Abstract
We have recently gained unprecedented insight into genetic factors that determine risk for Barrett’s esophagus (BE) and progression to esophageal adenocarcinoma (EA). Next-generation sequencing technologies have allowed us to identify somatic mutations that initiate BE and track genetic changes during development of tumors and invasive cancer. These technologies led to identification of mechanisms of tumorigenesis that challenge the current multi-step model of progression to EA. Newer, cost-effective technologies create opportunities to rapidly translate the analysis of DNA into tools that can identify patients with BE at high risk for cancer, detect dysplastic lesions at earlier stages, and uncover mechanisms of carcinogenesis.
Keywords: esophagus, genome-wide association study, mutational signature, chromothripsis, Cytosponge
Over the last 40 years, the incidence of esophageal adenocarcinoma (EA) has increased more than 6-fold in western countries1–3. The overall age-adjusted incidence in the United States is 2.7 cases per 100,000—a figure that reaches 6 and 9.4 per 100,000 among American and British white men respectively2,3. Overall 5-year survival is approximately 20% and about half of patients die within a year of diagnosis1. However, less than 50% of patients diagnosed early enough for curative treatment (surgery and neoadjuvant chemo or chemoradiotherapy) survive for 5 years4,5.
Barrett’s Esophagus (BE) is a precursor to development of EA is. In the absence of dysplasia (NDBE), the risk for transformation of BE to invasive cancer is 0.3% per year (reviewed in 6). BE, which is found in about 1%–2% of the general population, is a squamocolumnar metaplasia that develops in response to gastroesophageal reflux (GER). BE pathogenesis involves a combination of anatomical (hiatus hernia), genetic, and lifestyle risk factors 6–10. Neoplastic transformation of NDBE usually occurs through progressive grades of dysplasia. Endoscopic treatment is recommended upon identification of dysplasia, which is associated with a risk of progression to cancer of 10% per year or higher11–15. Unfortunately, clinical strategies for BE, which focus on endoscopic surveillance and endoscopic therapy, have not reduced the incidence or mortality of EA in the general population 16–18. This is because most cases of EA present without a prior diagnosis of BE. It has been estimated that 40% of EA cases have no history of GER symptoms and an additional 52% of cases have a history of GER but did not receive a diagnosis or undergo endoscopic surveillance 7.
Even when diagnosed there are no systems to stratify patients with BE, based on cancer risk, for surveillance and endoscopic therapy. Limited sensitivity of current endoscopic imaging technologies and sampling bias causes many dysplastic lesions to be missed. There is also low inter-observer reproducibility among pathologists in grading dysplasia, leading to overdiagnosis or underdiagnosis. When patients with invasive EA are identified, there are few therapeutic options.
Some of these issues can be improved by increasing our understanding of molecular factors associated with development of EA, including inherited (germline) and acquired (somatic) genetic alterations (Figure 1, Figure 2, Supplementary table 2). Development of massively parallel and less costly sequencing techniques (next-generation sequencing) has led to a number of genome-wide datasets, which can be used to study the genomic features of EA. We review the germline and somatic variants identified in different stages of the NDBE to EA spectrum, and discuss the challenges to translating findings from genomic analyses into screening, diagnostic, and therapeutic strategies (Supplementary Table 1, Figure 1).
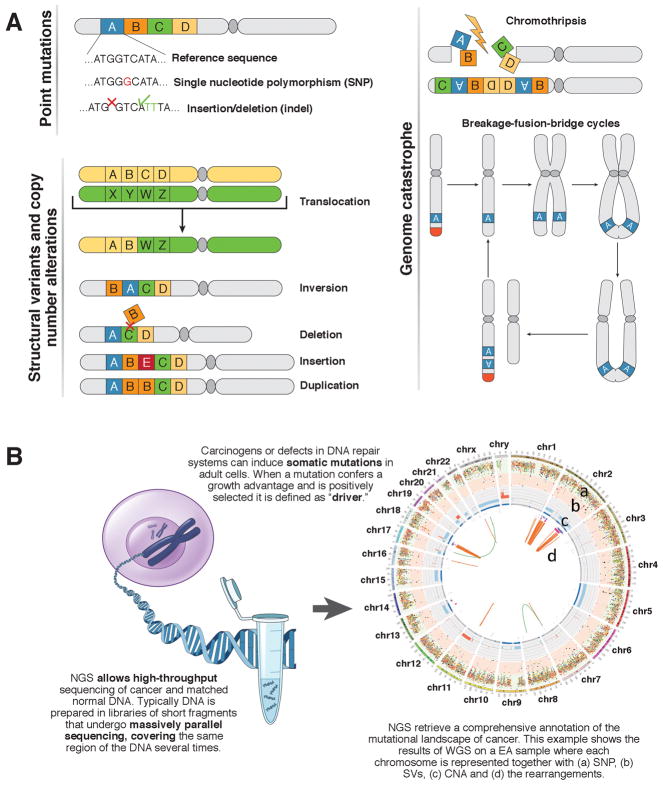
Figure 1. Somatic mutations and next-generation sequencing of cancer.
A) Tumor tissues can have point mutations, structural variations, copy number alterations, and genome catastrophes. Possible mechanisms of mutation are shown in a chromosome (2 arms linked by a dark gray centromere); these can involve a large segment of genome (lettered rectangles) or single DNA base pairs. Structural variations can cause loss or gain of genetic material and result in copy number changes. Complex structural variations occur in regions of genome catastrophes such as chromothripsis and breakage fusion bridge cycles 100,101,103,115. In cycles of breakage fusion bridge, an unprotected DNA end is generated following the loss of the telomeres (red) or a double-strand break115. During anaphase the broken chromatids can fuse (anaphase bridge) and then tear unevenly when the 2 chromatids are pulled apart. This event can be repeated through several cycles, leading to amplification of oncogenes. B) Next-generation sequencing of DNA extracted from cancer cells can identify somatic mutations that arise during carcinogenesis.
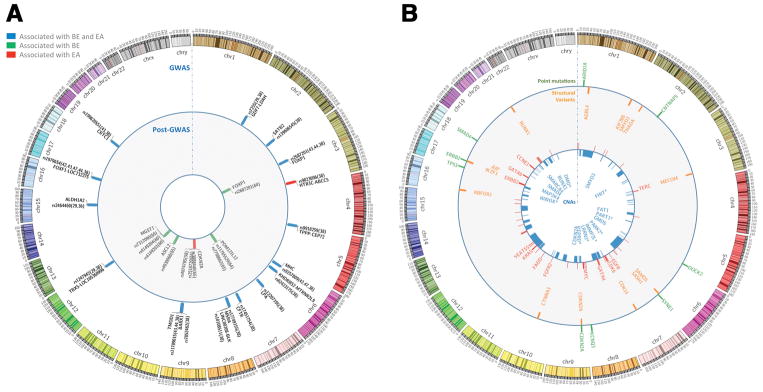
Figure 2. Variants that Increase Risk for BE and EA and Genomic Alterations Frequently Detected in EAs.
A) Circos plot of the loci associated with BE or EA risk in GWASs and in post-GWASs studies, reference to the first report followed by reference to confirmatory reports is shown in brackets. B) Circos plot of genomic alterations frequently detected in EAs. From the center of the circos to the outer ring: a) Significant regions of copy number losses (blue) according to the Gistic analysis (a tool to identify somatic copy number alterations; Broad Institute, US) reported by 78,85,91 on their respective cohorts; b) Copy number gains (red) according to the criteria above; c) Most frequent recurrent gene hits by SVs reported by 78, fragile sites were excluded; d) Recurrent point mutations in driver genes according to Mutsig and MutsigCV (bioinformatic tools to identify driver mutations; Broad Institute, US) in >/= 10% of cases by 54,78,91. * Common Fragile Site Genes. For an extended annotation of the data shown, see Supplementary Table 2.
Germline Variations and Susceptibility
Family studies
Evidence that germline mutations contribute to development of EA originated from reports of familial aggregation of this cancer19 and BE20–23. Orloff et al performed linkage analyses comparing 21 concordant affected sibling pairs (42 siblings with BE and/or EA) and 11 discordant sibling pairs using a 100K SNP set 24. Subsequent fine-mapping of regions of interest in an independent set of persons with BE or EA and controls, integration with publicly available gene expression data, and mutational analyses revealed 3 candidate genes for validation, performed in an independent set of 58 persons with BE or EA. Variants in MSR1, on chromosome 8p22, were significantly associated with BE or EA in the validation sample and in the pooled sample24. More recently, analyses of 42 multiplex pedigrees linked BE and EA with 3 chromosome regions (2q31, 4p14 and 12q23), and an additional region (15q26), in 18 female, affected pedigrees.25 The specific variants that mediate these associations have not been identified.
The extent to which BE or EA (including adenocarcinoma of the gastroesophageal junction) in siblings determines risk of BE or EA was examined using a training set data of 879 BE pedigrees and a validation set of data from 643 pedigrees, obtained from the Barrett’s Esophagus Translational Research Network 26. In male and female individuals, having a sibling with BE or EA associated with increased risk. For example, a 50-year old man with 1 unaffected brother was estimated to have a 3.2% baseline risk for BE or associated cancers. With 1 or 2 affected brothers, his risk increases 2.8-fold (to 9.1%) and 8.3-fold (to 26.6%), respectively. Similar increases in relative risk were estimated for a 50-year old woman, but applied to a much lower baseline risk (0.5%.) However, when the discrimination accuracy (determined from area under the curve) of a risk prediction model containing only demographic and clinical risk factors was compared to a model that contained family history, there was only minimal improvement (from 0.803 to 0.806). This likely reflects the relative rarity of a positive history in siblings in the general population, and the strength and higher prevalence of the other established risk factors that were included in the models.
Heritability
An estimate of heritability (genetic variance explained) of EA and BE among unrelated individuals was calculated using pooled genome-wide association study (GWAS, Supplementary figure 1) data from 1509 patients with EA, 2383 patients with BE, and 2170 control participants, contributed by 14 epidemiologic studies in the Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON). Using autosomal markers and genome-wide complex trait analysis, Ek et al estimated that 25% (standard error, 5%; one-sided P = 2 × 10−7) of EA cases and 35% of BE cases (standard error, 6%; one-sided P = 1 × 10−9) were determined by the composite effect of many common mutations of small individual relative risk 27,28. Furthermore, they demonstrated substantial polygenic overlap between EA and BE, indicating that shared genes influence the development of the 2 disorders. No other studies have reported on the EA genetic variance explained, nor on the overlap between EA and BE. However, Palles et al reported a lower figure for BE (10.0%; standard error,1.2%) for genetic variance explained. However this was based on the combined contributions of fewer single nucleotide polymorphisms (SNPs; 521,744 compared to 797,518 from the BEACON study)29.
A portion of the heritability of EA and BE may be explained by germline variants that affect development and severity of risk factors for these conditions, including symptomatic GER and obesity30–33. For example, a study based on self-administered questionnaires found that GER symptoms were substantially more prevalent among first-degree relatives of persons with BE or EA than among first-degree relatives of their spouses34. Twin studies of symptomatic GER support the concept of an important susceptibility component, with heritability estimates ranging from 13% to 41%35–37. Gharahkhani et al estimated heritability based on genotype arrays and reported that 7% of the variance in GER symptoms could be explained by genetic factors38. Furthermore, they found evidence for substantial genetic overlap between symptomatic GER and BE and EA. The heritability of obesity, measured by body mass index (BMI), waist circumference, and waist:hip ratio, appears to be even higher than for symptomatic GER, with estimates ranging from 40% to 70% from twin and family studies39. GWAS studies have identified close to 100 loci at the genome-wide level of significance (P<5 × 10−8), and estimated that more than 20% of variation in body mass index (BMI) can be accounted for by common variants 40. Using Mendelian randomization methods, researchers associated a risk score based on 29 BMI-associated variants was with a 12%–16% increase in risk of BE and EA, respectively, per 1 kg/m2 increase in BMI 41
GWASs
The first GWAS of BE was based on a discovery dataset of 1852 case and 5172 control participants from the Wellcome Trust Case Control Consortium42. After replication, researchers confirmed that 2 SNPs were associated with BE risk. One was located on chromosome 6p21, within the major histocompatibility complex, and the other on chromosome 16q24. A multi-phase extension of this study identified 3 additional loci, on chromosomes 2p24, 12q24, and 15q21 respectively, that were significantly associated with risk of BE 29.
A larger GWAS, which was the first to include EA cases (n=2390) in addition to BE cases (n=3175), was conducted by the BEACON consortium43. This study took advantage of the previous finding of extensive genetic overlap between EA and BE27, pooling BE and EA cases in the main analyses to increase statistical power. The researchers found 3 additional novel loci, on chromosomes 3p14, 9q22, and 19p13. They also observed that the previously reported association between BE and a locus on 16q24 also extended to risk of EA. Confirmatory evidence for an association between risk of EA and 3 of the 4 BEACON-reported SNPs (3p14, 9q22 and 16q24) was reported in a study from Germany using targeted genotyping44.
A meta-analysis of data from 4 GWASs, performed in 6 countries, included 4112 cases of EA, 6167 cases of BE, and 17,159 control participants of European ancestry45. The analysis confirmed associations between BE, EA, and the combined case group, with 7 of the 8 previously reported loci at the traditional level of statistical significance (P<5 × 10−8.) The 8th, on chromosome 9q22, narrowly missed this threshold (P=6.2 × 10−7.) This analysis also identified 9 additional loci, 8 of which were associated with BE and EA, and 1, on chromosome 3q27, which was associated with only EA.
In summary, a total of 17 independent loci associated with the development of BE and/or EA have been identified by traditional GWASs (Figure 2, Supplementary Table 2). One striking finding is that many of the identified SNPs are located in or near genes that regulate development and differentiation of the esophagus, stomach, and intestine (such as FOXP1, FOXF1, BARX1, GDDF1, and ABCC5)29,42,43,45–47. Given the importance of GER in development of BE and EA, and the fact that hiatal hernia substantially predisposes to GER, the findings identify mechanisms by which these variants might affect development of BE and EA. Support for this concept was provided by pathway analyses, which identified processes related to muscle cell differentiation as well as mesenchyme development and differentiation associated with these conditions45.
The large meta-analysis identified an intriguing association between a SNP on chromosome 7q31, located within the CFTR gene, and risk of BE and EA45. This gene is mutated in patients with cystic fibrosis—a condition characterized by severe dysfunction of the respiratory and gastrointestinal tract beginning in childhood, including a high prevalence of GER (in 35%–81% of patients) 48,49. The incidence of cystic fibrosis is about 6-fold higher in persons of European ancestry vs African ancestry, as are incidences of BE and EA.50 It was highlighted that CTFR and ABCC5 each encode proteins belonging in the same class of trans-membrane ion transporters (ATP-binding cassette), indicating an interesting area for research into pathogenic mechanisms of these disorders51.
After GWAS
Moving beyond GWAS, investigators have used a variety of analytic approaches to explore the influence of genetic factors on EA pathogenesis, including integrating knowledge of somatic mutation signatures with germline data, performing pathways-based analyses, and using epidemiologic data to examine genetic associations with risk factors. Somatic mutations occur at high frequencies in the CDKN2A and TP53 tumor suppressor genes in EAs (and other malignancies) 52–54; loss of heterozygosity at these loci has been associated with progression from BE to cancer (see section on somatic mutation analyses). 55–57 Reasoning that these loci may be implicated in susceptibility to cancer, investigators from the BEACON consortium tested 13 SNPs at the TP53 locus and 24 SNPs in CDKN2A, which were within 2-kb flanking regions and satisfied quality control constraints. While none of the SNPs in TP53 were associated with EA risk, 3 polymorphisms in CDKN2A were associated with a 10%–16% reduction in risk for EA (p<0.05) (Figure 2, Supplementary Table 2)58. The investigators then tested whether any of the variants predicted neoplastic progression in a separate prospective cohort of 408 patients with BE, and found that 2 of the variants (rs2518720; hazard ratio, 0.57 and rs3088440; hazard ratio, 0.34) were independently significantly associated with reduced risks of progression. Expression of one of the variants (rs3088440) in cell lines indicated that it reduces microRNA-mediated repression of the CDKN2A mRNA.
Systemic and local (esophageal) inflammation, caused by factors such as abdominal obesity, GER, and cigarette smoking, may be a common pathway in the development of BE and EA59. The role of genetic variation in inflammatory responses was investigated using a principal components-based approach in the BEACON GWAS. Variants in the cyclooxygenase (COX) pathway were significantly associated with risk of BE. Gene-level analyses identified an association with MGST1 (on chromosome 12p12), and a meta-analysis, which added BE and control participants from the Wellcome Trust GWAS, confirmed associations between 4 SNPs and risk of BE (Figure 2, Supplementary Table 2) 60. Analyses of GWAS data examining the role of germline variation in other pathways, including the biogenesis and activity of microRNAs61, androgens62, and the estrogen and oxytocin pathways63, also indicated associations, but these have not been replicated.
Gene–Environment Interactions
Some genetic factors affect susceptibility to BE or EA depending on other factors. Another approach to identifying so-called risk-modifying genes is therefore to test for differences in statistical associations across strata of exposure to those factors (BMI, sex, etc.) Using the well-annotated BEACON GWAS, Dai et al, examined the first 7 SNPs identified as associated with BE or EA at the genome-wide level of significance for interactions with BMI, GER symptoms, and smoking status.64 They found that the previously identified variant near FOXP1 (rs2687201) significantly modified the association between GER symptoms occurring at least weekly and risk of BE, such that the association was stronger (odds ratio, 6.2) among persons with 0 minor alleles, compared to those with 1 or 2 (odds ratios, 3.6 and 4.0, respectively,) (Pinteraction=0.0005; FDR=0.042.)
Dai et al developed a set of constrained testing methods to increase statistical power for tests of gene–environment interactions in settings in which several risk factors may act through a common pathway.65 Inflammation is a frequently accompanies cigarette smoking, abdominal obesity, and GER. When the constrained score statistics were applied to the BEACON dataset, 3 loci were identified that simultaneously interacted with smoking, obesity, and GER (Supplementary Table 2). Further explorations in this area will likely require much larger datasets that also include accurate annotation of key environmental risk factors.
Pleiotropic analysis of risk loci
To investigate whether risk-associated loci from GWAS of other cancer sites might also modify risk of BE or EA, Lee et al tested 387 candidate SNPs.66 None were found to be associated with risk of BE or EA, and there was no evidence for interactions with smoking, obesity, or GER symptoms.
Somatic Mutations that Affect BE Progression
With the advent of next-generation sequencing, mutations have been reported from hundreds of cases in studies of coding regions (whole-exome sequencing, WES) and the entire genome (whole-genome sequencing, WGS). These data can be obtained from 2 large pan-cancer consortia: the Cancer Genome Atlas (https://tcga-data.nci.nih.gov) and the International Cancer Genome Consortium (http://icgc.org). New data are being added every day.
Progression from pre-malignant BE to EA
There was a reasonable expectation that sequencing the genomes of BE or EA tissues would identify somatic alterations required for progression from BE to EA. This was expected to lead to biomarkers that could assist clinicians in identifying preneoplastic lesions at highest risk for progression to invasive cancer. In our current model, dysplasia progresses to invasive EA via early loss of CDKN2A, emergence of dysplastic clones with mutations in TP53 and/or additional somatic alterations, and increases in copy number 55,57,67–73. Although the basics of this model, largely characterized before the advent of NGS techniques, appear to hold true, sequencing studies have shown the BE genome to be highly complex—even when non-dysplastic for many years —and that progression can be non-linear74,75
It is now apparent that point mutations accumulate during early stages of disease and BE lesions often have a higher rate of mutation rate than many common, invasive cancers 52,70,75. At the time BE becomes dysplastic, the tissue has a mutation rate comparable to that of EA52,75. Mutations are found in a number of tumor suppressor genes important in chromatin remodeling, such as ARID1A and SMARCA475 (Supplementary table 3). Mutations in TP53 and SMAD4 are usually found only in tissues with high-grade dysplasia and EA, respectively. In contrast to patients with NDBE with no history of disease progression, mutations in TP53 are found in NDBE tissues adjacent to EA 67,70. This observation is consistent with the high allele fraction of TP53 mutations in many different cancer types, indicating that either this mutation appears early during tumorigenesis or it is able to promote expansion of a dominating clone 76.
Mutations in PIK3A and CTNNB1 have also been found in BE, although accumulation of activating mutations and amplifications in oncogenes is a marker of invasive EA 70. Similarities in mutation patterns provide evidence for the common origin of BE and EA(see Figure 3)53,70. However, fewer than 20% of specific variants overlap between adjacent BE and EA, so either the cancer clone diverged at an early stage or originated separately 53,70. Analysis BE patients suggested that the genetic diversity of different clones did not change significantly over time, but the extent of divergence of clones at baseline was the strongest predictor of progression77.
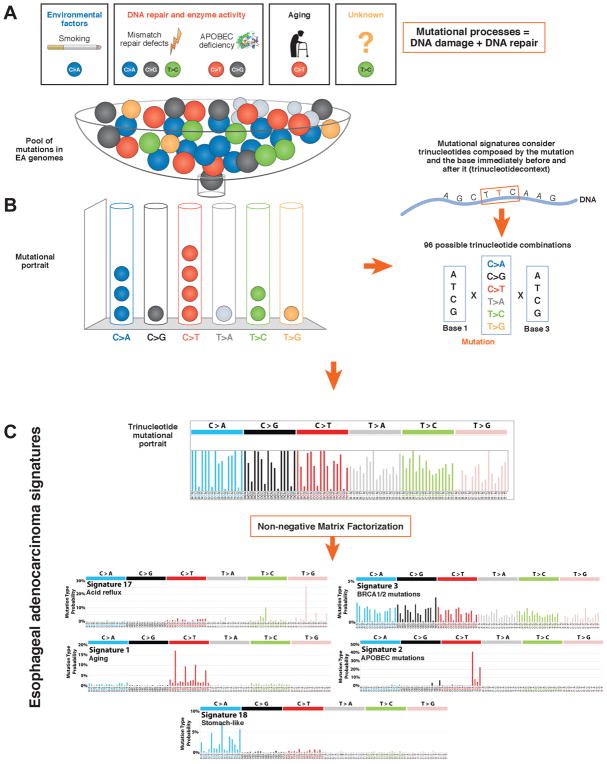
Figure 3. Mutational Signatures of Tumors.
A) Mutational processes are biological activities (e.g.: aging, smoking, UV light exposure, unknown carcinogens) that generate patter of mutations (mutational signatures) through a damage of the DNA sequence and its attempt to repair it by DNA repair mechanisms. B) The mutational portrait is the the total pattern of genetic changes in cancer cell that derive from the sum of all the mutational signatures occurring in a lifetime 86. C) Mathematical approaches, such as non-negative matrix factorization (NMF), can be used to extract mutational signatures from the mutational portraits of groups of patient’s cancer genomes. The pattern includes all base substitutions and flanking nucleotides (96 possible combinations shown in bar charts). NMF estimates the relative contribution of each signature to the mutational portrait and can highlight cancers that are predominantly driven by some mutational signatures. A comprehensive catalogue of the signatures identified by Alexandrov et al is available on the catalogue of somatic mutations in cancer (COSMIC, www.cancer.sanger.ac.uk). Mutation signatures associated with EA include a) S17, also called an acid signature—there are 2 forms, S17A and B; b) S3, associated with defects in the BRCA1/2-led homologous recombination pathway; c) S1, associated with aging; d) S2, caused by APOBEC mutations, and e) S18, detected in gastric cancer and neuroblastoma, arises via an unknown mechanism78,85.
The mutational lanscape found in BE and EA differs more dramatically at a chromosomal scale. For example, compared to BE epithelium, EAs have marked differences in genomic copy number profiles. Genomes of BE tissues are relatively stable compared to those of invasive tumors, in which almost 40% of the genome is non-diploid (median, range 2%–97%). The only common copy number alteration found in BE is 9p loss of heterozigosity (CDKN2A) 53,70,71. Invasive tumors have an increased number copy numbers of several oncogenes (GATA4, KLF5, MYB, PRKCI, CCND1, FGF3, FGF4, FGF19, and VEGFA) and loss of common fragile sites (FHIT, WWOX, PDE4D, PTPRD, and PARK2) 53,70,78–80
The stochastic and gradual accrual of copy number alterations fits into the linear multistep process of BE progression, but does not entirely account for the frequent whole-genome doubling observed by Stachler at al— particularly in EA tissues with TP53 mutations. The authors propose that following TP53 loss, whole-genome doubling occurs, which accelerates tumor progression and requires few other mutations 70. It is also observed that BE can progress to cancer via multiple different pathways, and suddenly accelerate, due to crises involving large regions of the genome (genomic catastrophes). Tumors with unstable genomes are more likely to progress rapidly 55,59,81, so the frequency of copy number changes is a good biomarker for development of EA. In the 24 months before a patient is diagnosed with esophageal cancer, biopsies from BE tissues show a marked increase in DNA content 69. These findings indicate that the time course and pathways to tumor development vary to a greater extent than previously appreciated (Figure 4).
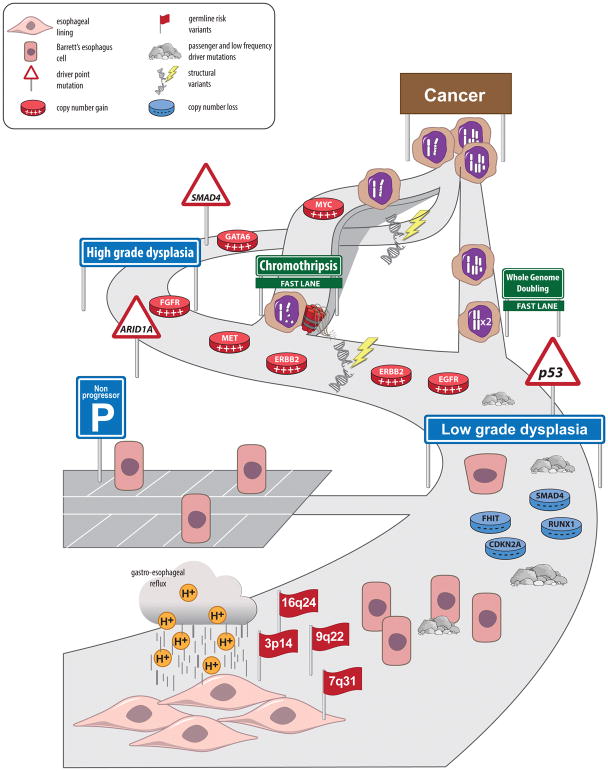
Figure 4. Paths of BE Progression to EA.
Findings from next-generation sequencing studies indicate BE progression can accelerate via genome doubling, genome catastrophes, and other unknown mechanisms—even at early stages of tumor progression. The main path represent the multistep progression of BE to EA through dysplasia. BE and EA pathogenesisis include genetic risk factors (each flag indicate GWAS identified regions), exposure to environmental risk factors (e.g. acid reflux) and the accumulation of different types of driver and passenger mutations. Genomic catastrophes such as chromothripsis and whole genome doubling can occour at any stange and dramatically accelerate progression of BE.
On a practical note, it is a challenge to predict the lifetime course of a patient’s BE progression. In the past, when esophagectomy was the only therapy available, patients were followed until it was clear they had invasive cancer. Now, intervention is appropriate earlier in the disease course 11,12, due to the availability of outpatient-based endoscopic techniques such as endoscopic mucosal resection and radiofrequency ablation. The agenda has therefore shifted towards identifying early genomic events that distinctly mark the presence of dysplasia, awaiting for more refined risk models for NDBE. The modality of tissue sampling is critical, because BE is a polyclonal disease and endoscopic biopsies have inherent sampling bias. Fortunately, several new modes of sample collection have been developed, which could overcome some of these limitations.
One of these approaches, the Cytosponge sampling device, collects cells from the entire length of the esophagus; it is simple to perform and inexpensive, allowing for repeated sample collection in a primary care setting 82,83. The diagnostic yield of the Cytosponge for new cases of BE in individuals with a history of reflux is being compared with standard of care in a cluster randomised clinical trial of 9,000 patients in primary care (registration number: REC 16-EE-0546). As well as diagnosing BE as noted previously risk stratification is essential. Analysis of a single Cytosponge sample was able to recapitulate the same sequencing results as samples collected from polyclonal lesions in multiple biopsies 53,75. Furthermore, a panel of biomarkers can be applied to BE cells from the same sample (identified as Trefoil Factor 3 (TFF3) positive cells at immunostaining), in order to stratify patients into three risk groups according to the following criteria: presence of glandular atypia, p53 abnormality and a ploidy measure (Aurora kinase A positivity), along with joint effects of major risk factors such as age, obesity and length of the Barrett’s segment (if known). Using this algorithm 35% patients fell into the low-risk category, and were eligible for a less-intense surveillance regimen and this was reliable in a validation cohort 84. (Figure 5).
Figure 5. Translating Findings from Genetic Studies Into Clinical Practice.
Genetic data can be used to determine an individual’s risk for developing BE or EA, and to manage patients at different stages of disease progression. Test are available for use in primary (pink) secondary (light blue), and tertiary (orange) care settings. For each group (left), we provide example of clinical applications. The most suitable technology for each test is presented in the bottom row. The left column indicates the group size relative to the general population.
In summary, it seems that regardless of the sampling method, more informative assays are required to identify genomic instability and increasing copy number in patients requiring endoscopic therapy; this would avoid reliance on detection of dysplasia as the basis for clinical decision making 55,69,77.
Whole-exome and whole-genome analyses of EA
Point mutations and indels
Based on sequencing studies, EAs have a high degree of inter-sample genomic heterogeneity and a high mutation burden. Each tumor genome has a median 8 mutations/Mb (range, 1.5–35 mutations/Mb)—one of the highest mutation rates observed in tumors, along with bladder, colorectal, and lung tumors and melanoma 85. Other tumor types, such as breast and ovarian tumors, have fewer than 2 mutations/MB respectively 76. EAs might have a high mutation rate depending on the esposure to environmental mutagens, the efficiency in DNA repair, the rate of proliferation, and ther inflammatory response. Although no mutagen has been convingingly proven to cause esophageal adenocarcinoma (EA), carcinogenesis is believed to involved acid and bile reflux. Little is known about the mechanisms by which these luminal constituents might cause DNA damage, inherited mismatch repair gene deficiencies are not commonly observed 78.
One method to identify and classify mutational processes is through the statistical analysis of the frequency of base-changes (A>C, T>G etc.) throughout the entire genome (mutational portrait) (Figure 3). This can be carried out by analysis of 1 base at a time or in the context of the base either side (so-called tri-nucleotide context). Analyses of a large number of normal and cancer tissue genomes have identified mutation signatures. These have, in some cases, be associated with mutagens such as ultraviolet radiation, cigarette smoke, or aging 86 (Figure 3). Alexandrov et al created catalogue of these signatures, using a non-negative matrix factorization algorithm. Tumors can therefore be characterized according to the most commonly occurring signatures (S,number), 87 (Figure 3).
One interesting aspect of EA is the frequency of T>G substitutions in a CTT context, called the S17 signature. This mutation signature has been associated with gastric acid reflux and often referred to as an acid-signature 78,85. Other signatures include one associated with aging (S1)—a complex pattern caused by defects in the BRCA1/2-regulated homologous recombination pathway (S3); C>T mutations in a TCA/TCT context, due to apolipoprotein B mRNA editing enzyme catalytic polypeptide-like(APOBEC)-mutations (S2); and C>A/T dominant in a GCA/TCT context (S18), also found in gastric carcinoma and neuroblastoma78,85. The APOBEC signature has been associated with characteristic clusters of localized hypermutation named kataegis, in which a single strand accumulates a high burden of C>T and C>G mutations 88. Further analysis of these signatures may help to elucidate mechanisms of carcinogenesis and to aid in classification and treatment 78 (Figure 3).
For a cancer to occur it is estimated that at least 3 driver genes mutations are required 89. Despite the large number of mutations found in EA tissues, they contain an average of 1.7 driver mutations per case. Bioinformatic tools can be used to identify driver mutations, such as MutSig and more recently MutSigCV. These have identified only 8 genes that are consistently mutated (in more than 10% of cases) 54,75,78,90,91 (Figure 2, Supplementary Table 2, Supplementary Table 3). TP53 is by far the most frequently mutated gene—more than 70% of samples contain loss of function mutations in TP53. Studies are needed to determine the combination of mutations required for EA tumorigenesis.
Copy number alterations and structural variants
Two WGS studies have highlighted that EA genomes are predominantly characterized by large scale genomic rearrangements (i.e. structural variants) and gain or losses of genomic regions (copy number alterations) 78,85 (Figure 1). Chromosome instability stands out as a hallmark of EA when compared to squamous esophageal cancer and gastric adenocarcinoma 10,91. Copy number alterations of genes encoding EGFR, ERBB2, MET, and FGFR2 and other receptor tyrosine kinases are also common in EA and show a high degree of redundancy with downstream targets 79,92,93 (Figure 2, Figure 4, Supplementary Table 2).
Rearrangements are variably distributed in the genomes of EA samples. Nones et al proposed a classification of EA genomes unstable (with 450 or more structural variations), scattered (fewer than 450 structural variations, evenly distributed across the genome) and complex localized (with a concentration of clustered structural variationsin a single or few chromosomes), based on the pattern of structural variations distribution 85.
Highly recurrent rearrangements have been mainly reported in common fragile sites but their biological significance is unclear. For instance, the fragile histidine triad gene (FHIT or FRA3B) and WW domain containing oxidoreductase gene (WWOX or FRA16D) contain rearrangements in up to 95% of cases. Despite evidence that these are tumor suppressor genes 94,95,94, their loci are frequently rearranged following perturbation of DNA replication and replication stress96,97. Beside common fragile sites, structural variations could be a common mechanism of recurrent mutation in EA. RUNX1, a gene translocated in acute myeloid leukemia, and SMYD3, are rearranged in 39% and 27% of cases of EA78. Although functional studies are needed to confirm a driver role in EA, these alterations are possibly the most common after TP53 mutations.
In addition, a peculiar class of structural variations is represented by mobile element insertions that occur as a consequence of the excision and re-insertion of repeated L1 and Alu sequences that are transposed as DNA or through the reverse transcription of an mRNA intermediate. In EA, L1 insertions have been reported in the coding sequence of several genes (ERBB4, CTNNA3, CTNNA2, CDH18, and SOX5). Mobile element activity represent the most relevant contributor to the total SV burden in several EA genomes but further work is required to clarify their functional consequences 78,98,99.
WGS has revealed that many EA samples have evidence of genomic catastrophes, which result in the accumulation of structural variants in specific areas of the genome. These can be single events (chromothripsis) or repeated breakage–fusion–bridge cycles100,101 (Figure 1B). There is evidence for chromothripsis in about 30% of EAs and breakage–fusion–bridge events in 25% of EAs. Genomic catastrophes could be a common mechanism through which preneoplastic lesions rapidly progress to invasive tumors 78,85,102–104 (Figure 1 and Figure 4). Crises or punctuated equilibria (as opposed to gradual mutational accrual) can alter cell phenotyes and overcome the oncogene stress (cell cycle arrest or cellular senescence due to activation of an oncogene).
Furthermore, shattered chromosome segments not incorporated into the derivative chromosome can be linked to form a double-minute (circular) chromosome 104. For example, MYC-containing double minutes have been convincingly described in chromothriptic EAs 85,105. The second type of genomic catastrophes, breakage–fusion–bridge cycles, are related to telomere shortening, observed in advanced EAs 85. Unprotected telomere ends and sister chromatids fuses and are subsequently torn apart during anaphase. This process can be repeated for several cell cycles resulting in inverted duplications increase copy numbers of genes including KRAS, MDM2, and VEGFA 85.
Application to Therapy
Most cases of EAs present de novo without any prior diagnosis of BE. Standard treatment for EA remains chemo-radiotherapy followed by surgery and only incremental gains in survival have been made in the last 20 years. Since loss of TP53 is the most common mutation, it would make sense to try to restore its function as a therapeutic strategy. Several agents designed to increase P53 activity are in development, but their clinical efficacy has not yet been demonstrated (reviewed in 106).
Most trials of patients with EA have targeted receptor tyrosine kinases. However, trials are often performed without information on the expression level of the targeted receptor within the tumor. Recent sequencing data indicate that tumors from each patient have dysregulations in multiple receptor tyrosine kinases and their signaling pathways, so multiple agents could be required 78,79,93. An approach to overcome the genomic heterogeneity of EA could be to identify broader pathways that may emerge by a combined analysis of low frequency somatic variants and transcriptome. The clinical translation of such an approach is an open challenge due to the exponential complexity of cross talks and primary or secondary resistance 107,108. Mutational signature analysis is an alternative method for identifying therapeutic vulnerability in subgroups of patients. EA genomes can be grouped into 3 categories according to their dominant mutation signatures: C>A/T dominant (associated with age, S18-like), DNA damage repair impaired (BRCA), and mutagenic (predominantly S17A or S17B) 78. EAs with a DNA-damage repair defect signature have significantly more defects in homologous recombination and chromosome segregation pathways and may therefore respond better to DNA-damaging agents or photon irradiation in combination with inhibitors of PARP. On the other hand, EAs with a mutagenic signature have a higher neoantigen load and are characterized by infiltration of CD8+ T cells; these are more likely to respond to blockade of PDL1 and CTLA4, as observed in studies of patients with non small-cell lung cancer and melanoma 109. More pre-clinical studies are needed to test these strategies.
An area of active investigation is whether genomic alterations detected in endoscopic biopsies, cytosponge or peripheral blood samples can be used as a tool to monitor disease during treatment or surveillance. In peripheral blood samples, circulating tumor DNA has been used successfully to monitor response to therapy, as well as to identify the emergence of novel alterations conferring secondary resistance to targeted therapies 110,111 (Figure 5). In addition, genomic alterations indicating locoregional recurrence might be detected in biopsies and Cytosponge samples earlier than currently available diagnostic tests. Clinical trials are evaluating close follow-up as an alternative to surgery in patients with complete pathological response to neoadjuvant radio-chemotherapy112. Early detection of recurrence could offer further margins for salvage surgery in those patients.
Future Directions
In the past few years there have been intense international collaborative efforts to increase our understanding of genetic factors associated with BE and EA. These have produced many important and exciting findings. Genome-wide susceptibility scans have been performed on more than 6000 patients with BE and more than 4000 patients with EA. Although such sample sizes might seem large from the perspective of gastroenterology, they are dwarfed by patient collections for other human traits such as obesity (~300,000)40 and breast cancer (~62,000 cases)113. Experience gained from studying those other conditions has shown that increasing study size brings greater ability to detect associations with rare genetic variants.
For now, however, several conclusions seem reasonable. BE and EA each have sizable heritable components, estimated at around 35% and 25% respectively; heritability is conferred by a combination of many genetic factors that each increase risk by a small amount. Almost all of the genetic variants discovered have been associated with BE and EA, indicating that EA arises via a metaplasia–dysplasia–carcinoma pathway.
Annotations of the genes indicate that aberrations in the musculature of the foregut, perhaps during embryogenesis, that likely contribute to GER. GER is associated with development of BE and EA. Understanding the underlying functional mechanisms through which these variants act to increase risk will be necessary for these discoveries to yield practical utility.
The value of exploring genomic data using a variety of approaches beyond straightforward association testing has been demonstrated already, but much work remains to done. More powerful methods for analyzing pathways and detecting gene-environment interactions need to be explored for germline as well as somatic variants. Well-annotated epidemiologic datasets with careful measurement of environmental and phenotypic factors will be essential to achieve these aims. For example, exploring the possible role of DNA repair genes in esophageal carcinogenesis would be enhanced by analyzing associations with genotype separately among ever smokers and never smokers. Functional studies will be needed to characterize the downstream effects of genetic variants identified through the association studies currently underway. In the absence of valid animal models for BE or EA, mechanistic data will be crucial for identifying potential targets for treatment or chemoprevention.
Genomes of BE and EA cells are both highly mutated and heterogeneous and contain well-defined mutational signatures. Analyses of germline and somatic mutations indicate that these disorders have a common etiology. Several pathways are involved in the progression from BE to EA but the final common feature is that of an abnormal copy number profile with large-scale structural variants, and with amplifications and complex rearrangements occurring to a variable extent. In some instances, disease progression can be very slow with a cumulative number of points mutations in tumor suppressor genes; other tumors have punctuated evolution resulting in rapid progression.
To generate a coherent picture of genomic alterations that accumulate during progression of EA, genomes of EA must be integrated with analyses of the epigenome, transcriptome, and proteome and compared with other cancers. Significant effort will be required to investigate the causes and consequences of mutations—especially with respect to rearrangements and the functional consequences of putative driver mutations.
There are many opportunities to translate the findings from genetic susceptibility and somatic sequencing studies to the clinic (Figure 5)114. Functional studies of susceptibility loci, to identify causal variants and the carcinogenic mechanisms, may eventually lead to the discovery of biomarkers of risk and targeted approaches to prevention and treatment. Data on susceptibility loci could be combined with clinico-demographic factors to generate a risk score, to identify individuals who might benefit from targeted screening for BE. Screening tests for BE could use newer non-invasive sampling methods more suitable for primary care. However, rather than relying solely on histopathological assessment, inclusion of biomarkers emerging from genome wide data could help objectively determine whether the epithelium is genomically unstable.
The most important criterion for determining the success of such tools is whether they accurately predict cancer risk; this needs to be assessed in large prospective studies. Ideally the term dysplasia would be superceded by a molecular readout detailing TP53 mutations and copy number alterations, so that a risk:benefit profile for endoscopic therapy can be determined. For patients with invasive cancer, increasing our understanding of the genomic landscape could improve tumor sub-classification and lead to personalized therapy. Tests might soon be available to detect DNA shed from tumors into blood; these could be used to monitor response to therapy or identify emergent clones for therapeutic targeting 110. With the advent of affordable and rapid sequencing technologies, we can move from discovery science into the clinic.
Supplementary Material
Techniques Used to Analyze Tumor Genomes
Data to Support Figure 2
A) Loci associated with BE or EA from GWASs and post-GWASs; B) significant GISTIC copy number gain regions detected by next-generation sequencing studies; C) significant GISTIC copy number loss regions detected by next-generation sequencing studies. (format xlsx file)
Point Mutations that Promote Development of EA.
These were identified by MutSig or MutSigCV, with a greater than 10% frequency in at least 1 next-generation sequencing study. * NCC: network of Cancer Genes, ncg.kcl.ac.uk 116.
GWAS analysis of germline DNA (from peripheral blood) can identify variants associated with the development of a condition
Table 1.
Whole-exome and Whole-genome Studies of EAC.
| Consortium | Sequencing technique | Cohort (Number of cases) | Sample Type | Main findings | Study |
|---|---|---|---|---|---|
| N/A | WES + sanger sequencing | 11 Chemonaive EAs, 12 Chemonaive esophageal squamous cell carcinoma, 2 Matched BE | Frozen biopsies |
|
(Agrawal et al. 2012) |
| TCGA | WES + WGS on selected samples | Chemonaive EAs(149) | Frozen biopsies |
|
(Dulak et al. 2013) |
| ICGC | WGS + Targeted sequencing | Chemonaive EAs (112), BE (84), HGDs (61) | Frozen biopsies, Cytosponge ® |
|
(Weaver et al. 2014) |
| ICGC | WGS +targeted sequencing | Paired BE and chemonaive EAs samples (23), longitudinal sampling of BE (1) | Frozen biopsies, paraffin, Cytosponge ® |
|
(Ross-Innes et al. 2015) |
| TCGA | WGS + targeted sequencing | Paired BEs and chemonaive EA samples (25), multiple sampling of BE and EAs (5) | Frozen biopsies, paraffin |
|
(Stachler et al. 2015) |
| N/A | WGS | Chemonaive EAs (22) | Frozen biopsies |
|
(Nones et al. 2015) |
| N/A | WES | Multiregion, paired pre+post chemotherapy (8) | Frozen biopsies |
|
(Murugaesu et al., 2015) |
| ICGC | WGS | Chemonaive EAs (129) | Frozen biopsies |
|
(Secrier et al. 2016) |
| TCGA | WES + SNParrays integrated with DNAmethylation and mRNA-sequencing | gastroesophageal samples (592) of which 171 Chemonaive EAs and 90 Chemonaive esophageal squamous cell carcinoma | Frozen biopsies |
|
(Kim et al. 2017) |
| ICGC | WGS | Chemonaive EAs (62), and chemotherapy treated EAs (58) and comparison of matched pre and post chemo EAs (10) | Frozen biopsies |
|
(Noorani et al., 2017) |
TSG, tumor suppressor gene.
Acknowledgments
Funding sources
G.C. is a National Institute for Health Research Lecturer as part of a NIHR professorship grant to R.C.F. TLV supported in part by NIH K05CA124911 and NIH R21 CA197502, DCW is supported by a Research Fellowship (APP1058522) from the National Health and Medical Research Council of Australia. R.C.F. is funded by an NIHR Professorship (RG67258), the Medical Research Council (RG74187), the Biomedical Research Centre and the Experimental Cancer Medicine Centre, and a programme grant from Cancer Research UK (RG66287).
Abbreviations
- EA
Esophageal adenocarcinoma
- BE
Barrett’s esophagus
- NDBE
Non-dysplastic Barrett’s esophagus
- GWAS
Genome wide association study
- GER
Gastro-esophageal reflux
- WGS
Whole genome sequencing
Footnotes
Conflict of interest
RCF is named on patents pertaining to the Cytopsonge and associated assays that have been licensed by the Medical Research Council to Covidien (now Medtronic). All the authors declare no other competing interests.
Author’s contribution
GC and RCF designed the review, GC, TLV, DW and RCF wrote the manuscript, GC designed figures and tables.
References
Author names in bold designate shared co-first authorship.
- 1.Hur C, Miller M, Kong CY, et al. Trends in esophageal adenocarcinoma incidence and mortality. Cancer. 2013;119:1149–1158. doi: 10.1002/cncr.27834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Cancer Institute. Cancer of the Esophagus - SEER Stat Fact Sheets. Cancer Stat. 2015 Available at: http://seer.cancer.gov/statfacts/html/esoph.html.
- 3.Cancer Resarch UK. Oesophageal cancer incidence statistics. Available at: cancerresearchuk.org/cancer-info/cancerstats/
- 4.Hirst J, Smithers BM, Gotley DC, et al. Defining cure for esophageal cancer: analysis of actual 5-year survivors following esophagectomy. Ann Surg Oncol. 2011;18:1766–74. doi: 10.1245/s10434-010-1508-z. [DOI] [PubMed] [Google Scholar]
- 5.van Hagen P, Hulshof MCCM, van Lanschot JJB, et al. Preoperative Chemoradiotherapy for Esophageal or Junctional Cancer. N Engl J Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 6.Anaparthy R, Sharma P. Progression of Barrett oesophagus: role of endoscopic and histological predictors. Nat Rev Gastroenterol Hepatol. 2014;11:525–534. doi: 10.1038/nrgastro.2014.69. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan TL, Fitzgerald RC. Precision prevention of oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:243–248. doi: 10.1038/nrgastro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spechler SJ, Souza RF. Barrett’s esophagus. N Engl J Med. 2014;371:836–845. doi: 10.1056/NEJMra1314704. Available at: http://dx.doi.org/10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 9.Spechler SJ, Fitzgerald RC, Prasad GA, et al. History, Molecular Mechanisms, and Endoscopic Treatment of Barrett’s Esophagus. Gastroenterology. 2010;138:854–869. doi: 10.1053/j.gastro.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett’s esophagus in the general population: An endoscopic study. Gastroenterology. 2005;129:1825–1831. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 11.Phoa KN, van Vilsteren FGI, Weusten BLaM, et al. Radiofrequency Ablation vs Endoscopic Surveillance for Patients With Barrett Esophagus and Low-Grade Dysplasia. Jama. 2014;311:1209. doi: 10.1001/jama.2014.2511. [DOI] [PubMed] [Google Scholar]
- 12.Shaheen NJ, Sharma P, Overholt BF, et al. Radiofrequency ablation in Barrett’s esophagus with dysplasia. N Engl J Med. 2009;360:2277–88. doi: 10.1056/NEJMoa0808145. [DOI] [PubMed] [Google Scholar]
- 13.Rastogi A, Puli S, El-Serag HB, et al. Incidence of esophageal adenocarcinoma in patients with Barrett’s esophagus and high-grade dysplasia: a meta-analysis. Gastrointest Endosc. 2008;67:394–398. doi: 10.1016/j.gie.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 14.NICE. Clinical Guidelines [CG106] Barrett ’s oesophagus: ablative therapy [Google Scholar]
- 15.Shaheen NJ, Falk GW, Iyer PG. ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. 2015:1–21. doi: 10.1038/ajg.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corley DA, Mehtani K, Quesenberry C, et al. Impact of endoscopic surveillance on mortality from Barrett’s esophagus-associated esophageal adenocarcinomas. Gastroenterology. 2013;145:312–9.e1. doi: 10.1053/j.gastro.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spechler SJ, Sharma P, Souza RF, et al. American gastroenterological association technical review on the management of Barrett’s esophagus. Gastroenterology. 2011;140:18–52. doi: 10.1053/j.gastro.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao Y, Hyder A, Bae SJ, et al. Surveillance in Patients With Barrett’s Esophagus for Early Detection of Esophageal Adenocarcinoma: A Systematic Review and Meta-Analysis. Clin Transl Gastroenterol. 2015;6:e131. doi: 10.1038/ctg.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jochem VJ, Fuerst PA, Fromkes JJ. Familial Barrett’s esophagus associated with adenocarcinoma. Gastroenterology. 1992;102:1400–1402. [PubMed] [Google Scholar]
- 20.Crabb DW, Berk MA, Hall TR, et al. Familial gastroesophageal reflux and development of Barrett’s esophagus. Ann Intern Med. 1985;103:52–54. doi: 10.7326/0003-4819-103-1-52. [DOI] [PubMed] [Google Scholar]
- 21.Chak A, Faulx A, Kinnard M, et al. Identification of Barrett’s esophagus in relatives by endoscopic screening. Am J Gastroenterol. 2004;99:2107–14. doi: 10.1111/j.1572-0241.2004.40464.x. [DOI] [PubMed] [Google Scholar]
- 22.Chak A, Lee T, Kinnard MF, et al. Familial aggregation of Barrett’s oesophagus, oesophageal adenocarcinoma, and oesophagogastric junctional adenocarcinoma in Caucasian adults. Gut. 2002;51:323–8. doi: 10.1136/gut.51.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X, Elston R, Barnholtz-Sloan J, et al. A segregation analysis of Barrett’s esophagus and associated adenocarcinomas. Cancer Epidemiol Biomarkers Prev. 2010;19:666–74. doi: 10.1158/1055-9965.EPI-09-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orloff M, Peterson C, He X, et al. Germline Mutations in MSR1, ASCC1, and CTHRC1 in Patients With Barrett Esophagus and Esophageal Adenocarcinoma. JAMA. 2011 Jul 27;306(4):410–419. doi: 10.1001/jama.2011.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Elston R, Falk GW, et al. Linkage and related analyses of Barrett’s esophagus and its associated adenocarcinomas. Mol Genet Genomic Med. 2016 doi: 10.1002/mgg3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Elston RC, Barnholtz-Sloan JS, et al. Predicting Barrett’s Esophagus in Families: An Esophagus Translational Research Network (BETRNet) Model Fitting Clinical Data to a Familial Paradigm. Cancer Epidemiol Biomarkers Prev. 2016;25:727–735. doi: 10.1158/1055-9965.EPI-15-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ek WE, Levine DM, D’Amato M, et al. Germline genetic contributions to risk for esophageal adenocarcinoma, Barrett’s esophagus, and gastroesophageal reflux. J Natl Cancer Inst. 2013;105:1711–8. doi: 10.1093/jnci/djt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palles C, Chegwidden L, Li X, et al. Polymorphisms near TBX5 and GDF7 are associated with increased risk for Barrett’s esophagus. Gastroenterology. 2015;148:367–378. doi: 10.1053/j.gastro.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook MB, Corley DA, Murray LJ, et al. Gastroesophageal Reflux in Relation to Adenocarcinomas of the Esophagus: A Pooled Analysis from the Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON) PLoS One. 2014;9:e103508. doi: 10.1371/journal.pone.0103508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corley DA, Kubo A, Levin TR, et al. Abdominal Obesity and Body Mass Index as Risk Factors for Barrett’s Esophagus. Gastroenterology. 2007;133:34–41. doi: 10.1053/j.gastro.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 32.Edelstein ZR, Farrow DC, Bronner MP, et al. Central adiposity and risk of Barrett’s esophagus. Gastroenterology. 2007;133:403–11. doi: 10.1053/j.gastro.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Hoyo C, Cook MB, Kamangar F, et al. Body mass index in relation to oesophageal and oesophagogastric junction adenocarcinomas: a pooled analysis from the International BEACON Consortium. Int J Epidemiol. 2012;41:1706–18. doi: 10.1093/ije/dys176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero Y, Cameron AJ, GRL, et al. Familial aggregation of gastroesophageal reflux in patients with Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 1997;113:1449–1456. doi: 10.1053/gast.1997.v113.pm9352846. [DOI] [PubMed] [Google Scholar]
- 35.Cameron AJ, Lagergren J, Henriksson C, et al. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology. 2002;122:55–59. doi: 10.1053/gast.2002.30301. [DOI] [PubMed] [Google Scholar]
- 36.Lembo A, Zaman M, Jones M, et al. Influence of genetics on irritable bowel syndrome, gastro-oesophageal reflux and dyspepsia: a twin study. Aliment Pharmacol Ther. 2007;25:1343–1350. doi: 10.1111/j.1365-2036.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohammed I, Cherkas LF, Riley SA, et al. Genetic influences in gastro-oesophageal reflux disease: a twin study. Gut. 2003;52:1085–1089. doi: 10.1136/gut.52.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gharahkhani P, Tung J, Hinds D, et al. Chronic gastroesophageal reflux disease shares genetic background with esophageal adenocarcinoma and Barrett’s esophagus. Hum Mol Genet. 2016;25:828–835. doi: 10.1093/hmg/ddv512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apalasamy YD, Mohamed Z. Obesity and genomics: role of technology in unraveling the complex genetic architecture of obesity. Hum Genet. 2015;134:361–374. doi: 10.1007/s00439-015-1533-x. [DOI] [PubMed] [Google Scholar]
- 40.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thrift AP, Shaheen NJ, Gammon MD, et al. Obesity and risk of esophageal adenocarcinoma and barrett’s esophagus: A mendelian randomization study. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Z, Gay LJ, Strange A, et al. Common variants at the MHC locus and at chromosome 16q24. 1 predispose to Barrett’s esophagus. Nat Genet. 2012;44:1131–6. doi: 10.1038/ng.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine DM, Ek WE, Zhang R, et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and {Barrett}’s esophagus. Nat Genet. 2013;45:1487–1493. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker J, May A, Gerges C, et al. Supportive evidence for FOXP1, BARX1, and FOXF1 as genetic risk loci for the development of esophageal adenocarcinoma. Cancer Med. 2015 doi: 10.1002/cam4.500. n/a--n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gharahkhani P, Fitzgerald RC, Vaughan TL, et al. Genome-wide association studies in oesophageal adenocarcinoma and Barrett’s oesophagus: a large-scale meta-analysis. Lancet Oncol. 2016;17:1363–1373. doi: 10.1016/S1470-2045(16)30240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nat Publ Gr. 2017 doi: 10.1038/nm.4333. Available at: http://dx.doi.org/10.1038/nm.4333. [DOI] [PMC free article] [PubMed]
- 47.Dura P, van Veen EM, Salomon J, et al. Barrett associated MHC and FOXF1 variants also increase esophageal carcinoma risk. Int J cancer. 2013;133:1751–5. doi: 10.1002/ijc.28160. [DOI] [PubMed] [Google Scholar]
- 48.Pauwels A, Blondeau K, Dupont LJ, et al. Mechanisms of Increased Gastroesophageal Reflux in Patients With Cystic Fibrosis. Am J Gastroenterol. 2012;107:1346–1353. doi: 10.1038/ajg.2012.213. [DOI] [PubMed] [Google Scholar]
- 49.Houghton LA, Lee AS, Badri H, et al. Respiratory disease and the oesophagus: reflux, reflexes and microaspiration. Nat Rev Gastroenterol Hepatol. 2016;13:445–460. doi: 10.1038/nrgastro.2016.91. [DOI] [PubMed] [Google Scholar]
- 50.Schrijver I, Pique L, Graham S, et al. The Spectrum of CFTR Variants in Nonwhite Cystic Fibrosis Patients: Implications for Molecular Diagnostic Testing. J Mol Diagnostics. 2016;18:39–50. doi: 10.1016/j.jmoldx.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Meltzer SJ. Leaky transporters and sphincters in Barrett’s oesophagus? Lancet Oncol. 2016;17:1336–1337. doi: 10.1016/S1470-2045(16)30365-5. [DOI] [PubMed] [Google Scholar]
- 52.Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross-Innes CS, Becq J, Warren A, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet. 2015 doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blount PL, Galipeau PC, Sanchez CA, et al. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–5. [PubMed] [Google Scholar]
- 57.Barrett MT, Sanchez CA, Galipeau PC, et al. Allelic loss of 9p21 and mutation of the CDKN2/p16 gene develop as early lesions during neoplastic progression in Barrett’s esophagus. Oncogene. 1996;13:1867–73. [PubMed] [Google Scholar]
- 58.Buas MF, Levine DM, Makar KW, et al. Integrative post genome-wide association analysis of CDKN2A and TP53 SNPs and risk of esophageal adenocarcinoma. Carcinogenesis. 2014 doi: 10.1093/carcin/bgu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reid BJ, Li X, Galipeau PC, et al. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buas MF, He Q, Johnson LG, et al. Germline variation in inflammation-related pathways and risk of Barrett’s oesophagus and oesophageal adenocarcinoma. Gut. 2016 doi: 10.1136/gutjnl-2016-311622. gutjnl-2016-311622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buas MF, Onstad L, Levine DM, et al. MiRNA-Related SNPs and Risk of Esophageal Adenocarcinoma and Barrett’s Esophagus: Post Genome-Wide Association Analysis in the BEACON Consortium. PLoS One. 2015;10:e0128617. doi: 10.1371/journal.pone.0128617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ek WE, Lagergren K, Cook M. Polymorphisms in genes in the androgen pathway and risk of Barrett’s esophagus and esophageal adenocarcinoma. Int J Cancer. 2015 doi: 10.1002/ijc.29863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagergren K, Ek WE, Levine D, et al. Polymorphisms in Genes of Relevance for Oestrogen and Oxytocin Pathways and Risk of Barrett’s Oesophagus and Oesophageal Adenocarcinoma: A Pooled Analysis from the BEACON Consortium. PLoS One. 2015;10:e0138738. doi: 10.1371/journal.pone.0138738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai JY, de Dieu Tapsoba J, Buas MF, et al. A Newly Identified Susceptibility Locus near FOXP1 Modifies the Association of Gastroesophageal Reflux with Barrett’s Esophagus. Cancer Epidemiol Biomarkers Prev A Publ Am Assoc Cancer Res Cosponsored by Am Soc Prev Oncol. 2015;24:1739–1747. doi: 10.1158/1055-9965.EPI-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai JY, de Tapsoba JD, Buas MF, et al. Constrained Score Statistics Identify Genetic Variants Interacting with Multiple Risk Factors in Barrett’s Esophagus. Am J Hum Genet. 2016;99:352–365. doi: 10.1016/j.ajhg.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee E, Stram DO, Ek WE, et al. Pleiotropic Analysis of Cancer Risk Loci on Esophageal Adenocarcinoma Risk. Cancer Epidemiol Biomarkers Prev. 2015;24:1801–1803. doi: 10.1158/1055-9965.EPI-15-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prevo LJ, Sanchez CA, Galipeau PC, et al. Advances in Brief p53 -Mutant Clones and Field Effects in Barrett’s Esophagus 1. 1999:4784–4787. [PubMed] [Google Scholar]
- 68.Wong DJ, Barrett MT, Stöger R, et al. p16 INK4a Promoter Is Hypermethylated at a High Frequency in Esophageal Adenocarcinomas Promoter Is Hypermethylated at a High Frequency in Esophageal. Cancer Res. 1997:2619–2622. [PubMed] [Google Scholar]
- 69.Li X, Galipeau PC, Paulson TG, et al. Temporal and spatial evolution of somatic chromosomal alterations: A case-cohort study of Barrett’s esophagus. Cancer Prev Res. 2014;7:114–127. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stachler MD, Taylor-Weiner A, Peng S, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–55. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galipeau PC, Prevo LJ, Sanchez CA, et al. Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett’s) tissue. J Natl Cancer Inst. 1999;91:2087–95. doi: 10.1093/jnci/91.24.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reid BJ, Haggitt RC, Rubin CE, et al. Barrett’s esophagus. Correlation between flow cytometry and histology in detection of patients at risk for adenocarcinoma. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- 73.Levine DS, Reid BJ, Haggitt RC, et al. Correlation of ultrastructural aberrations with dysplasia and flow cytometric abnormalities in Barrett’s epithelium. Gastroenterology. 1989;96:355–67. doi: 10.1016/s0016-5085(89)91559-x. [DOI] [PubMed] [Google Scholar]
- 74.Weaver JMJ, Ross-Innes CS, Fitzgerald RC. The “-omics” revolution and oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2014;11:19–27. doi: 10.1038/nrgastro.2013.150. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23982683. [DOI] [PubMed] [Google Scholar]
- 75.Weaver JMJ, Ross-Innes CS, Shannon N, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–43. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3927368&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez P, Timmer MR, Lau CT, et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat Commun. 2016;7:12158. doi: 10.1038/ncomms12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016 doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paterson AL, Shannon NB, Lao-Sirieix P, et al. A systematic approach to therapeutic target selection in oesophago-gastric cancer. Gut. 2012;62:1415–24. doi: 10.1136/gutjnl-2012-302039. [DOI] [PubMed] [Google Scholar]
- 80.Dulak AM, Schumacher SE, Van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8692948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kadri SR, Lao-Sirieix P, O’Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett’s oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. doi: 10.1136/bmj.c4372. Available at: http://dx.doi.org/10.1136/bmj.c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ross-Innes CS, Debiram-Beecham I, O’Donovan M, et al. Evaluation of a Minimally Invasive Cell Sampling Device Coupled with Assessment of Trefoil Factor 3 Expression for Diagnosing Barrett’s Esophagus: A Multi-Center Case-Control Study. PLoS Med. 2015;12:1–19. doi: 10.1371/journal.pmed.1001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross-Innes CS, Chettouh H, Achilleos A, et al. Risk stratification of Barrett’s oesophagus using a non-endoscopic sampling method coupled with a biomarker panel: a cohort study. Lancet Gastroenterol Hepatol. 2016;3:1–9. doi: 10.1016/S2468-1253(16)30118-2. [DOI] [PubMed] [Google Scholar]
- 85.Nones K, Waddell N, Wayte N, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2015;5:1–9. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tomasetti C, Marchionni L, Nowak MA, et al. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci U S A. 2015;112:118–23. doi: 10.1073/pnas.1421839112. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4291633&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong C, Guo Y, Yang H, et al. iCAGES: integrated CAncer GEnome Score for comprehensively prioritizing cancer driver genes in personal genomes. Genome Med. 2016;8:1–22. doi: 10.1186/s13073-016-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim JJ, Bowlby R, Mungall AJ, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017 doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paterson AL, Donovan MO, Provenzano E, et al. Characterization of the timing and prevalence of receptor tyrosine kinase expression changes in oesophageal carcinogenesis. J Pathol. 2013;230:118–128. doi: 10.1002/path.4044. [DOI] [PubMed] [Google Scholar]
- 93.Kim J, Fox C, Peng S, et al. Preexisting oncogenic events impact trastuzumab sensitivity in ERBB2-amplified gastroesophageal adenocarcinoma. J Clin Invest. 2014;124:5145–5158. doi: 10.1172/JCI75200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saldivar JC, Miuma S, Bene J, et al. Initiation of Genome Instability and Preneoplastic Processes through Loss of Fhit Expression. PLoS Genet. 2012:8. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iliopoulos D, Guler G, Han SY, et al. Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett. 2006;232:27–36. doi: 10.1016/j.canlet.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 96.LeTallec B, Millot G, Blin M, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013;4:420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 97.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 98.Paterson AL, Weaver JMJ, Eldridge MD, et al. Mobile element insertions are frequent in oesophageal adenocarcinomas and can mislead paired-end sequencing analysis. BMC Genomics. 2015;16:473. doi: 10.1186/s12864-015-1685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Helman E, Lawrence MS, Stewart C, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 101.Garsed DW, Marshall OJ, Corbin VDA, et al. The Architecture and Evolution of Cancer Neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 102.Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang C-Z, Spektor A, Cornils H, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang C-Z, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Storlazzi CT, Fioretos T, Surace C, et al. MYC-containing double minutes in hematologic malignancies: Evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet. 2006;15:933–942. doi: 10.1093/hmg/ddl010. [DOI] [PubMed] [Google Scholar]
- 106.Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol. 2015;5:1–13. doi: 10.3389/fonc.2015.00288. Available at: http://journal.frontiersin.org/article/10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Niroula A, Vihinen M. Variation Interpretation Predictors: Principles, Types, Performance, and Choice. Hum Mutat. 2016;37:579–597. doi: 10.1002/humu.22987. [DOI] [PubMed] [Google Scholar]
- 108.Samyn B, Samyn B, Sergeant K, et al. Pathway and network analysis of cancer genomes. Nat Methods. 2015;2:1–6. doi: 10.1038/nmeth.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mcgranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (80- ) 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med. 2014;6:224ra24–224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Noordman BJ, Shapiro J, Spaander MC, et al. Accuracy of Detecting Residual Disease After Cross Neoadjuvant Chemoradiotherapy for Esophageal Cancer (preSANO Trial): Rationale and Protocol. JMIR Res Protoc. 2015;4:e79. doi: 10.2196/resprot.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vaughan TL. From genomics to diagnostics of esophageal adenocarcinoma. Nat Genet. 2014;46:806–807. doi: 10.1038/ng.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–54. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.An O, Dall’Olio GM, Mourikis TP, et al. NCG 5. 0: Updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res. 2016;44:D992–D999. doi: 10.1093/nar/gkv1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Techniques Used to Analyze Tumor Genomes
Data to Support Figure 2
A) Loci associated with BE or EA from GWASs and post-GWASs; B) significant GISTIC copy number gain regions detected by next-generation sequencing studies; C) significant GISTIC copy number loss regions detected by next-generation sequencing studies. (format xlsx file)
Point Mutations that Promote Development of EA.
These were identified by MutSig or MutSigCV, with a greater than 10% frequency in at least 1 next-generation sequencing study. * NCC: network of Cancer Genes, ncg.kcl.ac.uk 116.
GWAS analysis of germline DNA (from peripheral blood) can identify variants associated with the development of a condition