Abstract
High-capacity adenoviral vectors (HCAdVs) devoid of all coding genes are powerful tools to deliver large DNA cargos into cells. Here HCAdVs were designed to deliver a multiplexed complete CRISPR/Cas9 nuclease system or a complete pair of transcription activator-like effector nucleases (TALENs) directed against the hepatitis B virus (HBV) genome. HBV, which remains a serious global health burden, forms covalently closed circular DNA (cccDNA) as a persistent DNA species in infected cells. This cccDNA promotes the chronic carrier status, and it represents a major hurdle in the treatment of chronic HBV infection. To date, only one study demonstrated viral delivery of a CRISPR/Cas9 system and a single guide RNA (gRNA) directed against HBV by adeno-associated viral (AAV) vectors. The advancement of this study is the co-delivery of multiple gRNA expression cassettes along with the Cas9 expression cassette in one HCAdV. Treatment of HBV infection models resulted in a significant reduction of HBV antigen production and the introduction of mutations into the HBV genome. In the transduction experiments, the HBV genome, including the HBV cccDNA, was degraded by the CRISPR/Cas9 system. In contrast, the combination of two parts of a TALEN pair in one vector could not be proven to yield an active system. In conclusion, we successfully delivered the CRISPR/Cas9 system containing three gRNAs using HCAdV, and we demonstrated its antiviral effect.
Keywords: chronic HBV infection, multiplexed CRISPR/Cas9, high-capacity adenovirus, cccDNA degradation
Introduction
Advanced therapy medicinal products belong to an emerging field in medical science. The field includes gene therapy, somatic cell therapy, tissue engineering, or combinations thereof. To develop such new approaches, several issues, such as target identification, determination and optimization of adequate methods, transport and implementation of new therapies, and advanced monitoring of the effect of the complete product, have to be addressed. For gene therapy, implementing the transfer of therapeutic nucleic acids, virus-based and non-viral delivery strategies have been explored.
Adenovirus (Ad) is a widely used vector for gene transfer in gene therapy, which has been thoroughly studied regarding the structure and replication cycle.1, 2 In terms of vector usage, Ads have experienced a long-time development that is still ongoing.3, 4 From the first attempts to exploit virtual wild-type Ad, progress led to the development of the third-generation Ad vector that lacks almost all original viral DNA sequence and is termed high-capacity adenoviral (HCAdV) or helper-dependent adenoviral (HDAdV) vector.5, 6 The vector production procedure, which is also applied in this study, is established to current good manufacturing practices standard (ClinicalTrials.gov: NCT01433133 and NCT00542568).7, 8, 9 This and other advantages like the ability to transduce a large variety of dividing and nondividing cell types, lack of viral protein expression, capacity of up to 36 kb of foreign DNA, and a very low risk of insertional mutagenesis, because of an episomal status of vector genome in transduced cells, make it a very attractive delivery system.8
Attractive genetic cargos to be delivered for gene therapeutic approaches are designer nucleases like zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), both acting as a protein dimer and requiring two separate expression cassettes, or the CRISPR/Cas9 system.10 Designer nucleases as tools for precise editing of DNA are handled as a new milestone in science, and efficient delivery is an inevitable issue to be solved. Several studies exemplified successful application of Ad vectors for the delivery of designer nucleases. For instance, ZFNs were delivered by adenoviral vectors.7, 11, 12, 13 Similarly, intact TALEN sequences could be maintained as individual expression cassettes in first- and second-generation adenoviral vectors,14, 15, 16 as well as a complete TALEN pair in HCAdV.7, 17 The CRISPR/Cas9 system was packed in early generation adenoviral vectors, too, including the expression cassettes of Cas9 and guide RNA (gRNA) apart18 or in combination.19, 20, 21, 22 The latest development in the field of hepatitis B virus (HBV) gene therapy was the use of recombinant single-stranded adeno-associated viruses (ssAAVs) for co-delivery of Cas9 expression units, which was derived from Staphylococcus aureus (SaCas9) in combination with one gRNA.23
Here, the manufacturing of a multi-compound CRISPR/Cas9 construct harbored in one HCAdV was established and tested on its feasibility. The chronic HBV infection serves as an exemplary disease for the application of designer nucleases targeting multiple DNA sequences in a virus genome (Figure 1A). The HBV is an enveloped DNA virus, and the replication includes the formation of covalently closed circular DNA (cccDNA), a persistent DNA species in the nucleus of infected cells.24 The cccDNA is the major hurdle in chronic HBV infection. If the immune system fails to eliminate the virus during acute infection, the virus nests itself within the infected cells as episomal cccDNA (Figure 1B), leading to a chronic state of infection. Current therapies of chronic HBV infection target the process of viral replication or inhibit the inflammatory process to prevent continuing liver damage. Both strategies have in common that they require long-lasting medication with strict adherence to prevent reactivation of the infection.25 In this sense, a strategy to attack the persistent HBV DNA using gene-editing tools to induce mutations or even complete destruction of the HBV genome would provide a basis for advanced therapeutics to cure chronic HBV infection.
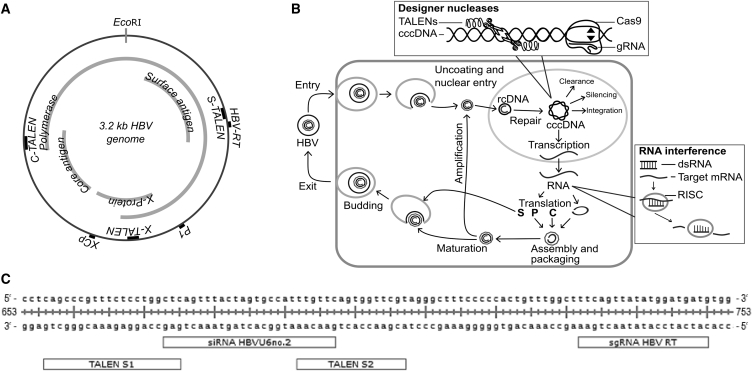
Figure 1.
Overview of HBV Target Sites and Mode of Action of Anti-HBV Strategy
(A) Schematic representation of the HBV genome with the unique EcoRI restriction enzyme site adjusted to the top, showing the location of the target sites of designer nucleases used in this study. The four viral open reading frames (ORFs) polymerase, surface antigen, X-protein, and core antigen are indicated in the inner circle. The S-/X-/C-TALEN labels on the outer circle mark the target sites designed for TALENs, which are based on No. 2, No. 5 and No. 6 RNAi target sites reported by McCaffrey et al.30 The target sites for the CRISPR/Cas9 system were adopted from Kennedy et al.26 (HBV-RT) and Lin et al.27 (P1 and XCp). (B) Illustration of the anti-HBV strategy against the background of the hepatitis B virus replication cycle. Upon entry of enveloped virions into liver cells, the capsid with the relaxed circular (rc) DNA is released into the cytoplasm and the DNA enters uncoated the nucleus. Here, the rc DNA is completed to form the covalently closed circular (ccc)DNA. The cccDNA can be eventually cleared out, silenced, or integrated into the host genome. Usually it is episomally maintained in the nucleus and is transcribed and translated by the host cell machinery. Subsequently, newly formed viral genomes are encapsulated together with the translated viral polymerase (P) through the translated capsid proteins (C). The nucleocapsid either migrates back to the nucleus to increase the pool of cccDNA or it is internalized by the endoplasmatic reticulum. In the latter process, it is enveloped with ER-membrane that already harbors translated viral surface proteins (S). Finally, it buds and is released from the cell. The cccDNA is the target for designer nucleases, which cut double-stranded DNA. The aim is to induce mutations or even degrade the cccDNA completely. RNAi acts on a later stage of replication, namely on the transcribed RNA. The RNA is marked for degradation through the host cell machinery, including the RNA-induced silencing complex (RISC), and, in this way, viral replication is shut down. (C) Enlargement of the target sites in the surface antigen ORF. TALEN-binding sites (TALEN S1 and TALEN S2), which were based on the previously published RNAi target site (HBVU6no.2) and Cas9 localization site (gRNA HBV RT), lie in close proximity and can be easily compared.
In this study, complete nuclease systems with newly conceived TALEN arrays targeting the HBV genome or published chimeric gRNA sequences for the CRISPR/Cas9 system from Kennedy et al.26 and Lin et al.27 were introduced into HCAdVs. Both elements of a TALEN pair or the CRISPR/Cas9 system, including three gRNAs expressed from the Pol-III U6 promoter and a Cas9 expression cassette, were cloned into a shuttle vector and then transferred into the HCAdV production plasmid. TALEN expression was driven from the liver-specific human alpha-1-antitrypsin (hAAT) promoter in combination with the HCR-1 (hepatic locus control region 1) of gene locus ApoE. Expression of the Cas9 endonuclease was controlled by a chicken β-actin short promoter (CBh).
Activity of nucleases was either tested on cells transiently transfected with the 1.3× HBV genome containing plasmid pTHBV2,28 in HepG2.2.15 cells with a stably integrated HBV genome,29 or in HBV-infected HepG2-sodium-taurocholate cotransporting polypeptide (NTCP) cells (C. Ko, A. Chakraborty, W.-M. Chou, J.M. Wettengel, D. Stadler, R.Bester, T. Asen, K. Zhang, J.A. McKeating, W.-S. Ryu, and U.P., unpublished data). Treated cells were evaluated in terms of viral protein production, viral DNA transcription, genome copy numbers by qPCR, and integrity by mutation detection assays.
Results
Characterization of Designer Nucleases
The TALENs used in this study were designed on the basis of RNAi target sequences published by McCaffrey and colleagues.30 The three target sites in the HBsAg-open reading frame (ORF) (S-TALEN), protein X-ORF (X-TALEN), and HBcAg-ORF (C-TALEN), respectively, were chosen based on the performance in the RNAi experiments on the one hand and on the other hand on the location far apart from each other on the HBV genome. Therefore, all HBV ORFs were affected at least once (Figure 1A). Appropriate target sites were found by applying the web-based software TAL Effector Nucleotide Targeter.31 The target sites were specifically designed for the HBV genome used in the experiments (subtype, ayw3; genotype, D3), but the conservation profile of the binding sites among the different HBV genotypes was also analyzed using representative genotypes (Figure S1). TALENs were cloned utilizing Voytas lab Golden Gate cloning method,31 and the respective expression vectors pCS2TAL3DD and pCS2TAL3RR32 were modified such as TALEN expression is driven from the hAAT liver-specific promoter. Besides TALENs, three CRISPR/Cas9 gRNA sequences were adopted from published studies by Kennedy et al.26 (HBV-RT) and Lin et al.27 (P1 and XCp), and then they were cloned into expression vectors following the Zhang lab cloning protocol.33 Furthermore, all three gRNA expression cassettes were combined into the expression vector pShV, also including the Cas9 expression cassette.34 The detailed localization of the TALEN, CRISPR/Cas9, and small interfering RNA (siRNA) target sites in the HBsAg-ORF is shown in Figure 1C. Because of the close proximity of these target sites, comparisons of efficacy in this study were mainly based on this location.
The functionality of the different constructs (Figures S2 and S3) was tested after transient co-transfection with the HBV replication-competent plasmid pTHBV2 into HEK293 cells. Induction of specific nuclease-mediated mutagenesis was detected by a mismatch-selective endonuclease assay (Figure 2A), and the estimated gene modification was calculated using the formula developed by Miller et al.35 (Table 1). The three different TALEN pairs induced mutations at a comparable rate, ranging from 5.7% induction through the TALEN pair targeting the HBcAg-ORF and 9.4% induction through the TALEN pair targeting the protein X-ORF. To conduct a comparative study to a previously published HCAdV-delivered small hairpin RNA (shRNA) coding sequence against HBV (HBVU6no.2, here shRNA-S) by Rauschhuber et al.,36 we continued the work in this study with the TALEN pair directed against the HBsAg-ORF, which achieved a targeted disruption of 7.3% of the corresponding ORF.
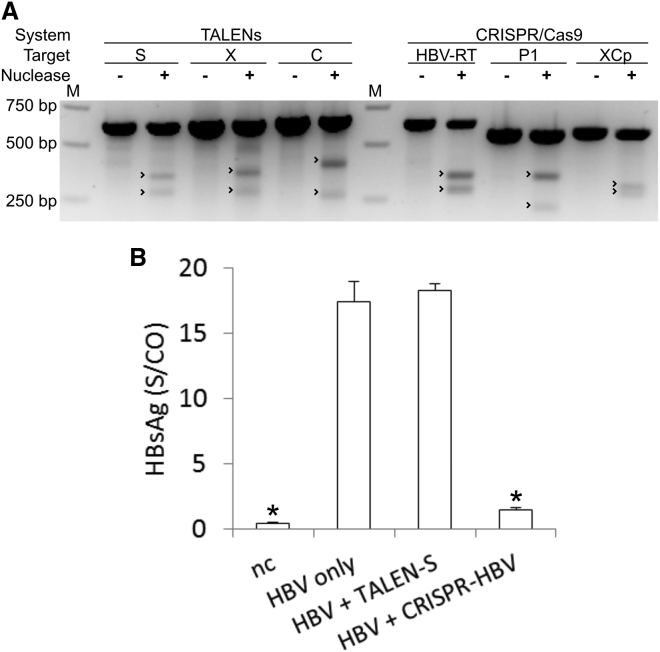
Figure 2.
Proof of Concept of Anti-HBV Efficacy of TALENs or CRISPR/Cas9
(A) HEK293 cells were co-transfected with an HBV expression plasmid pTHBV2 and either of the complete nuclease systems. DNA was isolated after 4 days and examined for mutagenesis at target sites by T7E1 assay. Undigested and digested products were separated on an agarose gel side by side for comparison. Expected cleavage product sizes are indicated by arrowheads. “M” indicates the molecular weight marker lane. (B) Huh7 cells were co-transfected with an HBV expression plasmid pTHBV2 and TALEN or CRISPR/Cas9 nuclease systems. HBsAg concentrations in the supernatant were measured after 4 days by ELISA. Data are represented as means of S/CO (sample to control ratio) values and error bars indicate SD of three replicates. Statistically significant differences to HBV only are indicated by an asterisk (*p < 0.01).
Table 1.
Genome-Editing Efficacy Estimated by T7E1 Digest and Sequence Trace Decomposition
Co-transfection of the CRISPR/Cas9 construct containing all three gRNA expression cassettes with pTHBV2 into HEK293 cells resulted in different target disruption efficiencies for the different target sites. The gRNA HBV-RT, which was directed to the coding sequence of the YYMD motif of the catalytic domain of HBV polymerase and at the same time also to the HBsAg-ORF, performed best with a targeted disruption of 14.0%. The gRNAs published by Lin et al. (P1 and XCp) were less efficient, with mutation induction percentages of 9.4% for P1 and 3.0% for XCp. The CRISPR/Cas9 genome-editing efficacy was additionally quantified by sequence trace decomposition37 (Table 1). Here, the targeted disruption percentage was confirmed for gRNA P1 with an efficiency of 9.2%. In contrast, the results for HBV-RT and XCp were about three times higher compared to the efficiencies determined by the mismatch-selective endonuclease assay. The gRNAs HBV-RT and XCp achieved 37.4% and 10.1% targeted disruption, respectively, as measured by sequence trace decomposition.
In a next step, the expression cassettes of the left and right TALEN subunits directed against the HBsAg-ORF were combined in one plasmid under the control of the liver-specific hAAT promoter.
After co-transfection of nuclease-expressing plasmids with pTHBV2 into the human liver cancer cell line Huh7, concentrations of HBsAg were measured with an enzyme immunoassay (Figure 2B). HBsAg concentrations were significantly reduced after treatment of the cells with the CRISPR/Cas9 system directed by the three gRNAs HBVRT, P1, and XCp in combination. In contrast, the TALEN pair directed against the HBsAg ORF caused no significant reduction of HBsAg concentration in the supernatant.
Characterization of HCAdV Vector Constructs
Vectors containing the triple-guide RNA construct or the double HBsAg-TALEN construct were produced using a well-established protocol9, 38 employing helper virus (HV) co-transductions for HCAdV amplification in the producer cell line 116.39 Production was finalized in purified large-scale batches of HCAdV, which were analyzed regarding concentration, purity, and vector integrity.
After purification of the HCAdV, a basic characterization was performed based on the concentration of the HCAdV by optical density (viral particles [vps]) and the level of infectious particles of HCAdV versus HV in final vector preparations quantified by qPCR (infectious units [IUs]). For the vp titer, viral DNA was extracted from the vector preparation, and the OD260 was measured and converted to number of particles per milliliter by use of the extinction coefficient of wild-type adenovirus, as determined by Maizel et al.40 The IUs were identified by virus-selective qPCRs on genomic DNA isolated from cells 3 hr post-transduction. In Table 2 the titers of different vector preparations are summarized, and parameters as particle-to-infectious unit ratio (vp:IU) for HCAdV, representing the infectivity of the HCAdV, and infectious unit-to-particle ratio (IU:vp) for HV, as an indication of HV contamination, are provided. The Food and Drug Administration has recommended that the infectivity of clinical grade Ad vector be <30:1,41 and this objective was met by the vector preparations. Furthermore, low HV contamination levels (<0.2%) could be achieved.
Table 2.
Characterization of Virus Preparations
| Total |
HCAdV |
Helper Virus |
|||
|---|---|---|---|---|---|
| vp/mL | IU/mL | vp:IU | IU/mL | IU/vp (%) | |
| HCAdV-FTC/CBh-Cas9-VAI-RT-P1-XCp | 1.20E+11 | 5.19E+09 | 23:1 | 1.44E+08 | 0.12 |
| HCAdV-FTC/hAAT-TALEN-S | 3.02E+11 | 1.07E+10 | 28:1 | 2.50E+07 | 0.01 |
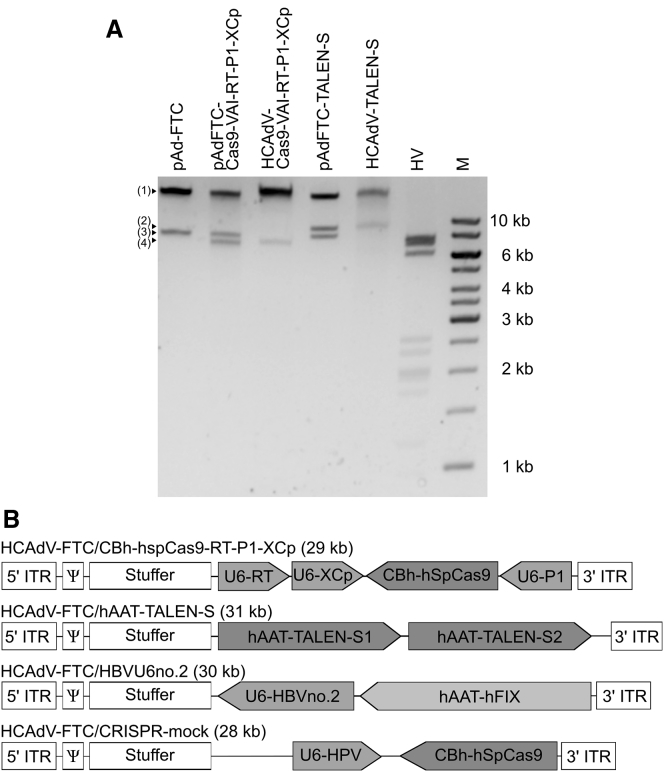
HCAdVs harboring TALEN sequences or the triple-gRNA construct were tested on genomic integrity by NotI and PspXI restriction enzyme co-digest of isolated viral DNA (Figure 3A). As controls, isolated HV DNA and isolated original empty and full vector plasmid DNA were digested and analyzed by agarose gel electrophoresis. The digested viral genomes revealed on the gel the band patterns that resembled their parental vector plasmids without the bacterial plasmid backbone. This was an indication that DNA rearrangements had been successfully avoided. In addition, the lanes containing the viral genomes clearly differed from the lane containing the digested HV genome, substantiating the titration outcomes. All vectors used in this study are summarized in Figure 3B.
Figure 3.
Characterization of Vector Genome Structures in Final Vector Preparations
(A) The results of NotI and PspXI restriction enzyme analyses of virion DNA of indicated vector preparations (HCAdV-Cas9-VAI-RT-P1-XCp and HCAdV-TALENs) and helper virus (HV) in comparison to parental plasmids (pAdFTC-Cas9-VAI-RT-P1-XCp and pAdFTC-TALENs) are shown. pAdFTC is the original plasmid containing the HCAdV genome without insert. Bands referenced with numbers on the left side of the gel represent the following: (1) the stuffer DNA, (2) the hAAT-TALEN-S insert, (3) the pAdFTC bacterial backbone, and (4) the Cas9-VAI-RT-P1-XCp insert. “M” indicates the molecular weight marker. (B) Schematic illustration of recombinant adenoviral vector genomes used in the study. HCAdV-Cas9-VAI-RT-P1-XCp, the CRISPR/Cas9 system was expressed from the CBh promoter; gRNAs RT, XCp, and P1 were expressed under the control of the U6 promoter; HCAdV-TALENs were expressed from the tissue-specific human alpha-1-antitrypsin (hAAT) promoter; HCAdV-HBVU6no.2, expresses a small hairpin RNA against HBsAg and the human coagulation factor IX (hFIX) under the control of the hAAT promoter; HCAdV-CRISPR-mock, instead of expressing gRNAs against HBV as shown in HCAdV-Cas9-VAI-RT-P1-XCp, this vector expresses an irrelevant gRNA (U6-HPV).
Inhibition of HBV Infection In Vitro
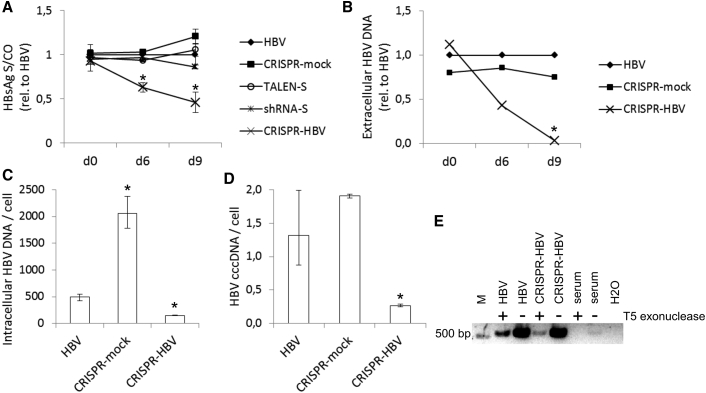
After producing HCAdVs that express either one of the nuclease systems, our vectors were challenged in infection models of HBV. Transduction of HepG2.2.15 cells with nuclease-expressing vectors for 9 days resulted in a reduction of HBsAg secretion of about 54% by the CRISPR/Cas9 system and about 14% by the previously published RNAi vector in relation to untreated HepG2.2.15 cells (Figure 4A). The TALEN vector did not induce any reduction of HBsAg secretion as well as the control CRISPR-mock vector. Measurement of secreted extracellular HBV DNA molecules by qPCR revealed a prominent decline at day 9 down to one-20th of the reference level in untreated cells (Figure 4B). In comparison, the HBV DNA levels in the medium of mock-treated cells ranged at three-quarters of the reference level. Intracellular HBV DNA levels were also analyzed by qPCR (Figures 4C and 4D). In untreated HepG2.2.15 cells, about 500 HBV DNA molecules were detected per cell. Interestingly, this number was raised about 4-fold in mock-treated cells. In contrast, the HBV-specific CRISPR/Cas9 system induced a decline to about 150 copies per cell. The HBV sequence amplified by qPCR was located outside of the nuclease-induced mutations, providing evidence for a mechanism of reduction of HBV DNA quantity. A corresponding outcome was found for cccDNA determination by cccDNA-selective qPCR from T5 exonuclease-digested samples. Interestingly, treatment with the CRISPR-HBV vector reduced the cccDNA content to 0.3 copies per cell, indicating a statistical clearance of 70% of the cells from cccDNA. To verify the required non-cccDNA elimination in the aforementioned assay, PCR products of T5 endonuclease-digested and undigested templates were separated on an agarose gel (Figure 4E). Reduction of PCR products after T5 endonuclease treatment proved the digestion of template DNA, which was not in a supercoiled circular state. In line with the premise that HBV-transgenic mouse serum does not contain cccDNA, there was no amplicon detected in T5 endonuclease-treated serum samples.
Figure 4.
Inhibition of HBV in HepG2.2.15 Cells
Either of the complete nuclease systems were delivered into cells by HCAdV and incubated for 9 days. (A) HBsAg concentrations in supernatant were measured at the indicated time points by ELISA. All measurements were performed with medium that has been on cells for 3 days. Data are represented as means of S/CO (sample to control ratio) values relative to HBV only, and error bars indicate SD of three replicates. Statistically significant differences to HBV only are indicated by an asterisk (*p < 0.01). (B) Extracellular HBV DNA was quantified by qPCR in extracted DNA from the same medium samples, which were examined by ELISA. HBV DNA was quantified relative to HBV-only samples using the ΔCt method. Data represent means of ΔCt ratios. Statistical differences in Ct values in comparison to HBV-only samples are indicated by an asterisk (*p < 0.01). (C) Intracellular HBV DNA was quantified by qPCR in extracted DNA from cells harvested on day 9 post-transduction. HBV DNA was quantified relative to the B2M reference gene using the 2−ΔΔCt method. Data represent means of HBV DNA copy numbers per cell, and error bars indicated SD of three replicates. Statistical differences in Ct values in comparison to HBV-only samples are indicated by an asterisk (*p < 0.01). (D) HBV cccDNA was quantified by qPCR in extracted and T5 exonuclease-digested DNA from cells harvested on day 9 post-transduction. cccDNA was quantified relative to the B2M reference gene in undigested but similarly treated templates using the 2−ΔΔCt method. Data represent means of cccDNA copy numbers per cell, and error bars indicate SD of three replicates. Statistical differences in Ct values in comparison to HBV-only samples are indicated by an asterisk (*p < 0.01). (E) PCR products from HBV templates of the transduction experiment or HBV-transgenic mouse serum with or without T5 exonuclease treatment prior to PCR to verify non-cccDNA elimination.
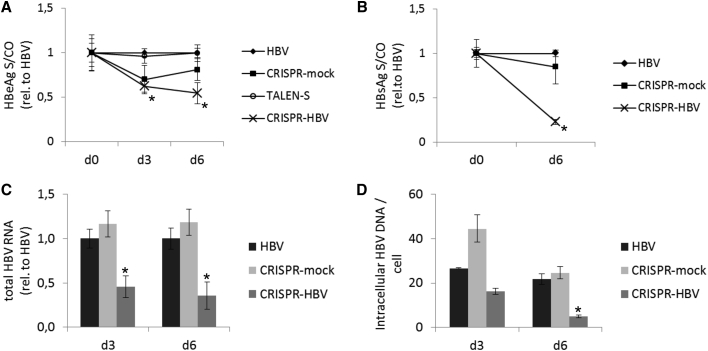
To further evaluate the effect of the designer nuclease-bearing vectors, HepG2-NTCP cells were infected with HBV and transduced with the respective constructs. This HBV model mirrors in a more accurate way the HBV infection, as the viral DNA is derived from the natural HBV and expression of viral genes and replication of the genome depend on the newly formed cccDNA.42 After infection with HBV and subsequent transduction with the therapeutic or control vectors, HBeAg (Figure 5A) and HBsAg (Figure 5B) concentrations were measured 3 and 6 days post-transduction as parameters of viral gene expression. After 3 days, the therapeutic CRISPR-vector induced a 37% reduction, and, after 6 days, a 45% reduction in HBeAg concentration relative to HBV-only-treated cells. The mock-CRISPR vector also lowered the HBeAg secretion, but not to a statistically significant extent. The TALEN vector did not alter the expression of HBeAg at all in comparison to HBV-only-treated cells. The HBsAg levels after treatment were only measured on day 6 in therapeutic CRISPR-treated and HBV-only samples. The reduction of 76% of HBsAg concentration in the supernatant relative to HBV-only-treated cells was more prominent than the measured loss of HBeAg. The mock treatment induced a similar effect for HBsAg as for HBeAg. All significant differences to the reference antigen level in HBV-only-treated samples are indicated by an asterisk in Figures 5A and 5B. The results are in accordance with the change in total HBV transcripts measured in isolated RNA of treated cells (Figure 5C). CRISPR/Cas9-treated cells with therapeutic gRNAs showed a significant reduction of total HBV transcripts of 54% on day 3 and 64% on day 6, respectively, relative to HBV-only-treated cells. In line with the previous results was the loss of total HBV DNA content in therapeutic CRISPR-treated cells of 39% on day 3 and 78% on day 6, respectively, relative to HBV-only-treated cells. Of note, here too the mock treatment resulted in an elevated HBV DNA level on day 3 but without statistical significance (Figure 5D).
Figure 5.
Inhibition of HBV in HepG2-NTCP Cells
Cells were infected with HBV 4 days prior to transduction of either of the complete nuclease systems by HCAdV and then incubated for 3 or 6 days. (A and B) HBeAg (A) and HBsAg (B) concentrations in supernatant were measured at the indicated time points by ELISA. All measurements were performed on medium that was on cells for 3 days. Data are represented as means of S/CO (sample to control ratio) values relative to HBV only, and error bars indicate SD of three replicates. Statistically significant differences to HBV only are indicated by an asterisk (*p < 0.01). (C) Total HBV RNA was measured by qRT-PCR from cells 3 and 6 days after transduction with the indicated vectors. RNA was quantified relative to the GAPDH reference gene using the 2−ΔΔCt method. Data represent means of HBV ratios relative to HBV-only treated samples, and error bars indicate SD of three replicates. Statistical differences in Ct values in comparison to HBV-only samples are indicated by an asterisk (*p < 0.01). (D) Intracellular HBV DNA was quantified by qPCR in extracted DNA from cells harvested on day 3 and 6 days after transduction. HBV DNA was quantified relative to the B2M reference gene using the 2−ΔΔCt method. Data represent means of HBV DNA copy numbers per cell, and error bars indicate SD of three replicates. Statistical differences in Ct values in comparison to HBV-only samples are indicated by an asterisk (*p < 0.01).
Discussion
Gene-editing techniques provide a whole new repertoire of tools to correct certain maladies for which no option of treatment or cure at all exists. A critical issue in the development of new treatment strategies is the delivery of the therapeutic agent. Here we present a delivery strategy based on packaging expression units into adenoviral vectors.
HCAdV vectors represent an advanced vehicle option for diverse and large genetic cargos. Their capacity and lower immunogenicity compared to earlier adenoviral vector generations belong to the main advantages.43, 44 The lowered immunogenicity of these vectors counts only for the acquired immune system native for the applied serotype, because the viral gene expression is omitted. However, the immunogenicity of the incoming capsid is still present. Moreover, the vector used in this study is derived from human adenovirus type 5 (Ad5), which is the most commonly used type in gene therapy but also has high prevalence of pre-existing immunity in human populations. It is possible to bypass the immune response induced by the incoming viral proteins, for example, by shielding the viral particle with polyethylene glycol (PEG).45 In addition, PEG modification counteracts vector sequestration in the bloodstream.46 Other approaches include capsid modification to ablate blood coagulation factor X binding to the hexon47, 48 or vector pseudotyping, in which Ad5 vectors are engineered with capsid parts from other serotypes with low pre-existing immunity to circumvent vector neutralization and, more importantly, acute toxicity.49, 50 A comparable strategy for vector optimization is to completely switch the vector serotype to a rare type.51 A study from Wang et al. hypothesized that the cargo itself might pose another challenge by induction of Cas9-specific immune responses.52 By using adenoviral vectors, the transferred genetic material is delivered in an episomal and transient way. On the one hand this can be advantageous with respect to limitation of the cargo immunogenicity, but on the other hand the dosage has to be carefully evaluated to guarantee the desired effect. Translational targeting as it was implemented here by using a liver-specific promoter is a strategy to counteract undesired side effects in cells other than the hepatocytes.53, 54 Another method for translational targeting is the post-transcriptional downregulation of transgenes.7 Such an approach would require the comparison of the microRNA (miRNA) expression profile in the optimal case from HCAdV- and HBV-infected primary hepatocytes against a panel of cells from various human cell types in order to find candidate miRNAs that downregulate the transgene expression in cells other than hepatocytes.
Chronic HBV infection is still a worldwide threatening disease with no complete reliable cure. Although the lifespan of patients can be extended to a normal level with current therapeutics, the quality of life is impeded by regular medication administration and side effects. In addition, the elevated risks for liver damage and liver cancer accompanying the HBV infection are not restored to normal risk levels because of the sustained impact of the suppressed infection. A potential solution to this problem might be achieved by designer nuclease-based therapy, which promises the complete eradication of latent HBV genomes. The effective eradication of the HBV genome from infected cells can also decrease the risk of a superinfection with the hepatitis delta virus, which is dependent on HBsAg production. This is in contrast to nucleos(t)ide analog RT inhibitors, which repress reverse transcription of the HBV genome but leave viral protein translation unaffected.
The approach applied in this study includes the delivery of expression units of three gRNAs along with one Cas9 in a one-vector system. In comparison to a recently published study, which utilizes ssAAVs to deliver one gRNA and one Cas9 expression unit from Staphylococcus aureus,23 this approach promises a more radical attack against the HBV genome. This study also confirmed the observation of others that the HBV cccDNA was rather degraded than mutated by CRISPR/Cas9 cleavage.55, 56 In theory, the probability of complete degradation of HBV cccDNA increases with every gRNA, and additionally multi-species targeting is possibly independent from consensus sequences. The presented reductions of HBV markers of replication in this study never went below the detection limit, which can be achieved with current therapeutic treatments. Possible reasons for this are that either a relatively short time frame was examined or data were collected in a model with high HBV content and cell turnover, which may lead to higher thresholds to asses significant activity. Nevertheless, the reductions in these experimental settings give reason to initiate further studies in a clinically relevant HBV infection animal model in vivo.
Comparison of TALENs with the CRISPR/Cas9 system in this study showed fewer efficacies of the TALENs in the context of the viral vector, although mutational activity was equal in transfection experiments. Of note, transfection experiments in a liver cell line also showed no effect from the TALEN system, but it was hypothesized that this was due to poor transfection efficiencies. Therefore, it was tried to enhance their potency by vectorization, which turned out unsuccessful in the later experiments. This suggests that there are difficulties with the vectorization of TALENs, which are not encountered with the CRISPR/Cas9 system, and we hypothesize that multiple factors may be responsible for the non-performance of HCAdV-TALEN vectors. It could be speculated that these issues are caused by the repeated sequences present in the repeat variable diresidues (RVDs). Although we did not observe major rearrangements in the analytic restriction enzyme digest, the occurrence of smaller deletions of, for instance, RVDs cannot be excluded. We performed Sanger sequencing only into the ends of the TALEN sequences, but not over the repetitive structures.
Another important issue could be the used promoter driving expression of designer nucleases. In plasmid-based experiments, we used the strong CMV promoter to drive expression of TALENs, and in HCAdV-mediated TALEN delivery, we applied the tissue-specific hAAT promoter. In contrast, Cas9 in the context of the CRISPR/Cas9 system was expressed from the strong CBh promoter in both non-viral and HVAdV approaches. The designer nuclease systems were additionally compared to a previously vectorized shRNA expression cassette.36 Here we could find equal to or superior suppression of antigen expression by the CRISPR/Cas9 system. Furthermore, because of the mutational effect of the designer nucleases, we believe the designer nuclease-mediated reduction to be a more permanent effect than the one by inhibition by RNAi. Here we found a reduction of cccDNA copy numbers to less than one copy per three HepG2.2.15 cells. In concordance with previous studies,23 we speculate that this may be due to definite loss of cccDNA caused by degradation. It is of note that HepG2.2.15 cells are considered to contain about 10 copies of cccDNA per cell,57, 58 but here only 1.3 copies per cell were measured in untreated samples, and, therefore, copy numbers are potentially underestimated. This may have a methodological reason but this phenomenon needs to be analyzed in further detail.
Furthermore, we observed a difference for quantified total intracellular and extracellular HBV DNA and the amount of cccDNA (Figure 4). Especially the intracellular HBV DNA level in CRISPR-mock-treated HepG2.2.15 cells was significantly higher than those in the untreated control. That adenovirus can increase HBV infection parameters, such as HBsAg levels, in mice shortly after infection was also observed in another study in mice.36 To further shed light on this phenomenon, it is of note that previous studies suggested that adenovirus types binding to αv integrins as co-receptor, such as Ad5 used in the present study, activate several signaling proteins, such as phosphoinositide-3-OH kinase (PI3K).59 PI3K plays a role in the autophagy pathway, which is also involved in the HBV DNA replication.60 Thus, circumventing activation of this signaling pathway by using alternative adenovirus types (e.g., serotypes 40 and 4161) may, therefore, further increase CRISPR activity in adenovirus-based approaches.
In summary, this study can be considered as a valuable improvement to the already known state of the art, which expands the possible use of the CRISPR/Cas9 system in the context of viral vectors by the inclusion of multiple gRNAs. Beyond that, the addition of further elements like immunomodulatory factors is imaginable to improve the system even more in the combat against chronic HBV infection. Future objectives are to test our vectors in animal models of HBV infection and eventually to optimize the vector for the needs of human application.
Materials and Methods
Cell Culture
The cell line HepG2.2.15 (stably transfected with the HBV genome) was cultured in William’s Medium E (PAN-Biotech, Aidenbach, Germany), supplemented with 10% fetal bovine serum (FBS, Standard Quality, EU approved, PAA, Pasching, Austria), 1× minimum essential medium (MEM) with non-essential amino acid (NEAA) solution (100×) without L-Glutamine (PAN-Biotech, Aidenbach, Germany), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Pen/Strep, PAN-Biotech, Aidenbach, Germany). Cell culture vessels were coated with collagen A (Biochrom, Berlin, Germany) prior to seeding. The human hepatoma cell line Huh7 was maintained in DMEM (PAN-Biotech, Aidenbach, Germany) supplemented with 10% FBS, 1× MEM NEAA, and Pen/Strep. HEK293 cells were grown in DMEM supplemented with L-glutamine, 10% FBS, and Pen/Strep. The HCAdV producer cell line 116, which is based on the HEK293 cell line stably expressing Cre recombinase,39 was cultured in MEM Eagle with Earle’s balanced salt solution (PAN-Biotech, Aidenbach, Germany) supplemented with 10% FBS, 100 μg/mL hygromycin B (PAN-Biotech, Aidenbach, Germany), and Pen/Strep. The FBS concentration in the medium was reduced to 5% and antibiotics were omitted during virus production. This cell line is obtainable from Philip Ng (Baylor College of Medicine, Houston, TX). HepG2-NTCP-K7 cells, which stably overexpress the NTCP as an entry receptor for HBV, were cultured on collagen-coated plates in full DMEM (10% fetal calf serum [FCS], 1% Pen/Strep, 1% sodium pyruvate, 1% NEAA, and 1% Glutamin; Life Technologies, Darmstadt, Germany). Before HBV infection, the culture medium was replaced with differentiation medium (full medium plus 2.5% DMSO; Sigma-Aldrich, Taufkirchen, Germany). Cells were infected with HBV 100 genome equivalents/cell (GEq/cell) in differentiation medium plus 4% PEG6000. All cell lines were maintained in a humidified incubator at 37°C and 5% CO2.
Plasmid Construction
TALEN pairs for HBsAg (S), HBcAg (C), and protein X (X) knockout were designed by the TALEN Targeter program (https://tale-nt.cac.cornell.edu/). The target sequences of the binding domains were for HBsAg 5′-T-CAGCCCGTTTCTCCTGGCTC and 5′-T-ACGAACCACTGAACAA with a 15-bp spacer, for HBcAg 5′-T-AGACGACGAGGCAGGTCCCC and 5′-T-GAGACCTTCGTCTGCGAGGC with a 17-bp spacer, and for protein X 5′-T-CTCTTTACGCGGACTCCCC and 5′-T-GCACACGGTCCGGCAGA with a 14-bp spacer sequence in between. TAL repeats were assembled by the Golden Gate TALEN and TAL Effector Kit 1.0 supplied by the Daniel Voytas laboratory (no longer available at Addgene, Cambridge, MA)31 and cloned into the TALEN expression cassette of pCS2TAL3DD or pCS2TAL3RR,32 which were before cloned into the multiple cloning site of a pBluescript-based vector.62 Promoters were exchanged with an hAAT liver-specific promoter by restriction enzyme cloning. Therefore, the hAAT promoter was amplified by PCR and ligated into the PstI restriction enzyme site of the respective vector. The resulting plasmids were named pBS-p/p-dNotI-hAAT-TALEN-DD-S1 and pBS-p/p-dNotI-hAAT-TALEN-RR-S2.
Three gRNA sequences were adopted from published sources26, 27 targeting either the ORF of HBV polymerase in the highly conserved reverse transcriptase domain (HBV-RT, TTCAGTTATATGGATGATG), in the RNase H domain (P1, GTTTTGCTCGCAGCAGGTCT, with one position adjusted to the HBV genome sequence used in this project), and the ORF of protein X (XCp, GGGGGAGGAGATTAGGTTAA). The HBV-specific gRNAs were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (42230, Addgene, Cambridge, MA), following the protocol from the Feng Zhang laboratory.33
The intermediate shuttle plasmid pShV34 was constructed by insertion of a synthetic DNA fragment into the pEX-K plasmid (Eurofins, Ebersberg, Germany). The synthetic DNA fragment is composed of a multiple cloning site (MCS), which is flanked by a 112-bp and a 116-bp non-coding random DNA sequence that do not share similarity with natural DNA sequences. They function as homology arms (HAs) for site-directed homologous recombination into the HCAdV genome contained in a bacterial artificial chromosome (BAC).63 The HA-flanked MCS is additionally flanked by the recognition sites for the homing endonucleases I-CeuI and PI-SceI, enabling the cloning into the HCAdV genome contained in the previously established plasmid pAdFTC.62 Outside of the I-CeuI and PI-SceI restriction sites, SwaI sites were added to release the synthetic DNA fragment from the pEX-K plasmid. The plasmid backbone was then digested with NotI to remove the original MCS of pEX-K, blunted, and ligated with the released synthetic DNA fragment. The resulting plasmid pShV served as the basis to create intermediate shuttle plasmids for cloning into the HCAdV genome.
To clone a TALEN pair-containing construct, the blunted SacII fragment from pBS-p/p-dNotI-hAAT-TALEN-DD-S1 containing the left TALEN targeting the HBsAg ORF was cloned into the HincII restriction site of pShV. Next, the blunted SacII fragment from pBS-p/p-dNotI-hAAT-TALEN-RR-S2 containing the right TALEN targeting the HBsAg ORF was cloned into the EcoRV restriction site of pShV-hAAT-TALEN-S1, generating the intermediate plasmid pShV-hAAT-TALEN-S.
To generate the intermediate plasmid containing the CRISPR/Cas9 system directed against HBV, the blunted XbaI/NotI fragment containing the hSpCas9 from pX330-U6-Chimeric_BB-CBh-hSpCas9 (Feng Zhang, plasmid 42230, Addgene, Cambridge, MA) was cloned into the EcoRV restriction site of pShV. The three gRNA expression cassettes were then subsequently cloned into pShV-CBh-hSpCas9. For this purpose, the expression units were amplified by PCR and restriction sites added by using primers with either NheI- (gRNA-NheI-fwd 5′-ATGCTAGCGAGGGCCTATTTCCCATGATTCC-3′; gRNA-NheI-rev 5′-ATGCTAGCCTGCAGAATTGGCGCACG-3′) or BamHI-restriction enzyme recognition sites (gRNA-BamHI-fwd 5′-ACTGGGATCCGAGGGCCTATTTCCCATGATTCC-3′; gRNA-BamHI-rev 5′-TGAGGATCCTGCAGAATTGGCGCACG-3′) at either end of the amplicon, respectively. First, the expression cassette for the HBV-RT gRNA was amplified using the NheI-primer set, digested with NheI, and cloned into the AvrII restriction enzyme site of pShV-CBh-hSpCas9, resulting in pShV-CBh-hSpCas9-RT. Second, the expression cassette for the P1 gRNA was amplified using the NheI-primer set, digested with NheI, and cloned into the NheI restriction enzyme site of pShV-CBh-hSpCas9-RT, resulting in pShV-CBh-hSpCas9-RT-P1. Third, the expression cassette for the XCp gRNA was amplified using the BamHI-primer set, digested with BamHI, and cloned into the BamHI restriction enzyme site of pShV-CBh-hSpCas9-RT-P1, resulting in pShV-CBh-hSpCas9-RT-P1-XCp. The I-CeuI/PI-SceI insert fragments of the different intermediate shuttle plasmids were cloned into the I-CeuI/PI-SceI of pAdFTC,62 resulting in the respective pAdV-FTC/insert plasmids pAdV-FTC/hAAT-TALEN-S and pAdV-FTC/CBh-hSpCas9-RT-P1-XCp.
Adenovirus Production
The pAdFTC/insert plasmids were digested with NotI to linearize the pAdFTC/insert vector DNA and transfected with FuGENE 6 Transfection Reagent (Promega, Madison, WI) into 116 cells.39 At 18 hr post-transfection, cells were transduced with the first-generation HV AdNG163R-239 containing a packaging signal flanked by loxP sites. To increase the titer of the HCAdV, five serial passages in 116 cells were performed. The virions containing the HCAdV-hAAT-TALEN-S or HCAdV-CBh-hSpCas9-RT-P1-XCp (in short HCAdV-CRISPR-HBV) vector genomes were purified by CsCl gradients. After one CsCl step gradient and one equilibrium gradient, the HCAdV vectors were purified at high titer (3E+09–1E+10 transducing particles per milliliter), with HV contamination lower than 0.2%.
The genome integrity of the adenovirus vector preparations was tested on virion DNA, which was isolated with a proteinase K-SDS lysis solution (1×TE, 0.5% SDS, and 0.1 μg/μL proteinase K) for 2 hr at 56°C, precipitated with ethanol, and then resuspended in 20 μL ddH2O. Analytical restriction enzyme digests, PCRs amplifying junctions in the vector construct, and Sanger sequencing were performed on viral DNA.
Transfection and Transduction Experiments to Test the Efficacy of Designed Plasmids and Vectors
Plasmid constructs were co-transfected with an HBV replication-competent plasmid, pTHBV228 with FuGENE 6 Transfection Reagent (Promega, Madison, WI), into HEK293 cells or Huh7 cells, according to the manufacturer’s instructions. Briefly, cells were plated in a 24-well plate 1 day before transfection to reach approximately 50%–80% confluency on the day of transfection. In total, 900 ng plasmid DNA was transfected per well using 2.7 μL transfection reagent. A molar target plasmid-to-nuclease plasmid ratio of 1:5 was used and adjusted to 900 ng with unrelated plasmid DNA as stuffer DNA. Transfected cells were incubated for 4 days, harvested for genomic DNA isolation with a proteinase K-SDS lysis solution (10 mM Tris-Cl [pH 8.0], 100 mM EDTA [pH 8.0], 50 mM NaCl, 0.5% SDS, 20 μg/mL RNase, and 100 μg/mL Proteinase K),64 and HBsAg was measured in the supernatant.
To analyze HBV knockdown by vector-delivered nucleases in the context of an established HBV infection, HepG2.2.15 cells were transduced for 9 days with HCAdV-FTC/CRISPR-mock (a CRISPR/Cas9 vector directed against HPV; E.E.-S., unpublished data), HCAdV-FTC/TALEN-S, HCAdV-FTC/HBVU6no.2 (an RNAi vector directed against HBV, which is here called shRNA-S36), or HCAdV-FTC/CRISPR-HBV at an MOI of 200. Medium was collected every third day and renewed from the day of transduction (d0). Therefore, cells were washed before new medium was added. Total DNA was isolated on day 9 post-transduction. HBsAg was measured in the cell culture supernatant of Huh7 and HepG2.2.15 cells in a 1:10 dilution in PBS using the Monolisa HBs Ag ULTRA kit (Bio-Rad, CA). HBV-infected HepG2-NTCP-K7 cells were transduced with the same vectors at an MOI of 300. Medium and cells were harvested on days 3 and 6 post-transduction with HCAdVs. The medium was tested for HBeAg concentrations (Siemens Molecular Diagnostics, Marburg, Germany) and HBsAg concentrations (Abbott Laboratories, Chicago, IL). DNA and RNA were isolated from cells with the NucleoSpin Tissue Kit (Macherey-Nagel, Düren, Germany) and the NucleoSpin RNA Kit (Macherey-Nagel, Düren, Germany), respectively. Cell viability of NTCP-HepG2 cells was monitored before harvest with the CellTiter-Blue Reagent (Promega, Madison, WI), according to the manufacturer’s instructions (Figure S4).
Mutation Detection
For the T7 endonuclease 1 (T7E1) (New England Biolabs, MA) mutation detection assay, DNA sequences that span the target sites were amplified from purified genomic DNA by PCR using standard conditions for the OneTaq 2X Master Mix with Standard Buffer (New England Biolabs, MA), with the following primer sets: S/HBV-RT, S-T7 for 5′-ttcctcttcatcctgctgct-3′ and S rev 5′-tgtaaaaggggcagcaaaac-3′; X, No. 5-forw 5′-actcctagccgcttgttttg-3′ and No.5-rev 5′-ataagggtcgatgtccatgc-3′; C, C-T7 for 5′-ctgggtgggtgttaatttgg-3′ and No.6-rev 5′-tacccgccttccatagagtg-3′; P1, P1_F 5′-gctttcactttctcgccaac-3′ and P1_R 5′-accttgggcaatatttggtg-3′; and XCp, XCp_F 5′-actctctcgtccccttctcc-3′ and XCp_R 5′-gcctgagtgcagtatggtga-3′. The PCR products were desalted by ethanol precipitation and resuspended in 1× NEB2 buffer (New England Biolabs, MA). Next, the PCR products were denatured and annealed to promote heteroduplex formation using the following program: 95°C-2 min, 95°C→85°C (Δ 2°C/s), 85°C→25°C (Δ 0.1°C/s), and 16°C. T7E1 was added to the reaction and incubated for 15 min at 37°C. The reaction was stopped using a gel loading dye containing SDS and then resolved by electrophoresis on a 2% agarose gel for 1 hr at 100 V. The gel was analyzed with the Gel Doc EZ System (Bio-Rad, CA) and Image Lab Software Version 5.2 (Bio-Rad, CA). The gene modification rates were estimated using the following approximation: fractional modification = (1- (1- (fraction of cleaved bands))1/2), as introduced by Miller et al.35
For quantification of the editing efficacy by a method named TIDE (tracking of indels by decomposition), the same PCR products were analyzed by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany) and the chromatograms analyzed using the TIDE online tool.37
Quantification of HBV Total DNA, cccDNA, and Total RNA
Total extracellular and intracellular DNA was quantified in duplicates from three separate samples by qPCR using My-Budget 5x EvaGreen QPCR-Mix II (Bio-Budget Technologies, Krefeld, Germany) in a 10-μL reaction volume, including 5 μL sample volume, according to the manufacturer’s instructions, with the following primer set: C_qRT-for 5′-tagaagaagaactccctcgcctcg-3′ and C-T7 rev 5′-cccagtaaagttccccacct-3′. qPCR was performed on the CFX96 Touch System (Bio-Rad, CA) using the following program: 95°C-15 min; 40× (95°C-15 s, 59°C-20 s, 72°C-20 s, plate read); melt curve 65°C→98°C (Δ 0.5°C/5 s). Ct values were determined by use of the Bio-Rad CFX Manager 3.1 software using the single-threshold mode. Extracellular HBV DNA was isolated from 150 μL medium using an SDS-lysis buffer. Samples were normalized against same input volume and quantified relative to HBV-only samples using the ΔCt method. Intracellular HBV DNA was isolated from approximately 1E+05 cells using an SDS-lysis buffer. The same qPCR protocol and primer set were used as for extracellular HBV DNA quantification, and, additionally, HBV-Ct values were normalized by an B2M qPCR65 using the 2−ΔΔCt method.
For cccDNA detection, 8.5 μL total cellular DNA was subjected to 30 min of T5 exonuclease (New England Biolabs, MA) digestion, as described in Xia et al.,66 with the exception that the DNA was precipitated with ethanol and resuspended in 12 μL ddH2O before performing the qPCR protocol, as described for total HBV DNA with the following primer set: P1_F 5′-gctttcactttctcgccaac-3′ and P1_R 5′-accttgggcaatatttggtg-3′. Ct values were normalized against B2M from samples receiving the same treatment but without T5 exonuclease digestion. A control reaction was based on DNA isolated from HBV-transgenic mouse serum, as described previously,67 which is considered negative for cccDNA.
Total cellular RNA was converted to HBV-specific cDNA and quantified with the Luna Universal One-Step RT-qPCR (New England Biolabs, MA). HBV total transcripts were amplified with primers binding on the common 3′ end, published by Yan et al.,42 and GAPDH qRT-PCR was taken as the reference gene.68 The qRT-PCR was performed on the CFX96 Touch System (Bio-Rad, CA, USA) using the following program: 55°C-10 min; 95°C-1 min; 45× (95°C-10 s, 60°C-30s, plate read); melt curve 60°C→95°C (Δ 0.2°C/5 s). Ct values were determined by use of the Bio-Rad CFX Manager 3.1 software using the single-threshold mode. Expression ratios were calculated in relation to HBV-only samples using the 2−ΔΔCt method.
Statistical Analysis
Statistical calculations were performed on technical replicates using an unpaired two-tailed Student’s t test, and differences were considered statistically significant when p was less than 0.01.
Author Contributions
Conceptualization, M.S., E.E.-S., T.B., and A.E.; Methodology and Investigation, M.S.; Resources, A.O., T.M., U.P., and A.E.; Writing – Original Draft, M.S.; Writing – Review & Editing, M.S., E.E.-S., A.O., T.M., U.P., and A.E.; Funding Acquisition, M.S., U.P., and A.E.; Supervision, A.E.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Philip Ng (Baylor College of Medicine, Houston, TX, USA) for providing 116 cells and the helper virus for HCAdV production and Jan Postberg (HELIOS Medical Centre Wuppertal, Witten/Herdecke University, Germany) for providing HepG2.2.15 cells. This work was supported by the Else Kröner-Fresenius-Foundation (grant 2012_A303) and German Liver-Foundation (grant S163/10137/2017) to M.S. and the German Research Foundation (DFG) to U.P. via TRR 179 and TP18.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.006.
Supplemental Information
References
- 1.Russell W.C. Adenoviruses: update on structure and function. J. Gen. Virol. 2009;90:1–20. doi: 10.1099/vir.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 2.Hoeben R.C., Uil T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a013003. doi: 10.1101/cshperspect.a013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crystal R.G. Adenovirus: the first effective in vivo gene delivery vector. Hum. Gene Ther. 2014;25:3–11. doi: 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonçalves M.A., de Vries A.A. Adenovirus: from foe to friend. Rev. Med. Virol. 2006;16:167–186. doi: 10.1002/rmv.494. [DOI] [PubMed] [Google Scholar]
- 5.Alba R., Bosch A., Chillon M. Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. 2005;12(Suppl 1):S18–S27. doi: 10.1038/sj.gt.3302612. [DOI] [PubMed] [Google Scholar]
- 6.Alonso-Padilla J., Papp T., Kaján G.L., Benkő M., Havenga M., Lemckert A., Harrach B., Baker A.H. Development of Novel Adenoviral Vectors to Overcome Challenges Observed With HAdV-5-based Constructs. Mol. Ther. 2016;24:6–16. doi: 10.1038/mt.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saydaminova K., Ye X., Wang H., Richter M., Ho M., Chen H., Xu N., Kim J.S., Papapetrou E., Holmes M.C. Efficient genome editing in hematopoietic stem cells with helper-dependent Ad5/35 vectors expressing site-specific endonucleases under microRNA regulation. Mol. Ther. Methods Clin. Dev. 2015;1:14057. doi: 10.1038/mtm.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitrani E., Pearlman A., Stern B., Miari R., Goltsman H., Kunicher N., Panet A. Biopump: Autologous skin-derived micro-organ genetically engineered to provide sustained continuous secretion of therapeutic proteins. Dermatol. Ther. (Heidelb.) 2011;24:489–497. doi: 10.1111/j.1529-8019.2012.01457.x. [DOI] [PubMed] [Google Scholar]
- 9.Jager L., Hausl M.A., Rauschhuber C., Wolf N.M., Kay M.A., Ehrhardt A. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat. Protoc. 2009;4:547–564. doi: 10.1038/nprot.2009.4. [DOI] [PubMed] [Google Scholar]
- 10.Chandrasegaran S., Carroll D. Origins of Programmable Nucleases for Genome Engineering. J. Mol. Biol. 2016;428(5 Pt B):963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Wang D., Liu S., Zheng X., Ji H., Xia H., Mao Q. Multiple copies of a linear donor fragment released in situ from a vector improve the efficiency of zinc-finger nuclease-mediated genome editing. Gene Ther. 2014;21:282–288. doi: 10.1038/gt.2013.83. [DOI] [PubMed] [Google Scholar]
- 12.Maier D.A., Brennan A.L., Jiang S., Binder-Scholl G.K., Lee G., Plesa G., Zheng Z., Cotte J., Carpenito C., Wood T. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum. Gene Ther. 2013;24:245–258. doi: 10.1089/hum.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan J., Wang J., Crain K., Fearns C., Kim K.A., Hua K.L., Gregory P.D., Holmes M.C., Torbett B.E. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol. Ther. 2012;20:849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holkers M., Maggio I., Liu J., Janssen J.M., Miselli F., Mussolino C., Recchia A., Cathomen T., Gonçalves M.A. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holkers M., Cathomen T., Gonçalves M.A. Construction and characterization of adenoviral vectors for the delivery of TALENs into human cells. Methods. 2014;69:179–187. doi: 10.1016/j.ymeth.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Zhang S., Huang X., Orwig K.E., Sheng Y. Rapid assembly of customized TALENs into multiple delivery systems. PLoS ONE. 2013;8:e80281. doi: 10.1371/journal.pone.0080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K., Yu C., Qu J., Li M., Yao X., Yuan T., Goebl A., Tang S., Ren R., Aizawa E. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15:31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maggio I., Holkers M., Liu J., Janssen J.M., Chen X., Gonçalves M.A. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci. Rep. 2014;4:5105. doi: 10.1038/srep05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C., Guan X., Du T., Jin W., Wu B., Liu Y., Wang P., Hu B., Griffin G.E., Shattock R.J., Hu Q. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J. Gen. Virol. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 20.Cheng R., Peng J., Yan Y., Cao P., Wang J., Qiu C., Tang L., Liu D., Tang L., Jin J. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N., Cowan C.A., Rader D.J., Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Mol. Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott T., Moyo B., Nicholson S., Maepa M.B., Watashi K., Ely A., Weinberg M.S., Arbuthnot P. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci. Rep. 2017;7:7401. doi: 10.1038/s41598-017-07642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dienstag J.L. Hepatitis B virus infection. N. Engl. J. Med. 2008;359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy E.M., Bassit L.C., Mueller H., Kornepati A.V.R., Bogerd H.P., Nie T., Chatterjee P., Javanbakht H., Schinazi R.F., Cullen B.R. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin S.R., Yang H.C., Kuo Y.T., Liu C.J., Yang T.Y., Sung K.C., Lin Y.Y., Wang H.Y., Wang C.C., Shen Y.C. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol. Ther. Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marion P.L., Salazar F.H., Liittschwager K., Bordier B.B., Seeger C., Winters M.A., Cooper A.D., Cullen J.M. A transgenic mouse lineage useful for testing antivirals targeting hepatitis B virus. In: Schinazi R.F., Sommadossi J.-P., Rice C.M., editors. Frontiers in Viral Hepatitis. Elsevier; Amsterdam: 2003. pp. 197–210. [Google Scholar]
- 29.Sells M.A., Chen M.L., Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 31.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., Grunwald D.J. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrke-Schulz E., Schiwon M., Leitner T., Dávid S., Bergmann T., Liu J., Ehrhardt A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 2017;7:17113. doi: 10.1038/s41598-017-17180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 36.Rauschhuber C., Xu H., Salazar F.H., Marion P.L., Ehrhardt A. Exploring gene-deleted adenoviral vectors for delivery of short hairpin RNAs and reduction of hepatitis B virus infection in mice. J. Gene Med. 2008;10:878–889. doi: 10.1002/jgm.1207. [DOI] [PubMed] [Google Scholar]
- 37.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehrke-Schulz E., Zhang W., Schiwon M., Bergmann T., Solanki M., Liu J., Boehme P., Leitner T., Ehrhardt A. Cloning and Large-Scale Production of High-Capacity Adenoviral Vectors Based on the Human Adenovirus Type 5. J. Vis. Exp. 2016;(107):e52894. doi: 10.3791/52894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer D., Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Maizel J.V., Jr., White D.O., Scharff M.D. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 41.Simek S., Byrnes A., Bauer S. FDA perspectives on the use of the adenovirus reference material. Bioprocess. J. 2002;1:40–42. [Google Scholar]
- 42.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunetti-Pierri N., Ng P. Helper-Dependent Adenoviral Vectors for Cell and Gene Therapy. In: Brunetti-Pierri N., Ng P., editors. Therapeutic Applications of Adenoviruses. CRC Press; 2016. pp. 59–84. [Google Scholar]
- 44.Weaver E.A., Nehete P.N., Buchl S.S., Senac J.S., Palmer D., Ng P., Sastry K.J., Barry M.A. Comparison of replication-competent, first generation, and helper-dependent adenoviral vaccines. PLoS ONE. 2009;4:e5059. doi: 10.1371/journal.pone.0005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croyle M.A., Le H.T., Linse K.D., Cerullo V., Toietta G., Beaudet A., Pastore L. PEGylated helper-dependent adenoviral vectors: highly efficient vectors with an enhanced safety profile. Gene Ther. 2005;12:579–587. doi: 10.1038/sj.gt.3302441. [DOI] [PubMed] [Google Scholar]
- 46.Danielsson A., Elgue G., Nilsson B.M., Nilsson B., Lambris J.D., Tötterman T.H., Kochanek S., Kreppel F., Essand M. An ex vivo loop system models the toxicity and efficacy of PEGylated and unmodified adenovirus serotype 5 in whole human blood. Gene Ther. 2010;17:752–762. doi: 10.1038/gt.2010.18. [DOI] [PubMed] [Google Scholar]
- 47.Alba R., Bradshaw A.C., Parker A.L., Bhella D., Waddington S.N., Nicklin S.A., van Rooijen N., Custers J., Goudsmit J., Barouch D.H. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood. 2009;114:965–971. doi: 10.1182/blood-2009-03-208835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alba R., Bradshaw A.C., Coughlan L., Denby L., McDonald R.A., Waddington S.N., Buckley S.M., Greig J.A., Parker A.L., Miller A.M. Biodistribution and retargeting of FX-binding ablated adenovirus serotype 5 vectors. Blood. 2010;116:2656–2664. doi: 10.1182/blood-2009-12-260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts D.M., Nanda A., Havenga M.J., Abbink P., Lynch D.M., Ewald B.A., Liu J., Thorner A.R., Swanson P.E., Gorgone D.A. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 50.Solanki M., Zhang W., Jing L., Ehrhardt A. Adenovirus hexon modifications influence in vitro properties of pseudotyped human adenovirus type 5 vectors. J. Gen. Virol. 2016;97:160–168. doi: 10.1099/jgv.0.000328. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W., Fu J., Liu J., Wang H., Schiwon M., Janz S., Schaffarczyk L., von der Goltz L., Ehrke-Schulz E., Dörner J. An Engineered Virus Library as a Resource for the Spectrum-wide Exploration of Virus and Vector Diversity. Cell Rep. 2017;19:1698–1709. doi: 10.1016/j.celrep.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Wang D., Mou H., Li S., Li Y., Hough S., Tran K., Li J., Yin H., Anderson D.G., Sontheimer E.J. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum. Gene Ther. 2015;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hafenrichter D.G., Ponder K.P., Rettinger S.D., Kennedy S.C., Wu X., Saylors R.S., Flye M.W. Liver-directed gene therapy: evaluation of liver specific promoter elements. J. Surg. Res. 1994;56:510–517. doi: 10.1006/jsre.1994.1082. [DOI] [PubMed] [Google Scholar]
- 54.Hafenrichter D.G., Wu X., Rettinger S.D., Kennedy S.C., Flye M.W., Ponder K.P. Quantitative evaluation of liver-specific promoters from retroviral vectors after in vivo transduction of hepatocytes. Blood. 1994;84:3394–3404. [PubMed] [Google Scholar]
- 55.Ramanan V., Shlomai A., Cox D.B., Schwartz R.E., Michailidis E., Bhatta A., Scott D.A., Zhang F., Rice C.M., Bhatia S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Hao R., Chen S., Guo D., Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J. Gen. Virol. 2015;96:2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- 57.Sells M.A., Zelent A.Z., Shvartsman M., Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rabe B., Glebe D., Kann M. Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J. Virol. 2006;80:5465–5473. doi: 10.1128/JVI.02303-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li E., Stupack D., Klemke R., Cheresh D.A., Nemerow G.R. Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J. Virol. 1998;72:2055–2061. doi: 10.1128/jvi.72.3.2055-2061.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sir D., Tian Y., Chen W.L., Ann D.K., Yen T.S., Ou J.H. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. USA. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madisch I., Hofmayer S., Moritz C., Grintzalis A., Hainmueller J., Pring-Akerblom P., Heim A. Phylogenetic analysis and structural predictions of human adenovirus penton proteins as a basis for tissue-specific adenovirus vector design. J. Virol. 2007;81:8270–8281. doi: 10.1128/JVI.00048-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehrhardt A., Kay M.A. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood. 2002;99:3923–3930. doi: 10.1182/blood.v99.11.3923. [DOI] [PubMed] [Google Scholar]
- 63.Mück-Häusl M., Solanki M., Zhang W., Ruzsics Z., Ehrhardt A. Ad 2.0: a novel recombineering platform for high-throughput generation of tailored adenoviruses. Nucleic Acids Res. 2015;43:e50. doi: 10.1093/nar/gkv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 65.Goff L.K., Neat M.J., Crawley C.R., Jones L., Jones E., Lister T.A., Gupta R.K. The use of real-time quantitative polymerase chain reaction and comparative genomic hybridization to identify amplification of the REL gene in follicular lymphoma. Br. J. Haematol. 2000;111:618–625. doi: 10.1046/j.1365-2141.2000.02352.x. [DOI] [PubMed] [Google Scholar]
- 66.Xia Y., Stadler D., Ko C., Protzer U. Analyses of HBV cccDNA Quantification and Modification. Methods Mol. Biol. 2017;1540:59–72. doi: 10.1007/978-1-4939-6700-1_6. [DOI] [PubMed] [Google Scholar]
- 67.Bloom K., Ely A., Mussolino C., Cathomen T., Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol. Ther. 2013;21:1889–1897. doi: 10.1038/mt.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim S., Kim T. Selection of optimal internal controls for gene expression profiling of liver disease. Biotechniques. 2003;35 doi: 10.2144/03353bm03. 456–458, 460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.