Abstract
Background and Aims
Anisotropic cell elongation depends on cell wall relaxation and cellulose microfibril arrangement. The aim of this study was to characterize the molecular function of AtDICE1 encoding a novel transmembrane protein involved in anisotropic cell elongation in Arabidopsis.
Methods
Phenotypic characterizations of transgenic Arabidopsis plants mis-regulating AtDICE1 expression with different pharmacological treatments were made, and biochemical, cell biological and transcriptome analyses were performed.
Key Results
Upregulation of AtDICE1 in Arabidopsis (35S::AtDICE1) resulted in severe dwarfism, probably caused by defects in anisotropic cell elongation. Epidermal cell swelling was evident in all tissues, and abnormal secondary wall thickenings were observed in pith cells of stems. These phenotypes were reproduced not only by inducible expression of AtDICE1 but also by overexpression of its poplar homologue in Arabidopsis. RNA interference suppression lines of AtDICE1 resulted in no observable phenotypic changes. Interestingly, wild-type plants treated with isoxaben, a cellulose biosynthesis inhibitor, phenocopied the 35S::AtDICE1 plants, suggesting that cellulose biosynthesis was compromised in the 35S::AtDICE1 plants. Indeed, disturbed cortical microtubule arrangements in 35S::AtDICE1/GFP-TuA6 plants were observed, and the cellulose content was significantly reduced in 35S::AtDICE1 plants. A promoter::GUS analysis showed that AtDICE1 is mainly expressed in vascular tissue, and transient expression of GFP:AtDICE1 in tobacco suggests that AtDICE1 is probably localized in the endoplasmic reticulum (ER). In addition, the external N-terminal conserved domain of AtDICE1 was found to be necessary for AtDICE1 function. Whole transcriptome analyses of 35S::AtDICE1 revealed that many genes involved in cell wall modification and stress/defence responses were mis-regulated.
Conclusions
AtDICE1, a novel ER-localized transmembrane protein, may contribute to anisotropic cell elongation in the formation of vascular tissue by affecting cellulose biosynthesis.
Keywords: Arabidopsis, anisotropic cell elongation, cellulose biosynthesis, cell wall integrity, endoplasmic reticulum, isoxaben, poplar, secondary wall thickening, vascular tissue formation
INTRODUCTION
Cell elongation is a fundamental process for optimal growth and development of plants by directing proper cell morphology. Cell elongation is initiated by vacuolar turgor pressure and limited by a strong but flexible cell wall that permits directional cell elongation. Plant cell walls are essential not only for protecting cells against external stresses but also the cell elongation process (Baskin, 2005). The cell wall is composed mainly of polysaccharides (e.g. cellulose, hemicelluloses and pectins) interwoven among a relatively small amount of highly glycosylated protein (Somerville et al., 2004; Carpita, 2011). The strongest component in the cell wall is a network of cellulose microfibrils. While cellulose microfibrils are synthesized by cellulose synthase (CesA) complexes (CSCs) at the plasma membrane (PM) and are extruded directly into the cell wall (Geisler et al., 2008; McFarlane et al., 2014), other polysaccharides are synthesized in the endoplasmic reticulum (ER) or Golgi and delivered to the cell wall through the intracellular secretory pathway (Gibeaut and Carpita, 1994). CesA subunits are synthesized in the ER, and CSC rosettes are assembled in the Golgi, transported via the trans-Golgi network (TGN) and inserted into the PM through cortical microtubule-assisted vesicle trafficking (Paredez et al., 2006; McFarlane et al., 2014; Zhang et al., 2016).
Cellulose microfibrils, as load-bearing components of the cell wall, are important in controlling anisotropic cell elongation in plants. Cellulose microfibrils are aligned perpendicular to the axis of elongation, thus forming a spring-like structure that reinforces the cell crosswise and favours longitudinal expansion in most growing cells (Baskin, 2005; Lei et al., 2014). Furthermore, because the newly synthesized cellulose microfibrils are aligned by the cortical microtubules through microtubule–CSC interacting proteins, such as CSI1/POM2, CSI3 and CC, it has been suggested that the shape of plant cells is determined by the orientation of cortical microtubules (Green, 1962; Bringmann et al., 2012; Endler et al., 2015). Thus, perturbation of either microtubules or cellulose microfibril organization leads to defects in cell elongation (Fagard et al., 2000; Lane et al., 2001; Schindelman et al., 2001; Pagant et al., 2002; Caño-Delgado et al., 2003; Somerville, 2006; Wang et al., 2006). The dwarf phenotype of cellulose-deficient mutants is therefore commonly interpreted as resulting from the loss of anisotropic cell elongation by weakened cell walls.
Many genes involved in either cell expansion or cellulose biosynthesis have been identified through genetic screening and subsequent cloning. For example, mutation of RSW1/CesA1, RSW5/CesA3, QUILL/PROCUSTE/CesA6, POM-POM1/CHITINASE LIKE1 (POM1/CTL1), KORRIGAN/LION’S TAIL1/RADIALLY SWOLLEN2 (KOR/LIT/RSW2), COBRA and KOBITO/ELONGATION DEFECTIVE1 (KOB1/ELD1) was associated with epidermal cell swelling, restricted elongation of root and hypocotyl, and reduced cell wall cellulose content (Hauser et al., 1995; Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000; Lane et al., 2001; Schindelman et al., 2001; Pagant et al., 2002; Wang et al., 2006). During cell expansion, the integrity of the cell wall is vital. Once the integrity of the cell wall is disrupted, a series of compensatory reactions, such as ectopic accumulation of lignin and callose, modification of cell wall composition, and enhanced stress/defence responses, follow. For example, mutations or treatment with chemical inhibitors of cellulose biosynthesis lead to ectopic lignification and/or callose deposition, and change the composition of the cell wall matrix (Desprez et al., 2002; Pagant et al., 2002). In both eli1 (ECTOPIC LIGNIFICATION1) (Caño-Delgado et al., 2000) and cev1 (CONSTITUTIVE EXPRESSION OF VSP1) (Ellis and Turner, 2001) mutants, which were identified as different mutant alleles of a cellulose synthase gene CesA3, the jasmonic acid (JA) and ethylene-signalling pathways were highly activated and stress responsive genes were up-regulated. Also, cob-5, a loss of function mutant of COBRA, exhibits abnormal cell growth with a massive accumulation of stress response chemicals, such as anthocyanins and callose. Furthermore, cob-5 over-accumulates JA and induces defence and stress-related genes coordinately (Ko et al., 2006b). These reports suggest that plant cells initially sense biotic stresses at the level of the integrity of the cell wall (Ko et al., 2006b; Wolf et al., 2012).
In this study, we report the functional characterization of a gene named AtDICE1 (DEFECT IN CELL ELONGATION1) from Arabidopsis. A gain-of-function mutation of AtDICE1 caused severe dwarfism with defects in anisotropic cell elongation. Further anatomical, molecular, biochemical and whole transcriptome analyses suggest that AtDICE1 is a novel ER-localized transmembrane protein contributing to the proper anisotropic cell elongation process in the vascular tissue through participation in cell wall formation.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana, ecotype Columbia (Col-0), was used in both the wild-type (WT) and transgenic experiments. Plants were growth on soil or on MS-agar plates [0.5× MS, 2 % sucrose, 0.8 % (w/v) agar] in a growth chamber (16-h light/8-h dark) at 23 ± 2 °C, after stratification. For the growth measurement of young seedlings, MS-agar plates were vertically orientated. For isoxaben (36138, Sigma-Aldrich, St Louis, MO, USA) treatment, isoxaben was dissolved in methanol and added to the A. thaliana seedling cultures to final concentrations of 1, 5 or 10 nm. Seedlings were grown in 0.5× MS medium containing 2 % (w/v) sucrose, 0.8 % (w/v) agar and isoxaben at different concentrations and observed after 7 d.
Histological analysis
The stem area located immediately above the rosette (basal level) was cross-sectioned by hand and stained with 2 % phloroglucinol/HCl or 0.05 % toluidine blue O for 1 min. Microtome (Leica RM2025, Leica, http://www.leica.com) sectioning after paraffin embedding was used to observe the detailed structure of hypocotyls and stems. For confocal laser scanning microscopy, a Zeiss PASCAL microscope (Jena, Germany), with a 488-nm excitation mirror, a 560-nm emission filter and a 505–530-nm emission filter, was used to record images. Image analysis was performed using laser scanning microscope PASCAL LSM version 3.0 SP3 software. For scanning electron microscopy (SEM), a tabletop microscope (Hitachi TM3000, Tokyo, Japan) was used to visualize hypocotyls from 7-d-old seedlings of WT and 35S::AtDICE1 (7-5 line) plants using the default conditions without any pretreatments.
Plasmid vector construction and generation of transgenic Arabidopsis plants
The full-length cDNA of AtDICE1 (At2g41610) was amplified by PCR and inserted downstream of the 35S promoter in the pB2GW7 or pB7BWIWG2(II) vectors (Karimi et al., 2002) using the Gateway cloning system to produce 35S::AtDICE1 or 35S::AtDICE1-RNAi constructs, respectively. The N-terminal deletion fragment of AtDICE1 (ΔNT-AtDICE1) was amplified by PCR using the 35S::AtDICE1 construct as a template and inserted downstream of the 35S promoter in the pB2GW7 vector to produce the 35S::ΔNT-AtDICE1 construct. For the AtDICE1 promoter::GUS construct, the 1.0-kb XbaI/BamHI genomic fragment flanking the 5′ end of the AtDICE1 coding sequence was amplified using genomic DNA extracted from Arabidopsis leaf tissue as template, and subcloned into the pCB308 vector (Xiang et al., 1999). To construct the 35S::GFP:AtDICE1 plasmid, the Gateway destination vector pEarleyGate103 was used to translationally fused GFP:AtDICE1. All the vector constructs produced were verified by DNA sequencing and introduced into Agrobacterium tumefaciens (C58), which was used to transform Arabidopsis (Col-0) or the GFP-TuA6 background using the floral-dip method described by Clough and Bent (1998).
Extraction of alcohol-insoluble cell wall residue
Stem tissues (5 cm from rosette level) from 60-d-old WT and 35S::AtDICE1 plants were ground to a fine powder. The ground material (approx. 1 g) was washed in 15 mL of 70 % ethanol and heated for 15 min at 70 °C to inactivate endogenous enzymes and remove the cell contents. Samples were centrifuged for 10 min at 4000 g, and the pellets were washed twice with 100 % ethanol and once with 100 % acetone. The remaining pellet was considered to be the alcohol-insoluble cell wall residue (AIR) and was dried at 70 °C.
Analysis of cell wall monosaccharide composition
Cell wall sugars (as alditol acetates) were determined using the procedure reported by Hoebler et al. (1989). Briefly, AIRs (3 mg) were incubated with 70 % sulfuric acid at room temperature for 30 min, followed by the addition of inositol as the internal standard and dilution with water to a final concentration of 6 % sulfuric acid. After heating for 120 min at 105 °C, the solution was treated with 25 % ammonium solution. After reduction with sodium borohydride in dimethyl sulfoxide, the solution was heated for 90 min at 40 °C, followed by sequential treatment with glacial acetic acid, acetic anhydride, 1-methylimidazole, dichloromethane and water. The organic layer containing the alditol acetates of the hydrolysed cell wall sugars was washed three times with water, and sugars were analysed on a gas–liquid chromatograph (model 6890; Hewlett-Packard, http://www.hp.com) equipped with a 30 m × 0.25 mm (i.d.) silica capillary column DB 225 (Alltech Associates Inc., Deerfield, IL, USA).
Statistical analysis
Data were analysed with a two-tailed, unpaired t-test by using GRAPHPAD PRISM 7.03 (Graphpad Software, San Diego, CA, USA). P-values of <0.05 were considered significant.
Histochemical GUS staining
Histochemical GUS staining of transgenic plants was performed as described by Jefferson et al. (1987) with slight modifications as described by Nguyen et al. (2016). Briefly, samples were incubated at 37 °C for 18–24 h in GUS reaction buffer and visualized after removing chlorophyll by rinsing with 70 % ethanol.
Intracellular localization of AtDICE1
Each 35S::GFP and 35S::GFP:AtDICE1 construct was introduced into Agrobacterium tumefaciens (strain C58) by the freeze–thaw method (Höfgen and Willmitzer, 1988). Transient expression in Nicotiana benthamiana leaf epidermal cells was determined by the agroinfiltration method (Di Sansebastiano et al., 2004). The N. benthamiana leaves were further grown for 12 h, and the fluorescence was observed using a Zeiss PASCAL confocal laser scanning microscope as described above.
RNA extraction and semi-quantitative RT-PCR analysis
Total RNAs were extracted using Trizol reagent (Gibco-BRL, http://www.invitrogen.com) according to the manufacturer’s instructions. Total RNAs were reverse transcribed using Superscript II reverse transcriptase (Invitrogen) in 20-µL reactions. Semi-quantitative reverse transcriptase PCR (RT-PCR) was carried out using 1 µΛ of the reaction products as a template. Amplified DNA fragments were separated on a 1 % agarose gel and stained with ethidium bromide. The primers used for RT-PCR analysis are shown in Supplementary Data Table S3.
Whole transcriptome microarray analysis
Total RNAs were isolated from 7-d-old seedlings of 35S::AtDICE1 (7-5, 10-1, 11-6) and WT grown on MS agar plates. An Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA) was used to check RNA quality. Nucleic acid labelling was performed according to the manufacturer’s recommended procedures for single-colour arrays. Labelled RNA was hybridized to Agilent 4x44K Arabidopsis Gene Expression (V4) Microarrays (Agilent G2519F-021169). Arrays were scanned with an Agilent G2565B Array Scanner, and images were analysed using the Feature Extractor v9.5 default protocol GE1-v5_95_Feb07. The resulting hybridization intensity values (i.e. signal intensity) of each spot reflect the abundance of a given mRNA relative to the total mRNA population and were used in all subsequent analyses. All data normalization and selection of genes showing fold-changes were performed using GeneSpringGX 7.3.1 (Agilent). Genes were filtered by removing flag-out genes in each experiment. Intensity-dependent normalization [locally weighted scatterplot smoothing (LOWESS)] was performed, where the ratio was reduced to the residual of the LOWESS fit of the intensity versus ratio curve. The averages of normalized ratios were calculated by dividing the average of the normalized signal channel intensity by the average of normalized control channel intensity. Differentially expressed genes were identified using the Single t-test, which assumes unequal variances between groups. The criterion for significant genes was P < 0.05. The raw microarray data were deposited in the Gene Expression Omnibus database together with details of the protocol.
RESULTS
Overexpression of AtDICE1 results in a severe dwarf phenotype
Previously, using a global comparative transcriptome analysis, we found a gene network regulating secondary xylem development in Arabidopsis (Ko et al., 2006a). Among the genes in this network, we were interested in a completely unknown gene locus, At2g41610, which we named AtDICE1. AtDICE1 is specifically expressed in vascular tissues or cells (Supplementary Data Fig. S1) and the Potri.014G108300 gene (named as PtrDICE1), the closest homologue of AtDICE1 in poplar, is also preferentially expressed in the wood-forming tissues of poplar (Fig. S1C). Consistently, our promoter–GUS reporter analysis showed that AtDICE1 promoter activity was detected mainly in the vascular tissues of cotyledon and hypocotyl, and in the roots of young seedlings as early as 2 d old (Fig. S1D). In cross-sections of stems and roots, activity of the AtDICE1 promoter was found mainly in vascular bundles, especially in cambium and/or xylem parenchyma cells (Fig. S1E). Further sequence analysis identified that AtDICE1 is a plant-specific and transmembrane protein (Fig. S2). Both TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) analyses predicted that AtDICE1 has seven transmembrane domains with the N-terminus outside and C-terminus inside (Fig. S2B, C). Interestingly, the putative external N-terminal motif is highly conserved in all plant species examined (Fig. S2A).
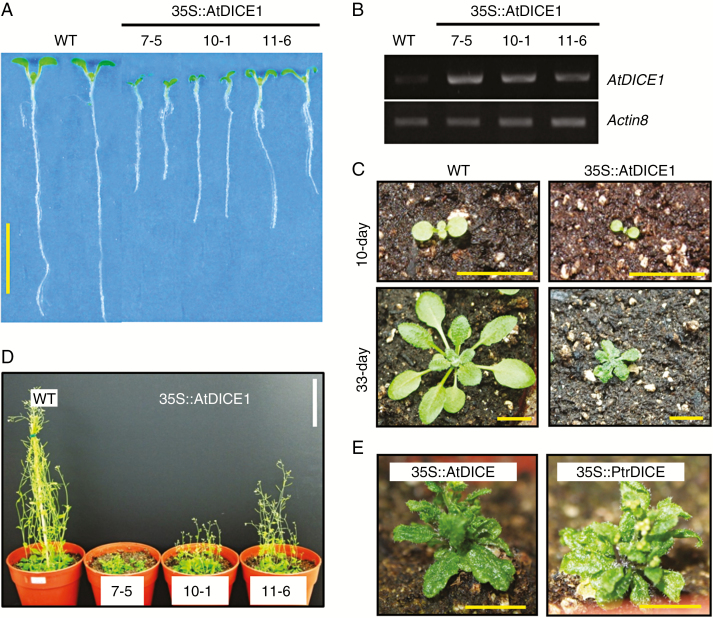
To characterize the molecular function of AtDICE1, we first generated gain-of-function transgenic Arabidopsis plants by overexpressing AtDICE1 under control of the cauliflower mosaic virus (CaMV) 35S promoter (i.e. 35S::AtDICE1 plants). This was done because T-DNA knock-out mutants were unfortunately not available. A total of 26 T1 transgenic plants were examined and we found that most of them were dwarfed compared to WT Arabidopsis plants (data not shown). To perform additional detailed analyses, three T3 homozygote transgenic plants (i.e. 7-5, 10-1 and 11-6 lines of 35S::AtDICE1) based on phenotypic severity were selected (Fig. 1). The dwarfing phenotypes of 35S::AtDICE1 plants were evident not only in young seedlings but also at the adult stage (Fig. 1; Table S1). Furthermore, the degree of dwarfing was consistent with the expression levels of the AtDICE1 gene (Fig. 1B). In adult plants, 35S::AtDICE1 plants showed extremely small rosette leaves, very short inflorescence stems, small flowers and reduced apical dominances (Figs 1C, D and S3). Furthermore, overexpression of PtrDICE1 reproduced almost the identical phenotype observed in 35S::AtDICE1 plants (Fig. 1E). This result suggests that PtrDICE1 is orthologous to AtDICE1.
Fig. 1.
Gain-of-function mutation of AtDICE1 resulted in a severe dwarfing phenotype. (A) Seven-day-old seedlings of wild-type (WT) and 35S::AtDICE1 plants. (B) Semi-quantitative RT-PCR analysis for AtDICE1 transcript accumulation in each of three T3 homozygote 35S::AtDICE1 plants. Actin8 was used as a control. (C) Growth features of 35S::AtDICE1 (line 7-5) and WT plants on soil at 10 d (upper) and 33 d (lower). (D) Forty-day-old WT and 35S::AtDICE1 (7-5) plants grown in pots. (E) Forty-day-old 35S::AtDICE1 (7-5) (left) and 35S::PtrDICE1 (right) plants. Scale bars represent 1 cm, except 10 cm in D.
We produced RNA interference (RNAi) suppression lines of AtDICE1 (i.e. 35S::AtDICE1-RNAi) to analyse the loss-of-function phenotype (Fig. S4). Among a total of seven independent T3 homozygous lines, two of them (7-1, 12-1) showed significantly decreased expression of AtDICE1 (Fig. S4A). However, we were unable to observe any significant alterations in phenotype in seedlings or adult plants (Fig. S4B, C).
35S::AtDICE1 plants exhibit defects in anisotropic cell elongation
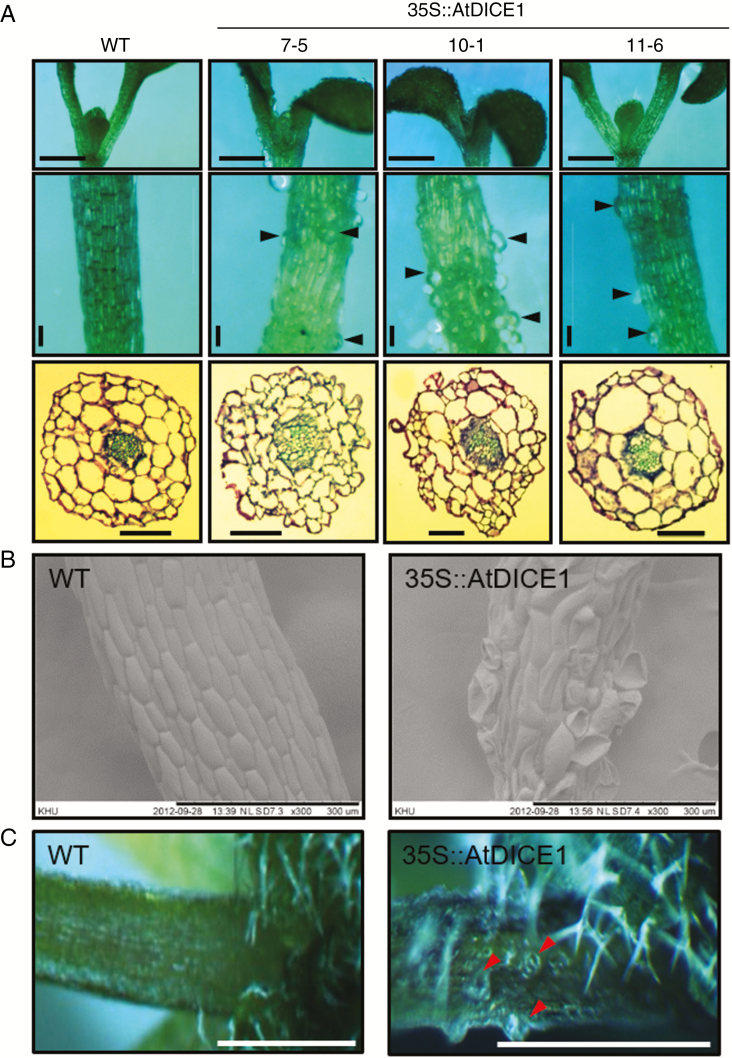
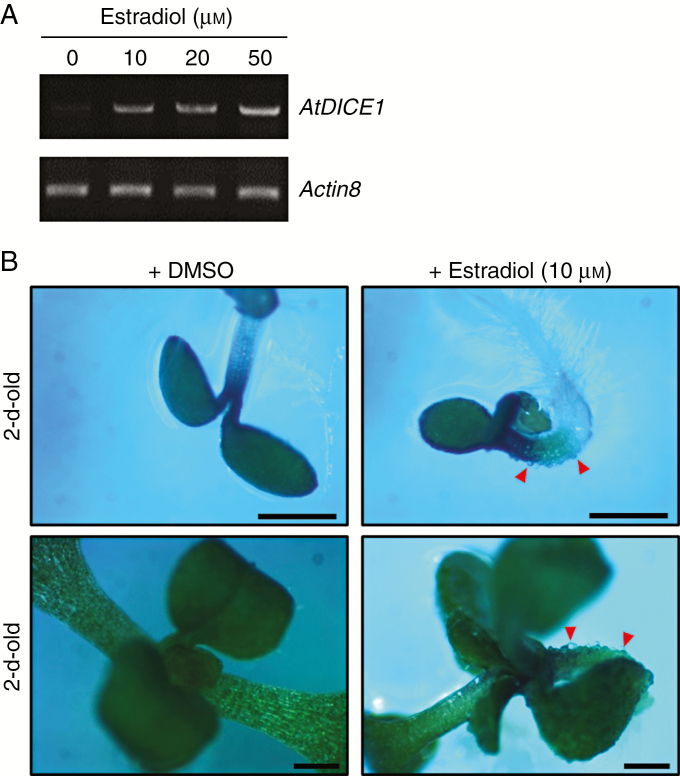
In young seedlings of 35S::AtDICE1 plants, epidermal cell swelling is evident in both hypocotyls and cotyledons; this was never seen in WT seedlings (Fig. 2). Analysis of a cross-section of hypocotyls from 7-d-old seedlings revealed that the cell swelling was found not only in epidermal cells but also in cells inside, which caused disarray of the cell arrangement in 35S::AtDICE1 plants (Fig. 2A). SEM clearly showed epidermal cell enlargements, which were distorted during the imaging process due to vacuum application (Fig. 2B). This isotropic cell expansion was found persistently in adult plants (Fig. 2C), which probably caused the severe dwarfism of 35S::AtDICE1 plants. To verify this phenotype, we produced additional transgenic Arabidopsis plants in which AtDICE1 expression can be induced. For inducible gene expression, the Gateway Destination vector pMDC7 (Curtis and Grossniklaus, 2003) was used. This vector contains the XVE inducible promoter for 17-β-estradiol inducible expression in plants (Fig. 3). Our results clearly demonstrated that treatment with 17-β-estradiol at concentrations as low as 10 αm successfully induced AtDICE1 expression (Fig. 3A) and resulted in epidermal cell swelling as early as in 2-d-old seedlings (Fig. 3B).
Fig. 2.
35S::AtDICE1 plants showed defects of anisotropic cell expansion. (A) Epidermal cell swelling in cotyledons (upper), hypocotyls (middle) and hypocotyl cross-sections (lower) of 7-d-old 35S::AtDICE1 seedlings compared to WT. Arrowheads indicate swollen cells. Scale bars represent 1 mm (top), 100 µm (middle and bottom). (B) Scanning electron micrographs of hypocotyls of 7-d-old plants. (C) Petioles of rosette leaves from 30-d-old plants. Arrowheads indicate swollen cells. Scale bars represent 0.5 cm.
Fig. 3.
Inducible expression of AtDICE1 reproduced the cell swelling phenotype of 35S::AtDICE1 plants. (A) Semi-quantitative RT-PCR analysis performed to confirm AtDICE1 induction by 17-β-estradiol treatment of 10-d-old seedlings of T3 homozygous AtDICE1/pMDC7 plants (line 8-4). (B) Estradiol treatment of AtDICE1/pMDC7 plants (line 8-4). AtDICE1/pMDC7 plants were grown on MS-agar plates with (+Estradiol) or without 17-β-estradiol (+DMSO). Red arrowheads indicate epidermal cell swelling. Scale bars represent 1 mm.
Cellulose biosynthesis is impaired in 35S::AtDICE1 plants
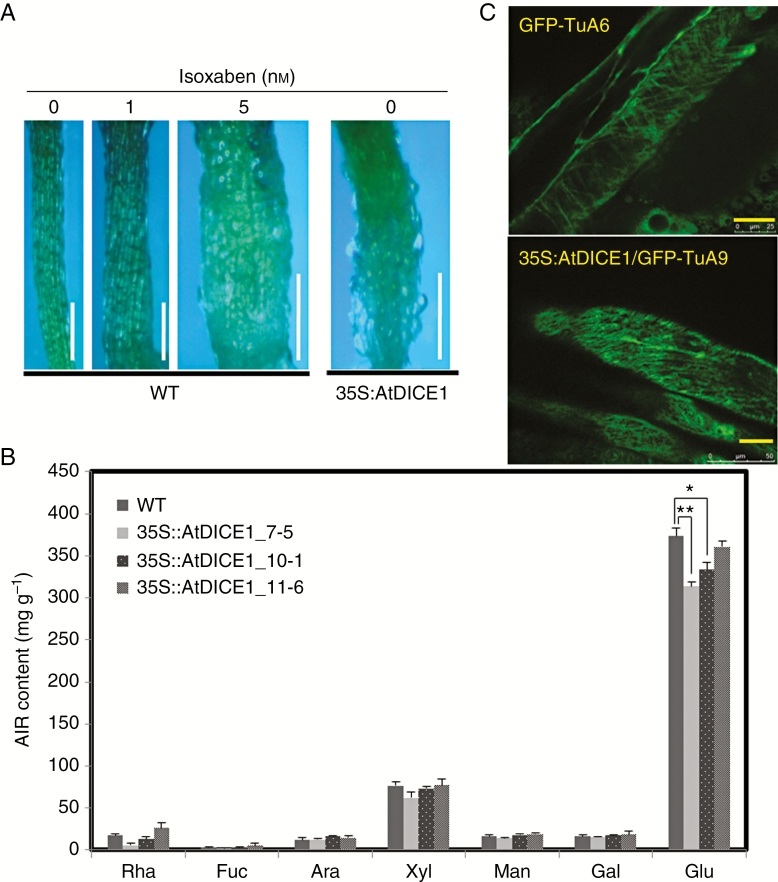
Perturbations in either cellulose biosynthesis or cortical microtubule arrangements are known to cause cell swelling (Fagard et al., 2000; Lane et al., 2001; Schindelman et al., 2001; Pagant et al., 2002; Caño-Delgado et al., 2003; Somerville, 2006; Wang et al., 2006). To test our hypothesis, isoxaben, a well-known inhibitor of cellulose biosynthesis, was applied to both WT and 35S::AtDICE1 plants. Treatment of WT plants with isoxaben successfully mimicked the epidermal cell swelling phenotype of 35S::AtDICE1 plants at concentrations as low as 5 nm (Fig. 4A). Because isoxaben treatment is known to prevent the insertion of CSCs into the PM (Paredez et al., 2006; Gutierrez et al., 2009), the cell swelling phenotype was probably caused by defects in cellulose biosynthesis in the cell wall.
Fig. 4.
35S::AtDICE1 resulted in defects in cellulose biosynthesis. (A) Treatment with isoxaben in WT plants. Plants were grown for 7 d on MS-agar plates containing isoxaben as indicated. Scale bars represent 0.5 mm. (B) Cell wall monosaccharide composition analysis. Inflorescence stems of 60-d-old soil-grown plants were used. AIR, alcohol-insoluble residue. Neutral monosaccharides in the sulfuric acid-soluble fraction were determined by quantification of their corresponding alditol acetates by gas chromatography. Rha, rhamnose; Fuc, fucose; Ara, arabinose; Xyl, xylose; Man, mannose; Gal, galactose; Glu, glucose. Error bars indicate s.e. using triplicate experiments (*P < 0.05; **P < 0.01, unpaired t-test). (C) Observation of cortical microtubule arrangement by confocal laser microscopy. Cortical microtubules were visualized in normal cells from GFP-TuA6 plants (upper) and swollen cells from 35S::AtDICE1/GFP-TuA6 plants (lower). Scale bars indicate 25 αµ.
To further confirm whether cellulose biosynthesis was affected in 35S::AtDICE1 plants, we analysed the monosaccharide composition of cell wall materials prepared from stem tissues of WT and 35S::AtDICE1 plants. To our surprise, the glucose content was significantly decreased in 35S::AtDICE1 plants (Fig. 4B). Because the AIRs of the cell wall were used in this analysis, most of the glucose content came from the degradation of cellulose. Thus, this result suggests that the cellulose content of the cell walls of 35S::AtDICE1 plants was reduced.
Next, to determine whether there were any changes in the cortical microtubule arrangement of 35S::AtDICE1 plants, we overexpressed AtDICE1 in GFP-TuA6 plants. These plants constitutively express a green fluorescent protein (GFP)-fused tubulin A6, and thus cortical microtubules can be visualized using confocal microscopy (Ueda and Matsuyama, 2000). Interestingly, we were able to observe abnormal arrangements of cortical microtubules in swollen cells from 35S::AtDICE1/GFP-TuA6 plants compared to GFP-TuA6 plants, which showed arrangements of microtubules perpendicular to the elongation direction (Fig. 4C).
AtDICE1 protein is probably localized in the ER
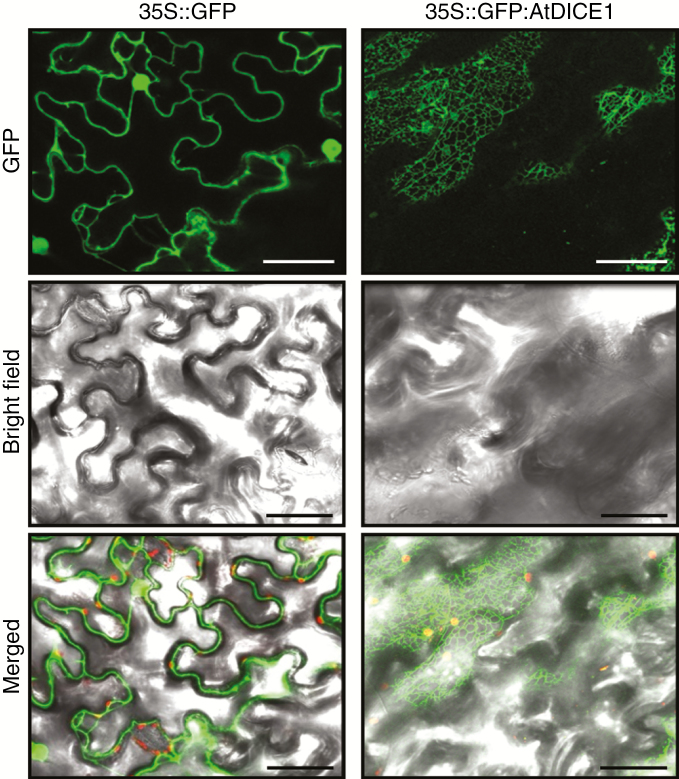
Our amino acid sequence analysis of AtDICE1 suggested that AtDICE1 is a putative transmembrane protein having seven transmembrane domains (Fig. S2). To investigate the intracellular localization of AtDICE1, a transient expression analysis was performed using tobacco epidermal cells. The vector construct expressing the AtDICE1 protein fused to the C-terminus of GFP driven by the CaMV 35S promoter (35S::GFP:AtDICE1) was introduced into the leaves of tobacco using Agrobacterium and the epidermal cells were observed under a confocal laser microscope. As shown in Fig. 5, the fluorescent signal of GFP:AtDICE1 was observed exclusively in the reticulate tubular networks and in sheets typical of the ER in the cortical regions of cells, indicating that AtDICE1 is probably localized in the ER. The same result was obtained by using the 35S:: AtDICE1:GFP construct (data not shown). By contrast, in a control experiment, the fluorescent signal of GFP alone was found in the plasma membrane, cytoplasm and nucleus of the cells (Fig. 5).
Fig. 5.
Intracellular localization of the AtDICE1 protein. Intracellular localization of either GFP alone (left) or GFP:AtDICE1 (right) driven by the 35S promoter was observed by confocal imaging of transiently transformed tobacco epidermal leaf cells. Single confocal optical sections visualizing images of GFP fluorescence (500–520 nm) were photographed using dark field (upper) and bright field (middle) and then merged (lower). Scale bars represent 50 αµ.
The N-terminal conserved domain is necessary for AtDICE1 function
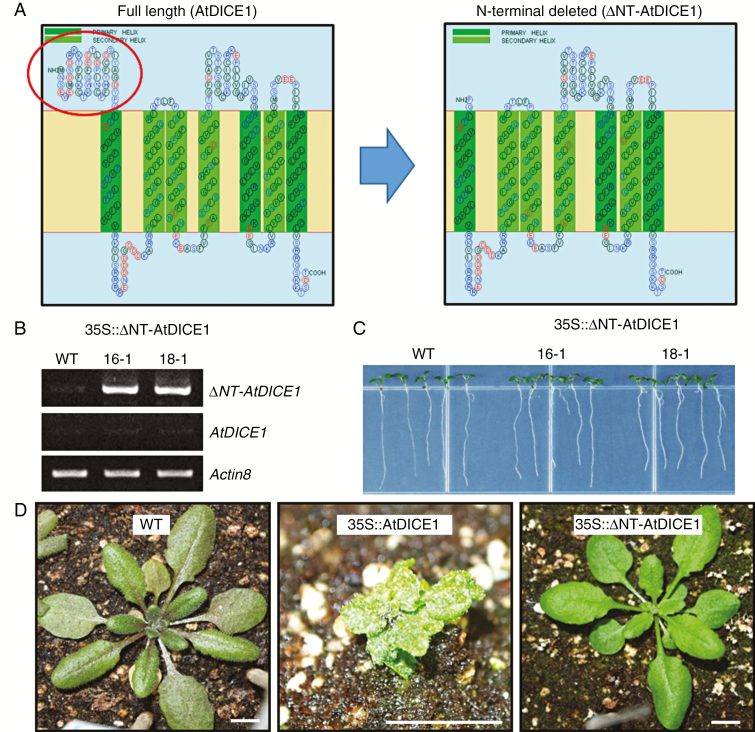
Because AtDICE1 has a putative external N-terminal motif, which is highly conserved in the plant species examined (Fig. S2A), we attempted to determine the functional significance of the motif using deletion analysis. We generated transgenic Arabidopsis plants overexpressing an N-terminal deleted AtDICE1 gene (35S::ΔNT-AtDICE1) (Fig. 6A, B). However, we did not observe any phenotypic changes in either the young seedlings or adult plants of the 19 independent T3 homozygous transgenic plants we examined (Fig. 6C, D). This suggests that the highly conserved N-terminal external domain of AtDICE1 might play an important role in AtDICE1 function.
Fig. 6.
The N-terminal conserved domain is necessary for AtDICE1 function. (A) Schematic diagram of full length (left) and N-terminal conserved domain-deleted (right; ΔNT-AtDICE1) AtDICE1. The N-terminal conserved domain is indicated by the red circle. (B) Semi-quantitative RT-PCR analysis to confirm the expression of ΔNT-AtDICE1 in 35S::ΔNT-AtDICE1 plants. (C) Seven-day-old 35S::ΔNT-AtDICE1 seedlings compared to WT. (D) Phenotypic consequences in a 33-d-old 35S::ΔNT-AtDICE1 plant compared to both WT and 35S::AtDICE1 plants. Scale bars indicate 1 cm.
Transcription phenotype of 35S::AtDICE1 plants is highly correlated with its physiological phenotype
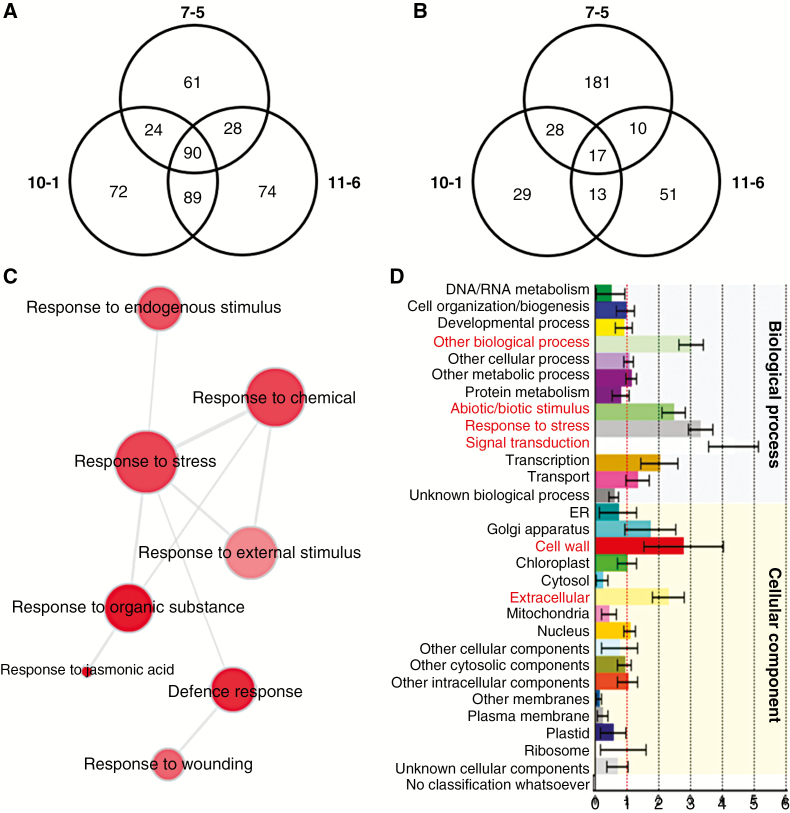
To examine the transcription phenotype of 35S::AtDICE1 plants, a whole-transcriptome analysis was performed using total RNAs extracted from 7-d-old seedlings of three independent T3 homozygous lines of 35S::AtDICE1 (i.e. 7-5, 10-1, 11-6) and WT plants grown on MS agar plates (see Materials and Methods). Gene expression profiles were obtained using Agilent’s Whole Arabidopsis Gene Expression Microarray (G2519F-021169, V4, 4x44K), which contains a total of 43 505 probe sets covering the entire Arabidopsis transcriptome (http://www.genomics.agilent.com). In the subsequent gene expression analysis, we found a total of 90 probe sets (representing 72 genes) which were significantly upregulated (three-fold as a threshold) in all three lines of 35S::AtDICE1 plants compared to WT plants. Meanwhile only 17 probe sets (representing 16 genes) were downregulated (Fig. 7A, B; Table S2).
Fig. 7.
Whole transcriptome analyses of 7-d-old 35S::AtDICE1 plants compared to WT. (A and B) Venn diagrams showing differentially expressed probe sets from three independent lines (i.e. 7-5, 10-1, 11-6) of 35S::AtDICE1 plants compared to WT. The number of probe sets in each line is displayed within a circle. The number of common probe sets is shown in the intersections between lines. Upregulated (A) or downregulated (B) probe sets in each line compared to WT using a three3-fold change as the threshold. (C) Gene Ontology (GO) analysis performed using REVIGO (http://revigo.irb.hr/). The representative GO terms of the 90 upregulated probe sets of A are visualized in semantic similarity-based scatterplots. (D) Functional classification generated using the Classification SuperViewer (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi). The Classification SuperViewer generates an overview of functional classification of the 90 upregulated probe sets of A based on the GO database. The input set was bootstrapped 100 times to provide an idea as to over- or underrepresentation reliability. Gene categories are shown on the left, and their normalized frequencies are found on the x-axis and are calculated as follows: (Number_in_Classinput_set/Number_Classifiedinput_set)/(Number_in_Classreference_set (25k)/Number_Classifiedreference_set).
To gain functional insight into the 72 upregulated genes in 35S::AtDICE1, we performed Gene Ontology (GO) analyses (Fig. 7C, D). In the REVIGO (http://revigo.irb.hr) analysis, which provides a visual summary of the long list of GO terms as a scatterplot by removing redundant GO terms, the responses to stress, response to organic substance (e.g. JA) and defence response categories plotted most clearly (Fig. 7C). In addition, using the Classification SuperViewer tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi), genes related to the abiotic/biotic stimulus, response to stress and cell wall categories were found to be over-represented (Fig. 7D). Accordingly, more than half of the 72 upregulated genes (37 genes, 51.4 %) were involved in the stress and/or defence mechanisms (Table 1). In addition, many cell wall formation and modification-related genes were significantly upregulated, which probably reflects the defects in the cell elongation phenotype of 35S::AtDICE1 plants (Fig. 7D and Table 1). These results clearly show that the transcription phenotype of 35S::AtDICE1 plants is highly correlated with its physiological phenotype.
Table 1.
List of genes upregulated in 35S::AtDICE1 plants
| AGI* | WT | F† | 7-5 | F | 10-1 | F | 11-6 | F | 7-5/WT‡ | 10-1/WT | 11-6/WT | cob-5 /WT§ | Gene description | St/Def¶ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT1G72260 | 6.7 | A | 7978.0 | P | 2015.0 | P | 2907.6 | P | 1185.5 | 299.3 | 431.9 | 106.4 | thionin 2.1 | YES |
| AT4G15100 | 15.9 | A | 1239.8 | P | 156.3 | P | 88.0 | P | 78.2 | 9.9 | 5.5 | 12.5 | serine carboxypeptidase- like 30 | |
| AT2G39330 | 267.0 | P | 8145.8 | P | 4897.3 | P | 4446.0 | P | 30.5 | 18.3 | 16.7 | 43.1 | jacalin-related lectin 23 | YES |
| AT2G41610 | 408.1 | P | 8791.5 | P | 6760.8 | P | 3807.9 | P | 21.5 | 16.6 | 9.3 | 0.7 | AtDCE1 | |
| AT1G54020 | 166.3 | P | 3426.4 | P | 1788.4 | P | 1949.7 | P | 20.6 | 10.8 | 11.7 | 53.9 | GDSL esterase/lipase | YES |
| AT1G52030 | 805.8 | P | 14800.8 | P | 9016.5 | P | 10847.8 | P | 18.4 | 11.2 | 13.5 | n/a | myrosinase-binding protein 2 | YES |
| AT1G52040 | 181.9 | P | 2666.8 | P | 2117.9 | P | 2019.6 | P | 14.7 | 11.6 | 11.1 | 27.6 | myrosinase-binding protein 1 | YES |
| AT1G33760 | 41.1 | P | 525.4 | P | 394.8 | P | 295.1 | P | 12.8 | 9.6 | 7.2 | 20.2 | ERF022 | YES |
| AT4G01985 | 16.9 | A | 215.6 | P | 298.5 | P | 118.7 | P | 12.7 | 17.6 | 7.0 | n/a | hypothetical protein | |
| AT3G28220 | 3471.2 | P | 38 894.9 | P | 28 177.1 | P | 36 552.9 | P | 11.2 | 8.1 | 10.5 | 13.6 | TRAF-like family protein | |
| AT1G66370 | 43.3 | P | 478.5 | P | 254.8 | P | 224.4 | P | 11.1 | 5.9 | 5.2 | 7.4 | MYB113 | YES |
| AT4G27160 | 17.4 | A | 167.7 | P | 53.5 | P | 162.2 | P | 9.6 | 3.1 | 9.3 | 0.3 | seed storage albumin 3 | |
| AT1G35140 | 1734.1 | P | 15 869.4 | P | 13 207.7 | P | 17 206.2 | P | 9.2 | 7.6 | 9.9 | 3.5 | PHI-1/EXL1 | YES |
| AT5G28237 | 4.9 | A | 41.3 | P | 51.1 | P | 26.7 | P | 8.4 | 10.3 | 5.4 | n/a | tryptophan synthase-like protein | |
| AT4G27170 | 18.9 | A | 157.0 | P | 57.5 | P | 103.7 | P | 8.3 | 3.0 | 5.5 | 0.5 | seed storage albumin 4 | |
| AT3G27810 | 7.2 | A | 53.6 | P | 57.1 | P | 45.0 | P | 7.4 | 7.9 | 6.2 | 1.6 | MYB21 | YES |
| AT4G17920 | 25.1 | A | 184.6 | P | 247.3 | P | 222.6 | P | 7.3 | 9.8 | 8.9 | n/a | RING-H2 finger protein ATL29 | |
| AT1G61120 | 21.4 | A | 150.9 | P | 180.1 | P | 321.5 | P | 7.0 | 8.4 | 15.0 | 71.5 | terpene synthase 04 | YES |
| AT1G17380 | 222.5 | P | 1471.2 | P | 982.5 | P | 1118.6 | P | 6.6 | 4.4 | 5.0 | 21.7 | JAZ5/TIFY 11A | YES |
| AT4G34410 | 35.5 | P | 230.8 | P | 121.3 | P | 148.0 | P | 6.5 | 3.4 | 4.2 | 0.3 | ERF109 | YES |
| AT1G52400 | 11461.6 | P | 72 721.5 | P | 44 118.3 | P | 51 794.5 | P | 6.3 | 3.8 | 4.5 | 3.1 | BGLU18/beta glucosidase 18 | YES |
| AT1G52410 | 3933.8 | P | 23 332.2 | P | 16 344.8 | P | 15 134.4 | P | 5.9 | 4.2 | 3.8 | 21.9 | TSK-associating protein 1 | YES |
| AT3G25180 | 11.3 | A | 66.3 | P | 70.7 | P | 131.2 | P | 5.9 | 6.3 | 11.7 | 2.9 | CYP82G1 | YES |
| AT1G76640 | 174.4 | P | 999.9 | P | 660.1 | P | 551.5 | P | 5.7 | 3.8 | 3.2 | 19.2 | calcium-binding protein CML39 | |
| AT2G34600 | 244.6 | P | 1319.1 | P | 798.1 | P | 1062.2 | P | 5.4 | 3.3 | 4.3 | 1.7 | JAZ7/TIFY 5B | YES |
| AT5G41750 | 622.9 | P | 3355.5 | P | 2809.3 | P | 2847.5 | P | 5.4 | 4.5 | 4.6 | n/a | TIR-NBS-LRR disease-resistant protein | YES |
| AT5G59310 | 1746.9 | P | 9175.7 | P | 24 203.0 | P | 17 251.5 | P | 5.3 | 13.9 | 9.9 | 1.7 | non-specific lipid-transfer protein 4 | YES |
| AT5G41740 | 354.1 | P | 1794.6 | P | 1501.2 | P | 1671.3 | P | 5.1 | 4.2 | 4.7 | 4.7 | TIR-NBS-LRR disease-resistant protein | YES |
| AT1G66610 | 16.7 | A | 83.5 | P | 91.2 | P | 54.5 | P | 5.0 | 5.4 | 3.3 | 1.5 | E3 ubiquitin-ligase SINA-like 1 | |
| AT1G66860 | 6.8 | A | 33.6 | P | 48.6 | P | 30.9 | P | 5.0 | 7.2 | 4.6 | 2.6 | Class I glutamine amidotransferase-like | |
| AT4G28790 | 21.5 | A | 100.4 | P | 172.9 | P | 175.4 | P | 4.7 | 8.1 | 8.2 | n/a | transcription factor bHLH23 | |
| AT5G59580 | 17.2 | A | 77.4 | P | 66.9 | P | 66.7 | P | 4.5 | 3.9 | 3.9 | 0.8 | UDP-glucosyl transferase 76E1/UGT76E1 | |
| AT5G24780 | 7476.2 | P | 33 245.3 | P | 35 412.6 | P | 39 064.4 | P | 4.4 | 4.7 | 5.2 | 4.8 | vegetative storage protein 1 | YES |
| AT4G11911 | 15.3 | A | 67.4 | P | 132.8 | P | 84.5 | P | 4.4 | 8.7 | 5.5 | n/a | hypothetical protein | |
| AT5G13220 | 260.0 | P | 1124.2 | P | 815.6 | P | 886.6 | P | 4.3 | 3.1 | 3.4 | 11.0 | JAZ10/TIFY 9 | YES |
| AT1G77640 | 338.2 | P | 1433.9 | P | 1595.4 | P | 1283.1 | P | 4.2 | 4.7 | 3.8 | 2.4 | ERF013 | |
| AT3G27415 | 20.9 | A | 87.1 | P | 146.4 | P | 111.3 | P | 4.2 | 7.0 | 5.3 | n/a | hypothetical protein | |
| AT5G57560 | 5382.4 | P | 22 307.3 | P | 21 813.0 | P | 21 676.0 | P | 4.1 | 4.1 | 4.0 | 1.3 | TCH4/XTH22 | |
| AT4G23215 | 17.7 | A | 73.1 | P | 89.4 | P | 74.1 | P | 4.1 | 5.0 | 4.2 | n/a | hypothetical protein | |
| AT4G29570 | 16.6 | A | 68.0 | P | 175.5 | P | 185.8 | P | 4.1 | 10.6 | 11.2 | 2.7 | cytidine/deoxycytidylate deaminase-like | |
| AT2G32200 | 259.6 | P | 1056.0 | P | 838.7 | P | 1025.1 | P | 4.1 | 3.2 | 3.9 | n/a | unknown protein | |
| AT1G03880 | 38.8 | P | 155.0 | P | 118.8 | P | 557.6 | P | 4.0 | 3.1 | 14.4 | 2.2 | cruciferin 2 | YES |
| AT4G10265 | 208.0 | P | 824.1 | P | 850.3 | P | 994.6 | P | 4.0 | 4.1 | 4.8 | n/a | putative wound-responsive protein | YES |
| AT5G24770 | 10375.6 | P | 40 351.9 | P | 65 197.1 | P | 55 171.0 | P | 3.9 | 6.3 | 5.3 | n/a | vegetative storage protein 2 | YES |
| AT4G12735 | 11.8 | A | 45.1 | P | 40.4 | P | 41.3 | P | 3.8 | 3.4 | 3.5 | n/a | hypothetical protein | |
| AT5G30341 | 9.5 | A | 36.2 | P | 43.5 | P | 40.7 | P | 3.8 | 4.6 | 4.3 | n/a | hypothetical protein | |
| AT5G11410 | 447.9 | P | 1698.6 | P | 1865.2 | P | 1458.5 | P | 3.8 | 4.2 | 3.3 | 18.9 | protein kinase family protein | |
| AT1G52000 | 482.5 | P | 1782.7 | P | 1549.3 | P | 1480.3 | P | 3.7 | 3.2 | 3.1 | 7.1 | jacalin-like lectin domain protein | YES |
| AT4G16590 | 33.7 | P | 124.4 | P | 247.1 | P | 397.6 | P | 3.7 | 7.3 | 11.8 | 4.2 | cellulose synthase-like A01 | |
| AT5G52760 | 107.3 | P | 395.5 | P | 503.6 | P | 354.6 | P | 3.7 | 4.7 | 3.3 | 7.9 | copper transport family protein | |
| AT1G21910 | 2375.9 | P | 8713.1 | P | 7190.5 | P | 7939.6 | P | 3.7 | 3.0 | 3.3 | 2.5 | ERF012/DREB26 | YES |
| AT5G58400 | 52.8 | P | 193.5 | P | 184.9 | P | 170.7 | P | 3.7 | 3.5 | 3.2 | 1.2 | peroxidase 68 | YES |
| AT1G69880 | 1654.5 | P | 6054.5 | P | 6756.3 | P | 6254.5 | P | 3.7 | 4.1 | 3.8 | 11.6 | thioredoxin H8 | YES |
| AT1G19610 | 305.4 | P | 1104.3 | P | 1089.4 | P | 1019.9 | P | 3.6 | 3.6 | 3.3 | 9.3 | PDF1.4/defensin-like protein 19 | YES |
| AT2G38250 | 121.0 | P | 435.5 | P | 681.0 | P | 363.1 | P | 3.6 | 5.6 | 3.0 | 3.6 | DNA-binding protein | |
| AT5G49520 | 284.6 | P | 998.3 | P | 991.5 | P | 900.4 | P | 3.5 | 3.5 | 3.2 | 3.7 | WRKY48 | YES |
| AT3G57260 | 74.7 | P | 260.4 | P | 800.4 | P | 1107.0 | P | 3.5 | 10.7 | 14.8 | 8.4 | BGL2/β-1,3-glucanase 2/PR2 | YES |
| AT1G68290 | 172.0 | P | 599.1 | P | 532.4 | P | 625.9 | P | 3.5 | 3.1 | 3.6 | 2.4 | endonuclease 2 | |
| AT2G17040 | 1983.1 | P | 6898.2 | P | 6486.4 | P | 5957.2 | P | 3.5 | 3.3 | 3.0 | 5.0 | ANAC036 | YES |
| AT4G02170 | 45.9 | P | 157.5 | P | 188.2 | P | 172.0 | P | 3.4 | 4.1 | 3.7 | 2.3 | hypothetical protein | |
| AT4G02330 | 4488.5 | P | 15 318.2 | P | 18 712.9 | P | 21 081.7 | P | 3.4 | 4.2 | 4.7 | 10.0 | pectinesterase/ATPMEPCRB | |
| AT3G50770 | 504.3 | P | 1692.4 | P | 1544.9 | P | 2176.5 | P | 3.4 | 3.1 | 4.3 | 2.1 | calcium-binding protein CML41 | |
| AT1G72520 | 438.8 | P | 1431.8 | P | 1316.7 | P | 2118.1 | P | 3.3 | 3.0 | 4.8 | 8.2 | lipoxygenase 4 | YES |
| AT2G35290 | 468.8 | P | 1491.9 | P | 1571.7 | P | 1476.1 | P | 3.2 | 3.4 | 3.1 | 0.8 | SAUR79 | |
| AT3G02550 | 111.9 | P | 351.8 | P | 411.3 | P | 461.1 | P | 3.1 | 3.7 | 4.1 | 1.2 | LOB41 | YES |
| AT4G32060 | 15.2 | A | 47.6 | P | 54.6 | P | 86.8 | P | 3.1 | 3.6 | 5.7 | 0.7 | calcium-binding EF hand protein | |
| AT4G25810 | 2394.0 | P | 7487.3 | P | 8327.9 | P | 7563.7 | P | 3.1 | 3.5 | 3.2 | 5.2 | XTH23 | |
| AT1G63750 | 1816.6 | P | 5667.8 | P | 6791.9 | P | 7526.2 | P | 3.1 | 3.7 | 4.1 | 1.7 | TIR-NBS-LRR disease-resistant protein | YES |
| AT2G14560 | 72.0 | P | 222.7 | P | 521.3 | P | 253.2 | P | 3.1 | 7.2 | 3.5 | 3.8 | LURP1 | YES |
| AT2G05580 | 32.8 | P | 100.1 | P | 130.3 | P | 115.9 | P | 3.1 | 4.0 | 3.5 | 0.2 | glycine-rich protein | |
| AT4G14365 | 591.9 | P | 1784.3 | P | 2416.5 | P | 2205.6 | P | 3.0 | 4.1 | 3.7 | 4.9 | E3 ubiquitin-protein ligase XBAT34 | YES |
| AT2G23100 | 19.5 | A | 58.7 | P | 61.5 | P | 87.2 | P | 3.0 | 3.2 | 4.5 | 1.5 | cys/his-rich C1 domain protein |
*AGI, Arabidopsis Gene Index.
†F, flag calling (presence, absence, marginal).
‡Fold change.
§Data from Ko et al. (2006b).
¶Stress/defence-related genes.
To verify the microarray results, we performed a semi-quantitative RT-PCR analysis of 15 selected genes, including six that were upregulated, four that were downregulated and five that had no significant changes in gene expression in 35S::AtDICE1 plants. The band intensities of the PCR products were quite comparable to the microarray data (Fig. S5).
Abnormal secondary wall thickenings in pith cells of the inflorescent stem in 35S::AtDICE1 plants
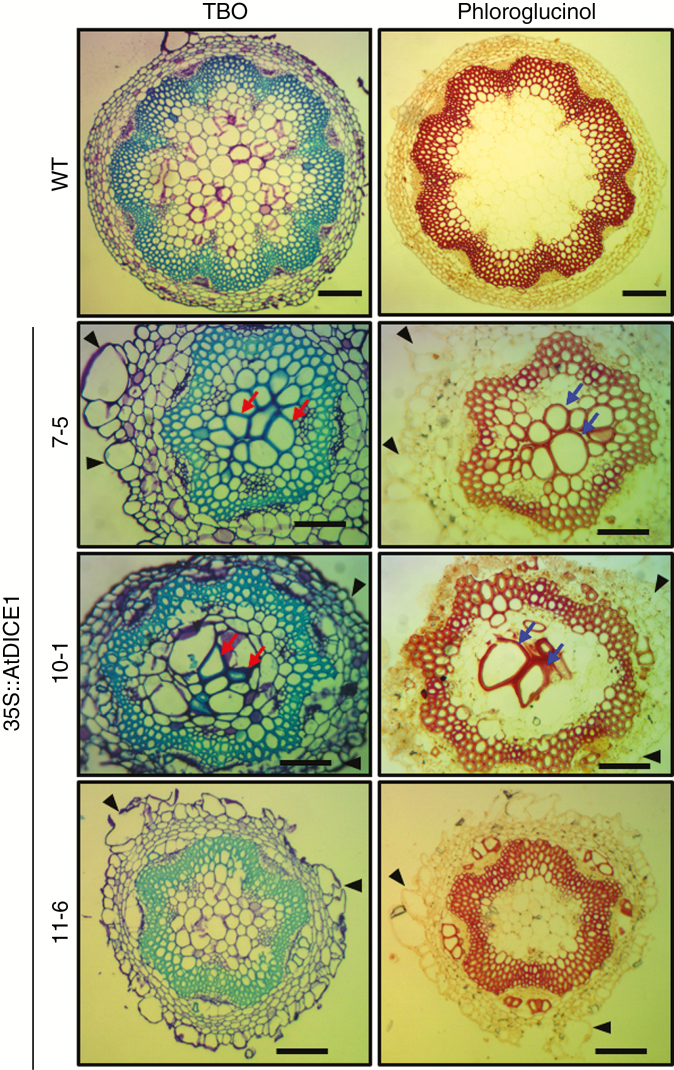
Because the growth of inflorescent stems in 35S::AtDICE1 plants was severely inhibited and the surface of the stems was bumpy, the anatomy of the inflorescent stems of 35S::AtDICE1and WT plants was examined using microtome sectioning. Surprisingly, we found abnormal secondary wall thickenings in the pith cells of 35S::AtDICE1 plants in addition to the epidermal cell swelling/enlargements (Fig. 8). This phenotype was also observed in the stem sections of 35S::PtrDICE1 plants (Fig. S7). Cross-sections along the stem length indicate that these abnormal secondary wall thickenings are regional or sporadic (Fig. S7B). Interestingly, this result is reminiscent of the eli1 mutant, which has defects in cell expansion due to mutation of the CesA3 gene (Caño-Delgado et al., 2000, 2003).
Fig. 8.
35S::AtDICE1 plants showed abnormal secondary wall thickenings in pith of inflorescent stems. Stem cross-sections of 50-d-old 35S::AtDICE1 and WT plants stained with 0.05 % toluidine blue O (TBO) and 2 % phloroglucinol/HCl (Phloroglucinol) as indicated. Pith cells having secondary wall thickenings are indicated by arrows and swollen cells by arrowheads. Scale bars represent 100 µm.
DISCUSSION
Precise regulation of cell expansion is essential for cellular morphogenesis and differentiation and consequently directs typical plant growth and development. It is well known that cellulose microfibrils strengthen cell walls and define the direction of anisotropic cell growth (Cosgrove, 2005; Lloyd and Chan, 2006). In this report, we attempted to characterize a gene locus, AtDICE1, which encodes a novel transmembrane protein probably localized to the ER.
Upregulation of AtDICE1 caused abnormal cell expansion
Upregulation of AtDICE1 caused severe growth and developmental defects at the whole plant level (Figs 1, 2 and S3, Table S1). The prominent cell swelling phenotype, caused by defects in anisotropic cell expansion, probably accounted for the dwarf phenotype of 35S::AtDICE1 plants (Figs 2 and 3). This phenotypic consequence was reproduced by overexpression of PtrDICE1, a poplar homologue of AtDICE1 (Fig. 1E) and was further verified by inducible expression of AtDICE1 which resulted in a clear phenotype as early as in 2-d-old Arabidopsis seedlings (Fig. 3).
We hypothesized that the cell swelling phenotype of 35S::AtDICE1 plants may result from alterations in cell wall integrity, especially the decrease in cellulose content estimated from our cell wall monosaccharide analysis (Fig. 4). This is further supported by the following results: (1) treatment with isoxaben, a well-known cellulose synthase inhibitor, mimicked the cell swelling phenotype of 35S::AtDICE1 plants in WT plants (Fig. 4A), and (2) cortical microtubule arrangements, which direct proper cellulose deposition in cell walls via an association with CesA interacting1/CSI1 (Paredez et al., 2006; Bringmann et al., 2012; Li et al., 2012), were severely altered in 35S::AtDICE1/GFP-TuA6 plants (Fig. 4C). Previously, loss-of-function mutants in several genes, including RSW1/CesA1, RSW5/CesA3, QUILL/PROCUSTE/CesA6, POM-POM1/CHITINASE LIKE1 (POM1/CTL1), KORRIGAN/LION’S TAIL1/RADIALLY SWOLLEN2 (KOR/LIT/RSW2), COBRA and KOBITO/ELONGATION DEFECTIVE1 (KOB1/ELD1), involved in cellulose biosynthesis or cell wall modification were reported to have cell swelling and dwarfing phenotypes similar to that observed in 35S::AtDICE1 plants (Hauser et al., 1995; Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000; Lane et al., 2001; Schindelman et al., 2001; Pagant et al., 2002; Wang et al., 2006). The korrigan mutant showed abnormal cell expansion phenotypes in roots and hypocotyls, and was found to be defective in an endo-1,4-d-glucanase (Nicol et al., 1998). The cobra mutant exhibited abnormal cell expansion and cellulose deposition in root cells resulting from a failure of orientated cell expansion (Roudier et al., 2002).
Furthermore, our whole transcriptome analysis revealed the relevant transcription profile of 35S::AtDICE1 plants responsible for the defects in anisotropic cell expansion (Table 1). For example, expression of the MYB21 and ANAC036 transcription factors, which are known to negatively regulate cell elongation, were up-regulated up to 7.9-fold in 35S::AtDICE1 plants. Overexpression of an MYB21-GR fusion protein resulted in decreased hypocotyl cell expansion, shorter stems, and smaller and narrower leaves (Shin et al., 2002). Overexpression of ANAC036 also resulted in a semi-dwarf phenotype (Kato et al., 2010). In addition, many cell wall formation/modification-related genes were upregulated, including xyloglucan endotransglucosylase/hydrolase (XTH22/TCH4 and XTH23), pectinesterase, cellulose synthase-like A01, lipid transfer proteins and peroxidase (Passardi et al., 2004; Nieuwland et al., 2005) (Table 1).
Upregulation of AtDICE1 invokes a cellular defence mechanism
Interference with cell wall integrity results in various compensatory reactions, such as the reactive oxygen species (ROS) response, ectopic lignin deposition (Caño-Delgado et al., 2003; Denness et al., 2011), altered pectin methyl-esterification status (Manfield et al., 2004), elevated callose deposition (Desprez et al., 2002), and increased JA and ethylene production with an associated resistance to pathogens (Ellis et al., 2002). Thus, the finding that a high proportion (51.4 %) of the upregulated genes in 35S::AtDICE1 plants were genes involved in stress and defence responses can be explained as the response of plant cells to changes in cell wall integrity and abnormal cell shape (Fig. 7 and Table 1). In particular, JA biosynthesis, signalling and responsive genes were massively upregulated. For example, Thionin 2.1 (AT1G72260), a cysteine-rich protein with antimicrobial properties, was dramatically upregulated (up to 1185-fold). Myrosinase-binding proteins (MBP1, AT1G52040; and MBP2, AT1G52030), involved in metabolizing defence compounds to protect against herbivory, were upregulated (up to 18-fold). Additionally, vegetative storage proteins (VSP1, AT5G24780; and VSP2, AT5G24770), known to be induced by JA, were upregulated (up to six-fold).
It has been reported that COBRA, a glycosyl phosphatidyl inositol (GPI)-anchored protein, is targeted to the plasma membrane and involved in orientated cell expansion in Arabidopsis (Roudier et al., 2005). cob-5, a loss-of-function mutant of COBRA with an epidermal cell swelling phenotype similar to 35S::AtDICE1, over-accumulates JA and coordinately induces stress- and defence-related genes (Ko et al., 2006b). Interestingly, our whole transcriptome analysis revealed that the upregulated genes in 35S::AtDICE1 plants overlapped dramatically with those upregulated in cob-5. Specifically, 32 of the 72 upregulated genes were also upregulated more than three-fold in cob-5 and, among them (i.e. 32 genes), 23 genes (~72 %) were involved in the stress/defence response (Table 1). This suggests that the changes in cell wall integrity caused by cell swelling in 35S::AtDICE1 plants induce cellular defence responses as in cob-5. Taken together, these results further support the hypothesis that plant cells initially perceive biotic stress at the cell surface (Ko et al., 2006b).
While the overexpression of AtDICE1 induced conspicuous phenotypes, the suppression of AtDICE1 resulted in no distinguishable phenotypic changes compared to WT (Fig. S4). However, expression of Thionin 2.1, which was dramatically upregulated in 35S::AtDICE1 plants, was significantly downregulated in AtDICE1_RNAi plants (Fig. S6). Thus, it is possible that the suppression of AtDICE1 in AtDICE1_RNAi plants was not strong enough to cause phenotypic changes. Generating a loss-of-function mutant using CRISPR/Cas9 technology may be required to address this hypothesis.
Functional significances of AtDICE1
Amino acid sequence analysis predicted that AtDICE1 is a membrane protein having seven transmembrane domains, and containing a putative di-lysine (KK) motif at the C-terminus, which is required for ER targeting (Fig. S2; Teasdale and Jackson, 1996). Indeed, the results of our transient expression analysis of GFP:AtDICE1 suggest that AtDICE1 is probably localized to the ER (Fig. 5). The ER is a cytoplasmic network of flattened, membrane-enclosed sacs or tube-like structures known as cisternae that play important roles in the folding of protein molecules and the transport of synthesized proteins in vesicles to the Golgi apparatus (Chen et al., 2012). Correct folding of newly synthesized proteins is made possible by several ER chaperone proteins and only properly folded proteins are transported from the rough ER to the Golgi apparatus. We hypothesize that AtDICE1 might be involved in the processing of some proteins responsible for cell wall formation, such as CesA proteins, in the ER via a currently unknown mechanism. Previously, mutants of the PEANUT1 gene encoding an ER-localized mannosyltransferase required for synthesis of the GPI anchor showed radially swollen embryos and decreased crystalline cellulose in the cell wall (Gillmor et al., 2005). Thus, it might also be plausible that the overly increased AtDICE1 protein population in the ER might disturb other proteins such as PEANUT1. If this is the case, the N-terminal external domain is obligatory because overexpression of ΔNT-AtDICE1 was not associated with any phenotypic changes (Fig. 6). In addition, because AtDICE1 was identified as a member of a gene network regulating secondary xylem development in Arabidopsis (Ko et al., 2006a) and confirmed to be expressed specifically in vascular tissues (Fig. S1), it might be plausible that AtDICE1 may function in vascular tissue formation by regulating anisotropic cell elongation.
It remains uncertain how cell elongation is integrated with secondary wall formation, which occurs irreversibly in some specialized cells such as xylem cells and fibres after cessation of cell elongation (Boerjan et al., 2003; Déjardin et al., 2010). Previously, the eli1 mutant exhibited a stunted phenotype and xylem cell disorganization in stem pith, which was caused by altered cell expansion with inappropriate initiation of secondary wall formation (Caño-Delgado et al., 2000). Because ectopic secondary wall formation or lignification phenotypes were observed in many other cell expansion mutants (e.g. rsw1, korrigan1/lit, det3), a mechanism that senses cell size and induces subsequent secondary wall formation may exist (Caño-Delgado et al., 2000). Subsequently, it was shown that eli1 mutants occur in the cellulose synthase gene CesA3 (Caño-Delgado et al., 2003). Recently, AtC3H14, a plant-specific tandem CCCH zinc-finger protein, was suggested to be a coordinate regulator of cell elongation and secondary wall formation during xylogenesis because overexpression of AtC3H14 or PtC3H17, a poplar homologuw of AtC3H14, suppressed cell elongation but increased secondary wall thickening (Kim et al., 2014; Chai et al., 2015). In this report, we found abnormal secondary wall thickenings in the pith cells of 35S::AtDICE1 and 35S::PtrDICE1 plants (Figs 8 and S7). This result further supports the hypothesis that a cell size sensing mechanism exists, leading to secondary wall formation. However, because these abnormal secondary wall thickenings in stem tissues of 35S::AtDICE1 plant are patchy (Fig. S7B), we assumed that the overall cellulose contents in adult stems of 35S::AtDICE1 plants were lower than in the WT, as shown in Fig. 4B. Furthermore, expression of AtC3H14 was not significantly altered in 35S::AtDICE1 plants and, furthermore, no changes in AtDICE1 were observed in 35S::AtC3H14 plants (data not shown), suggesting that it is unlikely that a mechanistic connection exists between them.
In conclusion, our data suggest that AtDICE1 is a novel putative ER-localized transmembrane protein contributing to a proper anisotropic cell elongation process through participation in cell wall formation via a currently unknown mechanism. Because AtDICE1 was identified as a member of a gene network regulating secondary xylem development in Arabidopsis (Ko et al., 2006a) and confirmed to be expressed specifically in secondary wall-forming tissues (Fig. S1), it is possible that AtDICE1 might be involved in the anisotropic cell elongation during xylogenesis. Further in-depth investigations, including loss-of-function mutation employing CRISPR/Cas9 technology, immunohistochemistry and protein–protein interactions with cell-wall-forming proteins (e.g. AtDICE1 with CESA complexes), will be required to elucidate AtDICE1 function in detail.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Growth features of 35S::AtDICE1. Table S2: List of genes differentially expressed in 35S::AtDICE1. Table S3: Primers used in this study. Figure S1: AtDICE1 (At2g41610) is specifically expressed in vascular tissues/cells. Figure S2: AtDICE1 is a plant-specific transmembrane protein. Figure S3: Floral organs of 35S::AtDICE1 plants are smaller than in WT. Figure S4: Phenotypic characterization of AtDICE1_RNAi plants. Figure S5: Confirmation of the microarray data using semi-quantitative RT-PCR analysis. Figure S6: Expression of the Thionin 2.1 gene was significantly downregulated in AtDICE1_RNAi plants. Figure S7: 35S::PtrDICE1 resulted in abnormal secondary wall thickenings in pith cells as in 35S::AtDICE1 plants.
ACKNOWLEDGEMENTS
We are grateful to Prof. Kim (Youngrok) at Kyung Hee University for technical assistance with SEM analysis. This work was supported by the Forest Resources Genome Project (2014071G10-1722-AA04) and by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF-2015R1D1A1A01060807).
LITERATURE CITED
- Arioli T, Peng L, Betzner AS, et al. . 1998. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720. [DOI] [PubMed] [Google Scholar]
- Baskin TI. 2005. Anisotropic expansion of the plant cell wall. Annual Review of Cell and Developmental Biology 21: 203–222. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S. 2012. POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado AI, Metzlaff K, Bevan MW. 2000. The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado AI, Penfield S, Smith C, Catley M, Bevan M. 2003. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. The Plant Journal 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Carpita NC. 2011. Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1→4)-β-d-glycans. Plant Physiology 155: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai G, Kong Y, Zhu M, et al. . 2015. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. Journal of Experimental Botany 66: 2595–2609. [DOI] [PubMed] [Google Scholar]
- Chen J, Doyle C, Qi X, Zheng H. 2012. The endoplasmic reticulum: a social network in plant cells. Journal of Integrative Plant Biology 54: 840–850. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin A, Laurans F, Arnaud D, Breton C, Pilate G, Leplé JC. 2010. Wood formation in Angiosperms. Comptes Rendus Biologies 333: 325–334. [DOI] [PubMed] [Google Scholar]
- Denness L, McKenna JF, Segonzac C, et al. . 2011. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiology 156: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, et al. . 2002. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiology 128: 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano GP, Renna L, Piro G, Dalessandro G. 2004. Stubborn GFPs in Nicotiana tabacum vacuoles. Plant Biosystems 138: 37–42. [Google Scholar]
- Ellis C, Turner JG. 2001. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. 2002. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, et al. . 2015. A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, et al. . 2000. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler DA, Sampathkumar A, Mutwil M, Persson S. 2008. Laying down the bricks: logistic aspects of cell wall biosynthesis. Current Opinion in Plant Biology 11: 647–652. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. 1994. Biosynthesis of plant cell wall polysaccharides. The FASEB Journal 8: 904–915. [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, et al. . 2005. Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PB. 1962. Mechanism for plant cellular morphogenesis. Science 138: 1404–1405. [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. 2009. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nature Cell Biology 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. 1995. Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebler C, Barry JL, David A, Delort-Laval J. 1989. Rapid acid hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. Journal of Agricultural and Food Chemistry 37: 360–367. [Google Scholar]
- Höfgen R, Willmitzer L. 1988. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kato H, Motomura T, Komeda Y, Saito T, Kato A. 2010. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. Journal of Plant Physiology 167: 571–577. [DOI] [PubMed] [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kang H, Kim J, Han KH. 2014. AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mRNAs in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. The Plant Journal 80: 772–784. [DOI] [PubMed] [Google Scholar]
- Ko JH, Beers EP, Han KH. 2006a Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Molecular Genetics and Genomics 276: 517–531. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim JH, Jayanty SS, Howe GA, Han KH. 2006b Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. Journal of Experimental Botany 57: 2923–2936. [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, et al. . 2001. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiology 126: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Li S, Bashline L, Gu Y. 2014. Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Frontiers in Plant Science 5: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y. 2012. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proceedings of the National Academy of Sciences USA 109: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Chan J. 2006. Not so divided: the common basis of plant and animal cell division. Nature Reviews Molecular Cell Biology 7: 147–152. [DOI] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, et al. . 2004. Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. The Plant Journal 40: 260–275. [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Döring A, Persson S. 2014. The cell biology of cellulose synthesis. Annual Review of Plant Biology 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Nguyen VP, Cho JS, Choi YI, Lee SW, Han KH, Ko JH. 2016. Evaluation of a novel promoter from Populus trichocarpa for mature xylem tissue specific gene delivery. Plant Physiology and Biochemistry 104: 226–233. [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. 1998. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. The EMBO Journal 17: 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, et al. . 2005. Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17: 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, et al. . 2002. KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. 2004. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends in Plant Science 9: 534–540. [DOI] [PubMed] [Google Scholar]
- Roudier F, Schindelman G, DeSalle R, Benfey PN. 2002. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiology 130: 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, et al. . 2005. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, et al. . 2001. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes & Development 15: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B, Choi G, Yi H, et al. . 2002. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. The Plant Journal for Cell and Molecular Biology 30: 23–32. [DOI] [PubMed] [Google Scholar]
- Somerville C. 2006. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology 22: 53–78. [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, et al. . 2004. Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Jackson MR. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annual Review of Cell and Developmental Biology 12: 27–54. [DOI] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T. 2000. Rearrangement of cortical microtubules from transverse to oblique or longitudinal in living cells of transgenic Arabidopsis thaliana. Protoplasma 213: 28–38. [Google Scholar]
- Wang J, Howles PA, Cork AH, Birch RJ, Williamson RE. 2006. Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiology 142: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H. 2012. Growth control and cell wall signaling in plants. Annual Review of Plant Biology 63: 381–407. [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation. Plant Molecular Biology 40: 711–717. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nikolovski N, Sorieul M, et al. . 2016. Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nature Communication 7: 11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.