Abstract
The mountain pine beetle (Dendroctonus ponderosae) is an insect native to western North America; however, its geographical range has recently expanded north in BC and east into Alberta. To understand the population structure in the areas of expansion, 16 gene‐linked microsatellites were screened and compared to neutral microsatellites using outlier analyses of F st and F ct values. One sex‐linked gene, inhibitor of apoptosis (IAP), showed a strong signature of positive selection for neo‐X alleles and was analyzed for evidence of adaptive variation. Alleles of IAP were sequenced, and differences between the neo‐X and neo‐Y alleles were consistent with neutral evolution suggesting that the neo‐Y allele may not be under functional constraints. Neo‐Y alleles were amplified from gDNA, but not effectively from cDNA, suggesting that there was little IAP expression from neo‐Y alleles. There were no differences in overall IAP expression between males and females with the common northern neo‐X allele suggesting that the neo‐X allele in males compensates for the reduced expression of neo‐Y alleles. However, males lacking the most common northern neo‐X allele thought to be selected for in northern populations had reduced overall IAP expression in early October—at a time when beetles are preparing for overwintering. This suggests that the most common allele may have more rapid upregulation. The reduced function of neo‐Y alleles of IAP suggested by both sequence differences and lower levels of expression may foster a highly selective environment for neo‐X alleles such as the common northern allele with more efficient upregulation.
Keywords: inhibitor of apoptosis, microsatellites, neo‐X/neo‐Y evolution, selection
1. INTRODUCTION
The mountain pine beetle (Dendroctonus ponderosae, Figure 1) is native to North American forests, but during epidemics this insect can devastate pine forests at the landscape level (Safranyik & Carroll, 2006). The frequency, duration, and severity of outbreaks are influenced by the availability of susceptible host trees and by the suitability of the climate (Taylor, Carroll, Alfaro, & Safranyik, 2006). Climate has traditionally been considered the main constraint on the range of the beetles in North America because under bark temperatures of less than −40°C are fatal to the mountain pine beetle (Carroll, Taylor, Regniere, & Safranyik, 2003). As epidemics can have severe ecological and economic impacts, it is important to model and understand mountain pine beetle outbreaks to inform future forest management practices.
Figure 1.

A teneral adult mountain pine beetle (Dendroctonus ponderosae)
The devastation caused during an epidemic is no longer limited to the historic range of D. ponderosae, but is now also a concern in new habitats where the beetles have spread in recent outbreaks. This includes movement into higher elevations in the United States, north and east in Canada, and even into a novel host, that is, jack pine (Pinus banksiana), in northern Alberta (Cullingham et al., 2011). As the mountain pine beetle has moved into these new areas, surveying the genetic diversity of beetles throughout their range has been useful in identifying both the spatial genetic structure (Samarasekera et al., 2012) and the genes potentially under selection in the expanding range (Janes et al., 2014). The identification of genes that are under selection at the northeastern limits of the beetles range may provide insights into how they have adapted to overcome the defenses of a novel host as well as the cold climates of these environments.
In looking for adaptive variation, microsatellites or Simple Tandem Repeats (STRs) are often overlooked in favor of other types of variation such as Single Nucleotide Polymorphisms (SNP's). However, microsatellites can be a useful tool to identify genes under selection that may provide local adaptation. This is especially true of Expressed Sequence Tags (ESTs) which have variable repeat lengths within the transcript which may directly result in functional differences and are easy to track through genotyping. Comparing variation in gene‐linked microsatellites to the variation in neutral microsatellites within the same populations can effectively identify outliers. These outliers may be under selection in populations where certain alleles of the associated gene provide a local adaptation causing allele frequencies to be different than would be expected based on the distribution of neutral markers (Holderegger et al., 2008; Meier, Hansen, Bekkevold, Skaala, & Mensberg, 2011).
In addition to being useful markers to identify genes under selection, microsatellites may provide local adaptation by creating variation in the expression or function in different alleles of the gene (Li, Korol, Fahima, & Nevo, 2004). The expression of a gene can be greatly affected by the number of repeat units in a microsatellite (Gemayel, Cho, Boeynaems, & Verstrepen, 2012; Gymrek et al., 2016). This is particularly true if the microsatellite is in a promoter region or transcription factor binding site. Furthermore, if the microsatellite is in between two transcription factor binding sites, variation in the repeat number may interrupt or promote interactions between transcription factors (Li et al., 2004). Finally, microsatellites have been shown to inhibit nucleosome formation resulting in an open chromatin structure around microsatellites thereby promoting transcription (Gemayel et al., 2012). Variation in repeat number of a microsatellite in the coding sequence may also affect the proper formation or function of the resulting protein. In particular, microsatellites can introduce frameshift mutations, amino acid repeats can result in protein aggregation, and variable numbers of amino acids can affect protein binding interactions (Gemayel et al., 2012).
In a previous study, Samarasekera, Keeling, Bohlmann, and Murray (2011) identified 50 polymorphic gene‐linked microsatellites as a resource to investigate local adaptations in mountain pine beetle populations. These were used to identify outliers showing signatures of selection. If these genes do provide a selective advantage, the location and repeat motif of the microsatellite may be able to inform what type of advantage is being conferred. As the population is expanding northeastwardly into ranges with colder climates, we hypothesize that alleles under positive selection may facilitate cold tolerance. Our screen identified the sex‐linked gene for inhibitor of apoptosis (IAP) which has a microsatellite in the coding sequence as a gene showing signatures of positive selection. This gene was previously found to be upregulated overall during overwintering by mountain pine beetle larvae (Robert et al., 2016). Subsequent analysis of variation in expression levels among beetles with different genotypes during early overwintering in a northern population suggests that there are elevated levels of transcription in early overwintering in larvae with certain genotypes which may be driving this positive selection. Furthermore, the male‐specific alleles of this sex‐linked gene were not detected in cDNA and the sequences were divergent from neo‐X linked alleles in a manner consistent with neutral evolution. Mountain pine beetle possesses a relatively recently evolved neo‐X/Y sex chromosome shared with only one other species, the Jeffrey pine beetle, in the genus Dendroctonus (Zúñiga, Cisneros, Hayes, & Macias‐Samano, 2002). Our findings suggest that the male‐specific allele was no longer functional. Therefore, having one functional allele may have resulted in a stronger selective pressure on the neo‐X alleles in male beetles. This is one example of local adaptation that may be facilitating the expansion of the range of mountain pine beetle to colder climates.
2. MATERIALS AND METHODS
2.1. Developing and screening a gene‐linked microsatellite database in western Canada
Individual beetles collected from six sampling locations representing at least two subpopulations of MPB in Western Canada (North: Houston, BC; Mackenzie, BC; Grande Prairie, AB, and South: AB; Green Lake, BC; Banff, AB; Nancy Greene, BC [Samarasekera et al., 2012]) were genotyped at 16 gene‐linked microsatellites (Table 1). Each of these microsatellites is associated with a unique scaffold (Supporting Information Table S1), and there is no evidence of linkage disequilibrium in either the neutral microsatellites (Samarasekera et al., 2012) or the gene‐linked microsatellites used (Samarasekera et al., 2011). M13 tailed primers were used to amplify polymorphic microsatellites with VIC, NED, PET, or FAM fluorescent labels on the amplicons (Life Technologies Inc., Burlington, ON, Canada). The primer sequences were those developed by Samarasekera et al. (2011) with the appropriate 5′ tails. The loci were amplified using either the touchdown PCR protocol described by Samarasekera et al. (2011) or by multiplex PCR using Qiagen Multiplex PCR kit (Qiagen Inc., Toronto, ON, Canada; Table 1). The thermocycling conditions used for multiplexed loci were: 95°C for 15 min, 27 cycles of 94°C for 30 s, 58°C for 90 s, and 72°C for 60 s, and a final extension step at 60°C for 30 min. Fragment analysis was performed on PCR products using ABI 3130xL and GeneScan 500 LIZ size standard following the manufacturer's protocol (Life Technologies). Amplicon size was scored using GeneMapper v4.0 (Life Technologies). Genotypes were added to a database of 14 neutral microsatellite markers (Samarasekera et al., 2012) and used to determine allele frequencies, F st values, F ct values, and to identify outliers as candidates for selection using GenAlEx v6.5 (Peakall & Smouse, 2006, 2012), Arlequin v3.5 (Excoffier & Lischer, 2010), and Bayescan v2.1 (Foll & Gaggiotti, 2008). A neutral microsatellite previously identified as being sex‐linked, Dpo486, was used for comparison to identify sex‐linked microsatellites (Davis et al., 2009).
Table 1.
A description of each gene‐linked microsatellite including the repeat motif, location within the transcript, and the PCR method used for each of the gene‐linked microsatellites screened for signatures of selection. The gene with the closest alignment from BLASTx and the organism the aligned gene is from are listed
| Microsatellite | Repeat motif | Location | PCR method | Description (BLASTx) – Predicted similar to |
|---|---|---|---|---|
| MPBC8_7725 | TAA | 5′ | Multiplex | MOB kinase activator‐like 4 (Pseudomyrmex gracilis) |
| MPBC5_6124 | AGG | CDS | Touchdown | WD repeat‐containing protein 55 homolog (Tribolium castaneum) |
| MPBC5_811 | CTC | CDS | Multiplex | Uncharacterized protein LOC111506581 (Leptinotarsa decemlineata) |
| MPBC6_675 | TAG | CDS | Multiplex | Death‐associated inhibitor of apoptosis 2 isoform X2 (Anoplophora glabripennis) |
| MPBC6_7245 | TGC | CDS | Multiplex | Prefoldin subunit 5 (Leptinotarsa decemlineata) |
| MPBC7_548 | GTG | CDS | Multiplex | Obscurin isoform X9 (Leptinotarsa decemlineata) |
| MPBC8_2778 | CAG | CDS | Touchdown | No significant hit |
| MPBC8_4511 | TCA | CDS | Touchdown | PREDICTED: zinc fin ger protein OZF‐like isoform X2 (Branchiostoma belcheri) |
| MPBC8_6649 | TCA | CDS | Multiplex | No significant hit |
| MPBC8_9094 | CAT | CDS | Multiplex | PREDICTED: WAS protein family homolog (Nicrophorus vespilloides) |
| MPBC8_9385 | CTA | CDS | Multiplex | No significant hit |
| MPBC5_6823 | AAT | CDS | Touchdown | Ornithine decarboxylase antizyme 1‐like protein (Tribolium castaneum) |
| MPBC8_884 | GTA | 3′ | Touchdown | F‐actin‐capping protein subunit alpha (Anoplophora glabripennis) |
| MPBC8_12800 | AAC | 3′ | Multiplex | Antennae‐rich cytochrome P450 (Tribolium castaneum) |
| MPBC5_4357 | (TA)2TTC(TA)7 | 3′ | Touchdown | Ring finger protein 141 (Anoplophora glabripennis) |
| MPBC7_24 | (TA)3TG(TA)8 | 3′ | Touchdown | Chitin deacetylase 1 precursor (Tribolium castaneum) |
2.2. Analysis of North American database
A database of 14 neutral microsatellites and one gene‐linked microsatellite, MPBC6_675 (inhibitor of apoptosis, IAP), was developed using beetles from 52 populations across western North America (Boone and Murray, unpublished data). This database was then divided into a female‐only and male‐only database because there were distinct neo‐X and neo‐Y linked microsatellites. The female‐only database was examined for F st and F ct outliers and to determine the distribution of neo‐X allele frequencies in North America. The male database was used to determine the neo‐Y allele frequencies in the same 52 populations. Analysis of the genotypic data was conducted using GenAlEx v6.5 (Peakall & Smouse, 2006, 2012), Arlequin v3.5 (Excoffier & Lischer, 2010), and BayeScan v2.1 (Foll & Gaggiotti, 2008).
2.3. Sequencing of IAP alleles
DNA was extracted from individual beetles using the Qiagen AllPrep DNA/RNA Mini Kit following the manufacturer's protocols (Qiagen), and IAP was amplified in a 10‐μl reaction using the Qiagen Multiplex PCR kit (Qiagen): 1X Qiagen Multiplex PCR Master Mix; 200 nM of forward primer (94F: 5′‐AGCGAGACAGACAACAGCAA‐3′); 200 nM of one of two reverse primers (1209R: 5′‐GGGGAGAAAAGTTCATGTTGA‐3′ or 1320R: 5′‐GCTCGGACCGTGGCTTCT‐3′); and 1 μl of template DNA. Thermocycling conditions were 95°C for 15 min, 30 cycles of 94°C for 30 s, 52°C for 90 s, and 72°C for 90 s, and a final extension step at 72°C for 10 min. Amplicons were run on a 0.8% agarose gel and extracted using GeneJET Gel extraction kit following the manufacturer's protocol (Life Technologies).
Amplified DNA was quantified using the Qubit fluorometric assay for dsDNA (Life Technologies), cloned into a pGEM‐T Easy vector system (Promega, Madison, WI, USA), and transformed into DH5α E. coli (Life Technologies). The plasmids were extracted from these using a QiaPrep Miniprep Kit following the manufacturers protocol (Qiagen). Plasmids were screened for the neo‐X and neo‐Y alleles of IAP using the primers for the MPBC6_675 microsatellite and the same PCR conditions described earlier for multiplex PCR. Selected plasmid inserts were sequenced using an ABI 3130xL with T7 and SP6 primers (Life Technologies). Sequences were aligned using CodonCode Aligner (v4.2.4), and the dN/dS ratios between the sequences of the neo‐X and neo‐Y alleles were determined using MEGA6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013).
2.4. Sample collection for IAP expression study
Larvae collected from Tête Juane Cache, BC in the autumn of 2008 as part of another study (Fraser, Bonnett, Keeling, & Huber, 2017) were used for expression studies of IAP. From this collection, we used larvae collected on October 3 and October 10 as these represent the time‐points immediately before and after the temperature dropped below 0°C. Briefly, upon removal from the tree, larvae were immediately frozen in 1.5 ml tubes using liquid nitrogen, transported to UNBC on dry ice, and stored at −80°C until use. Samples used solely for comparison between neo‐X and neo‐Y expression were collected from four populations: Utah, California, Cypress Hills, and Arizona, and sent to UNBC live. Utah samples were larvae collected in June 2012, California samples were larvae collected in July 2012, Cypress Hills samples were larvae collected in November 2010, and Arizona samples were larvae, pupae, and adults collected in June 2012.
2.5. Total RNA and DNA extraction and cDNA synthesis
The RNA and DNA from individual larvae were extracted using a Qiagen AllPrep DNA/RNA Mini Kit following the manufacturer's animal tissue protocol with optional DNase treatment (Qiagen). The amount and purity of RNA was evaluated using a Nanodrop ND‐100 (Nanodrop Technologies Inc., Wilmington, DE, USA). The integrity of RNA was visually inspected using Experion™ RNA StdSens Analysis Kit following the manufacturer's protocol (Bio‐Rad, Hercules, CA, USA). All RNA and DNA samples were stored at −80°C after extraction until required for downstream applications. The RNA extracted from each individual larva was converted to cDNA using a High Capacity cDNA Reverse Transcription Kit following the manufacturer's without RNase inhibitor protocol (Life Technologies). The total amount of RNA used per cDNA synthesis reaction was normalized to be 100 ng of RNA per reaction based on the concentration estimate from the Nanodrop ND‐100. The cDNA was stored at −80°C until required for downstream applications.
2.6. Genotyping of IAP and prefoldin subunit 5
The microsatellite regions of the IAP and another sex‐specific microsatellite region in prefoldin subunit 5 were amplified to determine the genotype of each larva and confirm its sex. These two loci were amplified in a duplex PCR using Qiagen Multiplex PCR kit (Qiagen). The final concentrations of reagents in a total volume of 10 μl were as follows: 1X Qiagen Multiplex PCR Master Mix; 160 nM IAP‐F, IAP‐R, and prefoldin‐F; 320 nM prefoldin‐R and VICtail fluorescent tag; and 1 μl of template DNA. PCR was performed twice for each larva, once using gDNA and once using cDNA as the template. Thermocycling conditions consisting of 95°C for 15 min, 30 cycles of 94°C for 30 s, 56°C for 90 s, and 72°C for 60 s, and a final extension step at 60°C for 30 min.
Fragment analysis was performed on each of the PCR products using ABI 3130xL with GeneScan 500 LIZ size standard, scored using GeneMapper v4.0 (Life Technologies) and visually inspected to determine any miscalled alleles. Many cDNA samples were manually scored because differences in peak heights between alleles resulted in miscalled alleles. The electropherograms produced by gDNA and cDNA from each individual were compared. The ratios of the peak heights representing the neo‐X and neo‐Y alleles of IAP were compared between amplicons produced from gDNA and cDNA.
2.7. Relative expression of IAP in different genotypes using RT‐qPCR
The cDNA synthesized from Tête Juane Cache larvae was used as the template for RT‐qPCR using an ABI 7300 system. IAP was the target gene, and actin, porphobilinogen deaminase (PBD), and RNA polymerase II (RPII) were used as reference genes. These reference genes were previously identified by Fraser et al. (2017) as effective for normalizing gene expression in D. ponderosae. Each time‐point was used as a separate experiment to account for any temporal changes in expression. The 3 October 2008 time‐point experiment was conducted using 26 larvae and two reference genes, actin and RPII. Reactions contained final concentrations of 1X TaqMan Environmental Master mix, 0.4 μM of each primer, 0.1 μM of TaqMan hydrolysis probe, and 1 ng of cDNA in a final volume of 25 μl. The 10 October 2008 time‐point experiment was conducted using 28 larvae and all three reference genes. Reactions contained final concentrations of 1X TaqMan Fast Advanced Master Mix, 0.5 μM of each primer, 0.25 μM TaqMan hydrolysis probe, and 1 ng of cDNA template in a total volume of 20 μl (Table 2).
Table 2.
The sequences for the primers and probes for the target gene (IAP) and the three reference genes (Actin, PBD, and RPII) used for RT‐qPCR of overwintering D. ponderosae larvae. For each probe, the reporter dye and the quencher used are reported
| Gene | Primer/probe | Primer sequence (5′‐3′) |
|---|---|---|
| Target gene | ||
| Inhibitor of apoptosis | IAP‐F | CCGAGAGATCGCCAAGGA |
| IAP‐R | CGCACACTGGACAATATCACTTTC | |
| IAP‐probe | VICN‐TGCTGGCAAAGGCTGGATTCTATTACAAAA‐TAMRA | |
| Reference genes | ||
| Actin | Actin‐F | AAATTTTAACCGAACGTGGATATTC |
| Actin‐R | CATCTCCTGTTCAAAGTCCAGAG | |
| Actin‐probe | JOEN‐TCACCACCACTGCCGAAAGGGAA‐BHQ‐1 | |
| Porphobilinogen deaminase (PBD) | PBD‐F | GGCTTCAATGTGTGTCCAGTG |
| PBD‐R | CACCAAACCAACGAAAAGATGTTC | |
| PBD‐probe | JOEN‐CGCCAATCTTATCACCGTTGCCG‐BHQ‐1 | |
| RNA polymerase II (RPII) | RPII‐F | GACGTTGGAGCAGTTCAAAGAG |
| RPII‐R | GGAAGAACACGAACATCTGGTC | |
| RPII‐probe | JOEN‐CGGCAAACCCAGCGAGAAGAGGC‐BHQ‐1 | |
For each target and reference gene, a standard curve was produced using three technical replicates of serially diluted pooled cDNA and used to optimize the conditions to obtain high R2 values, efficiencies between 90 and 100%, and to determine concentrations of cDNA template within the linear dynamic range as recommended by Bustin et al. (2009). Therefore, the thermocycling conditions were different for some of the genes used; however, they all had 45 cycles of denaturation and annealing/extension (Table 3).
Table 3.
The thermocycling conditions after optimization for the target gene inhibitor of apoptosis (IAP), and the three reference genes actin, porphobilinogen deaminase (PBD), and RNA polymerase II (RPII) all using 45 cycles of denaturation and anneal/extension
| Date | October 3, 2008 | October 10, 2008 | ||||||
|---|---|---|---|---|---|---|---|---|
| Step | All genes | IAP and actin | PBD | RPII | ||||
| Temp (°C) | Time | Temp (°C) | Time (s) | Temp (°C) | Time (s) | Temp (°C) | Time (s) | |
| UNG incubation | 50 | 5 min | 50 | 120 | 50 | 120 | 50 | 120 |
| Polymerase activation | 95 | 10 min | 95 | 20 | 95 | 20 | 95 | 20 |
| Denature | 95 | 30 s | 95 | 10 | 95 | 10 | 95 | 10 |
| Anneal/extension | 55 | 1 min | 56 | 30 | 58 | 45 | 56 | 60 |
Individuals were divided according to genotype into four groups: females homozygous for 162, females heterozygous with a 162 allele, males with a 162 neo‐X allele, and males lacking a 162 neo‐X allele. The data obtained from RT‐qPCR were analyzed using REST2009 software (Qiagen; Pfaffl, Horgan, & Dempfle, 2002) to determine the relative expression of IAP between different genotypes and also between males and females.
3. RESULTS
3.1. Gene‐linked microsatellite databases
3.1.1. Sex linkage of gene‐linked microsatellites
In the six Western Canadian populations genotyped, sex linkage was noted for three of the 16 gene‐linked loci when compared to a previously identified genetic marker for sex linkage (Davis et al., 2009). Two of these loci, MPBC6_675 (IAP) and MPBC6_7245 (prefoldin subunit 5) had distinct sets of neo‐X and neo‐Y alleles. The third locus, MPBC8_12800 (antennae‐rich cytochrome P450), only amplified from male samples and was therefore excluded from further analysis. The sex linkage of IAP was consistent in beetles from 52 North American populations genotyped.
3.1.2. Signatures of selection
The Western Canada microsatellite database with six sampling locations (Supporting Information Table S1) was divided into male and female datasets to eliminate any influence that the sex linkages of microsatellites could have on the identification of outliers. The female‐only dataset identified the trinucleotide repeat in the IAP coding sequence as the only significant outlier among 29 loci based on both the F st values and F ct values after accounting for population structure (Figure 2). IAP also showed a signature of diversifying selection in Western Canada after analysis using Bayescan (α = 1.553; q‐value = 0.000). In a second dataset containing neo‐X IAP and 14 neutral microsatellite loci in 52 North American population, IAP was again found to be a significant outlier based on F st values (F st = 0.437; p = 1e‐7), F ct values (F ct = 0.358, p = 1e‐7) and show a signature of diversifying selection using BayeScan (α = 0.110; q‐value = 0.048).
Figure 2.
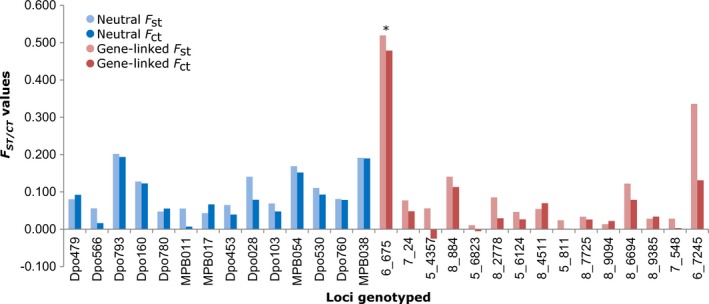
The F st and F ct values for neutral and gene‐linked microsatellites genotyped in mountain pine beetles collected in Western Canada. * indicates an outlier based on both F st and F ct values (p < 0.01)
3.1.3. Analysis of spatial distribution of IAP alleles
The frequencies of both neo‐X and neo‐Y IAP alleles in North American sampling locations were determined and trends were inspected. The 162‐bp allele, corresponding to a 14 serine repeat, was the most common neo‐X allele in the north and was near fixation in some northern populations. In the central and southern ranges, the 156‐bp and 159‐bp alleles are the most common neo‐X alleles and there is more of a mixture of genotypes; however, there are locations in South Dakota and Arizona where a 162‐bp allele is common (Figure 3). The neo‐Y specific alleles show more variation in the general clusters of allele frequencies than the neo‐X alleles. Some populations have a greater mix of neo‐Y alleles, and isolated populations such as Cypress Hills, Arizona, and South Dakota have otherwise uncommon neo‐Y alleles (Figure 3).
Figure 3.
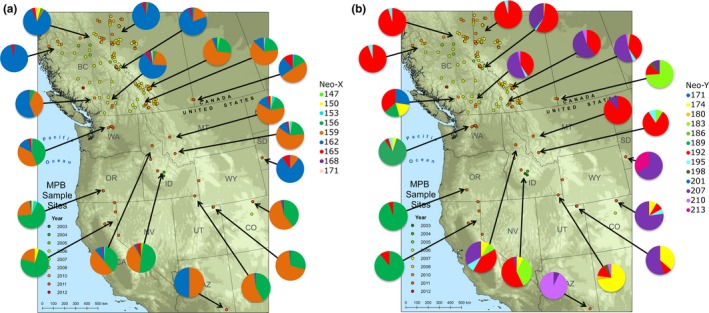
Spatial distribution of (a) neo‐X alleles and (b) neo‐Y alleles of IAP genotyped in mountain pine beetles collected from the indicated sampling locations across western North America
3.1.4. Neo‐X and neo‐Y allele sequences
Among the commonly observed neo‐X alleles of IAP sequenced (156, 159, and 162), there was no amino acid variation outside of the microsatellite region (Figure 4). Variation was greater among the neo‐Y alleles and between the neo‐X and neo‐Y alleles. In particular, there were two indels in the alignment of the neo‐X and neo‐Y sequences. The first indel region is associated with the trinucleotide (serine) repeat and lies in a region between two predicted BIR domains. A second indel region lies between the predicted BIR2 and RING domains. The dN/dS ratio between the 162‐bp neo‐X and 192‐bp neo‐Y alleles was 0.515 and a codon‐based Z‐test failed to reject neutral evolution (z = −1.44, p = 0.152). Four of the nonsynonymous changes resulting in amino acid changes were in the BIR2 and RING predicted domains.
Figure 4.

An alignment of the translated sequences of the inhibitor of apoptosis (IAP)156, 159, and 162‐bp neo‐X alleles; and 189‐, 192‐, and 207‐bp neo‐Y alleles. The amino acids differing from the 156‐bp allele are indicated, whereas the conserved amino acids are represented as dots and gaps as dashes. The conserved domains are indicated as follows: the two BIR domains are boxed with a solid line, whereas the RING domain is boxed with a dashed line
3.2. Expression of different IAP Alleles
3.2.1. Genotyping of individuals
Prior to analysis of expression, individual beetles were genotyped from both gDNA and cDNA using the sex‐linked microsatellites in IAP and prefoldin subunit 5 and were assigned into four groups based on their genotype. They were genotyped from both gDNA and cDNA to verify results because in some males from Utah, California, and Arizona the peak height of the neo‐Y allele of IAP was not discernible above background when genotyped from cDNA alone due to a lack of expression.
3.2.2. Qualitative relative expression of IAP alleles
The electropherograms from the amplification of prefoldin subunit 5 produced peaks with equal neo‐X to neo‐Y peak height ratios from gDNA and cDNA. In contrast, the electropherograms from the amplification of IAP from cDNA produced peaks with higher neo‐X to neo‐Y peak height ratios than those from gDNA (Figure 5). Based on the electropherograms from 18 male samples from Tête Juane Cache, BC the mean ratio of neo‐X to neo‐Y allele peak heights from cDNA was an average of 9.55 times higher than the ratio from gDNA (p < 0.005). A similar trend was found in all male individuals from Utah, California, Arizona, and Cypress Hills. In each location, the peak produced by the neo‐Y allele was reduced in the cDNA compared to the gDNA. However, the ratio was not quantifiable since the peak corresponding to the neo‐Y allele was not visible above background in many of the electropherograms.
Figure 5.
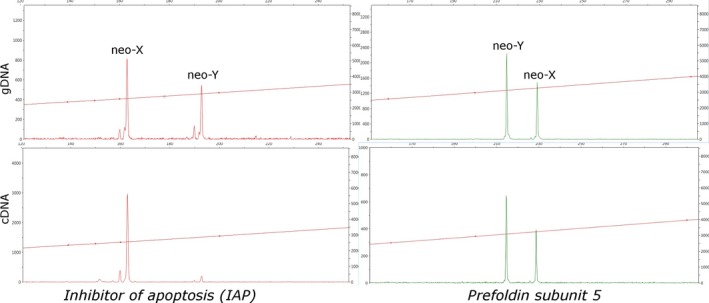
The peak heights of neo‐X and neo‐Y alleles of both IAP and prefoldin subunit 5 amplified from gDNA compared to the same alleles amplified from cDNA from a single individual larva. The neo‐Y allele of IAP amplified from cDNA is greatly reduced compared to the allele amplified from gDNA, but no difference is observable for the neo‐Y allele of prefoldin subunit 5
3.2.3. Quantitative relative expression of IAP in larvae
The optimized conditions for RT‐qPCR using cDNA from Tête Juane Cache resulted in standard curves having high R2 values and efficiencies over 90%. The relative expression of IAP in males lacking the 162‐bp neo‐X allele was reduced to 0.577‐fold of expression in females homozygous for the 162‐bp allele at the 3 October 2008 time‐point (p = 0.038). At the 10 October 2008 time‐point, there were no significant differences in IAP expression between any of the other genotypes compared to the females heterozygous for the 162‐bp allele (Figure 6). None of the female beetles sampled lacked the 162‐bp allele and so we were unable to make any comparisons with females lacking the 162‐bp allele altogether. The relative expression of IAP was also not significantly different between males and females at the 3 October 2008 time‐point (expression ratio = 0.883; p = 0.289) or the 10 October2008 time‐point (expression ratio = 0.958; p = 0.785). Finally, the overall IAP expression among all larvae was found to increase significantly 1.331‐fold (p = 0.013) from the 3 October to 10 October time‐point.
Figure 6.
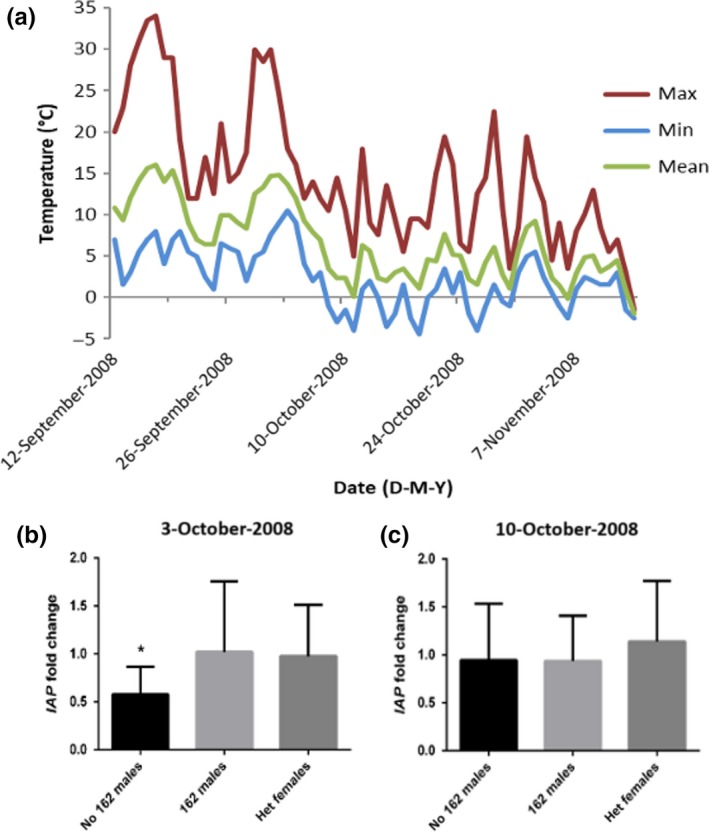
(a) The temperature data from September to November of the year larvae were sampled is shown. (b, c) The relative expression of IAP in larvae with different genotypes: males lacking the 162‐bp allele (no 162 males); males with the 162‐bp allele (162 males), and females heterozygous for the 162‐bp allele, compared to the expression of IAP in females homozygous for the 162‐bp allele. At the 3 October 2008 time‐point corresponding to a drop in temperature the male larvae lacking the 162‐bp allele had significantly less IAP expression than the females homozygous for the 162‐bp allele (b). By 10 October 2008, the differences in expression were no longer significant (c)
4. DISCUSSION
The inhibitor of apoptosis (IAP) gene was identified as a candidate for diversifying selection using a database of neutral and gene‐linked microsatellites and multiple outlier analyses. The neo‐X alleles of IAP were consistently identified as outliers based on F st values, F ct values (Figure 2), and Bayesian approaches. Upon inspection of the allele frequencies of the neo‐X IAP alleles across North America, the 162‐bp allele was noted to be the most common allele in the northernmost range as well as in some of the southernmost regions (Figure 3). Therefore, we hypothesized that the 162‐bp allele may be providing a local adaptation to beetles experiencing harsh conditions, including extreme cold in the north. Assuming that the southern 162‐bp allele is orthologous to the northern 162‐bp allele, this allele may be helping not only with cold tolerance, but more generally with adapting to stresses at the limits of D. ponderosae's geographical range.
A wide range of neo‐Y alleles was genotyped across North America and the distribution of alleles did not closely follow the distribution of neo‐X alleles (Figure 3) suggesting an independent evolution of neo‐Y alleles. Recent findings suggest that the neo‐Y chromosome in D. ponderosae forms three distinct geographically isolated SNP haplogroups (Bracewell, Bentz, Sullivan, & Good, 2017; Dowle et al., 2017). Using this single gene, we find consistency with the populations from the western United States being similar; however, in Canada, eastern US, and central US, we find more differences among neo‐Y alleles within populations than would be predicted by geographic range of the SNP haplogroups (Figure 3b). This suggests that the inclusion of neo‐Y microsatellite markers with SNPs (Bracewell et al., 2017; Dowle et al., 2017) will provide a finer resolution of within haplogroup variation which can inform evolution of the neo‐Y chromosome in these areas.
The independent evolution of neo‐X and neo‐Y alleles in D. ponderosae is of particular interest because there has been a recent fusion between an ancestral autosomal and X chromosome as well as the loss of the ancestral Y chromosome (Keeling et al., 2013; Lanier & Wood, 1968). This is informed by comparison with other Dendroctonus species which have 12 autosomal chromosomes and two sex chromosomes (X and Y), whereas D. ponderosae and D. jeffreyi have 11 autosomal chromosomes and two sex chromosomes (neo‐X and neo‐Y) (Zúñiga et al., 2002). It is expected that over time sequence divergence will occur between the ancestral autosomal portions of the neo‐X and neo‐Y chromosomes explaining the distinct IAP alleles observed. However, it is still early in this divergence so we were able to amplify and study both the neo‐X and neo‐Y alleles with common primer sets.
Analysis of sequence variation between the neo‐X and neo‐Y IAP alleles failed to reject neutral evolution. This variation suggests that the neo‐Y IAP may no longer be evolving under functional constraints indicating that it may not contribute substantially to overall IAP production. Furthermore, when male samples were genotyped the electropherograms for IAP amplified from cDNA had a reduced peak height produced by the neo‐Y allele. The peak height ratio of the IAP neo‐X allele compared to the neo‐Y allele was consistently larger in the electropherograms produced from amplification of cDNA than those from gDNA across all populations, representing seven different neo‐Y alleles (Figure 5). As the ratio of neo‐X to neo‐Y alleles in genomic DNA is one to one, this suggests that there were more neo‐X allele transcripts of IAP than neo‐Y allele transcripts to acts as templates for cDNA synthesis due to reduced expression of the neo‐Y allele. Reduced expression may have led to the loss of functional constraints which resulted in sequence variation between the neo‐Y and neo‐X alleles being consistent with neutral evolution.
In other insects, such as D. miranda with neo‐X/neo‐Y sex chromosomes, the neo‐Y chromosome is degenerating (Bachtrog & Charlesworth, 2002). Based on the sequence variation between the IAP neo‐X and neo‐Y alleles presented in this study, there is likely degeneration in the D. ponderosae neo‐Y chromosome. This is supported by recent findings that there has been degeneration in the neo‐Y chromosome of D. ponderosae. Bracewell et al. (2017) report large deletions in the neo‐Y chromosome suggesting degeneration, whereas we find divergence usually associated with a pseudogene (equal number of synonymous and nonsynonymous changes) within a single gene between the neo‐X and neo‐Y chromosomes. These pieces of evidence point to neo‐Y degeneration which may provide a highly selective environment for the orthologous neo‐X alleles. Outside of the variable number of serine repeats, there was no sequence variation in the coding sequences of neo‐X alleles analyzed, suggesting that differences in protein activity of different neo‐X alleles are unlikely. Therefore, potential expression differences among neo‐X alleles were examined.
Overall IAP expression among all larvae was found to increase significantly from 3 October to 10 October consistent with an overall trend of increasing IAP expression in larvae preparing for overwintering which had a 3.13‐fold increase from 26 September to 7 November 2008 in the same collection (Robert et al., 2016). This increase in expression as the larvae prepare for overwintering suggests that IAP may play a role in cold tolerance. This potential role in cold tolerance is supported by the role of IAP as a caspase inhibitor as apoptosis involving caspases has been shown to be inducible by cold shock (Fransen, Dieker, Hildebrands, Berden, & van der Vlag, 2011). Furthermore, in Drosophila melanogaster, an inhibitor of apoptosis Bcl‐2 was shown to be upregulated at the protein level during rapid cold‐hardening to compensate for dysregulated apoptosis which is common during cold shock (Yi, Moore, & Lee, 2007) Therefore, elevated levels of IAP expression may help the larvae to survive overwintering.
The relative expression of IAP in overwintering larvae was compared for beetles with varying genotypes. Individuals lacking a 162‐bp neo‐X allele were of interest as this allele was most common in the north. Among the larvae sampled on 3 October males lacking the 162‐bp allele had reduced IAP expression compared to females homozygous for the 162‐bp allele. However, among the 10 October samples, the expression of IAP was not significantly different in any group of larvae compared to females homozygous for the 162‐bp allele (Figure 6). The reduced expression of IAP in males lacking the 162 allele at the 3 October time‐point, but equal expression at the 10 October time‐point might reflect a more efficient or rapid upregulation of the 162‐bp IAP allele. Earlier expression of this IAP allele may give those larvae increased survival during early extreme cold events which would be more likely to occur earlier in the northeastern part of the beetle's range thereby providing a selective advantage. Unfortunately, due to the high frequency of the 162‐bp allele in this population, it was not possible to examine all allele combinations and compare with females completely lacking this allele.
The lack of expression differences between males and females suggests that dosage compensation may be occurring. As discussed, it appears that the neo‐Y alleles have reduced expression based on the electropherograms from cDNA. Therefore, dosage compensation would be necessary to allow equal expression of IAP in males and females. Although some form of dosage compensation is known to occur in many species with both neo‐X/neo‐Y and X/Y sex chromosomes, it has not been reported in bark beetles. D. miranda has a neo‐X/neo‐Y sex chromosome system which has dosage compensation partly developed for neo‐X alleles in males (Bachtrog & Charlesworth, 2002). In particular, there is increased expression of genes on the neo‐X chromosome which have degenerated orthologous genes on the neo‐Y chromosome (Marín, Siegal, & Baker, 2000). In D. ponderosae, the sequence variation and reduced expression suggests that the neo‐Y IAP allele may be degenerating. Furthermore, another sex‐linked gene, prefoldin subunit 5, did not show variation in the peak heights of neo‐X and neo‐Y alleles. This is also consistent with the neo‐X/neo‐Y dosage compensation in D. miranda which is not chromosome wide (Bone & Kuroda, 1996).
Although there is little known about dosage compensation in Coleoptera, Prince, Kirkland, and Demuth (2010) found that the majority of X‐linked genes in the red flour beetle, Tribolium castaneum, have increased expression in females. As the X‐linked genes in male flour beetles were expressed at approximately the same levels as autosomal genes, the increased expression of X‐linked genes in females is believed to be an evolutionary side effect of an imperfect dosage compensation mechanism. This hyperexpression of X‐linked genes shows that differential expression of sex chromosomes occurs in Coleoptera. However, the equal expression of IAP in male and female D. ponderosae suggests that, in contrast to T. castaneum, it is unlikely that neo‐X linked genes are globally hyperexpressed in females. It is possible that D. ponderosae is more similar to Xenos vesparum in which the chromosomal segment corresponding to the ancestral X shows dosage compensation but dosage compensation in the recently added segment of the X chromosome is incomplete (Mahajan & Bachtrog, 2015). However, the expression of more neo‐X linked genes particularly those in the ancestral X segment would need to be determined to make a definite conclusion on the extent and mode of dosage compensation in D. ponderosae.
Despite recent studies on sex chromosome evolution in D. ponderosae (Bracewell et al., 2017; Dowle et al., 2017), there is still little known about dosage compensation and much to be explored on a finer scale regarding sex chromosome evolution in bark beetles, so studying variation in both the sequence and expression of neo‐X and neo‐Y linked genes can help determine how the sex chromosomes are evolving and predict how they will continue to evolve. The sequence variation between the IAP neo‐X and neo‐Y alleles suggests that the neo‐Y chromosome is degenerating in D. ponderosae. Such degeneration fosters a highly selective environment for neo‐X alleles as they must compensate for an orthologous neo‐Y allele with a reduced or lost function. In the case of IAP, the 162‐bp allele appears to have faster upregulation compared to the other neo‐X alleles, which could provide a selective advantage allowing mountain pine beetle larvae in the north to survive early cold snaps.
The sequence variation and expression differences in mountain pine beetle IAP suggest that the neo‐Y allele is not contributing substantially to the overall production of the protein. However, relative expression of IAP in larvae preparing for overwintering shows that males do not have lower expression levels except for males lacking the northern 162‐bp allele at the earlier time‐point. This suggests that some mechanism of dosage compensation may be controlling the expression of this sex‐linked gene in mountain pine beetle. Further, the single functional copy of IAP in males would provide a highly selective environment for any alleles with earlier upregulation. Future research should investigate more sex‐linked genes that may be providing local adaptation and expression differences between males and females. This line of research could refine when expression differences occur in genes important for overwintering, provide insights into the mechanism of dosage compensation, and potentially explain the female skewed sex ratio in the adult population. This could help identify loci under strong selection that may explain how populations are expanding successfully into novel habitats northeast of the historic range.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
The experiments were designed and performed by LCH, CKB, GS, GKK, and BWM. Analysis of the data for this manuscript, preparation of the figures, and the manuscript were conducted by LCH and BWM. CKB, GS, and GKK critically reviewed the manuscript.
Supporting information
ACKNOWLEDGMENTS
We thank Dr. Dezene Huber, Dr. Jeanne Robert, and Jordie Fraser for access to the samples collected in Tête Juane Cache. We additionally thank Dr. Dezene Huber for access to the Experion (Bio‐Rad) and for useful discussions about these experiments. This research was supported by a Natural Sciences and Engineering Research Council (NSERC) of Canada Strategic Network grant (BWM Co‐investigator, grant no. NET GP 434810‐12) to the TRIA Network, with contributions from Alberta Agriculture and Forestry, fRI Research, Manitoba Conservation and Water Stewardship, Natural Resources Canada—Canadian Forest Service, Northwest Territories Environment and Natural Resources, Ontario Ministry of Natural Resources and Forestry, Saskatchewan Ministry of Environment, West Fraser and Weyerhaeuser. We also thank USDA Forest Service forest health professionals in Region 1‐Northern (MT, northern ID), Region 2‐Rocky Mountain (CO, SD), Region 3‐Southwestern (AZ, NM), Region 4‐Intermountain (MT, southern ID, UT, western WY), Region 5‐Pacific Southwest (CA) and Region 6‐Pacific Northwest (OR, WA), and Nevada Department of Forestry for assistance in sample collection. Additional research support was also received from an NSERC Undergraduate Summer Research Award and UNBC Research Project Award to LCH and UNBC Undergraduate Research Experience Awards to LCH and GKK.
Horianopoulos LC, Boone CK, Samarasekera GDNG, Kandola GK, Murray BW. Selection of the sex‐linked inhibitor of apoptosis in mountain pine beetle (Dendroctonus ponderosae) driven by enhanced expression during early overwintering. Ecol Evol. 2018;8:6253–6264. https://doi.org/10.1002/ece3.4164
REFERENCES
- Bachtrog, D. , & Charlesworth, B. (2002). Reduced adaptation of a non‐recombining neo‐Y chromosome. Nature, 416, 323–326. https://doi.org/10.1038/416323a [DOI] [PubMed] [Google Scholar]
- Bone, J. R. , & Kuroda, M. I. (1996). Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila . Genetics, 144(2), 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracewell, R. R. , Bentz, B. J. , Sullivan, B. T. , & Good, J. M. (2017). Rapid neo‐sex chromosome evolution and incipient speciation in a major forest pest. Nature Communications, 8(1), 1593 https://doi.org/10.1038/s41467-017-01761-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S. A. , Benes, V. , Garson, J. A. , Hellemans, J. , Huggett, J. , Kubista, M. , … Vandesompele, J. (2009). The MIQE guidelines: Minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry, 55(4), 611–622. https://doi.org/10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Carroll, A. L. , Taylor, S. W. , Régnière, J. , & Safranyik, L. (2003). Effect of climate change on range expansion by the mountain pine beetle in British Columbia In Shore T. L. et al. (Eds.), Mountain Pine Beetle symposium: Challenges and solutions, Oct. 30–31, 2003 (pp. 223–232). Kelowna BC: Natural Resources Canada, Information Report BC‐X‐399, Victoria. [Google Scholar]
- Cullingham, C. J. , Cooke, J. E. K. , Dang, S. , Davis, C. S. , Cooke, B. J. , & Coltman, D. W. (2011). Mountain pine beetle host‐range expansion threatens the boreal forest. Molecular Ecology, 20, 2157–2171. https://doi.org/10.1111/j.1365-294X.2011.05086.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. S. , Mock, K. E. , Bentz, B. J. , Bromilow, S. M. , Bartell, N. V. , Murray, B. W. , … Cooke, J. E. K. (2009). Isolation and characterization of 16 microsatellite loci in the mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Curculionidae: Scolytinae). Molecular Ecology Research, 9(3), 1071–1073. https://doi.org/10.1111/j.1755-0998.2009.02579.x [DOI] [PubMed] [Google Scholar]
- Dowle, E. J. , Bracewell, R. R. , Pfrender, M. E. , Mock, K. E. , Bentz, B. J. , & Ragland, G. J. (2017). Reproductive isolation and environmental adaptation shape the phylogeography of mountain pine beetle (Dendroctonus ponderosae). Molecular Ecology, 26, 6071–6084. https://doi.org/10.1111/mec.14342 [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , & Lischer, H. E. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3), 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Foll, M. , & Gaggiotti, O. (2008). A genome‐scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics, 180(2), 977–993. https://doi.org/10.1534/genetics.108.092221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen, J. H. , Dieker, J. W. , Hildebrands, L. B. , Berden, J. H. , & van der Vlag, J. (2011). Synchronized turbo apoptosis induced by cold‐shock. Apoptosis, 16, 86–93. https://doi.org/10.1007/s10495-010-0546-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, J. D. , Bonnett, T. R. , Keeling, C. I. , & Huber, D. P. (2017). Seasonal shifts in accumulation of glycerol biosynthetic gene transcripts in mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Curculionidae), larvae. PeerJ, 5, e3284 https://doi.org/10.7717/peerj.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel, R. , Cho, J. , Boeynaems, S. , & Verstrepen, K. J. (2012). Beyond junk‐variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes, 3(3), 461–480. https://doi.org/10.3390/genes3030461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek, M. , Willems, T. , Guilmatre, A. , Zeng, H. , Markus, B. , Georgiev, S. , … Erlich, Y. (2016). Abundant contribution of short tandem repeats to gene expression variation in humans. Nature Genetics, 48, 22–29. https://doi.org/10.1038/ng.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderegger, R. , Herrmann, D. , Poncet, B. , Gugerli, F. , Thuiller, W. , Taberlet, P. , … Manel, S. (2008). Land ahead: Using genome scans to identify molecular markers of adaptive relevance. Plant Ecology & Diversity, 1(2), 273–283. https://doi.org/10.1080/17550870802338420 [Google Scholar]
- Janes, J. K. , Li, Y. , Keeling, C. I. , Yuen, M. M. S. , Boone, C. K. , Cooke, J. E. K. , … Sperling, F. A. H. (2014). How the mountain pine beetle (Dendroctonus ponderosae) breached the Canadian Rocky Mountains. Molecular Biology and Evolution, 31(7), 1803–1815. https://doi.org/10.1093/molbev/msu135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, C. I. , Yuen, M. M. , Liao, N. Y. , Docking, T. R. , Chan, S. K. , Taylor, G. A. , … Bohlmann, J. (2013). Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biology., 14(3), R27 https://doi.org/10.1186/gb-2013-14-3-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, G. N. , & Wood, D. L. (1968). Controlled mating, karyology, morphology, and sex‐ratio in the Dendroctonus ponderosae Complex12. Annals of the Entomological Society of America, 61(2), 517–526. https://doi.org/10.1093/aesa/61.2.517 [Google Scholar]
- Li, Y. C. , Korol, A. B. , Fahima, T. , & Nevo, E. (2004). Microsatellites within genes: Structure, function, and evolution. Molecular Biology and Evolution, 21(6), 991–1007. https://doi.org/10.1093/molbev/msh073 [DOI] [PubMed] [Google Scholar]
- Mahajan, S. , & Bachtrog, D. (2015). Partial dosage compensation in Strepsiptera, a sister group of beetles. Genome Biology and Evolution, 7(2), 591–600. https://doi.org/10.1093/gbe/evv008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín, I. , Siegal, M. L. , & Baker, B. S. (2000). The evolution of dosage‐compensation mechanisms. BioEssays, 22(12), 1106–1114. https://doi.org/10.1002/1521-1878(200012)22:12<1106::AID-BIES8>3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- Meier, K. , Hansen, M. M. , Bekkevold, D. , Skaala, Ø. , & Mensberg, K. D. (2011). An assessment of the spatial scale of local adaptation in brown trout (Salmo trutta L.): Footprints of selection at microsatellite DNA loci. Heredity, 106(3), 488–499. https://doi.org/10.1038/hdy.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research‐an update. Bioinformatics, 28, 2537–2539. https://doi.org/10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. W. , Horgan, G. W. , & Dempfle, L. (2002). Relative expression software tool (REST) for group‐wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Research, 30(9), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, E. G. , Kirkland, D. , & Demuth, J. P. (2010). Hyperexpression of the X‐chromosome in both sexes results in extensive female‐bias of X‐linked genes in the flour beetle. Genome Biology and Evolution, 2, 336–346. https://doi.org/10.1093/gbe/evq024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, J. A. , Bonnett, T. , Pitt, C. , Spooner, L. J. , Fraser, J. , Yuen, M. M. , … Huber, D. P. (2016). Gene expression analysis of overwintering mountain pine beetle larvae suggests multiple systems involved in overwintering stress, cold hardiness, and preparation for spring development. PeerJ, 4, e2109 https://doi.org/10.7717/peerj.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safranyik, L. , & Carroll, A. L. (2006). The biology and epidemiology of the mountain pine beetle in lodgepole pine forests In Safranyik L., & Wilson B. (Eds.), The mountain pine beetle: A synthesis of biology, management and impacts on lodgepole pine (pp. 3–66). Victoria, BC: Natural Resources Canada, Canadian Forest Service, Pacific Forestry Centre. [Google Scholar]
- Samarasekera, G. D. N. G. , Bartell, N. V. , Lindgren, B. S. , Cooke, J. E. K. , Davis, C. S. , James, P. M. A. , … Murray, B. W. (2012). Spatial genetic structure of the mountain pine beetle (Dendroctonus poderosae) outbreak in western Canada: Historical patterns and contemporary dispersal. Molecular Ecology, 21, 2931–2948. https://doi.org/10.1111/j.1365-294X.2012.05587.x [DOI] [PubMed] [Google Scholar]
- Samarasekera, N. G. , Keeling, C. I. , Bohlmann, J. , & Murray, B. W. (2011). Isolation and characterization of EST‐derived microsatellite markers for the mountain pine beetle (Dendroctonus ponderosae Hopkins). Molecular Ecology Research, 11, 586–589. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. W. , Carroll, A. L. , Alfaro, R. I. , & Safranyik, L. (2006). Forest, climate and mountain pine beetle outbreak dynamics in western Canada In Safranyik L., & Wilson B. (Eds.), The mountain pine beetle: A synthesis of biology, management and impacts on lodgepole pine (pp. 67–94). Victoria, BC: Natural Resources Canada, Canadian Forest Service, Pacific Forestry Centre. [Google Scholar]
- Yi, S. X. , Moore, C. W. , & Lee, R. E. (2007). Rapid cold‐hardening protects Drosophila melanogaster from cold‐induced apoptosis. Apoptosis, 12(7), 1183–1193. https://doi.org/10.1007/s10495-006-0048-2 [DOI] [PubMed] [Google Scholar]
- Zúñiga, G. , Cisneros, R. , Hayes, J. L. , & Macias‐Samano, J. (2002). Karyology, geographic distribution, and origin of the genus Dendroctonus Erichson (Coleoptera: Scolytidae). Annals of the Entomological Society of America, 95(3), 267–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


