Abstract
Despite increasing evidence that landscape features strongly influence the abundance and dispersal of insect populations, landscape composition has seldom been explicitly linked to genetic structure. We conducted a genetic study of the melon aphid, Aphis gossypii, in two counties of Beijing, China during spring migration using samples from watermelon. We performed aphid genetic analysis using restriction site associated DNA sequencing (2b‐RAD) and investigated the relationship between land cover and the genetic diversity. The percentage area of land cover (cropland, vegetable, orchard, grassland, woodland) was quantified in each particular scale (ranging from 0.5 km to 3 km) and was used as a predictor variable in our generalized linear models. We found a moderate level of genetic differentiation among nine sampled populations. Geographic distance and genetic distance were not significantly associated, indicating that geographic location was not a barrier to migration. These nine populations could be clustered depending on their level of genetic diversity (high and low). The genetic diversity (Shannon’s information index) was positively correlated with grassland at the spatial scales of 1 and 2 km and negatively with orchard and vegetable at 0.5 and 1 km. Genetic diversity was best predicted by the grassland + orchard + vegetable model at a spatial scale of 1 km. Based on the method of relative weights, orchard land had the greatest relative importance, followed by grassland and vegetable land, in that order. This study contributes to our understanding of the genetic variation of aphids in agricultural landscapes.
Keywords: 2b‐RAD sequencing, landscape genetics, SNP loci, spring migration, watermelon
1. INTRODUCTION
Understanding how genetic variation is partitioned across the genome, both within and among populations, is a fundamental issue in ecological and evolutionary genetics (Catchen et al., 2013). Landscape genetics is an important emerging research topic that integrates landscape patterns with population genetics (Holderegger & Wagner, 2006). Studies in landscape genetics primarily evaluate the role of habitat loss and fragmentation in shaping the population genetic structure of a particular species, for example, bird, frog, or bee (Herrmann, Westphal, Moritz, & Steffan‐Dewenter, 2007; Johansson, Primmer, Sahlsten, & Merilä, 2005; Lindsay et al., 2008). Landscape variables are typically analyzed as putative barriers to the movement of individuals (Hemme, Thomas, Chadee, & Severson, 2010; Keyghobadi, Roland, & Strobeck, 1999), but land cover data have been seldom used (Perez‐Espona, McLeod, & Franks, 2012). Moreover, analyses of the impact of agricultural landscape features on the population genetics of insect pests are sparse. Here, we assess the effect of land cover on the genetic diversity of an insect pest using the melon aphid, Aphis gossypii Glover, as a model.
The melon aphid is distributed worldwide and is extremely polyphagous. It causes serious damage to melon, cotton and hundreds of other plant species (Blackman & Eastop, 2007). This aphid also has a complex life history. In regions with severe winters, A. gossypii has a complete life cycle (or holocycle). It lays eggs in autumn on a primary host (e.g., hibiscus), and in the spring, it migrates to secondary host plants (Zhang & Zhong, 1982). In areas with mild winters, it reproduces asexually throughout the year (Agarwala & Das, 2007; Fuller, Chavigny, Lapchin, & Vanlerberghe‐Masutti, 1999). A. gossypii is an important pest species of melon crops in the Beijing area. In recent years, the forest coverage in Beijing has greatly increased, and its urban agriculture is developing rapidly. Land use has changed, and the areas of cropland have decreased while the areas of woodland have increased. It is important to understand the relationship between the population genetics of aphids and landscape features.
The genetic variation of melon aphids can be attributed to many factors. One of the main factors is host plant species. The population genetics of A. gossypii is well documented to be affected by host plant species (Carletto et al., 2009; Charaabi et al., 2008). The population of A. gossypii often shows some degree of host specialization. Several host races are recognized the world over on congeneric host plants (Agarwala & Choudhuri, 2014) and on cultivated plants (Guldemond, Tigges, & De Vrijer, 1994). Reproductive mode is another factor influencing genetic diversity. Aphids reproducing by parthenogenesis have low genetic diversity at local and worldwide scales (Carletto et al., 2009; Chen et al., 2013). In contrast to the populations with an incomplete life cycle (or anholocycle), holocycle populations sexually reproduce. Evidence for high genetic diversity associated with sexual reproduction of A. gossypii has been found in China (Wang et al., 2016), France (Thomas, Boissot, & Vanlerberghe‐Masutti, 2012), and Iran (Razmjou, Vorburger, Moharramipour, Mirhoseini, & Fathipour, 2010). In addition, other factors such as insecticide selection pressure and competition can also affect genetic diversity (Brévault, Carletto, Linderme, & Vanlerberghe‐Masutti, 2008; Brévault, Carletto, Tribot, & Vanlerberghe‐Masutti, 2011; Carletto, Martin, Vanlerberghe‐Masutti, & Brévault, 2010; Fuller et al., 1999).
The effects of landscape features on aphid dispersal have been documented (Angelella, Holland, & Kaplan, 2016; Gilabert et al., 2017). Thomas, Vanlerberghe‐Masutti, Mistral, Loiseau, and Boissot (2016) conducted demo‐genetic analyses of aphid populations on aphid‐resistant plants during growth seasons and found that plant resistance changed the genetic structure of the aphid populations. Factors that modify the dispersal of founders may also modify the genetic composition of the population. Overall, the host colonization of A. gossypii likely could affect their population genetics. It has been suggested that host colonization of A. gossypii often occurs locally (Agarwala & Choudhuri, 2014; Carletto et al., 2009).
The landscape genetics of A. gossypii would provide new insights into host plant colonization. We conducted a landscape scale study on the population diversity of A. gossypii based on SNP loci in the genome. The advantage of a SNP study compared to using microsatellite data is that one can obtain more information. SNP data have the potential to identify both historical population structure and recent colonization history (Catchen et al., 2013). We focused on the spring migration of A. gossypii (founding individuals) to address the following questions: 1) Can genetic analysis reveal landscape aspects of A. gossypii colonization? 2) Are landscape features associated with the genetic diversity of A. gossypii colonizing the same host?
2. MATERIAL AND METHODS
2.1. Sampling
Sampling was conducted in the Beijing area of northern China (Figure 1). There were nine sites located in Daxing and Shunyi counties. Daxing is located south of Beijing, and Shunyi is northeast of Beijing. These two counties are in plain areas dominated by cultivated land. The sampling protocol consisted of collecting populations from as many watermelon plants as possible from different sites with a variety of landscape complexity.
Figure 1.
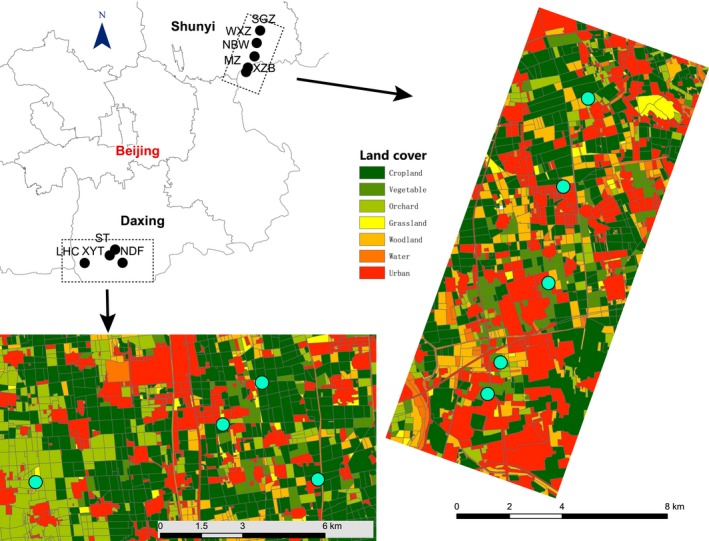
Sample localities of aphid populations in the vicinity of Beijing, China
Sampling was carried out in the early growth season, when aphids were migrating from primary hosts to the secondary host (melons). All melon aphids were sampled in greenhouses, because the spring melon crop is usually produced in greenhouses. We sampled 3–5 greenhouses at each location. In each greenhouse, we sampled five clones randomly by collecting leaves containing aphids. In each clone, only one alate aphid was used for further analysis. We randomly chose 10 individuals per location to represent the population of that location. Sample sizes for 2b‐RAD sequencing are listed in Table 1. A total of 90 aphids from nine locations were used for analysis of the population structure. All aphid samples were preserved in 95% ethanol at room temperature for DNA testing. We also obtained the sequence of the mitochondrial gene COI that had the best match with the COI sequence of A. gossypii in NCBI database. All samples (including those used for DNA analysis) in this study were deposited at our laboratory.
Table 1.
Sample collection data and genetic diversitya
| County | Sites (village) | Population | Sampling date | N | %poly | I | Ho | He | F |
|---|---|---|---|---|---|---|---|---|---|
| Daxing | Lihuacun | LHC | May 9‐26 | 10 | 25.96 | 0.156 | 0.180 | 0.107 | −0.441 |
| Daxing | Nandunfa | NDF | May 9‐26 | 10 | 69.43 | 0.306 | 0.153 | 0.181 | 0.385 |
| Daxing | Shitong | ST | May 9‐26 | 10 | 78.54 | 0.361 | 0.172 | 0.210 | 0.286 |
| Daxing | Xiyitang | XYT | May 9‐26 | 9 | 78.60 | 0.396 | 0.178 | 0.236 | 0.328 |
| Shunyi | Mazhuang | MZ | May 24‐31 | 7 | 60.78 | 0.335 | 0.214 | 0.209 | 0.126 |
| Shunyi | Nanbeiwu | NBW | May 24‐31 | 10 | 28.57 | 0.157 | 0.180 | 0.107 | −0.452 |
| Shunyi | Songgezhuang | SGZ | May 24‐31 | 10 | 28.26 | 0.165 | 0.179 | 0.112 | −0.452 |
| Shunyi | Wangxinzhuang | WXZ | May 24‐31 | 10 | 66.34 | 0.379 | 0.259 | 0.233 | 0.286 |
| Shunyi | Xiaozhubao | XZB | May 24‐31 | 10 | 62.42 | 0.298 | 0.169 | 0.182 | 0.149 |
N = sample size, %poly = percentage of polymorphic loci, I = Shannon’s information index, Ho = observed heterozygosity, He = expected heterozygosity, F = fixation index.
The distance between any two sample sites was at least 1 km. GPS coordinates were recorded using a Magellan® handheld GPS receiver, a latitude‐longitude projection and WGS84 datum. Sampling locations were further confirmed by consulting topographical maps of the study area and using Google Earth.
2.2. 2b‐RAD sequencing and genotyping
Genomic DNA was isolated using the DNeasy Tissue Mini Kit (Tiangen) following the manufacturer instructions. A total of 90 aphids were genotyped for SNP loci. 2b‐RAD sequencing and genotyping were outsourced to Shanghai OE Biotech Ltd. (Shanghai, China). The 2b‐RAD libraries were prepared following the protocol developed by Wang, Meyer, McKay, and Matz (2012). In brief, DNA from each sample was digested using BsaXI and then verified and separated on agarose gel. Next, library‐specific adaptors and the digestion products were linked with T4 DNA ligase. Ligation products were amplified by PCR, and the target band was excised from a 2% agarose gel. At last, sample‐specific barcodes were introduced by PCR with platform‐specific barcode‐bearing primers. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germany) and then pooled for sequencing using the Illumina Hiseq2500 platform (Illumina, San Diego, CA).
Quality filtering was conducted as follows: raw reads were trimmed to remove adaptors, and the terminal 2‐bp positions were discarded to eliminate artifacts that might have arisen by ligation. Reads contained no restriction site or long homo‐polymer regions (>10 bp). Ambiguous bases (N) or reads of low quality (>10 bp with quality less than Q20) were removed. SNPs were determined, and genotypes were called using a maximum‐likelihood statistical model implemented in the software Stacks v1.32 (Catchen, Amores, Hohenlohe, Cresko, & Postlethwait, 2011).
2.3. Data analysis
2.3.1. Population genetic analyses
The genotype data contained information for each locus and each individual. We used MEGA 7 to set the coverage at 95% and then output the selected loci for further analysis. All the genetic analyses were performed in GenALEx 6.5 (Peakall & Smouse, 2006, 2012). The genetic diversity of sampling locations was quantified using allele frequency, Nei’s unbiased heterozygosity, and Shannon’s information index. Shannon’s information index for information theory has often been used in population genetics. It offers statistical properties useful for measuring biological information (Peakall & Smouse, 2012; Sherwin, Jabot, Rush, & Rossetto, 2006). Therefore, we used Shannon’s information index to represent the genetic diversity. We estimated genetic variation (Fst) among all populations and between each pair of populations. To determine the proportion of genetic variation that could be attributed to differences between sampling sites, hierarchal analyses of molecular variance (AMOVAs) were performed. The correlation between genetic distance and geographic distance were analyzed using Mantel tests (10,000 permutations) which is the most widely used statistical method in landscape genetics’ studies. Principal coordinates analysis (PCoA) was used to find and plot the major pattern within a genetic distance matrix dataset.
2.3.2. Influence of landscape features on genetic diversity
Land cover maps of sampling sites were obtained from a digital map provided by the ZiYuan‐3 survey satellite (ZY‐3) with 5 m resolution. The landscape structure was analyzed using ArcGIS 10 (ESRI, Redlands, CA, USA). Land covers were classified into seven general categories, including cropland, vegetable, orchard, grassland, woodland, water, and urban. Cropland was categorized as land primarily being cultivated with crops such as winter wheat. The vegetable category included protected fields, for example, greenhouses, in which vegetables were usually planted. The orchard category had perennial crops, such as orchard trees. The grassland category was herb‐dominated land. Woodland included all forest and shrub land excluding the trees in residential sites. Water represented wetland and open water. Urban represented human architecture, roads, etc. For each of the sampling sites, landscape composition was estimated in four circular sectors (0.5, 1, 2, and 3 km radii), representing a nested set of landscape sectors at four spatial scales. The percentage of each category was measured within each landscape circle.
Generalized linear model regression (GLM) and correlation analyses were performed using the statistical program R version 3.4.0 (R Core Team 2017). Genetic data, such as Shannon’s information index, the percentage of polymorphic loci and expected heterozygosity were highly correlated. Only observed heterozygosity had a weak correlation with all the other indices. We used Shannon’s information index as a response variable and landscape features as predictor variables. The landscape variables were the percentages of cropland, vegetable, orchard, grassland, and woodland, all of which were belonging to vegetation cover. Each spatial scale was analyzed separately. We used the all‐subsets regression to select the predictor variables that best explained the model (Kabacoff, 2015). The all‐subsets regression was achieved using the package “leaps” (Thomas Lumley based on Fortran code by Alan Miller, 2017). We fit the models based on the all‐subset regression results. For all the scales, we chose the best model among different scales by adjusted r2, P‐value, and AIC. We used a new method called relative weights to assess the relative importance of predictors. The method closely approximates the average increase in R‐square obtained by adding a predictor variable across all possible submodels (Kabacoff, 2015).
3. RESULTS
3.1. Bioinformatics and filtering of data
Sequencing resulted in an average of 6,927,570 bp reads per individual. The percentage of high quality of reads was above 70% of the total reads in the libraries of the 90 individuals. The SNP loci number was 10,685, which could genotype at least 72 individuals (80% of samples). We removed four individuals (3 in MZ and 1 in XYT) because of their low‐quality reads and obtained a total of 6,280 loci, which scored 95% coverage.
3.2. Genetic diversity of aphid populations
The percentage of polymorphic loci ranged from 25.96% (LHC population) to 78.60% (XYT population). Shannon’s information index values across loci ranged from 0.156 (LHC population) to 0.396 (XYT population). The mean observed heterozygosity (Ho = 0.187) was similar to the mean expected heterozygosity (He = 0.175) in the total number of populations (Table 1). The reduction in genetic variation in the LHC and NBW populations was evident in their percentages of polymorphic loci, which were much lower than XYT and WXZ populations.
There was a moderate level of genetic differentiation among populations. Pairwise Fst values between the populations ranged from −0.042 to 0.310 (Table 2). The Mantel test result showed that no significant correlation was found between genetic and geographic distances across the nine populations (Z = −7.248, r = −0.074, p = 0.276).
Table 2.
Pairwise comparison of genetic distance (Fst) and geographic distance (km) among aphid populations
| LHC | NDF | ST | XYT | MZ | NBW | SGZ | WXZ | XZB | |
|---|---|---|---|---|---|---|---|---|---|
| LHC | 0.206 | 0.155 | 0.094 | 0.038 | 0.001 | 0.012 | 0.310 | 0.171 | |
| NDF | 10.306 | −0.042 | 0.001 | 0.119 | 0.170 | 0.125 | 0.155 | 0.030 | |
| ST | 9.027 | 4.088 | −0.020 | 0.075 | 0.123 | 0.086 | 0.131 | 0.011 | |
| XYT | 7.149 | 4.010 | 2.098 | 0.014 | 0.072 | 0.045 | 0.105 | 0.006 | |
| MZ | 67.910 | 61.698 | 59.940 | 62.024 | 0.035 | 0.032 | 0.175 | 0.080 | |
| NBW | 72.661 | 66.528 | 64.741 | 66.823 | 4.843 | −0.007 | 0.292 | 0.137 | |
| SGZ | 79.087 | 73.272 | 71.351 | 73.420 | 11.871 | 7.145 | 0.268 | 0.108 | |
| WXZ | 75.859 | 69.925 | 68.052 | 70.126 | 8.421 | 3.710 | 3.455 | 0.134 | |
| XZB | 69.116 | 62.943 | 61.169 | 63.251 | 1.268 | 3.586 | 10.605 | 7.154 |
Above diagonal; genetic distance among populations as measured by Fst.
Below diagonal; geographic distance between populations (km).
According to the results of AMOVA, the majority (89%) of the genetic variation was attributable to variation within populations, whereas approximately 11% of the variation was attributable to variation between populations. However, no variation was attributed to variation among regions (Daxing county and Shunyi county) (Table 3). The PCoA result (Figure 2) showed a high–low pattern of genetic structure. The first and second axes explained 69.04% and 23.95% of the overall variance, respectively. We, therefore, divided the populations into two groups: higher genetic diversity group (WXZ, XYT, ST, XZB, NDF), and lower genetic diversity group (MZ, SGZ, NBW, LHC). The AMOVA result showed that 11% of the variation was attributable to variation among these two groups (Table 3).
Table 3.
AMOVA analysis testing the partitioning of genetic variation across populations, geographic regions (Daxing and Shunyi) and two groups including a high genetic diversity group and a low genetic diversity group based on STRUCTURE analysis
| Source of variation | df | SS | % Variation | P‐value |
|---|---|---|---|---|
| Global analysis | ||||
| Among region | 1 | 1490.994 | 0 | 0.971 |
| Among population | 7 | 13498.768 | 11% | 0.001 |
| Among individual | 77 | 43315.367 | 0% | 0.001 |
| Hierarchical AMOVA (K = 2) | ||||
| Among group | 1 | 7216.514 | 11% | 0.001 |
| Among population | 7 | 7773.224 | 4% | 0.001 |
| Among individual | 77 | 43315.367 | 0% | 0.001 |
Figure 2.
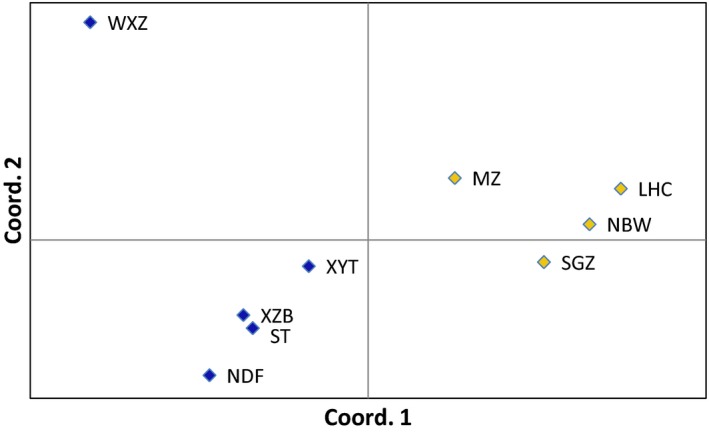
Principal coordinate analysis based on the genetic distance matrix of Fst values. Population codes are given in Table 1. Colors within the diamond: blue, high genetic diversity group; yellow, low genetic diversity group
3.3. Genetic diversity and their relationship with landscape feature
Shannon’s index was positively correlated with grassland at the spatial scales of 1 and 2 km and negatively with orchard at 0.5 and 1 km. Shannon’s index was best predicted by the grassland + orchard + vegetable model at a spatial scale of 1 km (Table 4), having the lowest AIC score of any model at the four spatial scales examined. The correlation between Shannon’s index with vegetable land was marginally significant in this model (p = 0.066). The total amount of variance accounted for by this model (R‐square = 0.796) was divided among the predictor variables. Orchard land accounted for 57.80% of the R‐square, grassland accounted for 27.72%, and vegetable land accounted for 14.48%. Based on the method of relative weights, orchard land had the greatest relative importance, followed by grassland and vegetable land, in that order.
Table 4.
The best model of GLM regression of genetic diversity (I, Shannon’s information index) related to landscape features. Landscape variables included percentage of cropland areas (cropland), percentage of vegetable areas (vegetable), percentage of orchard areas (orchard), percentage of grassland areas (grassland), and the percentage of woodland areas (woodland) at each scalea
| Scale | Model | Adjusted r 2 | p value | AIC |
|---|---|---|---|---|
| 0.5 km | −0.0021 × Orchard | 0.279 | 0.083 | −15.399 |
| 1 km | −0.0042 × Vegetable − 0.0033 × Orchard b + 0.036 × Grassland b | 0.674 | 0.035 | −21.576 |
| 2 km | 0.060 × Grasslandb | 0.353 | 0.054 | −15.399 |
| 3 km | −0.0028 × Orchard | 0.105 | 0.206 | −13.458 |
Bold indicates the best overall model. Intercept of models are omitted.
Significance of variables is indicated as follows: p < 0.05.
4. DISCUSSION
4.1. Genetic diversity of A. gossypii in melon growing areas
We completed genetic analysis of A. gossypii samples collected from watermelon in the Beijing area at landscape scales. We found evidence for moderate population differentiation at different sample sites with landscape features associated with the genetic variation.
Samples of A. gossypii populations were collected from watermelon on which reproduction was via apomictic parthenogenesis. The sampling period was during the establishment of the spring migration population. Early season individuals on melon were alates, and so the entire potential colonizing population was sampled. Host plant pressure on aphids occurs in melons and aphid‐resistant melon cultivars and could alter the genetic structure of A. gossypii (Thomas et al., 2012, 2016). In the present study, there was no known commercial aphid‐resistant melon varieties planted in the research area. Most of the watermelon cultivars in the study area were “Jingxin.” We therefore assume that any host plant pressure on the different A. gossypii samples was similar across the study area. We therefore excluded potential host plant effects and focused on the possibility that the different surrounding habitats could influence the genetic diversity of the melon aphid populations in greenhouses.
A recent genetic study of A. gossypii in China, based on microsatellite and mitochondrial DNA data, showed that aphids were genetically clustered into a western group and an eastern group (Wang, Yang, Lu, Zhou, & Wu, 2017). Their study had a large geographic scale, so a geographic effect was expected. In the smaller landscape scale of the present study, we found no geographic isolation. Most aphids are weak fliers, and their dispersal among secondary hosts is aided by wind (Dixon & Howard, 1986; Taylor, Woiwod, & Taylor, 1979). The genetic analysis showed that dispersal limitation only caused moderate differentiation. The colonization of aphids is potentially a local event.
The populations of A. gossypii tended to be clustered into two groups that could be most easily understood in terms of genetic diversity. The high diversity group, represented by WXZ, had a higher percentage of polymorphic loci and a high Shannon index. The low diversity group, represented by LHC, had a lower percentage of polymorphic loci and a low Shannon index. The genetic diversity pattern resembled a patchwork with high and low diversity. The presence of population structure suggests that one or more evolutionary forces may be operating. These forces include limited migration, small local population sizes, restricted mating, and intrapopulation selection (Vaughn & Antolin, 1998). Dispersal affects the distribution of genetic diversity across space. Gene flow mediated by dispersal may drive or inhibit local adaptation, modulate the effect of genetic drift, and ultimately regulate population persistence (Mazzi & Dorn, 2012). Our results indicate that geographic distance is not a barrier to A. gossypii gene flow in this area.
4.2. Relatedness of genetic structure and landscape features
The GLM results demonstrate that genetic diversity is significantly associated with landscape features. Populations within landscapes containing large expanses of grassland tended to have higher genetic diversity than that of populations within areas with less grassland. However, orchard influenced the genetic diversity in a manner opposite to that of grassland. The greater the extent of the orchard land area was, the lower the genetic diversity of A. gossypii became. The orchard had a more relative importance than grassland to predict the genetic diversity.
Many landscape ecology studies suggest that special habitat or land cover affects insect colonization (Angelella et al., 2016; Dong, Ouyang, Lu, & Ge, 2015; Sivakoff, Rosenheim, Dutilleul, & Carrière, 2013). As landscape composition can affect population establishment and expansion, it could also affect the genetic structure. There are several possible explanations for the genetic diversity of A. gossypii populations being affected by land cover.
First, aphids can move from an unsuitable host to a more suitable habitat. The landscape composition, therefore, could influence aphid colonization. We did not measure the nearest distance of primary host plants from where aphids migrated. It has been reported that many plant species serve as primary hosts in North China, such as Hibiscus syriacus (hibiscus), Punica granatum (pomegranate), and Zanthoxylum simulans (Chinese prickly ash) (Wang et al., 2016). Moreover, these primary hosts are seldom planted in large areas. The widespread and differing species of primary hosts may influence the genetic structure of immigrant populations of A. gossypii. Some of the alate migrants may come from much farther places even representing anholocyclic populations that overwintered as adults. It is possible that these aphids are selecting particular habitat types based on visual cues (Favret & Voegtlin, 2001; Smith, 1969). Perhaps long‐distance migrant aphids make a habitat choice and then increase the genetic diversity by mixing with local aphids.
Second, the vegetation cover in the present study could also simply reflect the host plant distribution. Grassland had a positive effect on aphid genetic diversity possibly because grassland supplies abundant aphid resources. Many weeds in the Capparidaceae, Convolvulaceae, and Euphorbiaceae are hosts of A. gossypii (Brévault et al., 2008). The host distribution affects the A. gossypii genetics through selection and clonal reproduction. We cannot rule out the possibility that A. gossypii also overwinters on weeds. By the time of watermelon emergence, the aphids may disperse between watermelon and grassland. Grassland plants might also act as transitional hosts to bridge the time gap between spring watermelons and overwintering hosts. The orchard area, however, contains perennial, deciduous crops such as orchard tree species. Most of these habitats do not contain suitable host plants for A. gossypii. As the orchard area increases, the melon crop areas harboring A. gossypii decrease. The low genetic diversity of A. gossypii occurs when the landscape is dominated with orchard land.
At last, the landscape scales also had effects on the dispersal and colonization of A. gossypii. We examined the impact of landscape variables on genetic diversity at four spatial scales, with landscape radii varying from 0.5 to 3 km. Landscape features at a radius of 1 km surrounding focal watermelon fields explained the highest proportion of the variation in genetic diversity. Although we know relatively little about the movement of A. gossypii, it is possible that a landscape of this size encompasses their ecological neighborhood (Addicott et al., 1987), containing a diversity of habitats utilized by this species. Bahlai, Sikkema, Hallett, Newman, and Schaafsma (2010) used landscape parameters to identify fields at greatest risk of becoming colonized by soybean aphids (Aphis glycines). The present study about landscape genetics of aphids may help improve our understanding of colonization of A. gossypii and the corresponding spatial scale. However, the limited sample size and location may reduce the statistical power. Measuring the distribution of primary hosts could be a way to investigate aphid colonization. An alternative analysis strategy used in some landscape studies (e.g., Gardiner et al., 2009) which focuses on landscape diversity and combined explanatory variables could also be helpful. Future work should test the effect of year and in particular should examine host plant species, which could act as reservoirs.
5. CONCLUSIONS
Our study clearly supports the hypothesis that landscape features can affect the genetic variation of A. gossypii. The populations were structured not by geography but by their surrounding habitats. They were divided into two groups (high and low) based on genetic diversity. The results suggest that the colonization of watermelon by A. gossypii occurs locally and is affected by the surrounding habitats. More grassland in the landscape tends to favor the maintenance of genetic diversity. Orchard and their perennial plants reduced the genetic diversity of A. gossypii. The land cover, such as grassland and orchard, had a strongest effect at the spatial scale of 1 km. By identifying features of the landscape that surrounds fields and can affect these dynamics, growers can develop more efficient crop protection strategies relying on habitat manipulation.
DATA ACCESSIBILITY
Raw 2b‐RAD sequence reads are available at NCBI PRJNA407691.
CONFLICT OF INTEREST
The authors have declared that no competing interests exist.
AUTHOR CONTRIBUTIONS
Z. Dong and Z. Zhang conceived and designed the study. Y. Li conducted sampling and isolated DNA. Z. Dong analyzed the data and prepared the manuscript.
ACKNOWLEDGMENTS
We thank Shuang Liu, Yuansheng Li, and Baofeng Mu for assistance with sampling and DNA extraction and the farmers for allowing the collection of aphids from their fields. This study was supported by National Natural Science Foundation of China (No. 31501646), National Key R&D Program of China (2017YFD0200300), Foundation for the Talents by the Beijing Municipal (2014000020124G080), and Scientific Research Project of Beijing Municipal Education Commission (KM201710020006).
Dong Z, Li Y, Zhang Z. Genetic diversity of melon aphids Aphis gossypii associated with landscape features. Ecol Evol. 2018;8:6308–6316. https://doi.org/10.1002/ece3.4181
REFERENCES
- Addicott, J. F. , Aho, J. M. , Antolin, M. F. , Padilla, D. K. , Richardson, J. S. , & Soluk, D. A. (1987). Ecological neighborhoods ‐ scaling environmental patterns. Oikos, 49, 340–346. https://doi.org/10.2307/3565770 [Google Scholar]
- Agarwala, B. K. , & Choudhuri, P. R. (2014). New host races of Aphis gossypii (Insecta: Hemiptera: Aphididae) from northeast India. Current Science, 107, 1172–1177. [Google Scholar]
- Agarwala, B. , & Das, K. (2007). Host plant‐based morphological, ecological and esterase variations in Aphis gossypii Glover populations (Homoptera: Aphididae). Entomon, 32, 89–95. [Google Scholar]
- Angelella, G. M. , Holland, J. D. , & Kaplan, I. (2016). Landscape composition is more important than local management for crop virus–insect vector interactions. Agriculture, Ecosystems & Environment, 233, 253–261. https://doi.org/10.1016/j.agee.2016.09.019 [Google Scholar]
- Bahlai, C. , Sikkema, S. , Hallett, R. H. , Newman, J. , & Schaafsma, A. (2010). Modeling distribution and abundance of soybean aphid in soybean fields using measurements from the surrounding landscape. Environmental Entomology, 39, 50–56. https://doi.org/10.1603/EN09127 [DOI] [PubMed] [Google Scholar]
- Blackman, R. , & Eastop, V. (2007). Taxonomic issues In Van Emden H., & Harrington R. V. (Eds.), Aphids as crop pests (pp. 1–29). London, UK: CAB International. [Google Scholar]
- Brévault, T. , Carletto, J. , Linderme, D. , & Vanlerberghe‐Masutti, F. (2008). Genetic diversity of the cotton aphid Aphis gossypii in the unstable environment of a cotton growing area. Agricultural and Forest Entomology, 10, 215–223. https://doi.org/10.1111/j.1461-9563.2008.00377.x [Google Scholar]
- Brévault, T. , Carletto, J. , Tribot, J. , & Vanlerberghe‐Masutti, F. (2011). Insecticide use and competition shape the genetic diversity of the aphid Aphis gossypii in a cotton‐growing landscape. Bulletin of Entomological Research, 101, 407–413. https://doi.org/10.1017/S0007485310000635 [DOI] [PubMed] [Google Scholar]
- Carletto, J. , Lombaert, E. , Chavigny, P. , Brévault, T. , Lapchin, L. , & Vanlerberghe‐Masutti, F. (2009). Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Molecular Ecology, 18, 2198–2212. https://doi.org/10.1111/j.1365-294X.2009.04190.x [DOI] [PubMed] [Google Scholar]
- Carletto, J. , Martin, T. , Vanlerberghe‐Masutti, F. , & Brévault, T. (2010). Insecticide resistance traits differ among and within host races in Aphis gossypii . Pest Management Science, 66, 301–307. https://doi.org/10.1002/ps.1874 [DOI] [PubMed] [Google Scholar]
- Catchen, J. M. , Amores, A. , Hohenlohe, P. , Cresko, W. , & Postlethwait, J. H. (2011). Stacks: Building and genotyping loci de novo from short‐read sequences. G3: Genes Genomes, Genetics, 1, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen, J. , Bassham, S. , Wilson, T. , Currey, M. , O’Brien, C. , Yeates, Q. , & Cresko, W. A. (2013). The population structure and recent colonization history of Oregon threespine stickleback determined using restriction‐site associated DNA‐sequencing. Molecular Ecology, 22, 2864–2883. https://doi.org/10.1111/mec.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charaabi, K. , Carletto, J. , Chavigny, P. , Marrakchi, M. , Makni, M. , & Vanlerberghe‐Masutti, F. (2008). Genotypic diversity of the cotton‐melon aphid Aphis gossypii (Glover) in Tunisia is structured by host plants. Bulletin of Entomological Research, 98, 333–341. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Vanlerberghe‐Masutti, F. , Wilson, L. J. , Barchia, I. , McLoon, M. O. , Smith, T. , & Herron, G. A. (2013). Evidence of superclones in Australian cotton aphid Aphis gossypii Glover (Aphididae: Hemiptera). Pest Management Science, 69, 938–948. https://doi.org/10.1002/ps.3455 [DOI] [PubMed] [Google Scholar]
- Dixon, A. F. G. , & Howard, M. T. (1986). Dispersal in aphids, a problem in resource allocation In Danthanarayana W. (Ed.), Insect flight (pp. 145–151). Berlin, Heidelberg: Springer; https://doi.org/10.1007/978-3-642-71155-8 [Google Scholar]
- Dong, Z. , Ouyang, F. , Lu, F. , & Ge, F. (2015). Shelterbelts in agricultural landscapes enhance ladybeetle abundance in spillover from cropland to adjacent habitats. BioControl, 60, 351–361. https://doi.org/10.1007/s10526-015-9648-5 [Google Scholar]
- Favret, C. , & Voegtlin, D. J. (2001). Migratory aphid (Hemiptera: Aphididae) habitat selection in agricultural and adjacent natural habitats. Environmental Entomology, 30, 371–379. https://doi.org/10.1603/0046-225X-30.2.371 [Google Scholar]
- Fuller, S. , Chavigny, P. , Lapchin, L. , & Vanlerberghe‐Masutti, F. (1999). Variation in clonal diversity in glasshouse infestations of the aphid, Aphis gossypii Glover in southern France. Molecular Ecology, 8, 1867–1877. https://doi.org/10.1046/j.1365-294x.1999.00782.x [DOI] [PubMed] [Google Scholar]
- Gardiner, M. M. , Landis, D. A. , Gratton, C. , DiFonzo, C. D. , O’Neal, M. , Chacon, J. M. , … Heimpel, G. E. (2009). Landscape diversity enhances biological control of an introduced crop pest in the north‐central USA. Ecological Applications, 19, 143–154. https://doi.org/10.1890/07-1265.1 [DOI] [PubMed] [Google Scholar]
- Gilabert, A. , Gauffre, B. , Parisey, N. , Le Gallic, J. F. , Lhomme, P. , Bretagnolle, V. , … Plantegenest, M. (2017). Influence of the surrounding landscape on the colonization rate of cereal aphids and phytovirus transmission in autumn. Journal of Pest Science, 90, 447–457. https://doi.org/10.1007/s10340-016-0790-3 [Google Scholar]
- Guldemond, J. A. , Tigges, W. T. , & De Vrijer, P. W. F. (1994). Host Races of Aphis gossypii (Homoptera: Aphididae) on Cucumber and Chrysanthemum. Environmental Entomology, 23, 1235–1240. https://doi.org/10.1093/ee/23.5.1235 [Google Scholar]
- Hemme, R. R. , Thomas, C. L. , Chadee, D. D. , & Severson, D. W. (2010). Influence of urban landscapes on population dynamics in a short‐distance migrant mosquito: Evidence for the dengue vector Aedes aegypti . PLoS Neglected Tropical Diseases, 4, e634 https://doi.org/10.1371/journal.pntd.0000634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, F. , Westphal, C. , Moritz, R. F. , & Steffan‐Dewenter, I. (2007). Genetic diversity and mass resources promote colony size and forager densities of a social bee (Bombus pascuorum) in agricultural landscapes. Molecular Ecology, 16, 1167–1178. https://doi.org/10.1111/j.1365-294X.2007.03226.x [DOI] [PubMed] [Google Scholar]
- Holderegger, R. , & Wagner, H. H. (2006). A brief guide to landscape genetics. Landscape Ecology, 21, 793–796. https://doi.org/10.1007/s10980-005-6058-6 [Google Scholar]
- Johansson, M. , Primmer, C. R. , Sahlsten, J. , & Merilä, J. (2005). The influence of landscape structure on occurrence, abundance and genetic diversity of the common frog, Rana temporaria . Global Change Biology, 11, 1664–1679. https://doi.org/10.1111/j.1365-2486.2005.1005.x [Google Scholar]
- Kabacoff, R. (2015) R in action: data analysis and graphics with R Manning Publications Co.
- Keyghobadi, N. , Roland, J. , & Strobeck, C. (1999). Influence of landscape on the population genetic structure of the alpine butterfly Parnassius smintheus (Papilionidae). Molecular Ecology, 8, 1481–1495. https://doi.org/10.1046/j.1365-294x.1999.00726.x [DOI] [PubMed] [Google Scholar]
- Lindsay, D. L. , Barr, K. R. , Lance, R. F. , Tweddale, S. A. , Hayden, T. J. , & Leberg, P. L. (2008). Habitat fragmentation and genetic diversity of an endangered, migratory songbird, the golden‐cheeked warbler (Dendroica chrysoparia). Molecular Ecology, 17, 2122–2133. https://doi.org/10.1111/j.1365-294X.2008.03673.x [DOI] [PubMed] [Google Scholar]
- Mazzi, D. , & Dorn, S. (2012). Movement of insect pests in agricultural landscapes. Annals of Applied Biology, 160, 97–113. https://doi.org/10.1111/j.1744-7348.2012.00533.x [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Resources, 6, 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics, 28, 2537–2539. https://doi.org/10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Espona, S. , McLeod, J. E. , & Franks, N. R. (2012). Landscape genetics of a top neotropical predator. Molecular Ecology, 21, 5969–5985. https://doi.org/10.1111/mec.12088 [DOI] [PubMed] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; URL https://www.R-project.org/. [Google Scholar]
- Razmjou, J. , Vorburger, C. , Moharramipour, S. , Mirhoseini, S. Z. , & Fathipour, Y. (2010). Host‐associated differentiation and evidence for sexual reproduction in Iranian populations of the cotton aphid, Aphis gossypii . Entomologia Experimentalis et Applicata, 134, 191–199. https://doi.org/10.1111/j.1570-7458.2009.00951.x [Google Scholar]
- Sherwin, W. B. , Jabot, F. , Rush, R. , & Rossetto, M. (2006). Measurement of biological information with applications from genes to landscapes. Molecular Ecology, 15, 2857–2869. https://doi.org/10.1111/j.1365-294X.2006.02992.x [DOI] [PubMed] [Google Scholar]
- Sivakoff, F. S. , Rosenheim, J. A. , Dutilleul, P. , & Carrière, Y. (2013). Influence of the surrounding landscape on crop colonization by a polyphagous insect pest. Entomologia Experimentalis et Applicata, 149, 11–21. https://doi.org/10.1111/eea.12101 [Google Scholar]
- Smith, J. (1969). Some effects of crop background on populations of aphids and their natural enemies on brussels sprouts. Annals of Applied Biology, 63, 326–330. https://doi.org/10.1111/j.1744-7348.1969.tb05497.x [Google Scholar]
- Taylor, L. , Woiwod, I. , & Taylor, R. (1979). The migratory ambit of the hop aphid and its significance in aphid population dynamics. Journal of Animal Ecology, 48, 955–972. https://doi.org/10.2307/4207 [Google Scholar]
- Thomas, S. , Boissot, N. , & Vanlerberghe‐Masutti, F. (2012). What do spring migrants reveal about sex and host selection in the melon aphid? BMC Evolutionary Biology, 12, 47. https://doi.org/10.1186/1471-2148-12-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Lumley based on Fortran code by Alan Miller (2017). leaps: Regression Subset Selection. R package version 3.0. https://CRAN.R-project.org/package=leaps
- Thomas, S. , Vanlerberghe‐Masutti, F. , Mistral, P. , Loiseau, A. , & Boissot, N. (2016). Insight into the durability of plant resistance to aphids from a demo‐genetic study of Aphis gossypii in melon crops. Evolutionary Applications, 9, 756–768. https://doi.org/10.1111/eva.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, T. T. , & Antolin, M. F. (1998). Population genetics of an opportunistic parasitoid in an agricultural landscape. Heredity, 80, 152–162. https://doi.org/10.1046/j.1365-2540.1998.00248.x [Google Scholar]
- Wang, S. , Meyer, E. , McKay, J. K. , & Matz, M. V. (2012). 2b‐RAD: A simple and flexible method for genome‐wide genotyping. Nature Methods, 9, 808–810. https://doi.org/10.1038/nmeth.2023 [DOI] [PubMed] [Google Scholar]
- Wang, X.‐Y. , Yang, X.‐M. , Lu, B. , Zhou, L.‐H. , & Wu, K.‐M. (2017). Genetic variation and phylogeographic structure of the cotton aphid, Aphis gossypii, based on mitochondrial DNA and microsatellite markers. Scientific Reports, 7, 1920 https://doi.org/10.1038/s41598-017-02105-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Zhang, S. , Luo, J.‐Y. , Wang, C. Y. , Lv, L. M. , Zhu, X. Z. , … Cui, J. J. (2016). Identification of Aphis gossypii Glover (Hemiptera: Aphididae) Biotypes from Different Host Plants in North China. PLoS ONE, 11, e0146345. https://doi.org/10.1371/journal.pone.0146345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , & Zhong, T. (1982). Study on life history of several species of aphids. Sinozoologia, 2, 7–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw 2b‐RAD sequence reads are available at NCBI PRJNA407691.


