Abstract
The casein kinase 1 (CK1) family of serine-threonine protein kinases are implicated in the regulation of many cellular processes, including the cell cycle, circadian rhythms, and Wnt and Hedgehog signalling. It is therefore critically important to understand how their activity is controlled in cells. Because these kinases exhibit constitutive activity in biochemical assays, it is likely that their activity in cells is controlled by subcellular localization, interactions with inhibitory proteins, targeted degradation, or combinations of these mechanisms. We identified members of the FAM83 family of proteins as partners of CK1 in cells. All eight members of the FAM83 family (FAM83A–H) interacted with the α and α-like isoforms of CK1; FAM83A, -B, -E, and -H also interacted with the δ and ε isoforms of CK1. We detected no interaction between any FAM83 member with the related CK1γ1, -2 and -3 isoforms. Each FAM83 protein exhibited a distinct pattern of subcellular distribution and colocalized with the CK1 isoform(s) to which it bound. The interaction of FAM83 proteins with CK1 isoforms was mediated by the conserved domain of unknown function 1669 (DUF1669) that characterises the FAM83 family. Mutations in FAM83 proteins that prevented them from binding to CK1 interfered with the proper subcellular localization of both the FAM83 proteins and their CK1 binding partners and interfered with the cellular functions of both families of proteins. Based on its function, we propose that DUF1669 be renamed the polypeptide anchor of CK1 (PACK1) domain.
Introduction
The eight members of the FAM83 family of proteins are conserved in vertebrates but are poorly characterised. They share a conserved N-terminal DUF1669 (domain of unknown function 1669) domain of ~300 amino acids, but each member possesses unique C-terminus of variable length (1, 2). The amino acid sequences of the FAM83 family members offer very few clues to their functions. The DUF1669 domain contains a putative phospholipase D–like (PLD-like) catalytic motif, which is characterized by the presence of an HxKxxxxD (HKD) sequence motif. Typically, two such motifs exist within each PLD protein, with the two HKD motifs coming together to form the catalytic core of the enzyme (3). FAM83 proteins, on the other hand, have only one HKD motif, and the histidine residue within the motif is absent from all but FAM83D (also known as CHICA) (fig. S1). No PLD activity has yet been demonstrated for any FAM83 member (4). Recent studies have implicated FAM83A and FAM83B in oncogenesis and resistance to tyrosine kinase inhibitors (4–6). FAM83D has been reported to localize to the mitotic spindle and interact with the chromokinesin kinesin family member 22 (KIF22, also called Kid), the microtubule-binding protein hyaluronan-mediated motility receptor (HMMR), and the light chain of the motor protein dynein (DYNLL1) to correctly orient the metaphase plate in mitosis (7, 8). FAM83G, also known as PAWS1 [protein associated with suppressor of mothers against decapentaplegic 1 (SMAD1)] interacts with the transcription factor SMAD1 and promotes the transcription of non-canonical bone morphogenetic protein (BMP) target genes (9). FAM83H mutations have been reported in both familial and spontaneous cases of amelogenesis imperfecta (AI), a genetic dental condition associated with soft enamel due to defective tooth mineralization (10–12). No functions have yet been reported for FAM83C, FAM83E, or FAM83F. Despite the increasing evidence that FAM83 proteins are involved in diverse biological processes, the precise molecular and biochemical roles of the FAM83 proteins, and in particular the DUF1669 domain that characterises them, remain undefined.
By taking a comprehensive proteomic approach to uncover potential roles of the FAM83 family and the DUF1669 domain, we identified many unique interactors of each of the FAM83 proteins, consistent with the diverse sequence composition of these related proteins. Nevertheless, the α, α-like, δ, and ε isoforms of casein kinase 1 (CK1) were identified as interacting with each of the FAM83 members, albeit with different affinities and specificities. CK1 enzymes in vertebrates include the α, α-like, δ, ε, γ1, γ2, and γ3 isoforms, all of which are serine-threonine protein kinases. CK1 isoforms consist of a highly conserved N-terminal kinase domain that has little homology outside this family (13, 14). Within the CK1 family, there is greater overall sequence homology between the α and α-like isoforms, between the δ and ε isoforms, and between the γ1, γ2, and γ3 isoforms (13, 14). CK1 isoforms play fundamental roles in many aspects of cellular homeostasis, including cell cycle progression (15), circadian rhythm (16–18), survival (19, 20), DNA damage repair (21), membrane trafficking, and integration of signalling processes (13–15). Increased catalytic activity of CK1 isoforms has been linked to cancer (14) and neurological pathologies (22). Due to their spontaneous in vitro kinase activity towards many substrates, CK1 isoforms are considered to be constitutively active kinases in cells (13). Consistent with the large number of cellular processes influenced by CK1 isoforms, they have been reported to localize to many subcellular compartments, including the plasma membrane, cytoplasm, nucleus, actin cytoskeleton, and mitotic spindle, and hundreds of putative substrates have been described (13, 15, 23). Although CK1 isoforms preferentially phosphorylate serine and threonine residues within the consensus sequence pS/pT-X-X-S/T, in many cases CK1 isoforms phosphorylate residues outside the context of the consensus motif, such as the phosphorylation of β-catenin on Ser45 (13). In some cases, acidic residues can substitute for the phosphoserine or phosphothreonine residues within the consensus motif (13). All of these studies indicate that the localization, activity, and substrate specificity of CK1 are tightly regulated in cells.
Interacting proteins that potentially control the subcellular localization, substrate accessibility, stability, or activity of CK1 isoforms remain elusive. Two scaffold proteins, the centrosomal and golgi N-kinase anchoring protein (CG-NAP, also known as AKAP450) and the DEAD-box RNA helicase DDX3, have been implicated in the centrosomal localization of CK1δ during the cell cycle and in Wnt-dependent phosphorylation of Dishevelled (DVL) by CK1ε, respectively (24, 25). The potential existence of CK1 scaffolds in cells is supported by an analogous role for the A-kinase anchoring proteins (AKAPs), which are established scaffolds that control the activity and substrate specificity of protein kinase A (PKA; also known as cAMP-dependent kinase) by interacting with PKA and tethering it to distinct subcellular compartments (26).
Our data suggest that the DUF1669 domain of the FAM83 family mediates the interaction of these proteins with CK1 isoforms. FAM83 members localized to different subcellular compartments and co-localized in cells only with the CK1 isoforms with which they interacted in vitro. Mutations within the DUF1669 domain that abolish the interaction with CK1 interfered with the localization of both the FAM83 members themselves as well as their CK1 binding partners. We hypothesise that FAM83 members, through their association with CK1 isoforms, restrict the function of CK1 enzymes in cells by directly controlling their subcellular localization, and perhaps their activity, stability, or substrate specificity.
Results
The FAM83 members interact with CK1 isoforms
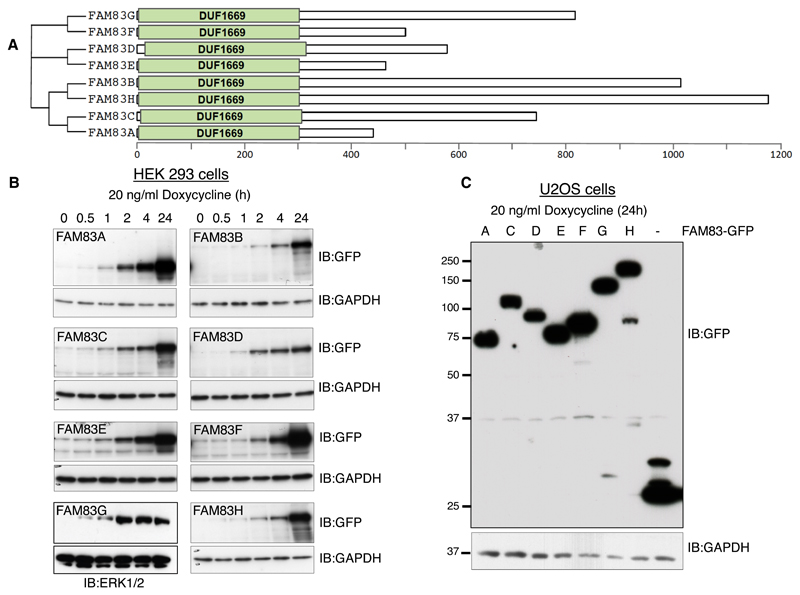
The FAM83 family of proteins is characterized by a conserved domain of unknown function, termed DUF1669, that is present at their N-termini, whereas the rest of the proteins vary in length and are not conserved between members (Fig. 1A; fig. S1). In order to investigate the roles of the FAM83 family of proteins, we generated transgenic human embryonic kidney HEK 293 cell lines each stably expressing a single copy of a FAM83 gene under the control of a tetracycline (Tet)-inducible promoter. All 8 of the transgenically-expressed FAM83 proteins were tagged at the N-terminus with green fluorescent protein (GFP). In these cell lines, doxycycline treatment induced the expression of the respective FAM83 protein in a time-dependent manner, with detectable amounts observed as early as 30 min after doxycycline treatment (Fig. 1B). Similarly, we generated U2OS osteosarcoma cells stably integrated with a single copy of each FAM83 gene tagged at the C-terminus with green fluorescent protein (GFP). All FAM83 proteins, except FAM83B, displayed robust expression following 24 h treatment with doxycycline (Fig. 1C). For both sets of cell lines (HEK 293 and U2OS), cells stably integrated with GFP alone under the Tet-inducible promoter were used as controls.
Fig. 1. Generation of HEK 293 and U2OS cells for tetracycline-inducible expression of FAM83 proteins.
A. Schematic representation of the human FAM83 family of proteins and the conserved domain of unknown function, DUF1669, that characterises them. B. A single copy of each FAM83 gene (A–H) tagged with GFP at the N-terminus was stably inserted downstream of a tetracycline-inducible promoter in HEK 293 cells. Cells were treated with doxycycline and lysed at the indicated times after treatment. Extracts were resolved by SDS-PAGE and subjected to immunoblotting (IB) for GFP. ERK1 and ERK2 (ERK1/2) and GAPDH are loading controls. This blot is representative of 2 independent experiments. C. A single copy of each FAM83 gene (A–H) tagged with GFP at the C-terminus was stably inserted downstream of a tetracycline-inducible promoter in U2OS cells. Extracts of doxycycline-induced cells were immunoblotted for GFP and the loading control GAPDH. FAM83B is not included on the blot because we were unable to detect FAM83B-GFP expression in U2OS cells. This blot is representative of 2 independent experiments.
Following induction with doxycycline, extracts from HEK 293 and U2OS cells expressing the GFP control and the FAM83 proteins tagged with GFP at either the N-terminus (HEK 293) or C-terminus (U2OS) were subjected to GFP immunoprecipitation and separation of the immunoprecipitated proteins by SDS-PAGE (fig. S2, A and B). The gel sections including the entire lane for each sample were excised and digested with trypsin (fig. S2, A and B). The resulting peptides were identified using mass spectrometry. In addition to confirming the identity of the respective FAM83 proteins in each lane, we identified SMAD isoforms in GFP-FAM83G immunoprecipitates (2) and DYNLL1 and HMMR in GFP-FAM83D immunoprecipitates (7, 8) (fig S2, A and B), consistent with previously reported observations of these protein-protein interactions. Under these conditions, at least one or more of the α, α-like, δ, and ε isoforms of CK1 were identified as interactors for every FAM83 family member, regardless of the positioning of the GFP tag and the cell line in which the fusion protein was expressed (Fig. 2A). Analysis of the top three precursor ion intensities of the individual CK1 isoforms bound to each GFP-FAM83 protein from HEK 293 extracts revealed that although all FAM83 members interacted with CK1α and α-like, only FAM83A, FAM83B, FAM83E, and FAM83H interacted with CK1δ and CK1ε (Fig. 2A). Similar patterns in spectral intensities were observed for CK1α, CK1δ, and CK1ε bound to FAM83-GFP proteins from U2OS cell extracts, whereas CK1α-like was not detected in FAM83C, FAM83D, and FAM83H immunoprecipitations (Fig. 2A). Although we observed differences in spectral intensities for each CK1 isoform associated with different FAM83 members, it is difficult to interpret these differences because the relative amount of FAM83 protein in each lane was quite different as judged by the intensity of Instant blue stains (fig S2, A and B).
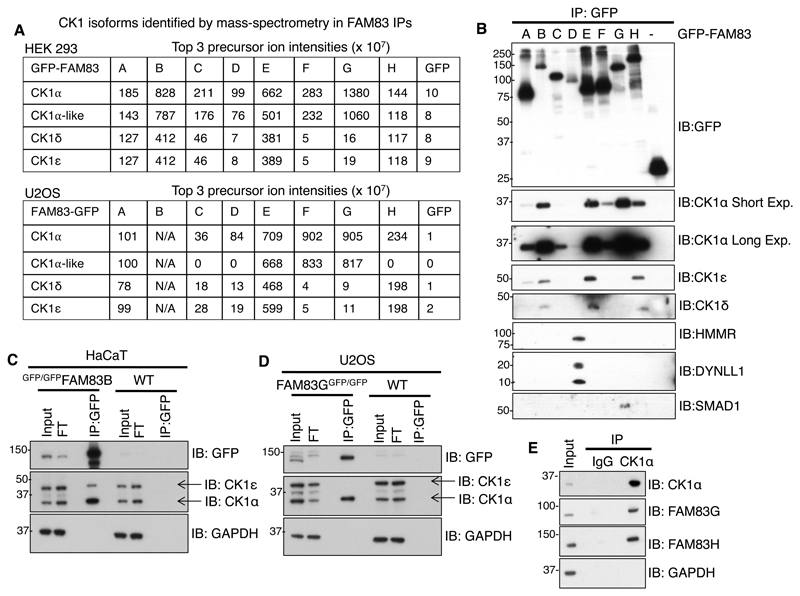
Fig. 2. FAM83 proteins interact with CK1 isoforms.
A. Mass fingerprinting of protein interactors of FAM83A–H proteins tagged N-terminally (HEK 293 cells) or C-terminally (U2OS cells) with GFP (fig. S2, A and B) identified one or more CK1 isoforms. These tables show the values for the top 3 precursor ion intensities of the indicated CK1 isoforms pulled down with each GFP-FAM83 protein (A–H) expressed in HEK 293 cells and each FAM83-GFP protein (A–H) expressed in U2OS cells. GFP expressed in each cell line was a negative control. Scaffold Q/Q+S V4.4.6 was used for analysis of the LC-MS/MS data from HEK 293 cells, and scaffold V4.3 was used for analysis of the LC-MS-MS data from U2OS cells. FAM83B-GFP did not express in U2OS cells. B. GFP Immunoprecipitates (IP) of GFP control or GFP-FAM83A–H proteins expressed in HEK 293 cells were immunoblotted (IB) with antibodies recognizing the indicated CK1 isoforms and other proteins known to interact with FAM83 family proteins. Short Exp., short exposure; Long Exp., long exposure. C. Extracts of wild-type (WT) or GFP-FAM83B knockin (GFP/GFPFAM83B) HaCaT cells were immunoprecipitated with GFP-TRAP A beads and immunoblotted to detect the indicated CK1 isoforms. GAPDH was used as a loading control. FT, flow through. D. As in C., except that proteins were immunoprecipitated from WT and FAM83G-GFP knockin (FAM83GGFP/GFP) U2OS cell extracts. E. U2OS extracts were immunoprecipitated using either pre-immune IgG or an antibody recognizing CK1α coupled to Protein-G sepharose beads and immunoblotted with antibodies recognizing the indicated FAM83 proteins and GAPDH. All blots are representative of 3 independent experiments.
To verify the interactions between FAM83 members and CK1 isoforms, GFP-FAM83A–H or control GFP immunoprecipitates from HEK 293 extracts were probed for co-precipitation of endogenous CK1α, δ, and ε isoforms. The relative amounts of FAM83 proteins in immunoprecipitates varied in that the amounts of FAM83B and FAM83D, both in extracts and immunoprecipitates, were lower as compared to other FAM83 members (Fig. 2B; fig. S3A). Under these conditions and in agreement with the mass spectrometry data above, all GFP-FAM83 proteins interacted with CK1α, whereas GFP alone did not (Fig. 2B). We observed that FAM83B, FAM83E, FAM83G, and FAM83H appeared to interact more strongly with CK1α than did FAM83A, FAM83C, FAM83D, and FAM83F (Fig. 2B). Endogenous CK1δ and ε were mainly detected in FAM83B, FAM83E, and FAM83H immunoprecipitates, although CK1ε was also observed in FAM83A immunoprecipitates (Fig. 2B). In line with the proteomic data, endogenous SMAD1 co-precipitated with only FAM83G, whereas endogenous HMMR and DYNLL1 co-precipitated exclusively with FAM83D (Fig. 2B).
We next sought to verify endogenous interactions between some FAM83 members and the α and ε isoforms of CK1. Given the absence of robust immunoprecipitating antibodies recognizing FAM83 members, we exploited CRISPR/Cas9 genome editing technology to introduce GFP-tag knock-ins at the FAM83B and FAM83G loci in HaCaT keratinocytes and U2OS cells respectively. GFP tags were thus inserted into the N-terminus of endogenous FAM83B and into the C-terminus of endogenous FAM83G. The disappearance of the endogenous FAM83B and FAM83G signals at the predicted molecular weights upon GFP-tag knock-ins and their concomitant appearance at higher molecular weights equivalent to the addition of GFP, in combination with genomic DNA sequencing, confirmed the insertion of the GFP tag at the appropriate loci (fig. S3B). Whereas we detected endogenous CK1α in GFP immunoprecipitates from extracts of cells expressing GFP-FAM83B and from cells expressing FAM83G-GFP, we detected CK1ε only in immunoprecipitates from cells expressing GFP-FAM83B (Fig. 2, C and D). We detected neither the α nor ε isoform of CK1 in GFP immunoprecipitates from wild-type cells (Fig. 2, C and D). Furthermore, we observed endogenous FAM83G and FAM83H proteins in CK1α immunoprecipitates from U2OS cell extracts but not in pre-immune IgG control immunoprecipitates (Fig. 2E). The CK1 branch of the human protein kinase family also includes γ1, γ2, and γ3 isoforms of CK1, tau-tubulin kinase (TTBK1), TTBK2, vaccinia-related kinase 1 (VRK1), VRK2, and VRK3 (27). In U2OS cell extracts, under co-expression conditions in which FAM83G interacted with CK1α, we were unable to detect interactions between FAM83G and either TTBK2 or CK1γ (fig. S4A). This, together with the proteomic data, suggests that FAM83 members interact only with one or more of the α, α-like, δ, and ε isoforms of CK1 but not with CK1γ1, CK1γ2, CK1γ3, or other members of the CK1 family.
The DUF1669 domain is sufficient to mediate the interaction of FAM83 proteins with CK1
Because all eight FAM83 family members contain the DUF1669 domain (Fig. 1A; fig S1), we postulated that this domain might mediate the observed interaction between FAM83 and CK1 proteins. To map the minimal domain within FAM83 proteins that can interact with CK1 isoforms, we co-expressed Myc-tagged FAM83G fragments with full-length hemagglutinin (HA)-tagged CK1α in FAM83G-/- U2OS cells (28) and performed coimmunoprecipitation experiments (Fig. 3A). HA-CK1α immunoprecipitates only included those FAM83G fragments that contained residues 165-307 within the DUF1669 domain (Fig. 3A). We asked whether the interaction between the DUF1669 domain and CK1 was direct by performing an in vitro binding assay with purified recombinant proteins: a His-tagged form of residues 124–304 of FAM83A [6xHis-FAM83A(124-304)] and the kinase domain of CK1ε. Following precipitation of His-FAM83A (124-304) with nickel resin and its elution using imidazole, we observed both CK1ε and FAM83A (124-304) in the eluate, suggesting a robust and direct interaction between the two (Fig. 3B). To probe the CK1 isoform–specific nature of the interactions of FAM83 members, we replaced the DUF1669 domain of FAM83G, which interacts only with CK1α, with that of FAM83H, which interacts with both CK1α and CK1ε. We expressed this chimeric protein (DUF1669H-FAM83G) in HEK 293 cells and tested whether they interacted with CK1α and CK1ε in cell extracts. The DUF1669H-FAM83G chimera interacted with both CK1α and CK1ε, much like FAM83H (fig. S5), suggesting that the DUF1669 domain of FAM83H is sufficient to confer selectivity for specific CK1 isoforms.
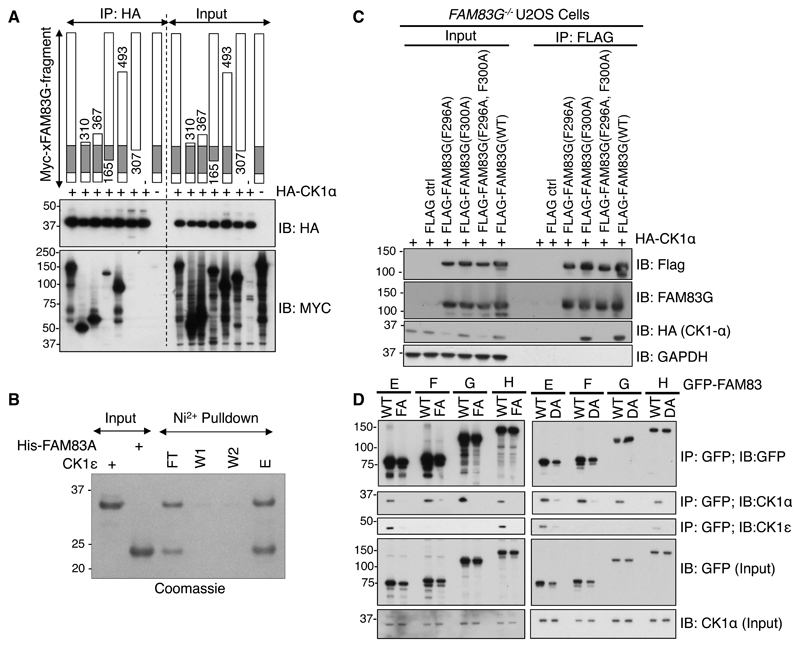
Fig. 3. The DUF1669 domain is sufficient to mediate the interaction of FAM83 proteins with CK1.
A. The indicated fragments of Myc-tagged Xenopus laevis FAM83G (Myc-xFAM83G) were co-expressed with HA-CK1α in FAM83G-/- U2OS cells, and then cell extracts or HA immunoprecipitates were subjected to immunoblotting (IB) with antibodies recognizing Myc or HA as indicated. This blot is representative of 3 independent experiments. B. A His-tagged fragment of FAM83A (amino acids122-304), which contains the DUF1669 and PLD-like domains, was mixed with recombinant CK1ε kinase domain (amino acids 1-294) in vitro. His-FAM83A(122-304) was then pulled down using Ni-sepharose (Ni2+) resin, which was washed twice before elution. The input, unbound flow-through (FT), wash solutions (W1 and W2), and eluate (E) were analysed by SDS-PAGE and stained with Coomassie blue. This gel is representative of 3 independent experiments. C. Empty Flag vector (ctrl) or the indicated FLAG-FAM83G mutant and wild-type (WT) proteins were overexpressed in FAM83G-/- U2OS cells. Cell extracts (input) and FLAG immunoprecipitates (IP) were subjected to immunoblotting for FLAG, CK1α, or GAPDH as indicated. This blot is representative of 3 independent experiments. D. WT and Phe→Ala (FA) and Asp→Ala (DA) mutant forms of GFP-FAM83E–H were transiently expressed in U2OS cells, immunoprecipitated (IP) from cell extracts with a GFP-specific antibody, and immunoblotted for GFP, CK1α, and CK1ε as indicated. This blot is representative of 3 independent experiments.
A CK1 docking motif that includes the amino acid sequence F-X-X-X-F was identified in nuclear factor of activated T cells 1 (NFAT1), Period 1 (PER1), and PER2, and mutation of either phenylalanine residue abolished CK1 interactions with these proteins (29). One such F-X-X-X-F motif is conserved within the DUF1669 domain of the FAM83A–G proteins, and FAM83H has four such motifs (30) (fig. S1). To determine whether mutations within this conserved motif were sufficient to disrupt the CK1 interaction, we tested the ability of FLAG-tagged wild-type and various mutant forms of FAM83G (FAM83GF296A, FAM83GF300A, and FAM83GF296A,F300A) to interact with HA-CK1 when coexpressed in FAM83G-/- U2OS cells. Whereas wild-type FAM83G interacted robustly with CK1α, the FAM83GF296A and FAM83GF296A,F300A mutants did not (Fig. 3C). Rather surprisingly, FAM83GF300A interacted with CK1α as robustly as did wild-type FAM83G (Fig. 3C), suggesting that the mode through which CK1 interacts with FAM83 proteins might differ from that through which it interacts with NFAT1, PER1, and PER2, which requires both phenylalanine residues (29). Consistent with this notion, mutational scanning of conserved residues within the 165–307 region of FAM83G uncovered another mutation, D262A, that also abolished the interaction with CK1α (31).
Armed with the knowledge that the D262A and F296A mutations both abolish the interaction of FAM83G with CK1α, we asked whether equivalent mutations in other FAM83 members also abolished their association with CK1α and CK1ε isoforms. We mutated the residues equivalent to FAM83G Asp262 and Phe296 in FAM83E (Asp243 and Phe277), FAM83F (Asp250and Phe284), and FAM83H (Asp236 and Phe270) to Ala. These substitutions are referred to as DA and FA, respectively. We individually expressed wild-type GFP-FAM83E–H, the DA mutants (GFP-FAM83E–HDA), and the FA mutants (GFP-FAM83E–HFA) in U2OS cells and tested their ability to coimmunoprecipitate endogenous CK1α or CK1ε isoforms. In comparison to wild-type FAM83E–H, both the DA and FA mutations attenuated the interaction of FAM83 proteins with CK1α and CK1ε isoforms (Fig. 3D). These observations suggest that the interaction between the DUF1669 domain and CK1 isoforms may be mediated through a conserved structural motif surrounding the residues equivalent to Asp262 and Phe296 in FAM83G. Consistent with previous observations (Fig. 2, B to D), although FAM83E and FAM83H bound to both CK1α and CK1ε, FAM83F and FAM83G bound only to CK1α (Fig. 3D).
FAM83 proteins and CK1α colocalize in cells
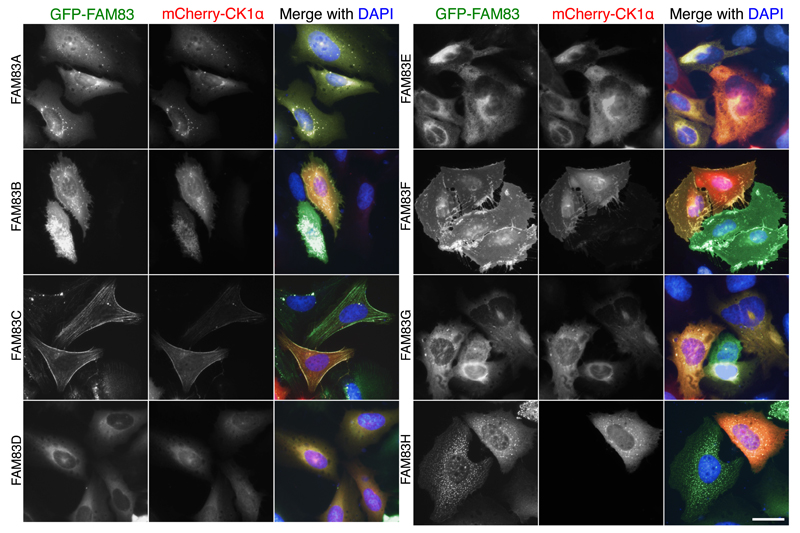
Given the interaction between all of the FAM83 members and CK1α in cell extracts, we sought to investigate whether FAM83 proteins also interact with CK1α in cells. We coexpressed mCherry-CK1α and an N-terminally GFP-tagged FAM83 family member (GFP-FAM83) under the control of a Tet-inducible promoter in U2OS cells and evaluated the localization of both proteins by fluorescence microscopy. We performed this experiment with each FAM83 family member (GFP-FAM83A–H). Upon induction of GFP-FAM83 expression, we observed overlapping colocalization of every GFP-FAM83 member with mCherry-CK1α (Fig. 4), with each FAM83 protein exhibiting a distinct pattern of subcellular localization. Pan-cellular staining was observed for both GFP-FAM83A and GFP-FAM83B, along with additional perinuclear punctate structures for FAM83A and membrane punctate structures for FAM83B (Fig. 4). GFP-FAM83C displayed distinct fibrous patterns of fluorescence in the cytoplasm and in the vicinity of membrane ruffles, suggesting possible colocalization with cortical actin stress fibres (Fig. 4). GFP-FAM83D displayed cytoplasmic staining, with some punctate staining in the nucleus (Fig. 4). FAM83D had previously been reported to localize to the spindle apparatus during mitosis (7, 8). GFP-FAM83E exhibited cytoplasmic and strong perinuclear staining (Fig. 4). GFP-FAM83F localized to the plasma membrane, with some staining also observed in the cytoplasm and nucleus (Fig. 4). As reported previously (2), GFP-FAM83G localized mainly to the cytoplasm, but some nuclear staining was also noted (Fig. 4). GFP-FAM83H displayed primarily cytoplasmic, and few nuclear, punctate fluorescence patterns (Fig. 4), like the patterns described previously for FLAG-FAM83H overexpressed in HCT116 cells (11). In cells expressing both mCherry-CK1α and the GFP tag (not fused to a FAM83 protein), the GFP signal was predominantly nuclear and did not overlap with the mCherry signal, which was present throughout the cell (fig. S6). When expressed alone, mCherry-CK1α displayed a pan-cellular staining pattern (fig. S6). These observations describe the subcellular localization profiles for all FAM83 members and demonstrate that each member colocalizes with CK1α.
Fig. 4. FAM83 proteins and CK1α colocalize in cells.
U2OS cells stably integrated with Tet-inducible expression of GFP-FAM83A–H were transfected with mCherry-CK1α. Cells were processed for fluorescence microscopy following 24h of doxycycline treatment. DNA was stained with DAPI. Images from one field of view representative of 3 independent experiments are shown. The number of cells that displayed staining patterns identical to the representative image were documented for each experiment: GFP-FAM83A (n=50); GFP-FAM83B (n=31); GFP-FAM83C (n=37); GFP-FAM83D (n=32); GFP-FAM83E (n=55); GFP-FAM83F (n=44); GFP-FAM83G (n=43); GFP-FAM83H (n=32). Fluorescence images for GFP-alone and mCherry-CK1α alone expressing cells are included in fig. S6. Scale bar, 20 µM.
To confirm whether endogenous CK1α also displayed similar overlapping subcellular distribution with FAM83 members, we examined the subcellular localization pattern of endogenous CK1α in U2OS cells stably expressing GFP, GFP-FAM83B, GFP-FAM83F, or GFP-FAM83H. No overlapping fluorescence was detected between endogenous CK1α and GFP, which was employed as a negative control (Fig. 5). Overlapping plasma membrane and perinuclear staining was observed for endogenous CK1α and GFP-FAM83B (Fig. 5). Likewise, strong overlapping plasma membrane staining was observed for endogenous CK1α and GFP-FAM83F (Fig. 5). GFP-FAM83H and endogenous CK1α displayed overlapping staining in cytoplasmic and nuclear speckles (Fig. 5). Collectively these observations demonstrate that upon overexpression, each FAM83 protein is capable of relocating endogenous CK1α to the distinct subcellular compartments in which they reside.
Fig. 5. FAM83 proteins colocalize with endogenous CK1α in cells.
U2OS cells stably integrated with Tet-inducible expression of GFP, GFP-FAM83B, GFP-FAM83F, or GFP-FAM83H were treated with doxycycline for 16 h prior to processing cells for fluorescence microscopy to detect GFP and endogenous CK1α (anti- CK1α). DNA was stained with DAPI. Images from one field of view representative of three independent experiments are shown. The number of cells displaying staining patterns identical to the representative image were documented for each experiment: GFP-FAM83B (n=56); GFP-FAM83F (n=60); GFP-FAM83H (n=48); GFP only (n=38); no transgene (n=82). Scale bars, 20 µm.
The association between FAM83 proteins and specific CK1 isoforms is selective in cells
The above data demonstrated that all eight FAM83 members (A–H) interacted and colocalized with both overexpressed and endogenous CK1α and that FAM83A, FAM83B, FAM83E, and FAM83H also interacted with the CK1δ and CK1ε isoforms in cell extracts. To determine whether the specificity of FAM83 proteins for binding to a specific subset of CK1 isoforms applied to cells as well as to cell extracts, we compared the subcellular distribution of CK1α and CK1ε with that of FAM83F, which interacted selectively with CK1α in cell extracts, and FAM83H, which interacted with both CK1α and CK1ε in cell extracts. GFP-FAM83F or GFP-FAM83H was coexpressed in U2OS cells with either mCherry-CK1α or mCherry-CK1ε. As observed earlier (Fig. 4 and 5), we found that both GFP-FAM83F and GFP-FAM83H displayed overlapping fluorescence with mCherry-CK1α, at the plasma membrane and in cytoplasmic speckles, respectively (Fig. 6A). In contrast, GFP-FAM83H, but not GFP-FAM83F, also colocalized with mCherry-CK1ε at these sites (Fig. 6A). Next, we sought to verify whether the binding specificity of GFP-FAM83F and GFP-FAM83H extended to endogenous CK1α and CK1ε. In wild-type U2OS cells, endogenous CK1α and CK1ε both displayed pan-cellular distributions, with some speckle-like structures also visible in the cytoplasm (Fig. 6B). In U2OS cells expressing GFP-FAM83F, we observed overlapping plasma membrane colocalization only with endogenous CK1α but not with CK1ε (Fig. 6B). In contrast, in U2OS cells expressing GFP-FAM83H, we observed overlapping cytoplasmic and nuclear speckle staining patterns with both endogenous CK1α and CK1ε (Fig. 6B). These data are consistent with our observations using overexpressed mCherry-CK1α and mCherry-CK1ε (Fig. 6A). Collectively, these data recapitulate in cells the distinct sets of interactions that we observed between FAM83 members and CK1 isoforms from the proteomic data (Fig. 2A).
Fig. 6. The association between FAM83 proteins and specific CK1 isoforms is selective in cells.
A. U2OS cells stably integrated with Tet-inducible expression of GFP-FAM83F or GFP-FAM83H were transfected with either mCherry-CK1α (α) or mCherry-CK1ε (ε). GFP-FAM83F and GFP-FAM83H expression was induced with doxycycline for 24 h prior to processing cells for fluorescence microscopy. DNA was stained with DAPI. Images from one field of view representative of three independent experiments are included. The number of cells that displayed staining patterns identical to the representative image were documented for each experiment: GFP-FAM83F + mCherry-CK1α (n=44); GFP-FAM83F + mCherry-CK1ε (n=40); GFP-FAM83H + mcherry-CK1α (n=32); GFP-FAM83H + mCherry-CK1ε (n=40). Scale bar, 20 µm. B. U2OS cells stably integrated with Tet-inducible expression of GFP-FAM83F or GFP-FAM83H were induced with doxycycline for 16 h prior to processing cells for fluorescence microscopy with CK1α (α) or CK1ε (ε) antibodies. Untransfected cells stained with CK1α or CK1ε antibodies were used as negative controls. Images from one field of view representative of three independent experiments are shown. The number of cells that displayed staining patterns identical to the representative image were documented for each experiment: GFP-FAM83F with CK1α (n=60); GFP-FAM83F with CK1ε (n=43); GFP-FAM83H with CK1α (n=48); GFP-FAM83H with CK1ε (n=35); no transgene with CK1α (n=82); no transgene with CK1ε (n=27). Scale bars, 20 µm.
Association with CK1 determines the subcellular localization of FAM83C
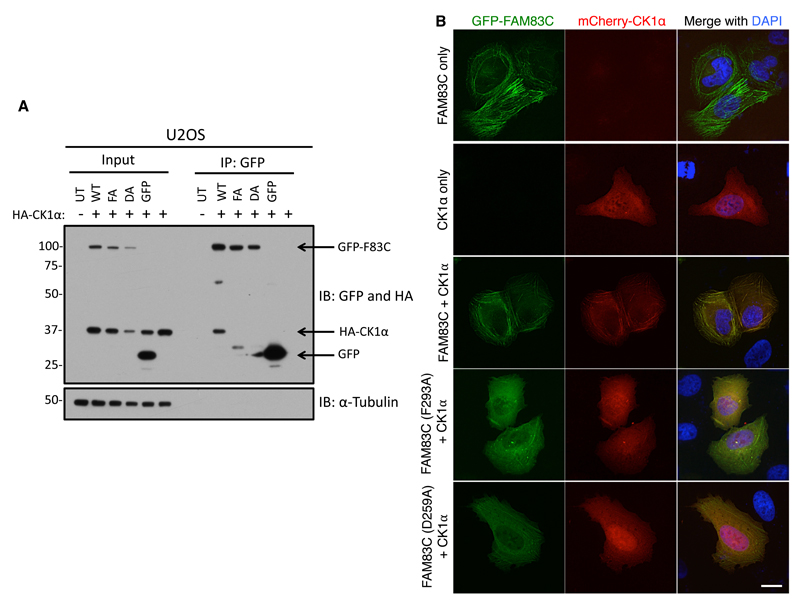
We next asked whether the interaction between CK1 and FAM83 proteins was important for their subcellular localizations. For this purpose, we chose GFP-FAM83C because of its distinct cortical fibre-like subcellular localization pattern (Fig. 4). First, we confirmed in cell extracts that only wild-type GFP-FAM83C coimmunoprecipitated HA-tagged CK1α and that the GFP-FAM83CD259A and GFP-FAM83CF293A mutants and control GFP did not (Fig. 7A). Next, we co-transfected U2OS cells with mCherry-CK1α and wild-type GFP-FAM83C, GFP-FAM83CD259A, or GFP-FAM83CF293A and examined their subcellular localization by fluorescence microscopy. Whereas GFP-FAM83C and mCherry-CK1α fluorescence colocalized along fibrous structures (Fig. 4 and 7B), GFP-FAM83CD259A and GFP-FAM83CF293A were predominantly found in the cytoplasm, in slightly distorted fibrous fluorescence patterns that did not overlap with mCherry-CK1α fluorescence (Fig. 7B). When it was expressed alone or with the FAM83CD259A and GFP-FAM83CF293A mutants, mCherry-CK1α was found in a diffuse cytoplasmic pattern, but adopted a fibrous appearance in the cytoplasm when it was co-expressed with wild-type FAM83C (Fig. 7B). These observations suggest that the interaction between FAM83C and CK1α determines the subcellular localization of both proteins.
Fig. 7. Association with CK1 determines the subcellular localization of FAM83C.
A. U2OS cells were cotransfected with plasmids encoding either GFP, GFP-FAM83C (WT), GFP-FAM83C(F293A) (FA), or GFP-FAM83C(D259A) (DA) plus a plasmid encoding HA-CK1α. Untransfected (UT) cells and cells transfected only with HA-CK1α were included as controls. Cell extracts (Input) or GFP-TRAP A Immunoprecipitates (IP) were immunoblotted (IB) with antibodies recognizing GFP and CK1α. α-Tubulin was used as a loading control. This blot is representative of 3 independent experiments. B. U2OS cells were transfected with plasmids encoding GFP-FAM83C, GFP-FAM83C(F293A), or GFP-FAM83C(D259A), together with mCherry-CK1α. Cells expressing GFP-FAM83C or mCherry-CK1α alone are negative controls. Cells were processed 24 h after transfection for fluorescence microscopy. DNA was stained with DAPI. Images from one field of view representative of three independent experiments are shown. The number of cells which displayed staining patterns identical to the representative image were documented for each experiment: GFP-FAM83C only (n=46); GFP-FAM83C + mCherry-CK1α (n=44); GFP-FAM83C (F293A) + mCherry-CK1α (n=41); GFP-FAM83C (D259A) + mCherry-CK1α (n=43); mCherry-CK1α only (n=45). Scale bar, 20 µm.
FAM83H co-localizes with and contributes to the subcellular localization of endogenous CK1α and CK1ε isoforms
We have shown that FAM83H displays a distinct punctate fluorescence pattern when overexpressed in U2OS cells (Fig. 4 to 6). We generated FAM83H-/- U2OS cells using CRISPR/Cas9 genome editing technology and asked whether endogenous CK1α colocalized with GFP-FAM83H that was transgenically expressed in these cells. In FAM83H-/- cells, CK1α was primarily cytoplasmic with few perinuclear puncta. When GFP-FAM83H was expressed transgenically, both GFP-FAM83H and endogenous CK1α adopted a pan-cellular punctate pattern (Fig. 8A). Although the majority of GFP-FAM83H puncta overlapped with endogenous CK1α staining, suggesting robust co-localization, the presence of some non-overlapping GFP-FAM83H and CK1α puncta suggest that FAM83H and CK1α may exist in complexes with other proteins (Fig. 8A). When the CK1-interaction deficient mutants, GFP-FAM83HD236A and GFP-FAM83HF270A, were expressed in FAM83H-/- U2OS cells, they displayed cytoplasmic, non-punctate fluorescence that did not overlap with endogenous CK1α (Fig. 8A). The intensity of CK1α punctate staining in FAM83H-/- cells and in FAM83H-/- cells expressing CK1-binding–deficient mutant forms of FAM83H was lower compared to that seen in cells expressing wild-type GFP-FAM83H (Fig. 8A), suggesting that the interaction with FAM83H determines the localization of CK1α to the punctate structures. Next, we quantified the co-localization correlation between CK1α and wild-type GFP-FAM83H, GFP-FAM83HD236A or GFP-FAM83HF270A. The localization of CK1α positively correlated with that of wild-type GFP-FAM83H (Pearson correlation coefficient 0.7523), whereas it did not correlate with the GFP-FAM83HD236A (0.001504) or GFP-FAM83HF270A (0.001504) mutants (Fig. 8B). Rescue of FAM83H-/- U2OS cells with wild-type or mutant GFP-FAM83H constructs was confirmed by Western blotting and suggested that the abundance of FAM83H in these cells was substantially higher than the amount of endogenous FAM83H in wild-type U2OS cells (Fig. 8C). In FAM83H-/- U2OS cells, endogenous CK1ε displayed similar immunostaining patterns to CK1α (fig. S7A) and displayed significant co-localization correlation with wild-type GFP-FAM83H but not the GFP-FAM83HD236A or GFP-FAM83HF270A mutants (fig. S7B). Overlapping fluorescence observed between CK1α immunostaining and mCherry-CK1α fluorescence (fig. S8A), and between CK1ε immunostaining and mCherry-CK1ε fluorescence (fig. S8B), confirmed the selectivity of the antibodies recognizing CK1α and CK1ε, respectively, for immunofluorescence applications.
Fig. 8. FAM83H colocalizes with and contributes to the subcellular localization of endogenous CK1α.
A. FAM83H-/- U2OS cells were transfected with plasmids encoding either GFP-FAM83H, GFP-FAM83H (D236A), or GFP-FAM83H (F270A). Untransfected knockout (FAM83H-/-) cells were used as controls. Cells were processed for fluorescence microscopy with antibody recognizing CK1α. DNA was stained with DAPI. Images from one field of view representative of 3 independent experiments are included. Scale bar, 10 µm. B. The boxplot shows the range, mean, and lower and upper quartiles of the Pearson’s correlation coefficients of GFP-FAM83H and endogenous CK1α intensities within above-background pixels in the cytoplasm. C. GFP-FAM83H constructs were transfected into FAM83H-/- U2OS cells, and extracts were immunoblotted (IB) with the indicated antibodies. Untransfected wild-type (WT) cells were used as controls. This blot is representative of three independent experiments.
The intrinsic catalytic activity of CK1 is not affected by or required for the association of CK1 with FAM83 proteins
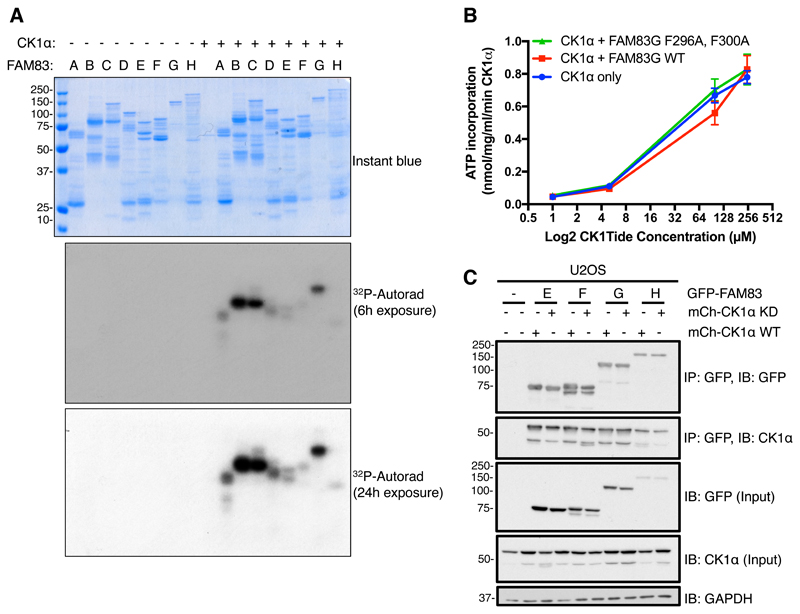
Because they associate with CK1 isoforms, it is possible that the FAM83 members could be substrates of CK1 or affect the intrinsic kinase activity of CK1. We tested whether CK1α phosphorylated FAM83 proteins in vitro using purified proteins. Whereas recombinant FAM83B, FAM83C, and FAM83G were robustly phosphorylated by CK1α, the other FAM83 members were phosphorylated poorly (Fig. 9A). The precise CK1 phosphorylation sites on most FAM83 proteins have not been mapped, and whether these phosphorylation events occur in cells and their potential functional consequences have not been investigated. The low activity of CK1α toward some of the FAM83 substrates in our in vitro kinase assay could reflect the poor purity of some of the recombinant FAM83 proteins (Fig. 9A) or their lack of any putative priming phosphorylation. The optimal CK1 phosphorylation motif is pS-x-x-S/T (13). We previously showed that CK1α phosphorylates FAM83G only on Ser614 in vitro, but this phosphorylation event does not appear to affect function of FAM83G in Xenopus laevis embryos, which requires its association with CK1 (31).
Fig. 9. The intrinsic catalytic activity of CK1 is not affected by or required for the association of CK1 with FAM83 proteins.
A. An in vitro kinase assay was performed in the presence of [γ32P]-ATP with recombinant GST-CK1α plus one of the following recombinant FAM83 fusion proteins: GST-FAM83A (A), MBP-FAM83B (B), GST-FAM83C (C), GST-FAM83D (D), GST-FAM83E (E), GST-FAM83F (F), GST-FAM83G-6His (G), or GST-FAM83H (H). After the reactions were stopped, samples were resolved by SDS-PAGE. The gel was stained with Instant blue, dried, and subjected to 32P autoradiography for the indicated times. Instant blue–stained gel and autoradiograph representative of 3 independent experiments are shown. B. An in vitro kinase assay was set up with recombinant GST-CK1α, and either recombinant GST-FAM83G-6His or the GST-FAM83G (F296A, F300A) double mutant in the presence of increasing amounts of the optimized CK1 peptide substrate CK1tide. GST-CK1α, without FAM83G addition, was used as a control. Data points represent the average from three independent experiments, each including three replicates. Error bars, SEM. C. U2OS cells were transiently co-transfected with GFP-FAM83E, GFP-FAM83F, GFP-FAM83G, or GFP-FAM83H and either WT CK1α or a catalytically inactive (kinase dead, KD) form of CK1α. After 24 h cell extracts (Input) were immunoprecipitated (IP) with GFP-TRAP A beads and immunoblotted (IB) with the indicated antibodies. This blot is representative of 3 independent experiments. GAPDH is a loading control.
In order to test whether the intrinsic catalytic activity of CK1α was affected by its association with FAM83, we performed an in vitro CK1α kinase assay with increasing concentrations of an optimized CK1 peptide substrate (CK1tide), and evaluated whether the addition of equimolar amounts of either wild-type FAM83G or a CK1-interaction–deficient FAM83G mutant (F296A, F300A) altered the rate of CK1α catalysis or its Michaelis constant (Km) toward the CK1tide substrate. The intrinsic CK1α catalytic activity against CK1tide was not significantly altered by the addition of either wild-type or the F296A, F300A mutant FAM83G at all CK1tide concentrations tested, suggesting that FAM83G does not affect the intrinsic kinase kinetics of CK1α (Fig. 9B). We also assessed whether the kinase activity of CK1α was required for its association with FAM83 members. For this, we transiently co-expressed GFP-FAM83E, GFP-FAM83F, GFP-FAM83G, or GFP-FAM83H with either mCherry-tagged wild-type CK1α or the catalytically inactive mutant CK1αN141A (32) in U2OS cells and performed co-immunoprecipitation assays. Equal amounts of both wild-type CK1α and the CK1αN141A mutant were detected in immunoprecipitates of FAM83E–H (Fig. 9C), suggesting that CK1 kinase activity is dispensable for the FAM83:CK1 interaction.
Discussion
The various CK1 isoforms are known to control a myriad of cellular processes, yet how their activities are regulated in cells remains poorly defined. In this report, we identified the FAM83 family of proteins as interactors of the α, α-like, δ, and ε isoforms of CK1 in mammalian cells. This interaction was mediated through the conserved DUF1669 domain of FAM83 proteins, with different family members exhibiting distinct affinities and isoform-selectivity for CK1. FAM83 proteins displayed unique subcellular distribution patterns that overlapped with the specific CK1 isoforms with which they associate. Point mutations within the DUF1669 domains of FAM83 proteins that abolished CK1 association disrupted not only the co-localization of FAM83 members with specific CK1 isoforms in cells, but also the subcellular localization of the respective FAM83 members themselves. Our findings imply that the DUF1669 domains of FAM83 proteins anchor CK1 α, α-like, δ, and ε isoforms in specific subcellular compartments and potentially mediate their association with substrates, perhaps similar to the A-kinase anchoring proteins (AKAPs) that streamline signal transduction by bringing protein kinase A into close proximity of its substrates (26).
Unlike AKAPs, which bind to the regulatory domain of PKA, the DUF1669 domain of FAM83 proteins appeared to associate directly with the kinase domain of CK1 isoforms and did so independently of CK1 catalytic activity. There are many other examples of the crucial roles that scaffolding and anchoring proteins play in organizing and streamlining signal transduction in cells (33–36). TPX2 (targeting protein for Xklp2) is a scaffold protein that recruits Aurora kinase A to the mitotic spindle and activates the kinase allosterically (37). However, FAM83G did not influence the intrinsic catalytic activity of CK1α in vitro. Because some FAM83 members were substrates for CK1 isoforms in vitro, future work will be required to establish whether there are roles for some FAM83 proteins as substrates of CK1 in cells. The DUF1669 domain contains a pseudo-PLD-like catalytic motif, yet FAM83 proteins do not exhibit phospholipase activity (5). Hence, there could very well be features within the DUF1669 domain that still harbour certain pseudo-PLD roles, such as binding to specific phospholipids, that might affect binding to CK1. Future work will aim to explain the specificity and affinity with which FAM83 members bind different CK1 isoforms.
Precisely how FAM83 members impact the diverse functions of CK1 isoforms in cells is largely unclear, but we are beginning to uncover some of these roles. We have established that FAM83G is a critical mediator of Wnt signalling in human cells and Xenopus embryos (31). Crucially, we showed that unlike wild-type FAM83G, two mutants incapable of interacting with CK1α are unable to activate Wnt signalling or induce axis duplication in Xenopus embryos (31). Similarly, a recent report suggested that FAM83H and the DNA binding protein SON recruit CK1 to nuclear speckles (38). From our proteomic data, it is evident that each FAM83 member interacts with unique proteins in addition to the CK1 isoforms. Future investigations will establish whether FAM83 proteins individually recruit distinct sets of substrates to specific CK1 isoforms. Furthermore, by controlling the localization of CK1 isoforms, different FAM83 proteins might be primed to streamline diverse signal transduction processes downstream of CK1. Future efforts will aim to establish precisely which CK1 substrates are affected by individual FAM83 members. Additionally, global phosphoproteomic approaches in cells devoid of individual FAM83 members generated by genome editing techniques will identify potential substrate maps for CK1 isoforms.
Given the involvement of CK1 isoforms in a wide range of cellular processes, it is no surprise that their misregulation has been linked to cancers and neurological disorders (15, 22). The pleiotropic nature of CK1 function in regulating many cellular processes, combined with poor understanding of its regulation, has limited the exploration of CK1 for therapeutics. Nonetheless, several potent inhibitors of individual CK1 isoforms have been developed, including CKI-7, IC261, D4476, PF-670462 and PF-4800567, although all suffer from selectivity issues, with off-target effects related to their inhibition of other CK1 isoforms and protein kinases (39–43). Our findings clearly place the FAM83 proteins at the helm of CK1 regulation in cells. Therefore, understanding the molecular bases for FAM83:CK1 associations may provide us with unique opportunities to target and disrupt this association with small molecules, which could prove to be useful in targeting specific CK1 isoforms in specific cellular compartments.
In light of our data that clearly demonstrates that the DUF1669 domain is responsible for facilitating the interaction between FAM83 members and CK1 isoforms, we propose that the DUF1669 domain be renamed polypeptide anchor of CK1 (PACK1) domain. Co-crystallization of the PACK1 domain with CK1 isoforms will potentially reveal the determinants of CK1 interaction specificity and affinity for each FAM83 member.
Materials and Methods
Plasmids
Recombinant DNA procedures were performed using standard protocols as described previously (2, 44). Human FAM83A-H and CK1 wild-type genes or appropriate mutants were sub-cloned into pcDNA5-FRT/TO vectors with a Green Fluorescence Protein (GFP) tag at either the N- or the C-terminus, or an mCherry tag at the N-terminus. All constructs are available to request from the MRC-PPU reagents webpage (http://mrcppureagents.dundee.ac.uk) and the unique identifier (DU) numbers indicated above provide direct links to the cloning strategy and sequence information. The following constructs were generated: pcDNA5-FRT/TO GFP-FAM83A (DU44235), pcDNA5-FRT/TO GFP-FAM83B (DU44236), pcDNA5-FRT/TO GFP-FAM83C (DU42473), pcDNA5-FRT/TO GFP-FAM83D (DU42446), pcDNA5-FRT/TO GFP-FAM83E (DU44237), pcDNA5-FRT/TO GFP-FAM83F (DU44238), pcDNA5-FRT/TO GFP-FAM83G (DU33272), pcDNA5-FRT/TO GFP-FAM83H (DU44239), pcDNA5-FRT/TO GFP-FAM83C (D259A) (DU28479), pcDNA5-FRT/TO GFP-FAM83C (F293A) (DU28480), pcDNA5-FRT/TO GFP-FAM83E (D243A) (DU28481), pcDNA5-FRT/TO GFP-FAM83E (F277A) (DU28482), pcDNA5-FRT/TO GFP-FAM83F (D250A) (DU28268), pcDNA5-FRT/TO GFP-FAM83F (F284A) (DU28488), pcDNA5-FRT/TO GFP-FAM83G (D262A) (DU28476), pcDNA5-FRT/TO GFP-FAM83G (F296A) (DU28477), pcDNA5-FRT/TO GFP-FAM83H (D236A) (DU28428), pcDNA5-FRT/TO GFP-FAM83H (F270A) (DU28487), pcDNA5-FRT/TO mCherry-CK1α (DU28407), pcDNA5-FRT/TO mCherry-CK1α (N141A) (DU28839), pcDNA5-FRT/TO GFP-FAM83H(M1-L284)-FAM83G(S311-P823) (DU28683), pcDNA5-FRT/TO GFP-FAM83G(M1-V310)-FAM83H(V285-K1179) (DU28688), pcDNA5-FRT/TO FAM83A-GFP (DU42864), pcDNA5-FRT/TO FAM83B-GFP (DU42833), pcDNA5-FRT/TO FAM83C-GFP (DU42825), pcDNA5-FRT/TO FAM83D-GFP (DU42835), pcDNA5-FRT/TO FAM83E-GFP (DU42826), pcDNA5-FRT/TO FAM83F-GFP (DU42832), pcDNA5-FRT/TO FAM83G-GFP (DU42816), pcDNA5-FRT/TO FAM83H-GFP (DU42865), pcDNA5-FRT/TO GFP only (DU41455), pcDNA5-FRT/TO GFP-FAM83H (F274A) (DU28658), pcDNA5-FRT/TO GFP-FAM83H (F270, 274A) (DU28182), pcDNA5-FRT/TO FLAG-FAM83G (DU33274), pcDNA5-FRT/TO FLAG-FAM83G (F296A) (DU28024), pcDNA5-FRT/TO FLAG-FAM83G (F296A, F300A) (DU28026), pcDNA5-FRT/TO FLAG-FAM83G (F300A) (DU28025), pCS2+ HA-CK1α (DU28216), pCMV5-FLAG TTBK2 (DU19028), pCMV-FLAG-CK1γ (DU5580), pCS2+ HA CK1δ (DU28189), pcDNA5-FRT/TO mcherry-CK1ε (DU28406). Myc-xFAM83G (Xenopus laevis FAM83G) constructs have been described previously (28). For CRISPR/Cas9 gene editing, pBABED P U6 FAM83H KO sense gRNA (DU52010), pX335-CAS9-D10A FAM83H KO antisense gRNA (DU52026), pBABED P U6 FAM83G KI sense gRNA (DU48528), pX335-Cas9-D10A FAM83G KI antisense gRNA (DU48529), pEX-K4 FAM83G Cter GFP donor (DU48585), pBABED P U6 FAM83B KI sense gRNA (DU54494), pX335-Cas9-D10A FAM83B KI antisense gRNA (DU54504), and pEX-K4 FAM83B Nter GFP donor (DU54547) were generated. Constructs were sequence-verified by the DNA Sequencing Service, University of Dundee (http://www.dnaseq.co.uk). For plasmid amplification, 1 μl of the plasmid was transformed into 10 μl of E. coli DH5α competent bacteria (Invitrogen) on ice, incubated at 42°C for 45 s, then on ice for 2 min, before plating on LB-agar medium plate containing 100 μg/ml ampicillin. Plates were inverted and incubated for 16 h at 37°C. A single colony was picked and used to inoculate 250 ml of LB medium containing 100 μg/ml ampicillin, and cultures were grown for 18 h at 37°C in a shaker (Infors HT). Plasmid DNA was purified using a Qiagen midi-prep kit as per the manufacturer’s instructions. The isolated DNA yield was subsequently analysed using a Nanodrop 1000 spectrophotometer (Thermo Scientific).
Antibodies
Rabbit anti-GAPDH (cat.: 2118, 1:5000), anti-CK1δ (cat.: 12417S, 1:1000), and anti-CK1ε (cat.: 12448, 1:1000) were from Cell Signalling Technology (CST). Rat anti-GFP for detection of endogenous GFP tags was from Chromotek (cat.: 3H9, 1:1000). Anti-CK1α (cat.: A301-991A, 1:1000 for immunoblotting, 5 µg antibody/mg of cell extract protein for immunoprecipitation) and anti-FAM83H (cat.: A304-327A, 1:1000) were from Bethyl. Anti-DYNLL1 (EP1660Y, 1:1000) and anti-FAM83B (cat.: 153829, 1:1000) were from Abcam. Anti-HMMR (cat.: ABC323, 1:1000) was from Millipore. Sheep anti-PAWS1/FAM83G (S876C, 3rd bleed, 1:1000), anti-FAM83H (SA273, 4th bleed, 1:1000), anti-GFP (S268B, 2nd bleed, 1:1000) and anti-SMAD1 (S618C, 3rd bleed, 1:1000) were generated by the Division of Signal Transduction Therapy (DSTT), University of Dundee (2, 45). anti-FLAG M2-Peroxidase (HRP) (cat.: A8592, 1:2000) and anti-c-Myc-HRP (cat.: A5598, 1:2000) were from Sigma and Anti-HA-HRP (cat.: 11667475001, 1:2000) was from Roche. For HRP-coupled secondary antibodies, goat anti-rabbit-IgG (cat.: 7074, 1:2500) was from CST, rabbit anti-sheep-IgG (cat.: 31480, 1:5000), goat anti-rat IgG (cat.: 62-9520, 1:5000) and goat anti-mouse-IgG (cat.: 31430, 1:5000) were from Thermo Fisher. For Immunofluorescence, anti-CK1-alpha (C-19 Santa Cruz Biotechnology, 1:100) and anti-CK1-epsilon (HPA026288 Sigma, 1:500) were used. For signal amplification, AlexaFluor-594 donkey anti-goat IgG (H+L) (A11058 Life Technologies, 1:300), AlexaFluor-594 goat anti-rabbit IgG (H+L) (A11012 Invitrogen™ Molecular Probes™, 1:500) and AlexaFluor-488 donkey anti-rabbit IgG (H+L) (A21206, Life Technologies, 1:500) were employed.
Cell Culture
Human osteosarcoma U2OS, human embryonic kidney HEK 293, human keratinocyte HaCaT, Flp-In T-Rex U2OS and HEK 293, and retroviral production HEK 293(FT) cell lines were grown in Dulbecco’s Modified Eagles Medium (DMEM; Gibco) containing 10% (v/v) Foetal Bovine Serum (FBS; Hyclone), penicillin (100 U/ml; Lonza), streptomycin (0.1 mg/ml; Lonza) and L-glutamine (2 mM; Lonza), and cultured at 37°C, 5% CO2 in a humidified incubator. For transient transfections, cells were transfected for 24 h with 2 μg (per 10 cm-dish), or 500 ng (per 6-well dish with coverslips) cDNA, in serum free OptiMem (Gibco) with the transfection reagent polyethylenimine (PEI) as described previously (44). Where applicable, Tet-inducible expression was achieved by adding doxycycline (20 ng/ml) for up to 24 h prior to cell lysis as indicated.
Generation of Stable Flp-In T-Rex Cell Lines
The Flp-In T-Rex U2OS or HEK 293 cells were transfected with the N- or C-terminal GFP-tagged FAM83A-H or GFP alone packaged into a pcDNA5-FRT/TO vector, together with the Flp recombinase pOG44 (Invitrogen) in a ratio of 1 μg : 9 μg as described previously (2, 46). Briefly, plasmids were diluted in 1 ml OptiMem (Gibco), 20 μl of 1 mg/ml polyethylenimine (PEI) was added, the mix vortexed and left at room temperature for 15 min and added dropwise to 10 cm dish of target cells in 10 ml complete medium. 24 h post-transfection, cells were selected in media containing hygromycin (50 μg/ml) and blasticidin (15 µg/ml). Resistant cells were grown up to confluency, tested for doxycycline-induced expression of GFP-tagged proteins and used in subsequent experiments.
Generation of FAM83GGFP/GFP and GFP/GFPFAM83B knock-in cells using CRISPR/Cas9
U2OS and HaCaT cells were transfected with vectors encoding a pair of guide RNAs (pBABED-Puro-sgRNA1 and pX335-Cas9-D10A-sgRNA2) targeting around the stop codon of FAM83G and the start codon of FAM83B (1 μg each), along with the respective donor plasmids carrying the GFP knockin insert and flanking homology arms (~500 bases) (3 μg each). 16 h post-transfection, cells were selected in puromycin (2 µg/ml) for 2 days. The transfection process was repeated one more time. GFP-positive cells were isolated by fluorescence-activated cell sorting (FACS) and single GFP-positive cell clones were plated on individual wells of two 96-well plates, pre-coated with 1% (w/v) gelatin as described previously (45). Viable clones were expanded, and the integration of GFP at the target locus was confirmed by Western blotting and genomic sequencing of the targeted locus. The DU identifier numbers for the plasmids listed above link to the sequences for gRNA and donors with homology arms for each target.
Generation of FAM83G-/- and FAM83H-/- cells using CRISPR/Cas9
CRISPR/Cas9 mediated deletion of FAM83G in U2OS cells was performed using Cas9 and a single gRNA targeting approach to delete exon 2 of the RefSeq gene for FAM83G (NM_001039999.2). Vectors containing the Cas9 and FAM83G targeting gRNA (ggaccgctccatcccgcagctgg) were transfected into 1x106 U2OS cells followed by selection with 2 ug/ml puromycin. Single cell sorting was used to isolate clone candidates, which were screened with Western blotting and confirmed by genomic sequencing. For FAM83H, U2OS cells were transfected with vectors encoding a pair of guide RNAs (pBABED-Puro-sgRNA1 and pX335-Cas9-D10A-sgRNA2) targeting the second exon of FAM83H (1 μg each). 16 h post-transfection, cells were selected in puromycin (2 µg/ml) for 2 days. The transfection process was repeated one more time. Cells were isolated by single-cell sorting and isolated clones were plated on individual wells of two 96-well plates, pre-coated with 1% (w/v) gelatin as described previously (45). Viable clones were expanded and successful knockout of FAM83H was confirmed by Western blotting and genomic sequencing of the targeted locus. The DU identifier numbers for the plasmids listed above link the sequences for gRNA for each target.
Cell Lysis and Immunoprecipitation
Cells were washed twice in ice-cold phosphate-buffered saline (PBS), before scraping on ice in lysis buffer (50 mM Tris-HCl pH 7.5, 0.27 M sucrose, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, and 1% Nonidet P40 substitute), supplemented with 1X protease inhibitor cocktail (Roche). Cell extracts were either cleared and processed immediately, or snap frozen in liquid nitrogen, before storing at -80°C. Protein concentrations were determined in a 96-well format using Bradford protein assay reagent (Pierce).
For immunoprecipitation, clarified extracts were diluted in lysis buffer to 1-5 mg/ml. Input aliquots were taken, and lysates were incubated overnight at 4°C with protein G-sepharose beads coupled to the antibody of interest, on a rotating wheel. For anti-GFP immunoprecipitations, GFP-TRAP A beads (Chromotek) were used; for anti-FLAG immunoprecipitations, anti-FLAG M2 affinity agarose gel (Sigma) were used. Following the incubation period, beads were pelleted and flow-through extracts collected. Beads were washed once in lysis buffer supplemented with 250 mM NaCl and 2-3 times in lysis buffer. Beads were eluted in 1X SDS sample buffer, at 95°C for 5 min.
For immunoprecipitations for mass-spectrometry, cells were lysed in DSP crosslinking lysis buffer (40 mM HEPES pH 7.4, 120 mM NaCl, 1 mM EDTA, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 1.5 mM sodium orthovanadate, 1% (v/v) triton, 1X protease inhibitor cocktail (Roche), and 2.5 mg/ml DSP as described previously (2). Following lysis, lysates were incubated for 30 mins at 4°C, before quenching the crosslinking reaction by adding 1 M Tris-HCl pH 7.4 in a ratio of 1:4 and incubating at 4°C for a further 30 min. Lysates were clarified by centrifugation at 15000 rpm for 20 min and filtered through 0.45 μM filters (BioRad). Extracts were pre-cleared by incubating with Protein-G sepharose beads for 1 h at 4°C on a rotating wheel. Pre-cleared lysates were quantified using the Bradford method, and extracts incubated with GFP-TRAP A beads (Chromotek) for 4 h at 4°C on a rotating wheel. Input and post-immunoprecipitation extract aliquots were taken for control blots. Beads were washed twice in lysis buffer supplemented with 250 mM NaCl, and three times in lysis buffer. 1X SDS sample buffer containing 0.1 M DTT was added to the beads (~50% slurry), and samples incubated at 37°C for 1 h. Samples were then boiled at 95°C for 5 min and eluted through SpinX columns (Corning).
Mass Spectrometry
The expression of GFP-tagged FAM83 proteins in stable Flp-In T-Rex HEK 293 and U2OS cells was induced with 20 ng/ml doxycycline for 24 h prior to lysis. Proteins were affinity purified from clarified extracts by GFP-TRAP A beads (ChromoTek) as described above. Purified proteins were resolved by 4-12% gradient SDS-PAGE, the gels were stained with Instant blue and gel slices covering each lane were excised and trypsin digested. The peptides were subjected to mass spectrometric analysis performed by LC-MS-MS on a Linear ion trap-orbitrap hybrid mass spectrometer (Orbitrap-VelosPro, Thermo) coupled to a U3000 RSLC Hplc (Thermo). Peptides were trapped on a nanoViper Trap column, 2 cm x 100 μm C18 5 μm 100 Å (Thermo, 164564) then separated on a 15 cm Thermo EasySpray column (ES800) equilibrated with a flow of 300 nl/min of 3% Solvent B. [Solvent A: 2% Acetonitrile, 0.1% formic acid, 3% DMSO in H2O; Solvent B: 80% acetonitrile, 0.08% formic acid, 3% DMSO in H2O]. The elution gradient was as follows; Time (min):Solvent B (%); 0:3, 5:3, 45:35, 47:99, 52:99, 55:3, 60:3. Data were acquired in the data-dependent mode, automatically switching between MS and MS-MS acquisition. Full scan spectra (m/z 400-1600) were acquired in the orbitrap with resolution R = 60,000 at m/z 400 (after accumulation to an FTMS Full AGC Target; 1,000,000; FTMS MSn AGC Target; 50,000). The 20 most intense ions, above a specified minimum signal threshold (2,000), based upon a low resolution (R=15,000) preview of the survey scan, were fragmented by collision induced dissociation and recorded in the linear ion trap (Full AGC Target; 30,000. MSn AGC Target; 5,000). Data files were analysed by Proteome Discoverer 2.0 (www.ThermoScientific.com), using Mascot 2.4.1 (www.matrixscience.com), and searching the SwissProt Human database. Scaffold Q/Q+S V4.4.7 (www.ProteomeSoftware.com) was also used to examine the Mascot result files. Allowance was made for the following fixed, Carbamidomethyl (C), and variable modifications, Oxidation (M), Dioxidation (M). Error tolerances were 10 ppm for MS1 and 0.6 Da for MS2. Scaffold Q/Q+S V4.3 (U2OS) or V4.4.6 (HEK 293) (www.ProteomeSoftware.com) was used to further analyse the data and obtain values for the Top 3 precursor ion intensities of each protein.
SDS-PAGE and Western Blotting
Reduced protein extracts (10–20 μg protein) or immunoprecipitates were resolved on either 8% (v/v) SDS-PAGE gels, or 4-12% NuPAGE bis-tris precast gradient gels (Invitrogen) by electrophoresis. Separated proteins were subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore), before membranes were blocked in 5% (w/v) non-fat milk powder (Marvel) in TBS-T (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.2% (v/v) Tween-20) and incubated overnight at 4°C in 5% milk TBS-T or 5% bovine serum albumin (BSA) TBS-T with the appropriate primary antibody. Membranes were then washed 3 X 10 min with TBS-T before incubating with HRP-conjugated secondary antibodies in 5% milk TBS-T for 1 h at room temperature. Membranes were then washed 3 X 10 min with TBS-T before detection with enhanced chemiluminescence reagent (Millipore) and exposure to medical-grade X-ray films (Konica Minolta), as described previously (2, 47, 48).
Fluorescence microscopy
Cells were plated on glass coverslips and treated/transfected as described above. Cells were washed twice in PBS, before being fixed either with methanol at -20°C for 2 min or 4% (w/v) paraformaldehyde (PFA) in 200 mM HEPES pH 7.4 for 20 min at RT. Cells fixed in methanol were washed three times in ice cold PBS after fixation, then blocked in 3% BSA/PBS + 0.01% Tween 20 on ice for 30 min. Cells fixed in PFA were washed twice with DMEM/10 mM HEPES followed by incubation in DMEM/10 mM HEPES for 10 min at RT. Cells fixed in PFA were washed once in PBS and permeabilised for 3 min in 1.5 ml 0.2% NP40. Cells were then washed twice in PBS containing 1-3% (w/v) BSA, followed by incubation in PBS/BSA for 15 min. Where appropriate, coverslips were then incubated with primary antibody in PBS/BSA (typically at 1:50-1:500 dilution as stated) at 30-37°C for 1-1.5 h. Cells were washed for a minimum of 3 X 10 min in PBS/BSA (PFA-fixed cells) or 3 X 5 min in PBS (methanol-fixed cells) on shaker. Coverslips were incubated with secondary Alexa-Fluor conjugated antibody in PBS/BSA (1:300-500 dilution) and DAPI (1:500) for 30 to 60 min at RT in the dark. Coverslips were then washed for 3 X 10 min in PBS/BSA (PFA-fixed cells) or 3 X 5 min in PBS (methanol-fixed cells), and mounted on glass microscopy slides using either ProLong® Gold anti-fade reagent with DAPI (Life Technologies) (if DAPI staining was not performed previously), or mounted using VECTASHIELD mounting solution (Vector Labs). Coverslips were sealed with clear nail varnish and left to dry overnight before imaging on either a Nikon TiS inverted microscope, or a DeltaVision Imaging Systems (GE Healthcare). Images were processed using either NIS Elements (Nikon) and Adobe Photoshop, or softWoRx (GE Healthcare) and Omero (49).
Colocalization was assessed using the Pearson’s correlation coefficient (PCC) as a measure of intensity correlation between the two channels. As explained by Adler and Parmryd (50), PCC is sensitive to the inclusion of background pixels. We therefore excluded background pixels by auto-thresholding each channel in the cytoplasmic region of interest using Otsu’s method (51). Thresholding and PCC calculation were implemented in an ImageJ macro developed by G. Ball (Dundee) and is included as a Supplementary File (File S1).
In vitro kinase assays
25 µl reactions were set up using 200 ng of kinase (GST-CK1α), 2 µg of substrate (GST-FAM83A, C, D, E, F, or H, MBP-tagged FAM83B, or GST-FAM83G-6XHis in a buffer containing 50 mM Tris pH 7.5, 0.1 mM EGTA, 10 mM Magnesium acetate, 2 mM DTT and 0.1 mM [γ32P]-ATP (~500 cpm/pmol). Assays were performed at 30°C for 30 min and stopped by addition of 9 μl of 4xSDS sample buffer with 5% β-mercaptoethanol and heating at 95°C for 5 min. The samples were resolved by SDS-PAGE and the gels were stained with Instant blue (Expedeon) and dried. Radioactivity was analysed by autoradiography. For peptide-based kinase assays, reactions were set up and performed as described previously (52), using an optimised CK1 peptide substrate (CK1tide (KRRRALS*VASLPGL), where S* indicates phosphoserine). Assays were performed in triplicates.
Protein expression and purification
The DUF1669 domain of FAM83A (a.a. 122-304) and the kinase domain of human CK1ε (a.a. 1-294) were expressed separately in E. coli strain BL21(DE3) R3-pRARE2 using the pNIC28-Bsa4 vector, which encodes for a N-terminal hexahistidine (6XHis) tag and TEV cleavage site. Cultures were grown at 37°C in LB medium supplemented with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol to an OD of 0.6, before expression at 18°C overnight by induction with 0.4 mM isopropyl 1-thio-β-D-galactopyranoside. Cells were harvested by centrifugation at 5000 g and pellets resuspended in binding buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM imidazole) supplemented with Calbiochem protease inhibitor set III. Cells were lysed by sonication before clarification of the lysate by centrifugation in a JA 25.50 rotor at 36,000 g. His-tagged proteins were immobilized on Ni-sepharose and bound proteins were eluted using step gradients of imidazole (50-250 mM). CK1ε protein was cleaved with TEV protease overnight at 4°C and both 6xHis-FAM83A and CK1ε were purified further by size exclusion chromatography using an S75 HiLoad 16/60 Superdex column equilibrated in buffer containing 50 mM HEPES pH 7.5, 300 mM NaCl, and 0.5 mM TCEP. Proteins were concentrated by centrifugal ultrafiltration using a 3 kDa molecular weight cut-off concentrator. Protein concentrations were determined by measuring absorbance at 280 nm. Protein purity of >95% was confirmed by SDS-PAGE and construct identities and tag cleavage were verified by mass spectrometry.
All other recombinant proteins used in the in vitro kinase assays were purified by the Division of Signal Transduction Therapy (DSTT; University of Dundee) and the identities of the expressed proteins verified by mass spectrometry. Each protein has a unique identification number to request from the MRC-PPU Reagents website (http://mrcppureagents.dundee.ac.uk) as follows: GST-CK1α (DU329),GST-FAM83A (DU24611), GST-FAM83C (DU28269), GST-FAM83D (DU28270), GST-FAM83E (DU28271), GST-FAM83F (DU28272), GST-FAM83H (DU28403) and GST-FAM83G (F296A, F300A) (DU28049). Briefly, the proteins were expressed in BL21(DE3) E. coli as described above and affinity purified using GSH-sepharose, Amylose-sepharose or Nickel-agarose columns as appropriate.
In vitro binding assay
For the in vitro binding assay, all proteins and Ni-sepharose were equilibrated in binding buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM imidazole) prior to use. 300 μg 6xHis-FAM83A (aa122-304) was immobilised onto 200 μl Ni-sepharose and washed before addition of 100 μg CK1ε. The Ni-sepharose was then washed with binding buffer and the flow through collected. Two 1 ml wash steps were performed using binding buffer before bound proteins were eluted with 1 ml binding buffer supplemented with 250 mM Imidazole. Fractions were run on a SDS-PAGE gel alongside the original protein solutions for molecular weight reference.
Statistical Analysis
For kinase assays, GraphPad (Prism) was used to generate plots and analyse data by two-way ANOVA and Tukey test to determine statistical significance, from 3 independent experiments, each containing 3 replicates. A p-value of <0.05 was deemed significant. For co-localization studies, GraphPad (Prism) was used to generate boxplots and analyze data by one-way ANOVA and Dunnett’s multiple comparison test to determine statistical significance. A p-value of <0.05 was deemed significant.
Supplementary Material
Acknowledgements
We thank G. Ball (Dundee) for analysing fluorescence images and developing an Image J Macro for Pearson correlation coefficient experiments. We thank L. Fin, J. Stark and A. Muir for help with tissue culture, the staff at the Sequencing Service (School of Life Sciences, University of Dundee, UK) for DNA sequencing, and the protein & antibody production and cloning teams at the Division of Signal Transduction Therapy (DSTT; University of Dundee) coordinated by H. McLauchlan and J. Hastie. We thank Dr. M. Gierliński (The Data Analysis Group, School of Life Sciences, University of Dundee, UK) for advice on appropriate statistical tests.
Funding: LJF, PB, TT-M are supported by the U.K. MRC PhD studentships. The Dundee Imaging Facility, which provided image analysis support, is funded by the 'MRC Next Generation Optical Microscopy' award [MR/K015869/1]. LJF also receives funding from the Queens College Scholarship, University of Dundee. KW and KD are supported by MRC Career Development Fellowships. GPS is supported by the U.K. Medical Research Council (grant number MC_UU_12016/3) and the pharmaceutical companies supporting the DSTT (Boehringer-Ingelheim, GlaxoSmithKline, Merck-Serono). JCS and KSD are supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001157), the UK Medical Research Council (FC001157), and the Wellcome Trust (FC001157). ANB, JCB and DMP are supported by the SGC, which is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, MSD, Merck KGaA, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda and Wellcome [106169/ZZ14/Z].
Footnotes
Author contributions: LJF, PB, TTM, KW, KD, TDC and SS performed experiments, collected and analysed data and contributed to the manuscript. TJM designed strategies and developed methodologies for, and generated, all CRIPSR/Cas9 knockin constructs. TJM, NTW and SW cloned genes and performed mutagenesis experiments. JV, RG and DGC performed mass-spectrometry experiments, collected and analysed data. JCB & DMP performed in vitro interaction between FAM83A-DUF1669 and the kinase domain of CK1ε and contributed to the composition of the manuscript. KSD, JCS & ANB contributed with data analysis and the composition of the manuscript. GPS conceived the project, analysed the data and wrote the manuscript.
Competing Interests: The authors declare that they have no competing interests.
Data and materials availability: The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository with the dataset identifier PXD009335.
References
- 1.Bartel CA, Parameswaran N, Cipriano R, Jackson MW. FAM83 proteins: Fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget. 2016 doi: 10.18632/oncotarget.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herhaus L, Al-Salihi MA, Dingwell KS, Cummins TD, Wasmus L, Vogt J, Ewan R, Bruce D, Macartney T, Weidlich S, Smith JC, et al. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open Biol. 2014;4 doi: 10.1098/rsob.140065. 140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Phospholipase D: enzymology, functionality, and chemical modulation. Chem Rev. 2011;111:6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, Bissell MJ. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122:3211–3220. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA, Brown HA, Stark GR, Jackson MW. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012;122:3197–3210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4:729–738. doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198:1039–1054. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr Biol. 2008;18:723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 9.Vogt J, Dingwell KS, Herhaus L, Gourlay R, Macartney T, Campbell D, Smith JC, Sapkota GP. Protein associated with SMAD1 (PAWS1/FAM83G) is a substrate for type I bone morphogenetic protein receptors and modulates bone morphogenetic protein signalling. Open Biol. 2014;4 doi: 10.1098/rsob.130210. 130210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Park JT, Seo BM, Hu JC, Simmer JP. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82:489–494. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuga T, Kume H, Kawasaki N, Sato M, Adachi J, Shiromizu T, Hoshino I, Nishimori T, Matsubara H, Tomonaga T. A novel mechanism of keratin cytoskeleton organization through casein kinase Ialpha and FAM83H in colorectal cancer. J Cell Sci. 2013;126:4721–4731. doi: 10.1242/jcs.129684. [DOI] [PubMed] [Google Scholar]
- 12.Lee SK, Lee KE, Jeong TS, Hwang YH, Kim S, Hu JC, Simmer JP, Kim JW. FAM83H mutations cause ADHCAI and alter intracellular protein localization. J Dent Res. 2011;90:377–381. doi: 10.1177/0022034510389177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venerando A, Ruzzene M, Pinna LA. Casein kinase: the triple meaning of a misnomer. Biochem J. 2014;460:141–156. doi: 10.1042/BJ20140178. [DOI] [PubMed] [Google Scholar]
- 14.Schittek B, Sinnberg T. Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer. 2014;13:231. doi: 10.1186/1476-4598-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Wurl PU, Eismann T, Stoter M. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie. 2005;28:508–514. doi: 10.1159/000087137. [DOI] [PubMed] [Google Scholar]
- 16.Camacho F, Cilio M, Guo Y, Virshup DM, Patel K, Khorkova O, Styren S, Morse B, Yao Z, Keesler GA. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- 17.Keesler GA, Camacho F, Guo Y, Virshup D, Mondadori C, Yao Z. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport. 2000;11:951–955. doi: 10.1097/00001756-200004070-00011. [DOI] [PubMed] [Google Scholar]
- 18.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 19.Stoter M, Bamberger AM, Aslan B, Kurth M, Speidel D, Loning T, Frank HG, Kaufmann P, Lohler J, Henne-Bruns D, Deppert W, et al. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene. 2005;24:7964–7975. doi: 10.1038/sj.onc.1208941. [DOI] [PubMed] [Google Scholar]
- 20.Zilian O, Frei E, Burke R, Brentrup D, Gutjahr T, Bryant PJ, Noll M. double-time is identical to discs overgrown, which is required for cell survival, proliferation and growth arrest in Drosophila imaginal discs. Development. 1999;126:5409–5420. doi: 10.1242/dev.126.23.5409. [DOI] [PubMed] [Google Scholar]
- 21.Bischof J, Randoll SJ, Sussner N, Henne-Bruns D, Pinna LA, Knippschild U. CK1delta kinase activity is modulated by Chk1-mediated phosphorylation. PLoS One. 2013;8:e68803. doi: 10.1371/journal.pone.0068803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev. 2011;31:924–954. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- 23.Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 25.Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol. 2002;322:785–797. doi: 10.1016/s0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- 26.Esseltine JL, Scott JD. AKAP signaling complexes: pointing towards the next generation of therapeutic targets? Trends Pharmacol Sci. 2013;34:648–655. doi: 10.1016/j.tips.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 28.Cummins TD, Wu KZL, Bozatzi P, Dingwell KS, Macartney TJ, Wood NT, Varghese J, Gourlay R, Campbell DG, Prescott A, Griffis E, et al. FAM83G/PAWS1 controls cytoskeletal dynamics and cell migration through association with the SH3 adaptor CD2AP. bioRxiv. 2017 doi: 10.1242/jcs.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol Cell Biol. 2004;24:4184–4195. doi: 10.1128/MCB.24.10.4184-4195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuga T, Sasaki M, Mikami T, Miake Y, Adachi J, Shimizu M, Saito Y, Koura M, Takeda Y, Matsuda J, Tomonaga T, et al. FAM83H and casein kinase I regulate the organization of the keratin cytoskeleton and formation of desmosomes. Sci Rep. 2016;6 doi: 10.1038/srep26557. 26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bozatzi P, Dingwell KS, Wu KZ, Cooper F, Cummins TD, Hutchinson LD, Vogt J, Wood NT, Macartney TJ, Varghese J, Gourlay R, et al. PAWS1 controls Wnt signalling through association with casein kinase 1alpha. EMBO Rep. 2018 doi: 10.15252/embr.201744807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barik S, Taylor RE, Chakrabarti D. Identification, cloning, and mutational analysis of the casein kinase 1 cDNA of the malaria parasite, Plasmodium falciparum. Stage-specific expression of the gene. J Biol Chem. 1997;272:26132–26138. doi: 10.1074/jbc.272.42.26132. [DOI] [PubMed] [Google Scholar]
- 33.Alexa A, Varga J, Remenyi A. Scaffolds are 'active' regulators of signaling modules. FEBS J. 2010;277:4376–4382. doi: 10.1111/j.1742-4658.2010.07867.x. [DOI] [PubMed] [Google Scholar]
- 34.Buday L, Tompa P. Functional classification of scaffold proteins and related molecules. FEBS J. 2010;277:4348–4355. doi: 10.1111/j.1742-4658.2010.07864.x. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, Neiswinger J, Zhang J, Zhu H, Qian J. Systematic Prediction of Scaffold Proteins Reveals New Design Principles in Scaffold-Mediated Signal Transduction. PLoS Comput Biol. 2015;11:e1004508. doi: 10.1371/journal.pcbi.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logue JS, Scott JD. Organizing signal transduction through A-kinase anchoring proteins (AKAPs) FEBS J. 2010;277:4370–4375. doi: 10.1111/j.1742-4658.2010.07866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 38.Kuga T, Kume H, Adachi J, Kawasaki N, Shimizu M, Hoshino I, Matsubara H, Saito Y, Nakayama Y, Tomonaga T. Casein kinase 1 is recruited to nuclear speckles by FAM83H and SON. Sci Rep. 2016;6 doi: 10.1038/srep34472. 34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, Holland J, Kleiman R, Nelson F, Reynolds L, St Germain K, et al. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther. 2007;322:730–738. doi: 10.1124/jpet.107.122846. [DOI] [PubMed] [Google Scholar]
- 40.Behrend L, Milne DM, Stoter M, Deppert W, Campbell LE, Meek DW, Knippschild U. IC261, a specific inhibitor of the protein kinases casein kinase 1-delta and -epsilon, triggers the mitotic checkpoint and induces p53-dependent postmitotic effects. Oncogene. 2000;19:5303–5313. doi: 10.1038/sj.onc.1203939. [DOI] [PubMed] [Google Scholar]
- 41.Chijiwa T, Hagiwara M, Hidaka H. A newly synthesized selective casein kinase I inhibitor, N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase I from bovine testis. J Biol Chem. 1989;264:4924–4927. [PubMed] [Google Scholar]
- 42.Rena G, Bain J, Elliott M, Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5:60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, Griffor M, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 44.Fulcher LJ, Macartney T, Bozatzi P, Hornberger A, Rojas-Fernandez A, Sapkota GP. An affinity-directed protein missile system for targeted proteolysis. Open Biol. 2016;6 doi: 10.1098/rsob.160255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas-Fernandez A, Herhaus L, Macartney T, Lachaud C, Hay RT, Sapkota GP. Rapid generation of endogenously driven transcriptional reporters in cells through CRISPR/Cas9. Sci Rep. 2015;5:9811. doi: 10.1038/srep09811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519. doi: 10.1038/ncomms3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruce DL, Macartney T, Yong W, Shou W, Sapkota GP. Protein phosphatase 5 modulates SMAD3 function in the transforming growth factor-beta pathway. Cell Signal. 2012;24:1999–2006. doi: 10.1016/j.cellsig.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Herhaus L, Perez-Oliva AB, Cozza G, Gourlay R, Weidlich S, Campbell DG, Pinna LA, Sapkota GP. Casein kinase 2 (CK2) phosphorylates the deubiquitylase OTUB1 at Ser16 to trigger its nuclear localization. Sci Signal. 2015;8:ra35. doi: 10.1126/scisignal.aaa0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, Macdonald D, Moore WJ, Neves C, Patterson A, Porter M, et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 51.Otsu N. Threshold Selection Method from Gray-Level Histograms. Ieee T Syst Man Cyb. 1979;9:62–66. [Google Scholar]
- 52.Hastie CJ, McLauchlan HJ, Cohen P. Assay of protein kinases using radiolabeled ATP: a protocol. Nat Protoc. 2006;1:968–971. doi: 10.1038/nprot.2006.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.