Abstract
Vaccination is one of the most successful immunology applications that has considerably improved human health. The DNA vaccine is a new vaccine being developed since the early 1990s. Although the DNA vaccine is promising, no human DNA vaccine has been approved to date. The main problem facing DNA vaccine efficacy is the lack of a DNA vaccine delivery system. Several studies explored this limitation. One of the best DNA vaccine delivery systems uses a live bacterial vector as the carrier. The live bacterial vector induces a robust immune response due to its natural characteristics that are recognized by the immune system. Moreover, the route of administration used by the live bacterial vector is through the mucosal route that beneficially induces both mucosal and systemic immune responses. The mucosal route is not invasive, making the vaccine easy to administer, increasing the patient’s acceptance. Lactic acid bacterium is one of the most promising bacteria used as a live bacterial vector. However, some other attenuated pathogenic bacteria, such as Salmonella spp. and Shigella spp., have been used as DNA vaccine carriers. Numerous studies showed that live bacterial vectors are a promising candidate to deliver DNA vaccines.
Keywords: lactic acid bacteria, antigen expression, oral delivery, carrier
1. Introduction
The DNA vaccine is a new vaccine with a bacterial plasmid as the antigen gene vector. Typically, the gene is expressed under host cell promoter regulation, and the expressed antigen is targeted to induce the host immune response [1]. DNA vaccine development is based on its advantages compared with conventional vaccines, such as the ability to induce a cellular immune response instead of only a humoral immune response [2]. The expressed antigen could be designed as an extracellular protein, which is detected by Major Histocompability Complex (MHC) class II or an intracellular protein that is recognized by MHC class I. DNA plasmid is also stable at room temperature, facilitating its production and distribution. DNA vaccine production does not directly use the pathogen organism, which is safer for the production system [1,2].
Some concerns about the DNA vaccine include potential genomic integration and auto-immune response [3]. However, extensive research found that the risk of integration is limited and significantly lower than the natural mutation rate [3,4,5]. Clinical trials also showed mild side effects after DNA vaccination [6,7,8].
Although promising, the DNA vaccine needs a superior delivering system to activate the potent immune system [9]. The DNA vaccine requires a large dose to effectively induce the immune system response [10]. At least 1–100 μg of DNA vaccine is needed to induce immune response [11]. Although some naked DNAs demonstrated efficacy [12,13,14], several delivery strategies have been studied to increase DNA vaccine efficiency. Chemical delivery systems, such as using micro particles [11,15] and nanoparticles [16,17], have successfully increased DNA vaccine delivery. Another potential delivery system to improve DNA vaccine delivery is the use of a live bacterial vector as the carrier.
Since being developed in the late 1970s, the idea of using live bacteria as the DNA carrier has been growing rapidly. The technique is also known as bactofection, in which the live bacteria are directly used to transfer the DNA to the target cells, tissues, or organs [18]. Bactofection has been widely used in the field of drug development research [19], such as cancer treatment [20], infections [21], inflammation diseases and other metabolic diseases [22,23]. As a DNA vaccine carrier, both native [24] or recombinant bacteria can be used. The live bacterial vector does not only deliver DNA inside the host cell but also induces a potent immune response due to its immunogenic features [25,26]. In this review, an overview of live bacterial vectors as DNA vaccine carriers and future prospects in this field is provided.
2. DNA Vaccine Components
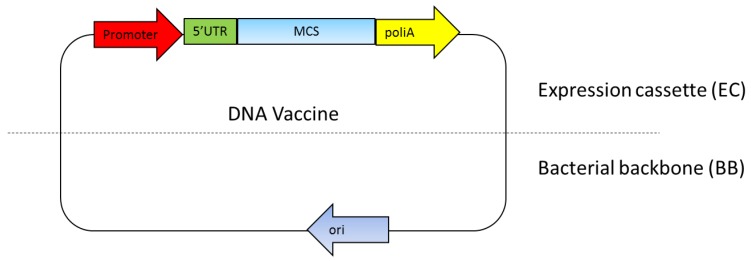
The DNA vaccine essentially includes two main parts: a mammalian expression cassette and bacterial backbone [27]. The mammalian expression cassette consists of a eukaryotic promoter for gene expression, 5′ untranslated region (5′UTR) including an intron and polyadenilation sequence (polyA) (Figure 1). The bacterial backbone consists of bacterial origin of replication (Ori) and an antibiotic resistance gene or other selection markers (Figure 1). Ideally, the multiple cloning sites (MCS) used to insert the target gene are located between the mammalian expression cassette and the bacterial backbone [27,28,29]. The mammalian expression cassette should be optimized so that the antigen gene is highly expressed to effectively generate the immune response [29]. The bacterial backbone should also be optimized so that a high yield of DNA plasmid can be produced with the fermentation production process [27,30]. The newest DNA vaccine version combines a component in the eukaryotic expression cassette while minimizing the bacterial backbone components, since it reduces antigen expression level [27,31].
Figure 1.
DNA vaccine components. The essential components in the DNA vaccines consist of a eukaryotic promoter, a multiple cloning site (MCS), a polyadenylation site (polyA), a selection marker and a bacterial origin of replication (ori). 5’UTR: 5′ untranslated region.
The Escherichia coli Ori ColE1 found, for instance, as OriV of the pUC vector, is still a prominent choice of bacterial origin of replication because of its high copy number of up to 500–700 copies per bacterial cell [32]. This OriV was used in the early generation DNA vaccines, such as in pVAK1 to the latest generation of DNA vaccines, such as NTC8385 [33]. This suggests that this OriV is still considered as an ideal ori for DNA vaccines [29,30].
Selectable markers are required for the maintenance of the plasmid inside the cells. Only the cells that contain plasmids with the appropriate selectable marker can survive under the selective conditions. Most DNA plasmids are based on antibiotic resistance genes as the selectable markers. However, use of antibiotic resistance genes has resulted in health concerns being expressed, such as the spreading of the resistance genes and the effect on the microbiota in the host system [34]. Thus, the European Medicines Agency (MEA, London, UK) recommends non-antibiotic resistance genes as the selection marker. The antibiotic resistance gene is replaced with other marker selections based on an auxotrophic strain, toxin-antitoxin systems, operator-repressor titration, RNA markers, or the minicircles [31,35,36,37].
The promoter recognized by the mammalian expression system plays significant roles in antigen gene expression. Cytomegalovirus (CMV), Simian virus 40 (SV40), and murine leukemia virus promoters are among the most prominent promoters used in DNA vaccines [38,39]. Although CMV promoter activity decreases under some conditions, such as when used in conjunction with cytokine treatment [29], the combination of CMV promoter with Intron A showed that the CMV promoter effectively activates gene expression. Some studies also combined a CMV promoter with other components, such as a modified chicken β-actin [39] or woodchuck hepatitis post-transcriptional regulatory element (WPRE), to create a hybrid promoter [40]. Other non-viral promoters have been demonstrated to have comparable efficacy to the CMV promoter, such as collagen [41] and keratinocytes [42] promoters. Another study indicated that the MHC class II promoter potently generated a transgene product that can be used in the DNA vaccine [43].
The polyadenylation sequence has a significant effect on transgene expression. The common polyadenylation sequences used in DNA vaccine construction are SV40, rabbit β-globin, and bovine growth hormone polyadenylation sequence [29,44]. 5′UTR is located upstream transgene and regulates transgene translation. Optimization of the regulatory element by inserting a sequence from the R region of the long terminal repeat from human T-cell leukemia virus type 1 (HTLV-1) to CMV enhancer/promoter markedly increased DNA vaccine immunogenicity in both mice and non-human primates [45].
3. Live Bacterial Vector as the DNA Vaccine Carrier
As a DNA vaccine delivery system, the live bacterial vector has several benefits. The bacteria typically used for a delivery system are recombinant bacteria that have been genetically modified so most of their pathogenicity components have been deleted to attenuate the bacteria and create a non-virulent organism, ensuring the safety of the host [26]. By using bacteria as the carrier, the vaccination can be delivered through mucosal routes, including intranasal, oral, or intravaginal routes. The mucosal route is favorable because it is non-invasive and more acceptable. Administration through oral routes also does not require special skills and is easier to manage. Vaccination through mucosal routes induces both mucosal and systemic immune system responses [46,47]. Bacteria protect the DNA vaccine from harsh environments and enzymatic reactions in the gut [48]. The intranasal route has also been thoroughly developed since it can hinder enzymatic reactions and withstand the high acidity conditions in the gut. Other studies showed that vaccines administered through the intranasal route induce the same or better immune response compared with oral route vaccinations [47,49].
The mucosal surface is the first location where the host and its environment contact; therefore, it has a prominent defense mechanism against pathogens. Mucosal route vaccine delivery systems are based on mucosa-associated lymphoid tissue (MALT), which is found on various mucosal surface areas. MALT is a lymphoid tissue in the nasopharynx, pharynx, salivary gland, and upper respiratory tract, which are known as nasal-associated lymphoid tissues (NALT). MALT is also found in the broncho epithelium and lower respiratory tract (BALT), gastrointestinal tract (GALT) and genital tract. MALT is composed of epithelial cells identified as follicle-associated epithelium or microfold cells (M cells) that act as the first mucosal barrier system and initiate the immune response [50].
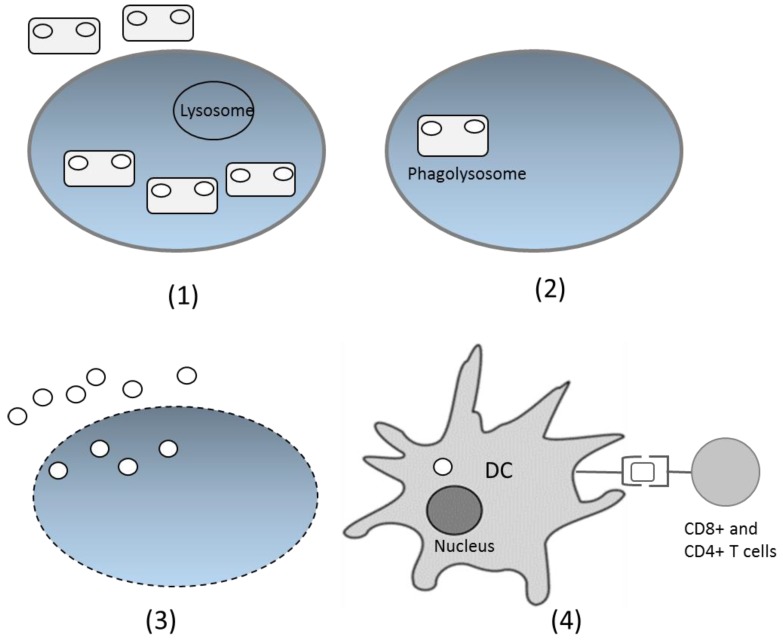
After being delivered through the oral route, bacteria with the DNA vaccine enter the digestion system. On the intestinal surface, the bacteria are recognized by M cells in Peyer’s patches and spread to the lamina propia [48]. The bacteria have specific characteristics in the form of microbe-associated molecular patterns (MAMPs), recognized by particular receptors, such as Toll-like receptors and Nod-like receptors. This introduction induces the native immune response and increases the adaptive immune response. Phagocytized bacteria shape phagolysosomes and trigger cell lysis, which further releases plasmids inside the bacteria. Cell components, including plasmids, are released and engulfed by dendritic cells (DCs) [51]. Inside DCs, plasmids enter the nucleus through special compartments, and the antigen gene is expressed by the host expression system. The expressed antigen is presented by class I MHC and activates the CD8+ T cells. The antigen can also be expressed as extracellular protein, presented by class II MHC, and activates antibody production and the T helper CD4+ cell response [25,52] (Figure 2). However, the precise mechanism of DNA transfer by live bacterial vectors is not yet fully understood for many species. The suggested mechanism is based on the bacterial invasion properties [53]. Thus, some invasive bacteria, such as Salmonella typhimurium and Listeria monocytogenes are preferable carriers for DNA vaccines.
Figure 2.
Proposed DNA vaccine delivery system using a live bacterial vector. (1) Bacteria are recognized by immune cells and phagocytized; (2) Inside the cells, bacteria fuse with the lysosome and form phagolysosomes; (3) Bacteria lyse and the DNA plasmids are released from cells; (4) DNA plasmids are engulfed by the dendritic cells (DCs), and inside the DCs, the antigen gene is expressed as protein, which will later be presented by class I or II MHC and delivered to CD4+ or CD8+ T cells.
Briefly, as a DNA vaccine carrier, bacteria are divided into two major groups: non-pathogenic bacteria and attenuated pathogen bacteria. The attenuated bacteria that have been studied as the DNA vaccine carrier include Salmonella spp. [4,23,47], Yersinia enterocolitica [54], Shigella spp. [55,56], and Listeria monocytogenes [57]. Pathogen bacteria target the mucous membranes as their infection route and as a result, they are suitable for mucosal administration. However, the main disadvantage includes the likelihood of causing infection, particularly in infants and immunocompromised patients [58]. Therefore, non-pathogen bacteria such as lactic acid bacteria (LAB) [59,60] may be preferable for development as DNA vaccine carriers. A comparison of the properties of several strains that are commonly used as DNA vaccine carriers is presented in Table 1.
Table 1.
Characteristics of live bacteria used as DNA vaccine carriers.
| Bacteria | Advantages | Limitations | Strategies | Ref. |
|---|---|---|---|---|
| Lactococcus lactis |
|
|
|
[50,60,61,62,63] |
| Salmonella spp. |
|
|
|
[64,65,66,67] |
| Listeria monocytogenes |
|
|
|
[25,68] |
| Shigella spp. |
|
|
[55,56] |
4. Lactic Acid Bacteria as the DNA Vaccine Carrier
LAB is an excellent candidate to be manipulated as a DNA vaccine carrier. LAB has been used in food fermentation for centuries and is a Generally Recognized as Safe (GRAS) organism [58,60]. LAB is also resistant to acidic conditions in the gastrointestinal (GI) system and is able to deliver the vaccine to the intestinal area [69,70]. Several LAB strains are famous as probiotic bacteria, such as Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus fermentum and Lactobacillus reuteri. Probiotic bacteria reduce lactose intolerance symptoms, such as diarrhea and flatulence, which appear in lactose intolerant patients who consume milk. Probiotic bacteria also increase the immune response toward pathogens by inhibiting pathogen colonization in the GI tract [59] and promote the mucosal immune system by activating plasma cells, inducing secretion of immunoglobulin A (IgA) and migration of T cells [71].
Lactococcus lactis is the most-studied LAB since its genome is easily manipulated, and many genetic tools have been engineered for L. lactis [54,64]. Notably, one of the main advantages of using L. lactis as a DNA vaccine carrier is its ability to pass through the intestinal tract without colonization [60].
Some studies confirmed that L. lactis is able to transfer DNA plasmid to the host cells [24,72,73,74]. A study conducted using native L. lactis showed its ability to deliver DNA plasmid into mammalian cells. The coincubation led to the expression and secretion of transgene products [24]. Yagnik et al. showed that L. lactis is capable of transferring DNA plasmid to Caco-2 cells in the absence of chemical treatment or other invasive proteins [72]. Another study demonstrated that glycine treatment escalates DNA transfer from L. lactis to Caco-2 cells [74]. The ability of L. lactis to transfer plasmid to mammalian cells in vivo was confirmed by the delivery of plasmid by L. lactis to murine epithelial membrane cells, and the protein was effectively expressed by the mammalian cell expression system [73,75].
Several attempts have been made to increase the efficacy of L. lactis delivery to the inside of DNA host cells. pValac, a new plasmid, was successfully constructed and demonstrated its ability to be delivered to the interior of porcine kidney cell lines [76]. The major advantage of the pValac vector is its small size (3.7 kb) compared with the previously used plasmid pLIG (10 kB) [24,75]. A smaller plasmid allows the cloning of larger DNA fragments with an easier transformation process.
To improve internalization capacity, L. lactis that expressed Listeria monocytogenes invasin internalin A (InlA) or Streptococcus pyogenes Fibronection-binding protein A (FnBPA) was developed. Internalin A mediates internalization through a binding interaction with the E-cadherin expressed on human epithelial and endothelial cells. Analysis showed that both strains effectively delivered the DNA plasmid to Caco-2 cells and the target protein was expressed inside the Caco-2 cells [62]. Another study demonstrated that the L. lactis expressing internalin A was able to deliver DNA to dendritic cells, either directly or by passing through the epithelial mono layer [52]. Moreover, the combination of an invasive L. lactis strain expressed In1A and pPERDBY, a reporter plasmid with immunostimulatory properties, successfully increased the expression level of the target gene in Caco-2 cells. This study demonstrated that the new combination increased the target gene expression three-fold compared with the expression in the non-invasive L. lactis strain [63].
5. Salmonella spp. as the DNA Vaccine Carrier
Salmonella spp. is a Gram-negative bacterium that causes salmonellosis through orofecal routes. As a DNA vaccine carrier, S. enterica serovars Typhimurium (S. typhimurium) is the most widely used Salmonella spp. [4,64,65]. This bacterium is suitable for oral administration as its natural infection route. However, it can induce both mucosal and systemic immune responses, activating the humoral and cellular immune systems [66]. As pathogenic bacteria, Salmonella spp. induce the immune response through their lipopolysaccharides (LPS) and flagellin content on their surface that is recognized as pathogen-associated molecular patterns (PAMPs). Flagellin induces the immune response by binding with Toll-like receptor 5 (TLR5), whereas LPS binds to TLR4. The binding activates nuclear factor-kappa B (NF-kB) and the mitogen-activated protein kinase (MAPK) pathway, which is followed by the release of cytokines [64,65]. S. enterica virulence genes are encoded in Salmonella pathogenicity islands (SPI). The two main SPIs are SPI1 and SP2, which participate in host cell invasion and intracellular host cell survival, respectively [64]. Due to their remarkable pathogenicity, mutant Salmonella have been developed as DNA vaccine carriers.
Mutant S. typhi and S. typhimurium that were developed as vaccine carriers have an aroA, aroC, or aroD mutation. Through these mutations, Salmonella is not able to produce aromatic substances; therefore, it cannot replicate inside the host. However, the bacteria still invade the host intestines and survive long enough to induce an immune system response. Other mutant types have also been established, such as mutants that cannot produce guanine and adenine bases, causing distress in cell wall production [77], the DNA repair system, or virulence gene regulation [26,51]. Kong et al. successfully constructed a recombinant attenuated Salmonella mutant strain that has a hyper invasive phenotype that can invade the host cell, escape the endosomes, and reduce the bacteria apoptosis. As a consequence, the DNA is allowed to efficiently enter the nucleus [77]. Several other manipulations have been studied to improve the ability of Salmonella spp. as a vaccine carrier, including the manipulation of lipid A, outer membrane vesicles and engineering the dual-plasmid system, as described briefly by Wang et al. [67].
The S. typhimurium mutant was demonstrated to be a remarkable candidate as an anti-atherosclerosis DNA vaccine carrier. S. typhimurium (aroA-, dam-) oral administration that contained a plasmid with the Flk-1 gene inhibited atherosclerosis and decreased aortic lesion size in atherosclerosis model mice. Flk-1 is a vascular epithelial growth factor receptor 2 (VEGRF2)-encoding gene in mice. The vaccination activated T cells and inhibited neoangiogenesis, which is involved in atherosclerosis. The cellular immune response was detected through decreasing the expression of VEGFR2 in endothelial cells [23,78]. This approach was also used in another study conducted by Hauer et al. by manipulating S. typimurium with a plasmid encoding for TIE2, which is an angiopoietin receptor in the endothelial surface that contributes to the development of atherosclerosis. Oral vaccination in atherosclerosis model mice induced a cellular immune response, decreased atherosclerosis lesion, and stabilized plaque. The cellular immune response was measured by the decrease in the number of endothelial cells that expressed TIE2 in vaccinated mice [79].
A similar study, using a DNA vaccine encoding for CD99, was also successfully conducted. CD99 is a protein expressed in leukocytes and endothelial cells involved in leukocyte recruitment in atherosclerosis lesion areas. The CD99 vaccine was consumed orally with S. typhimurium aroA- as the carrier. The vaccine generated CD8+ T cells that lysed CD99 expressing cells, so fewer leukocytes were detected in the lesion area. The vaccination also decreased lesion formation by 69% in carotid arterial [80].
Liang et al. confirmed Δasd/Δcrp mutant S. enterica successfully delivered the somatostatin gene in a mice model. The mutant lacks the antibiotic resistance gene and is based on the aspartate-semialdehyde dehydrogenase gene as the selection marker. Administered bacteria that contained a DNA vaccine effectively induced mucosal and systemic immune responses. The safety of the vaccine was shown by negligible integration of the plasmid gene into the host cellular genome [4].
6. Other Bacterial Live Vectors as DNA Vaccine Carriers
Other bacteria that were developed as DNA vaccine carriers include Listeria monocytogenes [65,81], Shigella spp. [51,82], and Yersinia enterolica [54].
L. monocytogenes is a Gram-positive bacterium that invades several cell types, such as mucosal epithelial cells, hepatocytes, macrophages, DCs and epithelial cells in the blood-brain barrier [25]. The bacteria invade and divide inside mammalian cells and consequently induce a high immune response. L. monocytogenes spread to other cells making it an effective DNA vaccine carrier against cancer. Given its ability to infect intestinal epithelium, L. monocytogenes has become an attractive candidate for oral vaccine delivery [57]. The disadvantage of this organism is the pathogenicity that leads to cholecystitis in human. Mice infected with L. monocytogenes showed bacterial colonization in the gall bladder. The mutant variant that was developed has mutations in the gene encoding for biotin metabolism (lmo0598) and ligase lipoate putative protein (lmo2566). The mutant variant was confirmed to induce an immune response, but did not replicate in the gall bladder [83]. Another study using mutant rsΔ2 L. monocytogenes showed that the bacterium, ingested orally, delivered a DNA vaccine containing the ovalbumin encoding gene. Increasing the antibody titer in a vaccinated mice serum demonstrated that the expressed antigen induced an immune response [57]. A different study indicated that attenuated recombinant L. monocytogenes, with a plasmid encoding for Mycobacterium tuberculosis antigen, was capable of inducing a robust cellular immune response in mice [84]. Conversely, a study conducted by Loeffler et al. revealed that L. monocytogenes induced a better immune response, when delivering recombinant antigen protein, than plasmid DNA. Antigens expressed by L. monocytogenes significantly increased the CD8+ T cell response, whereas the DNA vaccine carried by L. monocytogenes failed to induce a specific T cell response [85].
Shigella spp. are used as DNA vaccine carriers due to their ability to be retained in cytoplasm and evade endosomes, thus effectively delivering the DNA to the nucleus. Shigella also has a natural ability to target lymphoid tissues, triggering optimal mucosal and systemic immune systems. A study showed the efficacy of a DNA vaccine carried by mutant S. flexenery to attenuate human immunodeficiency virus (HIV) infection in a murine model. A single dose of the S. flexenery vaccine given intranasally induced a robust CD8+ T cell response [55]. A similar in vivo study demonstrated that a S. flexenery mutant successfully delivered the DNA vaccine encoding for the HIV gag gene. The DNA vaccine was intranasally delivered by recombinant bacteria inducing a cellular immune response comparable to a naked DNA vaccine given intramuscularly [56].
Y. enterocolitica is chosen as a DNA vaccine carrier because it can survive in host tissues for several days. During this period, the DNA vaccine is replicated in accordance with Y. enterocolitica proliferation, increasing the amount of DNA vaccine [54]. In a previous study, Y. enterocolitica was proven to deliver a DNA vaccine encoding for Brucella antigens bacterioferritin (BFR) and P39. The DNA vaccine induced antigen-specific antibodies and a Th1 response. The vaccinated mice showed resistance against Brucella infection [54].
7. Conclusions
DNA vaccines are facing challenges in terms of an effective delivery system that effectively targets the immune system. As live vectors, bacteria are new promising agents for vaccine delivery. Bacteria have unique natural characteristics that promote immune responses. However, the main concern about bacteria as vaccine carrier is patient safety. Although some attenuated recombinant strains have been developed, non-pathogenic bacteria such as LAB are considered more suitable DNA vaccine carriers.
Acknowledgments
The author would like to show gratitude to Kusworini Handono, Achmad Rudijanto, and Tri Yudani Mardining Raras for sharing their pearls of wisdom for the manuscript preparation. Comments by two anonymous reviewers greatly helped to improve an earlier version of this manuscript.
Conflicts of Interest
The author declares no conflict of interest.
References
- 1.Li L., Saade F., Petrovsky N. The future of human DNA vaccines. J. Biotechnol. 2012;162:171–182. doi: 10.1016/j.jbiotec.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coban C., Koyama S., Takeshita F., Akira S., Ishii K.J. Molecular and cellular mechanisms of DNA vaccines. Hum. Vaccines. 2008;4:453–457. doi: 10.4161/hv.4.6.6200. [DOI] [PubMed] [Google Scholar]
- 3.Ferraro B., Morrow M.P., Hutnick N.A., Shin T.H., Lucke C.E., Weiner D.B. Clinical applications of DNA vaccines: Current progress. Clin. Infect. Dis. 2011;53:296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang A., Riaz H., Dong F., Luo X., Yu X., Han Y. Evaluation of efficacy, biodistribution and safety of antibiotic-free plasmid encoding somatostatin genes delivered by attenuated Salmonella enterica serovar Choleraesuis. Vaccine. 2014;32:1368–1374. doi: 10.1016/j.vaccine.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Sheets R.L., Stein J., Manetz T.S., Duffy C., Nason M., Andrews C., Kong W., Nabel G.J., Gomez P.L. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile Virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol. Sci. 2017;91:610–619. doi: 10.1093/toxsci/kfj169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braat H., Rottiers P., Hommes D.W., Huyghebaert N., Remaut E., Remon J.P., van Deventer S.J.H., Neirynck S., Peppelenbosch M.P., Steidler L. A Phase I Trial with Transgenic Bacteria Expressing Interleukin-10 in Crohn’s Disease. Clin. Gastroenterol. Hepatol. 2006;4:754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Tavel J.A., Martin J.E., Kelly G.G., Enama M.E., Shen J.M., Gomez P.L., Andrews C.A., Koup R.A., Bailer R.T., Stein J.A., et al. Safety and Immunogenicity of a Gag-Pol Candidate HIV-1 DNA Vaccine Administered by a Needle-Free Device in HIV-1-Seronegative Subjects. J. Acquir. Immune Defic. Syndr. 2007;44:601–605. doi: 10.1097/QAI.0b013e3180417cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trimble C.L., Morrow M.P., Kraynyak K.A., Shen X., Dallas M., Yan J., Edwards L., Parker R.L., Denny L., Giffear M., et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: A randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386:2078–2088. doi: 10.1016/S0140-6736(15)00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurina V. DNA Vaccine: Mechanism of Action and factors which increase its efficacy. Vaccine. 2018;7:92–100. [Google Scholar]
- 10.Doria-Rose N.A., Haigwood N.L. DNA vaccine strategies: Candidates for immune modulation and immunization regimens. Methods. 2003;31:207–216. doi: 10.1016/S1046-2023(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 11.Denis-Mize K.S., Dupuis M., Singh M., Woo C., Ugozzoli M., Hagan D.T., Donnelly J.J., Ott G., McDonald D.M. Mechanisms of increased immunogenicity for DNA-based vaccines adsorbed onto cationic microparticles. Cell. Immunol. 2003;225:12–20. doi: 10.1016/j.cellimm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M., Obata C., Hisaeda H., Ishii K., Murata S., Chiba T., Tanaka K., Li Y., Furue M., Chou B., et al. A novel DNA vaccine based on ubiquitin-proteasome pathway targeting “self”-antigens expressed in melanoma/melanocyte. Gene Ther. 2005;12:1049–1057. doi: 10.1038/sj.gt.3302490. [DOI] [PubMed] [Google Scholar]
- 13.Fioretti D., Iurescia S., Rinaldi M. Recent advances in design of immunogenic and effective naked DNA vaccines against cancer. Recent Pat. Anticancer Drug Discov. 2014;9:66–82. doi: 10.2174/1574891X113089990037. [DOI] [PubMed] [Google Scholar]
- 14.Prather K.J., Sagar S., Murphy J., Chartrain M. Industrial scale production of plasmid DNA for vaccine and gene therapy: Plasmid design, production, and purification. Enzyme Microb. Technol. 2003;33:865–883. doi: 10.1016/S0141-0229(03)00205-9. [DOI] [Google Scholar]
- 15.Greenland J.R., Letvin N.L. Chemical adjuvants for plasmid DNA vaccines. Vaccine. 2007;25:3731–3741. doi: 10.1016/j.vaccine.2007.01.120. [DOI] [PubMed] [Google Scholar]
- 16.De Titta A., Ballester M., Julier Z., Nembrini C., Jeanbart L., van der Vlies A.J., Swartz M. A; Hubbell, J. Nanoparticle conjugation of CpG enhances adjuvancy for cellular immunity and memory recall at low dose. Proc. Natl. Acad. Sci. USA. 2013;110:19902–19907. doi: 10.1073/pnas.1313152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X., Yang X., Cai D., Mao D., Wu J., Zong L., Liu J. Intranasal immunization with chitosan/pCETP nanoparticles inhibits atherosclerosis in a rabbit model of Atherosclerosis. Vaccine. 2008;26:3727–3734. doi: 10.1016/j.vaccine.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 18.Gardlı R. Bacteria in gene therapy: Bactofection versus alternative gene therapy. Gene Ther. 2006;13:101–105. doi: 10.1038/sj.gt.3302635. [DOI] [PubMed] [Google Scholar]
- 19.Celec P. Gene therapy using bacterial vectors. Front. Biosci. 2017;22:4473. doi: 10.2741/4473. [DOI] [PubMed] [Google Scholar]
- 20.Gardlik R., Fruehauf J.H. Bacterial vectors and delivery systems in cancer therapy. IDrugs. 2010;13:701–706. [PubMed] [Google Scholar]
- 21.Chamcha V., Jones A., Quigley B.R., Scott J.R., Amara R.R. Oral Immunization with a Recombinant Lactococcus lactis-Expressing HIV-1 Antigen on Group A Streptococcus pilus Induces Strong Mucosal Immunity in the Gut. J. Immunol. 2015;195:5025–5034. doi: 10.4049/jimmunol.1501243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw Y.-W., Lin C.-Y., Lai Y.-S., Yang T.-C., Wang C.-J., Whang-Peng J., Liu L.F., Lin C.-P., Nieh S., Lu S.-C., et al. A Vaccine Targeted at CETP Alleviates high fat and high cholesterol diet-induced atherosclerosis and non-alcoholic Steatohepatitis in rabbit. PLoS ONE. 2014;9:e111529. doi: 10.1371/journal.pone.0111529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauer A.D., van Puijvelde G.H.M., Peterse N., de Vos P., van Weel V., van Wanrooij E.J.A., Biessen E.A.L., Quax P.H.A., Niethammer A.G., Reisfeld R., et al. Vaccination against VEGFR2 attenuates initiation and progression of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007;27:2050–2057. doi: 10.1161/ATVBAHA.107.143743. [DOI] [PubMed] [Google Scholar]
- 24.Guimarâes V.D., Innocentin S., Lefèvre F., Azevedo V., Wal J.M., Langella P., Chatel J.M. Use of native lactococci as vehicles for delivery of DNA into mammalian epithelial cells. Appl. Environ. Microbiol. 2006;72:7091–7097. doi: 10.1128/AEM.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor P., Becker P.D., Noerder M., Guzmán C.A., Becker P.D., Noerder M., Guzmán C.A. Delivery vehicles Bacteria as DNA vaccine delivery vehicles ND ES RIB. Hum. Vaccines Immunother. 2008;4:189–202. doi: 10.4161/hv.4.3.6314. [DOI] [PubMed] [Google Scholar]
- 26.Da Silva A.J., Zangirolami T.C., Novo-Mansur M.T.M., de Campos Giordano R., Martins E.A.L. Live bacterial vaccine vectors: An overview. Braz. J. Microbiol. 2014;45:1117–1129. doi: 10.1590/S1517-83822014000400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams J. Vector Design for improved DNA Vaccine efficacy, safety and production. Vaccines. 2013;1:225–249. doi: 10.3390/vaccines1030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pringle I.A., Hyde S.C. Progress and Prospects: The design and production of plasmid vectors. Gene Ther. 2009;16:165–171. doi: 10.1038/gt.2008.183. [DOI] [PubMed] [Google Scholar]
- 29.Garmory H.S., Brown K.A., Titball R.W. DNA vaccines: Improving expression of antigens. Genet. Vaccines Ther. 2003;1:2. doi: 10.1186/1479-0556-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J.A., Carnes A.E., Hodgson C.P. Plasmid DNA vaccine vector design: Impact on efficacy, safety and upstream production. Biotechnol. Adv. 2009;27:353–370. doi: 10.1016/j.biotechadv.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darquet M., Rangara R., Kreiss P., Schwartz B., Naimi S., Delaère P., Crouzet J., Scherman D. Minicircle: An improved DNA molecule for in vitro and in vivo gene transfer. Gene Ther. 1999;6:209–218. doi: 10.1038/sj.gt.3300816. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery D.L., Prather K.J. Design of plasmid DNA constructs for vaccines. Methods Mol. Med. 2006;127:11–22. doi: 10.1385/1-59745-168-1:11. [DOI] [PubMed] [Google Scholar]
- 33.Williams J., Luke J., Johnson L., Hodgson C. pDNAVACCultra vector family: High throughput intracellular targeting DNA vaccine plasmids. Vaccine. 2006;24:4671–4676. doi: 10.1016/j.vaccine.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 34.Glenting J., Wessels S. Ensuring safety of DNA vaccines. Microb. Cell Fact. 2005;4:26. doi: 10.1186/1475-2859-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams J. Improving DNA Vaccine performance through vector design. Curr. Gene Ther. 2014;14:170–189. doi: 10.2174/156652321403140819122538. [DOI] [PubMed] [Google Scholar]
- 36.Vandermeulen G., Marie C., Scherman D., Préat V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. 2011;19:1942–1949. doi: 10.1038/mt.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira P.H., Mairhofer J. Marker-free plasmids for biotechnological applications—Implications and perspectives. Trends Biotechnol. 2013;31:539–547. doi: 10.1016/j.tibtech.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Xia W., Bringmann P., McClary J., Jones P.P., Manzana W., Zhu Y., Wang S., Liu Y., Harvey S., Madlansacay M.R., et al. High levels of protein expression using different mammalian CMV promoters in several cell lines. Protein Expr. Purif. 2006;45:115–124. doi: 10.1016/j.pep.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Xu Z.L., Mizuguchi H., Ishii-Watabe A., Uchida E., Mayumi T., Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149–156. doi: 10.1016/S0378-1119(01)00550-9. [DOI] [PubMed] [Google Scholar]
- 40.Garg S., Oran A.E., Hon H., Jacob J. The hybrid cytomegalovirus enhancer/chicken beta-actin promoter along with woodchuck hepatitis virus posttranscriptional regulatory element enhances the protective efficacy of DNA vaccines. J. Immunol. 2004;173:550–558. doi: 10.4049/jimmunol.173.1.550. [DOI] [PubMed] [Google Scholar]
- 41.Kos S., Tesic N., Kamensek U., Blagus T., Cemazar M., Kranjc S., Lavrencak J., Sersa G. Improved Specificity of gene electrotransfer to skin using pDNA Under the control of collagen tissue-specific promoter. J. Membr. Biol. 2015;248:919–928. doi: 10.1007/s00232-015-9799-4. [DOI] [PubMed] [Google Scholar]
- 42.Itai K., Sawamura D., Meng X., Hashimoto I. Keratinocyte gene therapy: Inducible promoters and in vivo control of transgene expression. Clin. Exp. Dermatol. 2001;26:531–535. doi: 10.1046/j.1365-2230.2001.00883.x. [DOI] [PubMed] [Google Scholar]
- 43.Vanniasinkam T., Reddy S.T., Ertl H.C.J. DNA immunization using a non-viral promoter. Virology. 2006;344:412–420. doi: 10.1016/j.virol.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 44.Ertl P.F., Thomsen L.L. Technical issues in construction of nucleic acid vaccines. Methods. 2003;31:199–206. doi: 10.1016/S1046-2023(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 45.Barouch D.H., Yang Z., Kong W., Korioth-Schmitz B., Sumida S.M., Truitt D.M., Kishko M.G., Arthur J.C., Miura A., Mascola J.R., et al. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 2005;79:8828. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina E., Guzmán C.A. Use of live bacterial vaccine vectors for antigen delivery: Potential and limitations. Vaccine. 2001;19:1573–1580. doi: 10.1016/S0264-410X(00)00354-6. [DOI] [PubMed] [Google Scholar]
- 47.Cong H., Yuan Q., Zhao Q., Zhao L., Yin H., Zhou H., He S., Wang Z. Comparative efficacy of a multi-epitope DNA vaccine via intranasal, peroral, and intramuscular delivery against lethal Toxoplasma gondii infection in mice. Parasites Vectors. 2014;7:145. doi: 10.1186/1756-3305-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira V.B., Santiago M., Azevedo P., Diniz T., Saraiva L. Use of bacteria in DNA vaccine delivery. Vaccine. 2013;12:1993–2003. [Google Scholar]
- 49.Locht C. Live bacterial vectors for intranasal delivery of protective antigens. Pharm. Sci. Technol. Today. 2000;3:121–128. doi: 10.1016/S1461-5347(00)00256-X. [DOI] [PubMed] [Google Scholar]
- 50.Bermúdez-Humarán L.G. Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum. Vaccines. 2009;5:264–267. doi: 10.4161/hv.5.4.7553. [DOI] [PubMed] [Google Scholar]
- 51.Shata M.T., Stevceva L., Agwale S., Lewis G.K., Hone D.M. Recent advances with recombinant bacterial vaccine vectors. Mol. Med. Today. 2000;6:66–71. doi: 10.1016/S1357-4310(99)01633-0. [DOI] [PubMed] [Google Scholar]
- 52.De Azevedo M., Meijerink M., Taverne N., Pereira V.B., LeBlanc J.G., Azevedo V., Miyoshi A., Langella P., Wells J.M., Chatel J.M. Recombinant invasive Lactococcus lactis can transfer DNA vaccines either directly to dendritic cells or across an epithelial cell monolayer. Vaccine. 2015;33:4807–4812. doi: 10.1016/j.vaccine.2015.07.077. [DOI] [PubMed] [Google Scholar]
- 53.Lin I., Van T., Smooker P. Live-attenuated Bacterial vectors: Tools for Vaccine and therapeutic agent delivery. Vaccines. 2015;3:940–972. doi: 10.3390/vaccines3040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-mariri A., Tibor A., Lestrate P., Bolle X., Letesson J., Mertens P. Yersinia enterocolitica as a Vehicle for a Naked DNA Vaccine Encoding Brucella abortus Bacterioferritin or P39 Antigen Yersinia enterocolitica as a Vehicle for a Naked DNA Vaccine Encoding Brucella abortus Bacterioferritin or P39 Antigen. Infect. Immun. 2002;70:1915–1923. doi: 10.1128/IAI.70.4.1915-1923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shata M.T., Hone D.M. Vaccination with a Shigella DNA Vaccine Vector Induces Antigen-Specific CD8 + T Cells and Antiviral Protective Immunity Vaccination with a Shigella DNA Vaccine Vector Induces Antigen-Specific CD8+ T Cells and Antiviral Protective Immunity. J. Virol. 2001;75:9665–9670. doi: 10.1128/JVI.75.20.9665-9670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu F., Hong M., Ulmer J.B. Immunogenicity of an HIV-1 gag DNA vaccine carried by attenuated Shigella. Vaccine. 2003;21:644–648. doi: 10.1016/S0264-410X(02)00573-X. [DOI] [PubMed] [Google Scholar]
- 57.Sinha S., Kuo C.Y., Ho J.K., White P.J., Jazayeri J.A., Pouton C.W. A suicidal strain of Listeria monocytogenes is effective as a DNA vaccine delivery system for oral administration. Vaccine. 2017;35:5115–5122. doi: 10.1016/j.vaccine.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 58.Bermúdez-humarán L.G., Kharrat P., Chatel J.-M., Langella P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Fact. 2011;10:S4. doi: 10.1186/1475-2859-10-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pontes D.S., de Azevedo M.S.P., Chatel J.-M., Langella P., Azevedo V., Miyoshi A. Lactococcus lactis as a live vector: Heterologous protein production and DNA delivery systems. Protein Expr. Purif. 2011;79:165–175. doi: 10.1016/j.pep.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 60.Wyszyńska A., Kobierecka P., Bardowski J., Jagusztyn-Krynicka E.K. Lactic acid bacteria—20 years exploring their potential as live vectors for mucosal vaccination. Appl. Microbiol. Biotechnol. 2015;99:2967–2977. doi: 10.1007/s00253-015-6498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pontes D., Azevedo M., Innocentin S., Blugeon S., Lefévre F., Azevedo V., Miyoshi A., Courtin P., Chapot-Chartier M.P., Langella P., et al. Immune response elicited by DNA vaccination using Lactococcus lactis is modified by the production of surface exposed pathogenic protein. PLoS ONE. 2014;9:e84509. doi: 10.1371/journal.pone.0084509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Innocentin S., Guimarães V., Miyoshi A., Azevedo V., Langella P., Chatel J.M., Lefèvre F. Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl. Environ. Microbiol. 2009;75:4870–4878. doi: 10.1128/AEM.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yagnik B., Sharma D., Padh H., Desai P. Dual recombinant Lactococcus lactis for enhanced delivery of DNA vaccine reporter plasmid pPERDBY. Microbiol. Immunol. 2017;61:123–129. doi: 10.1111/1348-0421.12473. [DOI] [PubMed] [Google Scholar]
- 64.Hegazy W.A.H., Hensel M. Salmonella enterica as a vaccine carrier. Future Microbiol. 2012;7:111–127. doi: 10.2217/fmb.11.144. [DOI] [PubMed] [Google Scholar]
- 65.Shahabi V., Maciag P.C., Rivera S., Wallecha A. Live, attenuated strains of Listeria and Salmonella as vaccine vectors in cancer treatment. Bioeng. Bugs. 2010;1:235–239. doi: 10.4161/bbug.1.4.11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheminay C., Hensel M. Rational design of Salmonella recombinant vaccines. Int. J. Med. Microbiol. 2008;298:87–98. doi: 10.1016/j.ijmm.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Wang S., Kong Q., Curtiss R. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb. Pathog. 2013;58:17–28. doi: 10.1016/j.micpath.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruhn K.W., Craft N., Miller J.F. Listeria as a vaccine vector. Microbes Infect. 2007;9:1226–1235. doi: 10.1016/j.micinf.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Shigemori S., Watanabe T., Kudoh K., Ihara M., Nigar S., Yamamoto Y., Suda Y., Sato T., Kitazawa H., Shimosato T. Oral delivery of Lactococcus lactis that secretes bioactive HEME oxygenase-1 alleviates development of acute colitis in mice. Microb. Cell Fact. 2015;14:189. doi: 10.1186/s12934-015-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vesa T., Pochart P., Marteau P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 2000;14:823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 71.Silva V.D.O., Foureaux R.D.C., Araujo T.S., Peconick P., Zangeronimo M.G., Pereira L.J. Effect of Probiotic administration on the immune response: A Systematic review of experimental models in rats. Braz. Arch. Biol. Technol. 2012;55:685–694. doi: 10.1590/S1516-89132012000500007. [DOI] [Google Scholar]
- 72.Yagnik B., Padh H., Desai P. Construction of a new shuttle vector for DNA delivery into mammalian cells using non-invasive Lactococcus lactis. Microbes Infect. 2016;18:237–244. doi: 10.1016/j.micinf.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 73.Mancha-Agresti P., Drumond M.M., Carmo F.L.R., Santos M.M., Santos J.S.C., Venanzi F., Chatel J.-M., Leclercq S.Y., Azevedo V. A New broad range plasmid for DNA Delivery in Eukaryotic Cells using lactic acid Bacteria: In Vitro and in vivo assays. Mol. Ther. 2017;4:83–91. doi: 10.1016/j.omtm.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin T., Pavlovaa S.I., Ji X., Jin L., Spear G. A novel plasmid for delivering genes into mammalian cells with noninvasive food and commensal lactic acid bacteria. Plasmid. 2011;65:8–14. doi: 10.1016/j.plasmid.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatel J.-M., Pothelune L., Ah-Leung S., Corthier G., Wal J.-M., Langella P. In vivo transfer of plasmid from food-grade transiting Lactococci to murine epithelial cells. Gene Ther. 2008;15:1184–1190. doi: 10.1038/gt.2008.59. [DOI] [PubMed] [Google Scholar]
- 76.Guimarães V., Innocentin S., Chatel J.-M., Lefèvre F., Langella P., Azevedo V., Miyoshi A. A new plasmid vector for DNA delivery using Lactococci. Genet. Vaccines Ther. 2009;7:4. doi: 10.1186/1479-0556-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong W., Brovold M., Koeneman B.A., Clark-Curtiss J., Curtiss R. Turning self-destructing Salmonella into a universal DNA vaccine delivery platform. Proc. Natl. Acad. Sci. USA. 2012;109:19414–19419. doi: 10.1073/pnas.1217554109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrovan R.J., Kaplan C.D., Reisfeld R., Curtiss L.K. DNA vaccination against VEGF receptor 2 reduces atherosclerosis in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2007;27:1095–1100. doi: 10.1161/ATVBAHA.106.139246. [DOI] [PubMed] [Google Scholar]
- 79.Hauer A.D., Habets K.L.L., van Wanrooij E.J., de Vos P., Krueger J., Reisfeld R., van Berkel T.J.C., Kuiper J. Vaccination against TIE2 reduces atherosclerosis. Atherosclerosis. 2009;204:365–371. doi: 10.1016/j.atherosclerosis.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 80.Wanrooij E.J.A., Van Vos P., De Bixel M.G., Vestweber D., Berkel T.J.C., Van Kuiper J. Vaccination against CD99 inhibits Atherogenesis in low-density lipoprotein receptor-deficient mice. Cardiovasc. Res. 2008;78:590–596. doi: 10.1093/cvr/cvn025. [DOI] [PubMed] [Google Scholar]
- 81.Im E.-J., Borducchi E.N., Provine N.M., McNally A.G., Li S., Frankel F.R., Barouch D.H. An attenuated Listeria monocytogenes Vector primes more potent simian immunodeficiency virus-specific mucosal immunity than DNA Vaccines in mice. J. Virol. 2013;87:4751–4755. doi: 10.1128/JVI.03085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pasetti M.F., Barry E.M., Losonsky G., Singh M., Medina-Moreno S.M. Attenuated Salmonella enterica serovar Typhi and Shigella flexneri 2a strains mucosally deliver DNA vaccines encoding measles virus hemagglutinin, inducing specific immune responses and protection in cotton rats. J. Virol. 2003;77:5209–5217. doi: 10.1128/JVI.77.9.5209-5217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dowd G.C., Bahey-el-din M., Casey P.G., Joyce S.A., Hill C., Gahan C.G.M. Listeria monocytogenes mutants defective in gallbladder replication represent safety-enhanced vaccine delivery platforms. Hum. Vaccines Immunother. 2016;12:2059–2063. doi: 10.1080/21645515.2016.1154248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miki K., Nagata T., Tanaka T., Kim Y.H., Uchijima M., Ohara N., Nakamura S., Okada M., Koide Y. Induction of Protective cellular immunity against Mycobacterium tuberculosis by Recombinant attenuated self-destructing Listeria monocytogenes Strains harboring eukaryotic expression plasmids for antigen 85 complex and MPB/MPT51. Infect. Immun. 2004;72:2014–2021. doi: 10.1128/IAI.72.4.2014-2021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loeffler D.I.M., Schoen C.U., Goebel W., Pilgrim S. Comparison of different live vaccine strategies in vivo for delivery of protein antigen or antigen-encoding DNA and mRNA by virulence-attenuated Listeria monocytogenes. Infect. Immun. 2006;74:3946–3957. doi: 10.1128/IAI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]