Abstract
Intrinsically disordered proteins (IDPs)/intrinsically unstructured proteins are characterized by the lack of fixed or stable tertiary structure, and are increasingly recognized as an important class of proteins with major roles in signal transduction and transcriptional regulation. In this study, we report the identification and functional characterization of a previously uncharacterized protein (UPF0258/KIAA1024), major intrinsically disordered Notch2-associated receptor 1 (MINAR1). While MINAR1 carries a single transmembrane domain and a short cytoplasmic domain, it has a large extracellular domain that shares no similarity with known protein sequences. Uncharacteristically, MINAR1 is a highly IDP with nearly 70% of its amino acids sequences unstructured. We demonstrate that MINAR1 physically interacts with Notch2 and its binding to Notch2 increases its stability and function. MINAR1 is widely expressed in various tissues including the epithelial cells of the breast and endothelial cells of blood vessels. MINAR1 plays a negative role in angiogenesis as it inhibits angiogenesis in cell culture and in mouse matrigel plug and zebrafish angiogenesis models. Furthermore, while MINAR1 is highly expressed in the normal human breast, its expression is significantly downregulated in advanced human breast cancer and its re-expression in breast cancer cells inhibited tumor growth. Our study demonstrates that MINAR1 is an IDP that negatively regulates angiogenesis and growth of breast cancer cells.
Keywords: intrinsically disordered proteins, Notch2, angiogenesis, breast cancer, MINAR1
Introduction
Intrinsically disordered proteins (IDPs) are increasingly recognized as an important class of proteins with major biological functions. It was estimated that nearly 30% of proteins in eukaryotic cells lack a unique 3D structure (Gsponer and Babu, 2009), underscoring their ubiquitous functional importance in the biology of eukaryotic cells. Due to their intrinsically disordered nature, IDPs are dynamic and versatile. They interact with other proteins that are involved in key regulatory cellular events such as signal transduction and transcriptional regulation (Wright and Dyson, 2015; Chavali et al., 2017). It has been proposed that disordered sequences are essential for the function of transcriptional activators and copious other cell signaling proteins (Dyson and Wright, 2005). As IDPs have greater flexibility to interact with other proteins compared with the globular proteins, they rapidly form complexes with other proteins, and thus initiate a signaling cascade and dissociate quickly. These unique properties are critically important for highly complex processes of signal transduction and transcriptional regulation (Gsponer and Babu, 2009).
Angiogenesis, the formation of blood vessels, is a multistep and highly organized cellular process, which is largely initiated by the function of an intrinsically discorded transcription factor, hypoxia-inducible transcription factor1α (HIF1α) (Dames et al., 2002; De Guzman et al., 2005). HIF1α activates transcription of critical proteins such as vascular endothelial growth factor (VEGF), which is a key culprit of pathological and physiological angiogenesis (Rahimi, 2006a, b, 2012). Through its intrinsically disordered carboxyl-terminal activation domain (CAD), HIF1α associates with transcriptional coactivator protein (CREB)-binding protein (CBP)/p300 to activate transcription of VEGF (Dames et al., 2002). Initiation and maturation of angiogenesis is governed by a myriad of cell surface receptors such as VEGF receptors (VEGFR-1 and VEGFR-2), Tie and Notch receptors.
The Notch pathway is a highly evolutionary conserved intercellular signaling system and is activated by the interaction of transmembrane ligands, the Delta (Delta-like1 [Dll-1], Dll3, and Dll4) families and Jagged (Jagged1 and Jagged2) with Notch receptors (Notch1−4), which are usually expressed on the surface of neighboring cells. Ligand binding induces cleavage of the Notch receptors and subsequent nuclear translocation of the Notch intracellular domains (NICDs). In the nucleus, NICD binds to multiple DNA-binding proteins (Lu and Lux, 1996; Andersson et al., 2011; Chillakuri et al., 2012). Notch signaling plays a delicate role in angiogenesis. In the initial stages of angiogenesis, Notch activation is generally suppressed to allow endothelial cells to proliferate in response to VEGF stimulation, and its expression is subsequently upregulated when endothelial cells stop proliferating and the vessels begin to stabilize (Henderson et al., 2001; Taylor et al., 2002). Considering its overall function in angiogenesis, the Notch signaling pathway plays a negative role in angiogenesis. Interfering with the Notch pathway in a mouse resulted in the development of vascular tumors and lethal hemorrhage (Liu et al., 2011) and its activity in cell culture inhibited the angiogenic functions of endothelial cells such as capillary tube formation, migration, and proliferation (Leong et al., 2002; Itoh et al., 2004; Noseda et al., 2004; Williams et al., 2006). Notch receptors also play major roles in human cancers. They can function as either an oncogene or a tumor suppressor in human cancers. For example, Notch2 expression in breast cancer is associated with better survival, and high expression of Notch2 is correlated with well-differentiated breast tumors (Parr et al., 2004).
Here, we report the identification and functional characterization of a previously unknown protein, MINAR1. We demonstrate that MINAR1 is as an IDP, and have identified Notch2 as a MINAR1-binding protein. MINAR1 binds to Notch2 and gains stability and function. MINAR1 activity inhibits proliferation of breast cancer cells and angiogenesis and its expression is downregulated in human advanced breast cancer.
Results
Identification of major intrinsically disordered Notch2-binding receptor 1
We have recently identified multiple previously uncharacterized cell surface receptors through an in silico analysis of human genome (Rahimi et al., 2012; Arafa et al., 2015; Wang et al., 2016). Major intrinsically disordered Notch2-binding receptor 1 (MINAR1), which is encoded by a previously uncharacterized gene KIAA1024 located in chromosome 15 (GRCh38.p7), was among these putative cell surface receptors. MINAR1 consists of 916 amino acids with a predicted molecular weight of 103 kDa, and is composed of a large extracellular domain with multiple potential glycosylation sites, and a single transmembrane domain followed by a short intracellular domain (Supplementary Figure S1A). MINAR1 is highly conserved among the species ranging from human to rodents and Xenopus (Supplementary Figure S1B) and the sequence similarity of the human and mouse MINAR1 is 96.5% (Supplementary Figure S1C), suggesting a potentially evolutionarily conserved function for MINAR1.
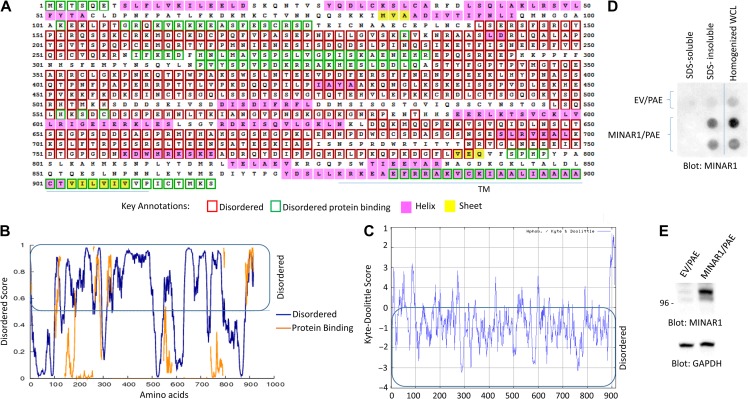
Uniquely, MINAR1 appeared to be a highly IDP, as its instability index was 58.24 (Guruprasad et al., 1990; Wilkins et al., 1999). Generally, proteins with higher stability have an instability index of less than 40, whereas unstable proteins have value above 40 (Guruprasad et al., 1990). IDPs commonly have a lower content of the order-promoting amino acids such as Cys, Trp, Tyr, Phe Ile, Leu, Val, and Asn, whereas they are enriched in the disorder-promoting residues (Pro, Arg, Gly, Gln, Ser, Glu, Lys, and Ala) (Campen et al., 2008). Consistent with these characteristics, MINAR1 is considerably low in the order-promoting amino acids and high in the disorder-promoting amino acids (Supplementary Figure S2). Furthermore, our analysis showed that majority of MINAR1 amino acids in Kyte–Doolittle scale are at the negative range (Figure 1C). Analysis of MINAR1 amino acid sequences via the online disorder prediction program DISOPRED3 (http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1) revealed that nearly 70% of MINAR1 is intrinsically disordered (Figure 1A and B). Similarly, other programs such as DISOclust (McGuffin, 2008; Roche et al., 2011), Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) (data not shown), and Metadisorder, which calculates disorder consensus from the results generated by 13 major different disorder programs such as DisEMBL, DISOPRED2, DISpro, Globplot, iPDA, IUPred, Pdisorder, Poodle-l, PrDOS, and Spritz, predicated MINAR1 as a highly IDP (Supplementary Figure S3).
Figure 1.
Identification of MINAR1 as an IDP. (A) The amino acid sequence of human MINAR1. Amino acids boxed in the red correspond to disordered segments of MINAR1. The data were generated using online DISOPRED3 (Disorder Prediction) program. (B) The graph is the representation of the data shown in A. (C) The Kyte–Doolittle hydrophobicity score of MINAR1. The graph was generated using online program http://web.expasy.org/protscale/. (D) PAE cells ectopically expressing empty vector (EV) or MINAR1 were lysed with lysis buffer containing Triton X-100. Whole-cell lysates (WCL) were centrifuged and Triton X-100 soluble fraction was separated from the insoluble fraction. The remaining insoluble fraction was further solubilized with 1% SDS. Both fractions were then blotted on cellulose acetate filter via a dot blot apparatus. The same cell groups were also homogenized in PBS plus 1% SDS and similarly blotted on cellulose acetate filter. The cellulose acetate filter was blotted for the presence of MINAR1 using anti-MINAR1 antibody (the MINAR1/PAE cell lysate group loaded in duplicate). (E) Western blot of the cell lysates from EV/PAE and MINAR1/PAE. Data in D and E are representative of at least three independent experiments.
Intrinsically disordered or unstructured proteins tend to aggregate and are typically insoluble (Hazeki et al., 2000). Therefore, we examined the tendency of aggregation of MINAR1 via the cellulose acetate filter trap assay, which is commonly used to study protein aggregation (Scherzinger et al., 1997). Initially, we lysed PAE cells ectopically expressing MINAR1 with a lysis buffer containing Triton X-100 and SDS and soluble and insoluble protein fractions were subjected to cellulose acetate filter trap assay. MINAR1 was detected only in the Triton X-100/SDS insoluble fraction (Figure 1D). However, when cells were homogenized and whole-cell homogenate was applied to the cellulose acetate filter trap assay, MINAR1 was retained on the cellulose acetate filter (Figure 1D), indicating that MINAR1 forms protein aggregates due to its intrinsically disordered characteristic. Intrinsically discorded proteins usually have short half-lives and are generally susceptible to degradation due to misfolding or lack of overall structure, which leaves them more accessible to proteases. Treatment of HEK-293 cells endogenously expressing MINAR1 with a chemical chaperone 4-phenylbutyrate (4BPA) increased the expression of MINAR1 (Supplementary Figure S4A) and decreased its sensitivity to partial trypsin digestion (Supplementary Figure S4B), demonstrating that in the presence of 4BPA, MINAR1 gains a partial secondary structure, and thus exhibits a reduced susceptibility to degradation by partial trypsin digestion.
MINAR1 is a cell surface receptor that is expressed in various human organs and tissues
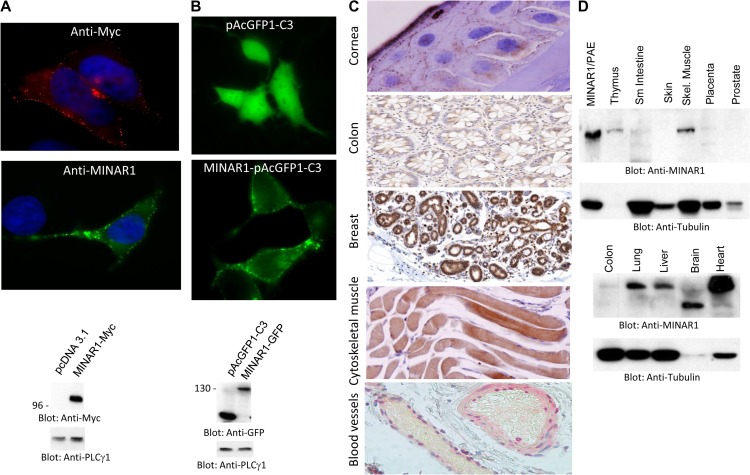
MINAR1 was predicated to be a cell surface protein (Figure 1). Therefore, we asked whether MINAR1 is expressed as a cell surface protein. When expressed in HEK-293 cells, Myc-tagged MINAR1 (MINAR1-Myc) was detected as a cell surface protein, as both anti-MINAR1 and anti-Myc antibodies located MINAR1 at the membranous region in HEK-293 cells (Figure 2A). Similarly, ectopic expression of GFP-tagged MINAR1 also showed that MINAR1 is expressed at the cell surface (Figure 2B). Furthermore, immunohistochemistry staining of human corneal epithelial cells showed that MINAR1 is expressed as a membrane-bound protein (Figure 2C). MINAR1 was also expressed in human colon, cytoskeletal muscle, breast, and endothelial cells of blood vessels (Figure 2C). Additionally, western blot analysis demonstrated that MINAR1 is highly expressed in heart, brain, liver, lung, skeletal muscle, and thymus (Figure 2D). Colon and placenta tissue cell lysates also were positive for MINAR1, although at significantly lower levels (Figure 2D). The apparent molecular weight of MINAR1 in human brain tissue was lower, which may be due to protein degradation. Furthermore, we used publically available human genome data (http://biogps.org) to examine the expression profile of MINAR1. MINAR1 mRNA is widely present in the majority of human organs; however, the highest of level of MINAR1 mRNA was present in heart, liver, skeletal muscle, cardiac myocytes, and adrenal cortex (Supplementary Figure S5). These results indicate that MINAR1 is a cell surface protein and widely expressed in human organs and tissues.
Figure 2.
MINAR1 is a cell surface receptor and is widely expressed in human organs and tissues. (A) HEK-293 cells expressing MINAR1-Myc were fixed in 4% PFA and stained with anti-MYC or anti-MINAR1 antibody. Pictures were taken under immunofluorescence microscope (40×). Expression of MINAR1 in HEK-293 cells is shown. (B) HEK-293 cells expressing GFP vector alone (pAcGFP1-C3) or GFP-MINAR1 were viewed under immunofluorescence microscope and pictures were taken (40×). Expression of MINAR1 in HEK-293 cells is shown. (C) Human cornea, breast, colon, and cytoskeletal tissues were subjected to immunohistochemistry analysis using anti-MINAR1 antibody. Slides were scanned and corresponding pictures are shown (40×). (D) Proteins extracted from various human organs were subjected to western blot analysis using anti-MINAR1 antibody or anti-tubulin as a loading control.
MINAR1 inhibits angiogenesis
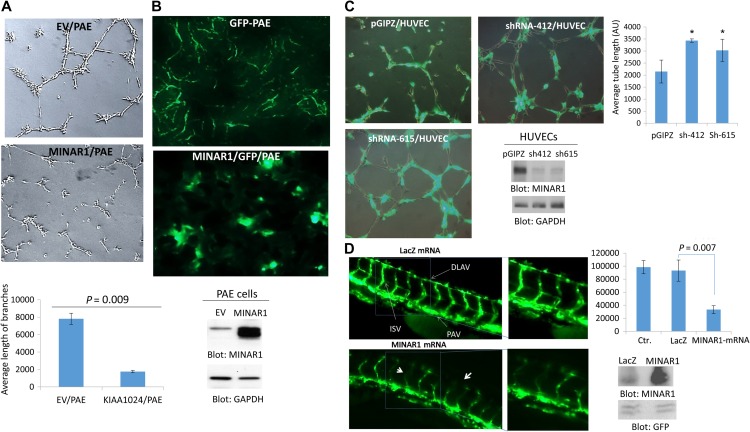
Considering its expression in the endothelial cells, we sought to examine possible biological activities of MINAR1 in angiogenesis. To this end, we examined the effect of overexpression of MINAR1 in porcine aortic endothelial (PAE) cells. Overexpression of MINAR1 in PAE cells inhibited capillary tube formation in an in vitro matrigel assay (Figure 3A). To corroborate the inhibitory effect of MINAR1 in angiogenesis further, we subjected PAE cells to a mouse matrigel plug angiogenesis assay. To visualize the tube formation of PAE cells, cells were also engineered to express GFP. Similar to in vitro matrigel assay, PAE cells expressing MINAR1 did not form capillary tubes; instead, they grew in clusters (Figure 3B). Next, we silenced expression of MINAR1 in human umbilical vein endothelial cells (HUVECs) by two different shRNAi and assessed their capillary tube formation. The knockdown of MINAR1 by shRNA increased capillary tube formation (Figure 3C). To further illustrate the in vivo function of MINAR1 in angiogenesis, we tested the role MINAR1 in zebrafish angiogenesis. Microinjection of in vitro-translated human MINAR1 mRNA into 1-cell-stage zebrafish embryos significantly disrupted intersegmental vessels (ISV) and dorsal longitudinal anastomotic vessel (DLAV) formation at 50 hpf (Figure 3D). Microinjection of LacZ mRNA, which was used as a control, had no effect on the blood vessel formation compared with control embryos with no injection (Figure 3D). Quantification of the blood vessel formation (ISV and DLAV) in response to microinjection of human MINAR1 mRNA is shown (Figure 3D). Western blot analysis was performed to confirm the overexpression of MINAR1 in microinjected fishes (Figure 3D). Altogether, the data obtained from cell culture, mouse, and zebrafish angiogenesis assays all demonstrated that MINAR1 negatively regulates angiogenesis.
Figure 3.
MINAR1 inhibits in vitro and in vivo angiogenesis. (A) PAE cells ectopically expressing empty vector (EV) or MINAR1 were subjected to in vitro angiogenesis assay and pictures were taken after 24 h. Pictures were taken and three randomly selected pictures from each group were selected and quantified using Image J program. (B) GPP-PAE cells expressing empty vector (EV) or MINAR1 were subjected to in vivo matrigel assay. Cells were mixed with growth factor-reduced matrigel and sub-dermally injected into mouse (three mice/group). After 7 days, the matrigel plugs were removed and were viewed under fluorescent microscope after cryo fixing and processing. The representative pictures are shown (40×). Expression of MINAR1 in PAE cells is shown. (C) HUVECs were transduced with an empty vector, pGIPZ, or two different MINAR1 shRNA. After 48 h, cells were subjected to in vitro angiogenesis assay. Cells were viewed under a fluorescent microscope and pictures were taken. Quantification of capillary tube formation was made by Image J program. Knockdown of MINAR1 is also shown. (D) Fli-eGFP transgenic fish embryos were injected with MINAR1 or LacZ mRNA at the 1-cell stage or 2-cell stage. The embryos were examined at 50 h post-fertilization (50 hpf) and representative immunofluorescence images are shown. Quantification of intersegmental vessels (ISV) and the dorsal lateral anastomosing vessel (DLAV) of 10 fish per group by ImageJ are shown in bar graph. Error bars represent SD. P = 0.007 for 10 ng MINAR1 mRNA compared with LacZ control. Western blot analysis of MINAR1 expression from tissue lysates of microinjected fish is performed with anti-MINAR1 antibody and protein loading control GFP.
MINAR1 expression is downregulated in human advanced breast cancer
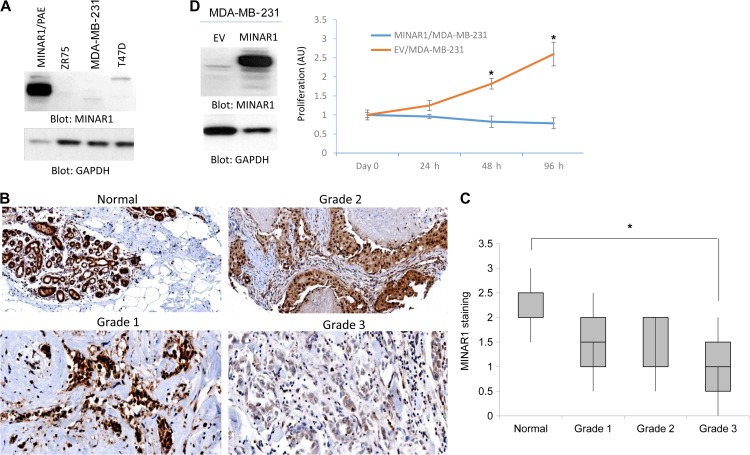
In addition to its expression in endothelial cells, MINAR1 is expressed in human epithelial cells of breast, colon, and other tissues (Figure 2C). Therefore, we examined the possible role of MINAR1 in human breast cancer. We initially examined several breast cancer cell lines, including MDA-MB-231, T47D, and ZR-75-1 cells for the expression of MINAR1 in western blot analysis. MINAR1 was hardly detectable in these breast cancer cell lines (Figure 4A).
Figure 4.
Expression of MINAR1 is downregulated in human advanced breast tumors and its expression in breast cancer cells inhibits tumor growth. (A) Cell lysates from human breast cancer cell lines, ZR-75-1, MDA-MB-231, and T47D, were prepared and subjected to western blot analysis using anti-MINAR1 antibody or anti-GAPDH antibody for loading control. (B) MINAR1 expression in human invasive ductal carcinoma breast cancer with different tumor grades. MINAR1 expression was scored for staining strength as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining) following a visual inspection of the cytoplasmic immunostaining for MINAR1. There is an inverse relation between MINAR1 expression and tumor grade (P < 0.05). (C) A tissue microarray consisting of 72 invasive ductal carcinoma tissues (16 cases of normal, 11 cases of grade 1, 32 cases of grade 2, and 12 cases of grade 3) for MINAR1 was carried out. Two pathologists assessed MINAR1 expression and tumor grades. The graph shows that MINAR1 expression is downregulated in advanced breast cancer (P < 0.05). (D) MDA-MB-231 cells expressing empty vector or MINAR1 were subjected to MTT assay. Expression of MINAR1 in MDA-MB-231 cells is shown.
This observation promoted us to examine the expression of MINAR1 in human in human breast cancer. IHC analysis showed that MINAR1 is highly expressed in normal/benign human breast (Figure 4B). However, the expression of MINAR1 was significantly reduced in poorly differentiated invasive ductal carcinoma (grade 3) of the breast (4 out 4 cases) (Figure 4B). Expression of MINAR1 in both well-differentiated (grade 1) and moderately differentiated (grade 2) breast carcinomas was also moderately reduced (Figure 4B). To confirm our observation, we stained a tissue microarray consisting of 72 invasive breast carcinoma tissues (16 cases of normal, 11 cases of grade 1, 32 cases of grade 2, and 12 cases of grade 3) for MINAR1 and two pathologists assessed MINAR1 expression and tumor grades. The result showed that MINAR1 expression has an inverse relation with tumor grade (Figure 4C). The higher the tumor grade, the lower was the MINAR1 expression (Figure 4C).
Given its observed downregulation in breast cancer, we ectopically expressed MINAR1 in MDA-MB-231 cells and assessed its effect in the proliferation of MDA-MB-231 cells. Proliferation of MDA-MB-231 cells expressing MINAR1 was significantly inhibited as measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (Figure 4D). Taken together, the data suggest that MINAR1 expression is downregulated in human breast cancer and its re-expression in breast cancer, MDA-MB-231 cells inhibits proliferation.
MINAR1 binds to Notch2 and gains stability and function
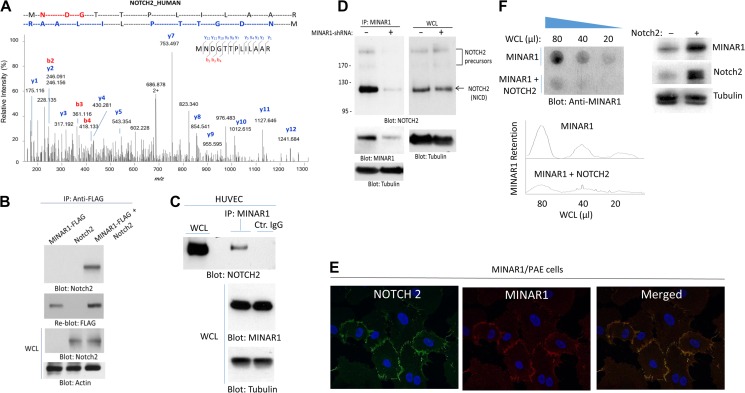
The unstructured or disordered segments in proteins often interact with other proteins and, in some cases, these regions form complexes with other proteins to gain secondary structure for functionality (Wright and Dyson, 1999; Tompa, 2002; Uversky, 2002). Given the intrinsically disordered nature of MINAR1, we hypothesized that MINAR1 could associate with other membrane-bound receptors or soluble proteins to gain structure and function. To identify possible MINAR1-binding proteins, we purified MINAR1 via immunoprecipitation from porcine aortic endothelial (PAE) cells ectopically expressing MINAR1 and analyzed the immunoprecipitated proteins by liquid chromatography–tandem mass spectrometry (LC-MS/MS). LC-MS/MS analysis identified Notch2 as a putative MINAR1-binding protein (Figure 5A). We confirmed the direct binding of MINAR1 with Notch2 in HEK-293 cells by co-transfection of MINAR1-FLAG and full-length Notch2. Cell lysates were immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-Notch2 antibody (Figure 5B). Moreover, we confirmed the binding of MINAR1 with Notch2 in human umbilical primary vein endothelial cells (HUVEC), which endogenously express both MINAR1 and Notch2. Cell lysates derived from HUVECs were immunoprecipitated with anti-MINAR1 antibody or with a control antibody, followed by immunoblotting with anti-Notch2 antibody. Notch2 was co-immunoprecipitated with MINAR1 (Figure 5C). Furthermore, knockdown of MINAR1 by shRNA in HUVECs abolished the co-immunoprecipitation of Notch2 with MINAR1 (Figure 5D). To demonstrate the Notch2 and MINAR1 co-localization in cells, we co-stained HUVECs with anti-MINAR1 and anti-Notch2 antibodies. The result showed that MINAR1 was co-localized with Notch2 (Figure 5E). These data further demonstrate that MINAR1 is a cell surface protein. To further corroborate the direct physical binding of MINAR1 with Notch2, we generated GST fusion MINAR1 encompassing the extracellular domain of MINAR1 (GST-E-MINAR1) without transmembrane and cytoplasmic domains (Supplementary Figure S6A and B). The purified GST-MINAR1 was used to examine whether GST-E-MINAR1 could interact with Notch2-GFP ectopically expressed in HEK-293 cells in an in vitro GST pull-down assay. The result showed that GST-E-MINAR1 binds to Notch2-GFP (Supplementary Figure S6C), indicating that MINAR1 directly interacts with Notch2.
Figure 5.
Identification of Notch2 as a binding partner of MINAR1. (A) PAE cells expressing empty vector or Myc-tagged MINAR1 were lysed, immunoprecipitated with anti-Myc antibody, resolved in SDS-PAGE. The corresponding band was cut and subjected to proteolytic digestion followed by LC-MS/MS analysis of the tryptic peptides. Shown here is the MS2 higher energy collisional dissociation (HCD) spectrum corresponding to Notch2 peptide ‘MNDGTTPLILAAR’, labeled with assignments for the detected N-terminal b-ion (red) and C-terminal y-ion (blue) fragments. (B) HEK-293 cells expressing FLAG-tagged MINAR1 alone or transfected with Notch2 were lysed and immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-Notch2 antibody. (C) Cell lysates from HUVECs were immunoprecipitated with anti-MINAR1 antibody or control IgG followed by immunoblotting with anti-Notch2 antibody. (D) HUVECs were transduced with two different MINAR1 shRNAs. After 48 h, cells were lysed and subjected to immunoprecipitation using anti-MINAR1 antibody, followed by immunoblotting with anti-Notch2 antibody. (E) PAE cells expressing MINAR1 were subjected to immunofluorescence staining for MINAR1 and Notch2. (F) Whole-cell lysates (WCL) from HEK-293 cells expressing MINAR1 alone or co-expressing MINAR1 with Notch2 were subjected to a filter trap assay as in Figure 1 and immunoblotted with anti-MINAR1 antibody. WCL of HEK-293 cells expressing MINAR1 alone or co-expressing MINAR1 with Notch2 were subjected to western blot analysis and blotted for MINAR1, Notch2, and the loading control tubulin.
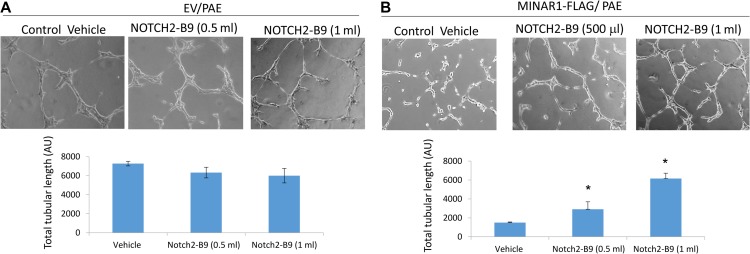
Next, we examined whether Notch2 is required for stability of MINAR1. Accordingly, we expressed MINAR1 alone or with Notch2 in HEK-293 cells. Cell homogenates were prepared and subjected to the cellulose acetate filter trap assay. The result showed that co-expression of MINAR1 with Notch2 significantly reduced aggregation of MINAR1, as was evident by the decrease in retention of MINAR1 on the cellulose acetate filter (Figure 5F). Western blot analysis of soluble MINAR1 and Notch2 expression is shown (Figure 5F). To assess whether the apparent effect of Notch2 in the stabilization of MINAR1 also affects the biological activity of MINAR1, we used a well-characterized recombinant monoclonal Notch2 blocking antibody, which binds to the extracellular region of Notch2 and inhibits its interaction with other proteins (Falk et al., 2012). Treatment of PAE cells, which express a very low amount of MINAR1, with the Notch2 blocking antibody (NOTCH2-B9) had no apparent effect on their capillary tube formation (Figure 6A). However, treatment of PAE cells expressing MINAR1 with NOTCH2-B9 antibody significantly reversed the inhibitory effect of MINAR1 on the capillary tube formation of PAE cells (Figure 6A). Taken together, we have identified Notch2 as a MINAR1-binding protein, which increases the stability of MINAR1.
Figure 6.
Blocking Notch2 antibody inhibits the effect of MINAR1 in tube formation of endothelial cells. PAE cells expressing empty vector or MINAR1 were subjected to a matrigel capillary tube formation assay treated with conditioned medium containing blocking anti-Notch2 antibody or control medium. Pictures were taken after overnight and quantified via Image J/angiogenesis assay.
Discussion
Due to their ability to interact with proteins involved in key cellular processes such as signal transduction and transcriptional regulation, IDPs/intrinsically unstructured proteins (IUPs) are considered a ‘protein–protein interaction hub’ (Wright and Dyson, 2015). Here we describe MINAR1, a previously uncharacterized protein, as an IDP/IUP. Remarkably, MINAR1 is a highly IDP with nearly 70% of its amino acid sequence predicted to contain no specific 3D structure or folding. Intrinsically disordered sequences in proteins are suggested to extend the interaction surface of proteins, providing flexibility and adaptability by allowing a given region of a protein to bind to a range of distinct protein partners (Dosztanyi et al., 2006; Singh et al., 2007; Cino et al., 2013). Such protein–protein interactions provide gain of 3D folding for IDP/IUPs that leads to stability and function (Berlow et al., 2015; Wright and Dyson, 2015).
We have identified Notch2 as a key MINAR1-binding protein. We propose that the Notch2 association with MINAR1 increases MINAR1 stability and function, as co-expression of Notch2 with MINAR1 reduced aggregation of MINAR1 as determined by a filter trap assay, consequently resulting in the increased detection of stable and solubilized MINAR by western blot. Moreover, Notch2 increased the level of endogenous MINAR1 in HEK-293 cells, likely due to binding to and inducing stability of MINAR1. This gain of stability of MINAR1, as demonstrated by 4PBA treatment in HEK-293 cells and subsequent trypsin digestion, likely increased its half-life by reducing its susceptibility to degradation. In a recent genome-wide data analysis on protein–protein interaction networks, it was demonstrated that IDP/IUPs generate more protein network activity than is the typical proteins with the conserved 3D structure (Demarest et al., 2002; Dosztanyi et al., 2006), a distinct feature that puts IDP/IUPs in an advantageous position in the context of protein–protein interaction, signal transduction, and biological functions. Our data point toward a critical role for MINAR1 in angiogenesis, as it regulates key angiogenic processes such as cell survival and capillary tube formation of endothelial cells. Interestingly, well-characterized IDP/IUPs such as p53, Mdm2, p300, BRCA1, or XPA have been associated with various human cancers. Our observation that MINAR1 expression is downregulated in advanced human breast cancer highlights the potential functional role of MINAR1 in regulating breast cancer progression.
Highly coordinated processes of capillary tube formation of endothelial cells constitute the most critical aspect of proper vessel formation in vivo. Our data demonstrate a significant role for MINAR1 in angiogenesis. Overexpression of MINAR1 in endothelial cells inhibited capillary tube formation, whereas silencing of its expression increased capillary tube formation in cell culture. MINAR1 also inhibited angiogenesis of endothelial cells in a mouse matrigel plug assay and vessel formation in zebrafish, underscoring its functional importance in angiogenesis. The underlying mechanism of MINAR1 function in angiogenesis likely requires Notch2, which binds strongly to Notch2 and appears to be necessary for stabilization of MINAR1. Remarkably, the biological effect of MINAR1 in endothelial cells overlaps with that of the Notch pathway in angiogenesis. Notch signaling is a well-known negative regulator of angiogenesis, as it inhibits angiogenesis (Leong et al., 2002; Itoh et al., 2004; Noseda et al., 2004; Williams et al., 2006). Notch2 has also been suggested to negatively regulate breast cancer growth (Parr et al., 2004). However, further studies are needed to establish the precise role of MINAR1 and its function, as an important participant in the Notch2 pathway.
Materials and methods
Plasmids, shRNAs, and antibodies
MINAR1/UPF0258 protein KIAA1024 cDNA (accession# BC160103 similar to accession# Q9UPX6) was purchased from Open biosystems/Dharmacon. MINAR1/KIAA1024 shRNAs cloned into pGIPZ lentivirus vector (three individual shRNA clones) were purchased from Dharmacon, Inc.: V3LHS_381948 (clone# 200306615), V3LHS_411743 (clone# 200253412), and V3LHS_411744 (clone# 200299719). The MINAR1 cDNA was subsequently cloned into multiple expression vectors including pAcGFP1-C3, which generates GFP-tagged MINAR1, mammalian and amphibian expression vector, pCS2+ vector with no tag, and pCDNA3.1 myc, which generates MINAR1-myc. MINAR1 was also cloned into retroviral vector pLNCX2. All the constructs were sequenced and the complete sequence of MINAR1 was verified. Rabbit polyclonal anti-MINAR1 antibody was generated against a 21-amino acid long peptide (KDGFLVEQVFSPHPYPASLKA) corresponding to the extracellular domain of MINAR1. The specificity of the antibody was validated in cells ectopically expressing MINAR1 or in cells endogenously expressing MINAR1. The immunoreactivity of anti-MINAR1 antibody was blocked by pre-incubation of anti-MINAR1 antibody with the corresponding peptide, which confirmed its specificity (data not shown). Anti-Nocth2 antibody was purchased from Cell Signaling. Notch2 blocking antibody (NOTCH2-B9) was purchased from Addgene (Falk et al., 2012). Full-length Notch2-GFP construct was kindly provided by Dr Carmela Abraham (Boston University). Full-length Notch2 cDNA with no tag was kindly provided by Dr Raphael Kopan (University of Cincinnati College of Medicine).
Cell lines
Porcine aortic endothelial cells (PAE) were kindly provided by Dr Carl-Henrik Heldin (Ludwig Cancer Research, University of Uppsala, Sweden). Human embryonic kidney epithelial cells (HEK-293) were kindly provided by Dr Vipul Chitalia (Boston University). PAE and HEK-293 were grown in DMEM medium supplemented with 10% FBS plus antibiotics. Human umbilical vascular endothelial cells (HUVECs) were purchased from Cell technologies (Frederick) and grown in the endothelial cell medium. Breast carcinoma cell lines, ZR75-1, MDA-MB-231, and T47D were grown in RPMI Medium 1640 supplemented with 10% FBS plus antibiotics. Retroviruses were produced in 293-GPG packaging cells as described (Rahimi et al., 2000). Lentiviruses were produced in 293T cells.
Immunoprecipitation and western blotting
Cells were prepared and lysed as described (Hartsough et al., 2013). Briefly, cells were washed twice with H/S buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, and 2 mM Na3VO4) and lysed in lysis buffer (10 mM Tris-HCl, 10% glycerol, pH 7.4, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM Na3VO4, and 20 μg/ml aprotinin). Whole-cell lysates were subjected to immunoprecipitation or were directly subjected to western blotting analysis as indicated in the figure legends.
In vitro GST pull-down assay
To generate the glutathione S-transferase (GST) fusion extracellular domain of MINAR1 (GST-E-MINAR1), the extracellular domain of MINAR1 was PCR amplified and cloned into pGEX-2T vector. The purified GST-MINAR1 protein was subsequently used for GST pull-down assay, which was performed as described (Rahimi et al., 2000).
Zebrafish angiogenesis assay
MINAR1 mRNA was prepared in vitro. Briefly, MINAR1 cloned into pCS2 vector were linearized with Not I restriction enzyme, treated with Proteinase K (Sigma) and extracted with phenol. Linearized plasmid DNA (1 μg/μl) in RNAse-free water was used for in vitro capped mRNA synthesis using the mMessage mMachine® SP6 kit (Ambion) according to manufacturer’s instructions and the RNA subsequently was used to injected into zebrafish embryos. Fli-eGFP-transgenic adult male and female zebrafish (Danio rerio) were housed in 14:19-h light-dark cycle at a temperature of (26.5°C) and a pH of (7.0–7.4) in a controlled multi-tank recirculating water system. A glass capillary needle attached to a Femtojet injector (Eppendorf) was used for injecting RNA (10 or 5 ng/μl in ~10 pl) into 1-cell or 2-cell-stage embryos. The embryos were grown at 28°C for 3 days. The embryos were examined after 50 hpf using an immunofluorescence microscope. The images of fish under same setting were obtained for 10 fish per group at every experiment and analyzed for the length of the vessels using Image J software.
Liquid chromatography–tandem mass spectrometry (LC-MS/MS)
MINAR1 was immunoprecipitated with anti-Myc antibody from PAE cells ectopically expressing MINAR1. The immunoprecipitated proteins were subjected SDS-PAGE, then gel bands were excised, followed by trypsin digestion (at 37°C overnight in 50 mM ammonium bicarbonate). Peptides were separated and analyzed using a 6550 Q-TOF MS with a 1200 series nanoflow HPLC-Chip ESI source with a HPLC-Chip consisting of a 360 nl trapping column and a 150 mM × 75 μM analytical column, both with Polaris C18-A 3 μm material (all from Agilent Corp.). After injection of the sample onto the trapping column, the column was washed at a rate of 2 μl/min with 2% acetonitrile and 0.1% formic acid in water for 4 min. Peptides were then separated on the analytical column at a flow rate of 0.3 μl/min using a gradient from 2% to 40% acetonitrile with 0.1% formic acid over a period of 25 min. The 6550 Q-TOF mass spectrometer was operated in positive mode using the high-resolution, extended dynamic range (2 GHz) setting. The instrument was operated in data-dependent mode; the 20 most abundant ions were selected for MS/MS. MS spectra were collected from m/z 285–1700, and MS2 spectra were collected from m/z 50–3000. The source gas temperature was set to 225°C, and the flow was set at 13 L/min, with a capillary voltage of 2100 V. Precursors ≥5000 counts and charge states ≥2 were selected for fragmentation, and the collision energy was set according to the equation y = mx + b, with y being the collision energy, slope m = 3.6, x representing the charge state, and the offset b = −4.8 for charge states 3 and 4. For charge state 2 peptides, slope m = 3.1, and the offset b = 1. Spectra were collected in centroid mode. The error for all peaks with signal-to-noise (S-to-N) ratio of >10 is within 5 ppm. MS/MS data were searched using a local copy of Mascot (www.matrixscience.com) using Uniprot database of human proteins. Parameters require a minimum of 2 peptides matching per protein with minimum probabilities of 95% at the peptide level.
Filter trap assay
Cells expressing MINAR1 were prepared and homogenized in phosphate buffered saline (PBS) as described (Scherzinger et al., 1997). Briefly, cells were washed twice in cold PBS and collected in filter trap buffer (PBS, 1 mM phenylmethylsulfonyl fluoride, 20 mg/ml aprotinin) and briefly sonicated using Thermo-Fisher Sonicator Dismembrator Model 1000. Proteins were solubilized by adding 1% SDS to the cell homogenates and these cell homogenates were subsequently blotted on a cellulose acetate filter via a dot blot apparatus. The homogenates were subjected to western blot analysis and blotted for MINAR1.
4-Phenylbutyric acid treatment and partial trypsin digest
HEK-293 cells were treated with 10 mM 4-phenylbutyric acid (4PBA) for 2 h and lysed in PBS (Wang et al., 2016). Cells designated for partial trypsin digest were homogenized by sonication and centrifuged to obtain supernatant. Trypsin (20 ng) was added to cell lysates (50 μl) and incubated at room temperature (26.5°C) for 0, 15, and 30 min. Trypsin digestion was terminated by adding sample buffer (5×), followed by incubation in the heat block at 95°C for 5 min. The samples were resolved on 10% SDS-PAGE and analyzed by western blot using anti-MINAR antibody.
In vitro capillary tube formation assay
PAE cells expressing pMSCV empty vector and MINAR (2 × 103 cells per well, triplicate wells per group) were seeded in each well of a 24-well plate coated with 200 μl growth factor-reduced Matrigel and allowed to adhere for 1 h. After 24 h, images of capillary tube formation were captured using a Zeiss microscope camera and Lumenera INFINITY ANALYZE Software. The length of capillary tube branching was analyzed with the Angiogenesis Analyzer via ImageJ software.
Ethical Approval and Consent to participate
The Boston University the Institutional Animal Care and Use Committee (IACUC) approved the use of mouse and zebrafish in this study. Boston University Medical Campus Institutional Review Board (IRB) approved the use of human tissues in this study, which is in accordance with the Declaration of Helsinki.
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported in part through grants from the NIH (R21CA191970 and R21CA193958 to N.R.) and P41GM104603 (to C.E.C.).
Conflict of interest: none declared.
References
- Andersson E.R., Sandberg R., and Lendahl U. (2011). Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612. [DOI] [PubMed] [Google Scholar]
- Arafa E., Bondzie P.A., Rezazadeh K., et al. (2015). TMIGD1 is a novel adhesion molecule that protects epithelial cells from oxidative cell injury. Am. J. Pathol. 185, 2757–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlow R.B., Dyson H.J., and Wright P.E. (2015). Functional advantages of dynamic protein disorder. FEBS Lett. 589, 2433–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campen A., Williams R.M., Brown C.J., et al. (2008). TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 15, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali S., Gunnarsson A., and Babu M.M. (2017). Intrinsically disordered proteins adaptively reorganize cellular matter during stress. Trends Biochem. Sci. 42, 410–412. [DOI] [PubMed] [Google Scholar]
- Chillakuri C.R., Sheppard D., Lea S.M., et al. (2012). Notch receptor–ligand binding and activation: insights from molecular studies. Semin. Cell Dev. Biol. 23, 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cino E.A., Killoran R.C., Karttunen M., et al. (2013). Binding of disordered proteins to a protein hub. Sci. Rep. 3, 2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dames S.A., Martinez-Yamout M., De Guzman R.N., et al. (2002). Structural basis for Hif-1α/CBP recognition in the cellular hypoxic response. Proc. Natl Acad. Sci. USA 99, 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman R.N., Wojciak J.M., Martinez-Yamout M.A., et al. (2005). CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry 44, 490–497. [DOI] [PubMed] [Google Scholar]
- Demarest S.J., Martinez-Yamout M., Chung J., et al. (2002). Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415, 549–553. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z., Chen J., Dunker A.K., et al. (2006). Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 5, 2985–2995. [DOI] [PubMed] [Google Scholar]
- Dyson H.J., and Wright P.E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208. [DOI] [PubMed] [Google Scholar]
- Falk R., Falk A., Dyson M.R., et al. (2012). Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods 58, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J., and Babu M.M. (2009). The rules of disorder or why disorder rules. Prog. Biophys. Mol. Biol. 99, 94–103. [DOI] [PubMed] [Google Scholar]
- Guruprasad K., Reddy B.V., and Pandit M.W. (1990). Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4, 155–161. [DOI] [PubMed] [Google Scholar]
- Hartsough E.J., Meyer R.D., Chitalia V., et al. (2013). Lysine methylation promotes VEGFR-2 activation and angiogenesis. Sci. Signal. 6, ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeki N., Tukamoto T., Goto J., et al. (2000). Formic acid dissolves aggregates of an N-terminal huntingtin fragment containing an expanded polyglutamine tract: applying to quantification of protein components of the aggregates. Biochem. Biophys. Res. Commun. 277, 386–393. [DOI] [PubMed] [Google Scholar]
- Henderson A.M., Wang S.J., Taylor A.C., et al. (2001). The basic helix-loop-helix transcription factor HESR1 regulates endothelial cell tube formation. J. Biol. Chem. 276, 6169–6176. [DOI] [PubMed] [Google Scholar]
- Itoh F., Itoh S., Goumans M.J., et al. (2004). Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 23, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.G., Hu X., Li L., et al. (2002). Activated Notch4 inhibits angiogenesis: role of β1-integrin activation. Mol. Cell. Biol. 22, 2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Turkoz A., Jackson E.N., et al. (2011). Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J. Clin. Invest. 121, 800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.M., and Lux S.E. (1996). Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc. Natl Acad. Sci. USA 93, 5663–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J. (2008). Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics 24, 1798–1804. [DOI] [PubMed] [Google Scholar]
- Noseda M., Chang L., McLean G., et al. (2004). Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol. Cell. Biol. 24, 8813–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C., Watkins G., and Jiang W.G. (2004). The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int. J. Mol. Med. 14, 779–786. [DOI] [PubMed] [Google Scholar]
- Rahimi N. (2006. a). Vascular endothelial growth factor receptors: molecular mechanisms of activation and therapeutic potentials. Exp. Eye Res. 83, 1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi N. (2006. b). VEGFR-1 and VEGFR-2: two non-identical twins with a unique physiognomy. Front. Biosci. 11, 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi N. (2012). The ubiquitin-proteasome system meets angiogenesis. Mol. Cancer Ther. 11, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi N., Dayanir V., and Lashkari K. (2000). Receptor chimeras indicate that the vascular endothelial growth factor receptor-1 (VEGFR-1) modulates mitogenic activity of VEGFR-2 in endothelial cells. J. Biol. Chem. 275, 16986–16992. [DOI] [PubMed] [Google Scholar]
- Rahimi N., Rezazadeh K., Mahoney J.E., et al. (2012). Identification of IGPR-1 as a novel adhesion molecule involved in angiogenesis. Mol. Biol. Cell 23, 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche D.B., Buenavista M.T., Tetchner S.J., et al. (2011). The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 39, W171–W176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., et al. (1997). Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558. [DOI] [PubMed] [Google Scholar]
- Singh G.P., Ganapathi M., and Dash D. (2007). Role of intrinsic disorder in transient interactions of hub proteins. Proteins 66, 761–765. [DOI] [PubMed] [Google Scholar]
- Taylor K.L., Henderson A.M., and Hughes C.C. (2002). Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc. Res. 64, 372–383. [DOI] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. (2002). Cracking the folding code. Why do some proteins adopt partially folded conformations, whereas other don’t? FEBS Lett. 514, 181–183. [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Meyer R.D., Bondzie P.A., et al. (2016). IGPR-1 is required for endothelial cell–cell adhesion and barrier function. J. Mol. Biol. 428, 5019–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Bairoch A., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. [DOI] [PubMed] [Google Scholar]
- Williams C.K., Li J.L., Murga M., et al. (2006). Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood 107, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., and Dyson H.J. (1999). Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J. Mol. Biol. 293, 321–331. [DOI] [PubMed] [Google Scholar]
- Wright P.E., and Dyson H.J. (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.