Abstract
PNRC2 (proline-rich nuclear receptor co-regulatory protein 2) was identified using mouse steroidogenic factor 1 (SF1) as bait in a yeast two-hybrid screening of a human mammary gland cDNA expression library. PNRC2 is an unusual coactivator in that it is the smallest coactivator identified so far, with a molecular weight of 16 kDa, and interacts with nuclear receptors using a proline-rich sequence. In yeast two-hybrid assays PNRC2 interacted with orphan receptors SF1 and estrogen receptor-related receptor α1 in a ligand-independent manner. PNRC2 was also found to interact with the ligand-binding domains of estrogen receptor, glucocorticoid receptor, progesterone receptor, thyroid receptor, retinoic acid receptor and retinoid X receptor in a ligand-dependent manner. A functional activation function 2 domain is required for nuclear receptors to interact with PNRC2. Using the yeast two-hybrid assay, the region amino acids 85–139 was found to be responsible for the interaction with nuclear receptors. This region contains an SH3 domain-binding motif (SEPPSPS) and an NR box-like sequence (LKTLL). A mutagenesis study has shown that the SH3 domain-binding motif is important for PNRC2 to interact with all the nuclear receptors tested. Our results reveal that PNRC2 has a structure and function similar to PNRC, a previously characterized coactivator. These two proteins represent a new type of nuclear receptor co-regulatory proteins.
INTRODUCTION
Nuclear receptors are transcription factors that modulate transcription of various cellular genes, either positively or negatively, by interacting with specific hormone-responsive elements located in the target gene promoters, thereby controlling diverse aspects of cell growth, development and homeostasis. The mechanisms by which nuclear receptors can regulate transcription from the target gene promoters are currently under intensive investigation. Recent data show that, in addition to contacting the basal transcriptional machinery directly, nuclear receptors enhance or inhibit transcription by recruiting an array of coactivator or corepressor proteins to the transcription complex. A number of these putative co-regulatory proteins for nuclear receptors have been identified and have been shown to act either as coactivators or corepressors (reviewed in 1,2). Most of these cofactors of nuclear receptors have molecular weights of ∼160 kDa and share a common motif containing a core consensus sequence LXXLL (L, leucine; X, any amino acid) (i.e. an NR box), which is necessary and sufficient to mediate the binding of these proteins to ligand-bound nuclear receptors. LXXLL is thus a defining feature of p160 coactivators (3).
Aromatase is an important enzyme that converts androgens to estrogen and its expression is elevated in human breast cancer tissue. Our laboratory is investigating the regulatory mechanism of aromatase expression in breast tissue. Steroidogenic factor 1 (SF1) is an orphan receptor that up-regulates the expression of aromatase by binding to a regulatory region, S1, in the human aromatase gene. In order to better understand the regulatory mechanism of SF1 on aromatase expression, we initiated a study to identify co-regulatory proteins that modulate the activity of the orphan receptor using the yeast two-hybrid screening method. Our laboratory has identified a new co-regulatory protein for nuclear receptors, proline-rich nuclear receptor co-regulatory protein (PNRC), that is expressed in human breast tissue and interacts with a significant number of nuclear receptors (4). This co-regulatory protein is unique in that it has a molecular weight of 35 kDa, much smaller than most of the known co-regulatory proteins, and it is proline-rich. Our site-directed mutagenesis studies have revealed that an SH3-binding motif (S-D-P-P-S-P-S) in PNRC is essential for its interaction with the ligand-binding region of nuclear receptors. PNRC contains a NR box sequence that is not required for its interaction with nuclear receptors. Recently, we have cloned a second member of the PNRC family, PNRC2. The molecular weight of this newly identified coactivator is only 16 kDa, one-tenth that of p160 coactivators. In this paper we report the nucleotide sequence of PNRC2 cDNA, compare the protein sequences of PNRC and PNRC2 and present the results from northern blot and RT–PCR analyses. A detailed functional characterization of PNRC2 has also been carried out. The coactivator function of PNRC2 was found not to be exactly identical to that of PNRC.
MATERIALS AND METHODS
Materials and reagents
The MatchMaker Two-Hybrid System kit, including a human mammary gland MatchMaker cDNA library, was purchased from Clontech (Palo Alto, CA). DNA sequencing kits were from US Biochemical (Cleveland, OH). Various restriction endonucleases were purchased from New England Biolabs (Beverly, MA) and Boehringer Mannheim (Indianapolis, IN). AmpiTaq polymerase was obtained from Perkin Elmer (Norwalk, CT). The luciferase assay system was purchased from Promega (Madison, WT). Oligonucleotide primers were synthesized in the DNA/RNA Chemistry Laboratory at the City of Hope. HeLa cells from ATCC (Rockville, MA) were maintained in McCoy’s 5A medium containing 10% fetal calf serum. 17β-Estradiol, progesterone, retinoic acid, deoxycorticosterone and testosterone were purchased from Sigma. l-3,3′,5′-Triiodothyronine (T3) was purchased from Calbiochem (La Jolla, CA). 9-cis-Retinoic acid was obtained from Biomol (Plymouth Meeting, PA). The yeast transformation kit was purchased from Bio 101 (La Jolla, CA) and the yeast culture media were purchased from Clontech. The mouse SF1 cDNA clone was kindly provided by Dr Keith L. Parker (Duke University Medical Center, Durham, NC).
Construction of plasmids
All recombinant DNA and plasmid constructs were prepared according to standard procedures, and the sequences and orientations of inserted DNA fragments in plasmid constructs were verified by standard DNA sequencing. The yeast expression plasmid for the Gal4 DNA-binding domain (DBD)–SF1 fusion protein (pGBT9-SF1) was prepared as previously described (4). The plasmid for overexpression of PNRC2 in mammalian cells was made by inserting a PCR-amplified fragment with BamHI sites at both ends into the BamHI site in vector pSG5 (Stratagene). The expression plasmids for mouse SF1 and human estrogen receptor-related receptor α1 (ERRα1), pSG5-SF1 and pSG5-ERRα1, were constructed as previously described (4). The plasmid for expression of wild-type human estrogen receptor α (ERα), pSG5-hERα, was constructed as follows. The full-length coding region of human ERα was generated by PCR using forward primer 5′-GCGGAATTCGCCGCCGCCATGACCATGACCCTCCACACCAAAGC-3′ and reverse primer 5′-GGGGTACTAGTAACCCGGGCGGATCCTCAGACTGTGGCAGGGAAACCCTC-3′ on the template DNA, pIC-ERF (ATCC). The PCR product was digested with EcoRI and BamHI and subcloned into vector pSG5 through the EcoRI and BamHI sites.
To construct the yeast expression plasmids for the NR box deletion mutants of PNRC2 and Gal4 activation domain (AD) fusion proteins, DNA fragments coding for PNRC285–130 were generated by PCR and inserted in-frame into the Gal4 AD vector pACT2 through the BamHI site. The PNRC285–139 fragment containing mutations 101P→A and 104P→A was generated by a PCR-based site-directed mutagenesis method described by Nelson and Long (5) and inserted into the pACT2 vector through the BamHI site. To construct the yeast expression plasmid pGBT9-hERα HBD, expressing a Gal4 DBD–hERα hormone-binding domain (HBD) fusion protein, the coding sequence from amino acid 274 to 595, the ligand-binding domain (LBD), of ERα was amplified by PCR on pSG5-hERα template DNA with sense primer 5′-GCGGAATTCATGGAAGTGGGGTCTGCTGGAGAC-3′ and antisense primer 5′-GGGGTACTAGTAACCCGGGCGGATCCTCAGACTGTGGCAGGGAAACCCTC-3′. The PCR product was digested with EcoRI and BamHI and the digested DNA fragment was subcloned in-frame into the EcoRI and BamHI sites of the yeast expression plasmid pGBT9. Several yeast expression plasmids coding for fusion proteins of Gal4 DBD with the HBDs of androgen receptor (AR), glucocorticoid receptor (GR), progesterone receptor (PR), thyroid receptor (TR), retinoic acid receptor (RAR) and retinoid X receptor (RXR) in vector pGBT9 were kindly provided by Dr Michael R. Stallcup (University of Southern California, Los Angeles, CA) (6).
A set of plasmids, termed pM-SF1, pM-ERRα1 HBD, pM-ERα HBD and pVP16-PNRC2, for mammalian two-hybrid assay were prepared as follows. The SF1 cDNA fragment was excised from pGBT9-SF1 and inserted in-frame into the Gal4 DBD vector pM (Clontech) through the EcoRI site. The cDNA fragments coding for the human ERα HBD was excised from pGBT9-ERα1 HBD and inserted in-frame into the vector pM through the EcoRI and SalI sites. PCR was used to generate the full-length PNRC2 coding region with BamHI sites at both ends. The PCR products were digested with BamHI and inserted in the proper reading frame into a pVP16 activation domain vector (Clontech) at the BamHI site.
The luciferase reporter plasmid, pGL3(ERE)3-Luciferase, which contains three copies of the ERE sequence, was constructed as follows. Two complimentary oligonucleotides for three random copies of the ERE consensus sequences, 5′-AATTCCAGGTCAGAGTGACCTGAGCTAAAATACCAGGTCAGAGTGACCTGAGCTAAAATACCAGGTCAGA-GTGACCTGAGCTAAAATAC-3′ and 5′-TCGAGTATTTTAGCTCAGGTCACTCTGACCTGGTATTTTAGCTCAGG-TCACTCTGACCTGGTATTTTAGCTCAGGTCACTCTGACCTGG-3′, were synthesized, phosphorylated, annealed, filled in and subcloned into the pGL3 promoter vector (Promega) through the SmaI site.
To study the function of PNRC2 on transcription mediated by SF1 and ERRα1, the reporter plasmid pGL3(SF1 site)3-Luciferase, which contains three random copies of the SF1-binding site, was constructed as follows. Two complimentary oligonucleotides containing three copies of the SF1-binding site (5′-CCAAGGTCAGAA-3′) from the human aromatase gene, 5′-CCAAGGTCAGAACCAAGGTCAGAACCAAGGTCAGAAC-3′ and 5′-TCGAGTTCTGACCTTGGTTCTGACCTTGGTTCTGACCTTGGAGCT-3′, were synthesized, phosphorylated, annealed and subcloned into pGL3 promoter vector through the SacI and XhoI sites.
Yeast two-hybrid screening
The yeast MatchMaker Two-Hybrid System (Clontech) was used to screen a MatchMaker mammary gland cDNA expression library according to the supplier’s protocol (protocol PT1030-1) using pGBT9-SF1 as the bait as previously described (4).
Isolation of putative full-length coding cDNA for PNRC2
The full-length PNRC2 cDNA was cloned by PCR amplification from the human mammary gland MatchMaker cDNA library DNA using one forward adapter primer corresponding to the sequence of the Gal4 AD domain (MatchMaker 5′ AD LD-insert Screening Amplimer; Clontech) and one reverse primer corresponding to the cDNA sequence encoding the C-terminus of PNRC2 (5′-TTATACCTGTACTTTAAGTAA-3′). The PCR products were isolated, subcloned into T-tailed pBluescript vector and five inserts with different lengths in the resulting plasmids were sequenced. The longest clone extended the PNRC2 sequence by 0.6 kb from the 5′-end of the longer original clone that was isolated from library screening.
RNA preparations, RT–PCR analysis and northern blot analysis
Total RNA was extracted from cultured cells according to the method of Chomczynski and Sacchi (7). The 0.25 kb PNRC2 cDNA probe was amplified and labeled with DIG with a DIG PCR Probe Synthesis Kit (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer’s instructions using primers 5′-GAGGACATGGTTATAACTCAG-3′ and 5′-GAAACAGGGACCCAGTGGCTTGGTGG-3′. RNA samples were electrophoresed on a 1.2% agarose/formaldehyde gel and capillary blotted onto a positively charged Zeta membrane (Bio-Rad Laboratories, Hercules, CA) in 20× SCC. The membrane was then subjected to prehybridization, hybridization and chemiluminescent detection according to the procedures provided in the standard DIG hybridization and detection kit (Roche Molecular Biochemicals).
Two micrograms of total RNAs from various cell lines were subjected to RT–PCR analysis using PNRC2-specific forward primer 5′-GAGGACATGGTTATAACTCAG-3′ and reverse primer 5′-CCTTATCTGAAGGATTAAAGGAAACAGGGA-3′. To verify the equal loading of total RNA in each sample, the transcripts of a housekeeping gene β-actin in 2 µg total RNA from cell lines were analyzed by RT–PCR using the sense primer 5′-AGGAGCCGTGCTGCTGA-3′ and the antisense primer 5′-CTAGAAGCATTTGCGGTGGAC-3′.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed as previously described (4) except that co-transformation of yeast strain Y187 was used in place of the yeast mating approach.
Mammalian two-hybrid assays
For mammalian two-hybrid assays, HeLa cells were transiently transfected with 1.5 µg reporter plasmid, pG5CAT (Clontech), and 1.0 µg pM-SF1 or pM-ERα HBD along with 1.0 µg pVP16-PNRC2 expression plasmid. Twenty-four hours after transfection, cells were harvested and CAT activity in the cell extract containing an equal amount of the protein from each sample was determined by the liquid scintillation counting (LSC) method as described previously (4).
Mammalian cell transfection and luciferase assays
HeLa cells were cultured in MEM Earle’s salts medium supplemented with 5% charcoal/dextran-treated fetal bovine serum. Cells were divided and cultured in 6-well plates until 80% confluent. The cells were transfected with 6 µg Lipofectin (Life Technologies Inc.), 1.25 µg plasmid DNA containing various amounts of the test plasmids indicated in each experiment and the appropriate amounts of empty vector (pSG5) to maintain the same overall amount of total DNA in all transfections. After 6 h incubation, the medium containing lipofectin and DNA was removed and the cells were cultured in growth medium containing 5% charcoal-treated fetal bovine serum with or without ligands. Twenty-four hours after transfection, the cells were treated with lysis buffer and harvested from the plates by scraping. The luciferase activities in the cell lysates (with the same amounts of protein) were measured according to the manufacturer’s instructions (Promega). All experiments were performed in triplicate and the representative examples of at least three independent experiments are presented.
RESULTS
Cloning of PNRC2 by its interaction with SF1 in the yeast two-hybrid screening
A Gal4-based yeast two-hybrid system was used to identify proteins encoded in a human mammary gland cDNA library that interact with mouse SF1. The mouse SF1 yeast expression plasmid, pGBT9-SF1, was used to transform yeast strain CG1945. The CG1945 transformants bearing pGBT9-SF1 plasmids were transformed again with a human mammary gland cDNA library (Clontech). Approximately 3.4 × 106 yeast transformants were screened in the absence of ligand since the ligand for SF1 is unknown. A total of 90 colonies appeared on histidine drop-out plates, 12 of which stained strongly positive when tested for expression of β-galactosidase. To test whether SF1 was required for interaction with the products of the isolated cDNAs, all of these plasmids were re-transformed into yeast CG1945. These transformants were used in yeast mating experiments to verify that the proteins identified from the library screen can only activate the reporter genes in the presence of Gal4 DBD–SF1 fusion protein. All hybrid proteins were found to interact with Gal4 DBD–SF1. The nucleotide sequences of these 12 cDNA inserts were determined. A database search revealed that four of the 12 sequences were PNRC, which was characterized recently (4). Another three clones were identical to a known nuclear receptor coactivator, receptor-interacting protein 140 (RIP140) (8,9). In addition to PNRC and RIP140, three clones encoding an unknown protein with regional homology to PNRC were also isolated. Since this protein contains an identical proline-rich region to PNRC (discussed below), we named it PNRC2.
Isolation of a putative full-length cDNA clone for PNRC2
All three cDNA clones isolated from library screening, with insert sizes ranging from 1.7 to 1.9 kb, contained a poly(A) tract but no putative translation start site. These results indicate that these cDNAs lacked the 5′-untranslated sequence. PCR amplification of human mammary gland MatchMaker cDNA expression library DNA was then performed with the MatchMaker 5′ AD LD-Insert Screening amplimer and a reverse primer corresponding to the 3′-end of the PNRC2 coding sequence, to obtain the 5′-end sequence of PNRC2 cDNA. Five clones with inserts of different lengths were sequenced. The longest clone extended the PNRC2 sequence by ∼0.6 kb from the 5′-end of the longer original clone isolated by library screening. This clone codes a mRNA of 2332 bp length (GenBank accession no. AF374386). It encodes a deduced 139 amino acid protein with a calculated molecular mass of 16 kDa. A search for PNRC2 cDNA and protein sequences against various databases revealed no high homology to other sequences except for the regional identity with PNRC protein (Fig. 1).
Figure 1.
Alignment of the amino acid sequences of PNRC and PNRC2. Identical residues are indicated by an asterisk (*). The SH3-binding motif-containing region and the NR box sequences are underlined.
Northern and RT–PCR analyses
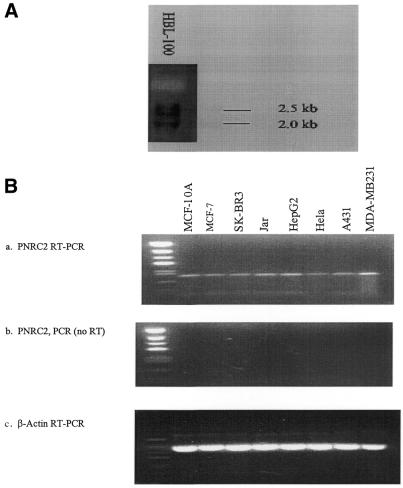
Our PNRC2 cDNA clones were isolated from a human mammary gland cDNA library. Figure 2A shows that two mRNA transcripts of ∼2.5 and 2.0 kb size were detected in the HBL-100 human non-cancer breast cell line. The size of the larger transcript, 2.5 kb, is consistent with the size of our longest PNRC2 cDNA clone, ∼2.4 kb. The two mRNA species are believed to be the result of differential processing. It is also possible that the 2.0 kb transcript is PNRC mRNA, since the 0.25 kb PNRC2 cDNA probe (used for northern hybridization) contains a 69 base region that is highly homologous between PNRC and PNRC2. We have also performed RT–PCR analysis to examine expression of PNRC2 in a series of human cell lines, using specific primers that are unique to PNRC2. As shown in Figure 2B(a), PNRC2 was expressed in all cell lines examined, including the non-cancer breast cell line MCF-10A and several breast cancer cell lines, MCF-7, SK-BR-3 and MDA-MB-231. The RT–PCR products obtained were not a result of contaminant DNA template in the RNA samples since no PCR product was detected when reverse transcriptase was excluded during the analysis [Fig. 2B(b)].
Figure 2.
(A) Northern analysis of PNRC2 in HBL-100 total RNA. Fifteen micrograms of total RNA from HBL-100 cells was subjected to northern blotting and hybridization as described in Materials and Methods using DIG-labeled PNRC2 cDNA probe. (B) RT–PCR analysis of PNRC2 mRNA in total RNA from various cell lines. (a) RT–PCR analysis of PNRC2 expression. Two micrograms of total RNA isolated from each cell line was subjected to RT–PCR and the products of RT–PCR reactions were analyzed on 1.5% agarose gels. A product with the expected size of 250 bp was detected in each reaction. (b) The reaction conditions were the same as in (a) except that no reverse transcription reaction was performed before the PCR reaction. (c) β-Actin RT–PCR. The conditions were the same as in (a) except that specific β-actin primers were used for the RT–PCR reaction.
Interactions between PNRC2 and multiple nuclear receptors including orphan receptors SF1 and ERRα1 in yeast
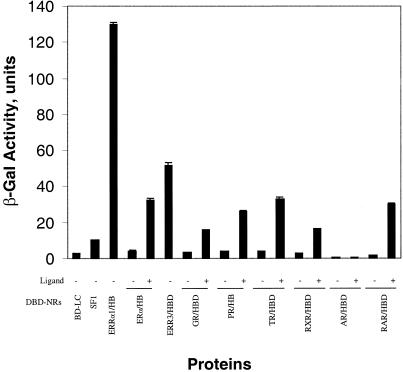
The protein–protein interactions between full-length PNRC2 or PNRC2 fragments and SF1, ERRα1 and several other nuclear receptors were analyzed by yeast two-hybrid assay. Yeast strain Y187 was co-transformed with plasmids for Gal4 DBD fusion proteins with the nuclear receptors and plasmids for Gal4 AD alone or Gal4 AD–PNRC2 fusion proteins. The expression of interacting hybrid proteins in yeast transformants was analyzed as LacZ expression, shown in Figure 3. Tests in the yeast two-hybrid assays indicated that PNRC2 interacted with orphan receptors SF1 and ERRα1 in the absence of any added activator or ligand. PNRC2 also interacted with six nuclear receptor HBDs, including ER, GR, TR, PR, RAR and RXR, and these interactions were completely ligand dependent. No interaction was detected between PNRC2 and AR HBD. In addition, no interaction was detected between PNRC2 and an irrelevant protein, human lamin C (Fig. 3), or between nuclear receptors and Gal4 AD alone (data not shown). These results suggest that the interaction occurred only in the presence of PNRC2 and nuclear receptors in the form of fusion proteins and that PNRC2 is a nuclear receptor-interacting protein with broad reactivity, with the exception of AR.
Figure 3.
Interaction between PNRC2 and nuclear receptors in yeast. Yeast strain Y187 was co-transformed with the appropriate pGBT9-receptor and pACT2-PNRC2 constructs and transformants harboring both plasmids were selected by growth on SD/–Leu/–Trp agar plates. The expression of interacting hybrid proteins in Y187 transformants was analyzed for induction of LacZ expression in the presence of the indicated amount of ligands as described in Materials and Methods. Gal4 AD was included as a control to monitor background transcriptional activity. Relative β-galactosidase activities in liquid cultures are expressed in Miller units as means ± SD of three independent assays.
Interaction of PNRC with SF1 and ERα in mammalian cells
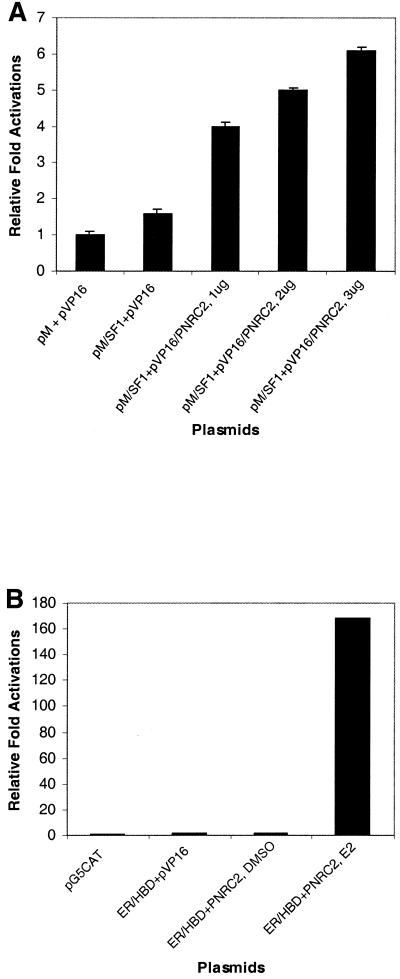
The mammalian MatchMaker two-hybrid assay system (Clontech) was used to confirm the interaction between PNRC2 and SF1 and ERα identified in yeast. Because the assays were performed in mammalian cells, the proteins are more likely to be in their physiological environment and the results are therefore more likely to represent biologically significant interactions. HeLa cells were co-transfected with three expression plasmids, including pM-SF1 for Gal4 DBD–SF1 fusion protein, pM-ERα HBD for Gal4 DBD–ERα HBD fusion protein or pVP16-PNRC2 for VP16 AD–PNRC fusion protein, and a third reporter plasmid, pG5CAT, which provides the Gal4 DBD, promoter and chloramphenicol acetyltransferase (CAT) reporter gene. As shown in Figure 4A and B, the CAT activities of cells transfected with the above three plasmids is ∼5–6-fold higher than that of cells transfected with the CAT reporter plasmid only. This PNRC2 interaction is specific for SF1 and ERα since no interaction was observed between PNRC2 and Gal4 DBD or between SF1 or ERα and VP16 AD. As shown in Figure 4B, the interaction between ERα and PNRC2 in the mammalian two-hybrid assay was also ligand dependent.
Figure 4.
PNRC2 interacts with SF1 (A) and ERα (B) in mammalian cells. HeLa cells were transiently co-transfected with 1.5 µg reporter plasmid (pG5CAT) and 1.0 µg each expression plasmid as indicated. The relative CAT activities are expressed as fold induction over the value obtained from co-transfection of reporter, pM and pVP16 vectors, which was given an arbitrary value of 1. The relative fold activations are means ± SD of three experiments.
Modulation of the transactivation activity of SF1, ERRα1 and ERα by PNRC2 in mammalian cells
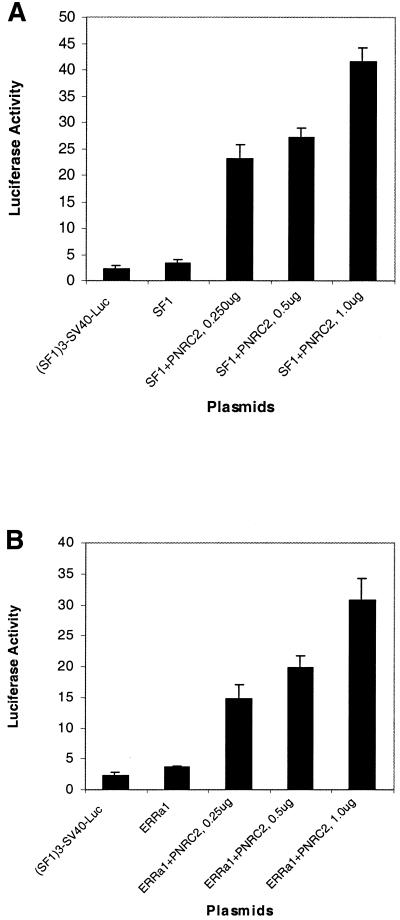
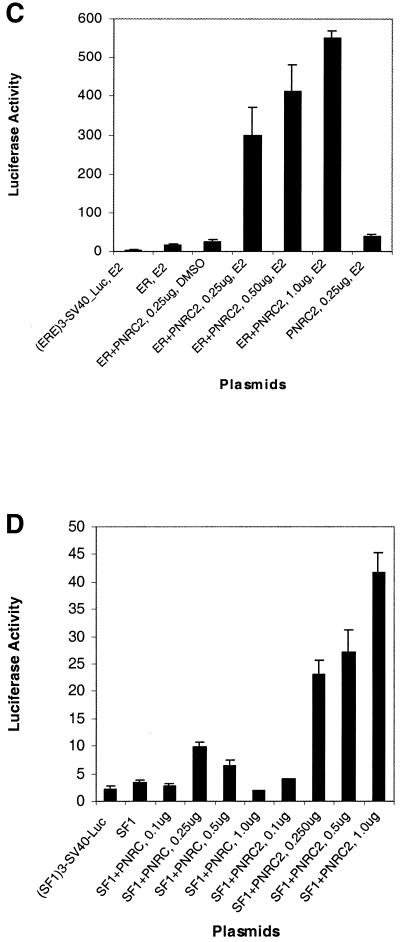
PNRC2 has been demonstrated to specifically interact with multiple members of the nuclear receptor family in both yeast and mammalian two-hybrid assays. To study the effect of PNRC2 overexpression on transcriptional activation of SF1 and ERRα1, the expression plasmid pSG5-PNRC2 was used to transiently transfect HeLa cells together with a reporter plasmid, pGL3(SF1 site)3-Luciferase, which contains three copies of the extended steroid hormone-binding half-site from the human aromatase gene (5′-CCAAGGTCAGAA-3′), the SV40 promoter and the luciferase reporter gene, along with a second expression plasmid for either mouse SF1 (pSG5-SF1) or human ERRα1 (pSG5-ERRα1). As shown in Figure 5A and B, co-transfection of PNRC2 enhanced SF1-stimulated and ERRα1-stimulated transcription from the SV40 promoter in a dose-dependent manner and this coactivation was ligand independent. PNRC2 was also found to act as a coactivator for chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) (results not shown). To test the function of PNRC2 on transcription mediated by human ERα, HeLa cells were transfected with a luciferase reporter plasmid, pGL3(ERE)3-Luciferase, which contains three tandem copies of the ER-binding site, the SV40 promoter and the luciferase reporter gene, along with the expression plasmid for human ERα (pSG5-hERα). As shown in Figure 5C, overexpression of PNRC2 enhanced ERα-stimulated transcription of luciferase in a dose-dependent manner and this coactivation was completely ligand dependent. Therefore, PNRC2 functions as a coactivator through its interaction with nuclear receptors.
Figure 5.
Coactivation functional analysis of PNRC2 on transcription mediated by multiple nuclear receptors in HeLa cells. HeLa cells were transfected with 0.25 µg pGL3(SF1 site)3-Luciferase reporter plasmid along with 10 ng expression plasmid for SF1 (pSG5-SF1) (A and D) or for ERRα1 (pSG5-hERRα1) (B) and increasing amounts of pSG5-PNRC2 expression plasmid as indicated. Appropriate amounts of empty vector pSG5 were included to maintain the 1.25 µg total DNA in all transfections. The luciferase activities in transfected cells were measured and are expressed as means ± SD of at least triplicate experiments. (A) Effect of PNRC2 overexpression on SF1-stimulated transcription of the luciferase gene directed by the SV40 promoter. (B) Effect of PNRC2 overexpression on ERRα1-stimulated transcription of the luciferase gene directed by the SV40 promoter. (C) Effect of PNRC2 overexpression on transcription mediated by human ERα. HeLa cells were transfected with pGL3(ERE)3-Luciferase reporter plasmid along with either pSG5-hERα expression plasmid alone or together with increasing amounts of PNRC2 expression plasmid (pSG5-PNRC2). All other features are as described for (A) and (B). (D) Comparison of coactivator activities of PNRC and PNRC2. The procedures for transfection and luciferase assay were the same as described for (A).
PNRC2 was found to be a stronger coactivator than PNRC (Fig. 5D). Moreover, unlike PNRC, which functions as a coactivator at low concentrations and functions as a repressor at higher concentrations, PNRC2 showed a constant coactivation function in a dose-dependent manner up to 1 µg pSG5-PNRC2 used (Fig. 5D).
Identification of the binding site on PNRC2 that is necessary and sufficient for its interaction with nuclear receptors
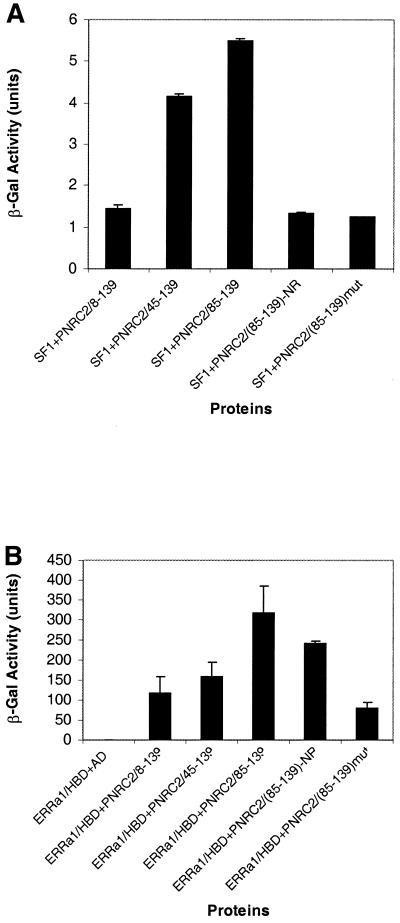
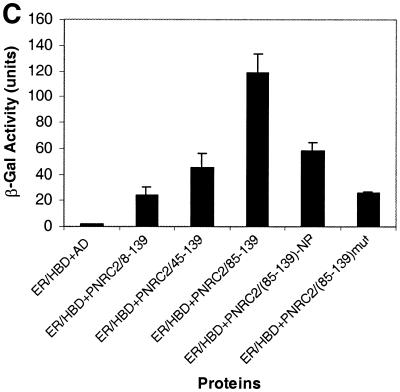
The three PNRC2 clones originally isolated from the yeast two-hybrid screen encoded three C-terminal peptides, amino acids 8–139, 45–139 and 85–139, respectively. As shown in Figure 6A, the shortest clone encoding amino acids 85–139 of PNRC2 showed the highest ability to bind SF1, suggesting that the region containing residues 85–139 was responsible for the interaction with SF1. Sequence comparisons revealed that there are two regions, a proline-rich 23 amino acid region (amino acids 92–114) and an NR box-like region (amino acids 131–138), with 96% identity between PNRC and PNRC2 (Fig. 1). The SH3 domain-binding motif (SDPPSPS) in the proline-rich 23 amino acid region, rather than the NR box-like sequence, has been demonstrated to be critical and sufficient for PNRC to interact with nuclear receptors. To test the importance of the SH3-binding motif and the NR box-like sequence in the interaction between PNRC2 and nuclear receptors, PNRC285–130, with the NR box deleted, and PNRC285–139, containing P101→A and P104→A mutations in the putative SH3-binding motif, were generated by PCR, expressed as fusion proteins with Gal4 AD and tested in the yeast two-hybrid assay for interaction with SF1, ERα HBD and ERRα1 HBD. Intriguingly, the NR box-containing sequence in PNRC2 is differentially required for its interaction with different nuclear receptors. As shown in Figure 6A, the intensity of the interaction between PNRC285–130 and SF1, as expressed by β-galactosidase reporter activity, was dramatically reduced when compared to the intensity of interaction between PNRC285–139 and SF1, indicating that the NR box region in PNRC2 plays a role in the interaction with SF1. This is in contrast to PNRC in that the NR box region is not necessary for PNRC to interact with SF1 (4). However, the intensities of interaction between ERRα1 HBD and PNRC285–139 and PNRC285–130 are about the same, indicating that the NR box region is not necessary for PNRC2 to interact with ERRα1 (Fig. 6B). Deletion of the NR box sequence also resulted in a remarkable reduction in the ability of PNRC2 to interact with ERα HBD, indicating that the NR box is required for interaction between PNRC2 and ERα (Fig. 6C).
Figure 6.

Localization of the interacting domain within PNRC2. Three PNRC2 clones with an N-terminal deletion, a NR box deletion and SH3-binding motif mutation were generated and tested for interaction in yeast two-hybrid assays with different nuclear receptors. The yeast expression plasmid for the NR box deletion [pACT2-PNRC285–139 (del NR)] and the plasmid for the PNRC2 mutation [pACT2-PNRC285–139 (P101A/P104A)] were prepared by PCR-based mutagenesis as described in Materials and Methods. Gal4 AD–PNRC2 deletion constructs were co-transformed with Gal4 DBD–nuclear receptor into yeast strain Y187 and transformants bearing both hybrid plasmids were selected and propagated in the presence of appropriate ligands. The β-galactosidase activities in yeast were determined and are expressed in Miller units as means ± SD of three independent assays. (A) Interaction between PNRC2 fragments and SF1. (B) Interaction between PNRC2 fragments and ERRα1. (C) Interaction between PNRC2 fragments and ERα. (D) Western blot of yeast protein extracts. Yeast protein extracts were prepared from log phase Y187 (grown in YPD medium) and Y187 harboring pGBT9-SF1 and pACT2-PNRC285–139 (wild type) or pACT2-PNRC285–139 (P101A/P104A) (grown in SD/–Leu/–Trp medium) using the TCA method according to the procedures provided by Clontech (PT3024-1). Protein extracts corresponding to 3 OD600 units of each type of yeast cultures were fractionated by 15% SDS–PAGE, transferred to a nitrocellulose membrane and probed by western blotting with a mouse monoclonal antibody against Gal4 AD (Clontech). The immunoprotein complex was detected with a 1:2500 diluted goat anti-mouse–horseradish peroxidase conjugate (Pierce) followed by the SupeSignal Substrate (Pierce).
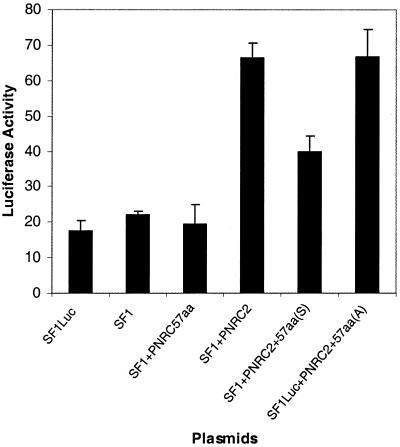
As shown in Figure 6, double mutation of P101→A and P104→A in the putative core site for SH3, i.e. SEPPSPS to SEAPSAS, in the amino acid 85–139 region of PNRC2 almost completely abolished the interactions between PNRC285–139 and the nuclear receptors tested, including SF1, ERRα1 and ERα. The dramatic reduction in the interaction between mutated PNRC285–139 and nuclear receptors was not due to a reduction in expression level of this mutant, as demonstrated by western blotting of yeast protein extract with anti-Gal4 AD antibody (Fig. 6D). This result strongly supports the hypothesis that, like PNRC, this SH3-binding motif in the region amino acids 85–139 is important for PNRC2 interaction with nuclear receptors. This conclusion was further supported by the fact that the C-terminal SH3-binding motif-containing domain of PNRC, but not the antisense version of this peptide, functioned as a dominant negative inhibitor of PNRC2 in transcription mediated by SF1 (Fig. 7). These results indicate that the peptide PNRC2270–327 can compete with full-length PNRC2 for binding to nuclear receptors while it does not have coactivator function.
Figure 7.
The C-terminal SH3-binding motif-containing domain of PNRC functions as a dominant negative inhibitor in the coactivation function of PNRC2. Expression plasmids (1.0 µg each) for wild-type PNRC2 (pSG5-PNRC2) and the C-terminal 57 amino acids of PNRC in the sense [pSG5-PNRC270–327 (S)] or antisense [pSG5-PNRC270–327 (A)] orientation along with 0.2 µg reporter plasmid [pGL3(SF1 site)3-Luciferase] and 10 ng SF1 expression plasmid (pSG5-SF1) were co-transfected into HeLa cells. The luciferase activities in the cell lysates from the transfected cells were assayed. Each result represents the mean ± SD of four experiments.
DISCUSSION
In this study we have obtained the full-length cDNA sequence of PNRC2, characterized its functions and determined the regions of PNRC2 that interact with nuclear receptors. The structural and functional analyses of PNRC2 indicate that it is a new member of the PNRC coactivator family. PNRC2 encodes a deduced 139 amino acid protein with a calculated molecular mass of 16 kDa. Since the C-terminus of PNRC2 contains regions highly homologous to that of PNRC, it was first thought that PNRC2 could be a splicing artifact resulting from cloning. To totally eliminate such a possibility, we performed RT–PCR analysis using tissue RNA as the template and two oligonucleotide probes derived from the N- and C-terminal regions of PNRC2, respectively. A RT–PCR product of correct size was produced and the nucleotide sequence of the product was found to be identical to that of the cloned cDNA (results not shown). To support the proposal that PNRC2 is a unique gene product, northern blot analysis was carried out, which revealed differences in tissue distribution between PNRC2 and PNRC. While PNRC is mainly expressed in liver, lung and fat tissue (Chen,B. Chen,S. and Zhou,D., unpublished results), PNRC2 transcript is found in heart, lung, muscle and different regions of brain (Zhou,D. and Chen,S., unpublished results).
We have performed a series of experiments to show that PNRC2 acts as a nuclear receptor coactivator. Physical interaction between PNRC2 and nuclear receptors has been demonstrated by GST pull-down assay (results not shown), yeast two-hybrid assay and mammalian two-hybrid assay. In transient transfection experiments PNRC2 was found to act as a coactivator for SF1, ERRα1 and ERα. Similar to other known coactivators, PNRC2 interacts with the activation function 2 (AF2) domain of nuclear receptors (results not shown). However, like PNRC, whose interaction with nuclear receptors depends on a SH3 domain-binding motif (4), the interaction between PNRC2 and nuclear receptors is also dependent on the SH3-binding motif (SEPPSPS) in a proline-rich sequence at the C-terminus, amino acids 85–139. This domain is not found in other nuclear receptor coactivators, including SRC1 (10), GRIP1 (6), RIP140 (8), TIFI (11), TIFII (12), ARA70 (13) and CBP/p300 (14,15) (using a BLAST sequence homology search). These two proteins belong to a new family of nuclear receptor co-regulatory proteins that interact with nuclear receptors through a different binding mechanism.
PNRC2 is the smallest nuclear receptor coactivator identified to date. The region amino acids 85–139 is the region that interacts with nuclear receptors as it has been shown to interact with nuclear receptors by yeast two-hybrid assay (Fig. 6), and the corresponding peptide in PNRC, i.e. PNRC270–327, can block the activity of PNRC2 by mammalian cell transfection assay (Fig. 7). These results, that the region amino acids 85–139 can interact with nuclear receptors but is not catalytically active, would also suggest that the N-terminal region of PNRC2 is important for coactivator function. A potential nuclear localization sequence is located at positions 40–45 (HKKKER). Interestingly, PNRC2 was found to be a significantly stronger coactivator than PNRC (Fig. 5D). The size of PNRC2 is approximately half that of PNRC, suggesting that the N-terminal region of PNRC contains sequence that down-regulates coactivator function. A detailed structure–function study of these proteins is underway in this laboratory.
It is worthwhile pointing out that although PNRC and PNRC2 are important coactivators that interact with SF1, PNRC2 does not interact with AR. Therefore, while PNRC and PNRC2 are nuclear receptor coactivators with broad reactivity, there are certain preferences as to which nuclear receptors they interact with. In addition, our results indicate that PNRC2 and PNRC bind with different affinities to different nuclear receptors. Analysis by yeast two-hybrid assay demonstrated that interaction of the orphan receptor ERRα1 was stronger with PNRC2 than PNRC. On the other hand, TR was found to have a stronger interaction with PNRC than with PNRC2. These two coactivators were isolated from a human breast tissue library. It is thought that they play important roles in breast tissue by modulating the activity of nuclear receptors. One mechanism is through interaction with nuclear receptors (such as ERRα1 and ERα) that bind to the S1 element in the promoter region of the aromatase gene, by which PNRC and PNRC2 can modify the levels of aromatase/estrogen biosynthesis in breast tissue.
While it has been shown that PNRC interacts with nuclear receptors mainly through the 23 amino acid fragment containing the SH3-binding domain, the NR box region of PNRC2 was found to be important for interaction with a few nuclear receptors. For example, a fragment lacking the NR box was unable to interact with SF1. However, this fragment had a similar affinity to the NR box-containing fragment for ERRα1. The NR box-negative fragment maintains some ability to interact with ERα. The structural differences, i.e. the region between the NR box and SH3-binding domain, may be important in modulating the ability of these two proteins to interact with various nuclear receptors.
Since both PNRC2 and PNRC contain an SH3 domain-binding motif, in addition to functioning as nuclear receptor coactivators they may also play a role in signal transduction, since most of the proteins possessing SH3 domains are involved in signal transduction (16). There are examples of nuclear receptor-binding proteins mediating the convergence of distinct signaling pathways. P62/ORCA is a ubiquitously expressed cytosolic protein identified through its binding of the SH2 domain of a member of the Src family of non-receptor tyrosine kinases, p56lck, in a phosphotyrosine-independent manner (17). Subsequently, p62/ORCA was identified as a coactivator of the orphan nuclear receptor COUP-TFII, suggesting that it mediates cross-talk between mitogenic and nuclear receptor signaling pathways (18). NRBP, a multidomain putative adapter protein has recently been reported to contain two putative NR boxes, a SH2-binding domain, a kinase-like domain and a nuclear localization signal (19). However, the function of NRBP has not yet been determined. Preliminary studies in our laboratory have confirmed an interaction between PNRC and Grb2 adapter protein. Experiments are currently underway to investigate the roles of PNRC in Grb2-mediated signaling pathways.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr Keith M. Quach for his help in the cloning of PNRC2 and Dr Bin Yu for his help in the northern analysis. This research was supported by NIH grants CA44735 and ES08258. D.Z. was also supported by the Third Military Medical University at Chongqing, China.
DDBJ/EMBL/GenBank accession no. AF374386
References
- 1.Horwitz K.B., Jackson,T.A., Bain,D.L., Richer,J.K., Takimoto,G.S. and Tung,L. (1996) Nuclear receptor coactivators and corepressors. Mol. Endocrinol., 10, 1167–1177. [DOI] [PubMed] [Google Scholar]
- 2.Glass C.K., Rose,D.W. and Rosenfeld,M.G. (1997) Nuclear receptor coactivators. Curr. Opin. Cell Biol., 9, 222–232. [DOI] [PubMed] [Google Scholar]
- 3.Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D., Quach,K.M., Yang,C., Lee,S.Y., Pohajdak,B. and Chen,S. (2000) PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors including orphan receptors SF1 (steroidogenic factor 1) and ERRα1 (estrogen related receptor α-1). Mol. Endocrinol., 14, 986–998. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R.M. and Long,G.L. (1989) A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem., 180, 147–151. [DOI] [PubMed] [Google Scholar]
- 6.Hong H., Kohli,K., Garabedian,M.J. and Stallcup,M.R. (1997) GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid and vitamin D receptors. Mol. Cell. Biol., 17, 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 8.Cavailies V., Dauvois,S., Danielian,P. and Parker,M.G. (1994) Interaction of proteins with transcriptionally active estrogen receptors. Proc. Natl Acad. Sci. USA, 91, 10009–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavailles V., Dauvois,S., l’Horset,F., Lopez,G., Hoare,S., Kushner,P.J. and Parker,M.G. (1995) Nuclear factor RIP 140 modulates transcriptional activation by the estrogen receptor. EMBO J., 14, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onate S.A., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- 11.Douarin B.L., Zechel,C., Garnier,J.M., Lutz,Y., Tora,L., Pierrat,B., Heery,D., Gronemeyer,H., Chambon,P. and Losson,R. (1995) The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptor, is fused to B-raf in the oncogenic protein T18. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voegel J.J., Heine,M.J.S., Zechel,C., Chambon,P. and Gronemeyer,H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J., 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh S. and Chang,C. (1996) Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl Acad. Sci. USA, 93, 5517–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamei Y., Xu,L., Heinzel,T., Torchia,J., Kurokawa,R., Gloss,B., Lin,S.C., Heyman,R.A., Rose,D.W., Glass,C.K. and Rosenfeld,M.G. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarti D., LaMorte,V.J., Nelson,M.C., Nakajima,T., Schulman,I.G., Juguilon,H., Montminy,M. and Evans,R.M. (1996) Role of CBP/p300 in nuclear receptor signalling. Nature, 383, 99–103. [DOI] [PubMed] [Google Scholar]
- 16.Pawson T. (1995) Protein modules and signaling network. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- 17.Joung I., Strominger,J.L. and Shin,J. (1996) Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl Acad. Sci. USA, 93, 5991–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus S.L., Winrow,C.J., Capone,J.P. and Rachubinski,R.A. (1996) A p56lck ligand serves as a coactivator of an orphan nuclear hormone receptor. J. Biol. Chem., 271, 27197–27200. [DOI] [PubMed] [Google Scholar]
- 19.Hooper J.D., Baker,E., Ogbourne,S.M., Sutherland,G.R. and Antalis,T.M. (2000) Cloning of the cDNA and localization of the gene encoding human NRBP, a ubiquitously expressed, multidomain putative adaptor protein. Genomics, 66, 113–118. [DOI] [PubMed] [Google Scholar]