://www.imedpub.com/articles/isolation-and-identification-of-ie-coliibacteria-for-the-synthesis-of-silver-nanoparticles-characterization-of-the-particles-and.pdf. Accessed 15 Sept 2017
- 29.Malarkodi C, Rajeshkumar S, Paulkumar K, Gnanajobitha G, Vanaja M, Annadurai G (2013) Bacterial synthesis of silver nanoparticles by using optimized biomass growth of Bacillus sp. Nanosci Nanotechnol 3:26–32 https://www.researchgate.net/profile/Rajeshkumar_Shanmugam/publication/236671464_Bacterial_synthesis_of_silver_nanoparticles_by_using_optimized_biomass_growth_of_Bacillus_sp/links/00b7d518d35968b7c9000000.pdf. Accessed 09 June 2017
- 30.Patil H, Borse S, Patil D, Patil U, Patil H (2011) Synthesis of silver nanoparticles by microbial method and their characterization. Arch Phys Res 2:153–158 https://www.researchgate.net/profile/Ulhas_Patil4/publication/268384142_Synthesis_of_Silver_Nanoparticles_by_Microbial_Method_and_Their_Characterization/links/54e337230cf2de71a71e58a0.pdf. Accessed 09 June 2017
- 31.Barapatre A, Aadil KR, Jha H. Synergistic antibacterial and antibiofilm activity of silver nanoparticles biosynthesized by lignin-degrading fungus. Bioresour Bioprocess. 2016;3:8. doi: 10.1186/s40643-016-0083-y. [DOI] [Google Scholar]
- 32.Phanjom P, Ahmed G. Biosynthesis of silver nanoparticles by Aspergillus oryzae (MTCC no. 1846) and its characterizations. J Nanosci Nanotechnol. 2015;5:14–21. [Google Scholar]
- 33.Devika R, Elumalai S, Manikandan E, Eswaramoorthy D. Biosynthesis of silver nanoparticles using the fungus Pleurotus ostreatus and their antibacterial activity. Sci Rep. 2012;1:1–5. [Google Scholar]
- 34.Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomedicine. 2010;6:103–109. doi: 10.1016/j.nano.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Rajeshkumar S, Malarkodi C. In vitro antibacterial activity and mechanism of silver nanoparticles against foodborne pathogens. Bioinorg Chem Appl. 2014;2014:1–10. doi: 10.1155/2014/581890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AbdelRahim K, Mahmoud SY, Ali AM, Almaary KS, Mustafa AE-ZM, Husseiny SM. Extracellular biosynthesis of silver nanoparticles using Rhizopus stolonifer. Saudi J Biol Sci. 2017;24:208–216. doi: 10.1016/j.sjbs.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey S, Thakur M, Shah R, Oza G, Mewada A, Sharon M. A comparative study of economical separation and aggregation properties of biologically capped and thiol functionalized gold nanoparticles: selecting the eco-friendly trojan horses for biological applications. Colloids Surf B. 2013;109:25–31. doi: 10.1016/j.colsurfb.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Okafor F, Janen A, Kukhtareva T, Edwards V, Curley M. Green synthesis of silver nanoparticles, their characterization, application and antibacterial activity. Int J Environ Res Publ health. 2013;10:5221–5238. doi: 10.3390/ijerph10105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cicek S, Gungor AA, Adiguzel A, Nadaroglu H. Biochemical evaluation and green synthesis of nano silver using peroxidase from Euphorbia (Euphorbia amygdaloides) and its antibacterial activity. J Chem. 2015;2015:1–7. doi: 10.1155/2015/486948. [DOI] [Google Scholar]
- 40.Song H, Ko K, Oh I, Lee B (2006) Fabrication of silver nanoparticles and their antimicrobial mechanisms. Eur Cells Mater 11:58 http://thesilveredge.com/pdf/Fabrication%20of%Silver%20Nanoparticles%20and%20Their%20Antimicrobial%20Mechanisms.pdf. Accessed 15 September 2017
- 41.Long Y-M, Hu L-G, Yan X-T, Zhao X-C, Zhou Q-F, Cai Y, Jiang G-B. Surface ligand controls silver ion release of nanosilver and its antibacterial activity against Escherichia coli. Int J Nanomedicine. 2017;12:3193. doi: 10.2147/IJN.S132327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.AshaRani P, Low Kah Mun G, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2008;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 43.Liao J, Mo AC, Wu HK, Zhang JC, Li YB, Lv GY. Antibacterial activity of silver-hydroxyapatite/titania nanoparticles on oral bacteria. Key Eng Mater. 2007;330–332:299–302. doi: 10.4028/www.scientific.net/KEM.330-332.299. [DOI] [Google Scholar]