Abstract
Type III polyketide synthases (PKSs) biosynthesize varied classes of metabolites with diverse bio-functionalities. Inherent promiscuous substrate specificity, multiple elongations of reaction intermediates and several modes of ring-closure, confer the proteins with the ability to generate unique scaffolds from limited substrate pools. Structural studies have identified crucial amino acid residues that dictate type III PKS functioning, though cyclization specific residues need further investigation. PKSIIINc, a functionally and structurally characterized type III PKS from the fungus, Neurospora crassa, is known to biosynthesize alkyl-resorcinol, alkyl-triketide- and alkyl-tetraketide-α-pyrone products. In this study, we attempted to identify residue positions governing cyclization specificity in PKSIIINc through comparative structural analysis. Structural comparisons with other type III PKSs revealed a motif with conserved hydroxyl/thiol groups that could dictate PKSIIINc catalysis. Site-directed mutagenesis of Cys120 and Ser186 to Ser and Cys, respectively, altered product profiles of mutant proteins. While both C120S and S186C proteins retained wild-type PKSIIINc product activity, S186C favoured lactonization and yielded higher amounts of the α-pyrone products. Notably, C120S gained new cyclization capability and biosynthesized acyl-phloroglucinol in addition to wild-type PKSIIINc products. Generation of alkyl-resorcinol and acyl-phloroglucinol by a single protein is a unique observation in fungal type III PKS family. Mutation of Cys120 to bulky Phe side-chain abrogated formation of tetraketide products and adversely affected overall protein stability as revealed by molecular dynamics simulation studies. Our investigations identify residue positions governing cyclization programming in PKSIIINc protein and provide insights on how subtle variations in protein cores dictate product profiles in type III PKS family.
Electronic supplementary material
The online version of this article (10.1007/s12088-018-0738-9) contains supplementary material, which is available to authorized users.
Keywords: Type III polyketide synthase, Neurospora crassa, PKSIIINc, Alkyl-resorcinol, Acyl-phloroglucinol
Introduction
Fungal genomes have revealed numerous polyketide synthase (pks) genes in several species [1–9]. Architecturally, PKSs are categorized into; type I modular/iterative PKSs [10–14] with several catalytic functions on a single polypeptide chain, type II PKSs [15, 16] with catalytic activities distributed on separate polypeptides and small homodimeric type III PKSs [17–21]. Type III PKSs display substrate promiscuity and catalyze repetitive decarboxylative condensations of a mono-carboxyl-coenzymeA (CoA) starter unit with di-carboxyl-CoA extender substrate. Repeated condensations elongate poly-β-keto linear intermediates that undergo enzyme-aided ring-closure through three distinct modes of cyclization [17]. Structural studies on type III PKSs have revealed residues that determine substrate preferences and number of chain length extensions [18]. However, efforts are on to delineate mechanisms that dictate intrinsic cyclization programming in these proteins [17]. Type III PKSs produce variety of molecules such as pyrones, chalcones, stilbenes, phloroglucinols, resorcinols, resorcinolic acids, acridones, quinones, etc. [22–25]. While C6–C1Claisen condensation of reaction intermediate generates chalcones and phloroglucinols, a C2–C7 aldol condensation produces stilbenes and resorcinols with their distinct biological activities. Type III PKSs in plants, bacteria, fungi, and algae have been implicated in several physiological functions like pigmentation [26], signaling [17], salinity resistance [27], stress adaptation [28], dehydration resistance [29], anti-viral agents [30], plant waxes [31] and cell wall remodeling factors [32]. Since first crystallographic study of CHS protein from Medicago sativa, several crystal structures and site-directed mutants have identified functionally and structurally crucial residues in type III PKS superfamily. These investigations highlight importance of conservation of physico-chemically equivalent amino acids and identify residue positions and motifs involved in substrate selection and/or preference, substrate binding, elongation of reaction intermediate and cyclization specificity [33–35]. In an extensive mutagenic investigation, CHS protein was engineered to 18xCHS mutant that functionally mimicked STS protein producing stilbene instead of chalcone [36]. Mutation at Thr197 position to Cys side-chain in CHS abolished chalcone production [18]. A G284W mutant of ArsC protein in Azotobacter, gained aldol condensation activity and produced alkyl-resorcinol products [37]. These studies suggest that steric interactions of amino acids at crucial residue positions control transition between lactonization, aldol and Claisen condensation chemistries. In this study, we attempted to investigate effects of substitutions at crucial residue positions, on cyclization specificity in PKSIIINc, a type III PKS from Neurospora crassa. PKSIIINc protein has been previously characterized to biosynthesize long-chain alkyl-resorcinols, alkyl-triketide- and alkyl-tetraketide-α-pyrone products. Three-dimensional crystallographic analyses and mutational studies established coordinated functioning of cyclization and substrate-binding pockets in the core of PKSIIINc protein [38, 39]. Our homology-based comparative sequence and structural analyses identified catalytically important amino acid motif and positions that could govern PKSIIINc functioning. Systematic site-directed mutagenesis and biochemical analyses of Cys120 and Ser186 residues corroborated our in silico findings and revealed altered product profile of mutant proteins. Active site comparisons and molecular dynamic simulations provided clues to the effect of amino acid substitutions on functionally important catalytic pockets and on overall stability of mutant proteins. Our analyses identify mechanistically crucial residue positions in PKSIIINc protein and provide new insights into the fundamental programming in type III PKS family of proteins.
Materials and Methods
Bacterial Strains and Materials
Escherichia coli XL-1 blue and BL21/(DE3) strains were used as cloning and expression strains, respectively. pET28c-PKSIIINc recombinant plasmid used as template for site-directed mutagenesis was kindly provided by Dr. Rajesh S. Gokhale (NII, India). Acyl-CoA substrates were purchased from Sigma. LB broth and LB agar were bought from Himedia. Ni2+–NTA was purchased from Qiagen. Analytical and HPLC grade solvent was purchased from Himedia and Merck. QuikChange® II Site-Directed Mutagenesis Kit was bought from Stratagene (La Jolla, CA).
Generation of PKSIIINc Mutants
Site-directed mutants were generated using QuikChange® II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The pET28c-PKSIIINc recombinant plasmid was used as template to generate C120S, C120F and S186C mutants. Details of the mutagenesis primers are given in Supplementary Table S1. Mutants were confirmed by DNA sequencing.
Expression and Purification of Wild-Type PKSIIINc and Mutant Proteins
Conditions used for expression and purification of wild-type protein were same as used for mutant proteins. Briefly, the pET28c-PKSIIINc, pET28c-PKSIIINc/C120S, pET28c-PKSIIINc/C120F and pET28c-PKSIIINc/S186C expression plasmids were transformed in Escherichia coli BL21/(DE3) strain and single colony was used to inoculate 10 ml of LB containing 100 μg ampicillin mL−1. The culture was incubated at 37 °C overnight and used to inoculate 1L LB containing 100 μg ampicillin mL−1 and grown at 30 °C until OD reached 0.5 at 600 nm. Protein expression was induced at 22 °C for 16 h by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation at 4000 rpm for 30 min and re-suspended in lysis buffer (50 mM Tris pH 8.0, 10% glycerol, 0.15 M NaCl). Cell disruption was carried out with 8 cycles of sonication at 30% amplitude and cell debris was removed by centrifugation (30 min, 13000 rpm). All proteins were purified by Ni2+−NTA affinity chromatography. 1 ml of Ni2+−NTA slurry per liter of culture was used and incubated at 4 °C for 1 h. Proteins were eluted using elution buffer (5/10/25/50/100/250 mM imidazole 50 Mm Tris pH 8.0, 10% glycerol, 0.15 M NaCl). Eluted protein fractions were analyzed on SDS-PAGE and appropriate fractions were pooled, estimated for protein concentration and used for in vitro product formation assays.
Cell-Free Enzymatic Assays and Product Characterization
A standard reaction mixture containing 100 mM starter substrate (C14/C18 acyl-CoA), 100 mM malonyl-CoA extender unit and 5 μg of purified wild-type/mutant protein was incubated at 30 °C for 60 min and reactions were quenched with 5% acetic acid. Products were extracted with 2 × 300 μl of ethyl acetate and dried under vacuum. Products were then dissolved in methanol and further resolved on C5-reverse phase-ultra fast liquid chromatography (UFLC) column using a 30–100% binary gradient of solvent B (99%CH3CN + 1% acetic acid) and solvent A (water + 1% acetic acid) for 60 min. Resolved products were detected at 280 nm using PDA detector in UFLC. UFLC resolved products were further characterized by high-resolution mass spectrometry (HRMS) using Sciex Triple-Time of Flight (TOF) 5600 mass spectrometer.
Homology-Based Comparative Studies
X-ray crystal structures of PKSIIINc (3E1H) [38], Esi-PKS1 (4B0N) [27], PKS18 (1TED) [40], 2-PS (1QLV) [35], CHS (1BQ6) [41], STS (1U0U) [36], 18 × CHS mutant (1U0V) [36] downloaded from Protein Data Bank (http://rcsb.org/pdb) and homology model of ArsB [42] generated using 72% identical 3VS9_A structure as template in Biovia Discovery Studio 4.5 were used for homology-based sequence and structural analyses. Chain A of PKSIIINc structure, 3E1H, was used as template for in silico substitutions to generate C120S, C120F and S186C in silico mutants for analyses in Biovia Discovery Studio 4.5 (Supplementary Table S2). The highest scoring predictions for the in silico mutations were used for further analyses.
Molecular Dynamics Simulations
Crystal structure of PKSIIINc (3E1H) and the in silico substituted C120S, C120F and S186C mutant proteins were used for molecular dynamics (MD) simulations in Discovery Studio 4.5 under CHARMM 36 force field. All proteins were solvated using solvation protocol with default parameters. The “standard dynamics cascade” protocol was utilized where equilibration was run for 20 ps and production was run for 100 ps. All other parameters were set to defaults (target temperature = 300 K; time step = 1 fs). The root-mean-square deviation (RMSD) was evaluated by “analyze trajectory” protocol.
Results
Homology-Based Sequence/Structure Comparisons of Type III Polyketide Synthases
Fungal PKSIIINc protein shares ~ 26% identity with plant and bacterial type III PKSs and biosynthesizes long-chain alkyl-resorcinols and alkyl-triketide- and alkyl-tetraketide α-pyrone products. Crystal structure of PKSIIINc has revealed conserved substrate-binding and cyclization pockets that work in coordination to determine cyclization specificity during PKSIIINc catalysis [38]. We attempted to identify additional amino acid positions in the catalytic pockets that could influence cyclization specificity in PKSIIINc protein. Multiple sequence alignment (MSA) was made for several functionally characterized type III PKSs with diverse product profiles. MSA of primary sequence of PKSIIINc (3E1H) [38], Esi-PKS1 (4B0 N) [27], PKS18 (1TED) [40], 2-PS (1QLV) [35], CHS (1BQ6) [41], STS (1U0U) [36], 18 × CHS mutant (1U0 V) [36] and ArsB [42] respectively, from N. crassa, E. siliculosus, M. tuberculosis, G. hybrida, M. sativa, P. sylvestris, P. sylvestris, and A. vinelandii can be found in Supplementary Fig. S1. Close examination of MSA of substrate-binding and cyclization pockets revealed a short motif of three contiguous residue positions that are primarily occupied by amino acids with functional hydroxyl or thiol group in each analyzed sequence. The motif in PKSIIINc corresponds to Thr119-Cys120-Thr121. While first and third motif positions are occupied by either serine or threonine (barring exceptional occurrence of alanine at third position in 2-PS), the second position shows conservation of threonine in all plant sequences and serine in microbial PKS18 and Esi-PKS1 proteins. Second position diverges to cysteine in PKSIIINc and ArsB proteins that exhibit similarity in substrate and cyclization specificities, and in product profiles. We probed possibility of involvement of second motif position in influencing product profiles of type III PKSs by superimposing crystal structures of the above eight proteins and observing spatial orientation of side-chain at this position in each structure. Based on cyclization chemistry, structures of proteins with similar mode of ring-folding were superimposed and the three-residue motif was observed for side-chain variations at second motif position. Structural superimpositions were carried out for; Esi-PKS1 and CHS proteins with Claisen condensation specificity (Fig. 1a); PKS18 and 2-PS proteins with lactonization specificity (Fig. 1b); and, STS and 18xCHS mutant plant proteins (Fig. 1c) and PKSIIINc and ArsB microbial proteins (Fig. 1d) with aldol condensation specificity. Interestingly, orientation of side-chain at second motif position differed with each cyclization chemistry and proteins with analogous specificities exhibited similar orientation of the side-chain. This analysis suggested that functional group and side-chain orientation of amino acid at second position in the three-residue motif facilitates ring-closure and determines product profile in type III PKSs. We speculated that structural orientation of thiol group at Cys120 position in PKSIIINc would be important in determining cyclization specificity of the protein. Thr135 at corresponding position in STS, hydrogen-bonds with a structured water molecule and facilitates aldol condensation [36]. However, STS mechanism of ring-folding has not been reported in structures of other aldol condensing type III PKSs. Cys106 at an equivalent position aids cyclization chemistry in THNS from S. erythraea [43].
Fig. 1.
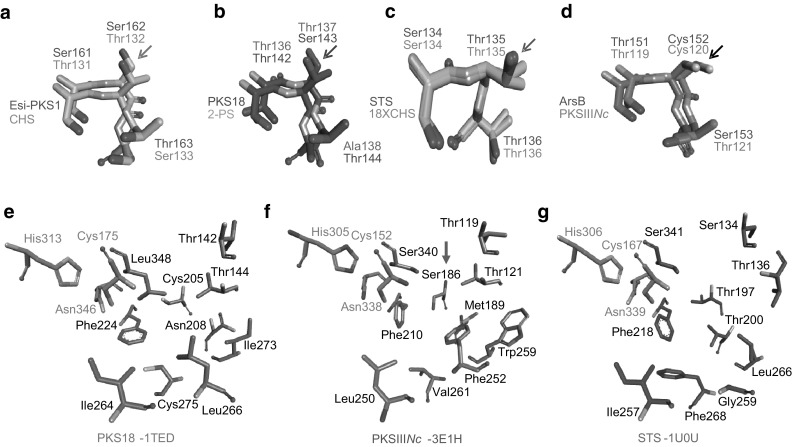
Superimposed structures of type III PKSs and analyses of conserved residues. Superimposition of structures of functionally equivalent a Esi-PKS1 and CHS, b PKS18 and 2-PS, c STS and 18xCHS mutant, and d PKSIIINc and ArsB, reveals a three-residue motif with conserved hydroxyl/thiol groups. A correlation can be observed between cyclization specificity of protein and spatial orientation of side-chain at second motif position (marked with an arrow in each panel). Structural comparison of active site architecture of e PKS18, f PKSIIINc, and g STS revealed conservation of hydroxyl/thiol group at Ser186 position (marked with an arrow) in PKSIIINc. Analogous positions in PKS18 and STS are Cys205 and Thr197, respectively
Structural comparison of active site cavities in PKSIIINc with cavities in functionally related aldol condensing STS and lactonization specific PKS18 revealed a structurally conserved position occupied by Ser186 in PKSIIINc (Fig. 1e–g). While the hydroxyl side-chain at this position is conserved in STS as Thr197, in PKS18 this position is occupied by a thiol as Cys205. This position in PKS18 and corresponding Thr197 residue in STS have been previously implicated in determining substrate specificity. Ser186 to phenylalanine substitution in PKSIIINc has previously shown to abrogate aldol condensation while retaining lactonization specificity. To probe the importance of conservation of hydroxyl/thiol functional groups at key positions in the protein core, we generated C120S, C120F and S186C site-directed mutants of PKSIIINc protein.
Biochemical Analysis of PKSIIINc Mutants
Systematic site-directed mutagenesis was performed to generate three mutant proteins of PKSIIINc, C120S, C120F and S186C. Purified C120S and C120F proteins were independently assayed for product formation using myristoyl-CoA (C14-CoA) as starter and malonyl-CoA as extender substrates. Figure 2a displays UFLC chromatogram of separated reaction products. As can be noted, reactions with mutant proteins resulted in altered peak profile as compared to product profile for wild-type PKSIIINc (wt-PKSIIINc). While C120F could not synthesize products with myristoyl-CoA, chromatographs for wt-PKSIIINc and C120S revealed two peaks, 1 and 2 at 280 nm. High-resolution mass spectrometric (HRMS) analysis revealed molecular ions of [M–H]− at m/z 293.2127 and m/z 335.2225 that could correspond to myristoyl-triketide α-pyrone (14a) (Fig. 2b) and myristoyl-tetraketide α-pyrone (14b) (Fig. 2c), respectively. Tandem MS/MS analysis confirmed identity of the molecules. Mutant proteins when assayed for product formation with stearoyl-CoA (C18–CoA) as starter substrate revealed additional chromatographic peaks (Fig. 2d). HRMS analysis of peak 2 from chromatogram of reaction products of C120F protein revealed existence of stearoyl-triketide α-pyrone (18a) with [M–H]− at m/z 349.2766 as the sole catalytic product (Fig. 2e). C120S protein, in contrast, displayed a peak profile similar to product profile for wt-PKSIIINc. A tandem MS/MS analysis of molecular ions from each peak confirmed existence of stearoyl-triketide α-pyrone (18a) in the five product peaks 1–5, obtained for both wt-PKSIIINc and C120S catalyzed reactions (Supplementary Fig. S2, S3). In addition to 18a, both the proteins catalyzed biosynthesis of tetraketide products (Fig. 2f,g, Supplementary Fig. S3), stearoyl-tetraketide α-pyrone (18b) with [M–H]− at m/z 391.2865 in peaks 1 and 2, and stearoyl-resorcinol (18c) with [M–H]− at m/z 347.2971 in peaks 3–5, of the chromatograms. Mass spectrometric identification of similar molecules has been previously reported for reaction products of PKS18 [34], PKSIIINc [30] and ArsB [35] proteins from M. tuberculosis, N. crassa and A. vinelandii, respectively. HRMS analysis revealed an additional ion of [M–H]− at m/z 391.2874 in peak 3 of chromatogram for C120S. While this molecular mass could be expected for ion of 18b, tandem MS/MS analysis, to our surprise, confirmed the product as stearoyl-phloroglucinol (18d) (Fig. 2h). MS/MS analysis of acyl-phloroglucinols has been previously reported for products of RppA from S. griseus [44]. The ability of C120S mutant to produce acyl-phloroglucinol in addition to alkyl-resorcinol has never been reported earlier for wt-PKSIIINc and is a unique observation in the family of fungal type III PKSs. A previous investigation reported C120S mutant of PKSIIINc to exhibit wild-type product profile albeit with diminished production of alkyl-resorcinol [45]. The study however failed to identify acyl-phloroglucinol product. Our homology-based structural comparisons in Fig. 1a–d revealed a correlation between cyclization specificity and orientation of side-chain at second motif position in type III PKSs. In an attempt to delineate structural basis for the unique catalytic potential of C120S mutant, we replaced Cys120 with Ser in PKSIIINc structure to generate an in silico C120S mutant. The in silico C120S mutant was compared with related structures previously aligned in Fig. 1a–d. Figure 2i shows structural overlay of the three-residue motif as present in in silico C120S mutant and the other proteins. Notably, side-chain of Ser120 in the mutant protein displayed a unique orientation and did not align with any of the compared orthologous chains. The new orientation of Ser120 side-chain upon switch of native thiol group to hydroxyl functionality at this position might be leading to new interactions with growing polyketide intermediates, and facilitating the additional Claisen condensation chemistry for ring-closure. This analysis provided clues to structural basis for unusual cyclization potential of C120S mutant of PKSIIINc, and substantiated the proposed importance of spatial positioning of side-chain at second motif position in dictating cyclization chemistry and product profile in type III PKSs.
Fig. 2.
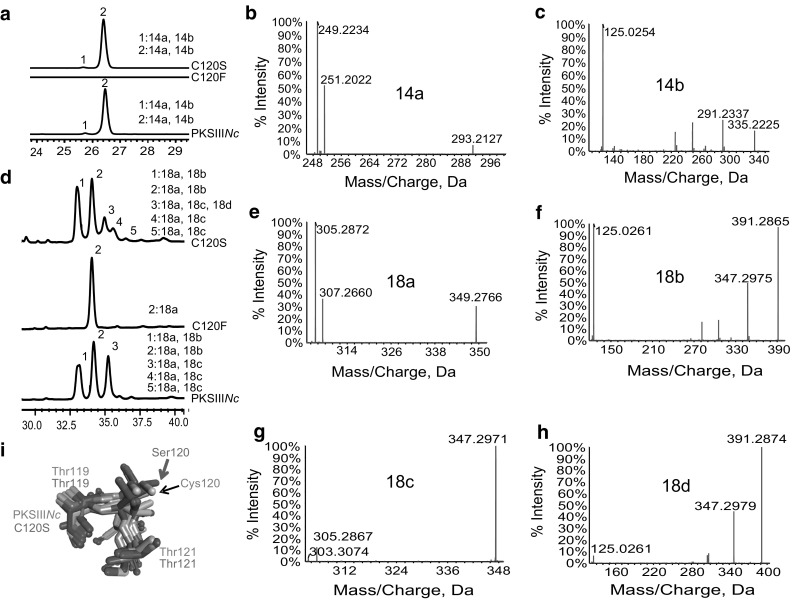
Characterization of C120S and C120F mutants of PKSIIINc. a UFLC profile of products from myristoyl-CoA primed reactions. Tandem MS/MS of C120S myristoyl-CoA primed products confirmed presence of b myristoyl-triketide α-pyrone (14a), and c myristoyl-tetraketide α-pyrone (14b). d UFLC profile of products from stearoyl-CoA primed reactions. MS/MS analysis of C120F product revealed e stearoyl-triketide α-pyrone (18a). MS/MS fragmentation analysis of C120S products with stearoyl-CoA confirmed presence of f stearoyl-tetraketide α-pyrone (18b), g stearoyl-resorcinol (18c), and h stearoyl-phloroglucinol (18d). i Structural superimposition of three-residue motif in Esi-PKS1, CHS, PKS18, 2-PS, STS, 18xCHS mutant, PKSIIINc, ArsB model and in silico C120S mutant of PKSIIINc reveals unique side-chain orientation of Ser120 in the mutant. Spatial orientations of Cys120 side-chain in PKSIIINc and Ser120 side-chain in C120S mutant are marked with arrows
Functional assays of S186C with myristoyl-CoA (Fig. 3a–c) or stearoyl-CoA (Fig. 3d–f, Supplementary Fig. S4) as starter and malonyl-CoA as extender substrates resulted in a product profile similar to that observed for wt-PKSIIINc. S186C though favored lactonization chemistry of ring-closure, resulting in increased biosynthesis of alkyl-α-pyrone products 14a, 14b, 18a and 18b. Previous substitution of Ser186 with bulky Phe residue in PKSIIINc was reported to abolish alkyl-resorcinol formation [38]. Our functional studies on mutant proteins, unambiguously establish Cys120 and Ser186 residues to play decisive roles in dictating cyclization specificity in PKSIIINc protein.
Fig. 3.
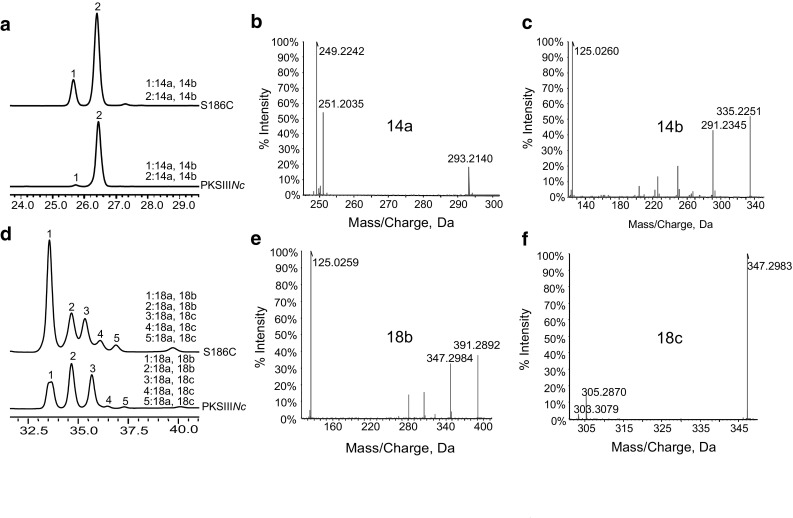
Biochemical analysis of S186C mutant of PKSIIINc. a UFLC profile of products from myristoyl-CoA primed reactions. Tandem MS/MS of S186C products from myristoyl-CoA revealed b myristoyl-triketide α-pyrone (14a), and c myristoyl-tetraketide α-pyrone (14b). d UFLC profile of products from stearoyl-CoA primed reactions. MS/MS analysis confirmed S186C catalyzed formation of products e stearoyl-tetraketide α-pyrone (18b) and f stearoyl-resorcinol (18c)
Architecture and Stability of Mutant Proteins
Our in vitro characterization of PKSIIINc mutants revealed that C120F protein could not accept myristoyl-CoA as a starter substrate. This mutant was also defective in chain elongation beyond two rounds of condensations of C18 starter with malonyl-CoA. Structurally, Cys120 in PKSIIINc occupies a position close to catalytic Cys152 in the active site pocket. We speculated that replacement of Cys120 with bulky Phe residue alters the active site architecture and affects protein catalysis. To understand effect of mutations on the active site architecture we independently mutated Cys120 to Phe/Ser, and Ser186 to Cys in PKSIIINc structure and compared the changes in active site cavity volume with each mutation. wt-PKSIIINc displayed a cavity volume of 524 Å3 (Fig. 4a). While C120S mutation lead to negligible increase in volume (Fig. 4b), there was substantial reduction with C120F mutation (Fig. 4c). Decrease in cavity volume due to bulky Phe120 at a position near catalytic Cys152 might account for the inability of C120F mutant to elongate polyketide intermediates and form tetraketide products. Mutation of Ser186 to Cys led to a small decrease in cavity volume (Fig. 4d). Replacing hydroxyl with thiol group at 186 residue position in PKSIIINc probably leads to stabilization of the intermediate in an orientation necessary for facile lactonization reaction while retaining aldol condensation mechanism of ring-closure. In order to probe effect of each independent mutation on overall stability, molecular dynamics (MD) simulations were performed for the wild-type and mutant proteins. Root mean square deviation (RMSD) was monitored to assess structural stability and plotted against time in Fig. 4e. Average RMSD values for C120S and S186C mutants were 0.909646 and 0.95665 Å2, respectively and were similar to RMSD value of 0.92255 Å2 for wt-PKSIIINc. In contrast, average RMSD of C120F mutant was 1.12497 Å2. wt-PKSIIINc attained equilibrium state at 30 ps, whereas, C120F showed steep peaks at 50 ps relating to instability of the mutant protein. Our MD simulation studies evidenced the importance of protein stability in dictating enzyme catalysis. We concluded that inability of C120F mutant to biosynthesize tetraketide products stems from the architectural differences in the catalytic pocket and overall structural instability of the mutant.
Fig. 4.
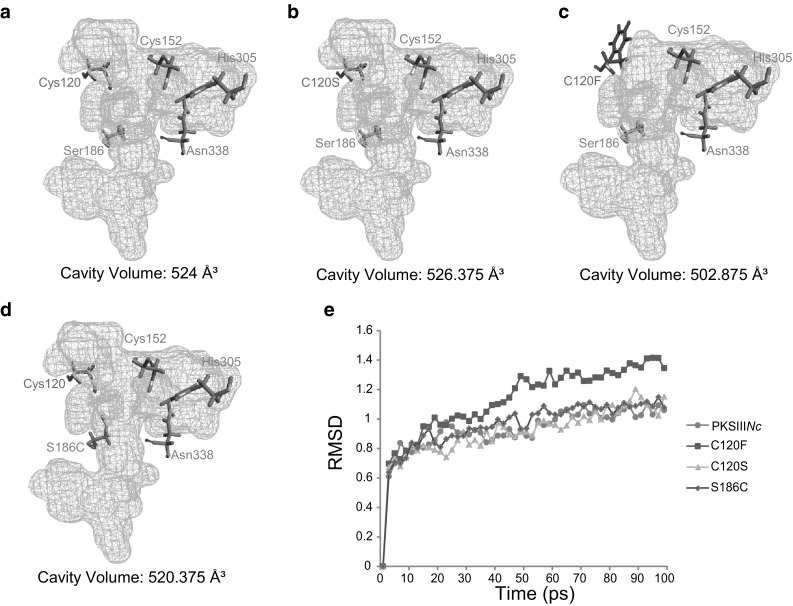
In silico active site analysis and molecular dynamics (MD) simulation studies. a wild-type PKSIIINc, b C120S in silico mutant, c C120F in silico mutant and d S186C in silico mutant were probed for differences in active site cavity volume. Cavity volumes are mentioned in Å3. e RMSD analysis of MD simulations for wild-type PKSIIINc and mutant proteins
Discussion
Structural characterization of several members of type III polyketide synthase (PKS) family has unraveled a plethora of information on structural features that dictate catalytic intricacies and mechanistic variations in these proteins. These studies reveal conserved αβαβα-fold architecture with buried substrate-binding, catalytic and elongation pockets in the protein structures. Little information, however, is available on the structural-basis and programming of ring-folding of reaction intermediates, and release of products. The study here, deals with delineation of residue positions that could play important roles in cyclization mechanisms of PKSIIINc protein from Neurospora crassa. Previous structural and mutational analyses of the protein had identified an inter-link between cyclization and substrate-binding pockets that facilitated their coordinated functioning in the core of the protein [38]. Our homology-based sequence analysis of structurally characterized type III PKSs, revealed a three-residue motif in all proteins with conservation of hydroxyl/thiol groups at the three positions in the motif. Hydroxyl/thiol functional groups, placed at crucial amino acid positions in the active sites have been earlier implicated to play important roles in repetitive elongation of the intermediates, positioning of intermediates in the catalytic pocket, and condensation reactions for specific ring-closures [18, 36, 43]. In silico comparative structural analysis of the motif, as present in structures of functionally equivalent proteins, revealed a correlation between cyclization specificity of the protein and spatial orientation of side-chain at second position in the motif. Site-directed mutagenesis of Cys120, occupying the second motif position in PKSIIINc, to a Ser amino acid residue revealed interesting results. C120S mutant exhibited unique mechanistic potential and biosynthesized acyl-phloroglucinol product in addition to alkyl-resorcinol, alkyl triketide- and alkyl tetraketide- α-pyrone products of wt-PKSIIINc protein. The tetraketide products, alkyl-tetraketide α-pyrone, alkyl-resorcinol and acyl-phloroglucinol are generated from a common intermediate that folds through three distinct cyclization chemistries. While a C2–C7 aldol condensation is responsible for production of alkyl-resorcinol, and C6–C1 Claisen condensation leads to acyl-phloroglucinol formation, alkyl-tetraketide α-pyrone is a product of lactonization of the tetraketide intermediate. It is interesting to note that lactonization reaction commonly accompanies aldol/Claisen chemistries of ring-closure, however, both aldol condensation and Claisen condensation in the same catalytic pocket is a unique observation, not known in fungal type III PKS family. A switch from thiol to hydroxyl side-chain at second position in the three-residue motif in PKSIIINc conferred unique mechanistic potentials and corroborated our predictions of involvement of this residue position in determining cyclization specificity in the protein. Mutation of Cys120 to bulky Phe residue abrogated formation of tetraketide products and resulted in alkyl-triketide α-pyrone as the sole catalytic product, specifically with the long-chain acyl-CoA. Structurally, Cys120 is positioned close to catalytic Cys152 residue in the active site of PKSIIINc. Comparative structural analysis revealed reduction in volume of active site cavity in C120F mutant as compared to cavity volume in wt-PKSIIINc. Time-dependent molecular dynamic (MD) simulations revealed inability of C120F mutant in attaining stable wild-type PKSIIINc conformation. Cys120 to bulky Phe side-chain in catalytic pocket of C120F leads to structural instability affecting overall protein catalysis. Structural comparisons of active site lining residues identified a conserved Ser186 amino acid in PKSIIINc placed at the juncture of substrate-binding and cyclization pockets. The hydroxyl at this position was replaced with thiol functional group by generating S186C mutant of PKSIIINc. Biochemical analysis of the mutant revealed a product profile similar to wild-type protein, albeit with a preference for lactonization chemistry of ring-closure of tetraketide intermediate. Substitution of Ser186 with Phe in PKSIIINc has been previously reported to abrogate aldol condensation specificity. Our investigations establish involvement of Cys120 and Ser186 in cyclization specificity of PKSIIINc and highlight the importance of spatial positioning of crucial amino acid side-chains in determining product profile in type III PKSs. The ability to confer additional cyclization capability and generate new polyketide scaffold by subtle variation of crucial amino acid in the protein core, opens possibilities of engineering the fungal type III PKS, PKSIIINc to generate novel type III polyketides with distinct chemical architectures and diverse biological activities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
P.S. gratefully acknowledges Innovative Young Biotechnologist Award (IYBA) of Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India and South Asian University (SAU) Startup Research Grant, SAU, India for providing funds and support for this study. A.P. acknowledges CSIR for Senior Research Fellowship. We are thankful to Dr. Rajesh S. Gokhale (NII, India) for kindly providing the pET28c-PKSIIINc recombinant expression plasmid. We thank Sciex, Gurugram, India for Mass Spectrometry data collection.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12088-018-0738-9) contains supplementary material, which is available to authorized users.
Amreesh Parvez and Samir Giri these authors contributed equally to this work.
References
- 1.Bond C, Tang Y, Li L. Saccharomyces cerevisiae as a tool for mining, studying and engineering fungal polyketide synthases. Fungal Genet Biol. 2016;89:52–61. doi: 10.1016/j.fgb.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakrabortti A, Li J, Liang Z-X. Complete genome sequence of the filamentous fungus Aspergillus westerdijkiae reveals the putative biosynthetic gene cluster of ochratoxin A. Genome Announcements. 2016;4:e00982-00916. doi: 10.1128/genomeA.00982-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Lee TA, Medema MH. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet Biol. 2016;89:29–36. doi: 10.1016/j.fgb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Noar RD, Daub ME. Bioinformatics prediction of polyketide synthase gene clusters from Mycosphaerella fijiensis. PLoS One. 2016;11:e0158471. doi: 10.1371/journal.pone.0158471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma KK. Fungal genome sequencing: basic biology to biotechnology. Crit Rev Biotechnol. 2016;36:743–759. doi: 10.3109/07388551.2015.1015959. [DOI] [PubMed] [Google Scholar]
- 6.Aylward J, Steenkamp ET, Dreyer LL, Roets F, Wingfield BD, Wingfield MJ. A plant pathology perspective of fungal genome sequencing. IMA fungus. 2017;8:1–45. doi: 10.5598/imafungus.2017.08.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Möller M, Stukenbrock EH. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol. 2017;15:756. doi: 10.1038/nrmicro.2017.76. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand RL, Abdel-Hameed M, Sorensen JL. Lichen biosynthetic gene clusters. Part I. Genome sequencing reveals a rich biosynthetic potential. J Nat Prod. 2018 doi: 10.1021/acs.jnatprod.7b00769. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Tang M-C, Tang S, Gao S, Soliman S, Hang L, Xu W, Ye T, Watanabe K, Tang Y. Genome mining and assembly-line biosynthesis of the UCS1025A pyrrolizidinone family of fungal alkaloids. J Am Chem Soc. 2018;140:2067–2071. doi: 10.1021/jacs.8b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Du L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl Microbiol Biotechnol. 2016;100:541–557. doi: 10.1007/s00253-015-7093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh H, Miura A, Matsui M, Arazoe T, Nishida K, Kumagai T, Arita M, Tamano K, Machida M, Shibata T. Knockout of the SREBP system increases production of the polyketide FR901512 in filamentous fungal sp. No. 14919 and lovastatin in Aspergillus terreus ATCC20542. Appl Microbiol Biotechnol. 2018;102:1393–1405. doi: 10.1007/s00253-017-8685-7. [DOI] [PubMed] [Google Scholar]
- 12.Ruocco M, Baroncelli R, Cacciola SO, Pane C, Monti MM, Firrao G, Vergara M, di San Lio GM, Vannacci G, Scala F. Polyketide synthases of Diaporthe helianthi and involvement of DhPKS1 in virulence on sunflower. BMC Genom. 2018;19:27. doi: 10.1186/s12864-017-4405-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson SA, Crossman L, Almeida EL, Margassery LM, Kennedy J, Dobson AD. Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. Isolates. Mar Drugs. 2018;16:67. doi: 10.3390/md16020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Xu M, Feng C, Hu C. Progress in fungal polyketide biosynthesis. Chin J Biotechnol. 2018;34:151–164. doi: 10.13345/j.cjb.170219. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, Ichinose K, Choi YH, van Wezel GP. Aromatic polyketide GTRI-02 is a previously unidentified product of the act gene cluster in streptomyces coelicolor A3 (2) ChemBioChem. 2017;18:1428–1434. doi: 10.1002/cbic.201700107. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Pan H-X, Tang G-L. New insights into bacterial type II polyketide biosynthesis. F1000Research. 2017 doi: 10.12688/f1000research.10466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu Y, Ogata H, Goto S. Type III polyketide synthases: functional classification and phylogenomics. ChemBioChem. 2017;18:50–65. doi: 10.1002/cbic.201600522. [DOI] [PubMed] [Google Scholar]
- 18.Lim YP, Go MK, Yew WS. Exploiting the biosynthetic potential of type III polyketide synthases. Molecules. 2016;21(6):806. doi: 10.3390/molecules21060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H-N, Liu X-Y, Gao S, Sun B, Zheng H-B, Ji M, Cheng A-X, Lou H-X. Structural and biochemical characterization of the plant type III polyketide synthases of the liverwort Marchantia paleacea. Plant Physiol Biochem. 2018;125:95–105. doi: 10.1016/j.plaphy.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 20.Kontturi J, Osama R, Deng X, Bashandy H, Albert VA, Teeri TH. Functional characterization and expression of GASCL1 and GASCL2, two anther-specific chalcone synthase like enzymes from Gerbera hybrida. Phytochemistry. 2017;134:38–45. doi: 10.1016/j.phytochem.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Xie L, Liu P, Zhu Z, Zhang S, Zhang S, Li F, Zhang H, Li G, Wei Y, Sun R. Phylogeny and expression analyses reveal important roles for plant PKS III family during the conquest of land by plants and angiosperm diversification. Front Plant Sci. 2016;7:1312. doi: 10.3389/fpls.2016.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taura F, Iijima M, Yamanaka E, Takahashi H, Kenmoku H, Saeki H, Morimoto S, Asakawa Y, Kurosaki F, Morita H. A novel class of plant type III polyketide synthase involved in orsellinic acid biosynthesis from Rhododendron dauricum. Front Plant Sci. 2016;7:1452. doi: 10.3389/fpls.2016.01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Zhuang Y, Bai Y, Bi H, Liu T, Ma Y. Biosynthesis of phlorisovalerophenone and 4-hydroxy-6-isobutyl-2-pyrone in Escherichia coli from glucose. Microb Cell Fact. 2016;15:149. doi: 10.1186/s12934-016-0549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto M, Koen T, Takahashi H, Suda C, Kitamoto K, Fujii I. Aspergillus oryzae CsyB catalyzes the condensation of two β-ketoacyl-CoAs to form 3-acetyl-4-hydroxy-6-alkyl-α-pyrone. J Biol Chem. 2014;289:19976–19984. doi: 10.1074/jbc.M114.569095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui T, Kodama T, Mori T, Tadakoshi T, Noguchi H, Abe I, Morita H. 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases. J Biol Chem. 2017;292:9117–9135. doi: 10.1074/jbc.M117.778977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS One. 2015;10:e011905. doi: 10.1371/journal.pone.0119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meslet-Cladière L, Delage L, Leroux CJ-J, Goulitquer S, Leblanc C, Creis E, Gall EA, Stiger-Pouvreau V, Czjzek M, Potin P. Structure/function analysis of a type III polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell. 2013;25:3089–3103. doi: 10.1105/tpc.113.111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sone Y, Nakamura S, Sasaki M, Hasebe F, Kim S-Y, Funa N. Identification and characterization of bacterial enzymes catalyzing the synthesis of 1, 8-dihydroxynaphthalene, a key precursor of dihydroxynaphthalene melanin, from Sorangium cellulosum. Appl Environ Microbiol: AEM. 2018 doi: 10.1128/AEM.00258-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Aslam M, Rabbi F, Vanderwel MC, Ashton NW, Suh D-Y. PpORS, an ancient type III polyketide synthase, is required for integrity of leaf cuticle and resistance to dehydration in the moss, Physcomitrella patens. Planta. 2018;247:527–541. doi: 10.1007/s00425-017-2806-5. [DOI] [PubMed] [Google Scholar]
- 30.Hou L, Huang H, Li H, Wang S, Ju J, Li W. Overexpression of a type III PKS gene affording novel violapyrones with enhanced anti-influenza A virus activity. Microb Cell Fact. 2018;17:61. doi: 10.1186/s12934-018-0908-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Wettstein-Knowles P. The polyketide components of waxes and the Cer-cqu gene cluster encoding a novel polyketide synthase, the β-diketone synthase, DKS. Plants. 2017;6:28. doi: 10.3390/plants6030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quadri LE. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit Rev Biochem Mol Biol. 2014;49:179–211. doi: 10.3109/10409238.2014.896859. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Li X, Chai T, Wang H. Outer-sphere residues influence the catalytic activity of a chalcone synthase from Polygonum cuspidatum. FEBS Open Biol. 2016;6:610–618. doi: 10.1002/2211-5463.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raja Abdul Rahman RNZ, Zakaria II, Salleh AB, Basri M. Enzymatic properties and mutational studies of chalcone synthase from physcomitrella patens. Int J Mol Sci. 2012;13:9673–9691. doi: 10.3390/ijms13089673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jez JM, Austin MB, Ferrer J-L, Bowman ME, Schröder J, Noel JP. Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol. 2000;7:919–930. doi: 10.1016/S1074-5521(00)00041-7. [DOI] [PubMed] [Google Scholar]
- 36.Austin MB, Bowman ME, Ferrer J-L, Schröder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 37.Satou R, Miyanaga A, Ozawa H, Funa N, Katsuyama Y, K-i Miyazono, Tanokura M, Ohnishi Y, Horinouchi S. Structural basis for cyclization specificity of two azotobacter type III polyketide synthases a single amino acid substitution reverses their cyclization specificity. J Biol Chem. 2013;288:34146–34157. doi: 10.1074/jbc.M113.487272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goyal A, Saxena P, Rahman A, Singh PK, Kasbekar DP, Gokhale RS, Sankaranarayanan R. Structural insights into biosynthesis of resorcinolic lipids by a type III polyketide synthase in Neurospora crassa. J Struct Biol. 2008;162:411–421. doi: 10.1016/j.jsb.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto M, Nonaka T, Fujii I. Fungal type III polyketide synthases. Nat Prod Rep. 2014;31:1306–1317. doi: 10.1039/C4NP00096J. [DOI] [PubMed] [Google Scholar]
- 40.Sankaranarayanan R, Saxena P, Marathe UB, Gokhale RS, Shanmugam VM, Rukmini R. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol. 2004;11:894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 41.Ferrer J-L, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Mol Biol. 1999;6:775. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 42.Funa N, Ozawa H, Hirata A, Horinouchi S. Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc Natl Acad Sci. 2006;103:6356–6361. doi: 10.1073/pnas.0511227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Austin MB, Izumikawa M, Bowman ME, Udwary DW, Ferrer J-L, Moore BS, Noel JP. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 44.Funa N, Ohnishi Y, Ebizuka Y, Horinouchi S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J Biol Chem. 2002;277:4628–4635. doi: 10.1074/jbc.M110357200. [DOI] [PubMed] [Google Scholar]
- 45.Rubin-Pitel SB, Zhang H, Vu T, Brunzelle JS, Zhao H, Nair SK. Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem Biol. 2008;15:1079–1090. doi: 10.1016/j.chembiol.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


