Abstract
Purpose
During retinal degeneration, Müller glia cells respond to photoreceptor loss by undergoing reactive gliosis, with both detrimental and beneficial effects. Increasing our knowledge of the complex molecular response of Müller cells to retinal degeneration is thus essential for the development of new therapeutic strategies. The purpose of this work was to identify new factors involved in Müller cell response to photoreceptor cell death.
Methods
Whole transcriptome sequencing was performed from wild-type and degenerating rd10 mouse retinas at P30. The changes in mRNA abundance for several differentially expressed genes were assessed by quantitative RT-PCR (RT-qPCR). Protein expression level and retinal cellular localization were determined by western blot and immunohistochemistry, respectively.
Results
Pathway-level analysis from whole transcriptomic data revealed the Hippo/YAP pathway as one of the main signaling pathways altered in response to photoreceptor degeneration in rd10 retinas. We found that downstream effectors of this pathway, YAP and TEAD1, are specifically expressed in Müller cells and that their expression, at both the mRNA and protein levels, is increased in rd10 reactive Müller glia after the onset of photoreceptor degeneration. The expression of Ctgf and Cyr61, two target genes of the transcriptional YAP/TEAD complex, is also upregulated following photoreceptor loss.
Conclusions
This work reveals for the first time that YAP and TEAD1, key downstream effectors of the Hippo pathway, are specifically expressed in Müller cells. We also uncovered a deregulation of the expression and activity of Hippo/YAP pathway components in reactive Müller cells under pathologic conditions.
Keywords: retina, Müller cells, Hippo/YAP pathway, photoreceptor degeneration
Müller cells are the major glial cell type of the vertebrate retina. Their cell body lies in the inner nuclear layer (INL) of the neural retina while their processes span the entire thickness of the tissue. Under normal conditions, Müller cells actively participate in the maintenance of retinal functions and homeostasis. In addition to their structural role, they release neurotrophic factors, regulate oxidative stress, form the retinal blood barrier, and participate in retinal synaptic activity by recycling neurotransmitters.1,2 Due to their important role, dysfunction or absence of Müller cells lead to retinal structure disorganization and loss of photoreceptor function.3,4 In response to pathologic conditions, these glial cells enter a state referred to as reactive gliosis with both detrimental and beneficial effects.5,6 Among the latter is neuroprotection via the increased release of neurotrophic and antioxidant factors.5,7 In addition, and under certain conditions, reactive Müller cells can dedifferentiate into cells displaying characteristics of retinal stem cells, proliferate and generate neurons.8,9 If such a process is very efficient in fish, it is however largely ineffective in mammals.10–12 Hence, increasing our knowledge of the molecular mechanisms underlying Müller cell function under normal and pathologic conditions is essential for developing new therapeutic strategies that take advantage of their neuroprotective and regenerative properties.
In various species, a large variety of pathways have been shown to influence Müller cell activation in response to retinal injury, including MAPK, Wnt, Notch, Hedgehog, glucocorticoid, Jak/Stat, TGFβ/Smad, or mTor.9,13 The Hippo/Yes-associated protein (YAP) signaling pathway is another key pathway that has recently brought a lot of interest as an important regulator of tissue homeostasis through its action on both cell proliferation and survival.14,15 So far, its participation in Müller cell reactivation has not been investigated.
The Hippo pathway regulates diverse biological processes including proliferation, differentiation, survival, and is conserved throughout evolution.16,17 The core of the Hippo pathway consists of a kinase cascade that phosphorylates the transcription coactivators, YAP and its homolog TAZ, at a specific serine residue of the amino-terminal region, leading to their sequestration within the cytoplasm. The Hippo pathway is regulated in response to diverse stimuli such as mechanical stress, DNA damage, or oxidative stress.15,18 When the pathway is off, YAP and TAZ translocate into the nucleus and interact with their DNA-binding partners of the TEAD (TEA domain transcription factor) family, driving gene regulatory networks involved in cell proliferation and differentiation.19 In the adult, the effector of the pathway, YAP, can stimulate regeneration of several injured mammalian organs such as the heart, liver, intestine20–22 or the tail and limb in Xenopus.23,24 Its function in the adult retina and in particular in Müller cells has hitherto not been investigated. On the other hand, studies in various species have implicated YAP in retinal development. In zebrafish, YAP controls the balance between proliferation and differentiation of retinal progenitors25,26 and is a key regulator of retinal pigment epithelium (RPE) genesis.27 In Xenopus tadpoles, YAP is required in retinal stem cells for postembryonic retinal growth.28 Yes-associated protein also positively regulates proliferation of mammalian retinal progenitors.29 Noteworthy, heterozygous YAP loss-of-function mutations in humans can result in autosomal dominant coloboma,30 and a mutation within the YAP-binding domain of TEAD131 causes Sveinsson's chorioretinal atrophy (SCRA), an autosomal dominant eye disease characterized by chorioretinal degeneration.32 However, the mechanisms underlying YAP/TEAD function in these diseases are so far unknown.
Meta-analysis using already published ChIP-Seq data,33 and whole transcriptome sequencing data (RNA-Seq) from retinas of the well-characterized degenerative mouse model of retinitis pigmentosa, rd10, led to the identification of a set of INL-enriched genes. Pathway-level analysis revealed the Hippo pathway as one of the main deregulated pathways. We thus undertook a detailed analysis of the expression of YAP and its potential partner TEAD1 in normal adult retina and during photoreceptor degeneration. We found that both are specifically expressed in Müller cells. Their expression, as well as that of their well-characterized direct target genes, Ctgf and Cyr61, is increased alongside photoreceptor loss. Thus, this work uncovers for the first time a link between the Hippo/YAP pathway and Müller cell reactivation in pathologic conditions.
Materials and Methods
Animals and Tissues
All mice were handled in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. C57BL6/J (Charles River, L'Arbresle, France) and rd10 mice (The Jackson Laboratory, Bar Harbor, ME, USA, kindly provided by Bo Chang) were kept at 21°C, under a 12-hour light/12-hour dark cycle, with food and water supplied ad libitum. For the chemical-induced retinal degeneration model, C57BL6/J adult mice were given a single intraperitoneal injection of 1-Methyl-1-nitrosourea (MNU) at a dose of 60 mg/kg body weight. The MNU solution (Ark Pharm, Libertyville, IL, USA) was freshly dissolved in sterile physiological saline immediately before use. Control animals received physiological saline.
After mouse euthanasia, the eyes were rapidly enucleated and processed for immunohistochemistry, western blot, RNA-Seq, and quantitative RT-PCR (RT-qPCR) as described in the following sections.
Whole Transcriptome Sequencing (RNA-Seq) and Data Analysis
Whole transcriptome analysis was performed on three independent biological replicates from wild-type (WT) and rd10 retina at postnatal stage 30 (P30). After harvesting, both retinas for each animal were collected and immediately frozen. RNA was extracted using Nucleospin RNA Plus kit, which includes DNase treatment (Macherey-Nagel, Düren, Germany). RNA quality and quantity were evaluated using a BioAnalyzer 2100 with RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). Stranded RNA-Seq libraries were constructed from 100 ng of high quality total RNA (RIN > 8) using the TruSeq Stranded mRNA Library Preparation Kit (Illumina, San Diego, CA, USA). Paired-end sequencing of 125 bases length was performed on a HiSeq 2500 system (Illumina). Pass-filtered reads were mapped using TopHat version 2.1.0 and aligned to UCSC mouse reference genome mm10.34 Count table of the gene features was obtained using HT-Seq.35 Normalization, differential expression analysis, and fragments per kilobase of exon per million fragments mapped (FPKM) values were computed using EdgeR.36 An FPKM filtering cutoff of two in at least one of the six samples was applied. A P value of less than or equal to 0.05 was considered significant, and a cutoff of a fold change (FC) of 1.2 was applied to identify differentially expressed isoforms. Pathways analysis was done using the Kyoto Encyclopedia of Genes and Genome (KEGG) and GO annotation obtained using DAVID Bioinformatics Resources 6.7.
RNA Extraction and Gene Expression Analysis by Real-Time PCR
Quantitative RT-PCR experiment was performed on two independent biological replicates from WT and rd10 retinas at each stage (P10, P20, P30, and P120). Each sample was a pool of retinas from at least three individuals. Total RNA was extracted from neural retina using RNeasy mini kit (Qiagen, Germantown, MD, USA) and treated with DNAse I according to the manufacturer's instructions. RNA quantity and quality were assessed using the NanoDrop 2000c UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and Experion automated electrophoresis system (Bio-Rad, Hercules, CA, USA). One microgram of total RNA was reverse transcribed in the presence of oligo-(dT)20 using Superscript II reagents (Thermo Fisher Scientific). For each RT-qPCR, 2 μL of a 10-fold dilution of the cDNA was used, and the reactions were performed in triplicates on a 7900HT Genetic Analyzer (Thermo Fisher Scientific) as previously described.37 Differential expression analysis was performed using the ΔΔCt method using the geometric average of Gak, Mrpl46, Srp72, and Tbp as the endogenous controls.38 For each gene, the relative expression of each samples was calculated using WT retina at each time point as the reference (1 arbitrary unit [a.u.]). Primers are listed in Supplementary Table S1.
Western Blotting
Western blot was performed on retina pools of at least three individuals unless otherwise specified in the figure legends. Eyes were removed and retinas were quickly isolated and frozen at −80°. Retinas were lysed in radioimmunoprecipitation assay (RIPA) (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Nonidet P-40, 0.5% sodium deoxycholate) and protease inhibitor cocktail (P-2714; Sigma-Aldrich Corp., St. Louis, MO, USA). Lysates concentration was determined using a Lowry protein assay kit (500011, DC Protein Assay; Bio-Rad). Equal amounts of proteins (20 μg of each sample) were loaded, separated by 7.5% SDS-PAGE and transferred onto nitrocellulose membranes. Western blots were then conducted using standard procedures. Primary and secondary antibodies are listed in Supplementary Table S2. An enhanced chemiluminescence kit (Bio-Rad) was used to detect the proteins. Each sample was probed once with tubulin for normalization. α-tubulin was used as the loading control for each sample. Quantification was done using ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA).
Acrylamide gels containing Phos-tag (40 μM final; SOBIODA, Montbonnot-Saint-Martin, France) were prepared according to the manufacturer's instructions. Immunoblotting was then performed according our standard protocol. As a control, and to allow discrimination between phosphorylated and nonphosphorylated forms of YAP, one rd10 P10 lysate was treated with 10 U of calf intestinal alkaline phosphatase (Thermo Fisher Scientific) for 1 hour at 37°C and processed thereafter for western blot as previously described.
Immunohistochemistry
Standard immunohistochemistry techniques on paraffin sections were applied with the following specificities: antigen unmasking treatment was done in boiling heat-mediated antigen retrieval buffer (10 mM sodium citrate, pH 6.0) twice for 20 minutes. Primary antibody was diluted in ready-to-use diluent (S0809, Dako, Glostrup, Denmark). The specificity of the anti-YAP and anti-TEAD1 antibodies has already been shown by other groups.39,40 Primary and secondary antibodies are listed in Supplementary Table S2. Sections were counterstained with 1:1000 4′,6-diamidino-2-phenylindole (DAPI) (1 mg/mL, 62248, Thermo Fisher Scientific).
Confocal Microscopy
Confocal images were acquired using a Zeiss LSM710 confocal microscope and Zen software (Zeiss, Thornwood, NY, USA). Images were taken near the optic nerve. The same magnification, laser intensity, gain, and offset settings were used across animals for any given marker. All experiments were done in triplicate. Image processing was performed using ImageJ software (National Institutes of Health).
Statistical Analysis
Results are reported as mean ± SEM. Nonparametric Mann-Whitney U test was used to analyze western blot data. P value ≤ 0.05 was considered significant.
Results
The Hippo Pathway Is One of the Main Pathways Altered in Response to Photoreceptor Degeneration
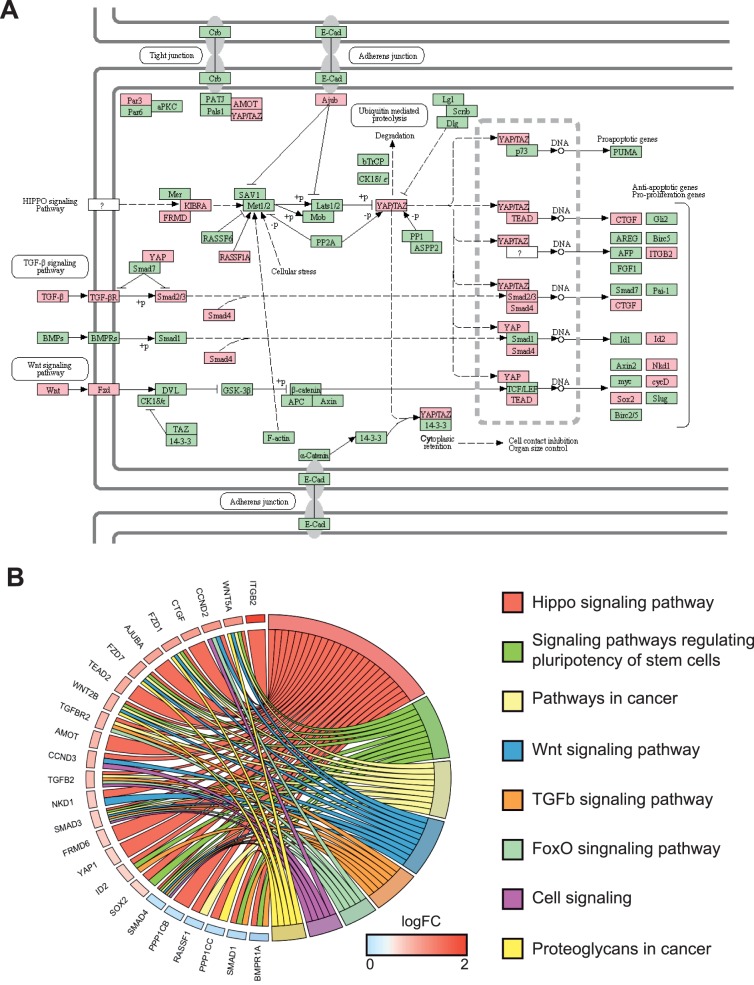
To gain insights into the molecular mechanisms triggered in Müller cells following photoreceptor cell death, we performed whole transcriptome sequencing of rd10 mouse retinas (Pde6brd10), and used C57Bl6/J WT mice as controls. In rd10 mice, degeneration is due to a mutation in the β subunit of the rod phosphodiesterase gene (Pde6b).41,42 In humans, mutations in the same gene cause retinitis pigmentosa.43 In rd10 mice, rod cell death starts around P16, and the vast majority of photoreceptors are lost by P60.41 We decided to perform RNA-Seq experiments on P30 retina in order to investigate the molecular response of Müller cells. By choosing this time point, we speculated that Müller cell response to injury would still be active since rod photoreceptor cell death is still occurring in contrast to a previous study performed at much later time points when all rods are already lost.44 Gene level analysis of the sequencing data allowed the identification of 3427 differentially expressed genes (DEGs) out of 24,062 genes using criteria of FC 1.2 with a P value cutoff of 0.05 and a minimum expression of two FPKMs in at least one sample (Fig. 1A). Such relaxed parameters allow the identification of several genes with small FC, which may be of importance if they are all part of the same pathway. With this objective in mind, the next step was the enrichment of the dataset with genes mostly expressed in the inner retina, where Müller cells are located. Therefore, we used neural retina leucine zipper (NRL) ChIP-Seq data to filter out photoreceptor-NRL target genes.33 Indeed, NRL is a rod-specific transcription factor required for rod photoreceptor development and homeostasis and is specifically expressed in photoreceptors.45 Putative photoreceptor-specific transcripts, as defined by at least one binding site for NRL, were discarded from our dataset, leading to 2643 DEGs named thereafter INL-enriched genes. Although among them, some are photoreceptor-expressed genes without NRL binding sites, such an approach circumvents the identification of a large number of genes expressed in rods and identified as downregulated due to photoreceptor cell death. In addition, based on PANTHER annotation, we kept only extrinsic factors and nuclear factors for further pathway analysis, which represent two of the main categories of signaling pathway regulators (extrinsic cues and effectors, respectively). This approach led to the identification of 293 DEGs that were then used for pathway analysis using KEGG (Fig. 1A). Among the top pathways, several were already known as deregulated during retinal degeneration such as PI3K-Akt signaling, Jak-STAT, and cytokine-related pathways (Supplementary Table S3). More interestingly, this analysis of the 293 DEGs belonging to extrinsic factors and nuclear factors categories revealed the Hippo pathway as a new signaling pathway deregulated in degenerative retina. Indeed, KEGG pathway analysis revealed a total of 11 Hippo signaling related genes as significantly differentially expressed in rd10 versus WT mouse retina (Fig. 1B) with nine of them upregulated and two downregulated. We then extended our analysis to our entire dataset of 2643 DEGs (regardless of the PANTHER annotation). Among genes identified in the KEGG database as related to the Hippo pathway, 25 were differentially expressed (Fig. 2A). Chord plot representation illustrates the relationship between those and other signaling pathways such as Wnt or TGFβ (Fig. 2B).
Figure 1.
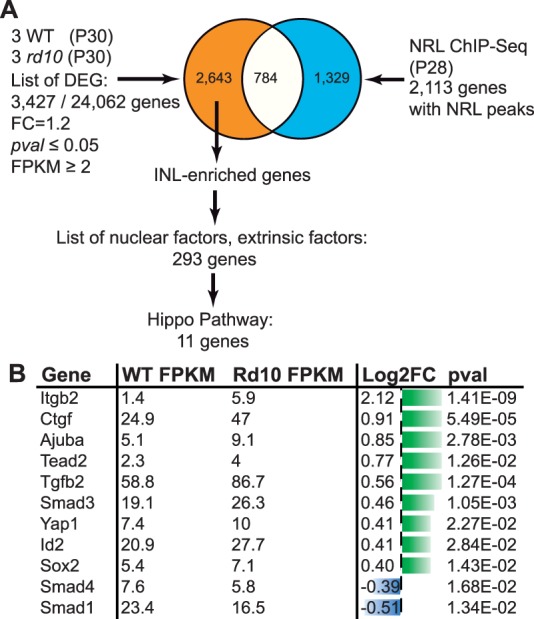
Identification of nuclear and extrinsic factors differentially expressed between rd10 and WT P30 retina. (A) Workflow of the meta-analysis used to identify nuclear and extrinsic factors differentially expressed in the degenerative rd10 retina at P30. (B) Table of the 11 nuclear and extrinsic factors differentially expressed and related to the Hippo pathway. Gene expression values in WT and rd10 are expressed in FPKM. Log2 of the FC and P value are also indicated. Green bars, upregulated genes; blue bars, downregulated genes. The length of the bar is proportional to the log2 of the FPKM value.
Figure 2.
Identification of 25 DEGs between rd10 and WT P30 retina related to the Hippo pathway and at the crossroad of other signaling pathways. (A) Schematic of the Hippo pathway from KEGG. Red boxes indicate 25 genes found differentially expressed in our dataset. Green boxes indicate unchanged genes. (B) ChordPlot representation shows the relationship between these 25 Hippo signaling components that are differentially expressed in rd10 retina, and other KEGG signaling pathways.
The Transcriptional YAP/TEAD Complex Is Upregulated in Response to Photoreceptor Degeneration
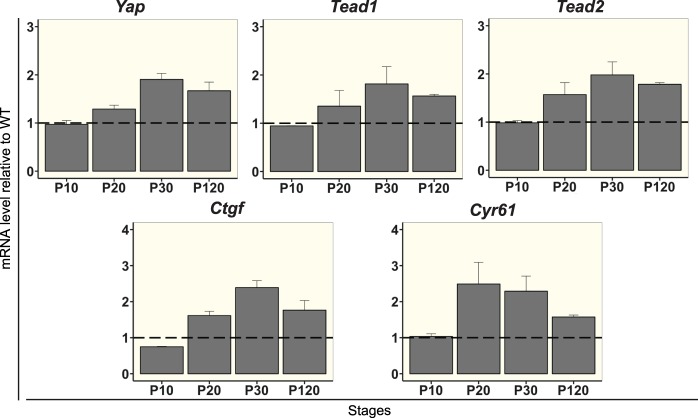
We then used RT-qPCR at P30 to validate some of our transcriptomic analysis findings. We also extended the study by analyzing gene expression at different stages (P10: before the degeneration; P20: after the onset of the degeneration; P30: stage corresponding to the transcriptomic analysis; P120: late onset of the degeneration when all rod photoreceptors are lost). We first focused on the terminal effector of the pathway, YAP. If Yap expression is not affected at P10 prior to degeneration, an increased expression in rd10 compared to WT was observed from P20 onwards (Fig. 3).
Figure 3.
RT-qPCR validation of selected Hippo pathway components identified by RNA-seq analysis during retinal degeneration. Differential expression analysis by RT-qPCR of Yap, Tead1, Tead2, Ctgf, and Cyr61 in rd10 retina at various stages as indicated, relative to WT levels. All values are expressed as the mean ± SEM from two biological replicates. The data were normalized against the geometric average ΔCt of four housekeeping genes: Gak, Mrpl46, Srp72, and Tbp.
As a cofactor, YAP mediates its transcriptional activity mainly by its association with TEAD transcription factors.46 We thus also focused our analysis on members of this gene family. Among the four members, only Tead2 was identified as differentially expressed in the RNA-Seq analysis and validated by RT-qPCR analysis (Fig. 3). Although RNA-Seq analysis did not show any significant difference in the expression of Tead1, RT-qPCR showed that it was also upregulated in rd10 retina (Fig. 3). The other two members of the Tead family (i.e., Tead3 and Tead4) were considered not expressed in mouse retinas (FPKM<1 by RNA-Seq and not detected by RT-qPCR).
Ctgf has been recognized as a direct YAP/TEAD target gene46 and identified as a DEG in our RNA-Seq analysis (Fig. 1B). We confirmed by RT-qPCR that Ctgf is differentially expressed following photoreceptor death in rd10 mouse versus WT (Fig. 3). Although Cyr61 is another bona fide YAP/TEAD target gene,47 it is not listed in the KEGG database on which our pathway analysis is based. Nevertheless, both our RNA-Seq and RT-qPCR analysis revealed a significant upregulation of Cyr61 in rd10 retina compared to WT (Fig. 3).
To support these conclusions in another model of retinal degeneration, we investigated the expression of these genes in the rd1 mouse model. The rd1 mouse line is another well-characterized model of retinitis pigmentosa caused by a mutation in the Pde6b gene.48 In rd1 animals, rod photoreceptors begin to degenerate at approximately P10. Microarray analysis retrieved from the KBaSS database (Knowledge Base for Sensory Systems: kbass.institut-vision.org/KBaSS)49 revealed an increased expression of Yap, Ctgf, and Cyr61, after the onset of the degeneration process from P13 onwards (Supplementary Fig. S1). We did not assess Tead family gene expression since the database did not give any signal level information for Tead probes.
Altogether, these data revealed an upregulation of key Hippo signaling factors during photoreceptor cell loss.
YAP Expression in Müller Cells
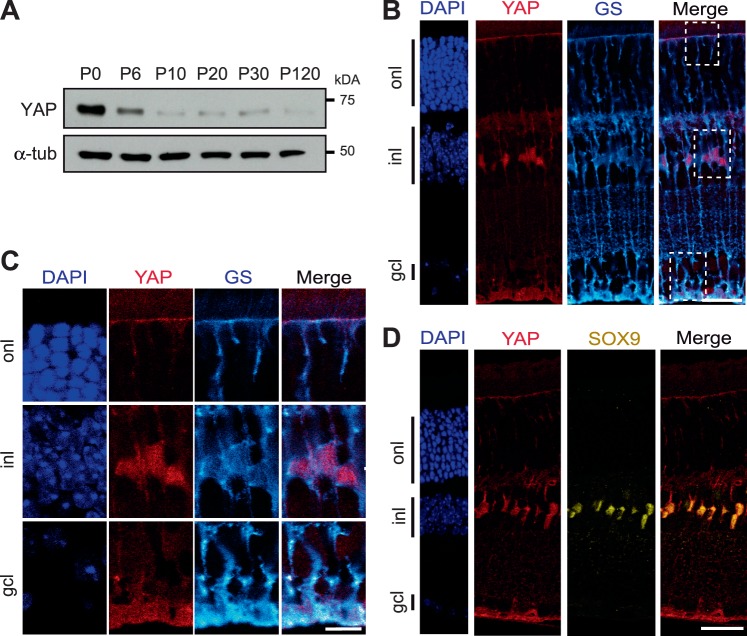
The finding that Hippo signaling effectors are upregulated at the transcriptional level in the INL of the degenerative retina prompted us to determine in which cells YAP is expressed in the adult retina, and if the upregulation could also be detected at the protein level. Western blot analysis was performed to evaluate the total amount of YAP protein in the retina at various postnatal time points from P0 to adult (Fig. 4A). A 65-kDa band corresponding to the predicted molecular weight of YAP was detected. As previously reported, a large amount of YAP protein was found in P0 retina, when retinal progenitors are still proliferating.29 Thereafter, the signal intensity strongly decreases up to P10, when all cells are postmitotic, and then remains at a steady-state level up to adulthood. To identify retinal cell types expressing YAP, we performed immunostaining on retinal sections of P60 WT mice. Although YAP is mostly known for being expressed in proliferative cells during development, we found YAP-positive cells in the INL, where postmitotic retinal cells reside. The presence of aligned and scattered labeled cells in the INL was already reported by our team in the Xenopus retina.28 Their position in this layer strongly suggests that YAP could be specifically expressed in Müller glial cells. This hypothesis was supported by our double staining analysis with anti-YAP and antiglutamine synthetase (GS, a Müller cell specific marker; Fig. 4B).50 Staining is detected in the nuclei as well as in the microvilli and the endfeet (Fig. 4C). The Müller cell specific expression was also confirmed by a colabeling with a nuclear marker of Müller cells, SOX9 (Fig. 4D).51 All YAP-positive cells were colabeled, demonstrating their Müller cell identity.
Figure 4.
YAP expression in Müller cells. (A) Representative western blots of retinal protein extracts from WT mice at different stages from P0 to P120 probed with anti-YAP antibody or anti-α-tubulin (α-tub) as a loading control. (B) Coimmunostaining with anti-YAP antibody (red) and anti-GS (cyan) antibody, on adult (P60) mouse retinal section. Nuclei are labeled with DAPI (blue). (C) Enlargement of framed areas (dashed lines) showing YAP expression in Müller cell microvilli (in the onl), nuclei (in the inl) and endfeet (in the gcl). (D) Colocalization of YAP immunostaining (red) with SOX9 (yellow). onl, outer nuclear layer; gcl, ganglion cell layer. Scale bars: 20 μm in B, D, and 10 μm in C.
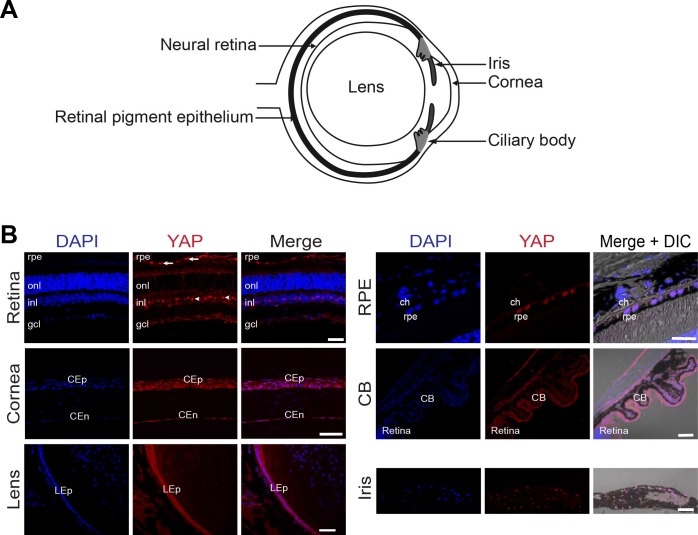
Of note, YAP immunolabeling was also detected in several nonneural ocular tissues, including the RPE, the cornea, the lens, the ciliary body, and the iris (Fig. 5). Expressed sequence tags (ESTs) retrieval from the BioGPS website supports this expression analysis.
Figure 5.
YAP expression in the adult mouse eye. (A) Schematic of a transverse section of an adult mouse eye, pointing to the different areas imaged in B. (B) Immunostaining with anti-YAP antibody (red) on adult (P60) eye sections. Different ocular regions are shown as indicated. Nuclei are DAPI counterstained (blue). Merge images plus phase contrast (DIC) are shown in the case of pigmented structures. YAP staining is detected in scattered cells within the INL and in the RPE but not in choroidal melanocytes (ch). YAP is also expressed in corneal epithelial (CEp) and corneal endothelial cells (CEd), in lens epithelial cells (LEp), in the ciliary body (CB), and the iris. onl, outer nuclear layer; gcl, ganglion cell layer. Scale bars: 50 μm except for RPE and iris where it is 20 μm.
YAP Expression in Müller Cells During Retinal Degeneration
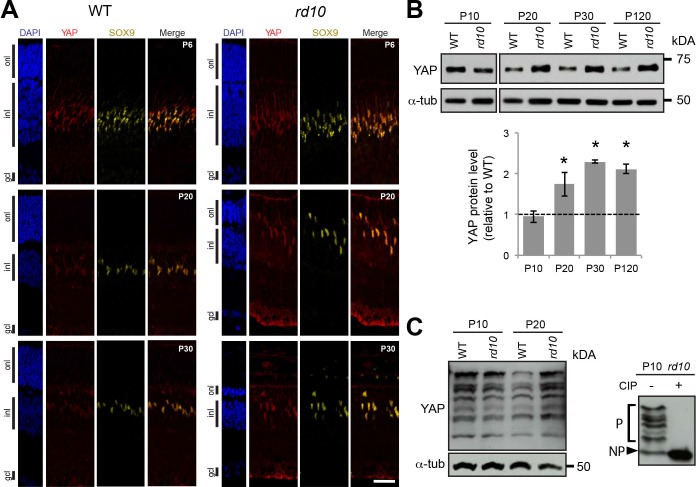
We next investigated whether the upregulation of Yap gene expression observed in our RNA-Seq and RT-qPCR analysis was also impacting YAP protein level and whether it was restricted in the retina to Müller glia cells. To address these questions, we performed immunostaining analysis and compared YAP expression in rd10 and WT mouse retina at different stages of photoreceptor degeneration (Fig. 6A). At P6, before the onset of photoreceptor cell death, YAP staining in Müller cells (SOX9-positive cells) was similar in rd10 and WT retinas (Fig. 6A). In contrast, we observed a stronger YAP staining intensity at P20 and P30 in the cytoplasm and the nucleus of Müller cells in rd10 retina compared to WT. This increase of YAP protein level correlates with a gliotic state of Müller cells, as inferred by immunostaining for GFAP, the gold standard marker of reactive gliosis (Supplementary Fig. S2).52 The increase in YAP protein levels in rd10 versus WT retina after the onset of retinal degeneration was strengthened by western blot analysis (Fig. 6B). A higher YAP protein level was maintained even after all photoreceptors died at P120.
Figure 6.
YAP expression during retinal degeneration. (A) Coimmunostaining with anti-YAP (red) and anti-SOX9 (yellow) antibodies on retinal sections of WT and rd10 mice at different stages from P6 to P30. Nuclei are DAPI counterstained (blue). (B) Representative western blots of retinal protein extracts from WT and rd10 mice at different stages from P10 to P120, probed with anti-YAP antibody or anti-α-tubulin (α-tub) as a loading control. Histogram representation of YAP quantification from western blot signals normalized to α-tubulin and relative to WT at each stage (dashed line). Mean values ± SEM from four independent western blot experiments are shown. Asterisk indicates P value ≤ 0.05 (Mann-Whitney U test). (C) Representative western blots of retinal protein extracts from WT and rd10 mice at P10 and P20 using Phos-tag gel and probed with anti-YAP antibody or anti-α-tubulin (α-tub; left panel). The shifted bands correspond to YAP phosphorylated isoforms since they are lost following Calf intestinal alkaline phosphatase (+CIP) treatment on rd10 P10 lysate (right panel). P, phosphorylated forms of YAP; NP, nonphosphorylated form of YAP; onl, outer nuclear layer; gcl, ganglion cell layer. Scale bars: 20 μm.
We wondered whether the protein level of YAP could be artificially increased because of the relative enrichment in inner retinal cells that occurs as photoreceptors die off in rd10 mice. α-tubulin may indeed not be a good reference protein if expressed in the entire retina. However, by immunostaining, we found that α-tubulin is mostly expressed in the INL/inner plexiform layer (IPL) and faintly in the photoreceptor layer (Supplementary Fig. S3), proving the pertinence of using this protein as a control, as previously shown for β-tubulin.53 To definitely assess the validity of our quantification approach, we also normalized YAP level with two INL-specific proteins, calbindin-D-28K (a marker of bipolar cells) and PKCα (a marker of horizontal cells). As with α-tubulin normalization, we found using these two markers as loading controls that YAP protein level in the retina is also significantly increased in rd10 compare to WT retinas (Supplementary Fig. S3).
Posttranslational modifications have been shown to play a critical role in the regulation of YAP activity. For instance, phosphorylation events by the Hippo pathway result in YAP cytoplasmic localization (cytoplasmic retention or poly-ubiquitination and degradation).19 To determine whether the upregulation of YAP in reactive Müller cells is associated with changes in its phosphorylation status, we performed Phos-tag-based western blots (Fig. 6C). This method allows greater separation of phosphorylated and nonphosphorylated protein and facilitates analysis at the isoform level.54 The efficiency of the method was validated following a phosphatase treatment. As expected in such conditions, only one form of nonphosphorylated YAP was detected while multiple bands are present in the control, reflecting various levels of phosphorylation. We observed that phosphorylated as well as nonphosphorylated forms of YAP are all increased at P20 in rd10 retina compared to WT retina. These data suggest that YAP phosphorylation overall profile is not markedly affected in reactive Müller cells.
In order to assess whether the increase in YAP expression also occurs in another degenerative context, we used the MNU-induced model, which is known to cause photoreceptor degeneration and reactive gliosis.55 In this model, we observed an increased level of YAP expression 72 hours after MNU injection, by both immunostaining and western blot analysis (Supplementary Fig. S4).
Altogether, using both a genetic and a chemical model of retinal degeneration, our results indicate that YAP level increases in reactive Müller cells upon photoreceptor degeneration.
TEAD1 Expression in Müller Cells During Retinal Degeneration
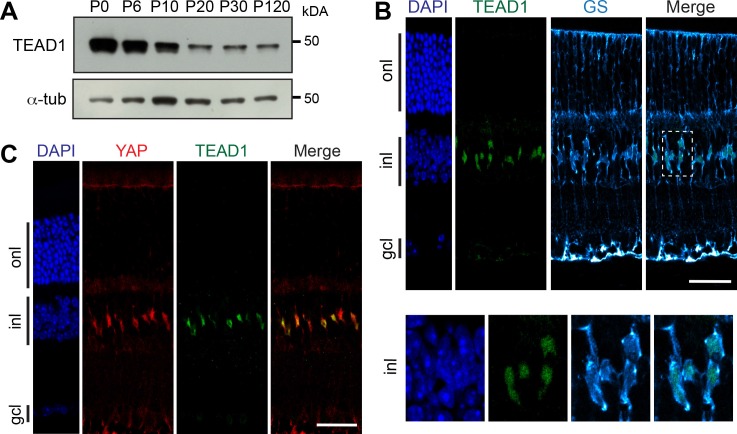
We next asked whether TEAD proteins were also expressed in adult Müller cells. By western blot, we observed that the level of TEAD1 protein in the retina decreases after P6, but that a band remains detectable at a roughly steady-state level from P10 to P120 (Fig. 7A), as previously shown for YAP (Fig. 4A). TEAD1 protein expression was restricted to Müller cells in adult retina, as demonstrated by coimmunostaining with GS antibody (Fig. 7B). We also confirmed that YAP and TEAD1 are coexpressed in Müller cell nuclei (Fig. 7C).
Figure 7.
TEAD1 expression in Müller cells. (A) Representative western blots of retinal protein extracts from WT mice at different stages from P0 to P120, probed with anti-TEAD1 antibody or anti-α-tubulin (α-tub) as a loading control. (B) Coimmunostaining with anti-TEAD1 (green) and anti-GS (cyan) on adult mouse (P60) retinal section. Nuclei are DAPI counterstained (blue). The framed area (dashed line) is enlarged showing TEAD1 expression in GS-labeled nuclei. (C) Coimmunostaining with anti-YAP (red) and anti-TEAD1 (green) antibodies on adult mouse retinal section. Nuclei are DAPI counterstained (blue). onl, outer nuclear layer; gcl, ganglion cell layer. Scale bars: 20 μm.
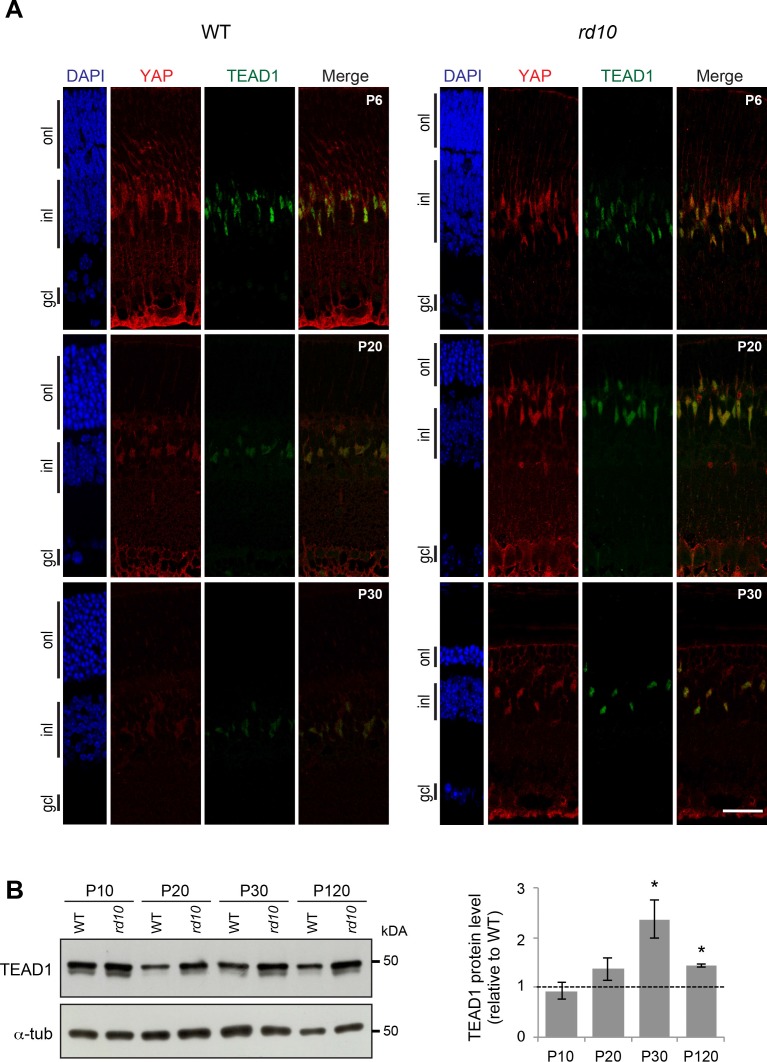
In order to assess whether TEAD1 protein level was affected in reactive Müller cells, we performed western blot and immunostaining analysis and compared TEAD1 expression levels between rd10 and WT retina. Similar to what was observed for YAP, we found that TEAD1 level was equivalent in rd10 and WT retina at P10 but was clearly upregulated in reactive Müller cells after the onset of photoreceptor degeneration (Figs. 8A, 8B). Similarly, in MNU-induced retinal degeneration model, we found a twofold increase of TEAD1 protein level in degenerating retinas compared to controls (Supplementary Fig. S5).
Figure 8.
TEAD1 expression during retinal degeneration. (A) Coimmunostaining with anti-YAP (red) and anti-TEAD1 (green) antibodies on retinal section of WT and rd10 mice at P20. Nuclei are DAPI counterstained (blue). (B) Representative western blots of retinal protein extracts from WT and rd10 mice at different stages from P10 to P120, probed with anti-TEAD1 antibody or anti-α-tubulin as a loading control. Histogram representation corresponds to the quantification of TEAD1 western blot signals normalized to α-tubulin (α-tub) and relative to WT at each stage (dashed line). Mean values ± SEM from two independent western blot experiments are shown. Asterisk indicates P value ≤ 0.05 (Mann-Whitney U test). onl, outer nuclear layer; gcl, ganglion cell layer. Scale bar: 20 μm.
Discussion
This study revealed profound changes in the expression of Hippo pathway components in degenerating retina. Specifically, we found that (1) YAP and TEAD1 are exclusively expressed in Müller cells in the adult neural retina; (2) the increase in their expression at the transcriptional and protein level strongly correlates with reactive gliosis occurring in Müller cells upon photoreceptor loss; (3) several other components of the Hippo pathway, including two of YAP/TEAD target genes Ctgf and Cyr61, are also upregulated in a mouse model of retinal degeneration; and (4) many of these genes are at the crossroad of other key signaling pathways.
We found that YAP is specifically expressed in Müller cells of the adult retina. Although they do not regenerate spontaneously in mammals, reactive Müller cells initiate a reprogramming event into progenitor-like cells through the upregulation of stem cell and cell cycle genes upon injury (upregulation of markers such as Nestin, Pax6, Cyclin D1, Cyclin D3, and Ki-678,9,56,57). Moreover, this limited mammalian retina regeneration can be stimulated by exogenous growth factors.11,58–62 Given these properties of Müller cells and the already described function of YAP in stem cells and regeneration, it is tempting to speculate that YAP may contribute to provide some stem-like characteristics to these glial cells. Indeed, the Hippo pathway has been proposed to act in numerous types of stem cells in a variety of organisms.63–65 YAP in particular was found enriched in multiple mouse stem and progenitor cells, including embryonic stem cells (ESCs).66 YAP overexpression was reported to regulate mouse ESCs self-renewal, to increase the efficiency of mouse induced pluripotent stem cells (iPS) generation, and more recently to promote the generation of naive human pluripotent stem cells.67–69 YAP also regulates the proliferation of stem and progenitor cells in a variety of organs including the intestine, liver, skin, retina, and brain28,70–72 and is involved in adult tissue repair and regeneration.20–22,73–75 Whether YAP could induce regenerative activity in Müller cells remains, however, to be investigated in species with regenerative properties such as zebrafish.
We found that the Hippo pathway, typically associated with growth control, is deregulated in a degenerating retina despite the absence of active proliferation. This might actually not be so surprising as Hippo signaling also emerged as a cellular stress response, important for maintaining cell and tissue homeostasis.15 What triggers the upregulation of multiple components of the pathway, in particular YAP/TEAD complex, following photoreceptor cell death? Several studies in mammalian cells revealed that YAP expression and subcellular distribution are regulated by mechanical cues, such as the stiffness of the extracellular matrix.76–79 For instance, substratum stiffness was shown to modulate the expression of YAP in human trabecular meshwork cells, known to stiffen in patients with glaucoma.80 Rearrangement of the cytoskeleton following photoreceptor loss in the retina may thus trigger, at least in part, the changes in YAP expression that we observed in Müller cells in both rd10 and MNU models.
Multiple lines of evidence also linked the Hippo pathway with oxidative stress.15,81,82 It is, however, difficult to draw a clear conclusion as studies on the effects of accumulated reactive oxygen species (ROS) on Hippo signaling are conflicting.81,83 Whether ROS in the degenerative retina is part of the triggering signal leading to Hippo pathway gene expression modulation is an attractive hypothesis that remains to be investigated.
Another related question is whether YAP increase in reactive Müller cells could mediate a cellular defense against oxidative stress. Indeed, several studies revealed that YAP has the ability to promote the expression of genes encoding proteins with antioxidant properties, resulting in decreased cellular ROS.20,84,85 In Drosophila, Hippo signaling has also recently been implicated in the transduction of cellular survival signals in response to chemical stress.86 Along this line, the secreted protein CYR61, a well-known direct target of YAP, which we found upregulated in reactive Müller cells, has recently been identified as a neuroprotective agent in organotypic retinal cultures of a mouse model of retinitis pigmentosa.87 It would thus be interesting to assess whether Cyr61 is a direct target of YAP in the retina, and thus whether Cyr61 upregulation in the rd10 model is a direct consequence of YAP upregulation. The aim would then be to investigate in vivo whether such YAP-Cyr61 axis in Müller cells has neuroprotective capacities in a degenerating retina. Regarding the other well-known YAP target gene Ctgf, it has been detected in the retina and was shown to be expressed by Müller cells. It was reported to induce the angio-fibrotic switch in diabetic retinopathy and to be involved in remodeling of the extracellular matrix in glaucoma.88–90
Our RNA-Seq analysis revealed that many of the deregulated genes of the Hippo pathway in rd10 retina are also related to other key signaling pathways, consistent with the idea that Hippo/YAP acts as a central node integrating a variety of signals. It is indeed well recognized that Hippo signaling establishes crosstalks with pathways such as Wnt, TGFβ, mTOR, or Notch.91–95 Among these pathways, some have been linked to retinal pathologies. For instance, uncontrolled Wnt signaling may cause familial exudative vitreoretinopathy, retinitis pigmentosa, and Norrie's disease.96 The mTOR pathway is affected during cone degeneration in retinitis pigmentosa and its inhibition results in the loss of red/green opsin.97 Therefore, it will be important to further explore the relationships in the retina between these pathways and Hippo signaling components identified in the present study.
Conclusions
As a whole, our work revealed differential expression of Hippo pathway components in a retinal degenerative model, which warrants further investigation to unveil its significance in terms of Müller cell reactivation and photoreceptor neuroprotection in diseased retina.
Supplementary Material
Acknowledgments
The authors thank Eric Jacquet and Prishila Ponien for their help with the RT-qPCR (Plateform qPCR of the ICSN, CNRS). They also thank Elodie-Kim Grellier for her help with the maintenance of mouse colonies and Megan Barnes for her technical assistance. The authors also thank Thierry Léveillard for providing the access to KBaSS database.
Supported by grants to MP from the FRM, Idex Paris-Saclay, Association Retina France and Fondation Valentin Haüy. They acknowledge NEI intramural research program for support as well as Anand Swaroop and the Neurobiology Neurodegeneration & Repair Laboratory (NNRL) for their support on whole transcriptome sequencing. AH was a Retina France association and an ARC fellow.
RNA-Seq data reported in this paper has been submitted to Gene Expression Omnibus (GEO), accession number GSE94534.
Disclosure: A. Hamon, None; C. Masson, None; J. Bitard, None; L. Gieser, None; J.E. Roger, None; M. Perron, None
References
- 1. Reichenbach A, Bringmann A. . New functions of Müller cells. Glia. 2013; 61: 651– 678. [DOI] [PubMed] [Google Scholar]
- 2. Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC. . Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. 2016; 51: 1– 40. [DOI] [PubMed] [Google Scholar]
- 3. Shen W, Fruttiger M, Zhu L,et al. Conditional Müller cell ablation causes independent neuronal and vascular pathologies in a novel transgenic model. J Neurosci. 2012; 32: 15715– 15727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byrne LC, Khalid F, Lee T,et al. AAV-mediated, optogenetic ablation of Müller glia leads to structural and functional changes in the mouse retina. PLoS One. 2013; 8: 1– 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bringmann A, Iandiev I, Pannicke T,et al. Cellular signaling and factors involved in Müller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009; 28: 423– 451. [DOI] [PubMed] [Google Scholar]
- 6. Belecky-Adams TL, Chernoff EC, Wilson JM, Dharmarajan S. . Reactive Müller glia as potential retinal progenitors. Neural Stem Cells New Perspect. 2013: 75. [Google Scholar]
- 7. Bringmann A, Pannicke T, Grosche J,et al. Müller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006; 25: 397– 424. [DOI] [PubMed] [Google Scholar]
- 8. Goldman D. . Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014; 15: 431– 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamon A, Roger JE, Yang XJ, Perron M. . Müller glial cell-dependent regeneration of the neural retina: an overview across vertebrate model systems. Dev Dyn. 2016: 1– 12. [DOI] [PMC free article] [PubMed]
- 10. Lenkowski JR, Raymond P. . Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014; 40 January: 94– 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh T. . Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A. 2008; 105: 19508– 19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wan J, Ramachandran R, Goldman D. . HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012; 22: 334– 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zelinka CP, Volkov L, Goodman ZA,et al. mTor-signaling is required for the formation of proliferating Muller glia-derived progenitor cells in the chick retina. Development. 2016; April: 1859– 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varelas X. . The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014; 141: 1614– 1626. [DOI] [PubMed] [Google Scholar]
- 15. Mao B, Gao Y, Bai Y, Yuan Z. . Hippo signaling in stress response and homeostasis maintenance. Acta Biochim Biophys Sin (Shanghai). 2015; 47: 2– 9. [DOI] [PubMed] [Google Scholar]
- 16. Yu FX, Zhao B, Guan KL. . Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015; 163: 811– 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meng Z, Moroishi T, Guan K-L. . Mechanisms of Hippo pathway regulation. Genes Dev. 2016; 30: 1– 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu C, Li L, Zhao B. . The regulation and function of YAP transcription co-activator. Acta Biochim Biophys Sin (Shanghai). 2014; 47: 16– 28. [DOI] [PubMed] [Google Scholar]
- 19. Zhao B, Li L, Lei Q, Guan KL. . The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010; 24: 862– 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Del Re DP, Yang Y, Nakano N,et al. Yes-associated protein isoform 1 (Yapl) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 2013; 288: 3977– 3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grijalva JL, Huizenga M, Mueller K,et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2014; 307: G196– G204. [DOI] [PubMed] [Google Scholar]
- 22. Johnson R, Halder G. . The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014; 13: 63– 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi S, Tamura K, Yokoyama H. . Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Dev Biol. 2014; 388: 57– 67. [DOI] [PubMed] [Google Scholar]
- 24. Hayashi S, Ochi H, Ogino H,et al. Transcriptional regulators in the Hippo signaling pathway control organ growth in Xenopus tadpole tail regeneration. Dev Biol. 2014; 396: 31– 41. [DOI] [PubMed] [Google Scholar]
- 25. Jiang Q, Liu D, Gong Y,et al. Yap is required for the development of brain, eyes, and neural crest in zebrafish. Biochem Biophys Res Commun. 2009; 384: 114– 119. [DOI] [PubMed] [Google Scholar]
- 26. Asaoka Y, Hata S, Namae M, Furutani-Seiki M, Nishina H. . The Hippo pathway controls a switch between retinal progenitor cell proliferation and photoreceptor cell differentiation in zebrafish. PLoS One. 2014; 9 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miesfeld JB, Gestri G, Clark BS,et al. Yap and Taz regulate retinal pigment epithelial cell fate. Development. 2015; 142: 3021– 3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabochette P, Vega-Lopez G, Bitard J,et al. Yap controls retinal stem cell DNA replication timing and genomic stability. Elife. 2015; 4(September 2015): 1– 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H, Deo M, Thompson RC, Uhler MD, Turner DL. . Negative regulation of Yap during neuronal differentiation. Dev Biol. 2012; 361: 103– 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williamson KA, Rainger J, Floyd JAB,et al. Heterozygous loss-of-function mutations in YAP1 cause both isolated and syndromic optic fissure closure defects. Am J Hum Genet. 2014; 94: 295– 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H. . Structural insights into the YAP and TEAD complex service. Genes Dev. 2010: 235– 240. [DOI] [PMC free article] [PubMed]
- 32. Fossdal R, Jonasson F, Kristjansdottir GT,et al. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration). Hum Mol Genet. 2004; 13: 975– 981. [DOI] [PubMed] [Google Scholar]
- 33. Hao H, Kim DS, Klocke B,et al. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012; 8: e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C, Pachter L, Salzberg SL. . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25: 1105– 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anders S, Pyl PT, Huber W. . HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31: 166– 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson MD, McCarthy DJ, . Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26: 139– 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brooks MJ, Rajasimha HK, Roger JE, Swaroop A. . Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes. Mol Vis. 2011; 17: 3034– 3054. [PMC free article] [PubMed] [Google Scholar]
- 38. Vandesompele J, De Preter K, Pattyn F,et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grove M, Kim H, Santerre M,et al. YAP/TAZ initiate and maintain Schwann cell myelination. eLife. 2017; 6: 1– 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X, Li H, Rajurkar M,et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016; 14: 1169– 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang B, Hawes NL, Pardue MT,et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 2007; 47: 624– 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Phillips MJ, Otteson DC, Sherry DM. . Progression of neuronal and synaptic remodeling in the rd 10 mouse model of retinitis pigmentosa. J Comp Neurol. 2010; 518: 2071– 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. . Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A. 1995; 92 April: 3249– 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uren PJ, Lee JT, Doroudchi MM, Smith AD, Horsager A. . A profile of transcriptomic changes in the rd10 mouse model of retinitis pigmentosa. Mol Vis. 2014; 20: 1612– 1628. [PMC free article] [PubMed] [Google Scholar]
- 45. Mears J, Kondo M, Swain PK,et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001; 29: 447– 452. [DOI] [PubMed] [Google Scholar]
- 46. Zhao B, Ye X, Yu J,et al. TEAD mediates YAP-dependent gene induction and growth control TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008: 1962– 1971. [DOI] [PMC free article] [PubMed]
- 47. Lai D, Ho KC, Hao Y, Yang X. . Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011; 71: 2728– 2738. [DOI] [PubMed] [Google Scholar]
- 48. Farber DB, Flannery JG, Bowes-Rickman C. . The rd mouse story: seventy years of research on an animal model of inherited retinal degeneration. Prog Retin Eye Res. 1994; 12: 31– 64. [Google Scholar]
- 49. Aït-Ali N, Fridlich R, Millet-Puel G,et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 2015; 161: 817– 832. [DOI] [PubMed] [Google Scholar]
- 50. Riepe RE, Norenberg MD. . Müller cell localization of glutamine synthetase in rat retina. Nature. 1977; 268: 654– 655. [DOI] [PubMed] [Google Scholar]
- 51. Muto A, Iida A, Satoh S, Watanabe S. . The group E Sox genes Sox8 and Sox9 are regulated by Notch signaling and are required for Müller glial cell development in mouse retina. Exp Eye Res. 2009; 89: 549– 558. [DOI] [PubMed] [Google Scholar]
- 52. Gargini C, Terzibasi E, Mazzoni F, Strettoi E. . Retinal organization in the retinal degeneration 10 (rd10) mutant mouse: a morphological and ERG study. J Comp Neurol. 2007; 500: 222– 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Srivastava P, Sinha-mahapatra SK, Ghosh A, Srivastava I. . Differential alterations in the expression of neurotransmitter receptors in inner retina following loss of photoreceptors in rd1 mouse. PLoS One. 2015; 10: e0123896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. . Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006; 5: 749– 757. [DOI] [PubMed] [Google Scholar]
- 55. Chen YY, Liu SL, Hu DP, Xing YQ, Shen Y. . N-methyl-N-nitrosourea-induced retinal degeneration in mice. Exp Eye Res. 2014; 121: 102– 113. [DOI] [PubMed] [Google Scholar]
- 56. Suga A, Sadamoto K, Fujii M, Mandai M, Takahashi M. . Proliferation potential of Müller glia after retinal damage varies between mouse strains. PLoS One. 2014; 9: e94556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kohno H, Sakai T, Kitahara K. . Induction of nestin, Ki-67, and cyclin D1 expression in Müller cells after laser injury in adult rat retina. Graefe's Arch Clin Exp Ophthalmol. 2006; 244: 90– 95. [DOI] [PubMed] [Google Scholar]
- 58. Ooto S, Akagi T, Kageyama R,et al. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004; 101: 13654– 13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Close JL, Liu J, Gumuscu B, Reh TA. . Epidermal growth factor receptor expression regulates proliferation in the postnatal rat retina. Glia. 2006; 54: 94– 104. [DOI] [PubMed] [Google Scholar]
- 60. Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. . Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007; 27: 4210– 4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fischer AJ, Bongini R. . Turning Müller glia into neural progenitors in the retina. Mol Neurobiol. 2010; 42: 199– 209. [DOI] [PubMed] [Google Scholar]
- 62. Löffler K, Schäfer P, Völkner M, Holdt T, Karl MO. . Age-dependent Müller glia neurogenic competence in the mouse retina. Glia. 2015; 63: 1809– 1824. [DOI] [PubMed] [Google Scholar]
- 63. Ramos A, Camargo FD. . The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 2012; 22: 339– 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hiemer SE, Varelas X. . Stem cell regulation by the Hippo pathway. Biochim Biophys Acta - Gen Subj. 2013; 1830: 2323– 2334. [DOI] [PubMed] [Google Scholar]
- 65. Yin MX, Zhang L. . Hippo signaling in epithelial stem cells. Acta Biochim Biophys Sin (Shanghai). 2014; 47: 39– 45. [DOI] [PubMed] [Google Scholar]
- 66. Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton D. . “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002; 298: 597– 600. [DOI] [PubMed] [Google Scholar]
- 67. Tamm C, Böwer N, Annerén C. . Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011; 124: 1136– 1144. [DOI] [PubMed] [Google Scholar]
- 68. Lian I, Kim J, Okazawa H,et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010; 24: 1106– 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qin H, Hejna M, Liu Y,et al. YAP induces human naive pluripotency. Cell Rep. 2016; 14: 2301– 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Camargo FD, Gokhale S, Johnnidis JB,et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007; 17: 2054– 2060. [DOI] [PubMed] [Google Scholar]
- 71. Avruch J, Zhou D, Fitamant J, Bardeesy N. . Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011; 104: 24– 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cao X, Pfaff SL, Gage FH. . YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008; 22: 3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bai H, Zhang N, Xu Y,et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012; 56: 1097– 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cai J, Zhang N, Zheng Y, De Wilde RF, Maitra A, Pan D. . The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010; 24: 2383– 2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. . Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature. 2015; 526: 715– 718. [DOI] [PubMed] [Google Scholar]
- 76. Dupont S, Morsut L, Aragona M,et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011; 474: 179– 183. [DOI] [PubMed] [Google Scholar]
- 77. Wada K-I, Itoga K, Okano T, Yonemura S, Sasaki H. . Hippo pathway regulation by cell morphology and stress fibers. Development. 2011; 138: 3907– 3914. [DOI] [PubMed] [Google Scholar]
- 78. Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. . Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012; 26: 54– 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. . YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014; 588: 2663– 2670. [DOI] [PubMed] [Google Scholar]
- 80. Thomasy SM, Morgan JT, Wood JA, Murphy CJ, Russell P. . Substratum stiffness and latrunculin B modulate the gene expression of the mechanotransducers YAP and TAZ in human trabecular meshwork cells. Exp Eye Res. 2013; 113: 66– 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lehtinen MK, Yuan Z, Boag PR,et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006; 125: 987– 1001. [DOI] [PubMed] [Google Scholar]
- 82. Ashraf A, Pervaiz S. . Hippo circuitry and the redox modulation of hippo components in cancer cell fate decisions. Int J Biochem Cell Biol. 2015; 69: 20– 28. [DOI] [PubMed] [Google Scholar]
- 83. Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, Igaki T. . Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012; 490: 547– 551. [DOI] [PubMed] [Google Scholar]
- 84. Wu H, Xiao Y, Zhang S,et al. The ETS transcription factor GABP is a component of the Hippo pathway essential for growth and antioxidant defense. Cell Rep. 2013; 3: 1663– 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shao D, Zhai P, Del Re DP,et al. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat Commun. 2014; 5: 3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Di Cara F, Maile TM, Parsons BD,et al. The Hippo pathway promotes cell survival in response to chemical stress. Cell Death Differ. 2015; 22: 1526– 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kucharska J, Del Río P, Arango-Gonzalez B,et al. Cyr61 activates retinal cells and prolongs photoreceptor survival in rd1 mouse model of retinitis pigmentosa. J Neurochem. 2014; 130: 227– 240. [DOI] [PubMed] [Google Scholar]
- 88. van Setten GB, Trost A, Schrödl F,et al. Immunohistochemical detection of CTGF in the human eye. Curr Eye Res. 2016; 41: 1571– 1579. [DOI] [PubMed] [Google Scholar]
- 89. Winkler JL, Kedees MH, Guz Y, Teitelman G. . Inhibition of connective tissue growth factor by small interfering ribonucleic acid prevents increase in extracellular matrix molecules in a rodent model of diabetic retinopathy. Mol Vis. 2012; 18: 874– 886. [PMC free article] [PubMed] [Google Scholar]
- 90. Junglas B, Kuespert S, Seleem AA,et al. Connective tissue growth factor causes glaucoma by modifying the actin cytoskeleton of the trabecular meshwork. Am J Pathol. 2012; 180: 2386– 2403. [DOI] [PubMed] [Google Scholar]
- 91. Barry ER, Camargo FD. . The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol. 2013; 25: 247– 253. [DOI] [PubMed] [Google Scholar]
- 92. Zhou D, Zhang Y, Wu H,et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci. 2011; 108: E1312– E1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. . A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012; 31: 1109– 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Saito A, Nagase T. . Hippo and TGF-β interplay in the lung field. Am J Physiol Lung Cell Mol Physiol. 2015; 309: L756– L767. [DOI] [PubMed] [Google Scholar]
- 95. Tumaneng K, Schlegelmilch K, Russell RC,et al. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012; 14: 1322– 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lad EM, Cheshier SH, Kalani MYS. . Wnt-signaling in retinal development and disease. Stem Cells Dev. 2009; 18: 7– 16. [DOI] [PubMed] [Google Scholar]
- 97. Punzo C, Kornacker K, Cepko CL. . Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009; 12: 44– 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.