Abstract
Visualizing pathologies in three dimensions can provide unique insights into the biology of human diseases. A rapid and easy-to-implement dibenzyl ether–based technique was used to clear thick sections of surgically resected human pancreatic parenchyma. Protocols were applicable to both fresh and formalin-fixed, paraffin-embedded tissue. The penetration of antibodies into dense pancreatic parenchyma was optimized using both gradually increasing antibody concentrations and centrifugal flow. Immunolabeling with antibodies against cytokeratin 19 was visualized using both light sheet and confocal laser scanning microscopy. The technique was applied successfully to 26 sections of pancreas, providing three-dimensional (3D) images of normal pancreatic tissue, pancreatic intraepithelial neoplasia, intraductal papillary mucinous neoplasms, and infiltrating pancreatic ductal adenocarcinomas. 3D visualization highlighted processes that are hard to conceptualize in two dimensions, such as invasive carcinoma growing into what appeared to be pre-existing pancreatic ducts and within venules, and the tracking of long cords of neoplastic cells parallel to blood vessels. Expanding this technique to formalin-fixed, paraffin-embedded tissue opens pathology archives to 3D visualization of unique biosamples and rare diseases. The application of immunolabeling and clearing to human pancreatic parenchyma provides detailed visualization of normal pancreatic anatomy, and can be used to characterize the 3D architecture of diseases including pancreatic intraepithelial neoplasia, intraductal papillary mucinous neoplasm, and pancreatic ductal adenocarcinomas.
Since the days of Virchow, pathologists have been studying diseases in two dimensions.1 For centuries, standard hematoxylin and eosin (H&E)-stained slides have framed our field of vision and therefore have narrowed the conceptual framework through which diseases are viewed. Although radiologists and experimental pathologists have broken through this framework with technologies such as three-dimensional (3D) computed tomography and organoid models, most morphologic studies of in situ human pathology still are viewed and conceptualized in two dimensions.2, 3
Visualizing pathologies in three dimensions provides insight into the biology of human diseases. Lesions can be measured accurately, the spatial relationships of various tissue and cell components can be defined, and morphologic changes can be put in the appropriate multidimensional context.2, 4 The addition of immunolabeling for protein and in situ hybridization for RNA would allow one to visualize protein and gene expression in 3D space, creating a true multidimensional landscape.
We present a simple clearing and labeling method to visualize intact thick sections of normal and diseased human pancreatic parenchyma in three dimensions.5, 6, 7 Organic solvents were selected to clear stroma-rich human tissues.8, 9 Proteins are preserved, allowing for immunofluorescent labeling of protein expression with an optimized protocol for penetration of antibodies in dense pancreatic tissue. In this study, cytokeratin 19 (CK19) was labeled to visualize the pancreatic ductal system, and the autofluorescent features of collagen and elastin were exploited to visualize blood vessels.
This methodology has a number of potential applications. The volume of small lesions, such as pancreatic intraepithelial neoplasia (PanIN) lesions, may be determined, the 3D relationships of cells (neoplastic, inflammatory, and so forth) to important structures (vessels, nerves, ducts, and so forth) may be defined, and changes in protein expression may be defined as cells interact, such as when neoplastic cells invade into tissues. When DNA and RNA studies are added in the future, the methodology may be used to define the drivers of 3D morphology.10, 11
Materials and Methods
This study was approved by our Institutional Review Board. Briefly, intact slabs (up to 2.0 × 2.0 × 0.5 cm) of excess normal or diseased surgically resected human pancreatic parenchyma were harvested. These included 17 slabs of grossly normal pancreatic parenchyma, 7 slabs of infiltrating pancreatic ductal adenocarcinoma with adjacent pancreatic parenchyma, and 2 slabs with a grossly dilated pancreatic duct, grossly suspicious for intraductal papillary mucinous neoplasm. Four of the 17 slabs of normal pancreatic parenchyma were first formalin-fixed and paraffin-embedded.
Fresh Samples
Fresh tissues were fixed as quickly as possible after surgical resection in 80% methanol/20% dimethyl sulfoxide (DMSO) to dehydrate the tissues and precipitate proteins to stop autolysis of the tissue by pancreatic enzymes. Fixation was performed overnight at room temperature. The following day, the tissues were rehydrated and fixed in neutral-buffered 10% formalin or 4% paraformaldehyde for 24 hours at room temperature. Next, the tissues were dehydrated with 70% methanol, 95% methanol, and 3 × 100% methanol, followed by chilling the tissue for an hour at 4°C in 100% methanol. Tissues then were incubated overnight in 66% dichloromethane/33% methanol at room temperature. Samples then were washed twice in 100% methanol and then 5% hydrogen peroxide was added to the 100% methanol for overnight incubation to oxidize endogenous pigments and autofluorescent proteins, resulting in increased tissue clearing and decreased tissue autofluorescence. Tissues were rehydrated in 1× phosphate-buffered saline (PBS) and then washed twice for an hour each in PBS/0.2% Triton X-100 (Millipore Sigma, St. Louis, MO). Next, the tissues were incubated for 4 days in a permeabilization solution of PBS/20% DMSO/0.2% Triton X-100/0.3 mol/L glycine at 37°C. Glycine was added to prevent background labeling.
Formalin-Fixed, Paraffin-Embedded Samples
After harvesting, four of the tissues were formalin-fixed and paraffin-embedded. The tissues were released from the block by dissolving the paraffin in xylene at 37°C. The tissues then were washed 3 times in 100% methanol and chilled for an hour at 4°C, followed by incubation overnight in 66% dichloromethane/33% methanol at room temperature. Samples were washed twice in 100% methanol. Five percent hydrogen peroxide was then added to the 100% methanol for overnight incubation. Tissues were rehydrated in PBS and washed twice for an hour each in PBS/0.2% Triton X-100, and then incubated for 2 days in PBS/20% DMSO/0.2% Triton X-100/0.3 mol/L glycine at 37°C. Glycine was added to prevent background labeling.9
Immunolabeling
Antigen blocking was performed by incubating the tissues in PBS/0.2% Triton X-100/10% DMSO/6% donkey serum for 2 days at 37°C. Samples then were washed twice in PBS/0.2% Tween-20 with 10 μg/mL heparin for 1 hour each at 37°C. Heparin was added to prevent background labeling.9 The extracellular matrix is a significant physical barrier for the penetration of antibodies.12, 13, 14 Two approaches were used to increase the penetration of the primary antibody, rabbit anti-human CK19. First, the antibody concentration was increased gradually over 4 days from a starting dilution of 1/800 to a final dilution of 1/200. Specifically, primary antibody (rabbit anti-human CK19; Abcam, Cambridge, UK) incubation was performed in PBS/5% DMSO/3% donkey serum/0.2% Tween-20 with 10 μg/mL heparin at a dilution of 1/800 on the first day. For the next 3 days, the antibody concentration was increased every day by 1/800 (1/800, 2/800, 3/800, and so forth) until a final dilution of 1/200. As a second approach, centrifugal flow (600 × g) was also used to promote antibody penetration.14 During these 4 days of antibody incubation, the tissues were centrifuged consecutively for 12 hours at 600 × g and shaken for 12 hours at 37°C. After the primary antibody was applied to the tissues, they were washed 5 times with PBS/0.2% Tween-20 with 10 μg/mL heparin for 1 hour each at room temperature. A pepsin-digested secondary antibody fragment with a smaller molecular weight than an intact IgG antibody was used to increase tissue penetration of the secondary antibody. The secondary antibody fragment (Alexa Fluor 488 AffiniPure F(ab’)2 fragment, donkey anti-rabbit IgG; Jackson ImmunoResearch, West Grove, PA) was incubated for 4 days and protected from light. During this time, the tissues were alternatively centrifuged for 12 hours at 600 × g and shaken for 12 hours at 37°C. The tissues were then washed 5 times with PBS/0.2% Tween-20 with 10 μg/mL heparin for 1 hour each at room temperature and protected from light.
Tissue Clearing
The tissues were dehydrated with 70% methanol, 95% methanol, and 3 × 100% methanol, followed by a 3-hour incubation in 66% dichloromethane/33% methanol at room temperature while shaking. Right before clearing in dibenzyl ether (DBE) for at least 48 hours, the tissues were incubated in 100% dichloromethane for 15 minutes.
Imaging
Immunolabeled tissues were visualized in three dimensions using either the Ultramicroscope II (Light Sheet Microscope; LaVision BioTec, Bielefeld, Germany) or the LSM800 (confocal laser scanning microscope; Carl Zeiss, Jena, Germany). The Ultramicroscope II is equipped with a Neo sCMOS camera (Andor Technology, Belfast, UK) and a 4× objective lens that was immersed in DBE in the imaging chamber. Alexa 488 signals of ductal cells were visualized with a bandpass filter set with an excitation range of 480/40 nm and an emission range of 525/50 nm. Although autofluorescence of the tissues was reduced as described in Fresh Samples and Formalin-Fixed, Paraffin-Embedded Samples, the elastic lamina and collagen of blood vessels still had detectable autofluorescence. This autofluorescence, combined with the unique morphology of elastic lamina, was used to identify vessels without the use of additional labeling. Autofluorescence was observed in an additional filter set of the LaVision Ultramicroscope II (excitation, 405/40 nm; emission, 460/50 nm). For the LSM800 microscope, the tissue was submerged in DBE in a petri dish to match the refractive index and to obtain a flat surface at the interface between materials of different refractive indices. We used both the 5× and 10× objectives. The 488-nm argon laser was used to excite the Alexa 488 fluorochrome, and the range of visualized emitted light was set at 510 to 615 nm. Autofluorescence was observed best in the LSM800 microscope when the 405-nm laser was used and the range of visualized emitted light was set between 410 and 470 nm. However, the autofluorescence signal also was observed while visualizing the Alexa 488 fluorochrome with both the Ultramicroscope II and the LSM800. 3D reconstructions were made with Imaris Software version 8.4 (Bitplane, Zurich, Switzerland).
Validation
Four of the cases were embedded in paraffin after labeling, clearing, and visualization. DBE was removed by washing it away with methanol (5 times for >1 hour), followed by rehydration with PBS. Tissues then were formalin-fixed and paraffin-embedded, and sectioned for routine H&E staining. The pathologies observed in the cleared tissues were compared with those in the H&E-stained slides.
Results
All 26 thick sections of human pancreatic parenchyma were cleared successfully (Figure 1). Normal parenchyma, fatty parenchyma, and even densely fibrotic parenchyma were cleared. All sections were labeled with an antibody targeting CK19. CK19 was selected as the first marker to examine in three dimensions because it allows visualization of the normal epithelial components of the pancreas, pancreatic cancer precursor lesions, and invasive pancreatic cancer, providing broad insights into normal and neoplastic processes in the human pancreas. Although the penetration of antibody into the dense fibrotic tissue of human pancreatic cancer was increased by applying centrifugal flow, the penetration obtained with fibrotic cancerous tissues was not as great as that observed with normal pancreatic parenchyma. CK19 labeling could be visualized from 0.7 to 1.5 mm into the normal tissue sections and 0.3 to 0.6 mm into the pancreatic ductal adenocarcinoma sections. Because depth-based limitations were not encountered when visualizing autofluorescent signals in the tissue, it can be concluded that the depth of visualization was limited by antibody penetration, not by efficacy of clearing or visualization capabilities of the light sheet microscope.
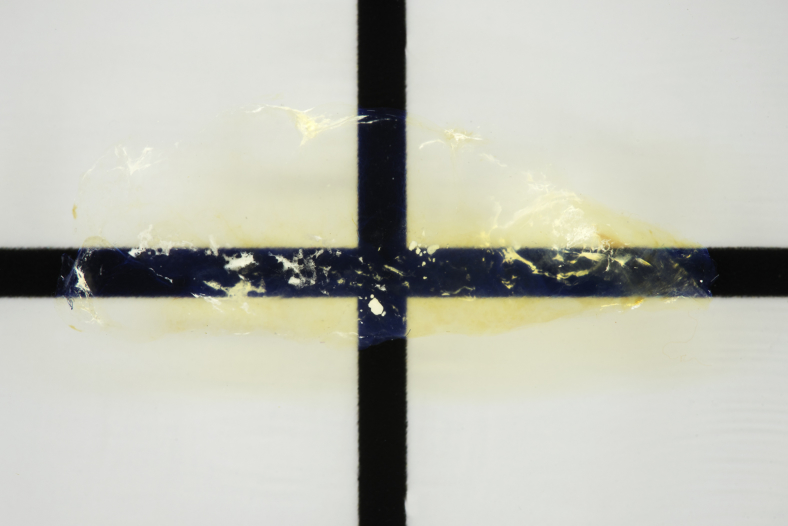
Figure 1.
Gross image of cleared human pancreatic parenchyma. In this 5-mm–thick cleared section of pancreatic tissue, the refractive index matching reduces light scattering and produces cleared tissue that enables fluorescent microscopy of deeper structures. Original magnification, ×2.5.
Normal Parenchyma
The 3D branching of ducts was visualized easily, and small periductal glands extending off of larger ducts into the stroma could be appreciated (Figure 2A and Supplemental Videos S1, S2, and S3). As has been described in rat pancreatic parenchyma, some of the ductules in the normal pancreas formed loops instead of linear branches (Figure 2A and Supplemental Video S2). Localized dilation of the ductules in a lobule was seen in one case, probably secondary to downstream obstruction (Supplemental Video S3).
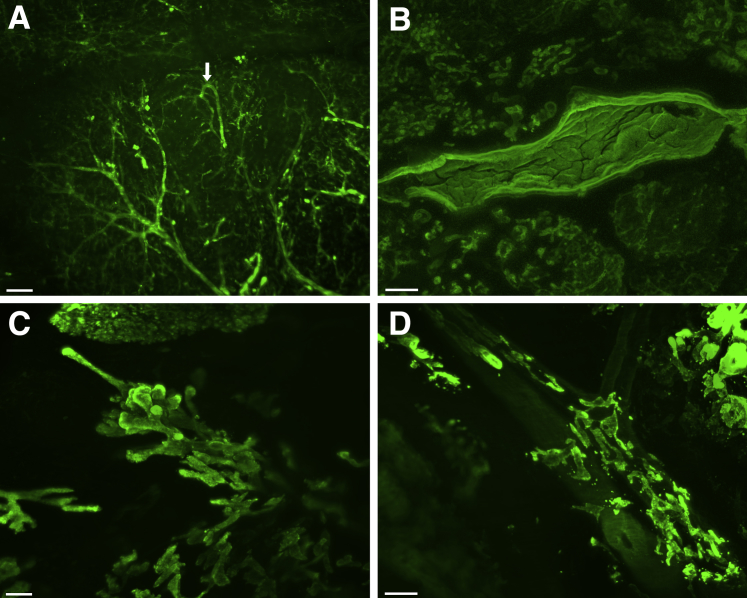
Figure 2.
Three-dimensional imaging of normal and neoplastic human pancreatic parenchyma. Sections of grossly normal and neoplastic human pancreas were immunolabeled with antibodies for cytokeratin 19 and then cleared. A: In the normal pancreas, the branching morphology at the edge of a lobule can be seen, including one of the ducts with looping (arrow). B: PanIN precursor lesions are identified in sections of grossly normal human pancreas—these lesions have intraluminal papillary projections of columnar epithelial cells. C: Imaging of grossly identified pancreatic ductal adenocarcinoma shows multiple patterns of invasion, including growth as invasive long and thin ductal structures. D: Analysis of pancreatic ductal adenocarcinoma shows a unique pattern of perivascular spread of malignant cells. The blood vessel passing from the upper left to the lower right is identifiable by its autofluorescence. In this tumor, invasive carcinoma grows in the perivascular space, parallel to the vessel. Scale bars: 200 μm (A, C, and D); 150 μm (B).
Precursor Lesions
Immunolabeling highlighted microscopic PanIN and intraductal papillary mucinous neoplasm lesions. The neoplastic papillary projections of the PanIN and intraductal papillary mucinous neoplasm lesions into the lumina of ducts was readily appreciated (Figure 2B and Supplemental Video S4). A PanIN in one case measured 2.8 mm × 0.5 mm × 0.5 mm.
Invasive Carcinoma
Invasive ductal adenocarcinomas were also well visualized (Figure 2C). The appearance of the neoplastic cells varied greatly, from sheets of individual cells embedded in the stroma (Supplemental Video S5), to large hollow globular clusters of cells with numerous blunt projections, to haphazardly arranged discrete long thin tubes (Figure 2C and Supplemental Video S6). Visualization in 3D allowed for the identification of cancerization of a duct (Supplemental Video S7), with bridging of neoplastic cells across the duct lumen. A remarkable finding was of invasive carcinoma growing in the connective tissue parallel to blood vessels (Figure 2D and Supplemental Video S8), as well as invasive carcinoma growing within blood vessels (Supplemental Videos S9 and S10). The blood vessels could be identified based on the autofluorescence and distinct morphology of the arterial elastic lamina. Specific CK19 labeling and nonspecific autofluorescence of the arterial elastic lamina could be distinguished reliably by careful visualization of the tissue and use of different excitation wavelengths and filters. The arterial elastic lamina produced a distinct narrow bright wavy line that was unique in fluorescent intensity, pattern, and thickness, distinguishing it from specific CK19 labeling of neoplastic epithelial cells. Moreover, the autofluorescence of the arterial elastic lamina remained in wavelengths and filters outside of the 488-nm Alexa Fluor filter that highlighted specific CK19 labeling.
Validation
With the exception of slight retraction of some epithelial cells into the lumina of ducts, tissue morphology was remarkably well preserved in the H&E sections taken after the tissue clearing. These H&E sections validated the interpretation of the 3D findings in all four cases (Supplemental Figure S1).
Discussion
The 3D visualization of cleared human solid organs provides an extraordinary opportunity to understand the true complexity of human diseases.2, 4, 5, 6, 7 The application of labeling and clearing to human pancreatic parenchyma provides detailed visualization of normal pancreatic anatomy, and can be used to characterize the 3D architecture of disease processes ranging from PanIN to invasive carcinoma. Importantly, with the addition of multicolor immunolabeling, the 3D relationships of normal ducts, precursor lesions, invasive carcinoma, immune cells, islets of Langerhans, vessels, and nerves should be visualizable.10, 15
Clearing methodologies have been described for 100 years.7 The more recent addition of immunolabeling using fluorescent antibodies has opened the door to 3D microscopy. These techniques were first applied to small pieces of tissue from experimental models, but technological improvements in large-scale microscopy have opened up this technology to the study of a broad array of human diseases.4 The basic principle underlying clearing is that light scattering, which occurs when photons transition through tissues with different refractive indices, can be reduced by matching refractive indices within tissue.8, 16 Lipid-containing membranes have a high refractive index, whereas water-based cytosol and interstitial fluid have a low refractive index. Dehydration and replacement of the cytosol and the interstitial fluid with a material with a high refractive index (such as DBE), followed by submersion of tissue in DBE during imaging, reduces light scattering, the main cause of tissue opaqueness. In contrast, other clearing techniques [eg, Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/in situ-hybridization-compatible Tissue hYdrogel (CLARITY)] exploit the removal of membranes to match the refractive indices.11 Unlike 3D reconstruction of histology from serially sectioned tissues, clearing avoids artifacts introduced when tissue is physically sectioned.
Poor antibody penetration can limit the application of clearing to dense tissues.14 This problem was addressed by gradually increasing the concentration of antibody and with centrifugal flow.14 A novel feature of our protocol is the combination of these strategies to increase antibody penetration with DBE-based tissue clearing to effectively visualize fibrotic human pancreatic tissue in three dimensions. In so doing, we were able to visualize processes such as cancerization of the ducts and cancer growing within a long segment of a vein. In addition, although it is known that the two-dimensional histologic finding of a gland next to a muscular vessel is suggestive of invasive cancer, here we show using 3D visualization that such proximity is not a random event.17, 18 Cords of neoplastic cells often run parallel to muscular blood vessels. Thus, this novel finding, observed in three dimensions, explains a long-known diagnostic aid in surgical pathology. For years, the presence of a gland next to a muscularized blood vessel in the pancreas has been recognized as a two-dimensional histologic feature of malignancy in the pancreas.18 Until our study, it was not clear whether this co-localization was simply owing to the haphazard growth of the neoplastic glands of pancreatic cancer or a more directed process. Our results show preferential invasion of malignant glands in the interstitium parallel to blood vessels. The mechanism for this preferential invasion remains to be elucidated—possibilities range from cancer growing within longitudinal perivascular interstitial tracks as well as chemical attractants.19, 20
We anticipate that this approach will provide new understandings of a wide range of processes, from the 3D anatomy of immune responses to neoplastic cells, to the events that take place at the leading edges of cancers as the neoplastic cells interact with normal parenchyma. In addition, this technique can also be applied to animal models of human disease (such as genetically engineered mouse models or xenograft models) to interrogate 3D anatomy at specific time points in disease progression.21, 22, 23 Importantly, just as the two-dimensional microscopic visualization of human diseases helped generate new hypotheses hundreds of years ago, so too is our hope that the 3D visualization of diseases of the pancreas will generate new hypotheses and new biological insights.
Acknowledgments
M.N., L.D.W., and R.H.H. designed the study; M.N. conducted technical experiments; N.R., K.A., M.S., V.P.G., and P.-H.W. provided input about the technical experiments; M.N., M.T.O., S.-M.H., S.J.K., G.J.A.O., L.A.A.B, D.W., L.D.W., and R.H.H. interpreted data; and all of the authors assisted with the writing and editing of the manuscript and approved the final version.
Footnotes
Supported by the Sol Goldman Pancreatic Cancer Research Center, Specialized Programs of Research Excellence (SPORE) in Gastro-Intestinal (GI) Cancer grant CA 62924, Susan Wojcicki and Dennis Troper, Rolfe Foundation for Pancreatic Cancer Research, NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant K08 DK107781, Buffone Family Gastrointestinal Cancer Research Fund, Kaya Tuncer Career Development Award in Gastrointestinal Cancer Prevention, American Gastroenterological Association (AGA)–Bernard Lee Schwartz Foundation Research Scholar Award in Pancreatic Cancer, Sidney Kimmel Foundation for Cancer Research Kimmel Scholar Award, American Association for Cancer Research (AACR) Incyte Corporation Career Development Award for Pancreatic Cancer Research, Joseph C. Monastra Foundation, and The Gerald O. Mann Charitable Foundation (Harriet and Allan Wulfstat, Trustees) (L.D.W.); The Dutch Digestive Foundation grant MLDS CDG 14-020 (L.A.A.B.); The Nijbakker-Morra Foundation and The Lisa Waller Hayes Foundation (M.N.); and NIH/National Cancer Institute grant R00 CA190889 (N.R.).
Disclosures: L.D.W. is a paid consultant for Personal Genome Diagnostics.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.04.002.
Contributor Information
Laura D. Wood, Email: ldwood@jhmi.edu.
Ralph H. Hruban, Email: rhruban@jhmi.edu.
Supplemental Data
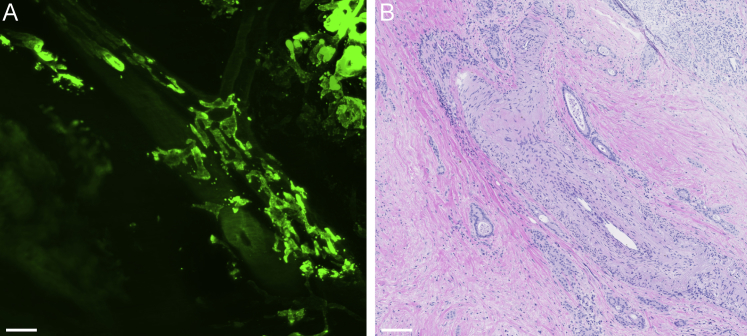
Supplemental Figure S1.
Validation of the three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels by hematoxylin and eosin (H&E) staining. After immunolabeling with antibodies for cytokeratin 19, clearing, and imaging, the tissue was formalin-fixed, paraffin-embedded and sectioned for routine H&E staining. A: Three-dimensional imaging with a Zeiss LSM800 confocal laser scanning microscope of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. B: A 4-μm thick section of the same area, confirming the presence of malignant ducts in the interstitium around a blood vessel. Scale bars: 200 μm (A); 100 μm (B). Original magnification, ×5 (A).
Three-dimensional imaging of normal human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. This highlights the branching structure of the pancreatic ductal system. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of normal human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. In addition to the branching structure of the pancreatic ductal system, looping of small ductules also can be seen. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of atrophic human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. An atrophic lobule with dilated lumina can be appreciated in the center of the cleared section. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of human pancreatic intraepithelial neoplasia (PanIN). A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Imaging shows a dilated ductule with intraluminal papillary projections of columnar epithelium, morphologically consistent with PanIN. Original magnification, ×10 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Sheet-like growth of the malignant cells can be seen in the bottom portion of the image, while the upper portion represents the lobular structure of non-neoplastic pancreatic parenchyma. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. In this tumor, the malignant cells grow as well-defined ductal structures. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of intraductal spread of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Malignant glands can be seen on the right portion of the image. In the center portion, what appears to be a pre-existing duct has been colonized by these malignant cells (so-called cancerization of the duct), resulting in bridges of malignant cells across the duct lumen. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. A muscular blood vessel can be identified by its autofluorescence. Malignant glands invade the perivascular space and can be seen running parallel to the blood vessel. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Malignant glands can be identified by their strong labeling. In addition, a blood vessel (likely an artery) can be identified in the middle of the image by its autofluorescent elastic lamina. To the left of this artery, a second blood vessel (likely a vein) also can be seen, although its autofluorescence is less prominent. A malignant gland extends across and invades into this second vessel. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A zoomed-in three-dimensional reconstruction of the same malignant duct growing into a blood vessel (likely a vein) as seen in Supplemental Video S9. Original magnification, ×10 (Zeiss LSM800 confocal laser scanning microscope).
References
- 1.Weller C.V. Rudolf Virchow–pathologist. Sci Mon. 1921;13:33–39. [Google Scholar]
- 2.Qiu H., Wild A.T., Wang H., Fishman E.K., Hruban R.H., Laheru D.A., Kumar R., Hacker-Prietz A., Tuli R., Tryggestad E., Schulick R.D., Cameron J.L., Edil B.H., Pawlik T.M., Wolfgang C.L., Herman J.M. Comparison of conventional and 3-dimensional computed tomography against histopathologic examination in determining pancreatic adenocarcinoma tumor size: implications for radiation therapy planning. Radiother Oncol. 2012;104:167–172. doi: 10.1016/j.radonc.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boj S.F., Hwang C.-I., Baker L.A., Engle D.D., Tuveson D.A., Clevers H. Model organoids provide new research opportunities for ductal pancreatic cancer. Mol Cell Oncol. 2016;3:e1014757. doi: 10.1080/23723556.2015.1014757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka N., Kanatani S., Tomer R., Sahlgren C., Kronqvist P., Kaczynska D., Louhivuori L., Kis L., Lindh C., Mitura P., Stepulak A., Corvigno S., Hartman J., Micke P., Mezheyeuski A., Strell C., Carlson J.W., Moro C.F., Dahlstrand H., Östman A., Matsumoto K., Wiklund P., Oya M., Miyakawa A., Deisseroth K., Uhlén P. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng. 2017;1:796–806. doi: 10.1038/s41551-017-0139-0. [DOI] [PubMed] [Google Scholar]
- 5.Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson D.S., Lichtman J.W. SnapShot: tissue clearing. Cell. 2017;171:496–496.e1. doi: 10.1016/j.cell.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Seo J., Choe M., Kim S.-Y. Clearing and labeling techniques for large-scale biological tissues. Mol Cells. 2016;39:439–446. doi: 10.14348/molcells.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azaripour A., Lagerweij T., Scharfbillig C., Jadczak A.E., Willershausen B., Van Noorden C.J.F. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem. 2016;51:9–23. doi: 10.1016/j.proghi.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Lin P.-Y., Peng S.-J., Shen C.-N., Pasricha P.J., Tang S.-C. PanIN-associated pericyte, glial, and islet remodeling in mice revealed by 3D pancreatic duct lesion histology. Am J Physiol Gastrointest Liver Physiol. 2016;311:G412–G422. doi: 10.1152/ajpgi.00071.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H., Park J.-H., Seo I., Park S.-H., Kim S. Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev Biol. 2014;14:48. doi: 10.1186/s12861-014-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flessner M.F., Choi J., Credit K., Deverkadra R., Henderson K. Resistance of tumor interstitial pressure to the penetration of intraperitoneally delivered antibodies into metastatic ovarian tumors. Clin Cancer Res. 2005;11:3117–3125. doi: 10.1158/1078-0432.CCR-04-2332. [DOI] [PubMed] [Google Scholar]
- 13.Calve S., Ready A., Huppenbauer C., Main R., Neu C.P. Optical clearing in dense connective tissues to visualize cellular connectivity in situ. PLoS One. 2015;10:e0116662. doi: 10.1371/journal.pone.0116662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E., Choi J., Jo Y., Kim J.Y., Jang Y.J., Lee H.M., Kim S.Y., Lee H.-J., Cho K., Jung N., Hur E.M., Jeong S.J., Moon C., Choe Y., Rhyu I.J., Kim H., Sun W. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang S.-C., Shen C.-N., Lin P.-Y., Peng S.-J., Chien H.-J., Chou Y.-H., Chamberlain C.E., Pasricha P.J. Pancreatic neuro-insular network in young mice revealed by 3D panoramic histology. Diabetologia. 2018;61:158–167. doi: 10.1007/s00125-017-4408-y. [DOI] [PubMed] [Google Scholar]
- 16.Becker K., Jährling N., Kramer E.R., Schnorrer F., Dodt H.-U. Ultramicroscopy: 3D reconstruction of large microscopical specimens. J Biophotonics. 2008;1:36–42. doi: 10.1002/jbio.200710011. [DOI] [PubMed] [Google Scholar]
- 17.Hruban R.H., Klimstra D.S. Adenocarcinoma of the pancreas. Semin Diagn Pathol. 2014;31:443–451. doi: 10.1053/j.semdp.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S., Green K.B. The pancreatic duct and its arteriovenous relationship: an underutilized aid in the diagnosis and distinction of pancreatic adenocarcinoma from pancreatic intraepithelial neoplasia. A study of 126 pancreatectomy specimens. Am J Surg Pathol. 2004;28:613–620. doi: 10.1097/00000478-200405000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Benias P.C., Wells R.G., Sackey-Aboagye B., Klavan H., Reidy J., Buonocore D., Miranda M., Kornacki S., Wayne M., Carr-Locke D.L., Theise N.D. Structure and distribution of an unrecognized interstitium in human tissues. Sci Rep. 2018;8:4947. doi: 10.1038/s41598-018-23062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X., Guadagni F., Hoffman R.M. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc Natl Acad Sci U S A. 1992;89:5645–5649. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., Tuveson D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Hiroshima Y., Maawy A.A., Katz M.H.G., Fleming J.B., Bouvet M., Endo I., Hoffman R.M. Selective efficacy of zoledronic acid on metastasis in a patient-derived orthotopic xenograph (PDOX) nude-mouse model of human pancreatic cancer. J Surg Oncol. 2015;111:311–315. doi: 10.1002/jso.23816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional imaging of normal human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. This highlights the branching structure of the pancreatic ductal system. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of normal human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. In addition to the branching structure of the pancreatic ductal system, looping of small ductules also can be seen. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of atrophic human pancreatic parenchyma. A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. An atrophic lobule with dilated lumina can be appreciated in the center of the cleared section. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of human pancreatic intraepithelial neoplasia (PanIN). A section of grossly normal human pancreas was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Imaging shows a dilated ductule with intraluminal papillary projections of columnar epithelium, morphologically consistent with PanIN. Original magnification, ×10 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Sheet-like growth of the malignant cells can be seen in the bottom portion of the image, while the upper portion represents the lobular structure of non-neoplastic pancreatic parenchyma. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. In this tumor, the malignant cells grow as well-defined ductal structures. Original magnification, ×4 (LaVision Ultramicroscope II Light Sheet Microscope).
Three-dimensional imaging of intraductal spread of human pancreatic ductal adenocarcinoma. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Malignant glands can be seen on the right portion of the image. In the center portion, what appears to be a pre-existing duct has been colonized by these malignant cells (so-called cancerization of the duct), resulting in bridges of malignant cells across the duct lumen. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. A muscular blood vessel can be identified by its autofluorescence. Malignant glands invade the perivascular space and can be seen running parallel to the blood vessel. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A section of grossly identified human pancreatic ductal adenocarcinoma was immunolabeled with antibodies for cytokeratin 19 (CK19) and cleared. Malignant glands can be identified by their strong labeling. In addition, a blood vessel (likely an artery) can be identified in the middle of the image by its autofluorescent elastic lamina. To the left of this artery, a second blood vessel (likely a vein) also can be seen, although its autofluorescence is less prominent. A malignant gland extends across and invades into this second vessel. Original magnification, ×5 (Zeiss LSM800 confocal laser scanning microscope).
Three-dimensional imaging of human pancreatic ductal adenocarcinoma and its relationship to blood vessels. A zoomed-in three-dimensional reconstruction of the same malignant duct growing into a blood vessel (likely a vein) as seen in Supplemental Video S9. Original magnification, ×10 (Zeiss LSM800 confocal laser scanning microscope).