Abstract
Sample preparation for matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) of DNA is critical for obtaining high quality mass spectra. Sample impurity, solvent content, substrate surface and environmental conditions (temperature and humidity) all affect the rate of matrix–analyte co-crystallization. As a result, laser fluence threshold for desorption/ionization varies from spot to spot. When using 3-hydroxypicolinic acid (3-HPA) as the matrix, laser fluence higher than the threshold value reduces mass resolution in time-of-flight (TOF) MS as the excess energy transferred to DNA causes metastable decay. This can be overcome by either searching for ‘hot’ spots or adjusting the laser fluence. However, both solutions may require a significant amount of operator manipulation and are not ideal for automatic measurements. We have added various sugars for crystallization with the matrix to minimize the transfer of excess laser energy to DNA molecules. Fructose and fucose were found to be the most effective matrix additives. Using these additives, mass resolution for DNA molecules does not show noticeable deterioration as laser energy increases. Improved sample preparation is important for the detection of single nucleotide polymorphisms (SNPs) using primer extension with a single nucleotide. During automatic data acquisition it is difficult to routinely detect heterozygous A/T mutations, which requires resolving a mass difference of 9 Da, unless a sugar is added during crystallization.
INTRODUCTION
The success of matrix-assisted laser desorption/ionization (MALDI) in analysis of oligonucleotides has introduced a direct procedure for examination of products of molecular biology assays. The specificity and ‘softness’ of MALDI combined with the simplicity and speed of time-of-flight (TOF) mass spectrometers has been particularly attractive to genomics endeavors where both reliability and throughput are critical. For example, MALDI has been used to detect PCR products for deletion/insertion (1,2) and for microsatellite analyses (3–5), to detect sequencing ladders (6–9) and to detect primer extension products for polymorphism analyses (10–13). Despite its success, continued efforts are being made to enhance sensitivity and resolution of oligonucleotides either by new instrument design (14–17) or by innovative MALDI sample preparation (18–26).
Improved sensitivity is important in achieving lower limits of detection, e.g. for multiplexed assays where product yields may not be high for all variable sites on the DNA template. Sample miniaturization was previously shown to directly influence detection sensitivity (20,25). The miniaturized matrix spot size concentrates analyte molecules in a much smaller region, thus increasing the amount of analyte in the laser interrogation area. As a result, MALDI-mass spectrometry (MS) detection sensitivity can be dramatically improved.
Improved resolution is highly desirable in assays where masses of diagnostic DNA products are very close or partially overlap (single base extension and multiplex assays) and in linear TOF mass spectrometers, where achievable resolution is modest. A variety of factors can lead to degradation of resolution in MALDI-MS analysis of nucleic acids. Most notable are formation of salt adducts with sodium or potassium ions and fragmentation of the molecular ion during MALDI ionization. Formation of cation adducts depends on oligonucleotide preparation, namely the effectiveness of the cleaning step, while DNA fragmentation during MALDI is largely dependent on the nature of the matrix used. The use of cation exchange beads (27) or ammonium salts (28–31) has proven to be very advantageous in decreasing the amount of salt adducts with DNA. Similarly, addition of basic organic species (32–34) to DNA samples has been somewhat effective in decreasing salt adducts and hence in increasing mass resolution. DNA fragmentation during MALDI results from excess energy deposited during laser irradiation. Some matrices are referred to as ‘hot’ if they result in excessive fragmentation, e.g. 2,4,6-trihydroxyacetophenone (29,35,36), while others are ‘cool’, e.g. 3-hydroxypicolinic acid (3-HPA) (37) and 6-aza-2-thiothymine (38), and result in very little fragmentation. The internal energy of the analyte ions may be reduced by collision with an inert gas as in an ion trap (39) or Fourier transform ion cyclotron resonance (FT-ICR) cell (16,40). Addition of a sugar to the matrix may also have a ‘cooling’ effect. Sugars have been reported to enhance ionization and/or to reduce metastable fragmentation of ions produced by MALDI. For example, Beavis et al. (41) used sucrose and glucose as a matrix for the multi-photon ionization of dipeptides in a reflectron TOF mass spectrometer. The use of sugars suppressed fragment ions resulting from sample pyrolysis due to IR laser irradiation.
Wilkins’ (42,43) research group used sugars as additives in MALDI ionization of peptides and small proteins. In their experiment, a mixture of analyte and 2,5-dihydroxybenzoic acid matrix in methanol/water/TFA was saturated with a selected sugar and air sprayed onto the probe tip of a 7 T FT-ICR mass spectrometer. The sugar additives in conjunction with optimization of electrostatic deceleration times and laser energies allowed successful coupling of the energetic MALDI ions to a FT-ICR mass spectrometer. Further experiments demonstrated that sugar additives enhanced the observed resolution, presumably by slowing metastable decay of the molecular ions and providing the requisite population of long-lived ions (43). Recently, Russell and co-workers (26) showed that addition of fructose to the MALDI matrix reduced the internal energies of desorbed DNA ions, reduced fragment ions and increased the yield of molecular ions analyzed in a MALDI-TOF mass spectrometer operating in reflected ion mode. The matrices tested consisted of α-cyano-4-hydroxycinnamic acid, 2,4,6-trihydroxyacetophenone and ferulic acid prepared as dried droplets and a novel fast evaporation-overlayer method.
In this report we describe the effect of sugar additives on the MALDI-TOF spectra of synthetic oligonucleotides as well as extension products of the single base extension assay (11–12), which is used for single nucleotide polymorphism (SNP) analysis. We are specifically interested in the effect of the sugar additive on the resolution of oligonucleotides for application to single base extension assays. In these assays, a primer is annealed upstream of the variable (mutation) site in the DNA sequence and is extended up to the mutation site by a polymerase in the presence of all four dideoxynucleoside triphosphates. Masses of the extension products are diagnostic of the base added to the variable site. Homozygous samples produce one diagnostic product while heterozygous samples produce two, one for each allele. The smallest mass difference between the extension products is 9 Da, corresponding to the difference between dideoxyadenosine (ddA) and dideoxythymidine (ddT). This mass difference is not easily resolvable with commercial linear TOF instrumentation, especially in automated analyses where an operator is not searching for ‘hot spots’ in the sample. Currently, in our hands, resolution values of 500–700 can be routinely achieved for DNA oligomers in the 4–8 kDa mass range during fully automated runs where 20 shots are acquired at 10 or 20 Hz. This resolution, although satisfactory for Mass Extend reactions (10,13) where the mass difference is at least 1 nt, is limiting for single base extension assays of A/T heterozygotes.
Our investigations started with screening of a number of sugars for their resolution-enhancing ability by comparing the resolution of synthetic DNA oligomers in mass spectra acquired under threshold (standard) and high laser intensity conditions from control and sugar-containing matrix samples. After suitable sugars were chosen, sample preparation was optimized for both manual and automatic modes of data acquisition. The utility of the sugar additive was further explored for resolving synthetic oligonucleotides (differing in mass by only 8 Da) in a clean background and subsequently put to test with actual single base extension assay reaction products differing in mass by 9 Da (A/T heterozygotes) in a real background of enzymes and buffers analyzed in both manual and automatic data acquisition modes. The results of these experiments are discussed below.
MATERIALS AND METHODS
Sample preparation
The sugars d(+)-trehalose, d(–)-ribose, N-acetyl-d-glucosamine, d(+)-glucosamine, d(+)-raffinose pentahydrate, α-cyclodextrin and stachyose hydrate were purchased from Fluka (Milwaukee, WI). l-Fucose and d-fructose were purchased from Aldrich Chemicals (Milwaukee, WI) and d(+)-glucose and sucrose were purchased from Sigma Chemical Co. (St Louis, MO). Individual sugar solutions were prepared in deionized water at concentrations ranging from 1.0 to 6.0 g/l. To remove residual Na+ and K+ ions from the sugar solutions, cation exchange columns were used. Each cation exchange column was prepared by packing a Fortuna 20 ml syringe, lined at the bottom with a Whatman 6 filter paper, with a 50 g/l slurry of Bio-Rad AG 50W-X8 hydrogen form cation exchange resin. Before use, the columns were conditioned with 1 M ammonium acetate solution.
A synthetic 28mer oligonucleotide (5′-CCA TCC ACT ACA ACT ACA TGT GTA ACA G-3′, [M + H]+ 8486.6 Da) was purchased from Operon Technologies (Alameda, CA). A synthetic 23mer oligonucleotide (5′-GTT TCC ATT TAG TCA GTC AAC TG-3′, [M + H]+ 7004.6 Da) was purchased from Integrated DNA Technologies (Coralville, IA). A second synthetic 23mer oligonucleotide (5′-ACA TTC TTC ATA GCA TTT TAG A*A-3′, * = ribose, [M + H]+ 7012.6 Da) was purchased from Tri Link Bio Tech (San Diego, CA). A stock solution of each oligonucleotide was prepared by dissolving the oligonucleotide in 1× TE buffer (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA) to a final concentration of 100 µM, and was kept at –20°C. Working solutions of the oligonucleotides were made by diluting the stock solutions with deionized water to a concentration of 10 µM. A mixture of the two 23mer oligonucleotides was prepared by diluting the oligonucleotides to 0.63 µM with either deionized water or a selected sugar solution (6 g/l). About 5 µg of cation exchange resin equilibrated with ammonium ions was added to the oligonucleotide mixture to prevent the formation of any Na+/K+ adducts.
Detection of SNP with single nucleotide extension
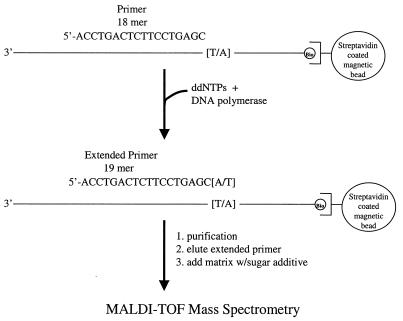
A selected region of human genomic DNA, namely Tissue Factor (TF), was first amplified using PCR. Genomic DNA was obtained from the Sequenom DNA banking facility. DNA samples were first amplified by PCR in a 50 µl volume containing 3 pmol reverse primer tagged with universal sequence at the 5′-end (13) (5′-AGC GGA TAA CAA TTT CAC ACA GGT TAT AAC TTG ACC GGG AAG AG-3′), 25 pmol forward primer (5′-CAC GTG TGG GAT TTA TCT TT-3′), 10 pmol 5′-biotinylated primer with the universal sequence (5′-biotin-AGC GGA TAA CAA TTT CAC ACA GG-3′), 2 µl of genomic DNA, 100 µM each dNTP, 1× Qiagen PCR buffer and 1 U Taq DNA polymerase, both purchased from Qiagen (Valencia, CA). The thermal cycling conditions of the PCR reaction, carried out in a Peltier Thermal 225 cycler (M.J. Research, Waltham, MA), were an initial incubation period of 15 min at 95°C and 45 subsequent cycles of denaturation at 95°C for 5 s, annealing at 56°C for 20 s and extension at 72°C for 30 s. The PCR products were analyzed by 1.5% agarose gel electrophoresis with ethidium bromide staining. An aliquot of 45 µl of biotinylated PCR product was then immobilized on 150 µg streptavidin-coated magnetic beads (Dynabeads M-280; Dynal, Lake Success, NY) following the manufacturer’s instructions. In brief, the beads were washed twice with 1 M NH4Cl and resuspended in 22.5 µl of 3 M NH4Cl. After mixing the beads with the PCR products, the mixture was incubated at room temperature for 20 min. After immobilization of the PCR products was complete, the double-stranded PCR products were denatured by incubating the beads in 50 µl of 100 mM NaOH solution at room temperature for 5 min. After capturing the beads on a magnet, the supernatant containing the non-biotinylated strand of the PCR products was removed and the beads were washed three times with 50 µl of 10 mM Tris–HCl, pH 8.0. A reaction cocktail for the single nucleotide extension (Fig. 1), which contained 260 mM Tris–HCl, pH 9.5, 65 mM MgCl2, 10 µM each ddNTP, 10 pmol specific primer and 2.5 U ThermoSequenase (Amersham, Piscataway, NJ), was added to the beads. The single nucleotide extension was carried out with an initial incubation period of 30 s at 80°C, followed by three cycles of 40°C for 15 s and 72°C for 30 s. After that, the beads were washed twice with 50 µl of 10 mM Tris–HCl, pH 8.0, to remove cations from the reaction and the extended primer was eluted in 5 µl of 50 mM NH4OH with heating at 60°C for 5 min.
Figure 1.
Schematic representation of single base extension products of an A/T heterozygote.
Mass spectrometry
The MALDI matrix, 3-HPA, was obtained from Fluka (Milwaukee, WI) and used without further purification. Standard matrix solutions were prepared by dissolving 35 mg 3-HPA and 7.14 mg diammonium citrate in 1 ml of 10% acetonitrile and water. Matrix solutions were filtered with syringe filters (0.2 µm i.d.; Alltech Inc., Deerfield, IL) prior to dispensing to remove any insoluble particles. Matrix and samples were either dispensed on silicon chips (96-well SpectroCHIPs) using a piezoelectric nano-plotter device (GeSim GmbH, Grosserkmannsdorf, Germany) or manually spotted on a stainless steel target. The volumes dispensed by the piezoelectric pipetter were ∼14 nl for both analyte and matrix, while 0.1 µl of both analyte and matrix were spotted manually on stainless steel targets.
All experiments were carried out in positive mode using both a Biflex III TOF-MS from Bruker (Bremen, Germany) and a Voyager DE TOF-MS from Applied Biosystems (Framingham, MA). Both instruments have a linear configuration and are equipped with a 337 nm N2 laser (Laser Science Inc., Franklin, MA). Instrument parameters on the Voyager DE were as follows: accelerating voltage +20 kV, grid voltage +18.9 kV, delay 400 ns, guide wire 0.001% of the accelerating voltage. The low mass ion gate was set at 3900 Da to reject ions with lower mass. The Voyager DE instrument is equipped with a 500 MHz LeCroy digitizer with a sample bin set at 5 ns, bandpass filter set at full (750 MHz) and sensitivity at 200 mV full scale. Each spectrum consisted of 5–50 laser shots processed with Data Explorer software (noise filter, correlation factor 0.7). Instrumental parameters used on the Biflex III TOF-MS were: accelerating voltage +20 kV, P2 lens voltage +18.9 kV, focusing lens voltage 9.4 kV, long delay >600 ns. The Biflex III is equipped with a LeCroy Waverunner digitizer; 6000 spectral points were acquired with the sample bin set at 5 ns. The bandwidth-limiting filter was on, corresponding to an input bandwidth of ∼250 MHz. The detector is gated so that ions below mass 2500 do not saturate the detector.
RESULTS AND DISCUSSION
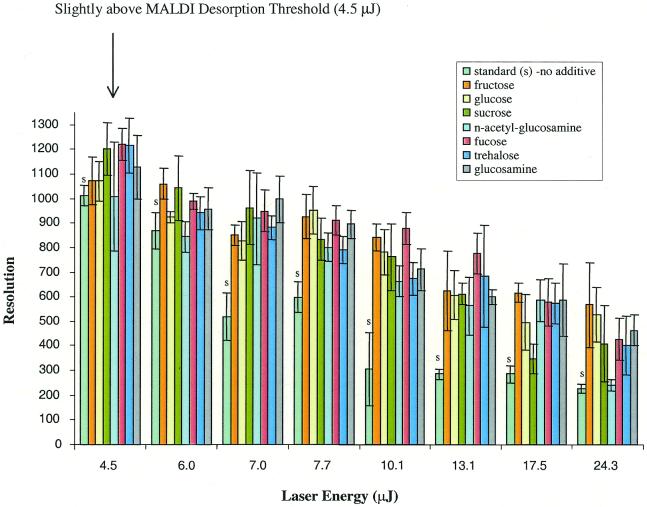
In this report we have examined the effects of sugars on the measurement of DNA using MALDI-TOF MS. In our initial experiments a pure 28mer oligonucleotide was used to represent the largest extended primer in the subsequent analysis of SNPs. Several sugars, including monosaccharides, disaccharides, trisaccharides and oligosaccharides, were selected to determine if appreciable differences in crystallization and improvements in mass resolution (m/Δm, where Δm is the peak width at half-maximum height) could be obtained. In these experiments the amount of sugar in the matrix solution was ∼4 g/l, slightly more than 10% in weight of the 3-HPA in the matrix solution. Mass spectra of the 28mer were manually acquired in standard matrix and sugar-doped matrices at laser intensities ranging from the desorption threshold to approximately six times the desorption threshold. The results of these experiments are shown in Figure 2. Based on these results all mono- and disaccharides tested appeared to improve the resolution of the 28mer over standard preparations, especially at higher laser intensities. The larger saccharides raffinose, α-cyclodextrin and stachyose, on the other hand, did not improve mass resolution of the 28mer (data not shown).
Figure 2.
Resolution of a synthetic 28mer (5′-CCA TCC ACT ACA ACT ACA TGT GTA ACA G-3′) as a function of laser energy (µJ) for selected sugar additives (4 g/l) added to the 3-HPA matrix (35 g/l). Each additive–matrix combination plus analyte was manually dispensed as five different spots and screened by manual data acquisition. From left to right are standard matrix without additive (S) and matrix with fructose, glucose, sucrose, n-acetylglucosamine, fucose, trehalose and glucosamine as additives, respectively.
It has been proposed that sugar additives might be degraded into smaller molecules such as CO2 and H2O during laser irradiation. This may create a dense environment that could act to collisionally cool the desorbed ions and slow the rate of decay (42,43), thus increasing resolution, since in FT MS resolution is directly proportional to the amount of time the transient ion signal is recorded. It was also reported that addition of sugars to the MALDI matrix increased the ion signal of peptide samples in a reflectron mode TOF instrument (44), presumably due to reduction of metastable fragmentation. The peptide matrices used in these studies are known to be ‘hot’ and may transfer excess energy to analytes. However, using a 3-HPA matrix we did not find any appreciable increase in ion intensity when sugar was added, even in a reflectron TOF instrument. This supports our finding that small oligonucleotides desorbed from a 3-HPA matrix are internally cool and metastable decay is not a major problem (16). The improvement in resolution found in the present study seems to support the assumption that there is a cooling effect (from gas phase collisions) that results in a narrower translational energy spread. It is also possible that the larger saccharides used in our experiments may not be as susceptible to decomposition as the smaller saccharides so that they could not provide enough small neutral molecules for collisional cooling, giving no improvements in mass resolution.
Another way to improve resolution in MALDI-TOF MS is through preparation of very flat samples. The flatness of sample preparations using sugar additives was visually inspected under a 10× microscope and compared to standard preparations. No appreciable differences were noted between crystals with and without sugar additives for samples deposited on the silicon chips (spot size 200 × 200 µm). Sugar additives also do not seem to provide a more homogeneous crystal distribution, nor do they improve DNA distribution in the matrix crystal. In fact, separate studies (45) have shown that DNA distribution in the matrix crystal is homogeneous while matrix distribution on the sample target is not. Similarly, microcrystals of miniaturized sample preparations still maintain variations in crystal distribution. However, the effect is minimized since the variation is now of the same order as the laser spot size (d ≈ 50–100 µm). These observations support our belief that the improvement in resolution is not related to sample flatness. The sugar-doped samples were only found to have a tougher crusted outer layer upon mass spectral analysis. Quality mass spectra were typically obtained after blasting the sample surface with a few laser shots at fluences higher than normally used for acquisition. This approach, known as matrix blasting, is commonly used to provide a clean sample surface before acquisition. Some attempts to remove the crusting using either accelerated drying with pressurized air or more volatile solvents to induce rapid evaporation were made. However, both methods were found to be unsuccessful as the matrix and sugar molecules did not dissolve well in organic solvents and accelerated drying was found to have no appreciable effect.
As noted in Figure 2, the highest resolution (>1000) was obtained at a laser energy (4.5 µJ) slightly above the MALDI desorption threshold in samples prepared with the sugar-doped matrix. In general, resolutions measured for the 28mer in the standard matrix were lower than those obtained in the sugar-doped matrix. With the use of sugar additives, the enhancing effect on resolution was more obvious at laser energies >13.1 µJ. Under these conditions the resolution was ∼400–800 and signal-to-noise (S/N) ratios ranged from 50 to 90 for the 28mer in the sugar-doped matrices, while the resolution was typically <325 and the S/N ratio <50 in the standard matrix. Although most monosaccharides produced similar results, the highest quality and most consistent mass spectra were obtained when fructose or fucose was present in the matrix. Therefore, these two sugars were used exclusively in the following studies.
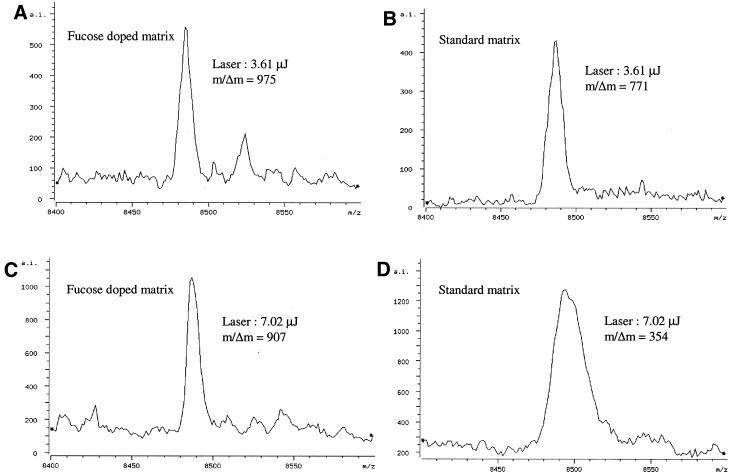
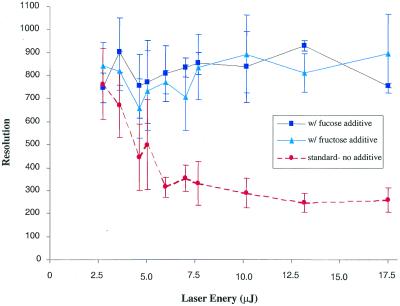
After the initial manual screening for suitable sugar additives, the amounts of sugar added to the matrix solution were varied in order to determine the effective sugar concentration for automatic data acquisition and to improve MALDI sample preparation. These studies were performed in automatic acquisition mode where five shots were summed after an initial blasting of the sample surface with eight high intensity laser shots (10.1 µJ). In automated acquisition the spectral peaks are evaluated for absolute intensity, resolution and S/N ratio using commercial fuzzy logic software. After the spectrum is saved, the sample translation stage in the ion source brings another sample spot into the focal point of the laser for interrogation and the process is repeated. It was found that high concentrations of sugar (>4 g/l) added to the matrix interfered with both matrix crystallization and desorption/ionization. The lowest sugar concentration tested was 1 g/l, a level below which little or no effect on resolution was observed. With 1 g/l sugar in the matrix solution, the following results were obtained for the 28mer. At threshold laser energy (3.6 µJ) the resolution of the 28mer in sugar-doped matrices was slightly higher than when using the standard matrix (Fig. 3A and B). However, as laser energies were increased to 7.0 µJ (Fig. 3C and D) the average resolution of the 28mer in the standard matrix measured from eight samples spots decreased drastically (m/Δm ≈ 353 ± 57), while the average resolution of the 28mer in the fucose-doped matrix remained high (∼834 ± 42). A plot describing the effects of increasing laser energies on resolution of the 28mer (averaged for eight different sample spots) in different matrices is shown in Figure 4. The resolution-enhancing effect of the sugar additives was more evident at higher laser energies. In summary, sugar concentrations ranging from 1 to 4 g/l were useful and significantly improved mass resolution over a standard matrix containing no sugar additives. The sugar concentration used in the remaining studies was 1–3 g/l for manual spotting. At high sugar concentrations, searching for ‘hot spots’ or spots with less crusting might be necessary, but most experienced users could easily accomplish this task. For automatic runs with samples deposited on silicon SpectroCHIPs as 200 × 200 µm spots, the lower sugar concentration of 1 g/l was more suitable because it reduced the time to locate ‘hot’ spots while still maintaining the resolution-enhancing effects. The matrix blasting option was always used during automated spectral acquisition in order to improve signal reproducibility. This is a straightforward option for the automatic mode settings of commercial MALDI-TOF instruments and only adds a fraction of a second to each acquisition. For example, usually eight laser shots at high fluence were used to blast the sample surface. At the laser repetition rate of 20 Hz, matrix blasting takes only an additional 0.4 s, which is only a fraction of the entire acquisition event, usually 3–4 s. By comparison, optimizing the laser energy for each sample spot during an automated run requires between three and ten times the standard acquisition time because the software evaluates several laser settings for each raster position.
Figure 3.
Mass spectra of a 28mer acquired in automatic mode. (B and D) Standard 3-HPA matrix (35 g/l); (A and C) fucose-doped (1 g/l) 3-HPA matrix. (A and B) At threshold energy (3.6 µJ); (C and D) above threshold energy (7.0 µJ).
Figure 4.
Average resolution as a function of laser energy (µJ) measured for a 28mer in: standard 3-HPA matrix (35 g/l) (filled circle); 3-HPA matrix (35 g/l) with fucose (1 g/l) (filled square); and 3-HPA matrix (35 g/l) with fructose (1 g/l) (filled triangle). All spectra were acquired in automatic mode. The resolution reported is an average of eight measurements from a single preparation dispensed as eight different spots.
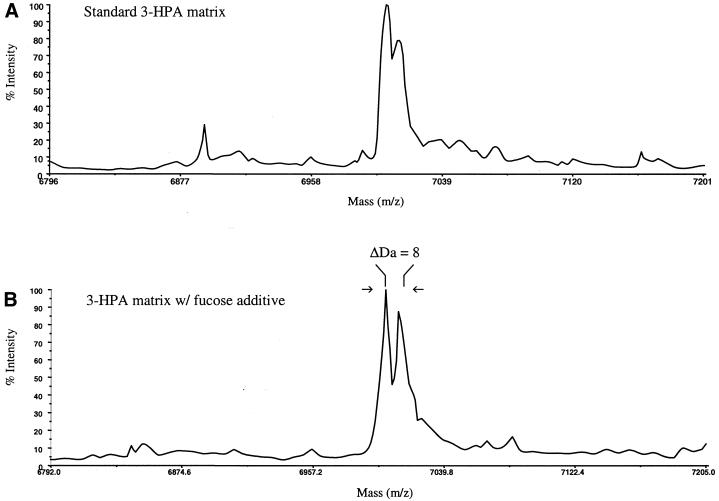
The utility of sugar additives in resolving closely spaced peaks was next tested with two pure synthetic oligonucleotides separated in mass by 8 Da (m/z 7004.6 and 7012.6 Da). The results obtained from manual data acquisition are shown in Figure 5. Laser energies around threshold conditions (∼3.0 µJ) were selected for standard 3-HPA matrix samples, while higher laser energy conditions (∼6.0 µJ) were selected for the sugar-doped matrix samples. These laser energies were chosen to directly compare the best possible resolution obtained for each matrix at its optimum laser energy level. Using the standard sample preparation (Fig. 5A), the two synthetic oligonucleotides could not be resolved at half height. The signal intensities for both 23mer oligonucleotides were 1.2 × 104 for m/z = 7004.6 Da and 9.4 × 103 for m/z = 7012.6 Da with 20 shots acquired per spectrum. However, the two oligonucleotide peaks were clearly better resolved by premixing samples with a fucose solution (final concentration of fucose ∼3 g/l). A resolution of 938 was obtained for the 7004.6 Da ion; the signal intensities were 1.9 × 104 for m/z = 7004.6 Da and 1.6 × 104 for m/z = 7012.6 Da when 20 shots were acquired per spectrum. For these samples, the sugar was premixed with the analyte and then dispensed onto the matrix, as this was found to further improve sample homogeneity.
Figure 5.
Manually acquired mass spectra of two synthetic 23mer oligonucleotides (5′-GTT TCC ATT TAG TCA GTC AAC TG-3′, [M + H]+ 7004.6 Da; 5′-ACA TTC TTC ATA GCA TTT TAG A*A-3′, * = ribose, [M + H]+ 7012.6 Da) separated in mass by only 8 Da. (A) Sample prepared using 3-HPA matrix without additives (laser energy at threshold, 3.0 µJ). (B) Sample prepared by addition of an equal volume of 6 g/l fucose solution to the analyte mixture (final fucose concentration 3 g/l) (laser energy above threshold, 6.0 µJ).
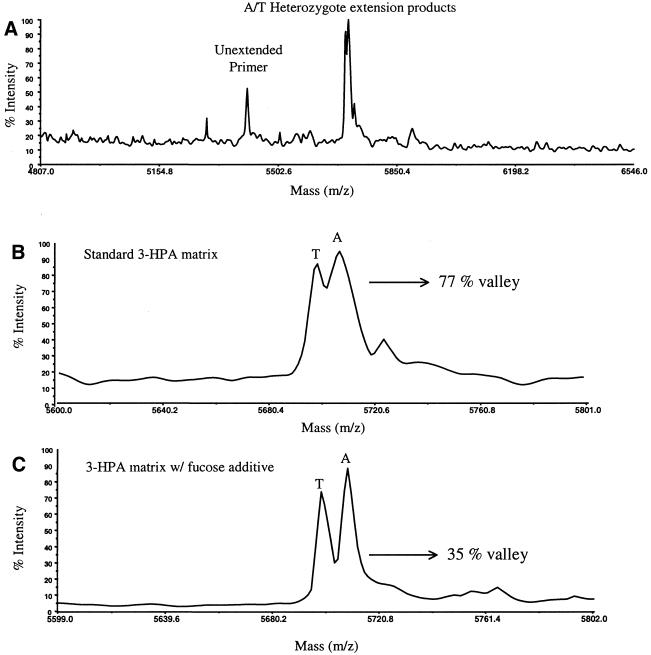
To further evaluate the utility of the sugar additives in the MALDI matrix, diagnostic products with a mass difference of 9 Da were analyzed. These products were obtained by extending a DNA primer with either ddA or ddT for a sample containing a heterozygous A/T mutation. Interference with sample–matrix co-crystallization by the chemical background of the sample makes it difficult to obtain mass spectra with high resolution from standard 3-HPA matrix samples. Figure 6 shows manually acquired mass spectra of the diagnostic products containing either A or T at the 3′-end using standard sample preparation with a laser energy slightly above threshold conditions. Again, the laser energies were chosen to directly compare the best resolution obtainable for each matrix at its optimum laser energy level. Laser energies at threshold conditions (3.0 µJ) were selected for standard 3-HPA matrix samples and a higher laser energy condition (∼6.0 µJ) was selected for the sugar-doped matrix sample. The unextended primer at 5410.6 Da was used to calibrate all the other peaks in the spectrum. The two diagnostic products were expected at 5698.8 (+ ddT) and 5707.8 Da (+ ddA). Figure 6A shows two peaks at m/z values of 5698.7 and 5707.2 Da, close to the expected values. The expanded view of this mass region displayed in Figure 6B shows these peaks to be poorly resolved (approximately to the 77% valley) with the standard 3-HPA matrix preparation. Searching the MALDI samples for ‘hot’ spots produced spectra with similar resolution for the two peaks. The ion signals were, respectively, 8.5 × 103 and 9.4 × 103 for m/z = 5698.7 and 5707.2 Da. An aliquot of the extension products was then premixed with an equal volume of 6 g/l fucose solution (final fucose concentration 3 g/l). The mass spectrum of this sugar-doped sample is shown in Figure 6C. As previously observed, the presence of the sugar in the sample greatly increased the resolution of the product peaks. The peaks corresponding to the A/T mutation were resolved to the 35% valley with resolutions of 845 and 888, respectively. The ion signals were 1.4 × 104 (m/z = 5699 Da) and 1.7 × 104 (m/z = 5707 Da) when the sugar additive was used. Results conclusively indicate that the presence of the sugar additive improves the resolution attained in MALDI-TOF mass spectra of these diagnostic products.
Figure 6.
Manually acquired mass spectra of A/T heterozygous products from a single base extension assay prepared (A) using a standard 3-HPA matrix and laser energy at the threshold level (3.0 µJ), with an expanded view of the A/T heterozygote products (B), and (C) by addition of an equal volume 6 g/l fucose solution to the analyte mixture (final fucose concentration 3 g/l) and using a laser energy above threshold (6.0 µJ).
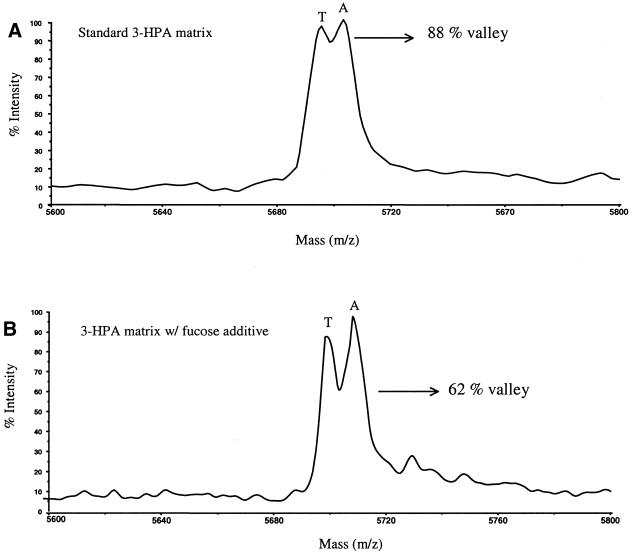
To determine if the resolution-enhancing effect could still be observed with automatic data acquisition, 14 nl of the single base extension products were dispensed with a piezo-electric pipetter onto SpectroCHIPs preloaded with either standard 3-HPA or fucose-doped 3-HPA matrix in an 8 × 12 array format. The fucose concentration used for automated runs was 1 g/l. The samples were measured in automatic mode with a set of pre-selected judging criteria comprising minimum signal intensity, S/N ratio and resolution in the mass region of interest. If these criteria were met, the mass spectrometer translation stage would move to the next sample for data acquisition. When these criteria were not met, the translation stage would move to a new raster position on the same sample spot and repeat the spectrum acquisition/judging step. After three attempts per sample spot the translation stage would move to a new spot for acquisition. The laser was set to threshold conditions (∼3.0 µJ) for standard matrix samples and well above threshold value (∼6.0 µJ) for sugar-doped matrix samples. These levels were previously found to be optimum for achieving high resolution spectra for each respective matrix. After the 96 sample spots were analyzed, all spectra were processed with Data Explorer software (noise filter, correlation factor 0.7). The mass spectra were analyzed and judged by the depth of separation (in percent valley) between the A/T heterozygote peaks. For example, Figure 7A and B shows mass spectra obtained from automatic measurement of the extension products in both matrices. The A/T peaks in the standard matrix (Fig. 7A) show the depth of separation of the two peaks to be at 88% valley. The same peaks analyzed with the fucose additive in the 3-HPA matrix (Fig. 7B) show the depth of separation of the two peaks to be at 62% valley. For the 96 samples analyzed the average depths of separation between the A/T peaks with both matrices are shown in Table 1. For the standard 3-HPA matrix it was found to be at 85.3 ± 5.1% valley, while for the 3-HPA matrix with fucose additive it was found to be lower at 67.3 ± 6.7% valley. These experiments showed that the fucose-doped matrices consistently produced spectra of the two peaks with higher resolution in automatic acquisition mode. In addition, it was also found that 23.9% of the standard matrix samples failed to resolve the two peaks, as only one broad peak was detected. That is in comparison to only 11.5% failure in fucose-doped matrix samples. In summary, our findings conclusively indicate that fucose-doped samples produce higher resolution spectra of A/T extension products compared to samples prepared in the standard matrix formulation in both manual and automated data acquisition modes. This is especially important in a high throughput environment where manual optimization and manipulation by an experienced user is not feasible. The ability to resolve oligonucleotides with very small mass differences demonstrates a viable application of this methodology to the analysis of diagnostic products and complex mixtures containing DNA oligomers with similar molecular weights.
Figure 7.
Mass spectra of the A/T heterozygous products from a single base extension assay acquired in automatic mode (A) with a standard 3-HPA matrix and laser energy set at threshold level (3.0 µJ) and (B) using fucose-doped matrix (1 g/l final fucose concentration) and a laser energy above threshold (6.0 µJ).
Table 1. Comparison of automatic data acquisition mode analysis of single base extension products of an A/T mutation with different 3-HPA matrices.
| Matrix |
Valley between heterozygote A/T peaks (%)a |
Failed sample spots (%)b |
| Standard 3-HPA | 85.3 ± 5.1 | 23.9 |
| Fucose-doped 3-HPA | 67.3 ± 6.7 | 11.5 |
96 MALDI samples were spotted on SpectroCHIPs and analyzed in automatic data acquisition mode.
a100% corresponds to no resolution between the two peaks (see Figs 6 and 7).
bPercentage of spots not resolved where the two peaks were detected as only one broad peak.
CONCLUSION
We have demonstrated that the quality of MALDI-TOF mass spectra is less dependent on laser fluence when selected sugars are added during matrix crystallization. The best results were produced by adding fructose or fucose to the MALDI matrix. Protocols for both manual and automatic modes of data acquisition were developed and used to obtain quality mass spectra. High resolution was maintained even with laser powers significantly higher than the desorption threshold. The improved formulation is important for routinely resolving oligonucleotides with small mass differences during automatic data acquisition, as shown in the selected single base extension assay for genotyping. The smallest mass change of 9 Da, representing an SNP of a heterogeneous A/T transition, was resolved and resolution could be maintained with a laser power higher than the desorption threshold. Therefore, during automatic data acquisition, a laser fluence higher than the desorption threshold could be chosen to guarantee ion generation without compromising mass resolution. This technique could be very useful for producing high quality mass spectra in cases where higher laser energy is needed to overcome low ion signals generated from weak assays. The increased resolution of the 3-HPA matrix with sugar additives could also prove to significantly improve the high throughput screening of DNA oligomers with similar masses, such as those in single base extension reactions or multiplex assays.
References
- 1.Ch’ang L.-Y., Tang,K., Schell,M., Ringelberg,C., Matteson,K.J., Allman,S.L. and Chen,C.H. (1995) Detection of ΔF508 mutation of the cystic fibrosis gene by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom., 9, 772–774. [DOI] [PubMed] [Google Scholar]
- 2.Bai J., Liu,Y., Liang,X., Zhu,Y. and Lubman,D.M. (1995) Procedures for detection of DNA by matrix-assisted laser desorption ionization mass spectrometry using a modified nafion film substrate. Rapid Commun. Mass Spectrom., 9, 1172–1176. [Google Scholar]
- 3.Ross P.L. and Belgrader,P. (1997) Analysis of short tandem repeat polymorphisms in human DNA by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem., 69, 3966–3972. [DOI] [PubMed] [Google Scholar]
- 4.Ross P.L., Davis,P.A. and Belgrader,P. (1998) Analysis of DNA fragments from conventional and microfabricated PCR devices using delayed extraction MALDI-TOF mass spectrometry. Anal. Chem., 70, 2067–2073. [DOI] [PubMed] [Google Scholar]
- 5.Taranenko N.I., Golovlev,V.V., Allman,S.L., Taranenko,N.V., Chen,C.H., Hong,J. and Chang,L.Y. (1998) Matrix-assisted laser desorption/ionization for short tandem repeat loci. Rapid Commun. Mass Spectrom., 12, 413–418. [DOI] [PubMed] [Google Scholar]
- 6.Roskey M.T., Juhasz,P., Smirnov,I.P., Takach,E.J., Martin,S.A. and Haff,L.A. (1996) DNA sequencing by delayed extraction-matrix-assisted laser desorption/ionizaton time of flight mass spectrometry. Proc. Natl Acad. Sci. USA, 93, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köster H., Tang,K., Fu,D., Braun,A., van den Boom,D., Smith,C.L., Cotter,R.J. and Cantor,C.R. (1996) A strategy for rapid and efficient DNA sequencing by mass spectrometry. Nat. Biotechnol., 14, 1123–1128. [DOI] [PubMed] [Google Scholar]
- 8.Fu D., Tang,K., Braun,A., Reuter,D., Darnhofer-Demar,B., Little,D.L., O’Donnell,M.J., Cantor,C.R. and Köster,H. (1998) Sequencing exons 5 to 8 of the p53 gene by MALDI-TOF mass spectrometry. Nat. Biotechnol., 16, 381–384. [DOI] [PubMed] [Google Scholar]
- 9.Kirpekar F., Nordhoff,E., Larsen,L.K., Kristiansen,K., Roepstorff,P. and Hillenkamp,F. (1998) DNA sequence analysis by MALDI mass spectrometry. Nucleic Acids Res., 26, 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun A., Little,D.P. and Köster,H. (1997) Detecting CFTR gene mutations by using primer oligo base extension and mass spectrometry. Clin. Chem., 43, 1151–1158. [PubMed] [Google Scholar]
- 11.Haff L. and Smirnov,I. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross P., Hall,L., Smirnov,I.P. and Haff,L. (1998) High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat. Biotechnol., 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 13.Tang K., Fu,D., Julien,D., Braun,A., Cantor,C.R. and Köster,H. (1999) DNA chip for genotyping by mass spectrometry. Proc. Natl Acad. Sci. USA, 96, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Y., Whittal,R.M., Weinberger,S.R. and Li,L. (1996) Accurate mass measurement of oligonucleotides using a time-lag focusing matrix-assisted laser desorption/ionization time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom., 10, 1792–1796. [DOI] [PubMed] [Google Scholar]
- 15.Juhasz P., Roskey,M.T., Smirnov,I.P., Haff,L.A., Vestal,M.L. and Martin,S.A. (1996) Applications of delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry to oligonucleotide analysis. Anal. Chem., 68, 941–946. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Tang,K., Little,D.P., Köster,H., Hunter,R.L. and McIver,R.T. (1996) High resolution MALDI Fourier transform mass spectrometry of oligonucleotides. Anal. Chem., 68, 2090–2096. [DOI] [PubMed] [Google Scholar]
- 17.Guo B., Wang,S. and Fan,Y. (2000) Improving the performance of MALDI-TOF in oligonucleotide analysis using a new SDIFA technology. Anal. Chem., 72, 5792–5797. [DOI] [PubMed] [Google Scholar]
- 18.Tang W., Zhu,L. and Smith,L.M. (1997) Controlling DNA fragmentation in MALDI-MS by chemical modification. Anal. Chem., 69, 302–312. [DOI] [PubMed] [Google Scholar]
- 19.Hunter J.M., Lin,H. and Becker,C.H. (1997) Cryogenic frozen solution matrixes for analysis of DNA by time-of-flight mass spectrometry. Anal. Chem., 69, 3608–3612. [DOI] [PubMed] [Google Scholar]
- 20.Little D.P., Cornish,T.J., O’Donnell,M.J., Braun,A., Cotter,R.J. and Köster,H. (1997) MALDI on a chip: analysis of arrays of low-femtomole to subfemtomole quantities of synthetic oligonucleotides and DNA diagnostic products dispensed by a piezoelectric pipet. Anal. Chem., 69, 4540–4546. [Google Scholar]
- 21.Hung K.C., Rashidzadeh,H., Wang,Y. and Guo,B.C. (1998) Use of paraffin wax film in MALDI-TOF analysis of DNA. Anal. Chem., 70, 3088–3093. [DOI] [PubMed] [Google Scholar]
- 22.Berkenkamp S., Kirpekar,F. and Hillenkamp,F. (1998) Infrared MALDI mass spectrometry of large nucleic acids. Science, 281, 260–262. [DOI] [PubMed] [Google Scholar]
- 23.Hung K.C., Ding,H. and Guo,B. (1999) Use of poly(tetrafluoroethylene)s as a sample support for the MALDI-TOF analysis of DNA and proteins. Anal. Chem., 71, 518–521. [DOI] [PubMed] [Google Scholar]
- 24.Kirpekar F., Berkenkamp,S. and Hillenkamp,F. (1999) Detection of double-stranded DNA by IR- and UV-MALDI mass spectrometry. Anal. Chem., 71, 2334–2339. [DOI] [PubMed] [Google Scholar]
- 25.Schuerenberg M., Luebbert,C., Eickhoff,H., Kalkum,M., Lehrach,H. and Nordhoff,E. (2000) Prestructured MALDI-MS sample supports. Anal. Chem., 72, 3436–3442. [DOI] [PubMed] [Google Scholar]
- 26.Koomen J.M., Russell,W.K., Hettick,J.M. and Russell,D.H. (2000) Improvement of resolution, mass accuracy and reproducibility in reflected mode DE-MALDI-TOF analysis of DNA using fast evaporation-overlayer sample preparations. Anal. Chem., 72, 3860–3866. [DOI] [PubMed] [Google Scholar]
- 27.Nordhoff E., Cramer,R., Karas,M., Hillenkamp,F., Kirpekar,F., Kristiansen,K. and Roepstoff,P. (1993) Ion stability of nucleic acids in infrared matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res., 21, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie G.J. and Yates,J.R. (1993) Analysis of oligodeoxynucleotides by negative-ion matrix-assisted laser desorption mass spectrometry. J. Am. Soc. Mass Spectrom., 4, 955–963. [DOI] [PubMed] [Google Scholar]
- 29.Pieles U., Zürcher,W., Schär,M. and Moser,H.E. (1993) Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a powerful tool for the mass and sequence analysis of natural and modified oligonucleotides. Nucleic Acids Res., 21, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y.F., Taranenko,N.I., Allman,S.L., Martin,S.A., Haff,L. and Chen,C.H. (1996) The effect of ammonium salt and matrix in the detection of DNA by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom., 10, 1591–1596. [DOI] [PubMed] [Google Scholar]
- 31.Li Y.C.L., Cheng,S.-W. and Chan,T.-W.D. (1998) Evaluation of ammonium salts as co-matrices for matrix-assisted laser desorption/ionization mass spectrometry of oligonucleotides. Rapid Commun. Mass Spectrom., 12, 993–998. [Google Scholar]
- 32.Simmons T.A. and Limbach,P.A. (1998) Influence of co-matrix proton affinity on oligonucleotide ion stability in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom., 9, 668–675. [Google Scholar]
- 33.Asara J.M. and Allison,J. (1999) Enhanced detection of oligonucleotides in UV MALDI MS using the tetraamine spermine as a matrix additive. Anal. Chem., 71, 2866–2870. [DOI] [PubMed] [Google Scholar]
- 34.Vandell V.E. and Limbach,P.A. (1999) Polyamine co-matrices for matrix-assisted laser desorption/ionization mass spectrometry of oligonucleotides. Rapid Commun. Mass Spectrom., 13, 2014–2021. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y.F., Chung,C.N., Taranenko,N.I., Allman,S.L., Martin,S.A., Haff,L.A. and Chen,C.H. (1996) The study of 2,3,4-trihydroxyacetophenone and 2,4,6-trihydroxyacetophenone as matrices for DNA detection in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom., 10, 383–388. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y.F., Taranenko,N.I., Allman,S.L., Taranenko,N.V., Martin,S.A., Haff,L.A. and Chen,C.H. (1997) Oligonucleotide sequencing by fragmentation in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom., 11, 897–903. [Google Scholar]
- 37.Wu K.J., Steding,A. and Becker,C.H. (1993) Matrix-assisted laser desorption time-of-flight mass spectrometry of oligonucleotides using 3-hydroxypicolinic acid as an ultraviolet-sensitive matrix. Rapid Commun. Mass Spectrom., 7, 142–146. [DOI] [PubMed] [Google Scholar]
- 38.Lecchi P., Le,H.M.T. and Pannell,L.K. (1995) 6-Aza-2-thiothymine: a matrix for MALDI spectra of oligonucleotides. Nucleic Acids Res., 23, 1276–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louris J.N., Cooks,R.G., Syka,J.E.P., Kelley,P.E., Stafford,G.C.Jr and Todd,J.F.J. (1987) Instrumentation, applications and energy deposition in quadrupole ion trap MS/MS spectrometry. Anal. Chem., 59, 1677–1685. [Google Scholar]
- 40.McIver R.T., Li,Y. and Hunter,R.L. (1994) High-resolution laser desorption mass spectrometry of peptides and small proteins. Proc. Natl Acad. Sci. USA, 91, 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beavis R.C., Lindner,J., Grotemeyer,J. and Schlag,E.W. (1988) Sample-matrix effect in infrared laser neutral desorption, multiphoton-ionization mass spectrometry. Chem. Phys. Lett., 146, 310–313. [Google Scholar]
- 42.Köster C., Castoro,J.A. and Wilkins,C.L. (1992) High-resolution matrix-assisted laser desorption/ionization of biomolecules by Fourier transform mass spectrometry. J. Am. Chem. Soc., 114, 7572–7574. [Google Scholar]
- 43.Castoro J.A. and Wilkins,C.L. (1993) Ultrahigh resolution matrix-assisted laser desorption/ionization of small proteins by Fourier transform mass spectrometry. Anal. Chem., 65, 2621–2627. [DOI] [PubMed] [Google Scholar]
- 44.Gusev A.I., Wilkinson,W.R., Proctor,A. and Hercules,D.M. (1995) Improvement of ion signal reproducibility and matrix/comatrix effects in MALDI analysis. Anal. Chem., 67, 1034–1041. [Google Scholar]
- 45.Horneffer V., Forsmann,A., Strupat,K., Hillenkamp,F. and Kubitscheck,U. (2000) Confocal laser scanning microscopy (CLSM) as a suitable imaging technique for studies of the analyte distribution in MALDI standard preparations. In Proceedings of the 48th ASMS Conference on Mass Spectrometry and Allied Topics, Long Beach, CA.