Abstract
Background and Aims
Clonal reproduction in polyploids is expected to exceed that in diploids, due to either the immediate direct effects of whole-genome duplication (WGD) or selection during establishment. The timing of polyploidy effects on clonality are largely unknown despite its hypothesized influence on polyploid success. This study tests the direction and timing of divergence in clonal traits in diploid and polyploid Chamerion angustifolium.
Methods
Root bud production and biomass allocation patterns were compared between diploids and synthesized tetraploids (neotetraploids), and between neotetraploids and naturally occurring tetraploids grown in a common environment.
Key Results
Neotetraploids produced more root buds and fewer sexual structures than diploids and natural tetraploids; diploids and natural tetraploids had similar root bud numbers and sexual investment. The root bud:inflorescence biomass ratio was 71 % higher in neotetraploids than in natural tetraploids. Root bud location suggests that ramet density in neotetraploid genets could be higher than in diploid genets.
Conclusions
WGD immediately increases investment in asexual vs. sexual reproduction in C. angustifolium, potentially promoting within-cytotype mating and establishment for neopolyploids. However, evolutionary change after the polyploidization event negates the direct effects of WGD. Natural polyploids and diploids have similar root bud production and biomass allocation patterns, probably resulting from habitat- and ploidy-mediated selection on polyploids to become more like diploids. These results highlight the value of studying the effects of polyploidization in young vs. established polyploids.
Keywords: Clonal reproduction, colchicine, Chamerion angustifolium (fireweed), neopolyploid, autopolyploidy, root buds, whole-genome duplication
INTRODUCTION
Polyploidy is considered to be a driving force behind adaptive divergence and diversification in the angiosperms (Otto and Whitton, 2000; Wood et al., 2009, Soltis and Soltis, 2016), although the mechanisms leading to the formation and establishment of polyploid populations are still poorly understood (Soltis et al., 2010). Autopolyploids often differ ecologically and phenotypically from their lower ploid parents (Stebbins, 1950; Levin, 1983; Ramsey and Schemske, 2002; Husband et al., 2016), but because studies are commonly performed on long-established cytotypes it is unclear whether differences are due to instantaneous changes associated with the whole-genome duplication (WGD) event (Stebbins, 1971; Otto and Whitton, 2000; Comai, 2005) or divergence through selection after the fact (Bretagnolle and Lumaret, 1995; Weiss-Schneeweiss et al., 2013). Newly produced, synthetic polyploids (i.e. neopolyploids) provide an opportunity to study the direct phenotypic effects of WGD separate from changes wrought by generations of selection in naturally occurring polyploid cytotypes. However, using neopolyploids to study the immediate ecological and evolutionary consequences of a polyploidization event is uncommon (Bretagnolle and Lumaret, 1995; Husband et al., 2008; Maherali et al., 2009; Baldwin and Husband, 2013; Oswald and Nuismer, 2010; Ramsey, 2011; Martin and Husband, 2012; Husband et al., 2016), and it frequently remains unknown how phenotypically similar neopolyploids are to diploids, or what processes contribute to differences between new and established polyploids.
The timing of phenotypic divergence can have a significant impact on the likelihood of polyploid establishment and persistence in sympatry with its progenitor (Ramsey, 2011; Husband et al., 2016). One of the most crucial barriers to establishment that neopolyploids must overcome is a frequency-dependent mating disadvantage, where rare polyploids experience a lack of same-cytotype mates resulting in a disproportionate number of between-cytotype fertilizations [minority cytotype exclusion (MCE); Levin, 1975]. Polyploid gametes are usurped by incompatible pollen, and subsequent odd-ploidy offspring have low viability and fertility, leading to low polyploid fitness and their eventual exclusion from the population (Husband, 2000; Baack, 2005). MCE may be weakened by increasing same-cytotype mating through shifts in mating system (Barringer, 2007; Husband et al., 2008), self-compatibility (Thompson and Lumaret, 1992; Robertson et al., 2011), flowering time (Husband and Sabara, 2004), flower morphology (Segraves and Thompson, 1999; Lim et al., 2008; Vallejo-Marin, 2012), eco-geographic differentiation (Martin and Husband, 2012; Thompson et al., 2014) and asexual reproduction via apomixis (Rodriguez, 1996; Otto and Whitton, 2000).
Asexual reproduction through clonal reproduction could also facilitate polyploid establishment (Stebbins, 1950; Thompson and Lumaret, 1992; Otto and Whitton, 2000; Weiss-Schneeweis et al., 2013). Both the evolutionary relationship between polyploidy and clonal reproduction and the underlying mechanisms behind any association remain understudied (Baldwin and Husband, 2013; Freeling, 2017; Herben et al., 2017; Kolář et al., 2017). Clonal reproduction can be broadly defined as vegetative asexual reproduction through the propagation of plant parts not involving seeds (Vallejo-Marin et al., 2010), such that a genetic individual (a genet) may consist of multiple, genetically identical, daughter ramets. Clonal reproduction can occur through a wide variety of modes (e.g. rhizomes, stolons, bulbils, plantlets and corms; Klimešová and De Bello, 2009).
Early researchers posited that a perennial life history and the ability to reproduce clonally may be direct consequences of a polyploidization event (Müntzing, 1936; Gustafsson, 1948), but it is more likely that WGD would quantitatively alter the clonal ability normally present in a diploid (Stebbins, 1950; Soltis et al., 2016). A WGD event can immediately alter phenotypic and reproductive patterns (Otto and Whitton, 2000; Ramsey and Schemske, 2002), and differences in the size or number of clonal propagules produced by a neopolyploid could be due to gene dosage effects (Guo et al., 1996; Ramsey and Schemske, 2002), or new gene interactions and functions (Osborn et al., 2003; Parisod et al., 2010; Roulin et al., 2013; Soltis et al., 2016). While the direction of genetic effects of WGD can be unclear, it is generally assumed that the genetic consequences of WGD will result in heightened rates of clonal reproduction in polyploids (Stebbins, 1950; Otto and Whitton, 2000; Herben et al., 2017). Neopolyploids could also experience modification to clonal traits as an artefact of the direct effects of WGD on other traits. Neopolyploids often have faulty sexual reproduction because of chromosome segregation irregularities (Comai, 2005; Cifuentes et al., 2010), resulting in sexual structures not being fully formed, or the abortion of malformed ovules (Stebbins, 1950; Levin, 2002). In this case, a re-allocation of resources towards clonal growth as an alternative form of reproduction could occur within the lifetime of a neopolyploid (Gustafsson, 1948; van Kleunen et al., 2002; Vallejo-Marin et al., 2010).
Enhanced clonal reproduction in neopolyploids could facilitate survival through the bottle-neck stage of establishment via physical competition and persistence even in the absence of sexual reproduction (Stebbins, 1950; Ramsey and Schemske, 2002; Husband et al., 2008). Moreover, since mating generally involves near neighbours (Levin and Kerster, 1974; Vallejo-Marin et al., 2010), producing spatially proximate clones will increase rates of geitonogamous pollen transfer between ramets in a genet (Charpentier, 2002; Baack, 2005), and increase the number of potential same-cytotype mates within a predominantly diploid population. If the costs of selfing are less than those of between-cytotype mating, the probability of polyploid establishment will increase as more same-cytotype matings occur and the negative effects of MCE are circumvented (Husband and Schemske, 1997; Barringer, 2007; Husband et al., 2008).
During the establishment process, we expect that selection will maintain or further increase clonal reproduction in polyploids to extend the associated advantageous conditions for spreading within the population until more sexually produced same-cytotype mates become available (Müntzing, 1936; Eckert, 2002; Honnay and Bossuyt, 2005). Moreover, since polyploids are expected to have lower inbreeding depression than diploids (Lande and Schemske, 1985; Barringer, 2007; Husband et al., 2008), geitonogamous selfing in polyploids should result in a smaller fitness reduction than in diploids. Thus, once fully established, naturally occurring polyploid cytotypes should be able to tolerate larger, more compact genets and have optimal fitness at higher rates of clonal reproduction than diploids (Baldwin and Husband, 2013).
Previous research quantifying clonal reproduction in polyploids has presented conflicting results. There is some support for the prediction that higher ploidies will have increased levels of clonality in comparison with diploids (Bruneau and Anderson, 1988; Hroudová and Zákravský, 1993; Schlaepfer et al., 2010), while other studies have found that polyploids produce fewer clonal propagules than diploids (Schulze et al., 2013; Baldwin and Husband, 2013; Hanzl et al., 2014; Martínková et al., 2015), and some have demonstrated no differences between ploidies (Keeler, 2004). The above sudies have all focused on naturally occurring ploidy levels, and there are no studies that have used neopolyploids in determining the immediate effects of WGD on clonal reproduction.
Here, we use the mixed-ploidy species Chamerion angustifolium to investigate the differences in clonal reproduction between diploids and tetraploids and whether such differences are the direct result of WGD or differential selection in natural populations. Baldwin and Husband (2013) previously tested for differences in size and spatial extent of clones between diploids and tetraploids in natural populations of this species, but did not assess the capacity for clonal reproduction in neotetraploid C. angustifolium. We investigate the following three questions. (1) Are polyploids more clonal than their diploid progenitors? (2) Do differences in clonal reproduction occur immediately following WGD? (3) Do differences arise through evolutionary modification in natural populations? To address these questions, we compare clonal reproduction via root bud production between diploid, naturally occurring tetraploid and synthetic neotetraploid C. angustifolium grown under common greenhouse conditions. Neotetraploids are contrasted with diploids to address the second question explicitly, and with naturally occurring tetraploids to address the third.
MATERIALS AND METHODS
Study species
Chamerion angustifolium is a perennial, self-compatible, insect-pollinated herb occurring widely across the northern hemisphere in predominantly open or disturbed habitat. This species exhibits ploidy variation with diploid (2n = 2x = 36), tetraploid (2x = 4x = 72) and infrequent triploid individuals occurring naturally in North America (Sabara et al., 2013). Tetraploids are autotetraploids, derived from the doubling of the diploid C. angustifolium genome (Roy, 2008). Typically, diploids occur at higher latitude and altitude than tetraploids, but mixed populations occur in a diffuse contact zone across southern parts of the boreal forest and in the Rocky Mountains in North America (Husband and Schemske, 2000; Sabara et al., 2013). Chamerion angustifolium reproduces both sexually through perfect flowers, and asexually through the production of independent plants via vertically growing adventitious root buds (Mosquin, 1966; Stocklin, 1992). Both diploids and tetraploids have high inbreeding depression and strongly outcrossing mating systems (Husband et al., 2008; Ozimec and Husband, 2011). In contrast, previous studies have found that newly synthesized neotetraploid C. angustifolium have a significantly lower cost of inbreeding than naturally occurring tetraploids (Husband et al., 2008). Consequently, fitness gains through sexual reproduction for diploids, naturally occurring tetraploids and neotetraploids can be greatly impacted by the spatial arrangement of ramets within a genet as determined by patterns of investment in clonal reproduction (Charpentier, 2002; Vallejo-Marin et al., 2010; Van Drunen et al., 2015).
Source material, neopolyploid synthesis and ploidy determination
Root bud production was compared between greenhouse-grown plants from three available cytotypes of C. angustifolium (diploid, naturally occurring tetraploid and newly synthesized tetraploids) in the summer of 2016. Diploid and natural tetraploid seed of C. angustifolium were the F1 progeny of within-population within-cytotype crosses performed during 2015 on plants collected from ten locations in the Rocky Mountains (four diploid populations, four tetraploid populations and two of mixed ploidy; Table 1). A total of 18 diploids and 18 natural tetraploids were used in this study, from three crosses within each of the six populations per naturally occurring cytotype (Table 1).
Table 1.
Source population locations for C. angustifolium within the Rocky Mountains and the number of individuals of each ploidy used in the current study
| Population | Ploidy | Latitude | Longitude | Number of individuals | ||
|---|---|---|---|---|---|---|
| 2x | 4xNeo | 4x | ||||
| Jasper Park Boundary | 2x | N 51°26.674 | W 116°12.532 | 3 | 3 | – |
| Marmot Basin | 2x | N 52°48.100 | W 118°04.956 | 3 | 2 | – |
| Wilcox Creek | 2x | N 52°13.082 | W 117°10.621 | 3 | – | – |
| Fortress Mountain | 2x | N 50°49.534 | W 115°12.052 | 3 | 4 | – |
| Moose Meadows | 4x | N 51°15.231 | W 115°52.336 | – | – | 3 |
| Powderface Low | 4x | N 51°01.318 | W 114°53.861 | – | – | 3 |
| Sibbald Mountain | 4x | N 51°03.160 | W 114°56.847 | – | – | 3 |
| Barrier Lake | 4x | N 51°01.785 | W 115°02.097 | – | – | 3 |
| Coleman Clearcut | Mixed | N 50°18.648 | W 114°36.761 | 3 | 2 | 3 |
| Rampart Creek | Mixed | N 52°02.511 | W 116°51.767 | 3 | 2 | 3 |
| Mount Kitchener | Mixed | N 52°16.461 | W 117°18.538 | – | 2 | – |
| Total | 18 | 15 | 18 | |||
Neotetraploids (4xNeo) are listed under the source population of their maternal parent.
To obtain newly synthesized tetraploid seed, diploid seedlings were treated with colchicine, resulting in genome duplication and conversion to tetraploids (i.e. neotetraploids, 4xNeo). The diploid seeds used originated from the diploid and mixed-ploidy populations indicated in Table 1. Seeds were sown in Petri dishes on moist filter paper and individually treated with 40 μL of 0.2 % colchicine solution after 10 d when cotyledons were expanded and the seedlings exhibited vertical growth. Eighteen hours after application, the colchicine was rinsed from the seedlings twice with deionized water, and seedlings were transplanted onto soil. Crosses were performed between 11 neotetraploid individuals surviving to maturity, but due to high rates of pollen sterility in the small number of successfully converted neotetraploids F1 crosses were performed both within and between population sources. A total of 15 neotetraploid individuals resulting from eight unique crosses were used in the current study. Neotetraploids are listed in Table 1 under the population source of their maternal parent.
Diploid, naturally occurring tetraploid and neotetraploid seeds were sown on moist filter paper in Petri dishes and kept in the dark at 4 ºC for 24 h before moving to a Percival growth cabinet for germination at 22 ºC over 2 weeks. Seedlings were transplanted into a mixture of 5:1 Sunshine Mix to turface for a further 4 weeks of growth for seedling establishment. Individuals were then transplanted into HML Elite 1000 pots (8.83 L) with the same soil composition, and randomly placed on a greenhouse bench. This final pot size is an approx. 40 % larger volume than those used in the previous study (Baldwin and Husband, 2013) in order to ensure that the root systems were not severely root bound (coiling roots tend to influence the location of root bud elongation, W. E.Van Drunen, pers. obs.).
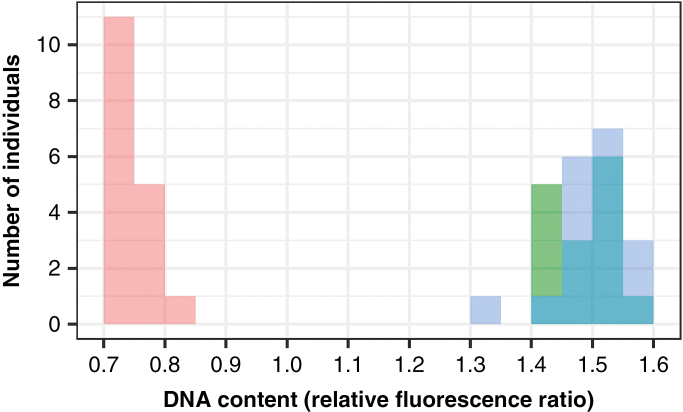
The ploidy of all plants was confirmed using estimates of DNA content via a FACSCalibur flow cytometer (BD Bioscience, San Jose, CA, USA). Approximately 1 cm2 of dried leaf tissue per plant was chopped with a clean razor blade along with an equal amount of internal standard (Solanum lycopersicum) in 0.7 mL of modified DeLaat’s buffer (Kron and Husband, 2009) with 50 μg mL–1 of the DNA-selective fluorochrome propidium iodide and 50 μg mL–1 RNase, and then passed through a 30 μm filter. Samples were stained for a minimum of 20 min. Relative fluorescence was measured with the FL2 detector (585/42 nm) and DNA content was quantified with FL2-area (integrated fluorescence). Ploidy was determined by estimating the mean relative fluorescence of the nuclei for each sample using ModFit LT (Verity Software House Inc., San Jose, CA, USA). Sample fluorescence peaks violating quality thresholds [coefficient of variation (CV) >8, nuclei count <500] were run twice and excluded if the peak was not identifiable as diploid or tetraploid. Fluorescence peaks are generally clearly recognizable in C. angustifolium, as the mean fluorescence values for diploid, tetraploid and the internal standard S. lycopersicum are non-overlapping. Figure 1 shows the distribution of fluorescence values for the three cytotypes in this study relative to those of the internal standard. The relative fluorescence values for 98 % of the individuals used in this study fell within 10 % of the mean fluorescence value for diploids (0.748 ± 10 % = 0.673–0.823), natural tetraploids (1.488 ± 10 % = 1.339–1.637) and neotetraploids (1.498 ± 10% = 1.348–1.648). The single sample that fell outside of these ranges was assigned to the nearest ploidy. Peak nuclei counts, CVs and relative fluorescence means for each cytotype are shown in Table 2.
Fig. 1.
Distribution of DNA content for diploids (2x, red), neotetraploids (4xNeo, green) and naturally occurring tetraploids (4x, blue), determined through flow cytometry on dried leaf tissue. Overlaps between neotetraploid and naturally occurring tetraploid distributions are darker blue. DNA content is the relative fluorescence ratio of each sample peak to the internal standard.
Table 2.
Flow cytometry on leaf tissue from diploids (2x), neotetraploids (4xNeo) and naturally occurring tetraploids (4x)
| Ploidy | n | Events per fluorescence peak | Peak CV | Relative fluorescence |
|---|---|---|---|---|
| 2x | 18 | 1278 ± 88 | 4.51 ± 0.27 | 0.748 ± 0.005 |
| 4xNeo | 15 | 2073 ± 213 | 3.42 ± 0.15 | 1.488 ± 0.012 |
| 4x | 18 | 1262 ± 138 | 3.91 ± 0.25 | 1.498 ± 0.012 |
Quality thresholds for peak CVs (CV <8) and nuclei counts (>500) were met for all samples. Values are means ± s.e.
Growth, biomass allocation and root bud traits
Plants were grown for 12 weeks under greenhouse conditions within the normal range experienced by C. angustifolium in source populations during the flowering season (Thompson et al., 2014), with a daylength of approx. 16 h and a day temperature of 22 ºC. Plants were watered biweekly and given fertilizer (Plant Prod pH Reducer, 18-9-18) on a weekly basis. After 12 weeks, plants were moved outside for 1 week to stimulate the beginning of senescence, then harvested. Above- and below-ground measurements were taken for all plants. Plant height (from the soil surface to the top of the primary stem) and the number of axillary branches per plant were assessed. The above-ground biomass of each plant was harvested and stored in paper bags. The above-ground biomass was separated into shoots and inflorescences, where an inflorescence was defined as all shoot mass above the lowest flower produced on a stem or branch. Inflorescence mass was used as a proxy for investment in sexual reproduction. Above-ground shoots and inflorescences were dried at 65 ºC for at least 72 h before being weighed.
After the above-ground biomass was harvested, root systems were gently excavated from the soil, washed, and stored at 4 ºC to inhibit further growth before root bud measurements could be taken. Root buds were identified by their light coloration, scaly appearance and vertical growth. Root buds were located along the roots, counted and their distances from the primary shoot recorded. Distances were measured on straightened roots, not on 3-D root topology as it grew in the pot, and thus are potential rather than realized distances. Root bud heights (base to tip) were measured using digital calipers. Root buds <1.5 mm in height have a similar appearance to small root initials, and as such were excluded from measurement. Root buds were removed from the roots and dried in silica for approx. 1 week before total root bud mass per plant was measured. Individual root bud mass was calculated as the total root bud mass divided by the number of root buds. Roots were placed in paper bags and dried at 65 ºC for at least 72 h before being weighed.
Statistical analysis
All analyses were performed in R (3.4.0; R Core Team, 2017). Linear mixed models were used to determine the effect of ploidy on plant growth, biomass allocation and root bud attributes. Models were formulated with the ‘lmer’ function in R (‘lme4’ package; Bates et al., 2015) and response variables were transformed where necessary in order to adhere to model assumptions (see Table 3). Plant growth traits included plant height and the number of axillary branches produced off the main stem of the plant. Total dry biomass was divided into four categories: above-ground shoot mass, inflorescence mass, root mass and root bud mass. Root bud attributes were the number of root buds, the average mass of an individual root bud, average root bud height, average bud distance along the root from the primary stem, and the minimum and maximum mean root bud distances per individual. The minimum and maximum root bud distances were included to provide information on the potential spatial extent and aggregation of a growing genet. To control for the effects of plant size on the measured traits, total biomass (growth and biomass attributes) or root mass (root bud attributes) were used as covariates in the models (see Table 3). To include variability between individual root buds more accurately, two additional models were used to quantify the effect of ploidy on root bud height and distance for all 1929 root buds identified. Significance testing for ploidy in all models was performed through likelihood ratio tests (LRTs) comparing full and reduced models for each response variable.
Table 3.
Results of linear mixed models for plant growth, biomass allocation and root bud characteristics
| TF | COV | Ploidy | Random effects | |||||
|---|---|---|---|---|---|---|---|---|
| LRT χ2 | d.f. | P | LRT χ2 | d.f. | P | |||
| Growth attributes | ||||||||
| Plant height (cm) | – | TM | 9.97 | 2 | 0.007 | 0 | 1 | 1 |
| Branches | – | TM | 13.66 | 2 | 0.001 | 0.01 | 1 | 0.917 |
| Biomass allocation | ||||||||
| Total mass (g) | – | – | 11.54 | 2 | 0.003 | 0 | 1 | 1 |
| Shoot mass (g) | – | TM | 19.14 | 2 | <0.0001 | 3.05 | 1 | 0.080 |
| Inflorescence mass (g) | – | TM | 17.03 | 2 | <0.001 | 3.95 | 1 | 0.047 |
| Root mass (g) | SR | TM | 0.85 | 2 | 0.652 | 0 | 1 | 1 |
| Root bud mass (mg) | SR | TM | 2.71 | 2 | 0.257 | 0.12 | 1 | 0.724 |
| Root bud mass/inflorescence mass (mg g–1) | FR | TM | 8.85 | 2 | 0.012 | 2.88 | 1 | 0.090 |
| Root bud attributes | ||||||||
| Bud number | SR | RM | 9.20 | 2 | 0.010 | 0.01 | 1 | 0.987 |
| Individual root bud mass (mg) | FR | RM | 2.49 | 2 | 0.288 | 0.01 | 1 | 0.904 |
| Average bud height (mm) | LOG | RM | 1.17 | 2 | 0.556 | 0 | 1 | 1 |
| Average bud distance (cm) | LOG | RM | 1.41 | 2 | 0.494 | 3.51 | 1 | 0.061 |
| Minimum bud distance (cm) | LOG | RM | 1.30 | 2 | 0.522 | 0.62 | 1 | 0.430 |
| Maximum bud distance (cm) | LOG | RM | 1.30 | 2 | 0.522 | 1.76 | 1 | 0.185 |
| All root buds (n = 1929) | ||||||||
| Bud height (mm) | LOG | – | 0.20 | 2 | 0.905 | 577.99 | 2 | <0.0001 |
| Bud distance (cm) | LOG | – | 237.39 | 3 | <0.0001 | 292.02 | 2 | <0.0001 |
Transformations (TFs) were performed on some variables to improve normality (SR – , FR – , LOG – log10). Total plant mass (TM) or root mass (RM) were used as covariates (COV) in the models. Test statistics from mixed models of the effect of ploidy and random effects on each characteristic are shown with likelihood ratio tests (LRTs) between full and reduced models along with the P-value associated with the LRT. Random effects for models for mean bud height and mean bud distance include population and plant nested within population, while all other models include population only. Significant test results are in bold.
All mixed models had source population as a random effect, and the two models, including all root bud height and distance measurements, contained both population and individual plant nested within population as random factors. To assess the contribution of random effects, the full model was compared with an analogous analysis of variance (ANOVA) model with no random component.
We tested for a relationship between root bud growth (height) and location along the root (distance from the primary stem) for all root buds identified using linear regression and linear mixed models in the R function ‘lmer’. Linear regression was performed on the raw root bud height and distance data across all plants to investigate their relationship for all cytotypes. Root bud height was then set as the response variable in a linear mixed model, while distance along the root, ploidy and their interaction were fixed effects. Both root bud height and distance were log transformed to improve model residual normality. Source population and individual plant nested within population were included in the model as random effects. Significance testing for fixed and random factors was performed through LRTs as described above.
RESULTS
Growth, biomass allocation and root bud traits
Cytotypes differed with respect to several growth traits, biomass allocation patterns and root bud attributes (Table 3; for raw variable distributions and means, see Supplementary Data Figs S1 and S2; Table S1). Tetraploids were significantly taller than both diploids and neotetraploids, while there was no difference in plant height between diploids and neotetraploids (Table 3). Neotetraploids had a significantly less branched growth pattern than diploids and tetraploids, while diploids and tetraploids did not differ in axillary branch number (Table 3).
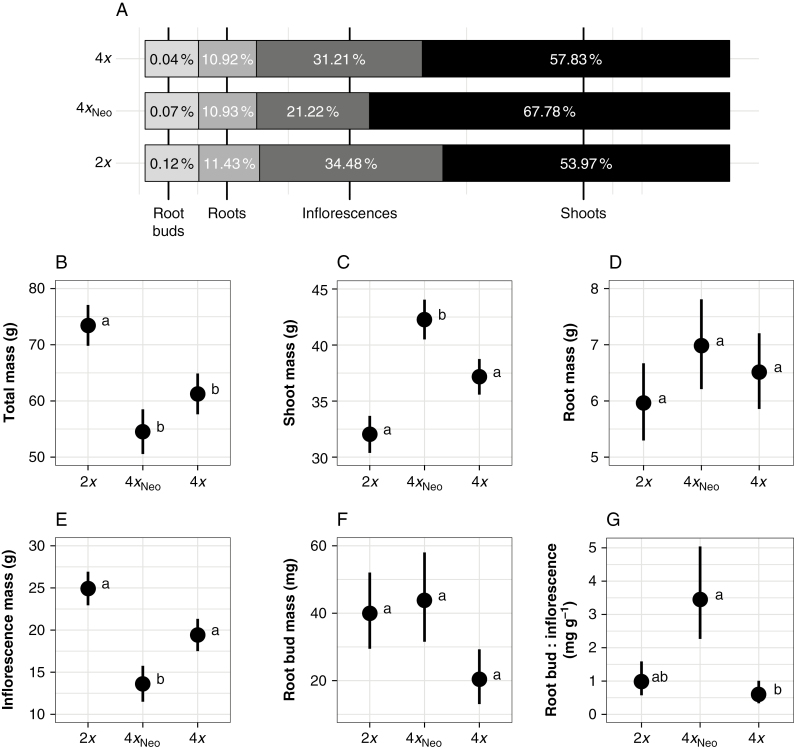
Diploids had a greater mean total biomass than both naturally occurring tetraploids and neotetraploids, whereas tetraploids and neotetraploids had comparable values (Table 3; Fig. 2B). Investment in shoot mass relative to total plant biomass was significantly higher in neotetraploids than in either diploids or tetraploids (68 % of total biomass vs. 54 and 58 %; Table 3; Fig. 2A, C). There were no differences in the relative investment in roots between cytotypes (Table 3; Fig. 2A, D). Mean inflorescence biomass in neotetraploids was significantly lower than in diploid and tetraploid plants (54 and 40 % lower), whereas tetraploid and diploid means were statistically indistinguishable (Table 3; Fig. 2E). Investment in inflorescence biomass relative to total plant biomass was lowest in neotetraploids; diploids and tetraploids invested 34 and 31 %, respectively, of their total biomass in inflorescences vs. only 21 % in neotetraploids (Fig. 2A). The raw mean root bud mass in diploids was greater than that in both neotetraploids and tetraploids (53 and 75 % higher; Supplementary Data Table S1), and neotetraploids had a higher mean total root bud mass than tetraploids (47 % higher; Supplementary Data Table S1), but no differences were significant due to high variability in total root bud mass (Table 3; Fig. 2F). The proportion of root bud mass relative to total plant biomass was similar across cytotypes (Fig. 2A).
Fig. 2.
Biomass allocation (A) for diploids (2x), neotetraploids (4xNeo) and naturally occurring tetraploids (4x). The percentage of the total biomass that a category represents for each cytotype is shown in white in each bar, and root bud biomass measures have been adjusted to the scale of the other biomass categories by adding 10 % of the average total biomass for each cytotype. (B–G) The adjusted least-square means (± s.e.) for each biomass category. Letters in (B–G) represent the Tukey’s HSD groupings for each cytotype according to the mixed models in Table 3.
Largely due to differences in allocation to sexual structures, there was a marked difference in the root bud:inflorescence biomass ratio between neotetraploids and tetraploids. Neotetraploids had a 71 % higher root bud mass to inflorescence mass ratio than tetraploids, but diploids did not differ from either neotetraploids or tetraploids (Table 3; Fig. 2G).
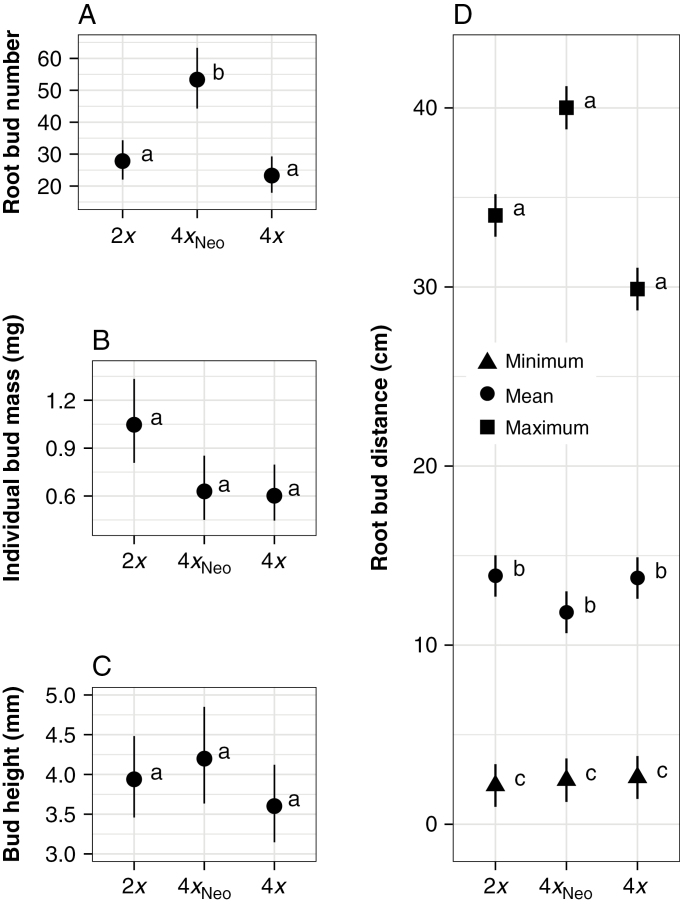
Neotetraploids produced a significantly higher mean number of root buds than both tetraploids and diploids (Table 3; Fig. 3A). Root bud production in neotetraploids was 29 % higher than in diploids and 49 % higher than in natural tetraploids (Supplementary Data Table S1). Diploids produced more root buds than tetraploids (27 % more; Supplementary Data Table S1), but this difference was not significant (Table 3; Fig. 3A).
Fig. 3.
Adjusted least-square mean values (± s.e.) of root bud attributes for diploids (2x), neotetraploids (4xNeo) and naturally occurring tetraploids (4x). (D) Minimum, mean and maximum root bud distances for each cytotype. Letters in (A–D) represent the Tukey’s HSD groupings for each cytotype according to the mixed models in Table 3.
Diploids had a mean individual root bud mass approximately twice that of both tetraploids and neotetraploids (Supplementary Data Table S1), but these differences were not significant due to large variances in mass among the buds per individual (Table 3; Fig. 3B). No differences were found between the cytotypes for mean root bud height per individual or for all root buds measured (Table 3; Fig. 3C), though diploids tended to have higher raw mean values in comparison with tetraploids and neotetraploids (Supplementary Data Table S1). The mean average root bud distance per individual was not significantly different between the cytotypes (Table 3; Fig. 3D). However, when the distance measurements from all 1929 root buds were included, diploids had a higher average distance than neotetraploids (Table 3). There were no cytotype differences in mean minimum and maximum root bud distances (Table 3; Fig. 3D).
Root bud height vs. distance
There was no significant relationship between root bud height and distance along the root for any of the three cytotypes. Fitting linear models to the raw distance and height data for each root bud measured revealed that the linear line of best fit for neotetraploids and tetraploids had slopes that were not different from zero (Supplementary Data Fig. S3; 4xNeo = 0.006 ± 0.019 s.e., P > 0.05; 4x = 0.031 ± 0.040, P > 0.05). The diploid model had a slightly negative slope estimate, though it was also not statistically different from zero (Supplementary Data Fig. S3; 2x = –0.039 ± 0.020, P > 0.05). The results of the linear mixed model with ploidy as a covariate showed that neither root bud distance nor ploidy had a significant relationship with root bud height (distance, LRT χ2 = 5.80, d.f. = 3, P = 0.122; ploidy, LRT χ2 = 5.80, d.f. = 4, P = 0.215). The interaction between root bud distance and ploidy was marginally significant (distance × ploidy: LRT χ2 = 5.61, d.f. = 2, P = 0.061), indicating that there may be weak differences in the relationship between root bud height and distance between the three cytotypes.
DISCUSSION
In this study, newly synthesized neotetraploid C. angustifolium produced more but smaller root buds than diploids, and invested fewer resources in sexual vs. clonal reproduction. Naturally occurring tetraploids produced fewer root buds than neotetraploids, and allocated more biomass towards sexual structures than root bud production. Consequently, naturally occurring tetraploids were generally more similar to diploids in their patterns of clonal and sexual reproduction than they were to neotetraploids. Overall, we find that WGD induces immediate increases in root bud production in C. angustifolium, supporting the expectation that clonal reproduction is higher in polyploids. However, there is also evidence that evolutionary processes acting after the WGD event continue to reshape resource allocation between sexual and clonal reproduction in established tetraploids. Selection appears to be operating in a direction opposite to predictions, resulting in naturally occurring tetraploids that are less clonal than neotetraploids and have levels of clonal reproduction comparable with diploids.
Immediate phenotypic shifts due to WGD
The neotetraploids used in this study were synthesized using colchicine, a technique that induces genome duplication by inhibiting chromosome segregation during mitosis (Caperta et al., 2006). The application of chemicals to induce WGD has the potential to create lasting side effects affecting growth, morphology and reproduction in new polyploids. In a recent study, Husband et al. (2016) examined the effects of colchicine in C. angustifolium by comparing the phenotypes of unexposed diploids with that of exposed diploids that did not convert into neotetraploids. They found significant phenotypic differences between unexposed and exposed diploids in the exposed generation, but no differences in the F1 seed generated from those plants. Studies on Fragaria and arabidopsis show similarly weak long-term effects of colchicine between treated and untreated diploids (Kwok, 2013; A. Green and B. C. Husband, University of Guelph, Canada, unpubl. res.). In contrast, Münzbergová (2017) found differences in plant performance between natural and second-generation synthetic polyploids, and evidence that selection during colchicine treatment could influence phenotype. While we cannot completely dismiss the possibility of selection in our synthetic polyploids, we have endeavoured to minimize its impact by treating a large number of diploid seedlings to maximize variation in successfully converted plants, and by using all converted plants to generate the individuals used in this study. Because previous studies on C. angustifolium have found no transgenerational effects of colchicine in F1 plants, it is likely that lingering effects are minimal and the differences observed here between diploids and neopolyploids can be directly attributed to genome duplication.
How might WGD directly affect root bud production? Adventitious root buds in C. angustifolium form endogenously from root tissue, often at the junction of lateral roots (Stocklin, 1992; Klimešová et al., 2009). Genetically regulated hormone production controls meristem growth and organogenesis in plants (Reinhardt et al., 2000; Horvath et al., 2003), and lateral root development is also governed by hormonal gradients (e.g. auxin and brassinosteroids; Vanstraelen and Benkova, 2012; Taylor-Teeples et al., 2016). The specific effects of genome duplication and polyploidy on these growth patterns are little researched (Levin, 2002), but a handful of studies demonstrate that meristem growth and the expression of hormone-controlling genes can be altered in polyploids (Hatano et al., 2012; Cheng et al., 2015; Dai et al., 2015). No studies have determined the immediate genetic or hormonal effects of WGD on adventitious root bud formation, or any other form of clonal reproduction. However, there is some evidence that WGD can cause abrupt changes in root size, root morphology and lateral root initiation (Kim et al., 2004; Tavan et al., 2015). It is plausible that the effect of WGD on gene expression, epigenetic interactions, chromosomal segregation and cell growth could have a large influence on across-plant hormonal balance and tissue development in C. angustifolium (Klimešová et al., 2009). If rates of lateral meristem formation below-ground increase due to altered hormone concentrations (Swarup et al., 2008), this could potentially result in an overproduction of adventitious root buds in neotetraploids. The precise developmental nature of root buds in C. angustifolium is unclear (Stocklin, 1992; Klimešová et al., 2009), but changes in gene expression could work either to stimulate higher root bud formation around lateral root sites (Baird et al., 1992; Sharma et al., 1993) or preferentially to ‘switch’ undetermined primordia into adventitious root buds vs. lateral root initials (Schirman and Zamora, 1978; Ellmore, 1981; Kirschbaum et al., 2004).
Root bud production in neotetraploids may also be the result of a trade-off with sexual effort. We see two key shifts above-ground in neotetraploids compared with diploids: (1) they have fewer lateral branches; and (2) they have approximately half the dry inflorescence biomass (Fig. 2E; Table 3; Supplementary Data Table S1). As in the root system, WGD could influence meristem and shoot growth above-ground by altering gene expression and hormone production. Interestingly, WGD has resulted in increased root bud production below-ground but decreased lateral branch growth above-ground, implying that hormone-regulated pathways for root vs. shoot systems may be affected by WGD in separate and opposing ways (Christianson and Warnick, 1983; Taylor-Teeples et al., 2016). Fewer lateral branches and fewer structures with the ability to bear flowers present a morphological constraint on sexual reproduction in neotetraploids, and they subsequently have a much lower investment in sexual reproduction than diploids. Additionally, though not assessed here, sex function in neopolyploids is often negatively impacted by meiotic irregularities leading to gamete sterility and seed abortion (Stebbins, 1950; Levin, 2002; Cifuentes et al., 2010). A decrease in sexual structures or non-functional sexual reproduction could lead to a restructuring of resource allocation and a compensatory increase in clonal reproduction (e.g. Geber et al., 1992; Vallejo-Marin et al., 2010; Van Drunen and Dorken, 2012). Studies that have found increases in clonal reproduction in extant polyploids often involve sexually sterile odd ploidies (Bruneau and Anderson, 1988; Hroudová and Zákravský, 1993; Holmes et al., 2009), where clonal reproduction may be offsetting the lack of sexual reproduction. Further study of the phenology of flowering and root bud production in C. angustifolium may reveal whether resource trade-offs between reproductive modes are occurring in neotetraploids.
Our data suggest that the reproductive shifts induced by WGD could considerably alter ecological interactions in neotetraploid C. angustifolium, and could impact their establishment potential. Diploid root buds elongated faster than neotetraploid root buds (Supplementary Data Figs S3 and S4), but were located further away from the parent shoot. Because neotetraploids produce more root buds, neotetraploid genets in natural populations may cover an area comparable with diploid genets but have a denser concentration of ramets closer to the parent stem. Large, aggregated genets would increase rates of geitonogamous selfing between ramets in neotetraploids (Vallejo-Marin et al., 2010) and, since inbreeding depression in neotetraploid C. angustifolium is much lower than that of diploids and naturally occurring tetraploids (Ozimec and Husband, 2011), we expect that the increase in same-cytotype mating will assist in overcoming MCE (Levin, 1975; Husband et al., 2008). Moreover, while fast growing diploid root buds could convey advantages in very competitive environments (e.g. old populations), higher root bud numbers could lead to rapid local habitat colonization for neotetraploids at population edges and contribute to polyploid persistence. Follow-up research conducted in natural populations to determine the consequences of higher root bud production in neotetraploid C. angustifolium is needed to understand further how phenotypic shifts in clonal investment and ramet location can impact the fitness and establishment dynamics of new polyploids.
Creating synthetic neopolyploids to explore the early stages of polyploid evolution requires that neopolyploids be created from extant diploid plants. Present-day diploids might differ both genetically and phenotypically from the diploids that gave rise to the established polyploids of the present day (Ramsey, 2011), and the neotetraploid C. angustifolium generated in this study may not reflect the phenotypes of historical neotetraploids. Though it is difficult to estimate the impact of these differences when hypothesizing how phenotypic shifts could have affected original polyploid establishment, they could be mitigated in young autopolyploids where presumably little change in diploids has occurred. Though the age of C. angustifolium polyploids is unknown, there is evidence that naturally occurring tetraploids in C. angustifolium have arisen recurrently and frequently (Roy, 2008), so the phenotypes of newly synthesized polyploids are still highly relevant to current population processes in this species.
Phenotypic change in established polyploids
The differences in root bud production and inflorescence biomass between neotetraploids and naturally occurring tetraploids indicate strong selection to suppress clonality and increase sexual reproduction after the WGD event. Furthermore, the phenotypic similarities between naturally occurring tetraploids and diploids is at odds with the prevalent hypothesis that polyploids are more clonal than their diploid relatives.
The habitat and ecological conditions that neotetraploid C. angustifolium would encounter in natural populations could play an important role in the shift away from clonal reproduction and towards sexual reproduction seen in naturally occurring tetraploids. Chamerion angustifolium is typically found in open areas with low competition (Mosquin, 1966), and is a rapid colonizer of disturbed areas (Myerscough and Whitehead, 1966; Stocklin, 1992). Diploids and naturally occurring tetraploids exhibit traits typical of such pioneers; sexual fruit and seed production are high (Stocklin, 1992), outcrossing is promoted (Husband and Schemske, 1997) and fast localized spread through clonal reproduction is an additional asset (Mosquin, 1966; Myerscough and Whitehead, 1966; Stocklin, 1992). While the skewed reliance on clonal reproduction seen in neotetraploids may initially aid in establishment, this allocation pattern may ultimately prove to be maladaptive in the common environment shared with their diploid parents. As tetraploids become naturalized, and perhaps sexual reproduction becomes more viable as chromosome segregation normalizes (Cifuentes et al., 2010; Hollister, 2015), we might expect to see a shift towards higher levels of sexual vs. clonal reproduction in order to optimize fitness.
The establishment period for new polyploids offers many opportunities for selection to drive phenotypic change. The data from this study indicate that neotetraploid genets have the potential to be large and dense, and may experience increased rates of geitonogamous selfing in the early stages of establishment. Due to the low inbreeding depression of neotetraploids (Husband et al., 2008), the high rate of self-fertilization is predicted to have relatively minor effects on overall seed production and viability. However, previous research has shown that neotetraploid C. angustifolium subject to several generations of self-fertilization have increased inbreeding depression at a level similar to that of diploids and naturally occurring tetraploids (Husband et al., 2008; Ozimec and Husband, 2011). Even with minimal sexual output in neotetraploids, the increasingly negative effects of inbreeding would probably cause selection pressure to decrease root bud production and genet size until clonal reproduction in tetraploids is comparable with that in diploids. Indeed, we find that total root bud production in naturally occurring tetraploids is generally lower than, but statistically similar to, that in diploids. This conclusion corroborates that of Baldwin and Husband (2013), where tetraploids produced fewer root buds and surveys of eight mixed- and single-ploidy populations of C. angustifolium found that tetraploid genets had fewer ramets than diploid genets.
CONCLUSIONS
A recent phylogenetic comparative study showed a strong evolutionary correlation between clonal reproduction and polyploidy in the central European flora (Herben et al., 2017), while early across-species surveys by Müntzing (1936) and Gustafsson (1948) revealed higher chromosome numbers and incidence of polyploidy in perennial or ‘root-wandering’ species. The population-level ecological and evolutionary mechanisms behind these large-scale patterns remain unclear, as researchers often do not find that extant polyploids are more clonal than close diploid relatives in mixed-ploidy species (Keeler, 2004; Baldwin and Husband, 2013; Schulze et al., 2013; Hanzl et al., 2014; Martínková et al., 2015; but see Schlaepfer et al., 2010). Clonal species may be pre-disposed towards generating successful polyploid lineages by facilitating polyploid establishment and persistence (Stebbins, 1950; Freeling, 2017; Herben et al., 2017), even if no quantitative differences in clonal reproduction are found between natural cytotypes. If, as we find in C. angustifolium, WGD immediately causes phenotypic shifts resulting in increased clonal output in neopolyploids, polyploid establishment may be even more rapidly accomplished.
This study is the first to measure the immediate influence of WGD and polyploidy on clonal reproduction using synthetic neopolyploids. To understand further how WGD can affect resource investment in clonality and sexual reproduction, and the implications of such changes on the evolutionary fate of neopolyploids, more research should be conducted on synthetic neopolyploids or polyploids of recent origin, and on species with different modes of clonal reproduction. Moreover, the realized consequences of clonality on the establishment potential of polyploids remains to be studied. Experiments determining how clonal reproduction affects selfing and outcrossing patterns in natural mixed-ploidy populations will provide valuable insight into how evolutionary interactions between clonal reproduction and polyploidy are operating within the angiosperms.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: biomass allocations with raw data distributions. Figure S2: root bud attributes with raw data distributions. Figure S3: regression of root bud height vs. distance for diploids, neotetraploids and naturally occurring tetraploids. Figure S4: distribution of root bud heights per ploidy for all measured root buds. Table S1: raw means of growth, biomass and root bud traits for diploids, neotetraploids and naturally occurring tetraploids.
ACKNOWLEDGEMENTS
The authors thank M. Mucci and T. Slimmon at the University of Guelph Phytotron for greenhouse support; and P. Kron, A. Abbruzzese and J. Seery for technical and data collection assistance. This work was supported by the Natural Science and Engineering Research Council of Canada (NSERC) with a Canada Graduate Scholarship (CGSD3-438830-2013) to W.E.V.D. and a Discovery Grant (400434) to B.C.H.
LITERATURE CITED
- Baack EJ. 2005. To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity 94: 538–546. [DOI] [PubMed] [Google Scholar]
- Baird JH, Dute RR, Dickens R. 1992. Ontogeny, anatomy, and reproductive biology of vegetative reproductive organs of Diodia virginiana L. (Rubiaceae). International Journal of Plant Sciences 153: 320–328. [Google Scholar]
- Baldwin SJ, Husband BC. 2013. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Molecular Ecology 22: 1806–1819. [DOI] [PubMed] [Google Scholar]
- Barringer BC. 2007. Polyploidy and self-fertilization in flowering plants. American Journal of Botany 94: 1527–1533. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Bretagnolle F, Lumart F. 1995. Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica: occurrence, morphological and genetic characteristics of first polyploids. Euphytica 84: 197–207. [Google Scholar]
- Bruneau A, Anderson GJ. 1988. Reproductive biology of diploid and triploid Apios americana (Leguminosae). American Journal of Botany 75: 1876–1883. [Google Scholar]
- Caperta AD, Delgado M, Ressurreição F, et al. . 2006. Colchicine-induced polyploidization depends on tubulin polymerization in c-metaphase cells. Protoplasma 227: 147–153. [DOI] [PubMed] [Google Scholar]
- Charpentier A. 2002. Consequences of clonal growth for plant mating. Evolutionary Ecology 15: 521–530. [Google Scholar]
- Cheng S, Zhu X, Liao T, et al. . 2015. Gene expression differences between high-growth Populus allotriploids and their diploid parents. Forests 6: 839–857. [Google Scholar]
- Christianson ML, Warnick DA. 1983. Competence and determination in the process of in vitro shoot organogenesis. Developmental Biology 95: 288–293. [DOI] [PubMed] [Google Scholar]
- Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. 2010. Genetic regulation of meiosis in polyploid species: new insights into an old question: research review. New Phytologist 186: 29–36. [DOI] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews. Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Dai F, Wang Z, Luo G, Tang C. 2015. Phenotypic and transcriptomic analyses of autotetraploid and diploid mulberry (Morus alba L.). International Journal of Molecular Sciences 16: 22938–22956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG. 2002. The loss of sex in clonal plants. Evolutionary Ecology 15: 501–520. [Google Scholar]
- Ellmore GS. 1981. Root dimorphism in Ludwigia peploides (Onagraceae): development of two root types from similar primordia. Botanical Gazette 142: 525–533. [Google Scholar]
- Freeling M. 2017. Picking up the ball at the K/Pg boundary: The distribution of ancient polyploidies in the plant phylogenetic tree as a spandrel of asexuality with occasional sex. The Plant Cell 29: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geber MA, Watson MA, Furnish R. 1992. Genetic differences in clonal demography in Eichhornia crassipes. Journal of Ecology 80: 329–341. [Google Scholar]
- Guo M, Davis D, Birchler JA. 1996. Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson Å. 1948. Polyploidy, life-form and vegetative reproduction. Hereditas 34: 1–22. [Google Scholar]
- Hanzl M, Kola F, Novakova D, Suda J. 2014. Nonadaptive processes governing early stages of polyploid evolution: insights from a primary contact zone of relict serpentine Knautia arvensis (Caprifoliaceae). American Journal of Botany 101: 935–945. [DOI] [PubMed] [Google Scholar]
- Hatano H, Mizuno N, Matsuda R, Shitsukawa N, Park P, Takumi S. 2012. Dysfunction of mitotic cell division at shoot apices triggered severe growth abortion in interspecific hybrids between tetraploid wheat and Aegilops tauschii. New Phytologist 194: 1143–1154. [DOI] [PubMed] [Google Scholar]
- Herben T, Suda J, Klimešová J. 2017. Polyploid species rely on vegetative reproduction more than diploids: a re-examination of the old hypothesis. Annals of Botany 120: 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD. 2015. Polyploidy: adaptation to the genomic environment. New Phytologist 205: 1034–1039. [DOI] [PubMed] [Google Scholar]
- Holmes GD, James EA, Hoffmann AA. 2009. Divergent levels of genetic variation and ploidy among populations of the rare shrub, Grevillea repens (Proteaceae). Conservation Genetics 10: 827–837. [Google Scholar]
- Honnay O, Bossuyt B. 2005. Prolonged clonal growth: escape route or route to extinction?Oikos 108: 427–432. [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. 2003. Knowing when to grow: signals regulating bud dormancy. Trends in Plant Science 8: 534–540. [DOI] [PubMed] [Google Scholar]
- Hroudová Z, Zákravsky P. 1993. Ecology of two cytotypes of Butomis umbellatus II. Reproduction, growth, and biomass production. Folia Geobotanica et Phytotaxonomica 28: 413–424. [Google Scholar]
- Husband BC. 2000. Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proceedings of the Royal Society B: Biological Sciences 267: 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Sabara HA. 2004. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytologist 161: 703–713. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. 1997. The effect of inbreeding in diploid and tetraploid populations of Epilobium angustifolium (Onagraceae): implications for the genetic basis of inbreeding depression. Evolution 51: 737–746. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. 2000. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. Journal of Ecology 88: 689–701. [Google Scholar]
- Husband BC, Ozimec B, Martin SL, Pollock L. 2008. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. International Journal of Plant Sciences 169: 195–206. [Google Scholar]
- Husband BC, Baldwin SJ, Sabara HA. 2016. Direct vs. indirect effects of whole-genome duplication on prezygotic isolation in Chamerion angustifolium: implications for rapid speciation. American Journal of Botany 103: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Keeler KH. 2004. Impact of intraspecific polyploidy in Andropogon gerardii (Poaceae) populations. American Midland Naturalist 152: 63–74. [Google Scholar]
- Kim YS, Hahn EJ, Murthy HN, Paek KY. 2004. Effect of polyploidy induction on biomass and ginsenoside accumulations in adventitious roots of ginseng. Journal of Plant Biology 47: 356–360. [Google Scholar]
- Kirschbaum DS, Cantliffe DJ, Shaw NL, Liu JR. 2004. Direct adventitious shoot formation on seedling radicles in seed cultures of strawberry. Journal of Plant Biology 47: 160–162. [Google Scholar]
- van K leunen M, Fischer M, Schmid B. 2002. Experimental life-history evolution: selection on the allocation to sexual reproduction and its plasticity in a clonal plant. Evolution 56: 2168–2177. [PubMed] [Google Scholar]
- Klimešová J, De Bello F. 2009. CLO-PLA: the database of clonal and bud bank traits of Central European flora. Journal of Vegetation Science 20: 511–516. [Google Scholar]
- Klimešová J, Pokorná A, Klimeš L. 2009. Establishment growth and bud-bank formation in Epilobium angustifolium: the effects of nutrient availability, plant injury, and environmental heterogeneity. Botany 87: 195–201. [Google Scholar]
- Kolář F, Čertner M, Suda J, Schönswetter P, Husband BC. 2017. Mixed-ploidy species: progress and opportunities in polyploid research. Trends in Plant Science 22: 1041–1055. [DOI] [PubMed] [Google Scholar]
- Kron P, Husband BC. 2009. Hybridization and the reproductive pathways mediating gene flow between native Malus coronaria and domestic apple, M. domestica. Botany 87: 864–874. [Google Scholar]
- Kwok A. 2013. The role of polyploidy in the evolution of gender dimorphism: an experimental approach using Fragaria vesca. MSc Thesis, University of Guelph, Canada. [Google Scholar]
- Lande R, Schemske DW. 1985. The evolution of self-fertilization and inbreeding depression in plants. I: genetic models. Evolution 39: 24–40. [DOI] [PubMed] [Google Scholar]
- Levin DA, Kerster HW. 1974. Gene flow in seed plants. Evolutionary Biology 7: 139–220. [Google Scholar]
- Levin DA. 1975. Minority cytotype exclusion in local plant populations. Taxon 24: 35–43. [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. American Naturalist 122: 1–25. [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. Oxford: Oxford University Press. [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, et al. . 2008. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS One 3: e3353. doi: 10.1371/journal.pone.0003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Martin SL, Husband BC. 2012. Whole genome duplication affects evolvability in natural populations of a flowering plant. PLoS One 7: e44784. doi: 10.1371/journal.pone.0044784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínková J, Klimešová J, Doležal J, Kolář F. 2015. Root sprouting in Knautia arvensis (Dipsacaceae): effects of polyploidy, soil origin and nutrient availability. Plant Ecology 216: 901–911. [Google Scholar]
- Mosquin T. 1966. A new taxonomy for Epilobium angustifolium L. (Onagraceae). Brittonia 18: 167–188. [Google Scholar]
- Müntzing A. 1936. The evolutionary significance of autopolyploidy. Hereditas 21: 263–378. [Google Scholar]
- Münzbergová Z. 2017. Colchicine application significantly affects plant performance in the second generation of synthetic polyploids and its effects vary between populations. Annals of Botany 120: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerscough PJ, Whitehead FH. 1966. Comparative biology of Tussilago farfara L., Chamaenerion angustifolium (L.) Scop., Epilobium montanum L. and Epilobium adenocaulon Hausskn. I. General biology and germination. New Phytologist 65: 192–210. [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, et al. . 2003. Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Oswald BP, Nuismer SL. 2010. Neopolyploidy and diversification in Heuchera grossulariifolia. Evolution 65-6: 1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Ozimec B, Husband BC. 2011. Effect of recurrent selfing on inbreeding depression and mating system evolution in an autopolyploid plant. Evolution 65: 2038–2049. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy: research review. New Phytologist 186: 5–17. [DOI] [PubMed] [Google Scholar]
- R Core Team 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; https://www.R-project.org/ [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics 33: 589–639. [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. 2000. Auxin regulates the initiation and radial position of plant lateral organs. The Plant Cell 12: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K, Goldberg EE, Igić B. 2011. Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution 65: 139–155. [DOI] [PubMed] [Google Scholar]
- Rodriguez DJ. 1996. A model for the establishment of polyploidy in plants: viable but infertile hybrids, iteroparity, and demographic stochasticity. Journal of Theoretical Biology 180: 189–196. [Google Scholar]
- Roulin A, Auer PL, Libault M, et al. . 2013. The fate of duplicated genes in a polyploid plant genome. The Plant Journal 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Roy Y. 2008. The evolutionary history of polyploidy in the herbaceous plant Chamerion angustifolium. MSc Thesis, University of Guelph, Canada. [Google Scholar]
- Sabara HA, Kron P, Husband BC. 2013. Cytotype coexistence leads to triploid hybrid production in a diploid–tetraploid contact zone of Chamerion angustifolium (Onagraceae). American Journal of Botany 100: 962–970. [DOI] [PubMed] [Google Scholar]
- Schirman R, Zamora BA. 1978. Bud development in excised roots of rush skeletonweed (Chondrilla juncea). Weed Science 26: 582–584. [Google Scholar]
- Schlaepfer DR, Edwards PJ, Billeter R. 2010. Why only tetraploid Solidago gigantea (Asteraceae) became invasive: a common garden comparison of ploidy levels. Oecologia 163: 661–673. [DOI] [PubMed] [Google Scholar]
- Schulze J, Erhardt A, Stoll P. 2013. Reduced clonal reproduction indicates low potential for establishment of hybrids between wild and cultivated strawberries (Fragaria vesca × F. × ananassa). Ecological Research 28: 43–52. [Google Scholar]
- Segraves KA, Thompson JN. 1999. Plant polyploidy and pollination: floral traits and insect visits to diploid and autotetraploid Heuchera grossulariifolia. Evolution 53: 1114–1127. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Yeung EC, Thorpe TA. 1993. Histology of shoot bud ontogeny from seedling root segments of Brassica napus L. Annals of Botany 71: 461–466. [Google Scholar]
- Soltis DE, Buggs RJ, Doyle JJ, Soltis PS. 2010. What we still don’t know about polyploidy. Taxon 59: 1387–1403. [Google Scholar]
- Soltis DE, Misra BB, Shan S, Chen S, Soltis PS. 2016. Polyploidy and the proteome. Biochimica et Biophysica Acta 1864: 896–907. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. Reading, UK: Addison-Wesley. [Google Scholar]
- Stocklin J. 1992. Differences in the location of subcotyledonary buds among Epilobium angustifolium L., E. dodonaei Vill. and E. fleischeri Hostst. (Onagraceae) and effects on architecture and population structure. Botanical Journal of the Linnean Society 108: 35–47. [Google Scholar]
- Swarup K, Benková E, Swarup R, et al. . 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology 10: 946–954. [DOI] [PubMed] [Google Scholar]
- Tavan M, Mirjalili MH, Karimzadeh G. 2015. In vitro polyploidy induction: changes in morphological, anatomical and phytochemical characteristics of Thymus persicus (Lamiaceae). Plant Cell, Tissue and Organ Culture 122: 573–583. [Google Scholar]
- Taylor-Teeples M, Lanctot A, Nemhauser JL. 2016. As above, so below: auxin’s role in lateral organ development. Developmental Biology 419: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Lumaret R. 1992. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology and Evolution 7: 302–307. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Husband BC, Maherali H. 2014. Climatic niche differences between diploid and tetraploid cytotypes of Chamerion angustifolium (Onagraceae). American Journal of Botany 101: 1868–1875. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín M. 2012. Mimulus peregrinus (Phrymaceae): a new British allopolyploid species. PhytoKeys 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marín M, Dorken ME, Barrett SCH. 2010. The ecological and evolutionary consequences of clonality for plant mating. Annual Review of Ecology, Evolution, and Systematics 41: 193–213. [Google Scholar]
- Van Drunen WE, Dorken ME. 2012. Trade-offs between clonal and sexual reproduction in Sagittaria latifolia (Alismataceae) scale up to affect the fitness of entire clones. New Phytologist 196: 606–616. [DOI] [PubMed] [Google Scholar]
- Van Drunen WE, van Kleunen M, Dorken ME. 2015. Consequences of clonality for sexual fitness: clonal expansion enhances fitness under spatially restricted dispersal. Proceedings of the National Academy of Sciences 112: 8929–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. 2012. Hormonal interactions in the regulation of plant development. Annual Review of Cell and Developmental Biology 28: 463–487. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.