Abstract
To investigate the mutation mechanism of purine transitions in DNA damaged with methoxyamine, a DNA dodecamer with the sequence d(CGCAAATTmo4CGCG), where mo4C is 2′-deoxy-N4-methoxycytidine, has been synthesized and the crystal structure determined by X-ray analysis. The duplex structure is similar to that of the original undamaged B-form dodecamer, indicating that the methoxylation does not affect the overall DNA conformation. Electron density maps clearly show that the two mo4C residues form Watson–Crick-type base pairs with the adenine residues of the opposite strand and that the methoxy groups of mo4C adopt the anti conformation to N3 around the C4–N4 bond. For the pair formation through hydrogen bonds the mo4C residues are in the imino tautomeric state. Together with previous work, the present work establishes that the methoxylated cytosine residue can present two alternate faces for Watson–Crick base-pairing, thanks to the amino↔imino tautomerism allowed by methoxylation. Based on this property, two gene transition routes are proposed.
INTRODUCTION
Genetic information is transmitted from generation to generation through DNA replication, in which the highest accuracy is achieved by forming Watson–Crick base pairs between adenine and thymine bases and between guanine and cytosine bases, as an absolute rule in every organism. When certain chemicals damage DNA, however, this rule is disturbed and errors are introduced into the synthesized DNA, which results in genetic mutations (1). Oxyamines such as hydroxyamine and methoxyamine are known to be mutagens (1) which predominantly attack and modify the exocyclic amino groups of adenine and cytosine residues (2). For the case of the former modification, we have reported the crystal structures of DNA duplexes containing N6-methoxyadenine (mo6A) paired with thymine (3) and with cytosine (4) residues of the opposite strand. For the latter modification too, it is expected that the N4-methoxycytosine (mo4C) moiety has chemical and physicochemical properties different from the unmodified base. It has been demonstrated that the methoxylated dCTP (mo4dCTP) is incorporated into the newly synthesized DNA strand for both templates, G and A, the latter template being preferable (5). On the other hand, when the template cytosine residue is methoxylated, RNA polymerase incorporates adenosine-5′-triphosphate (ATP) at the site opposite to the mo4C residue (6). In DNA replication, similar reaction will occur.
To investigate the interaction properties of mo4C with other nucleotides, several structural studies on oligonucleotides containing mo4C residue were performed as a molecular basis for understanding the purine transition. Meervelt et al. (7) reported the X-ray structure of a hexamer, in which the mo4C residue is in the imino tautomeric form with a syn methoxy group relative to N3, and forms a wobble pair with a guanine residue. In this duplex, the hexamer adopts a Z conformation. Such a conformation and wobble pairing are, however, not compatible with DNA polymerase binding because the enzyme accepts only Watson–Crick base pairs in B-form duplex (8). Recent X-ray study on a Dickerson–Drew type DNA dodecamer containing a mo4C residue (9) has revealed that one of the two mo4C residues forms a pair with a guanine residue of the opposite strand, the geometry being the canonical Watson–Crick type, and that the other mo4C residue forms a wobble pair with a guanine residue. These pairing modes are consistent with the results from an NMR study of a hepta-nucleotide containing mo4C (10). A similar situation was also found in the crystal structure of hexa-nucleotide, containing a pyrimidine analog 6H,8H-3,4-dihydropyrimido[4,5-c]-[1,2]oxazin-7-one (11).
On the other hand, in the case of interaction with adenine residue, NMR studies of oligonucleotides containing mo4C suggested that the mo4C residue in the imino tautomeric state formed pair with an adenine residue in the Watson–Crick geometry (12,13). To confirm this pairing and to visualize the exact geometry of mo4C paired with adenine residue in B-form duplex, a Dickerson–Drew type DNA dodecamer with the sequence of d(CGCAAATTmo4CGCG) has been synthesized and its crystal structure determined. The electron density maps clearly show that the mo4C residue indeed forms a Watson–Crick-type pair with the adenine residue of the opposite strand in the B-form duplex. For this pairing, the cytosine moiety must adopt the imino form. Together with the previous result, this work establishes that the mo4C residue has two faces in the amino and the imino tautomeric states and can form a stable base pair with either 2′-deoxyadenosine or 2′-deoxyguanosine. This property of mo4C makes it possible for us to propose two routes of gene transition mechanisms in DNA replication.
MATERIALS AND METHODS
Synthesis and crystallization
A DNA dodecamer with the sequence d(CGCAAATTmo4CGCG) containing 2′-deoxy-N4-methoxycytidine at the ninth position was synthesized by the reported method (14). Crystallization was carried out at 4°C by the hanging drop vapor diffusion method. Suitable crystals for X-ray analysis were obtained in a 4 µl droplet containing initially 0.5 mM DNA, 10 mM sodium cacodylate (pH 7.0), 4.5 mM spermine tetrahydrochloride, 18 mM magnesium acetate, 10% (v/v) 2-methyl-2,4-pentanediol, 40 mM sodium chloride and 0.1% n-octyl-β-d-glycoside, which was equilibrated to a 700 µl reservoir solution containing 25% 2-methyl-2,4-pentanediol and twice of other reagents in concentration, excluding DNA and n-octyl-β-d-glucoside. Crystals picked up in nylon cryoloops (Hampton Research) were immersed in a cryoprotectant containing 35% (v/v) MPD for few seconds and then rapidly transferred into liquid nitrogen.
Data collection
X-ray data were collected on a Quantum 4z CCD detector at 100 K with synchrotron radiation (λ = 1.00 Å) at BL18b of the Photon Factory in Tsukuba. Ninety frames of diffraction patterns, taken with 2.0° oscillation for each, were processed at 1.6 Å resolution by the program DPS/MOSFLM (15–18). Intensity data were put on a relative scale and merged into the independent reciprocal space using the program SCALA and TRUNCATE of the CCP4 suite (19). In total, 7855 independent reflections with Rmerge 5.8% were obtained from 40 352 observed reflections. Statistics of data collection and crystal data are summarized in Table 1.
Table 1. Crystal data, data collection and structure determination.
| Space group |
|
|
P212121 |
| Unit cell (Å) | a = 24.8, b = 40.3, c = 65.5 | ||
| Asymmetric unit (duplex) | 1 | ||
| Resolution (Å) | 100~1.6 | ||
| Unique reflections | 7855 | ||
| Completeness (%) | 86.1 | ||
| in the outer shell (%) | 69.7 (1.79~1.69 Å) | ||
| 38.4 (1.69~1.6 Å) | |||
| Multiplicity | 5.1 | ||
| Rmergea (%) | 5.8 | ||
| Structure refinement | |||
| Resolution range (Å) | 10~1.6 | ||
| Used reflection | 6515 (I/s >3) | ||
| R-factorb (%) | 21.8 | ||
| Rfreec (%) | 25.8 | ||
| DNA atoms | 488 | ||
| Water molecules | 111 | ||
| Magnesium atom | 1 | ||
| R.m.s. deviation from ideal geometry | |||
| Bond lengths (Å) | 0.004 | ||
| Bond angles (deg.) | 0.9 | ||
| Improper angles (deg.) | 1.4 | ||
| Average B-factors (Å2) | |||
| DNA | 27.6 | ||
| Waters | 39.4 | ||
| Magnesium atom | 29.7 |
aRmerge = 100 × ∑hklj|Ihklj – <Ihkl>|/∑hklj<Ihkl>
bR-factor = 100 × ∑||Fo|–|Fc||/∑Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
cCalculated using a random set containing 10% of observations that were not included during refinement (30).
Structure determination and refinement
The initial phase was derived by molecular replacement using the atomic coordinates of the original DNA dodecamer d(CGCGAATTCGCG) (20), with the program AMoRe (21). The molecular structure was constructed and modified on a graphic workstation by inspecting |Fo|–|Fc| omit maps at every nucleotide residue with the program QUANTA (Molecular Simulation Inc.). The |Fo|–|Fc| omit maps clearly showed that the two mo4C residues form base pairs with adenine residues and that the methoxy groups are in an anti conformation to the N3 atom around the N4–C4 bond. From the hydrogen-bonding scheme, the mo4C residues were assumed to adopt imino form. A combination of partial structure of unmodified 2′-deoxycytidine and the methoxy group of 3′,5′-di-O-acetyl-N4-methoxycytidine were used for the stereo-chemical parameters of mo4C residues (22). The atomic parameters were refined with the program CNS (23) through a combination of rigid body, simulated annealing, crystallographic conjugate gradient minimization refinement and B-factor refinement, followed by interpretation of omit map at every nucleotide residue. No restraints were applied between paired nucleotides during refinement. In the initial stages of refinement, all sugar puckering were assumed to be C2′-endo, but this restrain was released in the final refinements. One magnesium cation coordinated by 6 water molecules and 105 water molecules were included in the final refinements. Statistics of the structure determination are given in Table 1. All local helical parameters including all torsion angles and pseudorotation phase angles of 2′-deoxyribose rings were calculated by using the program NUPARAM (24).
RESULTS AND DISCUSSION
Effect of methoxylation on DNA conformation
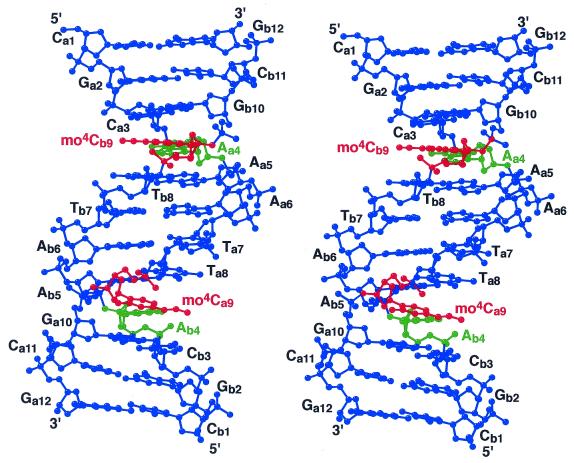
The crystal structure is isomorphous to that of the original Dickerson–Drew type dodecamer (20,25,26). Figure 1 shows a stereo-overview of two chains (a and b) of d(CGCAAATTmo4CGCG) which are associated to form a right-handed double helix. When the present and original duplexes are superimposed to each other with the corresponding atomic coordinates, the overall root-mean-square deviation is 0.48 Å, suggesting no significant discrepancy. The calculated local helical parameters, rise and displacement, and the sugar puckers (pseudorotation phase angles) fluctuate around their average values close to those of the typical B-form DNA. Their patterns, when plotted along with the nucleotide number, are similar to those of the original dodecamer, indicating that methoxylation gives no significant changes in the overall DNA conformation. The sugar puckering is more sensitive to local changes of DNA conformation. Pseudorotation phase angles of chains a and b also show a typical behavior similar to those of the corresponding residues of the original dodecamers. The mo4C residue at the ninth position of chain b has, however, a sugar pucker close to C2′-endo which is different from the intermediate state reported for the other Dickerson–Drew type DNA dodecamers. This difference may be due to the surrounding solvent structure. The terminal Ga12 residue of chain a is also different due to similar reason.
Figure 1.
A stereo-overview of the present DNA dodecamer with the sequence d(CGCAAATTmo4CGCG). This diagram was drawn with the program MOLSCRIPT (31). The mo4C residues are colored in red and the 2′-deoxyadenosine residues paired with mo4C are in green. The nucleotides are numbered from the 5′ end independently in the two strands a and b.
Base pair geometry of mo4C:A
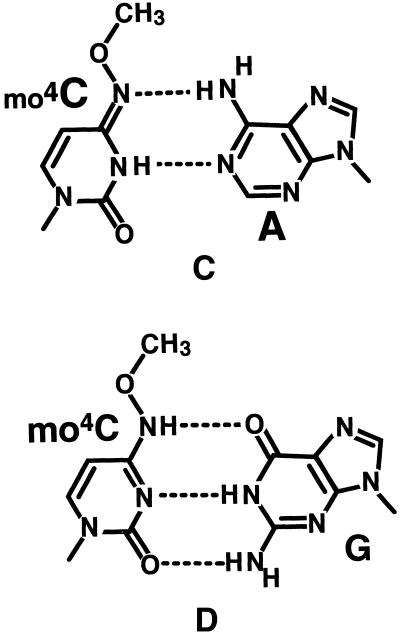
The base pairs in the duplex are all in the Watson–Crick types including the mo4C residues. Figure 2 shows the final 2Fo–Fc electron density maps at the two sites of the mo4C residues interacted with the opposite adenine residues and with the associated solvent molecules. These maps clearly indicate that both mo4C residues form base pairs with the opposite adenine residues. The pairing geometry at the two sites are very similar to the Watson–Crick T:A pairs found in the original Dickerson–Drew type dodecamer (26). Furthermore, in the mo4Ca9:Ab4 pair, the distances between N4(mo4Ca9) and N6(Ab4) (3.0 Å), and between N3(mo4Ca9) and N1(Ab4) (2.9 Å) suggest the hydrogen bond formation. For the mo4Cb9:Aa4 pair, the corresponding distances are 3.1 and 2.8 Å, respectively, suggesting also the formation of hydrogen bonds. To form these hydrogen bonds, the two mo4C residues must chemically adopt the imino tautomeric state. The methoxy groups of the two mo4C moieties take an anti conformation with respect to the N3 atom around the N4–C4 bond. This pairing mode is similar to the structure reported from an NMR study of hepta-deoxy-nucleotides containing mo4C (12). The present work is the first visualization of mo4C residues paired with adenine residues in the Watson–Crick-type geometry.
Figure 2.
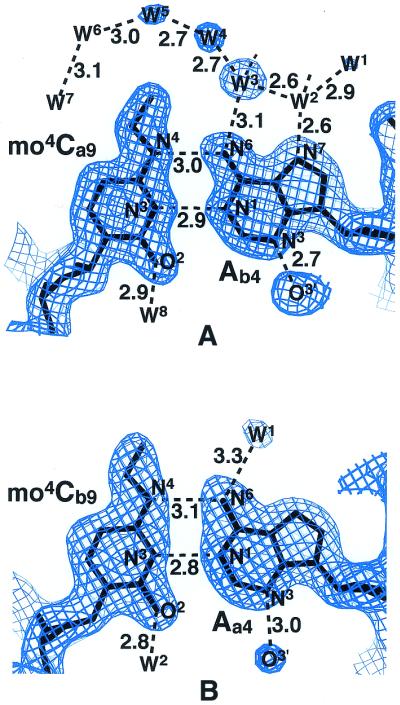
Final 2|Fo|–|Fc| electron density maps for the mo4Ca9:Ab4 (A) and mo4Cb9:Aa4 (B) base pairs, and the corresponding chemical structure (C). Maps are contoured at the 1.1σ level by the program O (32). The chemical structure of the Watson–Crick mo4C:G pair found in the Dickerson–Drew type dodecamer with the sequence d(CGCGAATTmo4CGCG) (9) is shown at the bottom (D). Broken lines indicate possible hydrogen bonds. W indicates a water molecule.
Crystal structure
Two duplexes related by crystallographic 21 symmetry along the c-axis make a direct contact to form an infinite column in a head-to-tail fashion. The two guanine residues, Ga12 and Gb2, located at one end of the duplex interact with the other two guanine residues, Ga2 and Gb12, respectively, at the other end of another duplex through double N2–H···N3 hydrogen bonds for each. As a common feature of Dickerson–Drew type dodecamer, a magnesium cation coordinated octahedrally by six water molecules is found in the major groove of the duplex. This cation takes a part for cementing the two duplex columns related by 21 symmetry along the b-axis. Three of the six water molecules form hydrogen bonds with the guanine bases at Ga2 and Gb10 of one duplex, and the other three are hydrogen bonded to the phosphate oxygen atoms of the Aa6 and the Ta7 residues of a neighboring duplex.
Water molecules found in the minor and the major grooves and around the phosphate groups occupy the positions similar to those of the original dodecamers, except around the methoxy groups. Other divalent and monovalent cations used for crystallization were not identified, but they may be disordered and involved in neutralization of the phosphate negative charges. Spermine may also contribute to such neutralization.
Biological implications
During replication of DNA, only the Watson–Crick-type base pair formation is acceptable in the active site of DNA polymerase, in which there is not enough space for a wobble type or other non-complementary base pairs. In addition, as the polymerase is bound in the minor groove of DNA, the methoxy group extruded into the major groove cavity does not seem to interfere with the binding of polymerase or the polymerase activity. Therefore, the Watson–Crick-type mo4C:A base pair is acceptable in the active site. For this pairing, the cytosine moiety must adopt the imino form. It is known that the unmodified cytosine residue always exists in the amino tautomeric state (27). However, methoxylation induces the cytosine moiety to occur tautomerization between the amino and imino forms, though the latter is predominant (28,29). In the imino form, the donor/acceptor sites for hydrogen bonding are similar to those of thymine moiety, so that the methoxylated cytosine can form a Watson–Crick-type base pair with adenine residue of the opposite strand. This mimicry is the origin of purine transition mutagenesis. The previous crystallographic study of a DNA dodecamer containing mo4C revealed that the two mo4C residues form two different types of base pairs with the opposite guanine residues in one duplex (9). One of the two mo4C residues, in the amino tautomeric state with a methoxy group in the anti conformation to N3 around the C4–N4 bond, forms a Watson–Crick base pair and the other mo4C residue in the imino tautomeric state with the syn methoxy group forms a wobble base pair with the guanine residue, as shown in Figure 2D. Taking both Watson–Crick-type mo4C:G and mo4C:A (present work) pairings into consideration, it is concluded that when cytosine residues of the template strand are methoxylated, they accept adenine residues as well as guanine residues in the synthesized DNA, and that when the reactant dCTP is methoxylated (mo4dCTP), it can be incorporated into the newly synthesized DNA at both sites of A and G residues of the template strand.
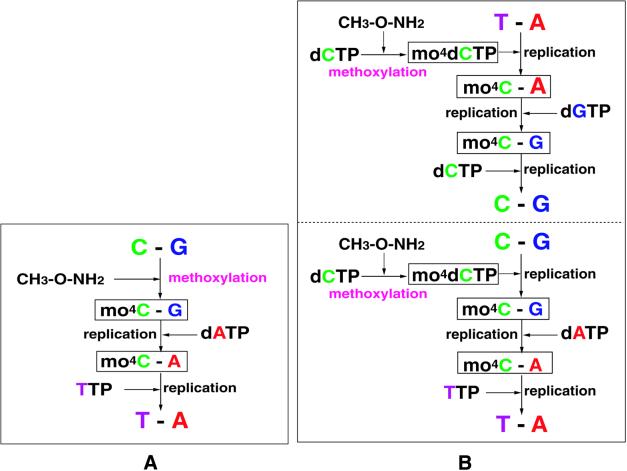
Based on the two cases of misincorporations, two routes of gene transition are possible, as shown in Figure 3. When a cytosine residue in DNA is methoxylated, the original C:G pair can be replaced with a T:A pair by two successive steps of replication at least. Suppose that dATP is incorporated instead of dGTP at the site opposite to the mo4C template in the first step, and then the synthesized strand is used for the second template, TTP will be incorporated at the A site in the second copy. In the case of mo4dCTP, T:A to C:G and C:G to T:A transition are possible to occur through three steps of replication at least. In the T:A→C:G case, if mo4dCTP is incorporated at a template A site by mimicking thymine base, the incorporated mo4C residue will then accept dGTP in the second step, and finally dCTP will be incorporated at the template G site. In the C:G→T:A case, if mo4dCTP is incorporated at the template G site by forming a Watson–Crick-type mo4C:G pair, then the incorporated mo4C can serve as the second template to accept dATP and, finally, TTP will be incorporated at the template A site of the second copy.
Figure 3.
Possible routes for gene transition mechanism. There are two ways that occur when template cytosine residues are methoxylated (A), and when reactant dCTP is methoxylated (B).
A similar situation was found in the case of methoxyadenine residue (3,4): the modified adenine base adopts the imino form and makes a pair with the opposite cytosine base in the Watson–Crick-type geometry. Methoxyamine attacks both adenine and cytosine residues to convert them to the methoxylated derivatives. From the present and the previous works, it has been established that these modifications allow adenine and cytosine moieties to form mis-pairs with the non-complementary nucleotides by mimicking a Watson–Crick-type geometry through tautomerization between the amino and imino forms. In addition the DNA polymerase is indifferent to such chemical modification in the major groove of DNA. These structural properties are the reason why oxyamines induce genetic mutation through purine and pyrimidine transitions.
Data Bank accession codes
The atomic coordinates and the structure factors have been deposited in the Protein Data Bank (PDB) with the ID code of 1J8L.
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Sakabe, M. Suzuki and N. Igarashi for facilities and help during data collection at Photon Factory (Tsukuba), and T. Simonson for proofreading of the original manuscript. This work was supported in part by a grant for the RFTF (97L00503) from the Japanese Society for the Promotion of Science and by the Structural Biology Sakabe Project and by Grants-in-Aid for Scientific Research (No. 12480177) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
PDB no. 1J8L
References
- 1.Singer B. and Kusmierek,J.T. (1982) Chemical mutagenesis. Ann. Rev. Biochem., 52, 655–693. [DOI] [PubMed] [Google Scholar]
- 2.Kochetkov N.K. and Budowsky,E.I. (1969) The chemical modification of nucleic acids. Prog. Nucl. Acid Res. Mol. Biol., 9, 403–438. [DOI] [PubMed] [Google Scholar]
- 3.Chatake T., Hikima,T., Ono,A., Ueno,Y., Matsuda,A. and Takénaka,A. (1999) Crystallographic studies on damaged DNAs II. N6-methoxyadenine can present two alternate faces for Watson–Crick base-pairing, leading to pyrimidine transition mutagenesis. J. Mol. Biol., 294, 1223–1230. [DOI] [PubMed] [Google Scholar]
- 4.Chatake T., Ono,A., Ueno,Y., Matsuda,A. and Takénaka,A. (1999) Crystallographic studies on damaged DNAs I. An N6-methoxyadenine residue forms a Watson–Crick pair with a cytosine residue in a B-DNA duplex. J. Mol. Biol., 294, 1215–1222. [DOI] [PubMed] [Google Scholar]
- 5.Reeves S.T. and Beattie,K.L. (1985) Base pairing properties of N4-methoxydeoxycytidine 5′-triphosphate during DNA synthesis on natural templates, catalyzed by DNA polymerase I of Escherichia coli. Biochemistry, 24, 2262–2268. [DOI] [PubMed] [Google Scholar]
- 6.Singer B. and Spengler,S. (1981) Ambiguity and transcriptional errors as a result of modification of exocyclic amino groups of cytidine, guanosine, and adenosine. Biochemistry, 20, 1127–1132. [DOI] [PubMed] [Google Scholar]
- 7.Meervelt L.V., Moore M.H., Kong Thoo Lin,P., Brown,D.M. and Kennard,O. (1990) Molecular and crystal structure of d(CGCGmo4CG): N4-MethoxycytosineûGuanine base-pairs in Z-DNA. J. Mol. Biol., 216, 773–781. [DOI] [PubMed] [Google Scholar]
- 8.Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalyticlly active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- 9.Hossain M.T., Chatake,T., Hikima,T., Tsunoda,M., Sunami,T., Ueno,Y., Matsuda,A. and Takénaka,A. (2001) Crystallographic Studies on Damaged DNAs III. N4-Methoxycytosine can form both Watson–Crick type and wobbled base pairs in a B-form duplex. J. Biochem., 130, 9–12. [DOI] [PubMed] [Google Scholar]
- 10.Fazakerley G.V., Gdaniec,Z. and Sowers,L.C. (1993) Base-pair induced shifts in the tautomeric equilibrium of a modified DNA base. J. Mol. Biol., 230, 6–10. [DOI] [PubMed] [Google Scholar]
- 11.Moore M.H., Meervelt,L.V., Salisbury,S.A., Kong Thoo Lin,P. and Brown,D.M. (1995) Direct observation of two base-pairing modes of a cytosine-thymine analogue with guanine in a DNA Z-form Duplex: Significance for base analogue mutagenesis. J. Mol. Biol., 251, 665–673. [DOI] [PubMed] [Google Scholar]
- 12.Gdaniec Z., Ban,B., Sowers,L.C. and Fazakerley,G.V. (1996) Methoxyamine-induced mutagenesis of nucleic acids A proton NMR study of oligonucleotides containing N4-methoxycytosine paired with adenine or guanine. Eur. J. Biochem., 242, 271–279. [DOI] [PubMed] [Google Scholar]
- 13.Stone M.J., Nedderman,A.N.R., Williams,D.H., Kong Thoo Lin,P. and Brown,D.M. (1991) Molecular basis for methoxyamine initiated mutagenesis. 1H nuclear magnetic resonance studies of base-modified oligonucleotides. J. Mol. Biol., 222, 711–723. [DOI] [PubMed] [Google Scholar]
- 14.Kong Thoo Lin P. and Brown,D.M. (1989) Synthesis and duplex stability of oligonucleotides containing cytosine-thymine analogues. Nucleic Acids Res., 17, 10373–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie A.G.W. (1992) Molecular data processing. In Moras,D., Pojarny,A.D. and Thierry,J.-C. (eds), Crystallographic Computing 5, From Chemistry to Biology. Oxford University Press, UK, pp. 50–61.
- 16.Steller I., Bolotovsky,R. and Rossmann,M.G. (1997) An algorithm for automatic indexing of oscillation images using Fourier analysis. J. Appl. Crystallog., 30, 1036–1040. [Google Scholar]
- 17.Rossmann M.G. and van Beek,C.G. (1999) Data processing. Acta Crystallog. D, 55, 1631–1653. [DOI] [PubMed] [Google Scholar]
- 18.Powell H.R. (1999) The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallog. D, 55, 1690–1695. [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallog. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 20.Shui X., McFail-Ison,L., Hu,G.G. and Williams,L.D. (1998) The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- 21.Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallog. A, 50, 157–163. [Google Scholar]
- 22.Meervelt L.V. (1991) Structure of 3′,5′-di-O-acetyl-N4-methoxy cytosine. Acta Crystallog. C, 47, 2635–2637. [Google Scholar]
- 23.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.-S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallog. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.Bansal M., Battacharyya,D. and Ravi,B. (1995) NUPARAM and NUCGEN: Software for analysis and generation of sequence dependent nucleic acid structures. Comput. Appl. Biosci., 13, 281–287. [DOI] [PubMed] [Google Scholar]
- 25.Dickerson R.E. and Drew,H.R. (1981) Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J. Mol. Biol., 149, 761–786. [DOI] [PubMed] [Google Scholar]
- 26.Sines C.C., McFail-Isom,L., Howerton,S.B., VanDerveer,D. and Williams,L.D. (2000) Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc., 122, 11048–11056. [Google Scholar]
- 27.Pieber M., Kroon,P.A., Prestegard,J.H. and Chan,S.I. (1973) Erratum. Tautomerism of nucleic acid bases. J. Am. Chem. Soc., 95, 3408. [DOI] [PubMed] [Google Scholar]
- 28.Brown D.M., Hewlins,M.J.E. and Schell,P. (1968) The tautomeric state of N(4)-hydroxy- and of N(4)-amino-cytosine derivatives. J. Chem. Soc. Sect. C, 1925–1929. [DOI] [PubMed] [Google Scholar]
- 29.Morzov Y.V., Savin,F.A., Chekhov,V.O., Budowsky,E.I. and Yakovlev,D.Y. (1982) Photochemistry of N6-methoxyadenosine and of N4-hydroxycytidine and its methyl derivatives I: Spectroscopic and quantum chemical investigation of ionic and tautomeric forms: syn-anti isomerization. J. Photochem., 20, 229–252. [Google Scholar]
- 30.Brünger A.T. (1992) Free-Rvalue: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 32.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cystallogr. A , 47, 110–119. [DOI] [PubMed] [Google Scholar]