Abstract
CRISPR/Cas9 has been confirmed as a distinctly efficient, simple-to-configure, highly specific genome-editing tool that has been used to treat monogenetic disorders. Epidermolytic palmoplantar keratoderma (EPPK) is a common autosomal dominant keratin disease resulting from dominant-negative mutation of the KRT9 gene, and it has no effective therapy. We performed CRISPR/Cas9-mediated treatment on a knockin (KI) transgenic mouse model that carried a small indel heterozygous mutation of Krt9, c.434delAinsGGCT (p.Tyr144delinsTrpLeu), which caused a humanized EPPK-like phenotype. The mutation within exon 1 of Krt9 generated a novel protospacer adjacent motif site, TGG, for Cas9 recognition and cutting. By delivering lentivirus vectors (LVs) encoding single-guide RNAs (sgRNAs) and Cas9 that targeted Krt9 sequence into HeLa cells engineered to constitutively express wild-type and mutant keratin 9 (K9), we found the sgRNA was highly effective in reducing expression of the mutant K9 protein in vitro. We injected the LV into the fore-paws of adult KI-Krt9 mice three times every 8 days and found that the expression of K9 decreased ∼14.6%. The phenotypic mitigation was revealed by restoration of the abnormal differentiation and aberrant proliferation of the epidermis. Our data are the first to show that CRISPR/Cas9 is a potentially powerful therapeutic option for EPPK and other PPK subtypes.
Keywords: CRISPR/Cas9, gene therapy, epidermolytic palmoplantar keratoderma, Krt9 gene (mouse), indel
Introduction
Hereditary palmoplantar keratoderma (PPK) is a heterogeneous group of genodermatoses characterized by abnormal thickening of the palms and soles. PPK arises from abnormal terminal differentiation and construction of the cornified envelopes.1, 2 Among the PPKs, epidermolytic PPK (EPPK; OMIM: 144200) inherited in an autosomal dominant pattern, with a worldwide incidence of 2.2–4.4 per 100,000 live newborns, is the commonest subtype. It manifests at birth or soon thereafter as diffuse yellow palmoplantar hyperkeratosis with distinct erythematous borders, sometimes combined with knuckle pads, digital mutilation, or camptodactyly. Histology may show epidermolytic areas in the upper stratum spinosum and granulosum.1, 2, 3 Currently, dominant-negative mutations in the KRT9 gene (encoding keratin 9 [K9]) on chromosome 17q12 are recognized as the molecular etiology in most cases.4, 5, 6, 7, 8 The precise molecular and cellular abnormalities underlying EPPK remain unknown, and there is still no effective therapy 117 years after its first description by Vorner in 1901. However, the unique pathological mechanism in which EPPK is exclusively caused by dominant-negative mutations of KRT9 makes it well suited for the development of gene-editing-based therapeutics that specifically inhibit the activity of the causative K9 allele.2, 6, 7, 8
To date, considerable numbers of reports have confirmed that the CRISPR/Cas9 is the most innovative genome-editing technique to treat genetic defects.7, 8, 9, 10, 11 In principle, CRISPR/Cas9 relies on a single-guide RNA (sgRNA) to recruit Cas9 nuclease to a specific locus by sequence complementarity and to induce double-strand breaks. These breaks are subsequently repaired either by non-homologous end joining or homology-directed repair in the presence of a donor template.12 By introducing the CRISPR/Cas9 system into a cell line, a zygote, or a specific tissue, it is possible to ameliorate the abnormal phenotype caused by a specific disease-causing gene mutation.11, 12, 13, 14, 15
Previously, using a gene-targeting technique, we constructed a knockin (KI) transgenic mouse model that carried a small insertion-deletion (indel) heterozygous mutation of the Krt9 gene, c.434delAinsGGCT (p.Tyr144delinsTrpLeu), corresponding to the human mutation KRT9/c.500delAinsGGCT (p.Tyr167delinsTrpLeu), and this caused a humanized EPPK-like phenotype.16 The indel mutation within exon 1 of Krt9 generated a novel protospacer adjacent motif (PAM) site, TGG, for Cas9 recognition and cutting. Thus, by delivering a lentivirus vector (LV) encoding sgRNAs and Cas9 that targeted the Krt9 DNA sequence into HeLa cells engineered to constitutively express either the wild-type (WT) or the c.434delAinsGGCT mutant allele, we developed an in vitro cell-based system to test the CRISPR potency. We then injected LV carrying sgRNA and Cas9 components into the right fore-paw of adult KI-Krt9 mice three times every 8 days. The phenotypic correction was demonstrated as ∼14.6% reduction of disease-associated K9 after the treatment compared with controls. The mitigation of the disease was also revealed by restoration of the abnormal differentiation and aberrant proliferation of the epidermis. Our results suggested that CRISPR/Cas9 is a potentially powerful therapeutic option for EPPK and other PPK subtypes.
Results
Construction of Krt9-Specific sgRNAs
The PAM (NGG sequence) is a critical feature of the CRISPR/Cas9 system, dictating its DNA target search mechanism.9, 10, 11 Based on the Krt9/c.434delAinsGGCT mutation, a novel PAM site (TAC→TGGCTC) within exon 1 of the Krt9 gene sequence was generated to design the sgRNA, and another novel PAM, two bases next to the mutation site, was also formed. Thus, two sgRNAs, sg1 and sg2, complementary to the sequence 20 nt adjacent to the PAM site were designed (Figure 1A). Assessed by the “Optimised CRISPR Design Tool” provided online by Dr. Feng Zhang (http://crispr.mit.edu/), sg1 and sg2 were calculated as having scores of 60 and 62, which were the highest-efficiency predictable sgRNAs that could be generated within a 10-base cut-to-mutation distance.13 Ten potential highest-ranking genomic off-target sites in the mouse genome were also predicted using this program (Table S1).
Figure 1.
Delivery of CRISPR/Cas9 Mediates Gene Editing In Vitro
(A) Experimental design. “A” in green indicates the WT base. Two sgRNAs (underlined) were designed according to the mutation site of four bases (red in the MUT lines). PAM is indicated with yellow frame; blue vertical lines mark the cut sites. Heterozygous KI-Krt9 mice harboring the indel mutant, c.434delAinsGGCT (p.Tyr144delinsTrpLeu), treated with sg1-Cas9-LV after in vitro experiments confirmed the efficacy of the LV vector. (B and C) FACS results showing the changes in FITC and mCherry fluorescence after treating HeLa cells with LV expressing both Krt9 exon1-EGFP and Krt9 exon1 MUT-mCherry (B). Thus, the HeLa Hez cell line was established. The reduction of EGFP fluorescence after transduction by sg1-Cas9-LV or sg2-Cas9-LV is shown in (C). See also Figure S2. (D and E) Immunoblotting of HeLa Hez cells against K9 (D) and semiquantitative immunoblotting analyses (relative to GAPDH and FLAG expression) (n = 3, *p < 0.05) (E). Each experiment was repeated at least three times. NC, untreated HeLa Hez cells; PC, HeLa Hez cells treated with Cas9-LV.
Specificity and Potency of Lentivirus-Delivered CRISPR/Cas9 System In Vitro
The allele specificity and potency of sg1 and sg2 were assessed in vitro. We established a heterozygous HeLa cell line (HeLa Hez) stably co-expressing WT allele-mCherry and mutant (MUT) allele-EGFP, i.e., heterozygous for Krt9 (Figure S1). HeLa Hez cells transduced by Cas9-LV without the sgRNA component served as the positive control, and cells without EGFP or mCherry fluorescence as the negative control. The fluorescence intensity of EGFP via flow cytometry (FACS) elucidated the efficacy of CRISPR/Cas9 on the target mutation sequence, and that of mCherry evaluated the possibility of off-target effects on the WT allele. Robust expression of FLAG demonstrated the transduction efficiency of LV and the high expression of Cas9, regardless of whether Cas9-LV, sg1-Cas9-LV, or sg2-Cas9-LV was transduced into cells (Figure S1).
After the transduction of sg1-Cas9-LV, potent reductions of 63.2% for total fluorescein isothiocyanate (FITC) fluorescence and 22.8% for mCherry fluorescence were observed in HeLa Hez cells, compared with lesser reductions of 32.1% and 7.0% measured in the cells infected with sg2-Cas9-LV (Figures 1B and 1C). These data indicated the specificity and potency of both sg1-Cas9-LV and sg2-Cas9-LV, but the vector sg1-Cas9-LV had a higher efficiency in vitro (Figure S2). The results of western blotting analysis were consistent with these observations. Using the expression levels of GAPDH and FLAG as references, we found reductions of 58.2% versus 45.8% and 43.5% versus 21.6% K9 protein in the different transduced HeLa Hez cells 72 hr after treatment, respectively (Figures 1A and 1E). Therefore, sg1-Cas9-LV was the first choice for the in vivo treatment.
Localized Subcutaneous Injection of the CRISPR/Cas9 System and Efficacy Assessment In Vivo
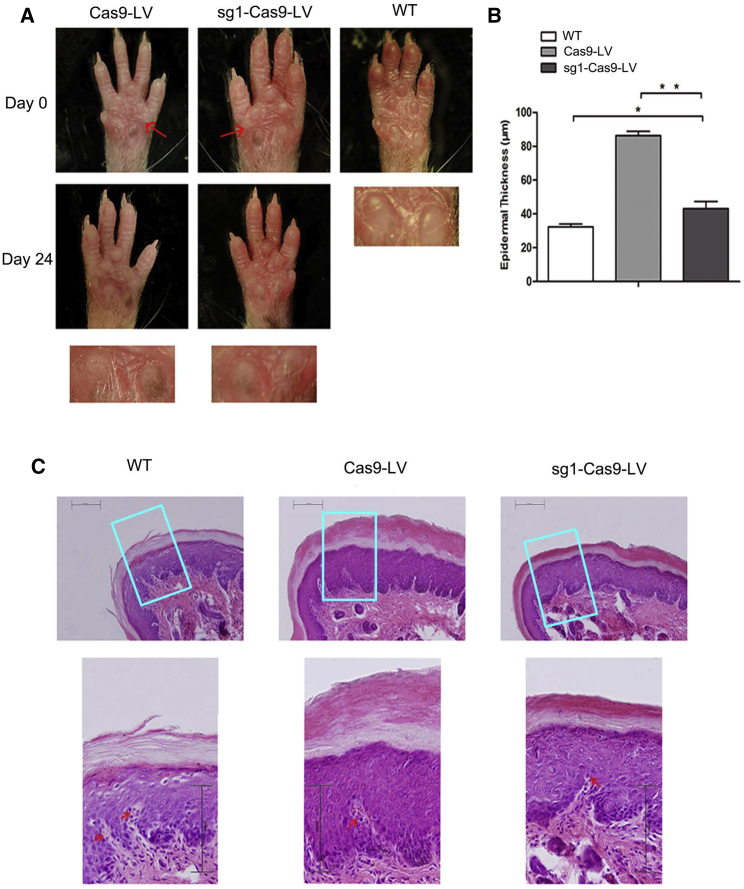
In the humanized EPPK-like mouse model, the phenotype developed on the major impact-dampening footpads of the main weight-bearing paws, the fore-paws. To determine whether sg1-Cas9-LV affects the process of hyperkeratosis in vivo, we treated 12-week-old KI-Krt9 mice (n = 4) with subcutaneous injections of 15 μL sg1-Cas9-LV into the right fore-paw three times every 8 days, each with a viral dose of 4 × 108 infectious units/mL. The left fore-paw of the model mice was injected with Cas9-LV as controls, and all mice were imaged before and after injection. Tissues from the paw pads were analyzed by H&E staining, immunofluorescent (IF) staining, transmission electron microscopy (TEM), and immunoblotting after injection for 24 days.
The hyperpigmented calluses of the right fore-paw pad regressed significantly after treatment, compared with those of the left pad (Figure 2A). H&E staining showed that the epidermal thickness of the right pad decreased, with less expansion of the epidermis and less massive hyperkeratosis. The average epidermal thickness of the right pad was ∼43 μm, half as thick as the left pad (86 μm), although it was still 1.34 times that of the normal pad of WT mice (32 μm) (Figure 2B). Usually, melanin covers the upper part of the nucleus of a keratinocyte to form a supranuclear cap to protect the nucleus from ultraviolet radiation. The melanin was increased in the epidermis of the pads of both K9 knockout and K9 mutant KI mouse models.16, 17 The results of H&E staining analysis showed reduced levels of melanin in the right pads similar to WT mice, compared with the left pads (Figure 2C).
Figure 2.
Fore-Paw Pad Abnormalities Are Partly Rescued in the sg1-Cas9-LV-Treated Adult KI-Krt9 Mouse
(A) Fore-paw pads of KI-Krt9 mice treated with Cas9-LV or sg1-Cas9-LV, and those of untreated WT mice. Red arrows indicate injection locations of Cas9-LV and sg1-Cas9-LV. (B) Analysis of epidermal thickness from the pads. (C) Representative images of H&E-stained epidermis showing that melanin (red arrows) and epidermal thickness decreased after sg1-Cas9-LV treatment. Each experiment was repeated at least three times. Scale bars, 100 μm. Mean ± SEM. *p < 0.05; **p < 0.01.
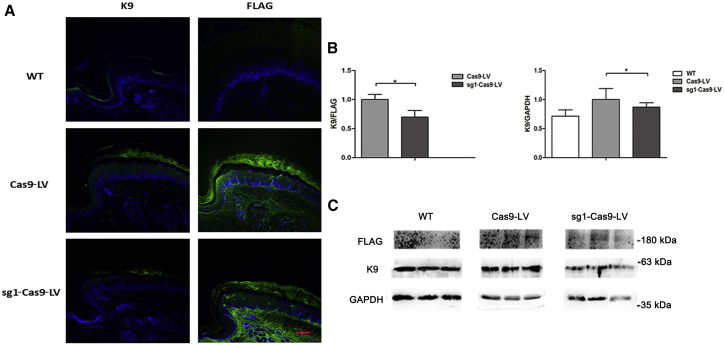
Analysis of both IF staining and immunoblotting against FLAG reflected the higher efficiency of sg1-Cas9-LV treatment (Figure 3). The expression levels of K9 were used for normalization. Defining the K9 expression in paw pads of WT mice as 100.0%, the ratio was 140.0% in the Cas9-LV-treated left pads of KI-Krt9 mice, but 120.0% in the sg1-Cas9-LV-treated right pads. Taking the K9 expression in the left pads of mutant mice as 100.0%, the ratio was only 71.4% for WT and 85.4% for the sg1-Cas9-LV-treated right pads of KI-Krt9 mice, i.e., an ∼14.6% decrease in K9 expression was found in the sg1-Cas9-LV-treated mice (Figure 3B). Relative to the expression level of FLAG, K9 expression was reduced 30.3% in the sg1-Cas9-LV-treated KI-Krt9 mice (Figure 3B).
Figure 3.
Delivery of sg1-Cas9-LV Decreases the Excessive Expression of K9
(A) IF staining against K9 and FLAG. Scale bar, 50 μm. (B) Semiquantitative immunoblotting analyses relative to GAPDH and FLAG (mean ± SEM; n = 3; *p < 0.05). (C) Immunoblotting against K9, GAPDH, and FLAG. Each experiment was repeated at least three times.
After injections for 24 days, the experimental mice were euthanized and genomic DNA (gDNA) prepared from the paw pads. PCR amplification targeted to the specific DNA region including exon 1 and intron 1 of Krt9, cloning, and Sanger sequencing were performed. In the humanized EPPK-like heterozygous mouse model, the mutant allele possessed a 121-bp KI fragment in intron 1 of Krt9 that differentiated it from the WT allele. Both the forward and reverse sequencing results of 72 clones established from the gDNA of the sg1-Cas9-LV-treated pads identified 31 mutant clones and 41 WT clones that maintained the intact Krt9 exon 1 and intron 1 sequence (Figure S3). DNA sequencing of the 10 potential highest-ranking genomic off-target sites in 200 clones showed only several single-nucleotide mutations at these 10 loci (Table S1). No abnormalities were observed in mice treated with either sg1-Cas9-LV or Cas9-LV.
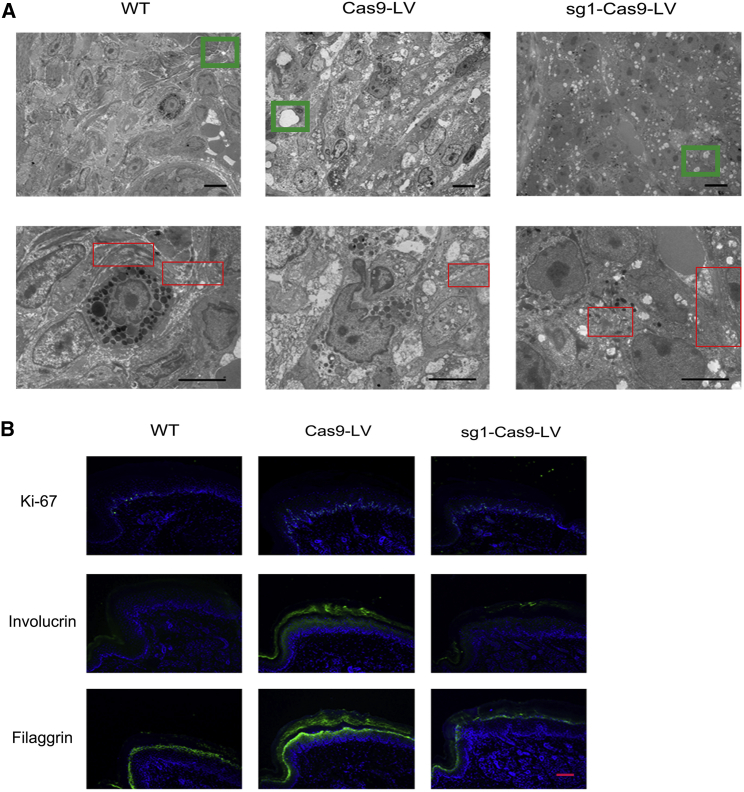
TEM analysis revealed more normal tonofilaments and fewer cytolysis-caused vacuoles in the suprabasal layers of the epidermis of the right pads, indicating the ameliorated phenotype of KI-Krt9 mice (Figure 4A).
Figure 4.
Disrupted Cytoskeletal Integrity and Abnormal Proliferation Are Inhibited
(A) Ultrastructural images showing the form of tonofilaments (red frames), cytolysis (green frames), and keratin filaments. Scale bars, 5 μm. (B) IF staining against Ki-67, involucrin, and filaggrin after treatment. Each experiment was repeated at least three times. Scale bar, 50 μm.
IF staining against the cellular proliferation markers Ki-67 and 5-bromo-2′-deoxyuridine (BrdU) and the keratinocyte differentiation markers involucrin and filaggrin demonstrated the partial rescue of abnormal proliferation after the sg1-Cas9-LV injection. Ki-67 is an excellent marker to determine the growth fraction of a given cell population (Figure 4B). BrdU, a thymidine analog that is incorporated into dividing cells during DNA synthesis, was also used to measure cell proliferation, and the data supported the result of Ki-67 staining (Figure S4).18 Although both involucrin and filaggrin appropriately localized to the granular layer in WT mice, we observed a decrease of involucrin and filaggrin staining intensity within the horny layer of the treated KI-Krt9 mice (Figure 4B).
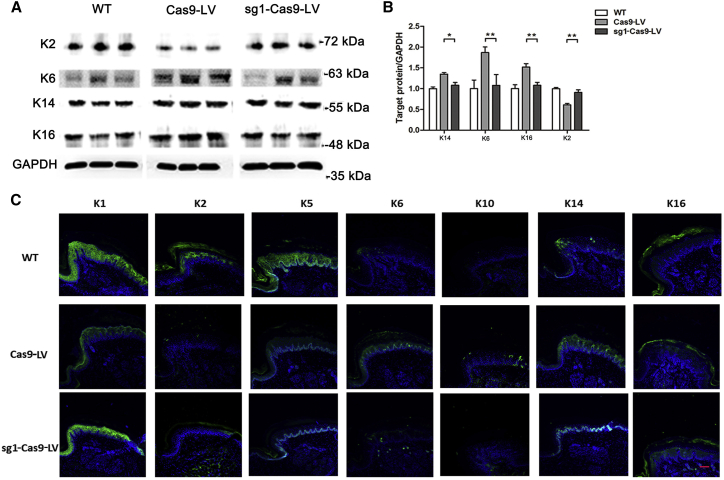
In the paw pads of KI-Krt9 mice, the dominant-negative mutation of K9 induces an imbalance in a subset of palmoplantar keratins (K1, K2, K5, K6, K10, K14, and K16).16 After sg1-Cas9-LV treatment, the results of both immunoblotting and IF staining showed that both the expression levels and localization of these keratins were rescued and similar to those in WT mice (Figure 5). Among them, the expression of K2 increased 48.9%; K6, 43.2%; K14, 19.7%, and K16, 28.9% (Figure 5B). Significant changes in the expression and localization of K1 and K14 were similar to those in WT mice, which also indicated that the differentiation of epidermis treated with sg1-Cas9-LV tended toward the normal state (Figure 5C).
Figure 5.
CRISPR/Cas9-Mediated Therapy Improves the Compensatory Expression of Other Palmoplantar Keratins
(A) Immunoblotting against K2, K6, K14, and K16. (B) Semiquantitative immunoblotting analyses relative to GAPDH (mean ± SEM; n = 3; *p < 0.05; **p < 0.01). (C) IF staining for K1, K2, K5, K6, K10, K14, and K16. Each experiment was repeated at least three times. Scale bar, 50 μm.
Discussion
The vast majority of mutations in inherited keratin disorders are either missense or small in-frame indel mutations, and the disease severity is often related to the position of the mutation. The dominant-negative mutant K9 protein interferes with the function of the normal allele. The treatment challenge is to specifically decrease the expression or impact of the mutant allele or its encoded mutant protein without disrupting the expression or function of the normal allele or its normal protein. Thus, the Krt9/c.434delAinsGGCT (p.Tyr144delinsTrpLeu) KI mouse is an excellent heterozygous model for gene therapy research.16 To the best of our knowledge, this study is the first to report CRISPR/Cas9 system delivery by LV, leading to in vivo gene therapy amelioration of the disease phenotype in a humanized EPPK-like mouse model.
Designing the optimal sgRNA is crucial for the CRISPR/Cas9 system to target a specific site in the genome, especially when the target site only has several bases different from the other DNA sequences. PAM-site mutations seem broadly effective, so sgRNA target mutations may have variable effects at different loci.19 Here, our data demonstrated that the sgRNA with the cut-site mutation (sg1) was more efficient than that with the PAM-site mutation (sg2) (Figures 1B and 1C).
Relative to FLAG and GAPDH, the reduction of K9 differed by 13.2% between in vitro and in vivo using semiquantitative analyses of immunoblotting (Figures 1D, 1E, 3B, and 3C). Because the expression of FLAG was positively correlated with the transduction efficiency of LV, this difference may be caused by disparities between the internal environment and in vitro. There may be some other regulatory pathways impacting the expression of K9 in the mouse. Although the efficiency of the CRISPR/Cas9 system has been evaluated on KRT12 by plasmid, on KRT14 in cells, and on recessive dystrophic epidermolysis bullosa cells from a patient’s skin biopsy and an embryo, no evaluation with a highly efficient vector in vivo has been carried out on genodermatoses.20, 21, 22, 23 Thus, the difference in efficiency between in vitro and in vivo should be considered when further CRISPR/Cas9 therapy is applied in vivo.
RNAi therapy has been used on humans with pachyonychia congenita, a genodermatosis with an autosomal dominant transmission that includes a disabling plantar keratoderma.24 During 100 days of treatment with 33 rounds of injections, clearing of the callus around the site of injection developed, but the level of pain during the injections is a significant concern, and the effect may not persist.24 Unlike the RNAi method, CRISPR/Cas9 targets DNA rather than RNA, ensuring that the effect is permanent.25 Compared with our previous shRNA therapy delivered by LV in the same Krt9/KI mouse model, the reduction of thickness of treated pads by the CRISPR/Cas9 system was 10% greater.16 Because CRISPR/Cas9 is derived from prokaryotes, it has less crosstalk with eukaryotic components than the RNAi pathway.9 The higher specificity of CRISPR/Cas9 may allow for fewer injections than RNAi therapy.9, 26 Because there are still no treatments for EPPK, the CRISPR/Cas9 system could provide an option for clinical trials.
Most researchers use adeno-associated virus (AAV) as a tool to deliver the CRISPR/Cas9 components. From the efforts reflected by the 2,463 clinical trials that have been conducted to date, 183 used AAV vectors and 158 used LV vectors in the gene therapy (October 2017; https://www.wiley.com/legacy/wileychi/genmed/clinical/). In contrast, LVs belong to the class of retroviruses that includes HIV, and are capable of DNA integration in non-dividing cells, including neurons.27, 28 Not showing preferential integration into any specific gene locus, LVs can thus reduce the chances of activating an oncogene in a large number of cells.29 In addition, LVs have a larger cargo capacity and a more stable long-term transgene expression than AAVs, which allows expression of the larger Cas9 proteins, enabling the targeting of more genomic sites.30, 31 So far, only a few studies have demonstrated in vivo gene editing following LV delivery of Cas9.31, 32, 33, 34 In this study, we chose LV as the delivery tool and suggested a better therapeutic efficacy over the renewal cycle of the epidermis. Then, the disrupted cytoskeletal integrity, abnormal differentiation, and irregular proliferation were ameliorated. Thus, we suggest that the CRISPR/Cas9-mediated genome editing tool is an effective means of gene modification in the epidermis and a therapeutic approach to correcting defective proteins.
The off-target effect is still of great concern for application of CRISPR/Cas9-mediated genome engineering.35 Rare “off-target” effects of a protein can be caused by DNA mutations in off-target genes and contribute to the development of certain human tumors.36 Identifying off-target cleavage is a formidable problem given the size and complexity of the eukaryotic genome.37 In this study, in the 49 potential highest-ranking genomic off-target sites predicted by the “Optimised CRISPR Design Tool” (Table S1), several off-target sites were located within the exons of functional genes, while only the top 24 of these sites were expressed in skin. The results of DNA sequencing from gDNA clones showed that the mismatched rate among the 10 potential highest ranking genomic off-target sites was only 3/200, indicating that there were no notable off-target effects (Table S2).
The CRISPR/Cas9 system has been widely used in spermatogonial stem cells, in various mouse models of human genetic disorders, and even in even age-related macular degeneration.38, 39, 40, 41, 42 There are 10 clinical trials under investigation based on the strategy of CRISPR/Cas9 (https://clinicaltrials.gov), two of which focus on the safety of CRISPR-Cas9. Although therapeutic safety and efficacy after transplantation remain to be evaluated clinically, CRISPR/Cas9-mediated treatment provides a new vision of cell therapy for many genetic diseases.26
After the genomic changes occur following the transduction of the CRISPR/Cas9 system, using cytokines as adjuvant therapy to accelerate regeneration could be a feasible means of enhancing the therapeutic efficacy.43, 44 Unlike the general therapeutic strategies for genetic diseases targeting either the genetic cause underlying a specific disorder or disease-specific pathophysiological pathways, the current standard care for EPPK and other keratin disorders still only has retinoids for treating the hyperkeratosis, focusing on symptomatic treatment. Although the CRISPR/Cas9 system had a significant effect on adult Krt9/KI mice after local injection, we predict that this therapy could be more effective with adjuvant treatment.
In conclusion, our research demonstrates the therapeutic benefits of LV-mediated CRISPR/Cas9 genome editing in an adult mouse model of EPPK, indicating that localized injection of the CRISPR/Cas9 system is a potentially powerful treatment for genetic defects that have local effects, such as hereditary PPKs.
Materials and Methods
Construction of Lentiviral Delivery for CRISPR/Cas9
Construction of the lentiviral delivery systems Cas9-LV, sg1-Cas9-LV, and sg2-Cas9-LV was based on the vector GV392. The EGFP fragment was replaced with the expression sequence FLAG. The sg1 strand 5′-CACCGCCGTCTGGCCTCTTGGCTCA-3′ and the sg2 strand 5′-CACCGTCAATTCCCGTCTGGCCTCT-3′ were synthesized by GeneChem (Shanghai, China). The Cas9-LV vector had no sgRNA strand.
Cell Culture and Transduction of HeLa Cells with Lentiviral Vectors
LVs containing Krt9 exon1 MUT-EGFP and Krt9 exon1-mCherry were produced as follows: 24 hr after plating, 293FT cells were co-transfected with 1.0 μg of Krt9 exon1 MUT-eGF or Krt9 exon1-mCherry (Figure S1A), 0.75 μg of psPAX2 (#VT1444; YouBio, Hunan, China), and 0.25 μg of pMD2.G (#VT1443; YouBio, Hunan, China) using Lipofectamine 2000 (#11668-019; Invitrogen). After incubation for 48 hr, the medium was replaced with fresh medium. The supernatant was collected, and reconstructed LVs were harvested by centrifugation and stored at −80°C for further experiments. HeLa cells were transduced with the two reconstructed LVs. After selection of cells expressing double fluorescence using BD FACSAria III (Becton Dickinson, Franklin Lakes, NJ, USA), a stable HeLa Hez cell line was established. Then, HeLa Hez cells were infected with CRISPR/Cas9-LV (1 × 107 infectious units/mL) at a dose of 10 μL for 3–5 × 102 cells. The cells were cultured at 37°C with 5% (v/v) CO2 and the medium replaced every 12 hr.
Flow Cytometry
The cells were prepared for FACS 60 hr after replacing the medium. Before FACS, the cells were suspended in PBS, pipetted, and filtered. The BD FACSAria III was operated using the standard protocol. Analysis was performed using FlowJo software.
Western Blotting
Epidermis from the fore-paw pads of WT and KI-Krt9 littermates was dissected and ground in liquid nitrogen. The next steps were the same for both cells and tissues. Radioimmunoprecipitation assay (RIPA) buffer (0.5% Nonidet P-40 [NP-40], 1% SDS, 50 mM Tris [pH 8.0], 0.1 mM EDTA, 150 mM NaCl, 100 μM Na3VO4, 1 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride, 3 μg/mL aprotinin, 2 μg/mL pepstatin, 1 μg/mL leupeptin) was used to lyse cells and tissues. The mixture was then centrifuged at 4°C for 20 min. After adding 4× loading buffer, the protein sample was denatured for 5 min at 95°C. The gel was run at 120 V for 90 min. After blotting the protein onto polyvinylidene fluoride membranes, the membranes were blocked for 60 min in skim milk and incubated overnight at 4°C with the primary antibody. The membranes were washed and incubated with the secondary antibody for 60 min. The bound antibodies were visualized with the Tanon 5200 chemiluminescent imaging system (Biotanon, Shanghai, China). The results of staining per area were generated by ImageJ with t test statistical analysis.
Animals
Mice on a C57BL/6 genetic background carrying Krt9/c.434delAinsGGCT were housed in groups with a 12-hr dark/light cycle and free access to food and water in accordance with the Regulations on Mouse Welfare and Ethics of Nanjing University, China. All procedures were conducted with the approval of Model Animal Research Center of Nanjing University. Mice between 12 and 16 weeks of age were used in all experiments.16
Localized Subcutaneous Injection
Mice were anesthetized by an intraperitoneal injection of chloral hydrate (40 mg/g). Localized subcutaneous injections of sg1-Cas9-LV into the right fore-paws of mice were performed using a microsyringe (location of injection is shown in Figure 2A).
BrdU Incorporation Assay
Each mouse received an intraperitoneal injection of 200 mg/kg BrdU twice every 1.5 hr, and epidermis from the fore-paw pads was dissected 2 hr after the second injection. BrdU (BrdU powder, No. E607203; Sangon Biotech, Shanghai, China) was prepared in PBS.
Histopathology and Immunofluorescence Analysis
Epidermis from the fore-paw pads of WT and KI-Krt9 littermates was dissected and fixed at 4°C for 4 hr in 4% paraformaldehyde/PBS. Then the tissues were dehydrated, embedded in paraffin, and cut at 8 μm. For histopathological analyses, the sections were deparaffinized and stained with H&E using standard protocols. Epidermal thickness was measured with t test statistical analysis using Photoshop software. For IF analysis, rehydrated paraffin-embedded sections were washed in PBS, blocked in 5% normal goat serum for 1 hr at room temperature, incubated with primary antibody (in 2.5% normal goat serum) at 4°C for 16 hr overnight, incubated with secondary antibody (in 2.5% normal goat serum) for 45 min, counterstained with DAPI, mounted, and imaged (the sections were washed three times in PBS before each step). Images were captured using a Leica SP5 Laser Scanning Confocal Microscope (Leica, Buffalo, IL, USA).
TEM and Barrier Function Assays
For TEM, tissues from the footpads of fore-paws were fixed in 2.5% glutaraldehyde in cacodylate buffer for 24 hr at 4°C. After rinsing twice with cacodylate buffer, samples were post-fixed in aqueous osmium tetroxide for 2 hr at 20°C. Samples were rinsed three times with cacodylate buffer and dehydrated through a graded series of ethanol concentrations. After dehydration through acetone, samples were infiltrated with complete embedding medium and dried in a 60°C oven for 48 hr. Then, the samples were cut at 70 nm and stained with 2% uranium acetate-saturated alcohol and lead citrate for 15 min each. Ultra-thin sections were examined on a TEM (HT7700; Hitachi, Tokyo, Japan).
T-A Cloning
To investigate the nature and frequency of the specific sg1-Cas9-LV-corrected indel, we performed T-A cloning and Sanger sequencing analysis using PCR amplicons from the gDNA at Krt9 exon 1 and 10 different loci. Epidermis from the fore-paw pads of WT and KI-Krt9 littermates treated with Cas9-LV or sg1-Cas9-LV was sampled for gDNA extraction. Then, PCR, T-A cloning (pMD 18-T Vector Cloning Kit, #6011; TaKaRa, Shiga, Japan), and Sanger sequencing were carried out according to standard protocols.
Author Contributions
Writing – Original Draft, X.-R.L., X.-L.C., and Y.-X.T.; Writing – Reviewing and Editing, X.-R.L., X.-L.C., Y.-X.T., J.-Y.Z., H.-P.K., Z.-Y.L., and X.-N.Z.; Supervision, X.G., H.-P.K., Z.-Y.L., and X.-N.Z.; Investigation, X.-R.L., X.-L.C., Y.-X.T., J.-Y.Z., and H.-P.K.; Methodology, X.-R.L., X.-L.C., Y.-X.T., and Z.-Y.L.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81450065), research funding of Zhejiang Chinese Medical University (711200F011), and the Ningbo Natural Science Foundation (2015A610187).
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.005.
Supplemental Information
References
- 1.Sybert V.P. Third Edition. Oxford University Press; 2017. Genetic Skin Disorders; pp. 34–53. [Google Scholar]
- 2.Has C., Technau-Hafsi K. Palmoplantar keratodermas: clinical and genetic aspects. J. Dtsch. Dermatol. Ges. 2016;14:123–139, quiz 140. doi: 10.1111/ddg.12930. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Valdez J., Rivera-Vega M.R., Gonzalez-Huerta L.M., Cazarin J., Cuevas-Covarrubias S. Analysis of the KRT9 gene in a Mexican family with epidermolytic palmoplantar keratoderma. Pediatr. Dermatol. 2013;30:354–358. doi: 10.1111/pde.12027. [DOI] [PubMed] [Google Scholar]
- 4.Reis A., Hennies H.C., Langbein L., Digweed M., Mischke D., Drechsler M., Schröck E., Royer-Pokora B., Franke W.W., Sperling K. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK) Nat. Genet. 1994;6:174–179. doi: 10.1038/ng0294-174. [DOI] [PubMed] [Google Scholar]
- 5.Du Z.F., Wei W., Wang Y.F., Chen X.L., Chen C.Y., Liu W.T., Lu J.J., Mao L.G., Xu C.M., Fang H., Zhang X.N. A novel mutation within the 2B rod domain of keratin 9 in a Chinese pedigree with epidermolytic palmoplantar keratoderma combined with knuckle pads and camptodactyly. Eur. J. Dermatol. 2011;21:675–679. doi: 10.1684/ejd.2011.1458. [DOI] [PubMed] [Google Scholar]
- 6.McLean W.H., Moore C.B. Keratin disorders: from gene to therapy. Hum. Mol. Genet. 2011;20(R2):R189–R197. doi: 10.1093/hmg/ddr379. [DOI] [PubMed] [Google Scholar]
- 7.Coulombe P.A. The molecular revolution in cutaneous biology: keratin genes and their associated disease: diversity, opportunities, and challenges. J. Invest. Dermatol. 2017;137:e67–e71. doi: 10.1016/j.jid.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.March O.P., Reichelt J., Koller U. Gene editing for skin diseases: designer nucleases as tools for gene therapy of skin fragility disorders. Exp. Physiol. 2018;103:449–455. doi: 10.1113/EP086044. [DOI] [PubMed] [Google Scholar]
- 9.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulei D., Berindan-Neagoe I. CRISPR/Cas9: a potential life-saving tool. What’s next? Mol. Ther. Nucleic Acids. 2017;9:333–336. doi: 10.1016/j.omtn.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ormond K.E., Mortlock D.P., Scholes D.T., Bombard Y., Brody L.C., Faucett W.A., Garrison N.A., Hercher L., Isasi R., Middleton A. Human germline genome editing. Am. J. Hum. Genet. 2017;101:167–176. doi: 10.1016/j.ajhg.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W.J., Zhu L.Y., Yan Z.Y., Xu Y., Wang Q.L., Lu X.J. CRISPR-Cas9 for in vivo gene therapy: promise and hurdles. Mol. Ther. Nucleic Acids. 2016;5:e349. doi: 10.1038/mtna.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyu Y.S., Shi P.L., Chen X.L., Tang Y.X., Wang Y.F., Liu R.R., Luan X.R., Fang Y., Mei R.H., Du Z.F. A small indel mutant mouse model of epidermolytic palmoplantar keratoderma and its application to mutant-specific shRNA therapy. Mol. Ther. Nucleic Acids. 2016;5:e299. doi: 10.1038/mtna.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu D.J., Thomson C., Lunny D.P., Dopping-Hepenstal P.J., McGrath J.A., Smith F.J.D., Irwin McLean W.H., Leslie Pedrioli D.M. Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J. Invest. Dermatol. 2014;134:754–763. doi: 10.1038/jid.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojtowicz J.M., Kee N. BrdU assay for neurogenesis in rodents. Nat. Protoc. 2006;1:1399–1405. doi: 10.1038/nprot.2006.224. [DOI] [PubMed] [Google Scholar]
- 19.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 20.Courtney D.G., Moore J.E., Atkinson S.D., Maurizi E., Allen E.H., Pedrioli D.M., McLean W.H., Nesbit M.A., Moore C.B. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108–112. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocher T., Peking P., Klausegger A., Murauer E.M., Hofbauer J.P., Wally V., Lettner T., Hainzl S., Ablinger M., Bauer J.W. Cut and paste: efficient homology-directed repair of a dominant negative KRT14 mutation via CRISPR/Cas9 nickases. Mol. Ther. 2017;25:2585–2598. doi: 10.1016/j.ymthe.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hainzl S., Peking P., Kocher T., Murauer E.M., Larcher F., Del Rio M., Duarte B., Steiner M., Klausegger A., Bauer J.W. COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa. Mol. Ther. 2017;25:2573–2584. doi: 10.1016/j.ymthe.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W., Lu Z., Li F., Wang W., Qian N., Duan J., Zhang Y., Wang F., Chen T. Efficient in vivo gene editing using ribonucleoproteins in skin stem cells of recessive dystrophic epidermolysis bullosa mouse model. Proc. Natl. Acad. Sci. USA. 2017;114:1660–1665. doi: 10.1073/pnas.1614775114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leachman S.A., Hickerson R.P., Schwartz M.E., Bullough E.E., Hutcherson S.L., Boucher K.M., Hansen C.D., Eliason M.J., Srivatsa G.S., Kornbrust D.J. First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol. Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgens D.W., Deans R.M., Li A., Bassik M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palfi S., Gurruchaga J.M., Ralph G.S., Lepetit H., Lavisse S., Buttery P.C., Watts C., Miskin J., Kelleher M., Deeley S. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: a dose escalation, open-label, phase 1/2 trial. Lancet. 2014;383:1138–1146. doi: 10.1016/S0140-6736(13)61939-X. [DOI] [PubMed] [Google Scholar]
- 29.Kotterman M.A., Chalberg T.W., Schaffer D.V. Viral vectors for gene therapy: translational and clinical outlook. Annu. Rev. Biomed. Eng. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- 30.Campochiaro P.A., Lauer A.K., Sohn E.H., Mir T.A., Naylor S., Anderton M.C., Kelleher M., Harrop R., Ellis S., Mitrophanous K.A. Lentiviral vector gene transfer of endostatin/angiostatin for macular (GEM) degeneration. Hum. Gene Ther. 2017;28:99–111. doi: 10.1089/hum.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue J., Gou X., Li Y., Wicksteed B., Wu X. Engineered epidermal progenitor cells can correct diet-induced obesity and diabetes. Cell Stem Cell. 2017;21:256–263.e4. doi: 10.1016/j.stem.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C., Akerstrom V., Baus J., Lan M.S., Breslin M.B. Comparative analysis of the transduction efficiency of five adeno associated virus serotypes and VSV-G pseudotype lentiviral vector in lung cancer cells. Virol. J. 2013;10:86. doi: 10.1186/1743-422X-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An H., Cho D.W., Lee S.E., Yang Y.S., Han S.C., Lee C.J. Differential cellular tropism of lentivirus and adeno-associated virus in the brain of cynomolgus monkey. Exp. Neurobiol. 2016;25:48–54. doi: 10.5607/en.2016.25.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmgaard A., Askou A.L., Benckendorff J.N.E., Thomsen E.A., Cai Y., Bek T., Mikkelsen J.G., Corydon T.J. In vivo knockout of the Vegfa gene by lentiviral delivery of CRISPR/Cas9 in mouse retinal pigment epithelium cells. Mol. Ther. Nucleic Acids. 2017;9:89–99. doi: 10.1016/j.omtn.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fruman D.A., O’Brien S. Cancer: a targeted treatment with off-target risks. Nature. 2017;542:424–425. doi: 10.1038/nature21504. [DOI] [PubMed] [Google Scholar]
- 37.White M.K., Kaminski R., Young W.B., Roehm P.C., Khalili K. CRISPR editing technology in biological and biomedical investigation. J. Cell. Biochem. 2017;118:3586–3594. doi: 10.1002/jcb.26099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., Xie Z., Bai M., Yin Q., Liang D. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25:67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakondi B., Lv W., Lu B., Jones M.K., Tsai Y., Kim K.J., Levy R., Akhtar A.A., Breunig J.J., Svendsen C.N., Wang S. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol. Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao H.K., Hatanaka F., Araoka T., Reddy P., Wu M.Z., Sui Y., Yamauchi T., Sakurai M., O’Keefe D.D., Núñez-Delicado E. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507.e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim K., Park S.W., Kim J.H., Lee S.H., Kim D., Koo T., Kim K.E., Kim J.H., Kim J.S. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–426. doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davar D., Tarhini A., Kirkwood J.M. Adjuvant therapy: melanoma. J. Skin Cancer. 2011;2011:274382. doi: 10.1155/2011/274382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirayama M., Tsubota K., Tsuji T. Bioengineered lacrimal gland organ regeneration in vivo. J. Funct. Biomater. 2015;6:634–649. doi: 10.3390/jfb6030634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.