Publisher's Note: There is a Blood Commentary on this article in this issue.
Key Points
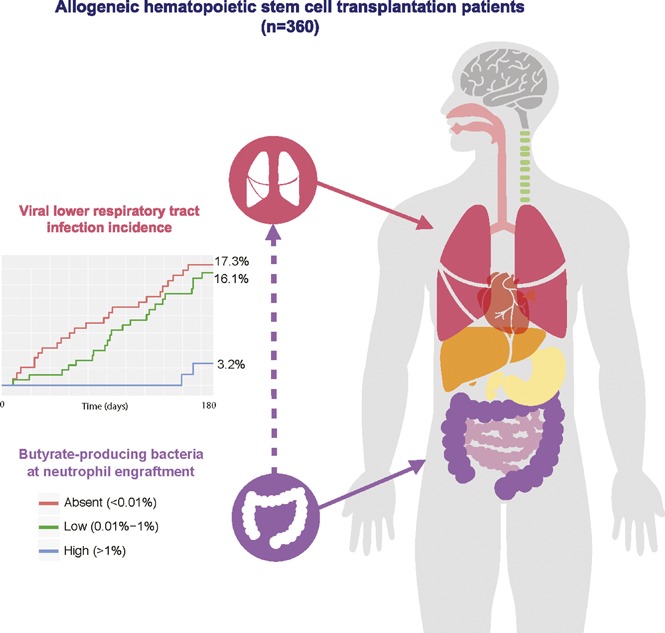
Butyrate-producing bacteria abundance is correlated with protection against viral LRTI following allo-HCT.
Abstract
Respiratory viral infections are frequent in patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HCT) and can potentially progress to lower respiratory tract infection (LRTI). The intestinal microbiota contributes to resistance against viral and bacterial pathogens in the lung. However, whether intestinal microbiota composition and associated changes in microbe-derived metabolites contribute to the risk of LRTI following upper respiratory tract viral infection remains unexplored in the setting of allo-HCT. Fecal samples from 360 allo-HCT patients were collected at the time of stem cell engraftment and subjected to deep, 16S ribosomal RNA gene sequencing to determine microbiota composition, and short-chain fatty acid levels were determined in a nested subset of fecal samples. The development of respiratory viral infections and LRTI was determined for 180 days following allo-HCT. Clinical and microbiota risk factors for LRTI were subsequently evaluated using survival analysis. Respiratory viral infection occurred in 149 (41.4%) patients. Of those, 47 (31.5%) developed LRTI. Patients with higher abundances of butyrate-producing bacteria were fivefold less likely to develop viral LRTI, independent of other factors (adjusted hazard ratio = 0.22, 95% confidence interval 0.04-0.69). Higher representation of butyrate-producing bacteria in the fecal microbiota is associated with increased resistance against respiratory viral infection with LRTI in allo-HCT patients.
Visual Abstract
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HCT) offers curative treatment of a variety of hematologic malignancies and immune disorders, but is associated with an array of complications, including severe infections.1 During the first 6 months after allo-HCT, ∼20% to 30% of patients develop viral respiratory tract infection, caused by influenza (A and B) virus, parainfluenza virus, respiratory syncytial virus, human metapneumovirus, coronavirus, adenovirus, and rhinovirus.2 Although these viral infections in healthy individuals are generally brief and limited to the upper respiratory tract, they can be severe and progress to lower respiratory tract infection (LRTI) in patients following allo-HCT, resulting in substantial morbidity and mortality. During respiratory virus infection, up to 18% to 44% of transplant patients develop LRTI, with associated mortality rates as high as 25% to 45%, according to some studies.2-4 Identifying patients at high risk of severe viral LRTI might provide opportunities for preemptive respiratory insufficiency.
Recently, it has been shown that the intestinal microbiota and its metabolites, such as short-chain fatty acids (SCFA), can modulate host inflammation and promote immune tolerance against a variety of bacterial and viral pathogens.5,6 For example, commensal-derived signals are thought to establish the activation threshold of the innate immune system required for optimal antiviral immunity7 and are associated with augmenting adaptive immune responses to respiratory influenza virus infection in mice.8,9 Butyrate, a microbiota-associated SCFA, serves as an important immunomodulatory compound. This product of microbial fermentation by anaerobic gut bacteria enhances the integrity of the intestinal epithelium10 and modulates enteric tolerance against microbial communities.11 In addition, butyrate contributes to host health in distant organs such as the lung. A recent study showed that administration of butyrate reduces lung inflammation and injury during pneumonia.12
The intestinal microbiota is severely compromised during the course of allo-HCT due to numerous factors, such as antibiotic treatment and disruption of the intestinal mucosa by conditioning chemotherapy and radiation. Altered intestinal microbiota composition, markedly reduced microbial diversity, and intestinal domination by bacterial pathogens during allo-HCT are associated with an increased risk of bacterial dissemination, pulmonary complications, and mortality.13-15 However, the relationship between alterations of the intestinal microbiota and risks of LRTI during respiratory viral infection is unclear.
In this study, we analyzed fecal samples collected from a prospective cohort of allo-HCT recipients and used bacterial metagenomic approaches to determine whether disruption of the intestinal microbiota composition, particularly depletion of butyrate-producing gut bacteria, is associated with the risk of LRTI associated with respiratory viral infection. We also assessed fecal volatile metabolite composition, using targeted metabolomics, in a nested subset of cases and controls.
Methods
Study design, participants, and sample collection
Study participants consisted of adult patients (≥18 years) undergoing allo-HCT at Memorial Sloan Kettering Cancer Center (MSKCC). Patients were enrolled in a prospective fecal collection protocol, where fecal samples were routinely collected during the initial transplant hospitalization and stored in a biospecimen bank, as described previously.13 We required that patients provide a minimum of 3 samples during hospitalization (1 pre- and 2 post-HCT). The study was approved by the institutional review board at MSKCC. All study patients provided written informed consent for biospecimen collection and analysis. The study was conducted in accordance with the Declaration of Helsinki.
We determined intestinal microbiota composition at the time of stem cell engraftment (first of 3 consecutive days where absolute neutrophil count remained ≥500 per milliliter), a pivotal time point during immune reconstitution when the intestinal microbiota would be anticipated, on the basis of experimental studies,13-15 to contribute to the differentiation of the recipients’ innate and adaptive immune system. Thus, in this study, we focused on the fecal sample collected closest in time to stem cell engraftment. Patients were excluded if no sample was collected within 3 days of this time point.
Data collection/variables
Our primary end point of interest was viral LRTI, which we defined as detection of a respiratory viral pathogen by polymerase chain reaction (PCR) assay (obtained in the setting of respiratory symptoms and/or abnormal vital signs), combined with new or changed radiographic findings that were consistent with lower respiratory tract involvement. We required that documented radiographic infiltrates occur between 7 days before to 14 days after diagnosis of respiratory viral infection by PCR assay. Two study physicians independently identified each pulmonary infiltrate, as further described in the supplemental Material, available on the Blood Web site. Imaging findings suggestive of cardiogenic pulmonary edema, atelectasis, or recurrent malignancy were not considered to be pulmonary infiltrates unless there was a specific documented concern for a superimposed infectious process. Respiratory viruses included influenza (A and B) virus, parainfluenza virus, respiratory syncytial virus, human metapneumovirus, coronavirus, adenovirus, and rhinovirus/enterovirus. Throughout the study period, these viruses were detected by multiplex PCR assay of nasopharynx or bronchoscopy specimens at our institution.
In addition to viral LRTI, the following clinical predictors were examined: age (at transplant), sex, underlying disease, pretransplant pulmonary function testing, conditioning regimen intensity,16 stem cell source, ex vivo T-cell depletion of stem cells, time to neutrophil engraftment, administration of any systemic corticosteroids, and administration of antibiotics (IV vancomycin, fluoroquinolones, metronidazole, and β-lactams).
Microbiota sequencing and assessment of butyrate producer abundance
The intestinal microbiota was assessed using the fecal sample collected at engraftment. DNA was extracted and purified from each fecal sample, and the V4 to V5 region of the 16S ribosomal RNA (rRNA) gene was PCR-amplified using modified universal bacterial primers.13-15 Purified PCR products were sequenced using the MiSeq Illumina platform.17 The 16S (V4-V5) paired-end reads were merged and demultiplexed. The UPARSE pipeline18 was used to (1) perform error filtering, using maximum expected error (Emax = 1),19 (2) group sequences into operational taxonomic units (OTUs) of 97% distance-based similarity, and (3) identify and remove potential chimeric sequences, using both de novo and reference-based methods. Prior to clustering, singleton sequences were removed as part of the protocol. Taxonomic assignment to species level was performed for representative sequences from each OTU; this was achieved by using a custom Python script incorporating nucleotide BLAST,20 with NCBI RefSeq21 as reference training set. We used a minimum E-value threshold of 1e-10 for assignments. Sequence designations and identity scores were manually inspected for quality and consistency in terms of taxonomic structure and secondary matches. Based on our testing and comparisons using mock community data, we have found this approach to yield good robust species-level approximations for our candidate sequences. Given prior observations demonstrating the potential benefits of butyrate as an immunomodulatory compound,11 we hypothesized that the presence of bacteria that produce butyrate and other SCFAs might be associated with protection against viral LRTI. In order to assess this feature in our cohort, we measured the abundance of 61 known bacterial species that produce butyrate, adapted from a comprehensive study that analyzed butyrate-producing pathways in 3184 sequenced bacterial genomes and the microbiomes of 15 healthy volunteers22 (listed in supplemental Table 1).
Measurement of SCFAs in selected samples
We also performed targeted metabolomics on a subset of samples, where samples from our cohort were selected in a nested case-control fashion. We selected samples from patients with microbiologically confirmed respiratory viral infection. Cases consisted of patients who developed evidence of viral LRTI, as defined in our primary end point, and controls consisted of patients who developed respiratory viral infection but no evidence of LRTI (ie, upper respiratory tract infection only).
The concentrations of 3 SCFAs were determined using targeted metabolomics methodology. Fecal samples were aliquoted into prechilled 2-mL microtubes with 2.8-mm ceramic beads (Omni International). Extraction solvent (80% MeOH; Fisher Scientific) containing deuterated internal standards: D-3 acetate, D-5 propionate, and D-7 butyrate (Cambridge Isotope Laboratories), was added to the tubes at a ratio of 100 mg fecal sample to 1 mL of extraction solvent. Fecal samples were homogenized using a bead ruptor homogenizer (Omni International) at 4°C followed by centrifugation for 15 minutes at 4°C and 20 000g. The supernatant was collected and derivatized similar to a previously described method.23 Briefly, 1 volume of supernatant was added to 1 volume of borate buffer, pH 11, and 4 volumes of 100 mM pentafluorobenzyl bromide (Thermo Scientific) in acetone (Fisher Scientific). Samples were then vortexed and heated to 65°C for 1 hour. The metabolites were then extracted into n-hexane, diluted 1:10 with additional n-hexane, and quantified by gas chromatography–mass spectrometry (Agilent 7890A GC system, Agilent 5975C MS detector) operating in negative chemical ionization mode, using methane as the reagent gas. Analysis was performed using Mass Hunter Quantitative analysis for gas chromatography–mass spectrometry software (B07.0; Agilent Technologies).
Statistical methods
We analyzed microbiota and clinical risk factors for viral LRTI within our prospective cohort, using survival methods. Analysis time began at the time of engraftment and continued until up to 180 days post-HCT. Observations were censored in the event of death due to any cause. Potential predictors were assessed using Cox proportional hazards modeling. All variables were first examined as univariate predictors of LRTI progression; variables with univariate P < .20 were included in a multivariate analysis. Kaplan-Meier estimates were calculated, and differences in survival were assessed using the log-rank test. All analyses were performed using R version 3.4.0 (R Development Core Team, Vienna, Austria). Data from this study are stored in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm. nih.gov/sra).
Results
We identified 360 patients who underwent allo-HCT at MSKCC between 15 May 2011 and 1 February 2017 and provided fecal samples at the time points outlined in “Methods.” Clinical characteristics of our study cohort were comparable with the case mix of the 553 patients who did not meet our inclusion criteria (supplemental Table 2). Sequencing of fecal samples taken at the time of hematopoietic stem cell engraftment yielded a total of 10 955 300 high-quality 16S rRNA gene sequences (average 30 431 per sample).
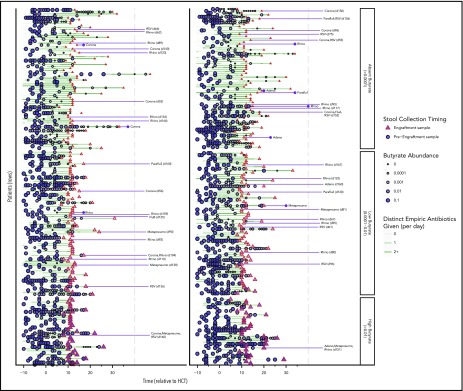
Clinical characteristics of the study cohort are shown in Table 1, grouped by butyrate producer abundance. High relative abundance of butyrate-producing bacteria (>1% of total sequences) was maintained in 69 (19.2%) patients; low butyrate-producing bacterial abundance (<1% of total sequences) was observed in 147 (40.8%) patients, and butyrate-producing bacteria were absent (0% of total sequences) in 144 (40.0%) patients. Patients with leukemia, receiving umbilical cord blood transplants, or experiencing longer time to engraftment were more likely to have lower abundances of butyrate-producing bacteria. Furthermore, patients who received β-lactams and/or metronidazole were also more likely to have a subsequent butyrate-producer deficiency. Figure 1 shows changes in the abundance of butyrate-producing bacteria for each patient over the course of time, prior to neutrophil engraftment. For the most part, abundances were high at the start of allo-HCT, but declined as antibiotics were administered.
Table 1.
Baseline characteristics of allogeneic hematopoietic cell transplantation (N = 360)
| Variable | Value | Butyrate group absent (<0.0001) (%) | Butyrate group low (0.0001-0.01) (%) | Butyrate group high (>0.01) (%) | Total (%) | Fisher P |
|---|---|---|---|---|---|---|
| Age, y | ≤29 | 14 (53.8) | 12 (46.2) | 0 (0.0) | 26 (100) | .156 |
| 30-39 | 23 (47.9) | 18 (37.5) | 7 (14.6) | 48 (100) | ||
| 40-49 | 19 (35.8) | 24 (45.3) | 10 (18.9) | 53 (100) | ||
| 50-59 | 39 (36.4) | 44 (41.1) | 24 (22.4) | 107 (100) | ||
| ≥60 | 49 (38.9) | 49 (38.9) | 28 (22.2) | 126 (100) | ||
| Sex | Female | 58 (44.3) | 57 (43.5) | 16 (12.2) | 131 (100) | .035 |
| Male | 86 (37.6) | 90 (39.3) | 53 (23.1) | 229 (100) | ||
| Underlying disease | Leukemia | 90 (48.1) | 69 (36.9) | 28 (15.0) | 187 (100) | .052 |
| Lymphoma | 20 (30.3) | 28 (42.4) | 18 (27.3) | 66 (100) | ||
| Multiple myeloma | 13 (36.1) | 14 (38.9) | 9 (25.0) | 36 (100) | ||
| Myelodysplastic syndrome | 18 (31.0) | 30 (51.7) | 10 (17.2) | 58 (100) | ||
| Other disorders | 3 (23.1) | 6 (46.2) | 4 (30.8) | 13 (100) | ||
| Conditioning intensity | Myeloablative | 63 (36.2) | 83 (47.7) | 28 (16.1) | 174 (100) | .071 |
| Reduced intensity | 72 (45.3) | 54 (34.0) | 33 (20.8) | 159 (100) | ||
| Nonmyeloablative | 9 (33.3) | 10 (37.0) | 8 (29.6) | 27 (100) | ||
| Stem cell source | Related identical | 32 (33.3) | 42 (43.8) | 22 (22.9) | 96 (100) | |
| Unrelated identical | 56 (40.6) | 51 (37.0) | 31 (22.5) | 138 (100) | ||
| Nonidentical | 18 (34.6) | 27 (51.9) | 7 (13.5) | 52 (100) | ||
| Umbilical cord | 38 (51.4) | 27 (36.5) | 9 (12.2) | 74 (100) | .105 | |
| Time to engraftment | <14 d | 97 (35.9) | 117 (43.3) | 56 (20.7) | 270 (100) | .027 |
| ≥14 d | 47 (52.2) | 30 (33.3) | 13 (14.4) | 90 (100) | ||
| Corticosteroid use | No | 58 (34.5) | 73 (43.5) | 37 (22.0) | 168 (100) | .122 |
| Yes | 86 (44.8) | 74 (38.5) | 32 (16.7) | 192 (100) | ||
| β−Lactam | No | 17 (23.9) | 29 (40.8) | 25 (35.2) | 71 (100) | .000 |
| Yes | 127 (43.9) | 118 (40.8) | 44 (15.2) | 289 (100) | ||
| Fluoroquinolones | No | 28 (45.2) | 19 (30.6) | 15 (24.2) | 62 (100) | .162 |
| Yes | 116 (38.9) | 128 (43.0) | 54 (18.1) | 298 (100) | ||
| Vancomycin (IV) | No | 3 (27.3) | 4 (36.4) | 4 (36.4) | 11 (100) | .399 |
| Yes | 141 (40.4) | 143 (41.0) | 65 (18.6) | 349 (100) | ||
| Metronidazole | No | 106 (35.3) | 129 (43.0) | 65 (21.7) | 300 (100) | .000 |
| Yes | 38 (63.3) | 18 (30.0) | 4 (6.7) | 60 (100) | ||
| Total | 144 (40.0) | 147 (40.8) | 69 (19.2) | 360 (100) |
Figure 1.
Timeline depicting longitudinal changes in butyrate-producing bacteria abundance and viral LRTI events following engraftment. Patients are sorted into 3 groups, depending on the abundance of butyrate-producing bacteria in the engraftment sample (triangle). For the most part, abundances were high at the start of allo-HCT, but declined as antibiotics (green lines) were administered. Viral lower respiratory tract events are depicted with their associated respiratory viruses by the purple lines following engraftment. Adeno, adenovirus; Corona, coronavirus; FluA, influenza virus type A; FluB, influenza virus type B; Metapneumo, human metapneumovirus; Paraflu1, parainfluenza virus type 1; Paraflu3, parainfluenza virus type 3; Rhino, rhino-/enterovirus; RSV, respiratory syncytial virus.
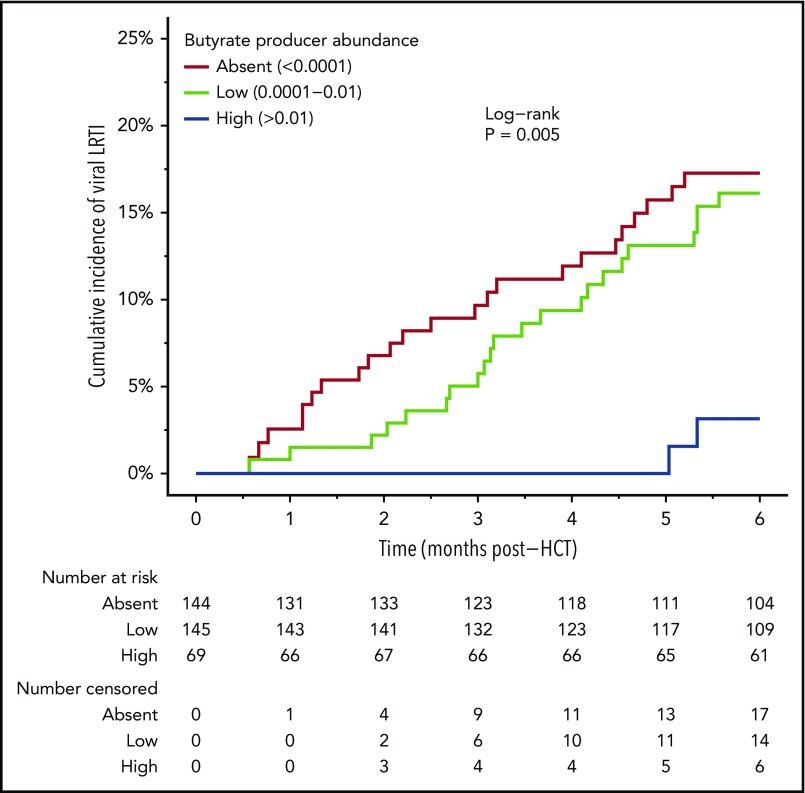
During the 6-month period following allo-HCT, respiratory viral infection occurred in 149 (41.4%) patients. Of those patients, 47 (31.5%, 13% overall) were associated with LRTI involvement. The breakdown of diagnosed virus pathogens is shown in Table 2. A total of 594 separate viral pathogens were identified from the 149 patients (through clinical testing of nasal/respiratory specimens). Rhinovirus/enterovirus was the most commonly encountered viral pathogen, occurring several-fold more frequently than all other viral pathogens. However, adenovirus, respiratory syncytial virus, and human metapneumovirus were most frequently associated with LRTI involvement (all ∼30%). Of the 47 cases of viral LRTI, 11 (23.4%) were diagnosed by lower tract specimens obtained by bronchoscopy (supplemental Table 3). In our analysis of clinical and microbiota predictors of viral respiratory infection, we found that patients with higher abundances of butyrate-producing bacteria were less likely to develop viral LRTI. The estimated incidence of viral LRTI at 180 days was 17.3% and 16.1% for the groups in which butyrate-producing bacteria were absent or low, respectively, and 3.2% for the high butyrate-producing abundance group (log-rank P = .005) (Figure 2). In addition, patients with the highest abundances of butyrate-producing bacteria (>1%) were independently associated with a fivefold decrease in risk of viral LRTI (Table 3). Other clinical variables that were independently predictive of viral LRTI were corticosteroid use and preengraftment macrolide use. Of note, other microbiome characterizations, including microbial diversity (inverse Simpson) and abundance of several notably beneficial taxonomic groups, were less associated with protection against viral LRTI (supplemental Table 4).
Table 2.
Respiratory viruses diagnosed in the prospective cohort (N = 360 subjects)
| Respiratory virus | Count | Developed LRTI (%) |
|---|---|---|
| Adenovirus | 22 | 7 (31.8) |
| Coronavirus 229E | 26 | 2 (7.7) |
| Coronavirus OC43 | 44 | 5 (11.4) |
| Coronavirus type HKU1 | 33 | 1 (3.0) |
| Coronavirus type NL63 | 25 | 3 (12.0) |
| Human metapneumovirus | 29 | 8 (27.6) |
| Human rhinovirus/enterovirus | 250 | 28 (11.2) |
| Influenza virus type A | 19 | 2 (10.5) |
| Influenza virus type B | 16 | 3 (18.8) |
| Parainfluenza virus type 1 | 12 | 2 (16.7) |
| Parainfluenza virus type 2 | 1 | 0 (0.0) |
| Parainfluenza virus type 3 | 47 | 4 (8.5) |
| Parainfluenza virus type 4 | 6 | 1 (16.7) |
| Respiratory syncytial virus | 64 | 18 (28.1) |
| Total | 594 | 84 (14.1) |
Figure 2.
Association between butyrate-producing bacteria abundance and the development of viral LRTI. Kaplan-Meier analysis starting at neutrophil engraftment. Log-rank P values are significant if P < .05.
Table 3.
Predictors of viral LRTI after allo-HCT
| Predictor | Univariate | Multivariate | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |
| Age, y | 1.00 (0.98-1.02) | .904 | ||
| Sex (female) | 1.01 (0.55-1.81) | .962 | ||
| Underlying disease (leukemia vs other) | 1.38 (0.78-2.49) | .271 | ||
| Conditioning intensity (myeloablative) | 1.00 | — | ||
| Conditioning intensity (reduced intensity) | 1.04 (0.58-1.85) | .904 | ||
| Conditioning intensity (nonmyeloablative) | 0.66 (0.13-2.02) | .509 | ||
| T-cell depletion (ex vivo) | 1.26 (0.71-2.27) | .435 | ||
| Stem cell source (related identical) | 1.00 | — | 1.00 | — |
| Stem cell source (unrelated Identical) | 0.80 (0.37-1.74) | .563 | 0.78 (0.36-1.71) | .524 |
| Stem cell source (nonidentical) | 1.79 (0.79-4.02) | .157 | 1.77 (0.78-3.99) | .172 |
| Stem cell source (cord blood) | 1.36 (0.60-3.05) | .454 | 0.89 (0.38-2.07) | .790 |
| Time to engraftment (≥14 d) | 1.27 (0.65-2.33) | .469 | ||
| Corticosteroid administration* | 2.36 (1.32-4.25) | .004 | 2.33 (1.27-4.32) | .007 |
| Abnormal DLCO (<80% predicted) | 1.10 (0.61-1.94) | .753 | ||
| Abnormal FEV1 (<80% predicted) | 1.09 (0.46-2.23) | .834 | ||
| Abnormal FVC (<80% predicted) | 1.08 (0.46-2.23) | .840 | ||
| Prior β-lactam use | 1.66 (0.78-4.19) | .201 | ||
| Prior fluoroquinolone use | 0.75 (0.39-1.56) | .420 | ||
| Prior metronidazole use | 1.10 (0.49-2.19) | .804 | ||
| Prior vancomycin (IV) use | 1.07 (0.29-9.46) | .935 | ||
| Prior macrolide use | 3.04 (1.11-6.80) | .033 | 2.79 (1.01-6.35) | .049 |
| Butyrate abundance: absent (<0.0001) | 1.00 | — | 1.00 | — |
| Butyrate abundance: low (0.0001-0.01) | 0.90 (0.50-1.61) | .722 | 0.92 (0.51-1.66) | .789 |
| Butyrate abundance: high (>0.01) | 0.20 (0.04-0.61) | .003 | 0.22 (0.04-0.69) | .006 |
Time-dependent predictor.
CI, confidence interval.
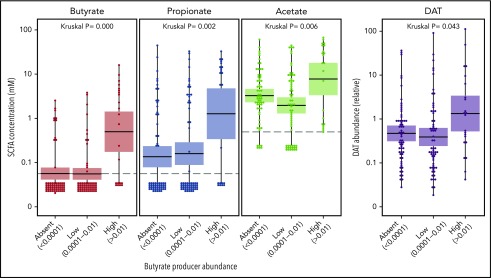
We performed targeted metabolomics analysis of volatile metabolites on 147 samples from patients with respiratory viral infection and found that measured fecal concentration of butyrate correlated with the abundance of butyrate-producing bacteria. The abundance of butyrate producers also correlated with production of other metabolites, including acetate, propionate, and desaminotyrosine (DAT) (Figure 3).
Figure 3.
Correlation between butyrate-producing bacteria abundance and absolute fecal concentrations of butyrate, acetate, and propionate, and relative abundance of DAT. Kruskal-Wallis P values are significant if P < .05.
Discussion
In this study, we examined clinical and microbiota predictors of viral LRTI in a cohort of 360 allogeneic HCT recipients. We found that patients who harbored a high abundance of butyrate-producing bacteria at the time of stem cell engraftment were less likely to develop viral LRTI in the 6 months following allo-HCT. In recent years, it has been shown that some of the bacterial species constituting the intestinal microbiota contribute to protection against infections through a variety of direct and indirect (immunomodulatory) mechanisms.5-9 In addition, the health benefits of microbiota-associated SCFA are increasingly appreciated, and several butyrate-specific mechanisms have recently been identified that suggest their association with protection from LRTI in our patient cohort. A recent study of mice showed that intraperitoneal butyrate administration reduced persistent lung inflammation during Klebsiella pneumoniae infection, leading to the suggestion that butyrate restores interleukin-10 (IL-10) levels in the lung by inhibiting histone deacetylase.12 Anti-inflammatory properties have been described in the intestine, where butyrate through the same IL-10-dependent mechanism was associated with heightened tolerance of macrophages toward the residing microbiota.11 Finally, as butyrate and butyrate-producing bacteria have been associated with promotion of T-regulatory cells and reduced incidence of graft-versus-host disease, a decrease of butyrate-producing bacteria might be associated with increased graft-versus-host disease and increased exposure to immunosuppressive agents such as steroids, which is a known risk factor for LRTI.24,25
Almost all major producers of butyrate are obligately anaerobic; our observation that β-lactams and metronidazole were associated with decreased abundances of these bacteria is not necessarily surprising, because these antibiotics exhibit potent antianaerobic activity. Furthermore, we did not observe significant associations with IV vancomycin and fluoroquinolones, which are less active against anaerobes in the gut lumen.
We found that high relative abundance of butyrate-producing bacteria in the gut was associated with elevated butyrate, propionate, and acetate concentrations in fecal samples, which underscores the close relationship between these 3 SCFAs. For example, a significant proportion of butyrate-producing bacteria harbors the butyryl-CoA:acetate CoA-transferase gene, which converts acetate into butyrate.26 Beyond SCFAs, we also found that DAT levels in feces were significantly elevated in patients with high abundance of butyrate-producing species. This degradation product of flavonoids is produced by Flavonifractor plautii (previously known as Clostridium orbiscindens) and has recently been associated with ameliorating influenza infection through augmentation of type I interferon signaling.9 These observations strengthen the notion that the potentially beneficial effects of butyrate-producing bacteria in the gut extend beyond butyrate alone, and that these communities might be reflected as a marker of immunological resilience.27,28
There are several limitations to this study. First, a broad clinical-radiographic definition of posttransplant viral LRTI was used to maximize detection of otherwise poorly defined events. We used a similar approach as taken previously by Harris and colleagues, which used abnormal radiographic patterns in conjunction with clinical information.14 However, the inclusion of all abnormal radiographic findings into 1 primary end point, regardless of severity of disease, respiratory viral infection, and causative pathogen, could misclassify and inflate or dilute the impact of butyrate-producing species in this cohort. In a sense, this partly mirrors our clinical experience, where it can be difficult to know whether symptoms and/or radiographic findings are related to a viral pathogen identified by clinical testing.
Second, despite that our findings show that the abundance of butyrate-producing bacteria is an independent predictor of viral LRTI, as depicted in Table 3, whether these bacteria or their metabolites can directly confer protection against LRTI is not known. It is possible that the loss of butyrate producers may reflect some underlying and undocumented differences between patients that also lead to increased susceptibility to viral LRTI. Third, the microbiota data were specifically studied during engraftment, and although we believe that this may be an immunologically crucial time point, subsequent microbiota time points may also be important but were not collected or captured. Finally, although this cohort was representative of the allo-HCT population at our center,13-15 it might not be fully comparable to other transplant centers, where exposure to microbial pathogens, antimicrobial prophylaxis, empiric treatment, and allo-HCT protocols differs.
In summary, high intestinal abundance of butyrate-producing bacteria at engraftment is an independent predictor of protection against viral LRTI in allo-HCT recipients. Recent studies of this patient group revealed the importance of the phylogenetic composition and diversity of the microbiota at the time of stem cell engraftment and support the notion that the microbiota contributes to optimization of immune reconstitution following allo-HCT.13-15 Interventions that protect the butyrate-producing compartment during allo-HCT, such as reduced exposure to antibiotics that target obligate anaerobic bacteria, such as some β-lactam antibiotics and metronidazole, might lead to improved outcomes. In addition, our study highlights a new opportunity for development of next-generation pre- or probiotics that enhance the abundance of butyrate-associated species as an adjuvant therapy during transplant hospitalization or following viral respiratory infection. Interestingly, a clinical trial assessing short-term administration of a dietary starch to increase levels of butyrate in patients subject to allo-HCT is currently ongoing (https://clinicaltrials.gov/ct2/show/NCT02763033). However, the underlying mechanisms by which butyrate-producing bacteria provide protection during allo-HCT should be investigated in greater detail, because it still remains unclear whether this relationship is directly causal. In addition, timing of potential treatment is of importance, because the anti-inflammatory effect of IL-10 has been shown to be detrimental in certain clinical situations.29 Experimental studies to define molecular mechanisms of antiviral protection followed by interventional studies that investigate the protective potential of intestinal reconstitution with butyrate-producing bacteria are key to developing microbiota-targeted interventions that could improve outcomes for patients following allogeneic HCT.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (UO1 AI124275; RO1 AI095706; RO1 AI42135) and National Cancer Institute (P30 CA008748; PO1 CA023766), Cycle for Survival to the MSK Center for Microbes, Inflammation and Cancer, and Parker Institute for Cancer Immunotherapy at MSKCC.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.W.H. conducted data collection, data analysis, data interpretation, and manuscript preparation; E.R.L. assisted in data analysis and interpretation and drafted the figures; J.-L.C., A.J.P., and J.R.C. conducted metabolomics analysis and assisted in manuscript preparation; E.F. conducted total 16s qPCRs and conducted microbiota sequencing; F.A. assisted in data collection and data interpretation; Y.G. and L.L. conducted the microbiota sequencing analysis; S.M.M., J.U.P., and M.R.v.d.B. were treating physicians of included patients and assisted in the manuscript preparation; A.I.G. assisted in data collection and data interpretation and in the manuscript preparation; E.G.P. and Y.T. drafted the study design and data collection protocol, assisted in data analysis and interpretation, and finalized manuscript preparation; and all authors critically reviewed and approved the manuscript.
Conflict-of-interest disclosure: J.U.P. receives research support and licensing fees from Seres Therapeutics. M.R.v.d.B. is an advisor for and receives research support from Seres Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Ying Taur, Infectious Diseases Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave #9, New York, NY 10065; e-mail: taury@mskcc.org.
REFERENCES
- 1.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813-1826. [DOI] [PubMed] [Google Scholar]
- 2.Peck AJ, Englund JA, Kuypers J, et al. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood. 2007;110(5):1681-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ison MG. Respiratory syncytial virus and other respiratory viruses in the setting of bone marrow transplantation. Curr Opin Oncol. 2009;21(2):171-176. [DOI] [PubMed] [Google Scholar]
- 4.Lee I, Barton TD. Viral respiratory tract infections in transplant patients: epidemiology, recognition and management. Drugs. 2007;67(10):1411-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuijt TJ, Lankelma JM, Scicluna BP, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abt MC, Osborne LC, Monticelli LA, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108(13):5354-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steed AL, Christophi GP, Kaiko GE, et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357(6350):498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111(6):2247-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty K, Raundhal M, Chen BB, et al. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat Commun. 2017;8:13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55(7):905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris B, Morjaria SM, Littmann ER, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med. 2016;194(4):450-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacigalupo A, Ballen K, Rizzo D, et al. Defining the intensity of conditioning regimens: working definitions. Biol Blood Marrow Transplant. 2009;15(12):1628-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996-998. [DOI] [PubMed] [Google Scholar]
- 19.Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31(21):3476-3482. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-410. [DOI] [PubMed] [Google Scholar]
- 21.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42(Database issue):D553-D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5(2):e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomcik K, Ibarra RA, Sadhukhan S, Han Y, Tochtrop GP, Zhang GF. Isotopomer enrichment assay for very short chain fatty acids and its metabolic applications. Anal Biochem. 2011;410(1):110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17(5):505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charrier C, Duncan GJ, Reid MD, et al. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology. 2006;152(Pt 1):179-185. [DOI] [PubMed] [Google Scholar]
- 27.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(suppl 1):S45-S56. [DOI] [PubMed] [Google Scholar]
- 28.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis. 2013;26(4):332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol. 2010;10(6):427-439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.