Abstract
“Musica universalis” is an ancient philosophical concept claiming the movements of celestial bodies follow mathematical equations and resonate to produce an inaudible harmony of music, and the harmonious sounds that humans make were an approximation of this larger harmony of the universe. Besides music, electromagnetic waves such as light and electric signals also are presented as harmonic resonances. Despite the seemingly universal theme of harmonic resonance in various disciplines, it was not until recently that the same harmonic resonance was discovered also to exist in biological systems. Contrary to traditional belief that a biological system is either at stead-state or cycles with a single frequency, it is now appreciated that most biological systems have no homeostatic “set point,” but rather oscillate as composite rhythms consisting of superimposed oscillations. These oscillations often cycle at different harmonics of the circadian rhythm, and among these, the ~12-hour oscillation is most prevalent. In this review, we focus on these 12-hour oscillations, with special attention to their evolutionary origin, regulation, and functions in mammals, as well as their relationship to the circadian rhythm. We further discuss the potential roles of the 12-hour clock in regulating hepatic steatosis, aging, and the possibility of 12-hour clock–based chronotherapy. Finally, we posit that biological rhythms are also musica universalis: whereas the circadian rhythm is synchronized to the 24-hour light/dark cycle coinciding with the Earth’s rotation, the mammalian 12-hour clock may have evolved from the circatidal clock, which is entrained by the 12-hour tidal cues orchestrated by the moon.
Keywords: 12h-clock, aging, chronotherapy, ER stress, mitochondria, NAFLD, Xbp1
We focus on 12-hour oscillations, with special attention to their evolutionary origin, regulation, and functions in mammals, as well as their relationship to the circadian rhythm.
1. The Concept of Harmonics in Physics and Their Biological Counterpart
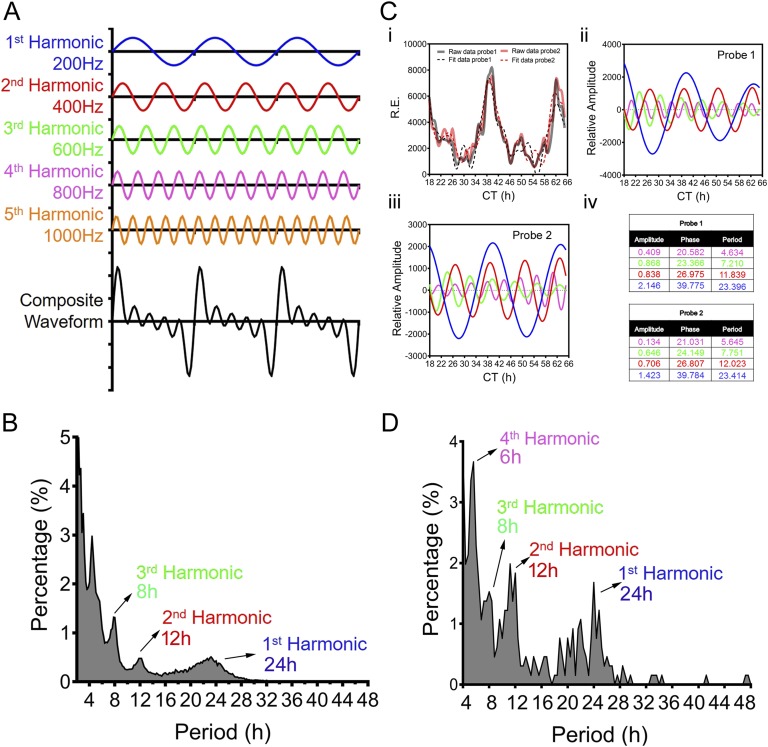
The term “harmonic” originates in musical instrument theory, where the wavelengths of the overtones of a vibrating string are derived from its fundamental wavelength. A harmonic of such a wave has a frequency that is a positive integer multiple of this fundamental frequency, which is also called the first harmonic [1, 2]. For instance, as shown in Fig. 1A, the fundamental, or the first, harmonic is a wave with a frequency of 200 Hz, whereas the second harmonic is 400 Hz (200 Hz × 2 = 400 Hz), and so on. Summing the harmonics generates a composite waveform that looks “distorted and noisy” (Fig. 1A, bottom) but is actually much closer to perception of music than the individual harmonics. This is because pure oscillation with a single frequency rarely exists in nature. Rather, physical rhythms often present as superpositions of basic waves such as the harmonic resonances found in music, light, and Keplers Law of planetary motion.
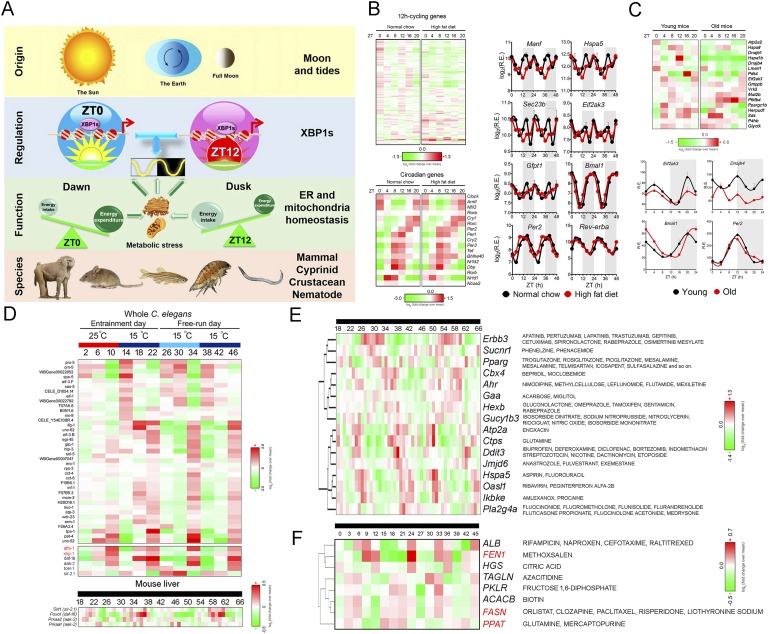
Figure 1.
The concept of harmonics in physics and their biological counterpart. (A) Diagram showing the individual harmonics (200 Hz, 400 Hz, 600 Hz, 800 Hz, and 1000 Hz) as well as the composite waveform resulting from adding up all of the harmonics. (B) Distribution of periods of all oscillations identified from the hepatic gene expression microarray dataset [8] via the eigenvalue/pencil approach. The vast majority of oscillations cycle at the first (24-hour), second (12-hour), or third (8-hour) harmonic of the circadian rhythm (24 hours). (C) Representative deconvolution of Gck gene mRNA expression by the eigenvalue/pencil method. Gck expressions detected by two different probe sets are analyzed by the eigenvalue/pencil method. (i) Raw microarray data (solid line) and model fit (dashed line) for Gck hepatic expression as reported in Ref. [8]. Superimposed harmonic oscillations revealed by the eigenvalue/pencil method for probe 1 (ii) and probe 2 (iii) are shown. (iv) Amplitudes, phases, and periods of different oscillations for the two probes with the color matching the different oscillations depicted in (ii) and (iii). (D) Distribution of periods of all oscillations identified from the hepatic metabolites dataset [11] via the eigenvalue/pencil approach. The vast majority of oscillations cycle at the first (24-hour), second (12-hour), third (8-hour), or fourth (6-hour) harmonic of the circadian rhythm (24 hours).
To uncover all superimposed oscillations in an unbiased manner, we used a recently developed eigenvalue/pencil method [3, 4–6] and disclosed that just as for other forms of physical oscillations, biological oscillations also are composite rhythms consisting of oscillations of various frequencies [3]. The eigenvalue/pencil method assumes that any oscillation can be approximated by a linear combination of exponentials (sine waves with a decay factor) plus noise and therefore can be applied to any situations where such assumptions are valid. Furthermore, unlike most of the current cycling transcript identification methodologies that require the user to define a narrow period range [7], the eigenvalue/pencil method does not preassign a period and thus permits the identification of all superimposed oscillations in an unbiased manner [3] (for a more detailed understanding of the strengths and limits of the eigenvalue/pencil method and its wider applications, please refer to Ref. [4]). Oscillations disclosed by this analysis often cycle at the harmonics of the circadian rhythm [3]. For instance, comprehensive analysis of a published mouse liver microarray dataset employing the eigenvalue/pencil method (where liver samples were taken every hour for a total of 48 hours under constant darkness condition) revealed that most oscillations cycle at periods close to 24 hours, 12 hours, 8 hours, and 4 hours, corresponding to the first, second, third, and sixth harmonics of the circadian rhythm (Fig. 1B) [3, 8]. Whereas the ~4-hour oscillations are near the Nyquist limit of sampling frequency and therefore may arise from technical artifacts [9], oscillations cycling with 12-hour and 8-hour periods are thought to be “real” biological oscillations [3]. One example is glucokinase (Gck), which encodes for a rate-limiting enzyme involved in both glycogenesis and glycolysis. Whereas Gck mRNA expression was originally thought to be solely under the circadian clock control [10], the new analysis revealed that it actually is comprised of superimposed major oscillations cycling at 24-hour, 12-hour, and 8-hour periods with comparable amplitudes (Fig. 1C) [3].
Similar to gene expression oscillations, mammalian metabolic rhythms also are composite oscillations cycling at different harmonics of the circadian rhythm. Analyzing a 1-hour resolution mouse hepatic metabolites profiling dataset [11] revealed that most metabolites consist of superimposed oscillations cycling at 24-hour, 12-hour, 8-hour, and 6-hour periods (Fig. 1D). This is consistent with the observed 24-hour, 12-hour, and 8-hour harmonic oscillations of the respiratory exchange ratio through indirect calorimetry measurement of energy expenditure for mice fed ad libitum [3]. Taken together, these unbiased analyses strongly indicate that as proposed thousands of years ago, harmonic resonance is a common “universal” theme that permeates from the physical world all the way to biological systems.
2. The Circatidal Rhythm: The Best Characterized ~12-Hour Cycling Oscillation
The wide prevalence of 12-hour gene expression and metabolic rhythms observed in mouse liver is intriguing. In fact, >20% of all mouse hepatic mRNA and metabolites cycle with an ~12-hour period, of which 4.2% and 11.5%, respectively, are dominant oscillations (~12-hour cycling amplitudes are the largest among all superimposed oscillations) [3]. The prevalent mammalian 12-hour rhythm of gene expression and metabolism is reminiscent of the ~12-hour circatidal rhythms of coastal and estuarine animals that modulate their behavior in tune to the ~12.4-hour ebb and flow of the tides [12]. (For the search related to circatidal rhythms in coastal and estuarine animals, the key words of “circatidal” and/or “tidal rhythms” were used and literatures between 1950 and the present were reviewed; For the 12-hour rhythms in mice, owing to the very limited number of publications on this topic, the authors are very acquainted with the handful of literatures and therefore no search was performed.) Prominent examples include numerous crustaceans such as the fiddler crab (Uca pugnax) [13, 14], mudflat crab (Chiromantes hematocheir) [15], green shore crab (Carcinus maenas) [16, 17], sea louse (Eurydice pulchra) [12, 18, 19], Dimorphostylis asiatica [20], Chelicerata horseshoe crab (Limulus polyphemus) [21], ragworm (Alitta virens) [22], Mollusca [23–26], and intertidal insects such as the Asian mangrove cricket (Apteronemobius asahinai) [27, 28]. These animals either emerge at low tides to forage, mate, and fight (such as U. pugnax and A. asahinai) or are more active during high tides when the incoming tide covers their forage and mating grounds (such as C. maenas and E. pulchra) [12]. More importantly, when removed to the laboratory and divorced from tidal cues, they can still maintain their circatidal locomotive activities at times of expected low water or high water under constant conditions [12–14, 16, 23, 27, 28]. Furthermore, their circatidal rhythms also can be entrained by artificial vibrations or inundation cycles [19, 23, 27]. This free-running behavior combined with entrainability indicates the presence of circatidal clocks in these animals, which under natural conditions are synchronized to the phase of the tidal cycle encountered on their home environment [12].
So how are these circatidal rhythms generated? Two mechanisms underlying tidal rhythms have been proposed. Some argue that a circatidal clock might be generated by two slightly longer (24.8-hour) circadian clocks acting antiphase to produce 12.4-hour peaks in behaviors such as swimming and foraging [29]. The second school of thought advocates that a dedicated circatidal clock with a period of ~12.4 hours is the basis for circatidal behavior. Superimposed on this tidal clock is a circadian oscillator that drives the day/night modulation observed in locomotor output of crustaceans and crickets [30]. One of the most convincing studies supporting the latter hypothesis was published in Current Biology in 2013 [19]. In this elegant study, Zhang et al. [19] used two orthogonal approaches (RNA interference against circadian clock genes and bright constant light to disrupt the circadian clock) to abolish the circadian clock in E. pulchra and tested their effects on the circatidal swimming rhythms of E. pulchra. Although both approaches disrupted the circadian clock as expected, neither influenced the tidal periodicity [19]. Additional studies performed on the American horseshoe crab Limulus polyphemes and mangrove cricket (A. asahinai) found similar results [30–33]. Collectively, these studies convincingly demonstrate the existence of a dedicated circatidal pacemaker in marine animals that is distinct from the circadian clock and responsible for the establishment of circatidal rhythms.
3. The Prevalence of Mammalian 12-Hour Rhythms
The first study that reported the presence of circadian harmonics of gene expression in mammalian systems, including 12-hour rhythms, was published by John Hogenesch’s group [8]. By comprehensively profiling temporal gene expression in mouse liver from animals maintained at constant darkness at high resolution (at 1-hour intervals for a total of 48 hours) using DNA microarray, the authors identified ~200 genes that cycle with a dominant 12-hour period using two statistical methods: a Fisher’s G-test and COSOPT [8]. Ingenuity pathway analysis of these 12-hour cycling hepatic genes revealed enriched gene ontology terms, including protein processing and endoplasmic reticulum (ER) homeostasis [8]. Contrary to the evenly distributed phases (the time of peak) of circadian genes, the acrophases of 12-hour cycling hepatic genes are more restricted to the dawn [circadian time (CT)0] and dusk (CT12) of a diurnal cycle, even though the underlying reason was then unknown [8] (By convention, the onset of activity of diurnal organisms defines CT0, whereas the onset of activity of nocturnal organisms defines CT12. Therefore, for nocturnal animals such as mice, they are active from CT12 to CT24). In addition to the liver, they also detected 12-hour cycling transcripts in many different tissues. For example, Hspa1b, a heat shock protein 70-kDa family member, showed clear 12-hour transcriptional rhythms in lung, kidney, adrenal gland, heart, and even in the hypothalamus of the central nervous system, indicating that the presence of circadian harmonics is not restricted to the liver [8]. Additionally, in all of these tissues, the phases of peak Hspa1b expression are nearly identical, again at dawn and dusk. Since then, a number of studies have confirmed the presence of these 12-hour rhythms of gene expression, especially in mouse liver, using different cycling transcript identification methods [34–37], even though the total repertoire of identified 12-hour cycling transcripts remained small.
We reasoned that the ~200 originally identified 12-hour cycling transcripts were a significant underestimation of the true prevalence of mammalian 12-hour rhythms of gene expression. Because the COSOPT/Fisher’s G-test methods used to identify 12-hour cycling transcripts in the original study require the user to preassign a period range, it favors the detection of genes with dominant 12-hour rhythms and is thereby biased against other 12-hour cycling transcripts having superimposed oscillations with larger amplitudes. As expected, when applying the eigenvalue/pencil method on the same mouse microarray dataset, we identified >3000 hepatic genes (~20% of all hepatic transcripts) with 12-hour rhythms [3]. As expected, most of them have superimposed 24-hour circadian rhythms and therefore evade detection by traditional methods. Among all ~3000 genes, 760 of them are dominant 12-hour cycling genes [3]. Gene ontology analysis of either all or dominant 12-hour cycling genes revealed that, in addition to the expected ER homeostasis and protein quality control pathways, various metabolism-related biological pathways are also highly enriched [3]. Whereas ER-related genes (such as Eif2ak3, Gfpt1, and Sec23b) have dominant 12-hour oscillations and lack superimposed circadian rhythms, 12-hour cycling metabolism genes often have superimposed circadian rhythms (including Gck, Acly, and Fasn) [3].
In addition to the widespread 12-hour cycling mature mRNAs in mouse liver, prevalent 12-hour cycling hepatic protein (~15% of hepatic proteome) also was observed from a time series mass spectrometry dataset [3, 38]. A limited ~35% overlap with 12-hour cycling mRNAs indicates that both transcriptional and posttranscriptional controls contribute to the regulation of 12-hour rhythms of hepatic gene expression [3]. Gene ontology analysis of 12-hour cycling proteins revealed strong enrichment of ER and, intriguingly, mitochondria-associated metabolism pathways, implicating potential coordinated actions of ER and mitochondria in maintaining systemic metabolic homeostasis and stress response [3]. Unlike mature mRNA, the phases of 12-hour protein oscillations are distributed evenly throughout the diurnal cycle, and a clear phase separation of proteins involved in opposing metabolic pathways was found [3].
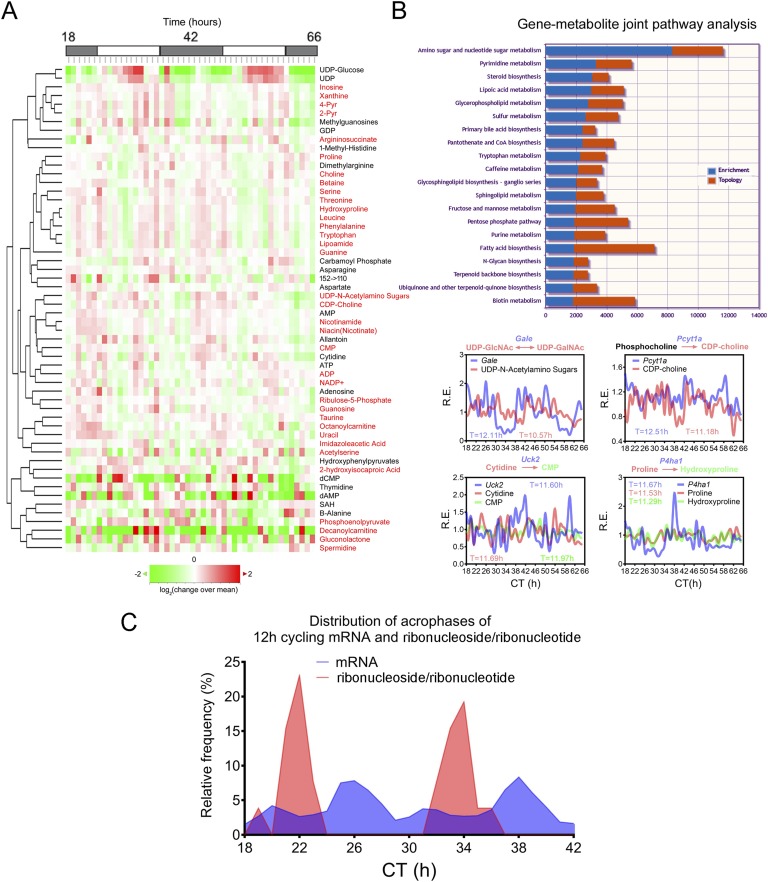
The prevalent hepatic 12-hour cycling mRNA and proteins involved in metabolic regulation predict a 12-hour rhythm for hepatic metabolites, which is confirmed by post hoc analysis of two recently published metabolomics datasets [39, 40]. Eigenvalue/pencil analysis revealed that >20% of hepatic metabolites profiled in these two studies exhibited 12-hour oscillations [3]. Furthermore, joint pathway analysis using MetaboAnalyst [41, 42] showed coordinated 12-hour rhythms of metabolites and gene expressions (Fig. 2A and 2B) [3]. For instance, top-enriched KEGG pathways exhibiting 12-hour rhythms of paired gene expression and metabolites include amino sugar and nucleotide sugar metabolism (Gfpt1/Gale: UDP-glucose/UDP-N-acetylamino sugars), pyrimidine metabolism (Uck2: cytidine/CMP), purine metabolism (Ppat/Gart/Atic/Adsl/Ampd2/Nt5e/Gmps: inosine/adenosine/AMP/ADP/ATP/xanthine/guanosine/guanine/GDP), polyamine metabolism (Srm/spermidine), glycerophospholipid metabolism (Pcyt1a: CDP-choline; Lpcat3: 2-Lyso-PC), sphingolipid metabolism (Sphk2: sphingosine), pentose phosphate pathway (Rbks: ribose), fatty acid biosynthesis (Fasn/Elovl6: palmitoleate/vaccinate), and amino acid metabolism (P4ha1: proline/hydroxyproline), among many others (Fig. 2A and AB) [3].
Figure 2.
Coupled 12-hour rhythms of hepatic gene expression and metabolism. (A) Heat map of mouse 12-hour cycling hepatic metabolites identified by the eigenvalue/pencil method from published metabolomic dataset [11] after hierarchical clustering. Metabolites with dominant 12-hour oscillations are highlighted in red. (B) (Top) Gene-metabolite joint pathway analysis using MetaboAnalyst [41, 42] reveals top enriched biological pathways. Both enrichment as well as topology scores are shown for each pathway. (Bottom) Representative data showing paired gene expression and metabolite oscillations. (C) Distribution of acrophases of all dominant hepatic 12-hour cycling mRNA and ribonuclesides/ribonucleotides.
These highly coupled 12-hour rhythms of hepatic metabolic gene expressions and metabolite oscillations strongly imply the presence of an endogenous 12-hour clock component that is responsible for the precisely timed orchestration of metabolic flux by temporally regulating the expressions of metabolic enzymes. This hypothesis was confirmed experimentally and is discussed in greater detail later. Of particular interest is the prevalent 12-hour hepatic ribonucleoside and ribonucleotide oscillations, which are building blocks for RNA synthesis (Fig. 2C). The acrophases of ribonucleoside and ribonucleotide oscillations are restricted at CT22 and CT34, preceding the two peaks of 12-hour mRNA transcription at CT26 and CT38, implying coordinated purine/pyrimidine metabolism and 12-hour mRNA transcription [3] (Fig. 2C). The 12-hour oscillations of metabolism are not restricted to liver tissues. Indirect calorimetry measurement of real-time mouse energy expenditure revealed the presence of 12-hour harmonics in respiratory exchange ratio oscillations [3], supporting the hypothesis that other peripheral tissues also possess a 12-hour clock that perhaps helps to synchronize 12-hour systemic metabolic homeostasis.
Although the prevalence of 12-hour rhythms of gene expression and metabolism in mice is undisputable, owing to the technical limitations and ethical constraint of obtaining time series tissue samples from humans, the evidence supporting the existence of 12-hour rhythms in human tissues in vivo is still largely anecdotal at this time. (For reported ~12-hour rhythms in humans, the key words of “circasemidian” and/or “12-hour rhythm” were searched against literatures published between 1950 and the present.) Nonetheless, in humans, using traditional spectrum analysis methods such as Fourier transform and wavelet analysis, rhythms of body temperature [43–48], blood pressure and heart rate variability [49–54], cognitive performance [48, 55–61], migraine onsets and subsequent cerebrospinal fluid sodium level [62], circulating serum and urine metal levels [63], circulating hormone levels [64–66], and sleep patterns [57, 58, 67–74] were all reported to exhibit a 12-hour rhythmic component.
Of these reported 12-hour rhythms in humans, daily variation of temperature and sleep–wake cycles are two of the most interesting. As discussed later, temperature is a strong Zeitgeber (a German word for time giver, which refers to a synchronizing agent) for 12-hour rhythms in C. elegans [3, 75]. Although it remains unknown whether temperature also can entrain the mammalian 12-hour clock, it is tempting to hypothesize that the 12-hour rhythm of mammalian gene expressions and metabolic rhythms are also in tune to the daily temperature oscillations, similar to what has been observed for the circadian clock [76–78]. As to the 12-hour rhythms of sleep–wake patterns, in addition to the well-characterized peak of sleepiness during the subjective night, a second smaller peak of sleepiness during the early afternoon is a widespread phenomenon (also known as “siesta” in Spanish-speaking countries). Moreover, because the afternoon sleep propensity occurred either under a constant routine [69, 79] or without having lunch [80], it is highly suggested that it is a part of an endogenous biological rhythm. Currently, sleep regulation is conceptualized by the popular “two-process” model that posits that homoeostatic and circadian drives control sleep [81]. The homoeostatic process is controlled by sleep pressure, which accumulates during the course of wakefulness and dissipates during sleep, and is tightly regulated by local changing levels of neuromodulators such as adenosine [82–84]. Alternatively, the sleep–wake cycle during the day and night is regulated by the circadian clock, independent of prior sleep or wake time [81]. Whereas it has long been thought that the afternoon sleepiness is under the homeostatic sleep control, several studies challenged this notion and suggested that the afternoon sleep propensity may instead reflect an endogenous 12-hour cycle of slow wave sleep (which is also referred to as nonrapid eye movement sleep or in more colloquial terms “deep sleep”), independent of duration of prior wakefulness [61, 69, 74]. Although it has been convincingly demonstrated that slow wave sleep can be strongly induced by local increased levels of adenosine in the basal forebrain through its adenosine A1 receptor and repressed by its antagonist caffeine [82, 85], the mechanism of how the concentration of adenosine is regulated in the brain remains elusive. Intriguingly, adenosine and three of its main precursors (ATP, AMP, and SAH) all exhibit robust 12-hour rhythms in mouse liver (Fig. 2A and 2B). Furthermore, xanthine, a close relative of caffeine (which is a methylxanthine) also revealed a 12-hour rhythm (Fig. 2A). Although it is premature to conclude that local intracellular and extracellular levels of adenosine and xanthine derivatives may also exhibit 12-hour rhythms in the central nervous system and is coupled to the 12-hour rhythms of slow wave activity cycle, these intriguing observations and correlations warrant future studies to either prove or refute this hypothesis.
4. Evidence Supporting the Existence of a Cell-Autonomous Mammalian 12-Hour Clock
Two pertinent questions arise regarding the mammalian 12-hour rhythms. First, are the 12-hour oscillations of gene expression and metabolism cell-autonomous? Second, are 12-hour rhythms established by a dedicated mammalian 12-hour clock or are they regulated by some variants of the well-characterized circadian clock? Theoretically, at least three possibilities exist. In the first scenario, the 12-hour rhythms are not cell-autonomous in nature. It is possible that one of the two peaks is established by the circadian clock and the second peak results from physiology-associated systemic cues only found in vivo. In the second scenario, the 12-hour rhythm is cell-autonomous. However, it is established by two circadian transcription activators or repressors appearing in antiphase and therefore is dependent on the circadian clock [36]. In the third scenario, the mammalian 12-hour rhythms are not only cell-autonomous, but also are established by a dedicated 12-hour clock “separate” from the circadian clock, a mechanism similar to the independent circatidal clock in certain crustaceans and intertidal insects [19, 32, 33].
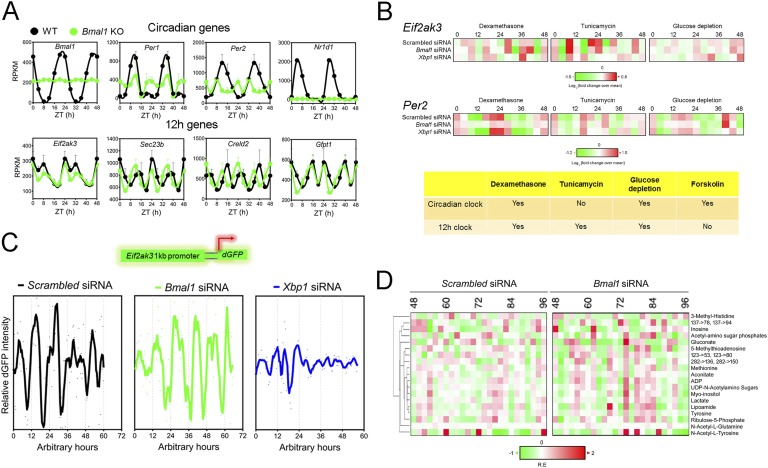
Early studies favor the first hypothesis (the 12-hour rhythms are not cell-autonomous and are established by the combined effects of circadian clock and fasting–feeding cues) due to the following three major observations. (1) No statistically significant 12-hour rhythms of gene expression were detected in forskolin-synchronized NIH3T3 cells, implying that mammalian 12-hour rhythms are not cell-autonomous (Fig. 3B) [8]. (2) Eight-hour daytime-restricted feeding substantially impairs the 12-hour rhythms of several hepatic gene expressions. These genes still maintained peak expression at approximately CT26, coinciding with the new feeding time; however, the subjective evening peak was largely absent. This evidence suggests that at least one component of the 12-hour rhythm is driven by feeding [8]. (3) Brain-specific rescue of ClockΔ19 mutant mice converted hepatic 12-hour rhythms into 24-hour rhythms, suggesting that signaling via the central circadian oscillator may be required to generate one of the two daily peaks of expression [37]. Whereas these results could suggest the lack of a cell-autonomous mammalian 12-hour clock, in light of strong new evidence supporting the existence of a cell-autonomous mammalian 12-hour clock [3], alternative interpretations of these data are offered and will be elaborated on in greater detail below.
Figure 3.
Twelve-hour rhythms of gene expression and metabolism are cell-autonomous, established by a dedicated 12-hour clock and evolutionarily conserved. (A) Representative reads per kilobase of transcript per million mapped reads (RPKM) of normalized hepatic circadian (top) as well as 12-hour cycling (bottom) gene expression from wild-type (WT) and conventional BMAL1 knockout (KO) mice under 12-hour/12-hour light/dark conditions as reported in Ref. [86]. Data are graphed as the mean ± SEM (n = 4) and double plotted for better visualization. (B) Heat map representation of oscillations of Eif2ak3 and Per2 mRNA level after dexamethasone, tunicamycin, or glucose depletion shock treatment in MEFs. The heat map is derived from quantitative PCR data reported in Ref. [3]. A summary of the conclusion is shown in the table below. (C) Representative recordings of single-cell time lapse microscopy analysis of Eif2ak3 promoter-driven dGFP oscillation in scrambled siRNA, Bmal1 siRNA, or Xbp1 siRNA transfected MEFs. (D) Heat map of 12-hour cycling metabolites in dexamethasone-synchronized human U2OS cells under both scrambled siRNA and Bmal1 siRNA transfection conditions compiled from Ref. [11]. Twelve-hour cycling metabolites are identified by the eigenvalue/pencil method [3].
An initial clue hinting at the possible existence of an independent mammalian 12-hour clock comes from the observed mathematical orthogonal relationship between superimposed 12-hour and circadian oscillations uncovered for most of the 12-hour cycling ER homeostasis and metabolism genes [3]. Subsequent post hoc analysis of hepatic RNA sequencing data in wild-type and whole-body BMAL1 knockout mice fed ad libitum (both conventional and adult BMAL1 deletion mice [86]) confirmed the independence of 12-hour rhythms of key ER and metabolism gene expressions from the circadian clock [3]. Specifically, for 12-hour cycling genes lacking superimposed circadian rhythms (such as Eif2ak3, Gfpt1, Creld2, and Sec23b), their 12-hour rhythms are almost identical between wild-type and BMAL1 knockout mice under either a 12-hour/12-hour light/dark schedule or a constant darkness condition [3, 86] (Fig. 3A). For 12-hour cycling genes with superimposed circadian rhythms (such as Gck, Fasn, Dnajb4, and Chka), more perceptible 12-hour rhythms were observed in BMAL1 knockout mice [3, 86]. The independent relationship between 12-hour and 24-hour rhythms for these ER homeostasis and metabolism genes also was verified in ClockΔ19 mutant mice [3, 87].
Can the 12-hour rhythm of gene expression be found in vitro or does its establishment require systemic cues only found in vivo as previously suggested [8, 35]? In light of the newly found independence of 12-hour cycling genes from the circadian clock, an alternative interpretation of the lack of observed 12-hour rhythms in forskolin-synchronzied NIH3T3 cells [8] is that forskolin is capable only of synchronizing the circadian clock but not the circadian clock-independent 12-hour clock. Consistent with this hypothesis, it was found that the circadian and 12-hour cycling genes are responding to a different repertoire of external cues (Fig. 3B) [3]. Whereas the cAMP inducer forskolin is only a strong synchronizer of the mammalian circadian clock [8], the ER stress inducer tunicamycin (which blocks UDP-GlcNAc–mediated UDP-N–linked glycosylation) can only synchronize the 12-hour clock [3] (Fig. 3B). This is consistent with observed dominant circadian and 12-hour rhythms of cAMP and UDP-N-acetylamino sugars levels, respectively, in mouse liver [11] (Fig. 2B). Contrary to the discriminatory nature of forskolin and tunicamycin, dexamethasone and glucose depletion can synchronize both the circadian and 12-hour clocks [3] (Fig. 3B). More importantly, whereas siRNA-mediated depletion of Bmal1 abolishes dexamethasone and glucose depletion–synchronized circadian Per2 expression, it does not affect dexamethasone, glucose depletion, or tunicamycin-synchronized 12-hour rhythms of Eif2ak3 expression in mouse embryonic fibroblasts (MEFs) [3] (Fig. 3B). Moreover, by cloning the Eif2ak3 promoter before a destabilized green fluorescence protein (dGFP) and performing time-lapse imaging of dGFP intensity in single cells without any prior perturbation, robust 12-hour oscillations were observed in MEFs [3], which also was not affected by Bmal1 siRNA knockdown (Fig. 3C) [3]. Lastly, 37 metabolites of the 137 examined also exhibited ~12-hour rhythms in dexamethasone-entrained human U2OS cells and 19 of them still maintained ~12-hour rhythms in the presence of Bmal1 knockdown [11] (Fig. 3D). Taken together, these observations strongly support the hypothesis that the 12-hour rhythm of gene expression, especially those involved in ER/metabolic pathways, and subsequent 12-hour rhythms of metabolism, are cell-autonomous and driven by a dedicated 12-hour pacemaker “distinct” from the circadian clock in mammalian cells [3].
Although the 12-hour rhythms of gene expression and metabolism are cell-autonomous, they can be disrupted by altered fasting–feeding cycles. Daytime-restricted feeding negatively influences 12-hour rhythms of hepatic gene expression [8]. Consistent with this finding, daytime-restricted feeding also substantially impairs 12-hour oscillations of respiratory exchange ratio in the mouse [3]. Thus, one also can envision how the circadian and 12-hour clocks may interact indirectly. For instance, manipulation/perturbation of the central circadian clock in the brain can lead to altered feeding–fasting and wake–sleep behaviors, which in turn may feed back to the 12-hour clock to alter 12-hour rhythms of gene expression and metabolism in peripheral tissues. In fact, this may very well account for the observed phase shift in several 12-hour cycling hepatic genes as well as the varied 12-hour cycling transcriptome reported in different circadian deficient mice models [3, 35, 86, 87]. Additionally, because many of the 12-hour cycling genes have superimposed circadian rhythms, alteration of the circadian clock can have a major influence on the overall gene oscillations, without necessarily affecting the superimposed 12-hour rhythms. This could be the case for the observed conversion of certain hepatic 12-hour rhythms into 24-hour rhythms upon brain-specific rescue of ClockΔ19 mutant mice [37].
Whereas these new data support the existence of a cell-autonomous mammalian 12-hour clock, they do not rule out the possibility that certain 12-hour rhythms are influenced by the circadian clock and/or the effects of certain external cues and are not cell-autonomous. Future studies are needed to profile the complete 12-hour transcriptome under the 12-hour clock control, as well as to investigate the physiologic conditions by which these distinct clocks can interact systemically in multiple model organisms.
5. The Spliced Form of XBP1 Transcriptionally Regulates the 12-Hour Clock
How is the mammalian 12-hour clock regulated? Observed prevalent 12-hour rhythms of hepatic nascent mRNA and enhancer RNA expression indicate that the mammalian 12-hour clock is regulated primarily at the transcriptional level [3, 88]. Consistent with the strong enrichment of ER stress and unfolded protein response (URP) pathways in 12-hour cycling hepatic transcriptome (for a review of mammalian UPR, see Refs. [89–91]), 12-hour cycling of UPR transcription factor spliced XBP1 (XBP1s) and ATF4 was found in mouse liver under both a 12-hour/12-hour light/dark schedule and constant darkness conditions with peak expressions found at CT10 to CT12 and CT22 to CT24, consistent with the acrophases of 12-hour cycling hepatic transcriptome [3, 35]. Furthermore, 12-hour cycling of XBP1s expression was found to be induced in tunicamycin-synchronized MEFs and is independent from the circadian clock [3]. Intriguingly, both the total level of Xbp1 mRNA (the combined level of Xbp1s and Xbp1us) and its splicing efficiency exhibited robust 12-hour rhythms both in mouse liver in vivo and in MEFs in vitro [3, 35]. The 12-hour rhythms of Xbp1 splicing efficiency and XBP1s expression in vivo is further supported by the observed 12-hour rhythms of its upstream endoribonuclease IRE1α phosphorylation at Ser724 [35].
Twelve-hour rhythms of nuclear XBP1s levels correlate with the 12-hour rhythms of chromatin recruitment of XBP1s to the promoters of several 12-hour cycling genes both in mouse liver in vivo and in tunicamycin-synchronized MEFs in vitro [3]. Knockdown of Xbp1 inhibits both tunicamycin and glucose depletion–synchronized 12-hour rhythms of ER homeostasis (such as Eif2ak3, Ddit3, Sec23b, and Herpud1) and metabolism (such as Gfpt1 and Acly) gene expression in MEFs [3] (Fig. 3B). Additionally, knockdown of Xbp1 abolished cell-autonomous 12-hour rhythms of Eif2ak3 promoter–driven dGFP intensity in a single MEF cell (Fig. 3C). In contrast, ablation of Xbp1 has no effects on dexamethasone and glucose depletion–synchronized circadian gene expression [3] (Fig. 3B). The transcriptional regulation of mammalian 12-hour transcriptome by XBP1s is further consistent with numerous past reports on the transcriptional regulation of the same repertoire of genes by XBP1s during pathological conditions of ER stress [92–95]. This evidence suggests that similar to the circadian clock, the mammalian 12-hour clock is also subject to major transcriptional control. Note that so far, the evidence supporting the role of Xbp1s in the regulation of the mammalian 12-hour clock is solely restricted to the study of a few 12-hour rhythmic genes in MEFs in vitro, and future studies using XBP1s knockout mice models are needed to comprehensively profile the XBP1s-regulated 12-hour transcriptome in an unbiased manner.
6. The Mammalian 12-Hour Clock Is Evolutionarily Conserved and May Be Circatidal in Origin
From where does the mammalian 12-hour clock originate? Given the distinctions of the circadian clock from the 12-hour clock/circatidal clock in both mouse and E. pulchra [3, 19] and the fact that mammals share common marine ancestors, it is logical to postulate that the mammalian 12-hour clock evolved from the ancient circatidal clock. Previous work revealed robust 12-hour circatidal mRNA rhythms for 10 mitochondrial DNA (mtDNA)–encoded protein-coding genes (Mt-Nd1~6, Mt-Cox1~3, and Cytb) that encode mitochondrial components of complexes I (NADH dehydrogenase) and IV (cytochrome c oxidase) in E. pulchra under free-running conditions, which are correlated with circatidal rhythms of oxygen consumption in the same animals [18, 96]. Post hoc analysis of two published mouse hepatic Gro-Seq [88] and Nascent-Seq [97] datasets that measure nascent RNA transcription rate revealed, surprisingly, conserved 12-hour rhythms of mtDNA-encoded gene transcription [3] These 12-hour rhythms of mtDNA-encoded gene expression were further confirmed at mature mRNA and protein levels in mouse liver [3, 38, 40]. Furthermore, cell-autonomous 12-hour rhythms of mtDNA-encoded gene expression were found in glucose depletion–synchronized MEFs in a BMAL1-independent manner [3]. Also supporting the hypothesis that the mammalian 12-hour clock evolves from a circatidal origin is the observed similar free-running periodicity of Eif2ak3 promoter–driven dGFP oscillation in MEFs (12.6 hours) with that of circatidal activity rhythms reported in E. pulchra after tidal cue entrainment (12.7 hours) and in the mangrove cricket, A. asahinai (12.6 hours to 12.9 hours) [19, 32, 33].
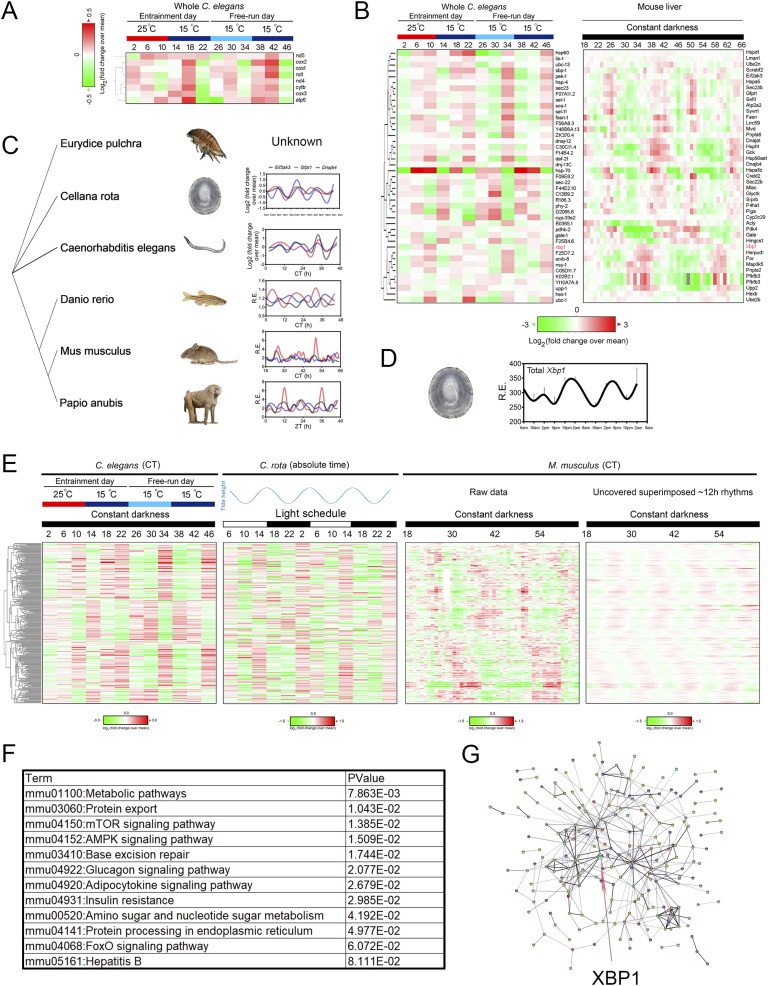
In addition to crustaceans and mammals, strikingly, mtDNA-encoded gene transcription was found in 12-hour/12-hour warm/cold temperature cycles-entrained as well as free-running C. elegans [75] (Fig. 4A). In fact, most of the C. elegans homologs of core 12-hour cycling mammalian transcriptome involved in ER homeostasis and metabolism revealed robust 12-hour rhythms under the same conditions [3, 75] (Fig. 4B). These include the C. elegans homologs of dominant 12-hour cycling mammalian genes such as total Xbp1, Eif2ak3, Hspa1b, Gfpt1, Hspa5, Sec23b, Creld2, Dnaja1, and P4ha1, with Hspa1b/hsp-70 being the strongest oscillating gene in both species (Fig. 4B). It is important that for mammalian 12-hour cycling genes with superimposed dominant circadian rhythms (such as Pfkfb3, Por, Map3k5, and Upp2), 12-hour oscillations are the dominant ones in C. elegans (Fig. 4B). This could be explained by the absence of oscillations of the canonical circadian clock genes in C. elegans [3, 75, 98]. Further to this point, these 12-hour rhythms oscillate more weakly or are completely absent in free-running C. elegans that are previously entrained by the 12-hour/12-hour light/dark cycles [75]. Taken together, this evidence indicates that the C. elegans 12-hour clock is responding to temperature, rather than light, cues, which is justified by the overall lack of exposure to light in their natural soil habitat. In addition to the nematode, 12-hour oscillations of ER homeostasis and metabolism gene expression also were found in light-entrained and free-running zebrafish [99] as well as in the liver of light-entrained baboons [100] (Fig. 4C). Most intriguingly, by thoroughly analyzing published time series database, we further identified a total of 280 genes (with a P value of 1.30 × 10−187 for observing the same number of conserved 12-hour genes by chance) that exhibited conserved ~12-hour rhythms in temperature-entrained C. elegans [75], naturally caught Cellana rota (a mollusc captured from its natural intertidal habitat and exhibiting a dominant circatidal clock) [23] and mouse liver [8] (Fig. 4E). As expected, gene ontology analysis of these 280 common ~12-hour cycling genes reveals enriched biological pathways in metabolic control and ER homeostasis (Fig. 4F).
Figure 4.
Twelve-hour rhythms of gene expression are evolutionarily conserved. (A) Heat map of temporal mtDNA-encoded gene expression in temperature-entrained as well as free-running C. elegans compiled from Ref. [75]. (B) Heat map of core ER homeostasis and metabolism related 12-hour cycling gene expression in both temperature-entrained as well as free-running C. elegans (left; compiled from Ref. [75]) and mouse liver under constant darkness (right; compiled from Ref. [8]). Xbp1 is highlighted in red. (C) Phylogenetic tree and relative mRNA expression of Eif2ak3, Gfpt1, and Dnajb4 in C. rota (second row; reported in Ref. [23]), Caenorhabditis elegans (third row; reported in Ref. [75]), Danio rerio (fourth row; reported in Ref. [99]), Mus musculus (fifth row; and reported in Ref. [8]), and Papio anubis (last row; reported in Ref. [100]) during a 48-hour interval. The status of the three genes was not reported for E. pulchra in the study [18]. The data for Papio anubis is double plotted for better visualization. (D) The mRNA level of Xbp1 ortholog in C. rota captured from different times of day from their natural intertidal habitat in the wild as compiled from Ref. [23]. (E) Heat map of all 280 evolutionarily 12-hour cycling gene expression in temperature-entrained as well as free-running C. elegans (left; compiled from Ref. [75]), naturally caught C. rota in tune to 12-hour cycling tidal cues under 12-hour/12-hour natural light condition (middle; compiled from Ref. [23]), and mouse liver under constant darkness (right; compiled from Ref. [8]). (F) Gene ontology analysis of enriched KEGG pathways from these 280 genes. (G) Predicted interactive network construction of these 280 proteins using STRING [102] with XBP1 highlighted by the arrow.
Although it is much appreciated that the molecular mechanisms for generating the circadian clock, namely, the transcriptional/translational feedback loop (TTFL), are conserved in multiple species, there is little overlap in the individual genes involved in the establishment of the circadian TTFL clockwork in different species. Therefore, many thought that the circadian clock evolved separately in different lineages [101]. However, the fact that the core 12-hour clock genes remained conserved even in divergent species that cross phylum boundaries (Fig. 4C) strongly implies that the 12-hour clock may even be more ancient and evolved earlier than the circadian clock. Further supporting this hypothesis is the conserved 12-hour oscillation of the proposed transcriptional regulator of the mammalian 12-hour clock, Xbp1 in C. rota, whose oscillation is also in phase with the general circatidal transcriptome peaking at rising tides [23] (Fig. 4D and 4E). Additionally, computational construction predicted a mammalian interactive network using these 280 genes via STRING [102] and put XBP1 in the center of the network connecting different peripheral subhubs (Fig. 4G), consistent with the experimental results demonstrating the essential role of Xbp1 in regulating the 12-hour clock in MEFs (Fig. 3). Notwithstanding the evidence, the causal relationships between the circatidal Xbp1/UPR-related gene oscillations and the circatidal behavior in these marine animals remain to be determined.
7. The Functions of the Mammalian 12-Hour Clock
It has been widely accepted that all organisms adjust their biochemical, physiological, and behavioral processes in the most advantageous manner, in phase with predictable environmental changes [103]. Alignment of endogenous clocks with environmental timing increases an organism’s overall fitness and survival, whereas misalignment between the two features considerably reduces an organism’s fitness and can be a strong predisposing factor to multiple morbidities in almost all species examined [104–111]. Whereas this conclusion was based mostly on studies on the circadian clock, it is no exception for the 12-hour clock. Intertidal animals exhibiting dominant circatidal clocks tune their tidal rhythmicity to ∼12-hour rhythmic cycles of inundation and exposure that give rise to rapid changes in temperature, salinity, hydrostatic pressure, osmotic pressure, food, and predation pressure [112].
The reasons for the existence of a 12-hour clock in mammals are, however, subtler. After all, it has been a long time since we “came out of the sea,” and mammals no longer experience 12-hour environmental changes of salinity, temperature, or hydrostatic pressure. Instead, we are constantly exposed to circadian changes of temperature, light, and food availability and, as a result, the circadian clock has long been thought to be the only clock present in higher organisms, including mammals. Nonetheless, if the theory that biological clocks are always in synchronization with environmental timing also holds true for the mammalian 12-hour clock, then we must have either overlooked some 12-hour cycling environmental factors to which the mammals are exposed to everyday, or there is “something else” cycling with a 12-hour period that is indirectly caused by the circadian environmental changes of temperature, light, or food commonly associated with land mammals.
We favor the latter hypothesis and conjecture that the mammalian 12-hour clock is in tune to a 12-hour cycling “stress cycle” or, more specifically, a 12-hour cycling “metabolic stress cycle” that results from the innate misalignment between energy intake and energy expenditure within a 24-hour window (Fig. 5A). The early clues suggesting the existence of such a 12-hour cycling metabolic cycle are the unique dawn and dusk phase distribution of 12-hour cycling mRNAs in mouse liver, both of which correspond to transition periods between fasting–feeding and sleep–wake (Fig. 2C) [3]. At the subjective dawn [Zeitgeber time (ZT)10 to ZT12 for the nocturnal mouse, where ZT0 is the time of light on and ZT12 is the time of light off), the prolonged absence of energy intake from the subjective night combined with reduced but still notable energy expenditure (during sleep the brain and other parts of the body still consume large amounts of energy to consolidate memory, dispose of the metabolic waste, and repair the body [113–115]) leads to a peak of energy “overdraft.” In contrast, at the subjective dusk (ZT0 to ZT2 for nocturnal mice), sufficient energy intake during the subjective day plus substantially reduced energy expenditure gives rise to a peak of energy “excess” (Fig. 5A). Both energy overdraft and energy excess create great metabolic stress for the mammalian cells. They activate the same stress response pathways to mitigate the original stress, including the two most well-known stress responses that evolved to cope with such metabolic stress: the classical ER-associated unfolded protein response (UPRER) [90] and a second stress response pathway reacting to unfolded proteins in the mitochondria coined “mitochondrial UPR” (UPRmt) [116–119].
Figure 5.
Mammalian 12-hour clock in diseases and chronotherapy. (A) Diagram summarizing the origin, regulation, function, and species conservation of 12-hour clock. Please see the main text for detailed description of each section. (B) High-fat diet disrupts the hepatic 12-hour cycling, but not circadian gene expression in mice. Heat map (left) and representative log2 normalized expression (right) of key 12-hour cycling and circadian genes under normal chow and high-fat diet conditions are compiled from Ref. [40]. The data are double plotted for better visualization. (C) Disrupted hepatic 12-hour rhythms are associated with aging. Heat map (top) and representative mRNA expression (bottom) of key 12-hour cycling and circadian gene in young and old male mice are compiled from Ref. [152]. (D) Aging-regulating genes are enriched for 12-hour rhythmicity in C. elegans. (Top) Heat map of aging-related 12-hour cycling gene expression in both temperature-entrained as well as free-running C. elegans as compiled from Ref. [75]. Of 62 genes that increase worm lifespan by 10% when knocked down postdevelopmentally [163], 38 (62%) of them showed 12-hour rhythm in temperature-entrained and free-running worms as shown in the heat map. Additionally, positive regulators of aging, including atfs-1, xbp-1, daf-16, aak-2, tcer-1, and sir-2.1, all exhibited 12-hour rhythms of gene expression. (Bottom) Heat map of mammalian ortholog of sir-2.1 (Sirt1), daf-16 (Foxo1), and aak-2 (Prkaa1 and Prkaa2) expression in mouse liver under constant darkness condition as compiled from Ref. [8]. Atfs1 and Xbp1 are highlighted in red. (E and F) Twelve-hour clock–based chronotherapy blueprint. Heat map of temporal mRNA (E) and protein (F) expression of hepatic 12-hour cycling genes with Food and Drug Administration–approved drug interactions as compiled from Refs. [8, 38]. The names of drug are indicated on the right. FEN1, FASN, and PPAT also exhibit 12-hour rhythms of mRNA expression and are highlighted in red.
For example, both fasting-associated hypoglycemia and feeding-associated hyperglycemia conditions can activate the UPRER [120–123]. Likewise, both increased and decreased mitochondria respiration capacity can activate UPRmt and other mitochondrial stress pathways [124]. Therefore, it is not surprising that both UPRER transcription factor xbp-1 and UPRmt transcription factor atfs-1 exhibit robust 12-hour rhythms of expression peaking at dawn (C10) and dusk (CT22), synchronized to the 12-hour rhythms of proposed cellular metabolic stress (Fig. 5A and 5D). Whereas the true mammalian homolog of atfs-1 remains elusive at this time, one potential candidate Ddit3/Chop [117] does exhibit a robust 12-hour rhythm in mice [3]. Based on these findings, we propose that the mammalian 12-hour clock may govern physiological 12-hour oscillations of UPRER and UPRmt to maintain metabolic homeostasis. These oscillations are further synchronized to the energy imbalance–induced 12-hour metabolic stress cycle occurring in the ER and mitochondria [3]. Therefore, we coined the term CREMA (coordinated rhythms of ER and mitochondria action) to reflect the coupled 12-hour rhythms of gene expression involved in both ER and mitochondria homeostasis [3]. Supporting the existence of such a 12-hour metabolic stress cycle are observed 12-hour cycling metabolites (such as ADP and inosine) in dexamethasone-synchronized human U2OS cells with or without BMAL1 knockdown (Fig. 3D) [11]. Nevertheless, more studies utilizing real-time imaging of stress-related metabolites in single cells are needed to substantiate that such a 12-hour metabolic stress cycle indeed exists in a fully cell-autonomous manner as predicted.
8. The Potential Roles of the 12-Hour Clock in Diseases and Therapy
In this section, we present early evidence supporting the potential roles of the 12-hour clock in regulating human diseases and further discuss how we may exploit this knowledge to better implement chronotherapy. Owing to the strong implications that the 12-hour clock is key to the controlling of hepatic metabolism and stress responses, we focus specifically on two conditions that are profoundly influenced by these two pathways: nonalcoholic fatty liver diseases (NAFLD) and aging [125–134].
A. Loss of a Functional 12-Hour Clock Is Strongly Associated With NAFLD Progression
It has been well established that both nutritional challenge and chronic ER/mitochondrial stress are strong contributing factors to the pathogenesis of NAFLD [125, 127, 128, 134–141]. Given the newly discovered housekeeping functions of the 12-hour clock in maintaining cycling metabolic stress response, we propose a new mechanism whereby pathological levels of metabolic stress impair the hepatic 12-hour clock and perturb the metabolic homeostasis that eventually results in NAFLD (Fig. 6A). This hypothesis is reminiscent of the well-established causal relationships among chronic “jet lag stress,” and the perturbed circadian rhythm and metabolic syndrome in both animal models and humans [142–145]. At least four pieces of evidence support this hypothesis. First, acute induction of severe ER stress by tunicamycin injection alters the 12-hour rhythmic gene expression in mouse liver [35]. Second, nutritional challenge by high-fat diet severely disrupts the 12-hour oscillation of UPR and metabolic genes, including Manf, Sec23b, Hspa5, Eif2ak3, and Gfpt1 in mouse liver. In contrast, circadian oscillations of all 17 core circadian clock genes, including Bmal1, Per2, and Rev-erbα, were affected either modestly or not at all by the high-fat diet challenge (Fig. 6B) [40]. Thirdly, gene set enrichment analysis revealed that the downregulation of 12-hour gene expressions is associated strongly with progression to hepatic steatosis and nonalcoholic steatohepatitis in humans [3, 146]. Lastly, using both genetic and pharmacological models, it has been convincingly shown that activating ER stress-sensing or ER quality control pathways, including Xbp1s, are protective against hepatic steatosis [147–150]. Although this evidence strongly supports an association of 12-hour clock disruption with progression to NAFLD, future studies using both genetic and pharmacological 12-hour clock–deficient animal models are needed to firmly establish the causal roles of the 12-hour clock in disease development (Fig. 6A).
Figure 6.
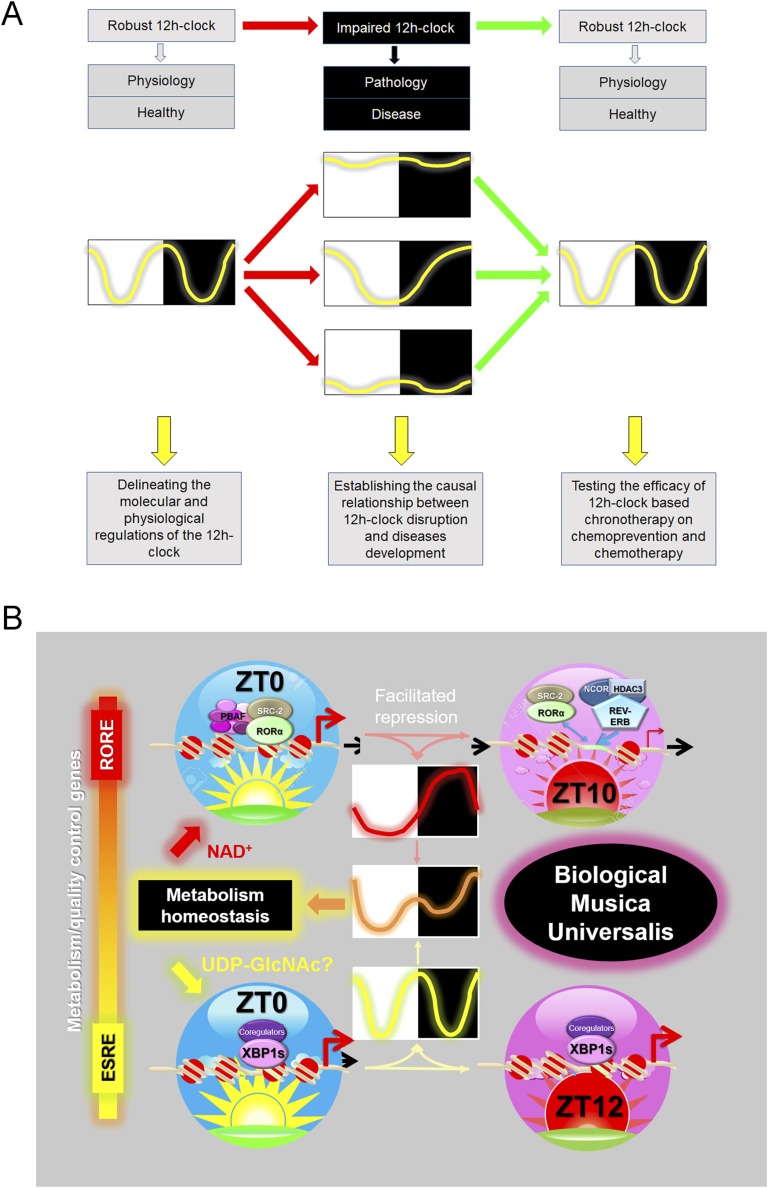
Framework of 12-hour clock study. (A) Diagram summarizing the potential causal roles of mammalian 12-hour clock in disease development with future directions outlined below. Twelve-hour rhythms can be disrupted either by the dampening of the amplitude or alteration of the period. (B) Diagram summarizing the basis for biological system being part of “musica universalis”. ERSE, ER response element; RORE, retinoic acid–related orphan receptor response element.
B. Is the 12-Hour Clock in Essence an Antiaging Hormetic Response?
Aging is a complex process characterized by a progressive loss of physiological integrity, leading to impaired function and increased vulnerability and eventually to death. In a comprehensive review published in 2013, a total of nine hallmarks representing the common denominator of the aging process in multiple organisms were proposed [133]. Of these hallmarks, loss of proteostasis, deregulated nutrient sensing, and mitochondria dysfunction are biological processes under strong 12-hour clock regulation, suggesting a potential role of the 12-hour clock in mediating aging, especially in the prevention of aging-related metabolic decline [3, 129, 133]. In fact, NAFLD itself is commonly thought to be an aging-related morbidity [134, 151]. Further supporting this hypothesis is the severely disrupted 12-hour rhythm (but not circadian rhythm) of hepatic gene expression in old mice (Fig. 5C) [152], reminiscent of similarly disrupted hepatic 12-hour rhythms in mice fed a high-fat diet (Fig. 5B). Intriguingly, genes known to regulate lifespan in C. elegans are highly enriched for 12-hour rhythms of gene expression (Fig. 5D) [75]. These include positive regulators of longevity such as sir2.1 (homolog of mammalian Sirt1) [153–157], daf-16 (homolog of mammalian Foxo1) [158, 159], aak-2 (homolog of mammalian Prkaa1/2, also known as Ampkα1/2) [160, 161], and tcer 1 (homolog of mammalian Tcerg1) [162] as well as a large number of negative regulators of C. elegans lifespan uncovered from an RNA interference screen [163] (Fig. 5D). More interestingly, Sirt1, Foxo1, Prkaa1, and Prkaa2 also exhibit 12-hour rhythms in mouse liver (Fig. 5D) [3].
The potential mechanisms of the 12-hour clock in mediating longevity and aging, in our opinion, may lie in the ancient concept of “hormesis,” which is the biological embodiment of the idiom “what does not kill you makes you stronger.” In scientific terms, hormesis is an adaptive cellular response whereby exposure to low doses of stress (but not a high dose) activates protective mechanisms that render the cell resistant to a subsequent challenge with higher doses of stress [164]. The exact molecular mechanisms underlying hormesis are still unclear but have been previously attributed to the actions of both ER and mitochondria, which are termed ER hormesis and mitohormesis, respectively [124, 165, 166]. In either case, the hormetic response is mediated either by xbp1-dependent UPRER (for ER hormesis) or atfs-1/hsp-60 (Hspd1 in mammals)–dependent UPRmt (for mitohormesis) [124, 165]. Both ER hormesis and mitohormesis are positively associated with organismal development, stress resistance, and, more importantly, longevity. For example, a study in C. elegans showed that expression of XBP1s in neurons was sufficient to increase longevity and induce ER hormetic response in distal, nonneuronal cell types via a cell-nonautonomous mechanism [167, 168]. Furthermore, mitochondrial ribosomal protein knockdown-mediated lifespan extension also requires hsp60-dependent UPRmt in both C. elegans and mammalian cells [169]. Considering the fact that both ER hormesis and mitohormesis master regulators xbp-1 and atfs-1/hsp-60 exhibit conserved 12-hour rhythms and the central roles of the 12-hour clock in stress response regulation, we conjecture that the hormetic response may exert its antiaging effects partially through the amplification/boosting of the endogenous 12-hour clock. Indeed, we found that only a low dose, but not a high dose, of tunicamycin (<30 ng/mL) is capable of synchronizing the 12-hour clock in MEFs, consistent with the concept of hormesis (Ref. [3] and unpublished data).
C. Twelve-Hour Clock–Based Chronotherapy
Chronotherapy is the concept that treatment of an illness or disorder should take the body’s natural rhythms and cycles into accounts [170]. It has been shown that drugs that target rhythmic, high-amplitude circadian gene products represent a potential pathway for mechanism-driven chronotherapy [171–175]. Herein we propose that similarly, 12-hour rhythmic transcriptome and proteome can function as a blueprint for future design of 12-hour clock–based chronotherapy. To this end, we probed Food and Drug Administration–approved drugs for potential interactions with high-amplitude 12-hour cycling hepatic transcripts and proteins using the Drug Gene Interaction Database [176, 177] and found a number of hits against both 12-hour cycling hepatic mRNA and/or their protein targets (Fig. 5E and 5F). For example, 6-mercaptopurine is a medication for treating a variety of blood cancer and autoimmune diseases. Among its mechanisms of action is the inhibition of de novo purine synthesis by directly inhibiting the rate-limiting enzyme phosphoribosyl pyrophosphate amidotransferase encoded by the Ppat gene [178, 179]. Because, as we discussed previously, coordinated 12-hour rhythms of hepatic purine metabolism with RNA transcription is a strong feature of the mammalian 12-hour clock (Fig. 2B and 2C) [3], it may be desirable to administer mercaptopurine (and other drugs targeting de novo purine and pyrimidine biosynthesis) at times of nadir purine/pyrimidine metabolism (at 3:00 pm to 4:00 pm) to minimize hepatic toxicity.
Owing to the very early stage of the 12-hour rhythm field (and ultradian rhythms field as a whole), very little is currently known about its definitive functions and implications in human diseases. To a large extent, we can only speculate on their functions and implications based on the limited literature available so far. We hope this review/preview can attract more scientists into this nascent field, because as outlined below, we think that only the tip of the iceberg has been uncovered thus far and that there is so much more to explore.
9. Future Directions
Future work could be directed toward three major aims: (1) delineating the molecular and physiological mechanisms of 12-hour clock regulation, (2) establishing the causal relationship between 12-hour clock disruption and disease progression, and (3) translating the above knowledge into chronotherapy-based medical practice (Fig. 6A). For example, at the molecular and cellular levels, is the 12-hour rhythm temperature-compensated? What is the full spectrum of the 12-hour cycling transcriptome, proteome, and metabolome under 12-hour clock control? What are the negative transcriptional regulators of the mammalian 12-hour clock? What are the coregulators for XBP1s-dependent 12-hour clock transcriptional control? Do some posttranscriptional and posttranslational mechanisms exist? If so, what are they? At the systemic and physiological level, how are the 12-hour rhythms in different tissues and organs coordinated and synchronized at the systemic level? Are there 12-hour cycling hormonal factors coordinating different tissues? Maybe a more tempting question is: does the 12-hour clock exist in the central nervous system and, if so, where? Furthermore, 12-hour clock–deficient animal models are needed in the future to rigorously establish the causal relationship between 12-hour clock dysfunction and multiple disease development. Our ultimate goal should be to successfully translate the newly learned knowledge into chronotherapy-based medicine and disease prevention.
10. Conclusion
Based on the current available literature, we propose a model where combined actions of both the circadian and 12-hour clock are required to ensure cellular homeostasis and promote organism integrity and overall fitness (Fig. 6B). Quite a number of genes in the mammalian genome are under dual circadian and 12-hour clock control, although through distinct molecular mechanisms. The circadian clock is transcriptionally regulated through a revised TTFL model that includes activation phase–facilitated repression mainly via the retinoic acid–related orphan receptor response element (Fig. 6B) [180], while the 12-hour clock is transcriptionally regulated by 12-hour rhythms of XBP1s chromatin recruitment to ER response element (Fig. 6B) [3]. Superimposition of both circadian and 12-hour rhythms of gene expression leads to a temporal gene expression pattern characterized by two nonsymmetrical peaks within a diurnal cycle (Fig. 6B), which is commonly observed in metabolic genes such as Fasn and Gck (Fig. 1C) [3]. Furthermore, the circadian clock and the 12-hour clock are synchronized to different environmental cues. The circadian clock is responding to the differential between the two opposite metabolic states, that is, fasting and feeding, whereas the 12-hour clock is entrained by the common denominator associated with both fasting and feeding: the peaking of metabolic stress (Fig. 6B).
In addition to the circadian and 12-hour clocks, it is very likely that additional clock components also exist in mammals, such as an 8-hour clock (Fig. 1B–1D). We reason that having multiple clock components endows organisms with more flexibility and the heightened ability to adapt to different environments, thus markedly increasing survival advantage. In theory, simply adjusting the relative phases and fine-tuning the amplitudes of different clocks can give rise to an infinite number of peak patterns to cope with a variety of daily environmental cycles. The coexistence of multiple harmonic biological rhythms in tune to the natural rhythms is reminiscent of the ancient philosophic concept of “musica universalis,” which was first proposed by the Greek philosopher Pythagoras more than 2000 years ago [181]. According to this theory, all natural appearing rhythms are in essence tuned to the rhythmic movements of celestial bodies [181]. Although the circadian rhythm is synchronized to the 24-hour light/dark cycle coinciding with the Earth’s rotation, our findings suggest that the 12-hour clock may have evolved from the ancient circatidal clock, which is in turn entrained by the 12-hour tidal cues orchestrated mainly by the moon. Thus, it appears that after 2000 years, we may have finally found evidence for the “biological musica universalis” (Fig. 6B).
Acknowledgments
We acknowledge all members of B.W. O’Malley’s laboratory for support for this project, with special thanks to Naomi Gonzalez, Dr. Brian York, and Dr. Maricarmen Delia Planas-Silva. We also thank Drs. Oren Levy and Yisrael Schnytzer from Bar-Ilan University for sharing the processed transcriptome data of C. rota. We apologize for omission of relevant works and citations due to space constraints.
Financial Support: This work was supported by funding from the Brockman Foundation to B.W.O. and C.C.D. and supported by funding from National Institute of Health HD07879 to B.W.O. and National Science Foundation 1703170 to C.C.D. and B.Z. This work also was supported by the Center for Advancement of Science in Space Grant GA-2014-136, National Science Foundation Grant 11703170, the Peter J. Fluor Family Fund, the Philip J. Carroll, Jr. Professorship, and the Joyce Family Foundation to C.C.D, as well as by American Diabetes Association junior faculty development award 1-18-JDF-025 to B.Z.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- CT
circadian time
- dGFP
destabilized green fluorescence protein
- Gck
glucokinase
- ER
endoplasmic reticulum
- MEF
mouse embryonic fibroblast
- mtDNA
mitochondrial DNA
- NAFLD
nonalcoholic fatty liver diseases
- TTFL
transcriptional/translational feedback loop
- UPR
unfolded protein response
- UPRER
endoplasmic reticulum–associated unfolded protein response
- UPRmt
mitochondrial unfolded protein response
- XBP1s
spliced XBP1
- Xbp1 us
unspliced Xbp1
- ZT
Zeitgeber time
References and Notes
- 1. Koff C, Fiveisky MM. Harmony From the Science of Acoustics. Studio City, CA: Koff Music Co.; 1975.
- 2. Pierce JR. The Science of Musical Sound. Revised ed. New York, NY: W.H. Freeman; 1992.
- 3. Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, O'Malley BW. A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell Metab. 2017;25(6):1305–1319.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antoulas AC, Zhu B, Zhang Q, York B, O’Malley BW, Dacso C. A novel mathematical method for disclosing oscillations in gene transcription: a comparative study. bioRxiv. 2017. https://doi.org/10.1101/151720. [DOI] [PMC free article] [PubMed]
- 5. Ionita AC, Antoulas AC. Parametrized model reduction in the Loewner Framework. In: Quarteroni A, Rozza G, eds. Reduced Order Methods for Modeling and Computational Reduction. Berlin, Germany: Springer; 2013:51–66.
- 6. Ionita AC, Antoulas AC. Data-driven parametrized model reduction in the Loewner Framework. SIAM J Sci Comput. 2014;36(3):A984–A1007. [Google Scholar]
- 7. Refinetti R, Lissen GC, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38(4):275–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shannon CE. Communication in the presence of noise. Proceedings of the IRE. 1949;37(1):10–21. [Google Scholar]
- 10. Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, Er U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM. Clock regulation of metabolites reveals coupling between transcription and metabolism [published correction appears in Cell Metab. 2017;25(5):1206]. Cell Metab. 2017;25(5):1206. [DOI] [PubMed] [Google Scholar]
- 12. Wilcockson D, Zhang L. Circatidal clocks. Curr Biol. 2008;18(17):R753–R755. [DOI] [PubMed] [Google Scholar]
- 13. Brown FA, Fingerman M, Sandeen MI, Webb HM. Persistent diurnal and tidal rhythms of color change in the fiddler crab, Uca pugnax. J Exp Zool. 1953;123(1):29–60. [Google Scholar]
- 14. Palmer JD. Comparative studies of tidal rhythms. VIII. A translocation experiment involving Circalunidian rhythms. Mar Behav Physiol. 1989;14(4):231–243. [Google Scholar]
- 15. Saigusa M. Hatching controlled by the circatidal clock, and the role of the medulla terminalis in the optic peduncle of the eyestalk, in an estuarine crab Sesarma haematocheir. J Exp Biol. 2002;205(Pt 22):3487–3504. [DOI] [PubMed] [Google Scholar]
- 16. Naylor E. Tidal and diurnal rhythms of locomotory activity in Carcinus maenas (L.). J Exp Biol. 1958;35:602–610. [Google Scholar]
- 17. Warman CG, Reid DG, Naylor E. Variation in the tidal migratory behaviour and rhythmic light-responsiveness in the shore crab, Carcinus maenas. J Mar Biol Assoc U K. 1993;73(02):355–364. [Google Scholar]
- 18. O’Neill JS, Lee KD, Zhang L, Feeney K, Webster SG, Blades MJ, Kyriacou CP, Hastings MH, Wilcockson DC. Metabolic molecular markers of the tidal clock in the marine crustacean Eurydice pulchra. Curr Biol. 2015;25(8):R326–R327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr Biol. 2013;23(19):1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akiyama T. Entrainment of the circatidal swimming activity rhythm in the cumacean Dimorphostylis asiatica (Crustacea) to 12.5-hour hydrostatic pressure cycles. Zool Sci. 2004;21(1):29–38. [DOI] [PubMed] [Google Scholar]
- 21. Chabot CC, Kent J, Watson WH III. Circatidal and circadian rhythms of locomotion in Limulus polyphemus. Biol Bull. 2004;207(1):72–75. [DOI] [PubMed] [Google Scholar]
- 22. Last KS, Bailhache T, Kramer C, Kyriacou CP, Rosato E, Olive PJ. Tidal, daily, and lunar-day activity cycles in the marine polychaete Nereis virens. Chronobiol Int. 2009;26(2):167–183. [DOI] [PubMed] [Google Scholar]
- 23. Schnytzer Y, Simon-Blecher N, Li J, Ben-Asher HW, Salmon-Divon M, Achituv Y, Hughes ME, Levy O. Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Sci Rep. 2018;8(1):4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balaparameswara Rao M. Studies on the oxygen consumption of a tropical intertidal limpet Cellana radiata (Born): effect of body size and tidal rhythm. Hydrobiologia. 1980;71(1-2):175–179. [Google Scholar]
- 25. Parpagnoli D, Pecchioli S, Santini G. Temporal determinants of grazing activity in the Mediterranean limpet Patella caerulea. Ethol Ecol Evol. 2013;25(4):388–399. [Google Scholar]
- 26. Gray DR, Hodgson AN. Endogenous rhythms of locomotor activity in the high-shore limpet, Helcion pectunculus (Patellogastropoda). Anim Behav. 1999;57(2):387–391. [DOI] [PubMed] [Google Scholar]
- 27. Satoh A, Yoshioka E, Numata H. Entrainment of the circatidal activity rhythm of the mangrove cricket, Apteronemobius asahinai, to periodic inundations. Anim Behav. 2009;78(1):189–194. [Google Scholar]
- 28. Satoh A, Yoshioka E, Numata H. Circatidal activity rhythm in the mangrove cricket Apteronemobius asahinai. Biol Lett. 2008;4(3):233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palmer JD. Review of the dual-clock control of tidal rhythms and the hypothesis that the same clock governs both circatidal and circadian rhythms. Chronobiol Int. 1995;12(5):299–310. [Google Scholar]
- 30. Naylor E. Crab clockwork: the case for interactive circatidal and circadian oscillators controlling rhythmic locomotor activity of Carcinus maenas. Chronobiol Int. 1996;13(3):153–161. [DOI] [PubMed] [Google Scholar]
- 31. Watson WH III, Bedford L, Chabot CC. Rhythms of locomotion expressed by Limulus polyphemus, the American horseshoe crab: II. Relationship to circadian rhythms of visual sensitivity. Biol Bull. 2008;215(1):46–56. [DOI] [PubMed] [Google Scholar]
- 32. Takekata H, Numata H, Shiga S, Goto SG. Silencing the circadian clock gene clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J Insect Physiol. 2014;68:16–22. [DOI] [PubMed] [Google Scholar]
- 33. Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol Lett. 2012;8(4):488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Veen DR, Gerkema MP. Unmasking ultradian rhythms in gene expression. FASEB J. 2017;31(2):743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11(1):47–57. [DOI] [PubMed] [Google Scholar]
- 36. Westermark PO, Herzel H. Mechanism for 12 hr rhythm generation by the circadian clock. Cell Reports. 2013;3(4):1228–1238. [DOI] [PubMed] [Google Scholar]
- 37. Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8(7):e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, Er U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM. Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 2017;25(4):961–974.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a Web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colquhoun WP, Blake MJ, Edwards RS. Experimental studies of shift-work I: a comparison of “rotating” and “stabilized” 4-hour shift systems. Ergonomics. 1968;11(5):437–453. [DOI] [PubMed] [Google Scholar]
- 44. Colquhoun WP, Paine MW, Fort A. Circadian rhythm of body temperature during prolonged undersea voyages. Aviat Space Environ Med. 1978;49(5):671–678. [PubMed] [Google Scholar]
- 45. Colquhoun WP, Paine MW, Fort A. Changes in the temperature rhythm of submariners following a rapidly rotating watchkeeping system for a prolonged period. Int Arch Occup Environ Health. 1979;42(3-4):185–190. [DOI] [PubMed] [Google Scholar]
- 46. Moore-Ede MC, Czeisler CA. Mathematical Models of the Circadian Sleep-Wake Cycle. New York, NY: Raven Press; 1984.
- 47. Kronauer RE, Jewett ME. The relationship between circadian and hemicircadian components of human endogenous temperature rhythms. J Sleep Res. 1992;1(2):88–92. [DOI] [PubMed] [Google Scholar]
- 48. Monk TH, Buysse DJ, Reynolds CF III, Kupfer DJ. Circadian determinants of the postlunch dip in performance. Chronobiol Int. 1996;13(2):123–133. [DOI] [PubMed] [Google Scholar]
- 49. Wan C, Wang Z, Cornélissen G, Halberg F. Age, gender and circadian or circasemidian blood pressure and heart rate variation of children. Chronobiologia. 1992;19(3–4):121–129. [PubMed] [Google Scholar]
- 50. Otsuka K, Murakami S, Kubo Y, Yamanaka T, Mitsutake G, Ohkawa S, Matsubayashi K, Yano S, Cornelissen G, Halberg F. Chronomics for chronoastrobiology with immediate spin-offs for life quality and longevity. Biomed Pharmacother. 2003;57(Suppl 1):1s–18s. [DOI] [PubMed]
- 51. Lee JS, Lee MS, Lee JY, Cornelissen G, Otsuka K, Halberg F. Effects of diaphragmatic breathing on ambulatory blood pressure and heart rate. Biomed Pharmacother. 2003;57(Suppl 1):87s–91s. [DOI] [PubMed]
- 52. Otsuka K, Oinuma S, Cornelissen G, Weydahl A, Ichimaru Y, Kobayashi M, Yano S, Holmeslet B, Hansen TL, Mitsutake G, Engebretson MJ, Schwartzkopff O, Halberg F. Alternating light-darkness-influenced human electrocardiographic magnetoreception in association with geomagnetic pulsations. Biomed Pharmacother. 2001;55(Suppl 1):63s–75s. [DOI] [PubMed]
- 53. Otsuka K, Cornelissen G, Furukawa S, Kubo Y, Hayashi M, Shibata K, Mizuno K, Aiba T, Ohshima H, Mukai C. Long-term exposure to space’s microgravity alters the time structure of heart rate variability of astronauts. Heliyon. 2016;2(12):e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Otsuka K, Cornélissen G, Halberg F. Circadian rhythmic fractal scaling of heart rate variability in health and coronary artery disease. Clin Cardiol. 1997;20(7):631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bjerner B, Holm A, Swensson A. Diurnal variation in mental performance; a study of three-shift workers. Br J Ind Med. 1955;12(2):103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mitler MM, Carskadon MA, Czeisler CA, Dement WC, Dinges DF, Graeber RC. Catastrophes, sleep, and public policy: consensus report. Sleep. 1988;11(1):100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dinges DF, Broughton RJ, eds. Sleep and Alertness:Chronobiological, Behavioral, and Medical Aspects of Napping. New York, NY: Raven Press; 1989.
- 58. Broughton RJ. SCN controlled circadian arousal and the afternoon “nap zone”. Sleep Res Online. 1998;1(4):166–178. [PubMed] [Google Scholar]
- 59. Reinberg A, Bicakova-Rocher A, Nouguier J, Gorceix A, Mechkouri M, Touitou Y, Ashkenazi I. Circadian rhythm period in reaction time to light signals: difference between right- and left-hand side. Brain Res Cogn Brain Res. 1997;6(2):135–140. [DOI] [PubMed] [Google Scholar]
- 60. Shub Y, Lewy H, Ashkenazi IE. Circadian pattern of simulated flight performance of pilots is derived from ultradian components. Chronobiol Int. 2001;18(6):987–1003. [DOI] [PubMed] [Google Scholar]
- 61. Iskra-Golec I, Smith L. Ultradian and asymmetric rhythms of hemispheric processing speed [published correction appears in Chronobiol Int. 2007;24(1):191]. Chronobiol Int. 2006;23(6):1229–1239. [DOI] [PubMed] [Google Scholar]
- 62. Harrington MG, Salomon RM, Pogoda JM, Oborina E, Okey N, Johnson B, Schmidt D, Fonteh AN, Dalleska NF. Cerebrospinal fluid sodium rhythms. Cerebrospinal Fluid Res. 2010;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kanabrocki EL, Sothern RB, Ryan MD, Kahn S, Augustine G, Johnson C, Foley S, Gathing A, Eastman G, Friedman N, Nemchausky BA, Kaplan E. Circadian characteristics of serum calcium, magnesium and eight trace elements and of their metallo-moieties in urine of healthy middle-aged men. Clin Ter. 2008;159(5):329–346. [PubMed] [Google Scholar]
- 64. Ayala DE, Hermida RC, Garcia L, Iglesias T, Lodeiro C. Multiple component analysis of plasma growth hormone in children with standard and short stature. Chronobiol Int. 1990;7(3):217–220. [DOI] [PubMed] [Google Scholar]
- 65. Tarquini R, Mazzoccoli G, Dolenti S, Gaudiano P, Comuni C, Laffi G, Perfetto F, Otsuka K, Cornelissen G, Halberg F. Circasemidian rather than circadian variation of circulating osteoprotegerin in clinical health. Biomed Pharacother. 2005;59(Suppl 1):225s–228s. [DOI] [PMC free article] [PubMed]
- 66. Francis SJ, Walker RF, Riad-Fahmy D, Hughes D, Murphy JF, Gray OP. Assessment of adrenocortical activity in term newborn infants using salivary cortisol determinations. J Pediatr. 1987;111(1):129–133. [DOI] [PubMed] [Google Scholar]
- 67. Broughton RJ. Biorhythmic variations in consciousness and psychological functions. Can Psychol Rev. 1979;16(4):217–239. [Google Scholar]
- 68. Broughton R, Mullington J. Circasemidian sleep propensity and the phase-amplitude maintenance model of human sleep/wake regulation. J Sleep Res. 1992;1(2):93–98. [DOI] [PubMed] [Google Scholar]
- 69. Hayashi M, Morikawa T, Hori T. Circasemidian 12 h cycle of slow wave sleep under constant darkness. Clin Neurophysiol. 2002;113(9):1505–1516. [DOI] [PubMed] [Google Scholar]
- 70. Billiard M, Carlander B, Besset A. [Circadian rhythm in normal and pathological sleep]. Pathol Biol (Paris). 1996;44(6):509–517. [PubMed] [Google Scholar]
- 71. Nobili L, Besset A, Ferrillo F, Rosadini G, Schiavi G, Billiard M. Dynamics of slow wave activity in narcoleptic patients under bed rest conditions. Electroencephalogr Clin Neurophysiol. 1995;95(6):414–425. [DOI] [PubMed] [Google Scholar]
- 72. Broughton R, Mullington J. Chronobiological aspects of narcolepsy. Sleep. 1994;17(8, Suppl):S35–S44. [DOI] [PubMed] [Google Scholar]
- 73. Broughton R, Dunham W, Newman J, Lutley K, Duschesne P, Rivers M. Ambulatory 24 hour sleep-wake monitoring in narcolepsy-cataplexy compared to matched controls. Electroencephalogr Clin Neurophysiol. 1988;70(6):473–481. [DOI] [PubMed] [Google Scholar]
- 74. De Koninck GC, Hébert M, Carrier J, Lamarche C, Dufour S. Body temperature and the return of slow wave activity in extended sleep. Electroencephalogr Clin Neurophysiol. 1996;98(1):42–50. [DOI] [PubMed] [Google Scholar]
- 75. van der Linden AM, Beverly M, Kadener S, Rodriguez J, Wasserman S, Rosbash M, Sengupta P. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8(10):e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bodenstein C, Heiland I, Schuster S. Temperature compensation and entrainment in circadian rhythms. Phys Biol. 2012;9(3):036011. [DOI] [PubMed] [Google Scholar]
- 77. Tamaru T, Hattori M, Honda K, Benjamin I, Ozawa T, Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One. 2011;6(9):e24521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Carskadon MA, Dement WC. Multiple sleep latency tests during the constant routine. Sleep. 1992;15(5):396–399. [DOI] [PubMed] [Google Scholar]
- 80. Stahl ML, Orr WC, Bollinger C. Postprandial sleepiness: objective documentation via polysomnography. Sleep. 1983;6(1):29–35. [DOI] [PubMed] [Google Scholar]
- 81. Borbély AA, Daan S, Wirz-Justice A, Deboer T. The two-process model of sleep regulation: a reappraisal. J Sleep Res. 2016;25(2):131–143. [DOI] [PubMed] [Google Scholar]
- 82. Bjorness TE, Kelly CL, Gao T, Poffenberger V, Greene RW. Control and function of the homeostatic sleep response by adenosine A1 receptors. J Neurosci. 2009;29(5):1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bjorness TE, Greene RW. Adenosine and sleep. Curr Neuropharmacol. 2009;7(3):238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huang ZL, Zhang Z, Qu WM. Roles of adenosine and its receptors in sleep–wake regulation. Int Rev Neurobiol. 2014;119:349–371. [DOI] [PubMed] [Google Scholar]
- 85. Ribeiro JA, Sebastião AM. Caffeine and adenosine. J Alzheimers Dis. 2010;20(Suppl 1):S3–S15. [DOI] [PubMed] [Google Scholar]
- 86. Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, FitzGerald GA. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8(324):324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28(1):251–277. [DOI] [PubMed] [Google Scholar]
- 90. Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69(2):169–181. [DOI] [PubMed] [Google Scholar]
- 91. Ho N, Xu C, Thibault G. From the unfolded protein response to metabolic diseases—lipids under the spotlight. J Cell Sci. 2018;131(3):131. [DOI] [PubMed] [Google Scholar]
- 92. Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, Criollo A, Luo X, Tan W, Jiang N, Lehrman MA, Rothermel BA, Lee AH, Lavandero S, Mammen PPA, Ferdous A, Gillette TG, Scherer PE, Hill JA. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156(6):1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, Liang G, Ye J, Lehrman MA, Hill JA, Horton JD, Scherer PE. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123(1):455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shao M, Shan B, Liu Y, Deng Y, Yan C, Wu Y, Mao T, Qiu Y, Zhou Y, Jiang S, Jia W, Li J, Li J, Rui L, Yang L, Liu Y. Hepatic IRE1α regulates fasting-induced metabolic adaptive programs through the XBP1s–PPARα axis signalling. Nat Commun. 2014;5(1):3528. [DOI] [PubMed] [Google Scholar]
- 95. Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320(5882):1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hastings MH. Semi-lunar variations of endogenous circa-tidal rhythms of activity and respiration in the isopod Eurydice pulchra. Mar Ecol Progr Ser. 1981;4(1):85–90.
- 97. Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Romanowski A, Garavaglia MJ, Goya ME, Ghiringhelli PD, Golombek DA. Potential conservation of circadian clock proteins in the phylum Nematoda as revealed by bioinformatic searches. PLoS One. 2014;9(11):e112871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li Y, Li G, Wang H, Du J, Yan J. Analysis of a gene regulatory cascade mediating circadian rhythm in zebrafish. PLOS Comput Biol. 2013;9(2):e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359(6381):eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Loudon AS. Circadian biology: a 2.5 billion year old clock. Curr Biol. 2012;22(14):R570–R571. [DOI] [PubMed] [Google Scholar]
- 102. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55(1):17–54. [DOI] [PubMed] [Google Scholar]
- 104. Roenneberg T, Merrow M. The circadian clock and human health. Curr Biol. 2016;26(10):R432–R443. [DOI] [PubMed] [Google Scholar]
- 105. Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. [DOI] [PubMed] [Google Scholar]
- 106. Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13(6):639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lambert G, Chew J, Rust MJ. Costs of clock-environment misalignment in individual cyanobacterial cells. Biophys J. 2016;111(4):883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. West AC, Smith L, Ray DW, Loudon ASI, Brown TM, Bechtold DA. Misalignment with the external light environment drives metabolic and cardiac dysfunction. Nat Commun. 2017;8(1):417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Krishnan HC, Lyons LC. Synchrony and desynchrony in circadian clocks: impacts on learning and memory. Learn Mem. 2015;22(9):426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30(3):333–349. [DOI] [PubMed] [Google Scholar]
- 111. O’Donnell AJ, Schneider P, McWatters HG, Reece SE. Fitness costs of disrupting circadian rhythms in malaria parasites. Proc Biol Sci. 2011;278(1717):2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Naylor E. Chronobiology of Marine Organisms. Cambridge, NY: Cambridge University Press; 2010.
- 113. Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156):373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Stickgold R. Sleep-dependent memory consolidation. Nature. 2005;437(7063):1272–1278. [DOI] [PubMed] [Google Scholar]
- 115. Dattilo M, Antunes HK, Medeiros A, Mônico Neto M, Souza HS, Tufik S, de Mello MT. Sleep and muscle recovery: endocrinological and molecular basis for a new and promising hypothesis. Med Hypotheses. 2011;77(2):220–222. [DOI] [PubMed] [Google Scholar]
- 116. Qureshi MA, Haynes CM, Pellegrino MW. The mitochondrial unfolded protein response: signaling from the powerhouse. J Biol Chem. 2017;292(33):13500–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21(17):4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217(Pt 1):137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response—synchronizing genomes. Curr Opin Cell Biol. 2015;33:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Iurlaro R, Püschel F, León-Annicchiarico CL, O’Connor H, Martin SJ, Palou-Gramón D, Lucendo E, Muñoz-Pinedo C. Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol Cell Biol. 2017;37(10):e00479-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. de la Cadena SG, Hernández-Fonseca K, Camacho-Arroyo I, Massieu L. Glucose deprivation induces reticulum stress by the PERK pathway and caspase-7- and calpain-mediated caspase-12 activation. Apoptosis. 2014;19(3):414–427. [DOI] [PubMed] [Google Scholar]
- 122. Mooradian AD, Haas MJ. Glucose-induced endoplasmic reticulum stress is independent of oxidative stress: a mechanistic explanation for the failure of antioxidant therapy in diabetes. Free Radic Biol Med. 2011;50(9):1140–1143. [DOI] [PubMed] [Google Scholar]
- 123. Zhong Y, Li J, Chen Y, Wang JJ, Ratan R, Zhang SX. Activation of endoplasmic reticulum stress by hyperglycemia is essential for Müller cell–derived inflammatory cytokine production in diabetes. Diabetes. 2012;61(2):492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19(5):757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhang XQ, Xu CF, Yu CH, Chen WX, Li YM. Role of endoplasmic reticulum stress in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20(7):1768–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ashraf NU, Sheikh TA. Endoplasmic reticulum stress and oxidative stress in the pathogenesis of non-alcoholic fatty liver disease. Free Radic Res. 2015;49(12):1405–1418. [DOI] [PubMed] [Google Scholar]
- 127. Gentile CL, Frye M, Pagliassotti MJ. Endoplasmic reticulum stress and the unfolded protein response in nonalcoholic fatty liver disease. Antioxid Redox Signal. 2011;15(2):505–521. [DOI] [PMC free article] [PubMed]
- 128. Pagliassotti MJ. Endoplasmic reticulum stress in nonalcoholic fatty liver disease. Annu Rev Nutr. 2012;32(1):17–33. [DOI] [PubMed] [Google Scholar]
- 129. López-Otín C, Galluzzi L, Freije JMP, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166(4):802–821. [DOI] [PubMed] [Google Scholar]
- 130. Brown MK, Naidoo N. The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol. 2012;3:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Martínez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell. 2017;16(4):615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Salminen A, Kaarniranta K. ER stress and hormetic regulation of the aging process. Ageing Res Rev. 2010;9(3):211–217. [DOI] [PubMed] [Google Scholar]
- 133. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gong Z, Tas E, Yakar S, Muzumdar R. Hepatic lipid metabolism and non-alcoholic fatty liver disease in aging. Mol Cell Endocrinol. 2017;455:115–130. [DOI] [PubMed] [Google Scholar]
- 135. Dara L, Ji C, Kaplowitz N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology. 2011;53(5):1752–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Nakamura A, Terauchi Y. Lessons from mouse models of high-fat diet-induced NAFLD. Int J Mol Sci. 2013;14(11):21240–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Polimeni L, Del Ben M, Baratta F, Perri L, Albanese F, Pastori D, Violi F, Angelico F. Oxidative stress: new insights on the association of non-alcoholic fatty liver disease and atherosclerosis. World J Hepatol. 2015;7(10):1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sunny NE, Bril F, Cusi K. Mitochondrial adaptation in nonalcoholic fatty liver disease: novel mechanisms and treatment strategies. Trends Endocrinol Metab. 2017;28(4):250–260. [DOI] [PubMed] [Google Scholar]
- 139. Ajith TA. Role of mitochondria and mitochondria-targeted agents in non-alcoholic fatty liver disease. Clin Exp Pharmacol Physiol. 2018;45(5):413–421. [DOI] [PubMed] [Google Scholar]
- 140. Simões ICM, Fontes A, Pinton P, Zischka H, Wieckowski MR. Mitochondria in non-alcoholic fatty liver disease. Int J Biochem Cell Biol. 2018;95:93–99. [DOI] [PubMed] [Google Scholar]
- 141. Sun WJ, Xiong J, Chen DF. [Cross-talk between endoplasmic reticulum and mitochondria in non-alcoholic fatty liver disease]. Zhonghua Gan Zang Bing Za Zhi. 2011;19(9):648–650. [DOI] [PubMed] [Google Scholar]
- 142. Arendt J. Shift work: coping with the biological clock. Occup Med (Lond). 2010;60(1):10–20. [DOI] [PubMed] [Google Scholar]
- 143. Fleet T, Stashi E, Zhu B, Rajapakshe K, Marcelo KL, Kettner NM, Gorman BK, Coarfa C, Fu L, O’Malley BW, York B. Genetic and environmental models of circadian disruption link SRC-2 function to hepatic pathology. J Biol Rhythms. 2016;31(5):443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106(3):447–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Froy O. Metabolism and circadian rhythms—implications for obesity. Endocr Rev. 2010;31(1):1–24. [DOI] [PubMed] [Google Scholar]
- 146. Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, Erhart W, Egberts J, Sipos B, Schreiber S, Häsler R, Stickel F, Becker T, Krawczak M, Röcken C, Siebert R, Schafmayer C, Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. [DOI] [PubMed] [Google Scholar]
- 147. Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21(17):2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15(6):829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30(7):1357–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Herrema H, Zhou Y, Zhang D, Lee J, Salazar Hernandez MA, Shulman GI, Ozcan U. XBP1s is an anti-lipogenic protein. J Biol Chem. 2016;291(33):17394–17404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bertolotti M, Lonardo A, Mussi C, Baldelli E, Pellegrini E, Ballestri S, Romagnoli D, Loria P. Nonalcoholic fatty liver disease and aging: epidemiology to management. World J Gastroenterol. 2014;20(39):14185–14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, Sassone-Corsi P. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 2017;170(4):664–677.e11. [DOI] [PMC free article] [PubMed]
- 153. Lee J, Kwon G, Park J, Kim JK, Lim YH. Brief communication: SIR-2.1-dependent lifespan extension of Caenorhabditis elegans by oxyresveratrol and resveratrol. Exp Biol Med (Maywood). 2016;241(16):1757–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. [DOI] [PubMed] [Google Scholar]
- 155. Wang X, Wang X, Li L, Wang D. Lifespan extension in Caenorhabditis elegans by DMSO is dependent on sir-2.1 and daf-16. Biochem Biophys Res Commun. 2010;400(4):613–618. [DOI] [PubMed] [Google Scholar]
- 156. Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9(5):605–615. [DOI] [PubMed] [Google Scholar]
- 157. Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125(6):1165–1177. [DOI] [PubMed] [Google Scholar]
- 158. Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. [DOI] [PubMed] [Google Scholar]
- 159. Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. [DOI] [PubMed] [Google Scholar]
- 160. Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18(24):3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5(2):119–126. [DOI] [PubMed] [Google Scholar]
- 162. Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009;5(9):e1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3(4):e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Mattson MP. Hormesis defined. Ageing Res Rev. 2008;7(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Mollereau B, Manié S, Napoletano F. Getting the better of ER stress. J Cell Commun Signal. 2014;8(4):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61(5):654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Martínez G, Duran-Aniotz C, Cabral-Miranda F, Hetz C. Commentary: XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Front Aging Neurosci. 2016;8:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kaur G, Phillips C, Wong K, Saini B. Timing is important in medication administration: a timely review of chronotherapy research. Int J Clin Pharm. 2013;35(3):344–358. [DOI] [PubMed] [Google Scholar]
- 171. Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci USA. 2017;114(20):5312–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Dakup PP, Porter KI, Little AA, Gajula RP, Zhang H, Skornyakov E, Kemp MG, Van Dongen HPA, Gaddameedhi S. The circadian clock regulates cisplatin-induced toxicity and tumor regression in melanoma mouse and human models. Oncotarget. 2018;9(18):14524–14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ye Y, Xiang Y, Ozguc FM, Kim Y, Liu CJ, Park PK, Hu Q, Diao L, Lou Y, Lin C, Guo AY, Zhou B, Wang L, Chen Z, Takahashi JS, Mills GB, Yoo SH, Han L. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 2018;6(3):314–328.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ozturk N, Ozturk D, Pala-Kara Z, Kaptan E, Sancar-Bas S, Ozsoy N, Cinar S, Deniz G, Li XM, Giacchetti S, Lévi F, Okyar A. The immune system as a chronotoxicity target of the anticancer mTOR inhibitor everolimus. Chronobiol Int. 2018;35(5):705–718. [DOI] [PubMed] [Google Scholar]
- 175. Paolino S, Cutolo M, Pizzorni C. Glucocorticoid management in rheumatoid arthritis: morning or night low dose? Reumatologia. 2017;55(4):189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wagner AH, Coffman AC, Ainscough BJ, Spies NC, Skidmore ZL, Campbell KM, Krysiak K, Pan D, McMichael JF, Eldred JM, Walker JR, Wilson RK, Mardis ER, Griffith M, Griffith OL. DGIdb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016;44(D1):D1036–D1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB, Eldred JM, Miller CA, Subramanian J, Govindan R, Kumar RD, Bose R, Ding L, Walker JR, Larson DE, Dooling DJ, Smith SM, Ley TJ, Mardis ER, Wilson RK. DGIdb: mining the druggable genome. Nat Methods. 2013;10(12):1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Dubinsky MC. Azathioprine, 6-mercaptopurine in inflammatory bowel disease: pharmacology, efficacy, and safety. Clin Gastroenterol Hepatol. 2004;2(9):731–743. [DOI] [PubMed] [Google Scholar]
- 179. Sahasranaman S, Howard D, Roy S. Clinical pharmacology and pharmacogenetics of thiopurines. Eur J Clin Pharmacol. 2008;64(8):753–767. [DOI] [PubMed] [Google Scholar]
- 180. Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell. 2015;60(5):769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Weiss P, Taruskin R. Music in the Western World: A History in Documents. 2nd ed. Belmont, CA: Thomson/Schirmer; 2008.