Abstract
The rice white tip nematode, Aphelenchoides besseyi, is widely distributed in rice planting areas worldwide and causes serious economic losses. Cathepsin genes have been demonstrated to have importance in studying the reproduction, development, pathogenicity, and control methods of plant nematodes. In this paper, a novel cathepsin B gene, Ab-cb-1, was found and cloned. The Ab-cb-1 gene was 1347 bp in length and encodes 369 amino acids. The Ab-CB-1 protein contains characteristic occluding loops but no signal peptide. A homology analysis showed that Ab-CB-1 had the highest identity value (64%) to the known amino acid sequence of cathepsin B-like cysteine protease 6 from Toxocara canis. When Ab-cb-1 was expressed in a prokaryotic system, the protein massed approximately 45 kDa and could decompose carrot callus. Ab-cb-1 mRNA was localized in the nematode intestine. The relative expression level of Ab-cb-1 in the A. besseyi Ab-S24 population, which had high reproductivity, was approximately 6.9 times that in the Ab-N10 population, which had low reproductivity, and the difference was significant (p<0.05). The Ab-cb-1 expression level was highest in females; the expression levels in males, juveniles and eggs were 30%, 12.2% and 5% of that in females, respectively, and the differences were significant among all four stages (p<0.05). Nematodes of the Ab-S24 population were treated with Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h, and their reproduction decreased with increasing time. These results demonstrated that Ab-CB-1 was a digestive enzyme with hydrolytic protease properties and that Ab-cb-1 played an important role in the reproduction of A. besseyi.
Background
Aphelenchoides besseyi is a migratory plant parasitic nematode with more than 200 host plants in over 35 genera. The main hosts are rice (Oryza sativa) and strawberry (Fragaria ananassa) [1]. A. besseyi is distributed over almost all the world’s rice planting areas, and it is one of the major rice seed-borne diseases that cause serious economic losses [2, 3, 4].
Cathepsins belong to the cysteine protease family and are commonly found in parasites as digestive enzymes. Cathepsins have hydrolytic protease properties [5, 6] and play important physiological and biochemical roles. In recent years, studies have shown that cathepsin genes are essential in the physiological and biochemical processes of parasites and insects, such as hatching, reproduction, development, infection, pathogenicity and immune evasion [7]. Several cathepsins including cathepsin L, cathepsin S, and cathepsin B have been demonstrated to have importance in plant parasitic nematodes, and thus far, cathepsin L has been the subject of greatest interest [8, 9]. So far, several cathepsin L genes have been successfully cloned from a variety of plant nematodes including Heterodera avenae (ACJ13100), H. glycines (Y09498), H. schachtii (ACJ13098), Globodera virginiae (ACJ13094), G. mexicana (ACJ13096) Meloidogyne incognita (CAD89795), Rotylenchulus reniformis (AAY45870), Bursaphelenchus xylophilus (ACH56225), and Ditylenchus destructor (GQ180107). Mi-cpl-1, a cathepsin L gene from M. incognita, has been shown to be correlated with parasite success and to be essential in the interaction between nematodes and plants [10], so this gene could be further developed for root-knot nematode control. Cathepsin S genes (cps) have also been successfully cloned from plant parasite nematodes including H. glycines [9], H. avenae [11] and Radopholus similis (EU659125) [12]. The cps gene in R. similis has been confirmed to be related to reproduction, parasitism and pathogenicity of the nematode [13]. However, cathepsin B genes (cb) in plant parasitic nematodes are seldom reported and have been cloned in only B. xylophilus (GU130153) and R. similis (GU360972). The Rs-cb-1 gene found in R. similis has been reported as a key gene that could affect its development and pathogenicity [14, 15].
In view of the important role and application of the cathepsin gene in the reproduction, development, parasitism and control of nematodes, a novel cathepsin gene was screened from the cDNA library of A. besseyi in this study and successfully cloned. Then, its biological characteristics and functions were identified for the first time.
Methods
Nematode and cultivation
The A. besseyi populations used in this study were the Ab-S24 population collected from strawberry (F. ananassa) in Longgang District, Shenzhen, Guangdong, China, and the Ab-N10 population collected from rice (O. sativa) in Luhe Town, Nanjing, Jiangsu Province, China. Nematodes were isolated, identified, preserved and cultured by the Laboratory of Plant Nematology, South China Agricultural University. The reproductivity was found to be higher in Ab-S24 than in Ab-N10 [16]. The preservation and cultivation of the nematodes were carried out using the method described by Cheng et al. [1].
Cloning of the full-length Ab-cb-1 gene from A. besseyi
Total RNA of approximately 20,000 mixed-stage nematodes of the Ab-S24 population was extracted using the Invitrogen TRIzol® Reagent kit (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA using RQ1 RNase-Free DNase (Promega, Madison, WI, USA). Both 3' RACE primers (CB-F1, CB-F2) and 5' RACE (CB-R1, CB-R2) (Table 1) primers were designed for cDNA amplification according to the conserved expressed sequence tag (EST) sequences of A. besseyi similar to the cathepsin B gene found in our previous study. The amplified products were purified and ligated with pMD 18-T vector (Takara, Japan) to obtain recombinant plasmid. Recombinant plasmid was transformed into Escherichia coli JM109 competent cells, and then positive clones were selected for sequencing (BGI Company). According to the sequencing results, primers QCCF and QCCR (Table 1) were designed for the full-length amplification of the Ab-cb-1 gene from A. besseyi.
Table 1. Primers used in this study.
| Primers | Sequences | Primer use |
|---|---|---|
| CB-F1 | 5′-TGCGGATTTGGTTGC-3′ | 3’ RACE |
| CB-F2 | 5′-CAAACCATGCCCAAAGGAACTATATC-3′ | 3’ RACE |
| CB-R1 | 5′-TTCCTTTGGGCATGGTTTGAAATGAG-3′ | 5’ RACE |
| CB-R2 | 5′-CTGGGAAAGTGTACGGTTG-3′ | 5’ RACE |
| QCCF | 5′-AAATGTTGGCGAAGTTAAGTGTAGC-3′ | Ab-cb-1 |
| QCCR | 5′-CAATCGAGCGAAATGTAAAATAAAA-3′ | Ab-cb-1 |
| CBfBamHI | 5′-CGGGATCCATGAAGACAAAAAACAATGATTTG-3′ | Ab-cb-1 plasmid |
| CBrXhoI | 5′-CCGCTCGAGTTAGAAAATATCATAGGAACTAGCC-3′ | Ab-cb-1 plasmid |
| qPCRC-F | 5′-TGAATGTTAGAAACCCAATCAAAG-3′ | qPCR |
| qPCRC-R | 5′-CACTACGACACATTGAACCCCA-3′ | qPCR |
| 18sF | 5′-CTCGTGGTGGCTGGTATGCTG-3′ | qPCR |
| 18sR | 5′-GTTTCCCGTGTTGAGTCAAATTAAG-3′ | qPCR |
| CB-IN-T7S1 | 5′-TAATACGACTCACTATAGGGATCTTGTGGCTCCTGTTGGTCAT-3′ | RNA probe |
| CB-IN-A1 | 5′-TCCGTTTGAGTGAATGCAGATGC-3′ | RNA probe |
| CB-IN-T7A1 | 5′-TAATACGACTCACTATAGGGTCCGTTTGAGTGAATGCAGATGC-3′ | RNA probe |
| CB-IN-S1 | 5′-ATCTTGTGGCTCCTGTTGGTCAT-3′ | RNA probe |
| CBRiF | 5′-CCGCAACATCAAACAACAAATTCGC-3′ | RNA probe |
| CBRiT7R | 5′-TAATACGACTCACTATAGGGCGGCTCCAAATGACCAACAGG-3′ | dsRNA template |
| CBRiT7F | 5′-TAATACGACTCACTATAGGGCCGCAACATCAAACAACAAATTCGC-3′ | dsRNA template |
| CBRiR | 5′-CGGCTCCAAATGACCAACAGG-3′ | |
| G-T7A | 5′-GGATCCTAATACGACTCACTATAGGGCGATGCGGTTCACCAGGGTGTCG-3′ | dsRNA template |
| G-S | 5′-CACAAGTTCAGCGTGTCCGGCG-3′ | dsRNA template |
| G-T7S | 5′-GGATCCTAATACGACTCACTATAGGGCACAAGTTCAGCGTGTCCGGCG-3′ | dsRNA template |
| G-A | 5′-CGATGCGGTTCACCAGGGTGTCG-3′ | dsRNA template |
Sequence analysis, alignment and phylogenetic analysis of Ab-CB-1
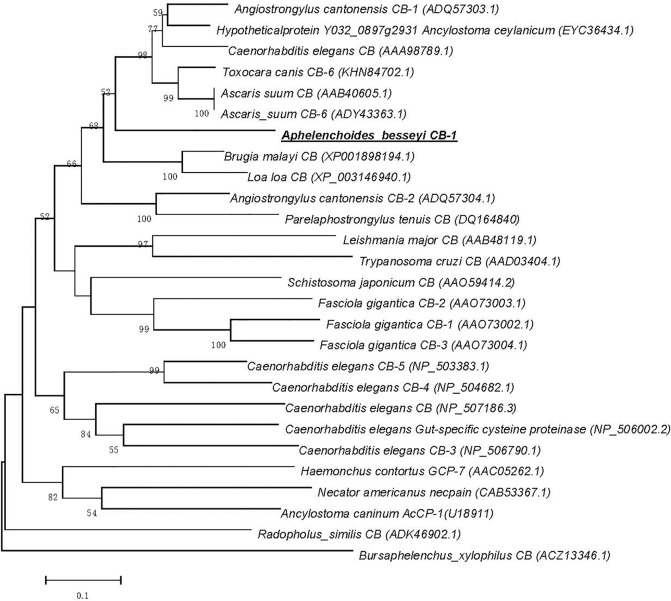
Sequence homology alignments were performed using BLASTN and BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein bioinformatic analysis was performed using Protein Machine software (http://www.expasy.ch/tools/), including predictions of protein transmembrane region, amino acid sequence, isoelectric point analysis, molecular weight and hydrophobicity analysis. Predictions of signal peptide and cleavage site were performed at http://www.cbs.dtu.dk/services/SignalP/. The phylogenetic tree was constructed using the neighbor-joining method [17] with the program MEGA (Molecular Evolutionary Genetics Analysis, USA) based on Ab-CB-1 and other 26 cathepsin B amino acid sequences from 17 species of representative nematodes in the NCBI database.
Prokaryotic expression and purification of recombinant Ab-CB-1
The full-length Ab-cb-1 gene was amplified from the plasmid with primers CBfBamHI and CBrXhoI (Table 1). The amplified product was purified by digestion with BamHI and XhoI (Takara DNA Fragment Purification Kit Ver 2.0), ligated to prokaryotic expression vector pET-28 (+) (Novagen, Madison, WI, USA), and subsequently introduced to E. coli JM109 for sequence confirmation. The recombinant plasmid thus obtained was introduced into E. coli BL21 (DE3) competent cells for prokaryotic expression using the method described by Cheng et al [1]. The recombinant fusion Ab-CB-1 protein with His-tag at the N-terminus was purified by affinity chromatography using Ni Sepharose High Performanc (GE Healthcare, Sweden) according to the manufacturer’s instructions. The purity of purified recombinant protein was confirmed by SDS-PAGE. Concentration of purified Ab-CB-1 was determined using Easy ⅡProtein Quantitative Kit (BCA) (Transgene, China).
Decomposition of carrot callus by recombinant Ab-CB-1
E. coli BL21 cells containing Ab-CB-1 constructs were collected from 10 mL bacterial suspension and induced to express the Ab-cb-1 gene by centrifugation (10,000 rpm, 1 min). These bacterial cells were broken ultrasonically for 30 min after mixing with PBS buffer, and the supernatant was obtained as a primary enzyme extract. Aliquots of 100 μL of each primary enzyme extract (from induced and non-induced E. coli cells containing Ab-CB-1 constructs), purified Ab-CB-1 (1.0 mg/ml) and sterile water were inoculated into carrot callus under sterile conditions, and then the callus was cultured at 25°C to observe changes in the carrot tissue.
Expression levels of Ab-cb-1 in Ab-S24 and Ab-N10
Total RNA was extracted from approximately 20,000 mixed-stage nematodes of Ab-S24 and Ab-N10 populations using the Invitrogen TRIzol® Reagent kit. RNA from different development stages of the Ab-S24 population was extracted from 500 each of females, males, juveniles and eggs using a MicroElute Total RNA kit (Omega, USA). The extracted RNA was reverse transcribed into cDNA using the RQ1 RNase-Free DNase (Promega) reverse transcription kit as described above. The expression levels of Ab-cb-1 in the Ab-S24 and Ab-N10 populations and in four different development stages of the Ab-S24 population were detected on a CFX-96 (Bio-Rad) qPCR machine with cDNA as a template using a SYBR Green Real-time PCR Master Mix Plus kit (Toyobo, Japan). Specific primers qPCRC-F and qPCRC-R (Table 1) were designed to detect Ab-cb-1 expression. A 140 bp sequence of 18S rRNA (AY508035) was amplified as a reference gene using the primers 18sF and 18sR (Table 1). The qPCR data were analyzed using the CFX Manager software provided by Bio-Rad. All experiments were performed in three biological replicates and each in three replicates.
In situ hybridization of Ab-cb-1
In situ hybridization was performed as described previously [1,18]. Approximately 10,000 nematodes of mixed stages of the Ab-S24 population were collected and concentrated into 30–50 μl. The nematodes were fixed in 3% paraformaldehyde for 18 h at 5°C and then at 22°C for 4 h. DIG-labeled sense and antisense RNA probes (Roche, Germany) were synthesized using sense primers (CB-IN-T7S1, CB-IN-A1) and antisense primers (RB-IN-TA1, CB-IN-S1) (Table 1) based on the full-length cDNA of Ab-cb-1. After adding the obtained DIG-labeled RNA probes, the hybridization solution containing nematodes was rotated for 12 h at 47°C. The results were examined and photographed by differential interference microscopy.
dsRNA synthesis and RNAi efficiency of Ab-cb-1
The RNAi knockdown of the Ab-cb-1 gene was performed by soaking the nematodes with double-stranded RNA (dsRNA) of Ab-cb-1 synthesized by in vitro transcription. Two primer pairs, CBRiF/CBRiT7R and CBRiT7F/CBRiR (Table 1), were designed to amplify the sense and antisense single-stranded RNA (ssRNA) products, respectively. Ab-cb-1 dsRNA was synthesized according to the instructions of the Script MaxTM Thermo T7 Transcription kit (Toyobo). The obtained dsRNA synthesis product was purified using the described method, then examined for integrity by electrophoresis, detected for concentration and quality by Nanodrop spectrophotometer, analyzed by 1.2% agarose gel electrophoresis, and finally stored at -80°C for later use. The non-endogenous control (green fluorescent protein gene, gfp) dsRNA (125 bp) was generated with the specific primers G-T7S, G-A and G-S (Table 1) [19]. Five hundred mixed-stage nematodes of the Ab-S24 population were separated from carrot callus and collected in a diethyl pyrocarbonate (DEPC)-treated centrifuge tube. A 50 μl aliquot of Ab-cb-1 dsRNA (2 μg/μL) was added into the tube to soak the nematodes at 25°C. The nematodes were soaked for 12 h, 24 h, 36 h, and 48 h, respectively. Non-endogenous gfp dsRNA solution 50 ml (2 μg/μL) was used as a control. In total, eight treatments were performed in triplicate. The RNA of the nematodes treated by soaking was extracted after washing the nematodes three times with DEPC-treated water. RNAi efficiency was examined by determining the Ab-cb-1 expression level by qPCR.
Effect of Ab-cb-1 RNAi on nematode reproduction
Females of the Ab-S24 population were treated with Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h and treated with gfp dsRNA as control. A total of 30 female nematodes were selected from each treatment and inoculated on carrot callus, and each treatment was repeated five times. Carrot callus dishes inoculated with nematodes were incubated at 25°C in the dark for 35 days, and then nematodes on carrot callus were separated and counted.
Statistical analysis
All data in this study were subjected to analysis of variance (ANOVA), and multiple comparisons of means were conducted by Duncan’s Multiple Range Test at p = 0.05 using SAS (Release 8.01).
Results
Cloning of the full-length Ab-cb-1 gene from A. besseyi
A 1347 bp full-length cDNA sequence from A. besseyi was amplified using the specific primers QCCF and QCCR (Table 1) and confirmed by sequencing. The cDNA sequence was named Ab-cb-1 and included a 1110 bp open reading frame (ORF) found with NCBI ORF finder, encoding 369 amino acids (GenBank accession number JQ686691). Ab-cb-1 began with an ATG initiation codon after 129 bp of upstream 5' untranslated region and ended with a TAA stop codon before 108 bp of downstream 3' untranslated region.
Sequence analysis, alignment and phylogenetic analysis of Ab-CB-1
The Ab-CB-1 protein sequence (S1 Fig) translated from the Ab-cb-1 gene sequence encoded 369 amino acids with a theoretical molecular mass of 41.8718 kDa. Sequence homology alignment analysis showed that Ab-CB-1 was a cathepsin B gene in the cysteine protease family. The results of protein bioinformatic analysis showed that Ab-CB-1 had the typical characteristics of the cysteine protease family: a cysteine protease catalytic triad of residues containing the sites cysteine (Cys, 122), histidine (His, 293) and asparagine (Asn, 313); two occluding loops (between residues 116 and 127 and between residues 291 and 301); glutamine oxyanion hole (116); predicted cleavage point between the mature and pro-domains (93); hemoglobinase motif (residues between 308 and 321); and protease S2 pocket (residues between 335 and 339).
The sequence homology alignment showed that the highest homology with the Ab-CB-1 sequence was found in cathepsin B-like cysteine protease 6 from Toxocara canis (GenBank accession: KHN84702.1, similarity 73%, identity 64%, E value: 3e-156), followed by two cathepsin B-like cysteine proteases from Ascaris suum (GenBank accessions: AAB40605.1 and ERG84961.1, similarity 73%, identity 62%, E value 2e-154) and hypothetical protein Y032_0897g2931 from Ancylostoma ceylanicum (GenBank accession: EYC36434.1, similarity 75%, identity 60%, E value 5e-153).
The phylogenetic tree was constructed based on Ab-CB-1 and other 26 cathepsin B amino acid sequences from 17 species of representative parasites in the NCBI database (Fig 1). The result showed that Ab-CB-1 had the closest overall relationship with cathepsin B-like cysteine protease 6 from Toxocara canis, which was consistent with the results of the alignment analysis obtained by blastx.
Fig 1. Phylogram constructed on the basis of amino acid sequences depicting evolutionary relationships among the cathepsin B genes of 16 parasitic species.
Aphelenchoides besseyi CB-1 is highlighted by the underline; accession numbers of the sequences are shown in brackets; distances on the X-axis correspond to the degree of homology between sequences; distances on the Y-axis are arbitrary.
Prokaryotic expression of recombinant Ab-CB-1 and purification
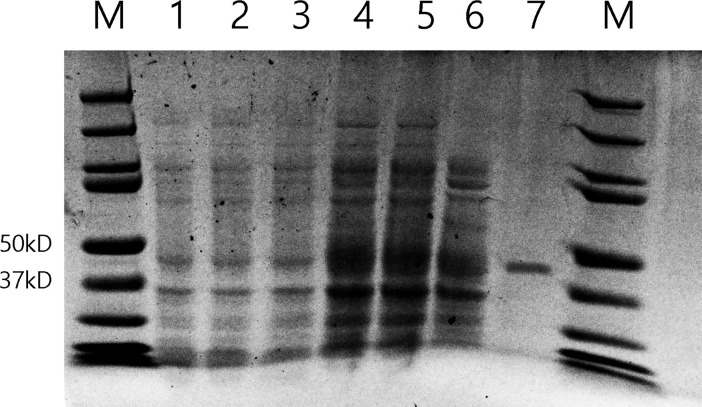
The expression of pET-28a (+)-Ab-CB-1 was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG), and the recombinant protein extract was examined by SDS-PAGE. Compared with non-induced E. coli containing the Ab-CB-1 construct and induced E. coli harboring the empty pET-28a (+) vector, only the IPTG-induced recombinant had an obvious band at 45 kDa (Fig 2). Prokaryotic expression of recombinant Ab-CB-1 was confirmed, and protein extract of Ab-CB-1 was obtained.
Fig 2. Sodium dodecyl sulfate polyacrylamide gel of lysate of Escherichia coli cells expressing Ab-CB-1 from the plasmid vector pET-28a (+).
M: PageRuler Prestained Protein Ladder (Thermo, USA); 1, 2: Protein extracts from induced E. coli cells harboring the empty pET-28a (+) vector by 0.5 and 1.0 mM IPTG; 3: Protein extract from non-induced E. coli cells containing the Ab-CB-1 construct; 4,5: Protein extracts from E. coli cells containing the Ab-CB-1 construct and induced by 0.5 and 1.0 mM IPTG; 6: Supernatant of cell lysate from E. coli cells containing the Ab-CB-1 construct; 7 Purified recombinant Ab-CB-1.
Decomposition of carrot callus by purified recombinant Ab-CB-1
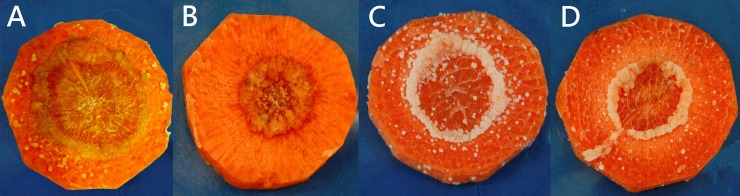
After inoculation with the purified protein Ab-CB-1, and the primary enzyme extract from induced E. coli cells containing the Ab-CB-1 construct, carrot callus changed color to yellow-brown and decomposed visibly (Fig 3A and 3B). In the treatments with sterile water (Fig 3C) and the primary enzyme extract from non-induced E. coli cells containing the Ab-CB-1 construct (Fig 3D), carrot callus remained white in color and was not visibly decomposed (Fig 3D). These results showed that the primary enzyme extract from prokaryotic expression of recombinant Ab-CB-1 could decompose carrot callus.
Fig 3. Inoculation of carrot callus with prokaryotically expressed products of Ab-cb-1.
A: carrot callus inoculated with the purified protein Ab-CB-1 in 1mg/ml; B: carrot callus inoculated with the primary enzyme extract from induced E. coli cells containing the Ab-CB-1 construct; C: carrot callus inoculated with distilled water; D: carrot callus inoculated with the primary enzyme extract from non-induced E. coli cells containing the Ab-CB-1 construct.
Ab-cb-1 expression in two populations and four developmental stages of A. besseyi
Ab-cb-1 expression levels in the mixed-stage nematodes of two populations and in four developmental stages including eggs, juveniles, females and males of the Ab-S24 population were detected by qPCR. The results showed that the relative expression level of Ab-cb-1 in the Ab-S24 population was approximately 6.9 times that in the Ab-N10 population, and the difference was significant (p<0.05) (Fig 4). The Ab-cb-1 expression level was highest in females, and the expression levels in males, juveniles and eggs were 30%, 12.2% and 5% of that in females. The differences among the four stages were significant (p<0.05) (Fig 5).
Fig 4. Expression levels of Ab-cb-1 in two populations of Aphelenchoides besseyi.
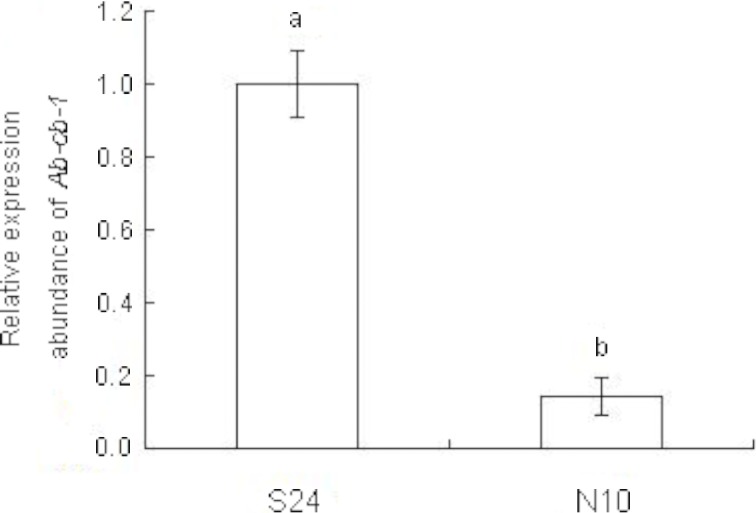
S24, N10: populations of A. besseyi collected from Fragaria ananassa and Oryza sativa, respectively; bars indicate standard errors of the mean (n = 3), and different letters indicate significant differences (p<0.05) between treatments.
Fig 5. Expression of Ab-cb-1 in different developmental stages of Aphelenchoides besseyi.
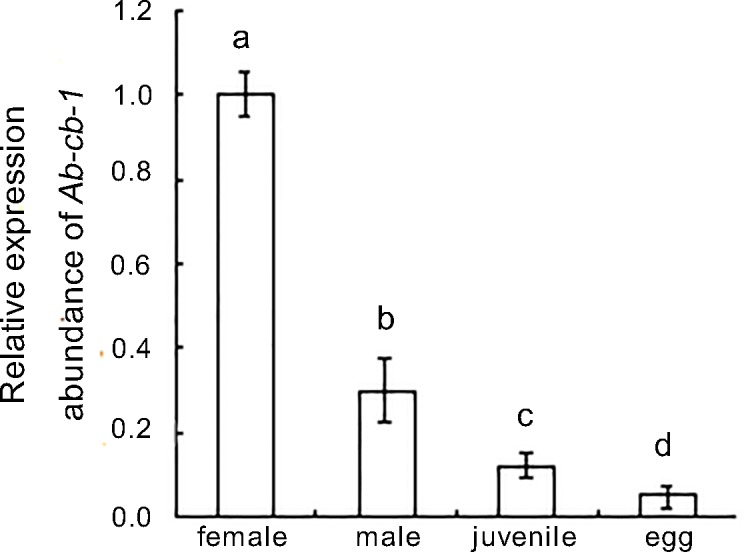
Bars indicate standard errors of the mean (n = 3), and different letters indicate significant differences (p<0.05) between treatments.
In situ hybridization of Ab-cb-1
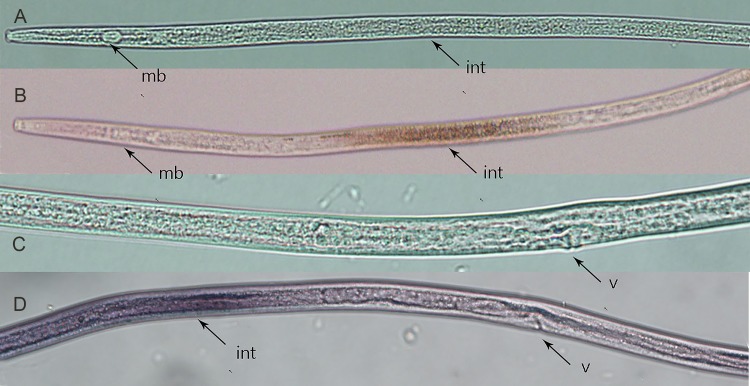
The results of in situ hybridization suggested that Ab-cb-1 was present in the intestine of females (Fig 6B and 6D). No hybridization signal was detected in nematodes when the control (sense Ab-cb-1 DIG-labeled RNA) probe was used (Fig 6A and 6C).
Fig 6. Tissue localization of Aphelenchoides besseyi cathepsin B gene Ab-cb-1 mRNA in females using in situ hybridization.
A, C: sense A. besseyi cb DIG-labeled cDNA probe; B, D: antisense A. besseyi cb DIG-labeled cDNA probe. Images suggested that Ab-cb-1 was present in the intestine of females. mb: medium bulb; v: vulva; int: intestine.
Ab-cb-1 RNAi
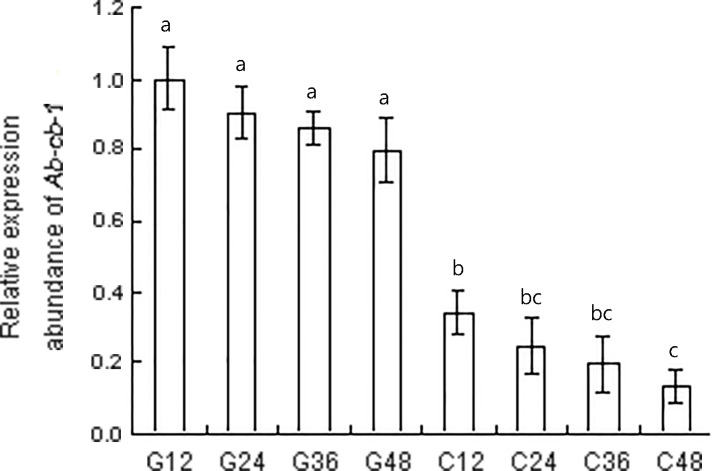
The RNAi efficiency of Ab-cb-1 was detected by qPCR after nematodes were treated with Ab-cb-1 dsRNA. Compared to the expression level of Ab-cb-1 in nematodes treated with the gfp dsRNA, the expression of Ab-cb-1 in these nematodes decreased significantly, by 65.8%, 65.6%, 66.5% and 66.3% when nematodes were soaked in Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h (p<0.05). Expression of Ab-cb-1 decreased with increasing dsRNA treatment time. The expression level of Ab-cb-1 was the lowest when nematodes were soaked for 48 h; this level was significantly lower than that of nematodes soaked for 12 h (p<0.05) but was not significantly different from those of nematodes soaked for 24h and 36 h (p>0.05). Nematode Ab-cb-1 expression was not significantly different among any of the gfp dsRNA treatments used as controls in this experiment (p>0.05) (Fig 7).
Fig 7. Expression levels of Ab-cb-1 mRNA in Aphelenchoides besseyi treated with Ab-cb-1 double-stranded RNA (dsRNA).
G12, G24, G36 and G48: expression of Ab-cb-1 mRNA in control nematodes soaked in non-endogenous gfp dsRNA solution for 12 h, 24 h, 36 h and 48 h, respectively; C12, C24, C36 and C48: expression of Ab-cb-1 mRNA in nematodes soaked in Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h, respectively. Bars indicate standard errors of the mean (n = 3), and different letters indicate significant differences (p<0.05) between treatments.
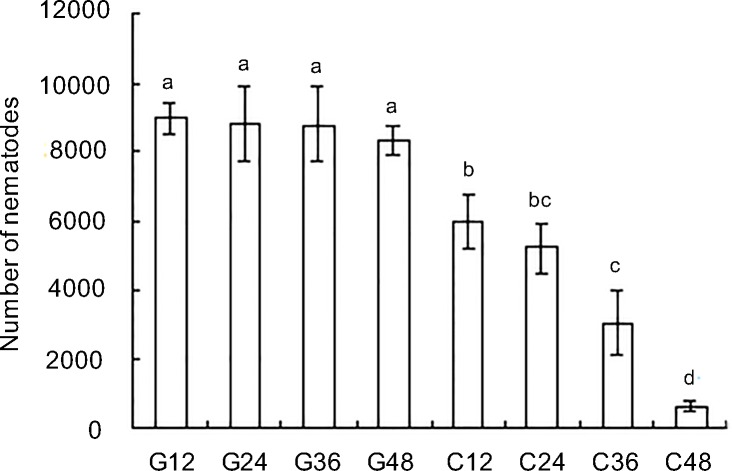
The effect of Ab-cb-1 RNAi on reproduction in A. besseyi was tested by inoculating the nematodes treated with Ab-cb-1 dsRNA on carrot disks. After culture on carrot disks for 35 d, nematodes treated with Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h had significantly lower reproduction than that of gfp dsRNA-treated nematodes (p<0.05) (Fig 8). In addition, reproduction of the nematodes treated with Ab-cb-1 dsRNA decreased with increasing treatment time; the differences were significant (p<0.05) among different RNAi treatment groups except between 12 h and 24 h and between 24 h and 36 h (p>0.05). No significant difference was found among the gfp dsRNA treatment groups in this experiment (p>0.05). Therefore, Ab-cb-1 expression in A. besseyi was effectively inhibited by soaking the nematodes in Ab-cb-1 dsRNA, and Ab-cb-1 RNAi depressed the reproduction of A. besseyi.
Fig 8. Number of Aphelenchoides besseyi on carrot callus after inoculating 30 females for 35days.
G12, G24, G36 and G48: the numbers of A. besseyi after inoculating nematodes treated with non-endogenous gfp dsRNA solution for 12 h, 24 h, 36 h and 48 h, respectively; C12, C24, C36 and C48: the numbers of A. besseyi after inoculating nematodes treated with Ab-cb-1 dsRNA for 12 h, 24 h, 36 h and 48 h, respectively. Bars indicate standard errors of the mean data (n = 5), and different letters indicate significant differences (p<0.05) among treatments.
Discussion
In this study, a 1347 bp full-length cDNA of the cathepsin B gene (Ab-cb-1) in A. besseyi was cloned based on an EST fragment that was highly similar to the cathepsin B gene in the cDNA library of A. besseyi. Ab-cb-1 was a differently expressed gene between A. besseyi populations Ab-S24 and Ab-N10 and the relative expression level of Ab-cb-1 in the Ab-S24 population was approximately 6.9 times that in the Ab-N10 population. The protein Ab-CB-1 consisted of 369 amino acids and had a molecular weight of 41.8718 kDa. Ab-CB-1 had the highest identity value (64%) to the known amino acid sequence of cathepsin B-like cysteine protease 6 from Toxocara canis, and this result was consistent with the results of a phylogenetic analysis. Ab-CB-1 contained the cysteine protease catalytic triad of residues (Cys, His, and Asn) and occluding loops. The triad of Cys, His, and Asn residues constitutes an active catalytic domain, which is highly functional and conserved among the cysteine proteases of peptidase family C1, including cathepsin B [20]. The occluding loop is characteristic of only cathepsin B; this loop is responsible for endopeptidase and exopeptidase activity, and its presence seems to indicate that cathepsin B possesses the ability to act [21, 22, 23] as a self-inhibitor by partially blocking the active site when the loop is inactive [24–28]. These structural features indicated that Ab-CB-1 had the functions and activities of cathepsin B. Ab-CB-1 was inferred to react with substrates directly without protein transportation after synthesis in cells because it did not contain any signal peptide sequence and because the transmembrane signal was weak in the putative transmembrane region.
Cathepsin B (CB) is a member of the cysteine protease family. Cysteine proteases from nematodes have mostly been found to express in gland or intestinal cells and act as the main "digestive enzymes" of nematodes [10], thus playing important roles in the nematode development and invasion process [20, 29, 30, 31]. Cathepsin B was also reported to be related to reproduction: cathepsin B mRNA increased during pregnancy in endometrial epithelia of the porcine uterus and placenta [32]; cathepsin B activity was a useful marker of oocyte quality in bovine oocytes [33]. In this study, we showed that Ab-CB-1 could decompose carrot callus, and the gene was expressed in the intestine of A. besseyi. Therefore, it could be confirmed that Ab-CB-1 acted as a digestive enzyme with hydrolytic protease properties and played an important role in the reproduction of A. besseyi. The method carrot callus tissue for culture of endoparasitic nematodes was established by Reise in 1987 [34]. It was used in several studies that Aphelenchoides besseyi can be cultured on carrot callus [1, 35]. A. besseyi also can be cultured on some fungi, for example, Botrytis cinerea. In this study we use carrot callus for nematode cultivation because carrot callus is also a kind of plant material and it is more similar to rice tissue comparing to fungi. And we suppose long time fungi culturing nematodes might lose its pathogenicity to plants.
The reproductivity of A. besseyi populations Ab-S24 and Ab-N10 are quite different and the reproductivity of Ab-S24 is approximately 19 times that of Ab-N10 [16]. Moreover, differences of Ab-cb-1 expression level were found between two A. besseyi populations with different reproductive rates. The Ab-cb-1 expression level of the high reproductive population Ab-S24 was significantly higher than that of the low reproductive population Ab-N10.Differences in Ab-cb-1 expression levels were found in the different developmental stages of the Ab-S24 population; the highest expression was in females, then males, juveniles and eggs, and this result was consistent with the biological characteristics and functions of the different developmental stages of A. besseyi. The females of A. besseyi bear the heaviest reproductive burden and represent the major nematode stage involved in infectivity; also, their bodies are larger than those of other stages, so they need to digest more food to obtain more nutrition and energy. The number and infectivity of the males were lower than those of the females, but compared to the juveniles, males had a well-developed intestine and reproductive functions. As expected, Ab-cb-1 expression in males was lower than that in females but higher than that in juveniles. Eggs have no functions related to feeding or digestion, and their Ab-cb-1 expression was the lowest among the four developmental stages. The expression levels of AC-cathB-1 in the different stages of Angiostrongy cantonensis [30] were similar to those of A. besseyi in this study. However, the expression of Rs-cb-1 in males of R. similis was significantly lower than that in other stages. The males of R. similis do not feed or infect, as the stylet and esophagus have degenerated; their main activity is mating with females. Furthermore, R. similis can reproduce through parthenogenesis, and the male stage is not necessarily required for the reproduction of the R. similis species [15].
After treatment with Ab-cb-1 dsRNA, the expression of Ab-cb-1 and the reproductivity of A. besseyi decreased with increasing time. The RNAi efficiency of Ab-cb-1 was the highest and nematode reproductivity was the lowest when the nematodes were treated with Ab-cb-1 dsRNA for 48 h. Although the RNAi effect of soaking with dsRNA was not persistent and hereditary, the Ab-cb-1 RNAi strongly affected the reproduction capacity of the first generation (F1) of A. besseyi. It resulted in a significant decrease in nematodes number of the second generation (F2) comparing to control. The life cycle of A. besseyi was about 6–7 days (25°C) and nematodes could complete approximately 5–6 generations during 35 days cultivation. The difference in nematodes number was started from the F2 generation between the number of A. besseyi in RNAi treatment and the control treatment. Furthermore, it was enlarged after 4–5 generations. This difference was caused by nematodes being soaked with Ab-cb-1 dsRNA comparing to control treatment. It was reported and demonstrated in many studies that soaking with dsRNA of target gene could affect nematodes in reproduction [1, 14, 15, 36, 37]. Our result agrees with that of a study reported in R. similis that showed the highest RNAi efficiency when nematodes were treated with Rs-cb-1 dsRNA for 48 h, and the reproductivity was significantly decreased on carrot callus [15].
Conclusion
This is the first work to clone a novel cathepsin gene from A. besseyi and then identify the biological characteristics and functions. These results showed that the cathepsin B gene played an essential role in the reproductivity of nematodes. Overall, the cb gene has important research and application value in exploring new methods to control plant nematodes based on biotechnological strategies.
Supporting information
Underscored: a predicted N-glycosylation site; shaded: the occluding loop; italics: the cysteine protease catalytic triad of residues (Cys, His, and Asn); bold: predicted cleavage points of mature and pro-domains; red: glutamine oxyanion hole; box: hemoglobinase motif; double underline: protease S2 pocket.
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by National Natural Science Foundation of China (No. 31371920 and 31071665 to H.X.).
References
- 1.Cheng X, Xiang Y, Xie H, Xun CL, Xie TF, Zhang C, et al. Molecular Characterization and Functions of Fatty Acid and Retinoid Binding Protein Gene (Ab-far-1) in Aphelenchoides besseyi. PLoS One. 2013; 8: e66011 doi: 10.1371/journal.pone.0066011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bridge J, Luc M, Plowright RA. Nematode parasites of rice In: Luc M, Sikora RA, Bridge J, editors. Plant parasitic nematodes in subtropical and tropical agriculture. Oxford: CABI Publishing; 1990: 69–76. [Google Scholar]
- 3.Rao Y S, Prasad J S, Panwar M S. Nematode pests of rice in India. Non insect pest and predators, 1985: 65–71. [Google Scholar]
- 4.Wang F, Li DL, Wang ZY, Dong AR, Wang BY, Chen QL, et al. Transcriptomic Analysis of the Rice White Tip Nematode, Aphelenchoides besseyi (Nematoda: Aphelenchoididae). PLoS One. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley C J, Urwin P E, McPherson M J, Atkinson HJ. Characterization of intestinally active proteinases of cyst-nematodes. Parasitology, 1996, 113(4): 415–424. [DOI] [PubMed] [Google Scholar]
- 6.Koiwa H, Shade RE, Zhu-Salzman K, D’Urzo MP, Murcock LL, Bressan R, et al. A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Letters, 2000, 471(1): 67–70; doi: 10.1016/S0014-5793(00)01368-5 [DOI] [PubMed] [Google Scholar]
- 7.Meemon K, Grams R, Vichasri-Grams S, Korge G, Viyanant V, Upatham ES, et al. Molecular cloning and analysis of stage and tissue-specific expression of cathepsin B encoding genes from Fasciola gigantica. Molecular and biochemical parasitology, 2004, 136(1): 1–10; https://doi.org/10.1016/j.molbiopara.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Shingles J, Lilley CJ, Atkinson HJ. Meloidogyne incognita: Molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Experimental Parasitology, 2007, 115(2): 114–120. doi: 10.1016/j.exppara.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 9.Wang GF, Peng DL, Sun JH, Huang WK, Peng H, Long HB. Cloning and sequence analysis of a new cathepsin L-like cysteine proteinase gene from Ditylenchus destructor. Chin J Biotech. 2011; 27 (1): 60−68 (In Chinese). [PubMed] [Google Scholar]
- 10.Dutta TK, Papolu PK, Banakar P, Choudhary D, Shirohi A, Rao U. Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Frontiers in Microbiology. 2015; 260: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilley CJ, Urwin PE, Atkinson HJ, McPherson MJ. Characterization of two cDNAs encoding cysteine proteinases from the soybean cyst nematode Heterodera glycines. Molecular and Biochemical Parasitology. 1997; 89: 195–207; https://doi.org/10.1016/S0166-6851(97)00116-3. [DOI] [PubMed] [Google Scholar]
- 12.Thakur PK, Kumar K, Kumar J, Gantasala NP, Rao U. Structural and functional analysis of cathepsin S of Heterodera spp: a promising candidate for its control. Indian Journal of Experimental Biology. 2014; 52 (3): 223–231. [PubMed] [Google Scholar]
- 13.Lv CH, Xiao L, Mao ZC, Chen GH, Yang YH, Xie BY. Molecular cloning and sequence analysis of cathepsin S-like cysteine proteinase genes of Radopholus similis. Plant Protection, 34(5):17–21 (In Chinese). [Google Scholar]
- 14.Wang K, Li Y, Huang X, Wang DW, Xu CL, Xie H. The cathepsin S cysteine proteinase of the burrowing nematode Radopholus similis is essential for the reproduction and invasion. Cell Biosci. 2016; 39, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Wang K., Xie H, Wang DW, Xu CL, Huang X, et al. Cathepsin B Cysteine Proteinase is Essential for the Development and Pathogenesis of the Plant Parasitic Nematode Radopholus similis. International Journal of Biological Sciences, 2015; 11(9): 1073–1087; doi: 10.7150/ijbs.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei YY, Luo AL, Xie H, Yang ZF, Cheng X, Xu CL. Reproductive fitness of 15 Aphelenchoides besseyi populations from China. Journal of Northwest A&F university (Nat. Sci.Ed), 2010, 38 (6): 165–170. (In Chinese) [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4: 406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 18.De BJM, Yan Y, Smant G, Davis EL, Baum TJ. In-situ hybridization to messenger RNA in Heterodera glycines. J Nematol. 1998; 30: 309–312. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Xie H, Xu CL, Cheng X, Li KM, Li Y. Differential expression of Rs-eng-1b in two populationsof Radopholus similis (Tylenchida: Pratylecnchidae) and its relationship to pathogenicity. European Journal of Plant Pathology, 2012, 133(4): 899–910. [Google Scholar]
- 20.Hashmi S, Britton C, Liu J, Guiliano David B, Oksov Y, Lustigman S. Cathepsin L is essential for embryogenesis and development of Caenorhabditis elegans. The Journal of Biological Chemistry, 2002, 277(5): 3477–3486. doi: 10.1074/jbc.M106117200 [DOI] [PubMed] [Google Scholar]
- 21.Rosenthal PJ. Cysteine proteases of malaria parasites. International Journal for Parasitology. 2004; 34, (1): 1489–1499. [DOI] [PubMed] [Google Scholar]
- 22.Illy C, Quraishi O, Wang J, Purisima E, Vernet T, Mort JS. Role of the occluding loop in cathepsin B activity. Journal of Biological Chemistry. 1997; 272, 1197–1202; doi: 10.1074/jbc.272.2.1197 [DOI] [PubMed] [Google Scholar]
- 23.Nomura H, Athauda SBP, Takase M, Ukai A, Azuma T, Hodotsuka K, Inoue H, et al. Cathepsin L-like cysteine proteases from Brugia malayi: cDNA cloning and comparison with Caenorhabditis elegans. Biomedical Research 25 (6) 287–293, 2004; doi: 10.2220/biomedres.25.287 [Google Scholar]
- 24.Turk D, Podobnik M, Kuhelj R, Dolinar M, Turk V. Crystal structures of human procathepsin B at 3.2 and 3.3 Angstroms resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Letters. 1996; 384 (3): 211–214. [DOI] [PubMed] [Google Scholar]
- 25.Bailly E., Jambou R., Savel J., Jaureguiberry G. Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and Pepstatin A (aspartyl protease inhibitor). J. Protozool. 1992: 39, 593–599; doi: 10.1111/j.1550-7408.1992.tb04856.x [DOI] [PubMed] [Google Scholar]
- 26.Bromme D, Nallaseth FS, Turk B. Production and activation of recombinant papain-like cysteine proteases. Methods 2004: 32, 199–206; https://doi.org/10.1016/S1046-2023(03)00212-3. [DOI] [PubMed] [Google Scholar]
- 27.Dluzewski AR, Rangachari K, Wilson RJ, Gratzer WB. Plasmodium falciparum: protease inhibitors and inhibition of erythrocyte invasion. Exp. Parasitol. 1986: 62, 416–422; doi: 10.1016/0014-4894(86)90050-0 [DOI] [PubMed] [Google Scholar]
- 28.Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Molecular and Biochemical Parasitology, 2002, 120(1): 1–21; doi: 10.1016/S0166-6851(01)00438-8 [DOI] [PubMed] [Google Scholar]
- 29.Cox GN, Pratt D, Hageman R, Boisvenue RJ. Molecular cloning and primary sequence of a cysteine protease expressed by Haemonchus contortus adult worms. Molecular and Biochemical Parasitology, 1990, 41(1): 25–34; https://doi.org/10.1016/0166-6851(90)90093-2. [DOI] [PubMed] [Google Scholar]
- 30.Carmona C, Dowd AJ, Smith AM, Dalton JP. Cathepsin L proteinase secreted by Fasciola hepatica in vitro prevents antibody-mediated eosinophil attachment to newly excysted juveniles. Molecular and Biochemical Parasitology, 1993, 62(1): 9–17; doi: 10.1016/0166-6851(93)90172-T [DOI] [PubMed] [Google Scholar]
- 31.Ni F, Wang Y, Zhang J, Yu L, Fang WZ, Luo D. Cathepsin B-like and hemoglobin-type cysteine proteases: stage-specific gene expression in Angiostrongy cantonensis. Experimental Parasitology, 2012, 131(4): 433–441; https://doi.org/10.1016/j.exppara.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Song G, Bailey DW, Dunlap KA, et al. Cathepsin B, Cathepsin L, and Cystatin C in the Porcine Uterus and Placenta: Potential Roles in Endometrial/Placental Remodeling and in Fluid-Phase Transport of Proteins Secreted by Uterine Epithelia Across Placental Areolae. Biology of Reproduction, 2010, 82: 854–864. doi: 10.1095/biolreprod.109.080929 [DOI] [PubMed] [Google Scholar]
- 33.Balboula AZ, Yamanaka K, Sakatani M. Cathepsin B activity is related to the quality of bovine cumulus oocyte complexes and its inhibition can improve their developmental competence. Molecular Reproduction & Development, 2010, 77 (5):439–448. [DOI] [PubMed] [Google Scholar]
- 34.Reise RW, Huettel RN, Sayre RM. Carrot Callus Tissue for Culture of Endoparasitic Nematodes. Journal of Nematology, 1987, 198(3): 387–9. [PMC free article] [PubMed] [Google Scholar]
- 35.Adnan Tülek, İlker Kepenekci, Sultan Çobanoğlu. A new culturing method for the rice white tip nematode, Aphelenchoides besseyi Christie, 1942, on carrot discs. Russian Journal of Nematology, 2009, 17 (2), 135–136 [Google Scholar]
- 36.Qiu XW, Wu XQ, Huang L, Ye JR. Influence of Bxpel1 Gene Silencing by dsRNA Interference on the Development and Pathogenicity of the Pine Wood Nematode, Bursaphelenchus xylophilus. International Journal of Molecular Science, 2016, 17, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng XY, Dai SM, Xiao L. Influence of cellulase gene knockdown by dsRNA interference on the development and reproduction of the pine wood nematode, Bursaphelenchus xylophilus. Nematology, 2010, 12 (2):225–233. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Underscored: a predicted N-glycosylation site; shaded: the occluding loop; italics: the cysteine protease catalytic triad of residues (Cys, His, and Asn); bold: predicted cleavage points of mature and pro-domains; red: glutamine oxyanion hole; box: hemoglobinase motif; double underline: protease S2 pocket.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.