Abstract
Circulating microRNAs (miRNAs) have been proposed as type 2 diabetes biomarkers, and they may be a more sensitive way to predict development of the disease than the currently used tools. Our aim was to identify whether circulating miRNAs, added to clinical and biochemical markers, yielded better potential for predicting type 2 diabetes. The study included 462 non-diabetic patients at baseline in the CORDIOPREV study. After a median follow-up of 60 months, 107 of them developed type 2 diabetes. Plasma levels of 24 miRNAs were measured at baseline by qRT-PCR, and other strong biomarkers to predict diabetes were determined. The ROC analysis identified 9 miRNAs, which, added to HbA1c, have a greater predictive value in early diagnosis of type 2 diabetes (AUC = 0.8342) than HbA1c alone (AUC = 0.6950). The miRNA and HbA1c-based model did not improve when the FINDRISC was included (AUC = 0.8293). Cox regression analyses showed that patients with low miR-103, miR-28-3p, miR-29a, and miR-9 and high miR-30a-5p and miR-150 circulating levels have a higher risk of disease (HR = 11.27; 95% CI = 2.61–48.65). Our results suggest that circulating miRNAs could potentially be used as a new tool for predicting the development of type 2 diabetes in clinical practice.
Keywords: type 2 diabetes mellitus, miRNAs, biomarkers, predictive models
Introduction
During the last decade, the prevalence of type 2 diabetes mellitus (T2DM) has been continuously on the rise; it has been estimated that the current number of T2DM patients worldwide will double in the next two decades1 and similar trends have been reported for obesity.2 It is, therefore, critical that there be early identification of individuals at high risk for developing diabetes in order to devise prevention strategies aimed at controlling the factors related to the development of the disease.3, 4
The traditional biomarkers used to identify T2DM and pre-DM patients include glucose-homeostasis-related parameters, lipid classes, and scores such as the Finnish Diabetes Risk Score (FINDRISC).5 In 2010, the American Diabetes Association (ADA) added glycated hemoglobin (HbA1c) as a diagnostic criterion for diabetes and prediabetes,6, 7 due to the strong association with the high risk of developing the disease. The use of HbA1c as a diagnostic test certainly has its advantages: convenience, less day-to-day variability, greater pre-analytical stability, and international standardization. Moreover, the FINDRISC has been successfully implemented as a practical screening test to assess the risk of diabetes and detect undiagnosed T2DM in European populations.8, 9 However, these parameters and tests also have their limitations and cannot precisely predict an individual’s risk of developing T2DM, mainly because certain medical conditions may affect HbA1c and cause falsely high or low readings.10, 11 In addition, previous studies have examined the performance of HbA1c in comparison with fasting plasma glucose (FPG) in diagnosis of dysglycemia in older adults, and the authors have demonstrated considerable discrepancy between FPG- and HbA1c-based diagnoses of T2DM and pre-T2DM, with differences accentuated by race and gender.12, 13 It has also been demonstrated that the variability of HbA1c could be influenced by biological determinants of hemoglobin glycation, including epidemiological, genetic, and physiological factors.12
It is, therefore, necessary to research into a new model that can use both HbA1c and the new biomarkers to improve sensitivity and specificity and increase the predictive value.
MicroRNAs (miRNAs) are autocrine and endocrine regulators of gene expression. In the latter case, it has been demonstrated that extracellular vesicles circulating in human plasma, which are a mixture of microparticles, exosomes, and other structures such as apoptotic bodies, contain miRNAs that can be considered as a novel class of signaling molecules mediating intercellular communication.14 Some of these circulating miRNAs have been linked to glucose metabolism,15 while others, such as miR-144, miR-146a, miR-150, miR-182, miR-192, mir-29a, miR-30d, and miR-320, seem to participate in the regulation of insulin signaling.16, 17 There are others, too, such as miR-375,18, 19, 20, 21 miR-96,22 and miR-124a,23 which have been associated with the development of T2DM and may also be involved in its progression.24 One previous study has shown that a set of miRNAs, including miR-15a, miR-29b, miR-126, miR-223, and miR-28-3p, deregulated prior to the development of T2DM.25 However, this study lacks a reliable predictive value, since it was performed in a reduced number of incident cases (n = 19), the T2DM diagnosis was not performed according to all the ADA diagnosis criteria,26 and a poor statistical analysis was conducted.25 Moreover, the idea about a circulating miRNA profile modulable by glucose homeostasis conditions is supported by two other studies, which related miRNA plasma levels with the grade of insulin resistance.27, 28
Overall, there is evidence to suggest that a specific profile of circulating miRNAs could become a valuable biomarker to identify those normoglycemic and prediabetic individuals at increased risk of developing T2DM. However, until now, no studies have been carried out in a large population at high risk of developing T2DM to test this hypothesis.
Based on this previous evidence, our aim was to identify whether circulating miRNAs, added to clinical and biochemical markers (HbA1c, glucose, insulin, glucose after 2 hr of oral glucose tolerance test [OGTT], hepatic insulin resistance index [HIRI], insulinogenic index [IGI], insulin sensitivity index [ISI], disposition index [DI], muscle insulin sensitivity index [MISI], homeostatic model assessment-insulin resistance [HOMA-IR], and FINDRISC), would have the potential to predict new incident cases of T2DM more accurately.
Results
Baseline Characteristics of the Patients
In the present work, from the full Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention (CORDIOPREV) study population (1,002 patients), we included the 462 patients who at baseline had not been clinically diagnosed with T2DM.
At baseline, we observed higher values of waist circumference, body mass index (BMI), triglycerides (TGs), glucose, insulin, and HbA1c in the subgroup of patients who developed T2DM in a median follow-up of 60 months (Incident-T2DM group), compared with patients who did not develop T2DM (non-T2DM group) (all, p < 0.05). Moreover, our results showed lower baseline ISI, IGI, and DI and higher baseline HIRI and HOMA-IR values in the Incident-T2DM subgroup than in the non-T2DM group (all, p < 0.05). Conversely, we did not observe any differences in the baseline MISI (Table 1).
Table 1.
Baseline Characteristics and Circulating miRNA Levels of Subjects Who Did Not Develop T2DM (Non-T2DM) versus Subjects Who Developed T2DM (Incident-T2DM) after the Follow-up Period
| Variable |
Non-T2DM Group |
Incident-T2DM Group |
p Value |
miRNAs |
Non-T2DM Group |
Incident-T2DM Group |
p Value |
q- Value |
|---|---|---|---|---|---|---|---|---|
| n | 355 | 107 | – | – | – | – | – | – |
| Age (years) | 57 ± 0.5 | 59 ± 0.9 | 0.171 | miR-103 | 103.9 ± 4.5 | 62.1 ± 4.8 | <0.001 | <0.001* |
| Waist circumference (cm) | 101.7 ± 0.6 | 105.3 ± 1.1 | 0.003* | miR-107 | 1.70 ± 0.09 | 1.65 ± 0.17 | 0.810 | 0.853 |
| BMI (kg/m2) | 29.9 ± 0.2 | 31.4 ± 0.5 | 0.002* | miR-223 | 79.0 ± 3.5 | 56.7 ± 3.6 | 0.016 | 0.032* |
| TG (mg/dL) | 119.5 ± 3.2 | 132.6 ± 6.6 | 0.041* | miR-29a | 68.1 ± 2.1 | 54.8 ± 4.0 | <0.001 | <0.001* |
| Total cholesterol (mg/dl) | 160.6 ± 1.6 | 164.9 ± 3.4 | 0.288 | miR-28-3p | 185.9 ± 9.6 | 101.1 ± 8.8 | <0.001 | <0.001* |
| c-LDL (mg/dL) | 91.1 ± 1.3 | 93.4 ± 2.7 | 0.473 | miR-126 | 31.4 ± 1.6 | 18.8 ± 1.4 | <0.001 | <0.001* |
| c-HDL (mg/dL) | 44.6 ± 0.5 | 43.5 ± 1.0 | 0.290 | miR-145 | 5.10 ± 0.27 | 4.11 ± 0.43 | 0.013 | 0.021* |
| Apo A1 (mg/dL) | 133.1 ± 1.2 | 135.2 ± 2.3 | 0.283 | miR-150 | 0.69 ± 0.04 | 1.07 ± 0.09 | <0.001 | <0.001* |
| Apo B (mg/dL) | 71.8 ± 1.0 | 76.3 ± 1.8 | 0.035* | miR-15a | 2.70 ± 0.13 | 2.08 ± 0.28 | <0.001 | <0.001* |
| hs-CRP (mg/L) | 2.51 ± 0.19 | 2.88 ± 0.29 | 0.031* | miR-182 | 58.7 ± 2.7 | 59.1 ± 4.9 | 0.7670 | 0.7887 |
| Glucose (mg/dL) | 92.6 ± 0.5 | 96.2 ± 1.1 | 0.002* | miR-30a-5p | 25.6 ± 1.3 | 35.2 ± 2.5 | <0.001 | <0.001* |
| HbA1c (%) | 5.8 ± 0.02 | 6.0 ± 0.03 | <0.001* | miR-320 | 23.2 ± 0.9 | 24.0 ± 1.4 | 0.434 | 0.419 |
| Insulin (mU/L) | 8.33 ± 0.31 | 10.51 ± 0.66 | 0.007* | miR-375 | 20.9 ± 0.9 | 15.4 ± 1.1 | 0.013 | 0.020* |
| HIRI | 1052 ± 37 | 1313 ± 90 | 0.002* | miR-7 | 6.38 ± 0.33 | 6.88 ± 0.58 | 0.398 | 0.425 |
| MISI (× 102) | 2.10 ± 0.11 | 1.89 ± 0.16 | 0.369 | miR-9 | 2.03 ± 0.15 | 0.93 ± 0.18 | <0.001 | 0.002* |
| DI | 1.01 ± 0.03 | 0.77 ± 0.04 | <0.001* | – | – | – | – | – |
| ISI | 4.30 ± 0.14 | 3.27 ± 0.18 | <0.001* | – | – | – | – | – |
| IGI | 1.14 ± 0.06 | 0.88 ± 0.07 | 0.025* | – | – | – | – | – |
| HOMA-IR | 2.58 ± 0.09 | 3.23 ± 0.22 | 0.002* | – | – | – | – | – |
Values expressed as mean ± SE. BMI, body mass index; c-HDL, high-density lipoprotein; c-LDL, low-density lipoprotein; TG, triglycerides; Apo A1, Apolipoprotein A1; Apo B, Apolipoprotein B; hs-CRP, high-sensitivity C-reactive protein; HbA1c, glycosylated hemoglobin; HIRI, hepatic insulin resistance index; MISI, muscle insulin sensitivity index; ISI, insulin sensitivity index; IGI, insulinogenic index; DI, disposition index; HOMA-IR, homeostasis model assessment-insulin resistance. Variables were calculated using one-way ANOVA through SPSS (now PASW Statistic for Windows, version 21.0) (IBM, Chicago, IL, USA). For data that did not fit the normal distribution, the Mann-Whitney U test was performed. *p < 0.05 in one-way ANOVA or after the false discovery rate (FDR) correction.
Bibliographic Identification of T2DM-Related Circulating miRNAs
Our bibliographic search of miRNAs associated with insulin sensitivity, insulin secretion and growth, and proliferation of beta cell yielded 24 miRNAs. When we measured the levels of miRNAs at baseline of the study (day 0 before dietary intervention), 17 were detected in the plasma of at least 80% of the patients, 4 were detected in less than 80% of the samples, and 3 were not detected at all in the plasma samples. (Table S1).
Multivariate Analysis by PCA and OPLS-DA
Whereas principal-components analysis (PCA) did not yield statistically significant results, the orthogonal partial least-squares discriminant analysis (OPLS-DA) showed differences between Incident-T2DM and non-T2DM (Q2 = 0.239; R2y = 0.270 and R2x = 0.575). Moreover, the variables importance projection (VIP) analysis identified 9 miRNAs (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223, and miR-375) with VIP scores > 1, which supports their relevance in differentiating between Incident-T2DM and non-T2DM patients (Figure 1).
Figure 1.
Orthogonal Partial Least-Squares Discriminant Analysis
Orthogonal partial least-squares discriminant analysis (OPLS-DA) was used to compare miRNA levels at baseline in order to analyze the differences between Incident-T2DM (gray circle) and non-T2DM patients (black circle) during follow-up. The quality of the models obtained by PCA and OPLS-DA was assessed by interrogation of the R2 and Q2 parameters. The miRNAs with VIP score > 1 were considered important for differentiating between groups (table within the image). Data were processed using SIMCA-P+ (version 14.0.0.1359; Umetrics, Umea, Sweden).
Baseline Levels of Circulating miRNAs
We observed that the baseline plasma levels of 9 miRNAs (miR-9, miR-15a, miR-28-3p, miR-29a, miR-103, miR-223, miR-126, miR-145, and miR-375) were significantly lower in Incident-T2DM patients than in non-T2DM patients. In contrast, the baseline plasma levels of 2 miRNAs (miR-30a-5p and miR-150) were significantly higher in Incident-T2DM as compared with non-T2DM patients. No significant differences between groups were found in the plasma levels of miR-7, miR-10, miR-182, and miR-320 (Table 1).
Baseline Circulating Levels of miRNAs According to the Year of T2DM Diagnosis
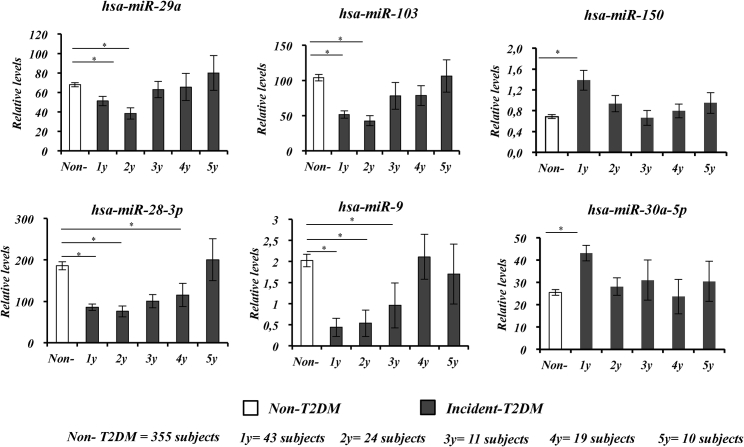
From the 462 subjects included in this study, a total of 107 subjects developed T2DM (Incident-T2DM group) during the follow-up period, of whom 43 were diagnosed after the first year of follow-up, 24 in the second year, 11 in the third year, 19 in the fourth year, and 10 in the fifth year. The remaining group of subjects (355 subjects) did not develop the disease (non-T2DM group). To evaluate the influence of the time before the T2DM diagnosis on the predictive value of each baseline miRNA to identify individuals at high risk of developing the disease, we analyzed the baseline circulating levels of miRNAs in each subgroup of patients according to the year in which the diagnosis of T2DM took place, as compared with patients who did not develop T2DM. Overall, we observed lower baseline circulating levels of miR-103, miR-28-3p, miR-9, and miR-29a in subjects diagnosed with T2DM in the first 3 years of follow-up and higher levels of miR-30a-5p and miR-150 in subjects diagnosed with T2DM in the first year of follow-up, compared with subjects diagnosed in later years and with the non-T2DM group (Figure 2).
Figure 2.
Circulating miRNA Levels at Baseline According to the Time Point at which Patients Were Diagnosed with T2DM
Data are represented as the mean ± SEM and correspond to the one-way ANOVA (in gray, subjects who developed T2DM [Incident-T2DM] and in white, subjects who did not develop T2DM [Non-T2DM]). The differences between groups were evaluated by Mann-Whitney U test. *p < 0.05.
Relationship between miRNA Plasma Levels and Insulin Sensitivity/Resistance and Beta Cell Function Indexes
We studied the relationship between the plasma levels of the 9 miRNAs, identified by OPLS-DA, and the insulin sensitivity, beta cell function, and the insulin resistance of peripheral tissues, such as liver and muscle, by lineal regression methods, adjusted by age, gender, BMI, TGs, and high-density lipoprotein cholesterol (c-HDL). We observed that the ISI, DI, and IGI were related with the plasma levels of miR-126, miR-103, and miR-150, respectively. Moreover, we also observed a relationship between miRNAs and fasting glucose (miR-28-3p, miR-30a-5p, miR-103, miR-126, and miR-150) and HbA1c (miR-28-3p, miR-103, miR-223, and miR-375) (Table 2).
Table 2.
Relationship between Circulating Levels of miRNAs and T2DM-Related Parameters
| hsa-miR103 | hsa-miR28-3p | hsa-mir-126 | hsa-miR150 | hsa-miR30a-5p | hsa-miR9 | hsa-miR375 | hsa-miR223 | hsa-miR29a | ||
|---|---|---|---|---|---|---|---|---|---|---|
| HbA1c (%) | r2 | −0.112 | −0.122 | −0.017 | 0.070 | 0.056 | −0.049 | −0.125 | −0.091 | −0.060 |
| p | 0.014* | 0.008* | 0.369 | 0.085 | 0.134 | 0.168 | 0.007* | 0.037* | 0.121 | |
| Glucosa (mg/dL) | r2 | −0.129 | −0.112 | −0.099 | 0.107 | 0.109 | −0.082 | 0.000 | −0.039 | 0.019 |
| p | 0.006* | 0.014* | 0.026* | 0.018* | 0.016* | 0.054 | 0.497 | 0.220 | 0.354 | |
| Insulina (mU/L) | r2 | 0.011 | −0.034 | −0.041 | 0.027 | 0.031 | −0.008 | −0.003 | 0.030 | 0.006 |
| p | 0.413 | 0.250 | 0.209 | 0.300 | 0.274 | 0.436 | 0.478 | 0.276 | 0.455 | |
| HOMA-IR | r2 | 0.016 | −0.026 | −0.013 | 0.010 | 0.001 | 0.059 | 0.027 | −0.006 | −0.040 |
| p | 0.380 | 0.306 | 0.396 | 0.424 | 0.490 | 0.123 | 0.301 | 0.452 | 0.217 | |
| MISI | r2 | −0.080 | 0.025 | 0.037 | −0.012 | 0.046 | −0.069 | −0.015 | 0.048 | −0.023 |
| p | 0.059 | 0.313 | 0.236 | 0.409 | 0.182 | 0.089 | 0.385 | 0.172 | 0.323 | |
| IGI | r2 | 0.053 | −0.016 | −0.015 | 0.102 | −0.021 | 0.080 | −0.031 | −0.013 | 0.030 |
| p | 0.148 | 0.377 | 0.382 | 0.022* | 0.340 | 0.060 | 0.269 | 0.398 | 0.280 | |
| ISI | r2 | 0.029 | 0.077 | 0.102 | −0.041 | 0.000 | −0.015 | 0.008 | 0.078 | 0.003 |
| p | 0.283 | 0.065 | 0.023* | 0.213 | 0.499 | 0.444 | 0.441 | 0.062 | 0.478 | |
| DI | r2 | 0.096 | 0.076 | 0.070 | 0.047 | −0.045 | 0.054 | −0.012 | 0.048 | 0.062 |
| p | 0.030* | 0.068 | 0.083 | 0.177 | 0.189 | 0.148 | 0.409 | 0.174 | 0.113 | |
| HIRI | r2 | 0.016 | −0.033 | −0.024 | 0.021 | 0.004 | 0.063 | 0.024 | −0.010 | −0.042 |
| p | 0.376 | 0.228 | 0.316 | 0.340 | 0.467 | 0.328 | 0.319 | 0.422 | 0.208 |
HbA1c, glycosylated hemoglobin; GLU, glucose; HOMA-B, homeostasis model assessment-beta cell function; HOMA-IR, homeostasis model assessment-insulin resistance; MIRI, muscle insulin resistance index; IGI, insulinogenic index; ISI, insulin sensitivity index; DI, disposition index; HIRI, hepatic insulin resistance index. Relationship was evaluated through a linear regression analysis adjusted by age, gender, body mass index (BMI), triglycerides (TG), and high-density lipoproteins (c-HDL), using SPSS (now PASW Statistic for Windows, version 21.0) (IBM, Chicago, IL, USA). *p < 0.05 in the linear regression analysis.
Received Operating Characteristic Curve Analysis
To evaluate the potential of miRNA plasma levels as a predictive biomarker of T2DM, the accuracy, sensitivity, and specificity were compared in 10 different models of received operating characteristic (ROC) analysis. We built models including all the miRNAs, measured together with the classic parameters (fasting glucose, 2-hr glucose, and HbA1c), beta cell function and insulin sensitivity/resistance indexes (IGI, ISI, DI, HIRI, and MISI), and the FINDRISC5 (Table S2).
First, we included the full miRNA dataset (15 miRNAs) as input variables in the model, and we obtained an area under the curve (AUC) = 0.849 (95% confidence interval [CI] = 0.808–0.889). Further, we reduced the number of miRNAs included in the model according to the VIP score in the OPLS-DA. Thus, a model built including the 9 miRNAs with a VIP value of over 1 (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223, and miR-375) yielded an only slightly reduced significance in the model AUC = 0.818 (95% CI = 0.771–0.864), as compared with the full 15 miRNA dataset.
Interestingly, the classic parameters, such as fasting glucose, insulin, 2-hr glucose (OGTT), and HbA1c, together with the insulin sensitivity indexes ISI, DI, IGI, HIRI, MIRI, HOMA-IR, and HOMA-beta yielded an AUC of 0.769 (95% CI = 0.713–0.826). In addition, the FINDRISC showed an AUC of 0.610 (95% CI = 0.550–0.671) (Figure 3A), which did not improve when we included insulin sensitivity indexes in the model AUC = 0.765 (95% CI = 0.709–0.820).
Figure 3.
ROC Analysis and Classification by Average Importance of Variables Included in a ROC Model Based on miRNAs and HbA1c
(A) The comparison among the AUC of ROC curves of three models: red line, miRNA- and HbA1c-based model; green line, clinical parameters model; blue line, FINDRISC model. (B) The average importance classification of the variables included in the ROC curve can be seen. Non-T2DM = 0; Incident-T2DM = 1. Analysis was carried out by MetaboAnalyst 3.0.
To obtain an implemented miRNA-based model, we combined the dataset of 9 miRNAs with the HbA1c, which was the classic clinical parameter with greater weight in the Average Importance of Variables classification. Thus, the HbA1c showed an AUC of 0.676 (95% CI = 0.605–0.737), which improved after inclusion of the 9 miRNAs (AUC of 0.834; 95% CI = 0.790–0.878, sensitivity = 0.766, specificity = 0.809, and accuracy = 80.0%) (Figure 3A). In addition, the ROC analysis showed 3 miRNAs (miR-9, miR-28-3p, and miR-29a) with the highest differentiation power from the Average Importance of Variables classification, followed by HbA1c and miR-150 (Figure 3B). Additionally, we built a model including the clinical parameters in the assessment of T2DM risk without miRNAs (fasting glucose, 2-hr glucose in OGTT, HbA1c, age, gender, BMI, c-HDL, TGs, diet, and waist circumference) showing an AUC = 0.7354 (Figure 3A).
Moreover, when we included the FINDRISC5 together with the dataset of the 9 miRNAs and HbA1c, we did not improve the model (AUC of 0.829; 95% CI = 0.785–0.874, sensitivity = 0.796, specificity = 0.774, and accuracy = 77.9%), as compared with the one that included the dataset of 9 miRNAs and HbA1c.
Also, the AUCs of all models were compared using the DeLong test, and we observed a significant difference between the model based on clinical parameters without miRNAs (fasting glucose, 2-hr glucose in OGTT, HbA1c, age, gender, BMI, c-HDL, TGs, diet, and waist circumference) and the model based on 9 miRNAs and HbA1c (adjusted by age, gender, BMI, c-HDL, TGs, diet, and waist circumference) (p = 0.01002).
Internal validation by bootstrap resampling of the original set (1,000 randomized samples) in the best model (9 miRNAs + HbA1c) showed a degree of over-optimism of 0.03, which represents the deviation from the mean SE in estimation of these 1,000 samples (bootstrapped AUC ROC = 0.796).
Cox Regression Analysis
To test the T2DM-predictive value of miRNAs by the Cox regression analysis, we selected those with the highest VIP values in the OPLS-DA multivariate analysis: 9 miRNAs (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223, and miR-375; Figure 1).
To perform the Cox regression analysis, we categorized subjects in tertiles according to the plasma levels for each miRNA as follows: low baseline plasma levels (T1), medium baseline plasma levels (T2), and high baseline plasma levels (T3). We compared the hazard ratio (HR) between T1 and T3 for each studied miRNA. We observed that low baseline plasma levels (T1) in 4 miRNAs (miR-9, miR-28-3p, miR-29a, and miR-103) and high plasma levels (T3) in 2 of 9 miRNAs (miR-30a-5p and miR-150) were associated with a high risk of T2DM development (HR T1 versus T3 ≥ 2.5) (Table 3).
Table 3.
Hazard Ratio Observed after Cox Regression Analysis for Each miRNA Included in the ROC Curve
| miRNA | HR T1 versus T1 (95% CI) | HR T1 versus T2 (95% CI) | HR T1 versus T3 (95% CI) |
|---|---|---|---|
| miR-150 | 1 (ref) | 2.40 (1.30–4.43) | 3.98a (2.21–7.17) |
| miR-103 | 1 (ref) | 2.07 (1.15–3.71) | 3.20a (1.85–5.55) |
| miR-28-3p | 1 (ref) | 2.83 (1.49–5.30) | 4.45a (2.40–8.24) |
| miR-126 | 1 (ref) | 1.82 (1.05–3.16) | 2.28 (1.34–3.90) |
| miR-9 | 1 (ref) | 0.99 (0.51–1.92) | 3.95a (2.34–6.65) |
| miR-30a-5p | 1 (ref) | 1.37 (0.79–2.40) | 2.53a (1.53–4.18) |
| miR-223 | 1 (ref) | 2.56 (1.47–4.47) | 1.90 (1.07–3.35) |
| miR-375 | 1 (ref) | 1.78 (1.05–3.00) | 1.54 (0.90–2.63) |
| miR-29a | 1 (ref) | 1.73 (0.97–3.07) | 2.51a (1.46–4.33) |
amiRNAs selected for multiple COX regression analyses.
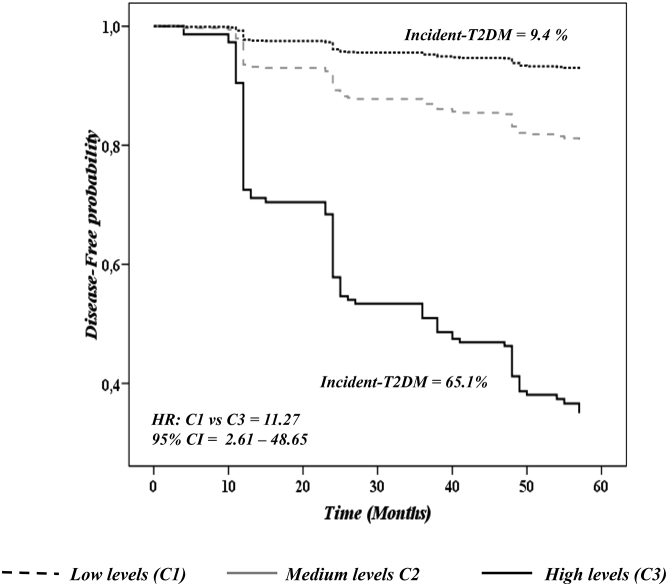
Accordingly, to study the accumulative predictive value of the six miRNAs together, we included these six miRNAs with HR T1 versus T3 ≥ 2.5 and performed a Cox regression multi-miRNA analysis (Figure 4).
Figure 4.
Disease-free Probability Analysis through a Cox Regression Model with the Six-miRNA Target
Data represent circulating levels for each miRNA by tertiles, low levels (T1), medium levels (T2), and high levels (T3). The analysis was carried out using SPPS (now PASW Statistic for Windows, version 21.0) (IBM, Chicago, IL, USA) and adjusted by diet, age, gender, BMI, TGs, c-HDL, HbA1c, and waist circumference.
Next, we categorized patients according to the levels of these six miRNAs as follows: C1 category was composed of patients with low levels in at least three of the four downregulated miRNAs (miR-9, miR-28-3p, miR-29a, and miR-103) and high levels in at least one of the 2 upregulated miRNAs (miR-150 and miR-30a-5p) (n = 46); C3 category was composed of patients with high levels in at least three of the four downregulated miRNAs and low levels in at least one of the two upregulated miRNAs (n = 32); and, finally, C2 was made up of patients with an intermediate miRNA deregulation profile (n = 356). We therefore obtained an HR between C1 and C2 of 2.34 (95% CI = 0.74–7.42) and an HR between C1 and C3 of 11.68 (95% CI = 3.56–38.34). Likewise, when we adjusted the Cox regression analysis by age, gender, BMI, diet, HbA1c, waist circumference, TGs, c-HDL, IGI, HOMA-IR, and DI, we observed a HR for C1 versus C2 of 2.66 (95% CI = 0.64–11.07) and for C1 versus C3 of 11.27 (95% CI = 2.61–48.65) (Figure 5).
Figure 5.
Disease-free Analysis through a Cox Regression Model Based on Multi-miRNAs, Including miR-103, miR-28-3p, miR-29a, miR-9, miR-150, and miR-30a-5p
The data represent circulating levels of all six miRNAs together; subjects were, therefore, classified into three categories as follows: C1 category was composed of patients with low levels in at least three of the four downregulated miRNAs (miR-9, miR-28-3p, miR-29a, and miR-103) and high levels in at least one of the 2 upregulated miRNAs (miR-150 and miR-30a-5p) (n = 46); C3 category was composed of patients with high levels in at least three of the four downregulated miRNAs and low levels in at least one of the two upregulated miRNAs (n = 32); and, finally, C2 was made up of patients with an intermediate miRNA deregulation profile (n = 356). The analysis was carried out through SPPS (now PASW Statistic for Windows, version 21.0) (IBM, Chicago, IL, USA) and adjusted by age, gender, BMI, diet, HbA1c, waist circumference, TGs, c-HDL, IGI, HOMA-IR, and DI.
To compare our miRNA-based predictive model with the FINDRISC,5 we categorized subjects into tertiles, according to the FINDRISC of our study as follows: low FINDRISC (T1), medium FINDRISC (T2), and high FINDRISC (T3). Next, we performed the Cox regression analysis and observed an HR between tertiles of FINDRISC T1 and T2 of 1.895 (95% CI = 1.131–3.174) and T1 and T3 of 2.362 (95% CI = 1.410–3.957) (Figure S1).
Discussion
Our study showed that plasma levels of 9 of the 17 miRNAs selected with detectable levels at baseline of the study in most of our subjects were significantly associated with glucose metabolism and type 2 diabetes risk. These 9 miRNAs, in combination with HbA1c plasma levels, were able to differentiate between the patients who developed type 2 diabetes (Incident-T2DM) after a median follow-up of 60 months and those patients who did not develop the disease (Non-T2DM), with an AUC in the model of 0.834. The Cox regression analysis showed that patients with low miR-103, miR-28-3p, miR-29a, and miR-9 and high miR-30a-5p and miR-150 plasma levels were at higher risk of developing T2DM.
The relevance of the T2DM prevalence figures lies in the fact that subjects with T2DM have higher cardiovascular morbidity and mortality compared with non-diabetic subjects.29 In addition, patients with acute myocardial infarction (AMI) and T2DM have a considerably higher risk of developing a new cardiovascular event than those without T2DM.30 It is, therefore, very important to identify early on individuals with a higher risk of developing T2DM, in order to prevent the onset of the disease by using powerful therapies and efficiently controlling the associated cardiovascular risk factors.
Nowadays, the standard tools for identifying patients with a risk of T2DM include HbA1c and scores such as the FINDRISC. However, these tools are not able to effectively predict the disease development. Previous studies have revealed a number of limitations of HbA1c as a diagnostic test in comparison with FPG in the elderly, in which the lower sensitivity and specificity of HbA1c was demonstrated.12, 13 Moreover, it has recently become evident that FINDRISC is not universally applicable among all ethnic groups and populations.31, 32
In addition, current evidence indicates that the blood miRNA profile may change under different pathophysiological conditions, such as cancer,33 cardiovascular diseases,34 and type 2 diabetes.35
In fact, two previous studies have related miRNA plasma levels with the grade of insulin resistance. One of them by Shah et al. identified 18 circulating miRNAs associated with plasma insulin and HOMA-IR levels,28 of which 4 were also identified as being deregulated in the study by Zampetaki et al., who identified 13 miRNAs associated with the incidence of T2DM.25 In our study, 9 miRNAs were identified as being important in the prediction model for T2DM, but only one matched with those identified by Zampetaki et al.25 and by Shah et al.28, suggesting that the differences in the phenotype between populations seem to be important in the deregulation of the miRNA profile associated with a pathological condition, such as glucose homeostasis.
In terms of T2DM prediction, the study by Zampetaki et al.25 investigated miRNA plasma levels before the diagnosis of T2DM, which identified 5 miRNAs (miR-15a, miR-28-3p, miR-29b, miR-126, and miR-223) associated with the incidence of T2DM. Nevertheless, the small number of incident cases (n = 19) reported in that study precluded the more extensive statistical predictive analysis that we have carried out in our study (ROC and Cox regression analyses). In contrast, our study included all 462 non-diabetic patients of the CORDIOPREV at baseline of the study, of whom 107 developed T2DM in a median follow-up of 60 months, and the statistical modeling (ROC and Cox regression models) allowed us to predict the onset of T2DM. In addition, in our study we followed ADA diagnosis criteria,26 whereas the study by Zampetaki et al.25 followed the World Health Organization guidelines, which do not take into account plasma levels of HbA1c as diagnosis criteria for diabetes and, therefore, did not produce a very accurate diagnosis.
The ROC analysis showed that HbA1c levels together with the set of 9 miRNAs were able to differentiate between Incident-T2DM and non-T2DM patients (AUC 0.834), and, furthermore, the model was more accurate than when only HbA1c (AUC 0.676) was used. In fact, the plasma levels of these 9 circulating miRNAs differentiate better between groups (Incident-T2DM versus non-T2DM) than the FINDRISC, on the basis of an AUC of 0.818 (9 miRNA plasma levels as input data) versus 0.610 obtained with the FINDRISC alone. It is worth noting that adding the FINDRISC to the 9-miRNA models (model 1: 9 miRNAs target + FINDRISC and model 2: 9 miRNAs target + FINDRISC + HbA1c) did not improve the accuracy in discriminating between the two groups of patients (AUC = 0.812 and AUC = 0.829, respectively).
Our results are in line with a recently published pilot cross-sectional study in which 8 miRNAs were able to discriminate between healthy people (n = 27), prediabetic patients (n = 12), type 1 diabetes patients (n = 16), and type 2 diabetes patients (n = 31),36 supporting the idea that miRNA plasma levels could be used as disease biomarkers. However, the study by Syehan et al.36 was performed in T2DM patients without a follow-up period, and, therefore, incidence was not analyzed, which means this study has no predictive value. More relevantly, our prospective study has shown the predictive value of measuring plasma miRNAs as biomarkers before T2DM development in patients at risk. Moreover, we studied different predictive models in a much larger population, including miRNAs for the first time as biomarkers. When these were added to the classic parameters, such as fasting glucose, insulin, 2-hr glucose (OGTT), HbA1c, insulin sensitivity indexes, and the FINDRISC, it allowed us to assess the risk of developing T2DM over the following 5 years.
Overall, after the multi-miRNA Cox regression analysis, we showed that the plasma levels of six miRNAs (miR-9, miR-28-3p, miR-29a, miR-103, miR-30a-5p, and miR-150) could potentially be used as predictive biomarkers, which is supported by their involvement in mechanisms related with the development of T2DM.
Circulating miRNAs are considered as a novel class of signaling molecules mediating intercellular communication,14 and the blood miRNA profile may change under different pathophysiological conditions, which allows for their potential use as biomarkers in certain diseases such as T2DM.25, 33, 35 However, circulating and intracellular miRNA levels may not match because, after the intracellular expression, miRNAs may be released,37 and, therefore, a reduction in release could lead to intracellular accumulation but low plasma levels. Additionally, previous studies demonstrated the miRNA levels in the T2DM-diagnosed status, in contrast with our study in which we evaluated the profile of miRNAs years before disease development and diagnosis. In fact, previous evidence has shown that upregulation of miR-29a in INS-β cell mediates β cell dysfunction and could contribute to the progression from impaired glucose tolerance to type 2 diabetes.38 Moreover, the inhibition of miR-103 leads to improved glucose homeostasis and insulin sensitivity in obese mice39 and an anti-inflammatory effect mediated by Cav1.40 In addition, the overexpression of miR-9 causes a decrease in glucose-stimulated insulin exocytosis by diminishing the expression of the transcription factor Onecut2, which represses the expression of granulophilin and negatively regulates insulin release.41
In the last few years, miRNA plasma levels have been shown to be useful as biomarkers for cardiovascular events34 and cancer.33 However, to the best of our knowledge, no studies have yet focused on the use of miRNA plasma levels as predictive biomarkers of T2DM development by combining a sample size large enough to obtain reliable results and robust statistical models. Thus, our study demonstrated for the first time that plasma levels of miR-9, miR-28-3p, miR29a, miR-103, miR-30a-5p, and miR-150 are also powerful predictive biomarkers that can discriminate between Incident-T2DM and non-T2DM patients. More importantly, our results showed that the alteration in miRNA levels precedes the development of T2DM by 3 years, and, therefore, our model has a predictive power in this period of time.
Nonetheless, our study has limitations. One limitation lies in the fact that we carried out a focused bibliographic search of the miRNAs associated with insulin sensitivity, insulin secretion and growth, and proliferation of beta cells, which meant that any potential miRNAs whose implication with T2DM has not yet been described were not included in our model. In addition, T2DM prevention was not the primary endpoint of the CORDIOPREV trial, but it was a secondary analysis conducted in the subgroup of cardiovascular patients without T2DM at baseline. Finally, the study included a large number of elderly patients with AMI, which limits our findings to people with these characteristics and precludes generalization to healthy individuals. In line with this, futures studies should focus the validation of this study on another population. Although diabetes prediction is very important as patients with AMI and T2DM have a considerably higher risk of developing a new cardiovascular event than those without T2DM,30 the validation in a cohort without cardiovascular disease and closer to the general population would allow us to adapt this method to the general population.
In conclusion, our results suggest that miRNA plasma levels added to HbA1c could become a valuable new tool for assessing the early risk of type 2 diabetes in clinical practice to prevent disease development.
Materials and Methods
Study Subjects
This work was conducted within the framework of the CORDIOPREV study. The rationale, methods, and baseline characteristics have been reported by Delgado-Lista et al.,42 and they are also provided in ClinicalTrials.gov (NTC00924937). Briefly, the CORDIOPREV study is an ongoing prospective, randomized, single blind, controlled dietary intervention trial in 1,002 patients with coronary heart disease (CHD), high cardiovascular risk, aged between 20 and 75 years old, who had their last coronary event more than 6 months prior to enrollment and had no severe diseases or a life expectancy of less than 5 years. In addition to conventional treatment for CHD, the subjects were randomized into two different dietary models (Mediterranean and low-fat diets). The intervention phase is still in progress and will have a median follow-up of 7 years. The patients were recruited from November 2009 to February 2012, mostly at the Reina Sofia University Hospital (Córdoba, Spain), but patients from other hospital centers from the Córdoba and Jaen provinces were also admitted. Written consent was obtained from all the subjects prior to recruitment, and the study protocol and all amendments were approved by the Ethics Committee of Hospital Reina Sofia, all of which follow the Helsinki Declaration and good clinical practices.
In the present work, all the subjects (n = 462) who had not been clinically diagnosed with T2DM at baseline in the CORDIOPREV-DIAB study were included.43 Of this group, 43 subjects were diagnosed as having T2DM after the first year of follow-up, 24 in the second year, 11 in the third year, 19 in the fourth year, and 10 in the fifth year, for a total of 107 subjects who developed T2DM (Incident-T2DM), according to all the ADA diagnosis criteria,26 evaluated on the basis of glucose tolerance tests performed each year during the median follow-up of 60 months (Figure S2). Of the 462 subjects included in the current work, 216 were randomized to consume a low-fat high-complex carbohydrate diet (LFHCC diet) and 246 to consume a Mediterranean diet (Med diet). The differences between diets were evaluated by Chi-square test, and we observed no statistical significance (Chi2 = 1.948; p = 0.163). The baseline characteristics of the subjects in the study are shown in Table 1.
Biochemical Measurements of Metabolic Parameters
Venous blood from the participants was collected in tubes containing EDTA after a 12-hr overnight fast. Lipid variables were assessed with the DDPPII Hitachi modular auto analyzer (Roche, Basel, Switzerland) using specific reagents (Boehringer-Mannheim, Mannheim, Germany). Measurements of total cholesterol (TC) and TG levels were performed by colorimetric enzymatic methods,44, 45 c-HDL was measured by colorimetric assay,46 and low-density lipoprotein (c-LDL) concentration was calculated by the Friedewald equation, using the following formula: c-LDL = CT − (c-HDL + TG/5). Glucose measurements were performed using the hexokinase method. The hs-C-Reactive Protein (hs-CRP) was determined by high-sensitivity ELISA (BioCheck, Foster City, CA, USA). Plasma insulin concentrations were measured by microparticle enzyme immunoassay (Abbott Diagnostics, Matsudo-shi, Japan). Non-esterified fatty acid concentrations were measured by enzymatic colorimetric assay (Roche Diagnostics, Penzberg, Germany). ApoA-1 and ApoB concentrations were determined by immunoturbidimetry.
Estimation of IR, Insulin Secretion, and Beta Cell Function Indexes and FINDRISC
Before starting the test, the patients had fasted (from food and drugs) for 12 hr, and they were asked to refrain from smoking during the fasting period and from alcohol intake during the preceding 7 days. They were also asked to avoid strenuous physical activity the day before the test was given. At 8:00 a.m., the patients were admitted into the laboratory to perform the OGTT (75 g dextrose monohydrate in 250 mL water, NUTER. TEC GLUCOSA 50), and 0-, 30-, 60-, and 120-min sampling was performed to establish plasma glucose and insulin levels.
The Matsuda ISI was calculated from the OGTT using the following formula: ISI = 10.000 ÷ √([fasting plasma glucose × fasting plasma insulin] × [mean glucose in OGTT × mean insulin in OGTT]).47 HOMA-IR was calculated as previously described by Song et al.48 Insulin secretion was measured by the IGI as follows: IGI = [30 min insulin − fasting insulin (pmol/L)]/[30 min glucose − fasting glucose (mmol/L)].49 Beta cell function was estimated by calculating the DI as follows: DI = ISI × [AUC30 min insulin/AUC30 min glucose], where AUC30 min is the AUC between baseline and 30 min of the OGTT for insulin (pmol/L) and glucose (mmol/L) measurements, respectively, calculated by the trapezoidal method.50 The indices used to determine tissue-specific IR were the HIRI and the MISI, which were calculated as described in previous work by our group,43 following the methods described by Matsuda and DeFronzo47 for HIRI and Abdul-Ghani et al. for MISI.51 The FINDRISC was calculated as defined by Lindström et al.5
Isolation of Circulating miRNAs from Plasma Samples
Total RNA was isolated from plasma using the miRNeasy Mini Kit (QIAGEN, Hilden, Germany). Briefly, venous blood from the participants was collected at baseline (day 0 before dietary intervention) in tubes containing EDTA and centrifuged at 2,000 × g for 10 min for plasma separation from blood cells. Further, 200 μL EDTA-plasma was mixed with 1 mL Qiazol, incubated for 5 min at room temperature, and subsequently mixed with 200 μL chloroform. We added 2 μg MS2 RNA carrier (Roche, Mannheim, Germany) before the chloroform protocol step. The organic and aqueous phases were separated by centrifugation at 12,000 × g for 15 min at 4°C. The aqueous phase was collected and the RNA was precipitated by adding 100% ethanol. The mixture was applied to an miRNeasy Mini spin column and was centrifuged at 8,000 × g for 2 min. Next, 700 μL RWT buffer was added to the RNeasy MinElute spin column at 8,000 × g for 2 min. It was then washed again with 500 μL RPE buffer and 500 μL 80% ethanol. RNA was eluted in 14 μL RNase-free water. RNA purity and concentration were evaluated by spectrophotometry using NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA).
miRNA Retrotranscription and Preamplification
The miRNA expression study was carried out on 24 miRNAs, which, based on a bibliographic search, were selected according to their association with insulin sensitivity, insulin secretion, inflammation, and growth and proliferation of beta cells (Table S1). The retrotranscription of RNA was carried out using the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA, USA). RT mix contained 2 μL RNA and 3 μL RT custom primer pool in a final volume of 7.5 μL. RT primer pool was customized selecting specific primers for our set of target miRNAs in the database (https://www.thermofisher.com/es/en/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/mirna-ncrna-taqman-assays.html). Plates were incubated in the iQ5 thermocycler (Bio-Rad, Hercules, CA, USA) at 16°C for 30 min, followed by 42°C for 30 min, and finally at 85°C for 5 min. In this step, the cDNA was stored at −20°C for a maximum of 1 week. Then, we prepared a mixture containing 10 μL customized PreAmp primer pool specific for our set of target miRNAs and 7.5 μL RT mix and 20 μL TaqMan PreAmp Master Mix (Life Technologies, Carlsbad, CA, USA) to a final volume of 40 μL. Next, the mixture was incubated in the iQ5 Thermocycler using the following steps: denaturation at 95°C for 10 min, 55°C for 2 min, and 72°C for 2 min; 20 cycles of amplification for 15 s at 95°C and 4 min at 60°C per cycle; and finally incubation at 99.9°C for 10 min. The pre-amplified products were then diluted with RNase-free water at a ratio of 1:40 and used for the real-time RT-PCR reactions.
Levels of Circulating miRNAs by Real-Time PCR
We measured the levels of miRNAs at baseline of the study with the OpenArray platform (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions. As a normalization method, we first selected the miRNAs that showed the least variability in their CT values in all samples. For this, we used the NormFinder Bioinformatic tool (MOMA-Department of Molecular Medicine, Aarhus University Hospital, Denmark),52 a software extensively used in expression studies.53, 54 The application showed that the most stable miRNAs were miR-143 and miR-144. Second, we used the BestKeeper method to calculate the geometric mean of the pairwise Ct values (Ct values of miR-143 and miR-144).55 The relative expression data were analyzed using OpenArray Real-Time qPCR Analysis Software (Life Technologies, Carlsbad, CA, USA).
PCA and OPLS-DA
Prior to the PCA, the data were normalized into a dataset suitable for analysis. Applying the procedures of mean-centering and unit variance (UV) scaling, the data of the miRNA levels were processed using SIMCA-P+ (version 14.0.0.1359; Umetrics, Umea, Sweden). PCA was applied to the dataset and the score plots were visually inspected for the detection of patterns and outliers. OPLS-DA was used to compare miRNA levels at baseline, in order to analyze the differences between Incident-T2DM and non-T2DM patients during follow-up. OPLS-DA validation was performed by cross-validation (CV) method in the SIMCA-P+ software using the default setting, which includes a procedure of 7-fold cross-validation where the dataset is split into 7 different subsets.56The quality of the models obtained by PCA and OPLS-DA was assessed by interrogation of the R2 and Q2 parameters.56, 57 Next, we selected those miRNAs with higher discriminatory power between groups from the VIP score obtained in the OPLS-DA model. The miRNAs with a VIP score > 1 were considered important for differentiating between groups.
Statistical Analysis
The quantitative data were evaluated for normal distribution by the Kolmogorov-Smirnov test, with the cutoff for normal distribution set at p > 0.05. For data that were not normally distributed, we used the Mann-Whitney U test. The relationship between miRNA plasma levels and insulin sensitivity/resistance and beta cell function indexes was evaluated through a linear regression analysis adjusted by age, gender, BMI, TGs, and c-HDL. We used the Cox proportional hazard regression analysis to test the potential predictive value of the miRNAs studied. The level values of each miRNA were categorized by tertiles: low levels (T1), medium levels (T2), and high levels (T3). The HR in the analysis of each miRNA studied was analyzed by comparing T1 versus T2 and T1 versus T3. Six miRNAs with HR T1 versus T3 ≥ 2.5 were selected for Cox regression multi-miRNA analysis. The subjects were, therefore, classified into three categories: C1 category was composed of patients with low levels in at least three of the four downregulated miRNAs (miR-9, miR-28-3p, miR-29a, and miR-103) and high levels in at least one of the 2 upregulated miRNAs (miR-150 and miR-30a-5p) (n = 46); C3 category was composed of patients with high levels in at least three of the four downregulated miRNAs and low levels in at least one of the two upregulated miRNAs (n = 32); and, finally, C2 was made up of patients with an intermediate miRNA deregulation profile (n = 356). This classification was called 6miRNAs-variable and was included in the Cox regression multi-miRNA analysis. The HR in the analysis between C1 versus C2 and C1 versus C3 was compared. The lineal regression and Cox regression analyses were adjusted by age, gender, diet, HbA1c, BMI, TGs, c-HDL, and waist circumference. p values ≤ 0.05 were considered statistically significant. We used ROC analysis to estimate the AUC, accuracy, specificity, and sensitivity of the variables for differentiation of Incident-T2DM from non-T2DM patients. The models were corrected for those covariables that were allowed, avoiding overestimating information, the set of covariables included: diet, age, gender, BMI, c-HDL, TGs, HbA1c, and waist circumference. For internal validation of the model, the degree of over-optimism was estimated using bootstrap resampling of the original set (1,000 randomized samples).
All the statistical analyses were carried out using SPSS (now PASW Statistic for Windows, version 21.0) (IBM, Chicago, IL, USA). Additionally, we used MetaboAnalyst 3.0 to classify the variables included in the ROC model according to the average importance of variables. Data normalization was performed using the auto-scaling method, based on mean centered and divided by the SD of each variable.58, 59 The DeLong test was performed to compare the ROC models as described in DeLong et al.60 for paired ROC curves using the software R.
Author Contributions
R.J.-L., O.A.R.-Z., A.C., and J. López-Miranda. conceived and designed the experiments. J.F.A.-D., J.D.-L., P.P.-M., J. López-Moreno, and J. López-Miranda participated in the recruitment and carried out the clinical and nutritional control of the volunteers. J.C.-V. was responsible for the management of samples and laboratory biochemical determinations. R.J.-L., A.C., and O.A.R.-Z. performed the experiments and collected the data. R.J.-L., O.A.R.-Z., J.F.A.-D., I.R.-R., E.M.Y.-S., A.C., and J. López-Miranda analyzed and interpreted the data. R.J.-L., O.A.R.-Z., A.C., and J. López-Miranda drafted the manuscript. J. López-Miranda conceived and designed the study. H.M.-A. supported the statistical analysis of data. J.P.C., J.M.O., and J. López-Miranda provided critical revision of the paper for the important intellectual content. J.D.-L., P.P.-M., A.C., and J. López-Miranda had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All the authors were involved in writing the paper and gave their final approval to the submitted and published versions.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
The CIBEROBN is an initiative of the Instituto de Salud Carlos III, Madrid, Spain. The CORDIOPREV study is supported by the Fundación Patrimonio Comunal Olivarero, Junta de Andalucía (Consejería de Salud; Consejería de Agricultura y Pesca; and Consejería de Innovación, Ciencia y Empresa), Diputaciones de Jaén y Córdoba, Centro de Excelencia en Investigación sobre Aceite de Oliva y Salud; and Ministerio de Medio Ambiente, Medio Rural y Marino, Gobierno de España. It was also partly supported by research grants from the Ministerio de Ciencia e Innovacion (AGL2009-122270 to J. López-Miranda); Ministerio de Economía y Competitividad (AGL2012/39615, PIE14/00005, and PIE 14/00031 to J. López-Miranda and AGL2015-67896-P to J.L.-M. and A.C.); Consejería de Innovación, Ciencia y Empresa, Proyectos de Investigación de Excelencia, Junta de Andalucía (CVI-7450 to J. López-Miranda); Fondo Europeo de Desarrollo Regional (FEDER); and U.S. Department of Agriculture-Agricultural Research Service (ARS), under Agreement 58-1950-4-003 (J.M.O.). A.C. is supported by an ISCIII research contract (Programa Miguel-Servet CP14/00114). H.M.-A. is partially supported by the VPPI of University of Seville. We would like to thank the Córdoba node of the Biobank of the Sistema Sanitario Publico de Andalucía (Andalucía, Spain) for providing the biological human samples. We would also like to thank the EASP (Escuela Andaluza de Salud Publica), Granada, Spain, which performed the randomization process for this study.
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.002.
Supplemental Information
References
- 1.Lau E., Carvalho D., Pina-Vaz C., Barbosa J.A., Freitas P. Beyond gut microbiota: understanding obesity and type 2 diabetes. Hormones (Athens) 2015;14:358–369. doi: 10.14310/horm.2002.1571. [DOI] [PubMed] [Google Scholar]
- 2.Ginter E., Simko V. Type 2 diabetes mellitus, pandemic in 21st century. Adv. Exp. Med. Biol. 2012;771:42–50. doi: 10.1007/978-1-4614-5441-0_6. [DOI] [PubMed] [Google Scholar]
- 3.Collins G.S., Mallett S., Omar O., Yu L.M. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 2011;9:103. doi: 10.1186/1741-7015-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penn L., White M., Lindström J., den Boer A.T., Blaak E., Eriksson J.G., Feskens E., Ilanne-Parikka P., Keinänen-Kiukaanniemi S.M., Walker M. Importance of weight loss maintenance and risk prediction in the prevention of type 2 diabetes: analysis of European Diabetes Prevention Study RCT. PLoS ONE. 2013;8:e57143. doi: 10.1371/journal.pone.0057143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindström J., Tuomilehto J. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 2003;26:725–731. doi: 10.2337/diacare.26.3.725. [DOI] [PubMed] [Google Scholar]
- 6.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Committee I.E., International Expert Committee International Expert Committee report on the role of the A1C assay in the diagnosis of diabetes. Diabetes Care. 2009;32:1327–1334. doi: 10.2337/dc09-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodovicz K.G., Dekker J.M., Rijkelijkhuizen J.M., Rhodes T., Mari A., Alssema M., Nijpels G., Williams-Herman D.E., Girman C.J. The Finnish Diabetes Risk Score is associated with insulin resistance but not reduced β-cell function, by classical and model-based estimates. Diabet. Med. 2011;28:1078–1081. doi: 10.1111/j.1464-5491.2011.03315.x. [DOI] [PubMed] [Google Scholar]
- 9.Saaristo T., Peltonen M., Keinänen-Kiukaanniemi S., Vanhala M., Saltevo J., Niskanen L., Oksa H., Korpi-Hyövälti E., Tuomilehto J., FIN-D2D Study Group National type 2 diabetes prevention programme in Finland: FIN-D2D. Int. J. Circumpolar Health. 2007;66:101–112. doi: 10.3402/ijch.v66i2.18239. [DOI] [PubMed] [Google Scholar]
- 10.Florkowski C. HbA1c as a Diagnostic Test for Diabetes Mellitus - Reviewing the Evidence. Clin. Biochem. Rev. 2013;34:75–83. [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao Y., Okumiya T., Saibara T., Park K., Sasaki M. Abnormally decreased HbA1c can be assessed with erythrocyte creatine in patients with a shortened erythrocyte age. Diabetes Care. 1998;21:1732–1735. doi: 10.2337/diacare.21.10.1732. [DOI] [PubMed] [Google Scholar]
- 12.Cohen R.M., Haggerty S., Herman W.H. HbA1c for the diagnosis of diabetes and prediabetes: is it time for a mid-course correction? J. Clin. Endocrinol. Metab. 2010;95:5203–5206. doi: 10.1210/jc.2010-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipska K.J., De Rekeneire N., Van Ness P.H., Johnson K.C., Kanaya A., Koster A., Strotmeyer E.S., Goodpaster B.H., Harris T., Gill T.M., Inzucchi S.E. Identifying dysglycemic states in older adults: implications of the emerging use of hemoglobin A1c. J. Clin. Endocrinol. Metab. 2010;95:5289–5295. doi: 10.1210/jc.2010-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guay C., Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013;9:513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 15.Tavintharan S., Chi L.S., Fang S.C., Arunmozhiarasi A., Jeyaseelan K. Riboregulators and metabolic disorders: getting closer towards understanding the pathogenesis of diabetes mellitus? Curr. Mol. Med. 2009;9:281–286. doi: 10.2174/156652409787847245. [DOI] [PubMed] [Google Scholar]
- 16.He A., Zhu L., Gupta N., Chang Y., Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 17.Karolina D.S., Armugam A., Tavintharan S., Wong M.T., Lim S.C., Sum C.F., Jeyaseelan K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE. 2011;6:e22839. doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Ouaamari A., Baroukh N., Martens G.A., Lebrun P., Pipeleers D., van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Xu X., Liang Y., Liu S., Xiao H., Li F., Cheng H., Fu Z. miR-375 enhances palmitate-induced lipoapoptosis in insulin-secreting NIT-1 cells by repressing myotrophin (V1) protein expression. Int. J. Clin. Exp. Pathol. 2010;3:254–264. [PMC free article] [PubMed] [Google Scholar]
- 20.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia H.Q., Pan Y., Peng J., Lu G.X. Over-expression of miR375 reduces glucose-induced insulin secretion in Nit-1 cells. Mol. Biol. Rep. 2011;38:3061–3065. doi: 10.1007/s11033-010-9973-9. [DOI] [PubMed] [Google Scholar]
- 22.Lovis P., Gattesco S., Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 23.Tang X., Muniappan L., Tang G., Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regazzi R. Diabetes mellitus reveals its micro-signature. Circ. Res. 2010;107:686–688. doi: 10.1161/CIRCRESAHA.110.228841. [DOI] [PubMed] [Google Scholar]
- 25.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 26.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2011;34(Suppl 1):S62–S69. doi: 10.2337/dc11-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones A., Danielson K.M., Benton M.C., Ziegler O., Shah R., Stubbs R.S., Das S., Macartney-Coxson D. miRNA Signatures of Insulin Resistance in Obesity. Obesity (Silver Spring) 2017;25:1734–1744. doi: 10.1002/oby.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah R., Murthy V., Pacold M., Danielson K., Tanriverdi K., Larson M.G., Hanspers K., Pico A., Mick E., Reis J. Extracellular RNAs Are Associated With Insulin Resistance and Metabolic Phenotypes. Diabetes Care. 2017;40:546–553. doi: 10.2337/dc16-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu K., Cowie C.C., Harris M.I. Diabetes and decline in heart disease mortality in US adults. JAMA. 1999;281:1291–1297. doi: 10.1001/jama.281.14.1291. [DOI] [PubMed] [Google Scholar]
- 30.Martín-Timón I., Sevillano-Collantes C., Segura-Galindo A., Del Cañizo-Gómez F.J. Type 2 diabetes and cardiovascular disease: Have all risk factors the same strength? World J. Diabetes. 2014;5:444–470. doi: 10.4239/wjd.v5.i4.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makrilakis K., Liatis S., Grammatikou S., Perrea D., Stathi C., Tsiligros P., Katsilambros N. Validation of the Finnish diabetes risk score (FINDRISC) questionnaire for screening for undiagnosed type 2 diabetes, dysglycaemia and the metabolic syndrome in Greece. Diabetes Metab. 2011;37:144–151. doi: 10.1016/j.diabet.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Omech B., Mwita J.C., Tshikuka J.G., Tsima B., Nkomazna O., Amone-P’Olak K. Validity of the Finnish Diabetes Risk Score for Detecting Undiagnosed Type 2 Diabetes among General Medical Outpatients in Botswana. J. Diabetes Res. 2016;2016:4968350. doi: 10.1155/2016/4968350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 34.Jansen F., Yang X., Proebsting S., Hoelscher M., Przybilla D., Baumann K., Schmitz T., Dolf A., Endl E., Franklin B.S. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J. Am. Heart Assoc. 2014;3:e001249. doi: 10.1161/JAHA.114.001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kantharidis P., Wang B., Carew R.M., Lan H.Y. Diabetes complications: the microRNA perspective. Diabetes. 2011;60:1832–1837. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seyhan A.A., Nunez Lopez Y.O., Xie H., Yi F., Mathews C., Pasarica M., Pratley R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci. Rep. 2016;6:31479. doi: 10.1038/srep31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roggli E., Gattesco S., Caille D., Briet C., Boitard C., Meda P., Regazzi R. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 40.Wang X.M., Kim H.P., Song R., Choi A.M. Caveolin-1 confers antiinflammatory effects in murine macrophages via the MKK3/p38 MAPK pathway. Am. J. Respir. Cell Mol. Biol. 2006;34:434–442. doi: 10.1165/rcmb.2005-0376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plaisance V., Abderrahmani A., Perret-Menoud V., Jacquemin P., Lemaigre F., Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 42.Delgado-Lista J., Perez-Martinez P., Garcia-Rios A., Alcala-Diaz J.F., Perez-Caballero A.I., Gomez-Delgado F., Fuentes F., Quintana-Navarro G., Lopez-Segura F., Ortiz-Morales A.M. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): Rationale, methods, and baseline characteristics: A clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am. Heart J. 2016;177:42–50. doi: 10.1016/j.ahj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco-Rojo R., Alcala-Diaz J.F., Wopereis S., Perez-Martinez P., Quintana-Navarro G.M., Marin C., Ordovas J.M., van Ommen B., Perez-Jimenez F., Delgado-Lista J., Lopez-Miranda J. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: the CORDIOPREV-DIAB randomised clinical trial. Diabetologia. 2016;59:67–76. doi: 10.1007/s00125-015-3776-4. [DOI] [PubMed] [Google Scholar]
- 44.Allain C.C., Poon L.S., Chan C.S., Richmond W., Fu P.C. Enzymatic determination of total serum cholesterol. Clin. Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 45.Bucolo G., David H. Quantitative determination of serum triglycerides by the use of enzymes. Clin. Chem. 1973;19:476–482. [PubMed] [Google Scholar]
- 46.Briggs C.J., Anderson D., Johnson P., Deegan T. Evaluation of the polyethylene glycol precipitation method for the estimation of high-density lipoprotein cholesterol. Ann. Clin. Biochem. 1981;18:177–181. doi: 10.1177/000456328101800309. [DOI] [PubMed] [Google Scholar]
- 47.Matsuda M., DeFronzo R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 48.Song Y., Manson J.E., Tinker L., Howard B.V., Kuller L.H., Nathan L., Rifai N., Liu S. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women’s Health Initiative Observational Study. Diabetes Care. 2007;30:1747–1752. doi: 10.2337/dc07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanson R.L., Pratley R.E., Bogardus C., Narayan K.M., Roumain J.M., Imperatore G., Fagot-Campagna A., Pettitt D.J., Bennett P.H., Knowler W.C. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am. J. Epidemiol. 2000;151:190–198. doi: 10.1093/oxfordjournals.aje.a010187. [DOI] [PubMed] [Google Scholar]
- 50.Tang W., Fu Q., Zhang Q., Sun M., Gao Y., Liu X., Qian L., Shan S., Yang T. The association between serum uric acid and residual β -cell function in type 2 diabetes. J. Diabetes Res. 2014;2014:709691. doi: 10.1155/2014/709691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdul-Ghani M.A., Matsuda M., Balas B., DeFronzo R.A. Muscle and liver insulin resistance indexes derived from the oral glucose tolerance test. Diabetes Care. 2007;30:89–94. doi: 10.2337/dc06-1519. [DOI] [PubMed] [Google Scholar]
- 52.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 53.Danese E., Minicozzi A.M., Benati M., Paviati E., Lima-Oliveira G., Gusella M., Pasini F., Salvagno G.L., Montagnana M., Lippi G. Reference miRNAs for colorectal cancer: analysis and verification of current data. Sci. Rep. 2017;7:8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Spiegelaere W., Dern-Wieloch J., Weigel R., Schumacher V., Schorle H., Nettersheim D., Bergmann M., Brehm R., Kliesch S., Vandekerckhove L., Fink C. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE. 2015;10:e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 56.Triba M.N., Le Moyec L., Amathieu R., Goossens C., Bouchemal N., Nahon P., Rutledge D.N., Savarin P. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015;11:13–19. doi: 10.1039/c4mb00414k. [DOI] [PubMed] [Google Scholar]
- 57.Worley B., Powers R. Multivariate Analysis in Metabolomics. Curr. Metabolomics. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia J., Broadhurst D.I., Wilson M., Wishart D.S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia J., Wishart D.S. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinformatics. 2011;Chapter 14:Unit 14.10. doi: 10.1002/0471250953.bi1410s34. [DOI] [PubMed] [Google Scholar]
- 60.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.