Abstract
The γ-tubulin complex is a large multiprotein complex that is required for microtubule nucleation at the centrosome. Here we report the purification and characterization of the human γ-tubulin complex and the identification of its subunits. The human γ-tubulin complex is a ring of ∼25 nm, has a subunit structure similar to that reported for γ-tubulin complexes from other species, and is able to nucleate microtubule polymerization in vitro. Mass spectrometry analysis of the human γ-tubulin complex components confirmed the presence of four previously identified components (γ-tubulin and γ-tubulin complex proteins [GCPs] 2, 3, and 4) and led to the identification of two new components, GCP5 and GCP6. Sequence analysis revealed that the GCPs share five regions of sequence similarity and define a novel protein superfamily that is conserved in metazoans. GCP5 and GCP6, like other components of the γ-tubulin complex, localize to the centrosome and associate with microtubules, suggesting that the entire γ-tubulin complex takes part in both of these interactions. Stoichiometry experiments revealed that there is a single copy of GCP5 and multiple copies of γ-tubulin, GCP2, GCP3, and GCP4 within the γ-tubulin complex. Thus, the γ-tubulin complex is conserved in structure and function, suggesting that the mechanism of microtubule nucleation is conserved.
INTRODUCTION
Microtubules are complex polymers composed of α/β-tubulin heterodimers assembled head-to-tail in protofilaments, which are arranged in a hollow cylinder (Tilney et al., 1973). Microtubules have a distinct polarity derived from the asymmetry of the α/β-tubulin heterodimer; one end of the microtubule is fast growing and is designated the plus end, whereas the other end is slow growing and is designated the minus end. In animal cells, most microtubules are nucleated by the microtubule organizing center, or centrosome. The centrosome is composed of a pair of centrioles that are surrounded by pericentriolar material. Microtubules are oriented with the minus ends at the centrosome and the plus ends growing away from the centrosome (Bergen et al., 1980; Heidemann and McIntosh, 1980). This results in microtubules being organized into a polarized radial array with the centrosome as its focus.
The pericentriolar material nucleates microtubules (Gould and Borisy, 1977) and contains a unique tubulin superfamily member, γ-tubulin, that does not assemble into microtubules. γ-Tubulin was discovered as a suppressor of a β-tubulin mutation in Aspergillus nidulans (Weil et al., 1986; Oakley and Oakley, 1989) and has now been found in all eukaryotes (reviewed by Jeng and Stearns, 1999). In animal cells, part of the total γ-tubulin is found at specific structures, including the centrosome (Stearns et al., 1991; Zheng et al., 1991), the mitotic spindle (Julian et al., 1993), and the midbody (Lajoie et al., 1994), and part is in the cytoplasm, not associated with any structures (Stearns and Kirschner, 1994). The cytoplasmic γ-tubulin is in the form of a large 32S complex (Stearns and Kirschner, 1994) that has a characteristic ring shape (Zheng et al., 1995). Zheng and coworkers have named this the γ-tubulin ring complex and have shown that it has a diameter of ∼25 nm (Oegema et al., 1999) and a defined subunit structure that is similar to a microtubule in cross section.
The cytoplasmic γ-tubulin complex can bind to microtubules (Stearns and Kirschner, 1994) and nucleate microtubule polymerization in vitro (Zheng et al., 1995). A simple model for centrosome-mediated microtubule nucleation is that cytoplasmic γ-tubulin complexes are recruited to the pericentriolar material of the centrosome, where they then can nucleate microtubule polymerization. This is supported by experiments showing that failure to recruit cytoplasmic γ-tubulin complex to the nascent sperm centrosome in egg cytoplasm abolishes microtubule nucleation (Felix et al., 1994; Stearns and Kirschner, 1994). Most importantly, ring-shaped structures containing γ-tubulin have been observed in the pericentriolar material and are thus likely to correspond to the centrosomal form of the γ-tubulin complex (Moritz et al., 1995; Vogel et al., 1997). The mechanism of attachment of the γ-tubulin complex to the centrosome is not understood, although it has been shown to require soluble factors (Moritz et al., 1998; Schnackenberg et al., 1998).
γ-Tubulin complexes from different organisms vary in size and complexity. The simplest known γ-tubulin complex is that of Saccharomyces cerevisiae, which is composed of two molecules of γ-tubulin and one molecule each of Spc97p and Spc98p (Geissler et al., 1996; Knop et al., 1997). The Spc97p and Spc98p proteins are related to each other (Murphy et al., 1998) and are both capable of binding to γ-tubulin (Knop et al., 1997; Nguyen et al., 1998). The γ-tubulin complexes from Xenopus (Zheng et al., 1995), Drosophila (Oegema et al., 1999), sheep (Detraves et al., 1997), pig (Fava et al., 1999), and mouse (Murphy et al., 1998) all contain the orthologues of Spc97p and Spc98p, as well as at least three additional proteins. We have adopted a standardized nomenclature for the proteins of the γ-tubulin complex, referring to them as GCPs (γ-tubulin complex proteins) (Murphy et al., 1998). The human homologues of Spc97p and Spc98p are GCP2 (Murphy et al., 1998) and GCP3 (Murphy et al., 1998; Tassin et al., 1998). The Drosophila (Oegema et al., 1999) and Xenopus (Martin et al., 1998) versions of these proteins have also been described. In Drosophila, a fraction of the γ-tubulin is in a small complex that, like the yeast complex, consists only of γ-tubulin and the fly orthologues of Spc97p/GCP2 and Spc98p/GCP3. This suggests that these three proteins form a conserved functional unit.
Although no γ-tubulin complex from an animal cell has been completely described, several of the individual components have been identified. Fava et al. (1999) reported the identification of p76, or GCP4, from human, Drosophila, and plant cells. Additional components of the γ-tubulin complex have been described in Drosophila (Dgrip128 and Dgrip163; Gunawardane et al., 2000) and Xenopus (Xgrip210; Zhang et al., 2000). Remarkably, each of these proteins is related to the GCP2 and GCP3 proteins, and like GCP2 and GCP3, each of these proteins has been shown to localize to the centrosome along with γ-tubulin.
In this study, we purified and characterized the human γ-tubulin complex and used mass spectrometry to identify the remaining components, GCP5 and GCP6. The size, structure, and activity of the human γ-tubulin complex are similar to those reported for the Drosophila and Xenopus γ-tubulin complexes. In addition, the GCP5 and GCP6 proteins are related to the other GCP components of the complex and with the others define a conserved protein superfamily. These results provide a framework for understanding the structure and function of the γ-tubulin complex and suggest that the molecular mechanism of microtubule nucleation is highly conserved.
MATERIALS AND METHODS
Cell Culture
HEK293 (human embryonic kidney) cells, U2OS (human osteosarcoma) cells, and BALB/c 393 subclone A31 (mouse fibroblast) cells were grown as monolayers at 37°C with 5% CO2. HEK293 cells were grown in DMEM medium (Life Technologies, Rockville, MD) with 10% fetal calf serum. U2OS cells were grown in McCoy's medium with 10% fetal calf serum. A31 cells were grown in DMEM media with 5% newborn calf serum and 5% fetal calf serum. A31γMycHis, A31GCP2MycHis, and A31GCP3MycHis (Murphy et al., 1998) were grown in DMEM medium with 5% newborn calf serum, 5% fetal calf serum, and 0.2 mg/ml geneticin (Life Technologies).
Cell lines stably expressing GCP4, GCP5, or GCP6 with C-terminal myc epitope and 6× histidine (mh) tags under the control of the cytomegalovirus (CMV) promoter were generated by transfecting U2OS cells with pTS1124 (CMV-GCP4mh), pTS1024 (CMV-GCP5mh), or pTS1090 (CMV-GCP6mh). U2OS cells were transfected with these plasmids with the use of the Effectene transfection protocol (Qiagen, Valencia, CA) followed by clonal selection in the presence of 0.4 mg/ml geneticin. Clonal cell lines were selected for further analysis based on the expression level of the transgene as judged by immunoblot analysis and immunofluorescence with the use of monoclonal anti-myc antibodies (9E10; Evan et al., 1985).
Purification of the γ-Tubulin Complex
HEK293 cells were grown to confluence, washed twice in HBS (50 mM HEPES, pH 7.4, 150 mM NaCl) at 4°C and collected by scraping. The cell pellet was resuspended in 3 volumes of lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM EGTA, 1 mM MgCl2, 0.25 mM GTP, 0.5% Triton X-100, aprotinin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride [PMSF]) and incubated for 5 min on ice. The resulting cell lysate was centrifuged at 12,000 × g for 15 min at 4°C, and then the supernatant was further clarified by centrifugation at 100,000 × g for 30 min at 4°C. The γ-tubulin complex was precipitated from the clarified lysate by adding an equal volume of 9% polyethylene glycol (Mr 3300) in buffer B (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, and 0.25 mM GTP), incubating the mixture at 4°C for 30 min and then centrifuging at 12,000 g for 15 min. The resulting pellet was resuspended in buffer B, clarified by centrifuging at 100,000 × g for 30 min at 4°C, and then desalted on a Sephadex G-25 medium (Amersham Pharmacia Biotech, Piscataway, NJ) column.
The desalted protein was applied to an anti-GCP2 affinity column at a flow rate of 0.02 ml/min or bound in batch format to the column. The anti-GCP2 affinity column was made by binding affinity-purified anti-GCP2 antibody to protein A-Sepharose and then cross-linking the antibody to protein A with dimethylpimelimadate (Pierce Chemical, Rockford, IL) as described by Harlow and Lane (1988). The column was washed with 10 column volumes of buffer B plus 0.5% Triton X-100, 10 column volumes of buffer B with 250 mM NaCl, and 10 column volumes of buffer B. The column was then incubated for 1 h in the presence of 1 column volume of buffer B with 300 μg/ml GCP2 antigenic peptide and then washed with 10 column volumes of buffer B. For mass spectrometry analysis, the column was washed with 10 column volumes of buffer B with 10 mM HEPES, pH 7.4, and then eluted by incubating in the presence of 100 mM glycine, pH 2.5, for 10 min. The elution was neutralized by adding 0.1 column volume of 1.5 M Tris, pH 8.8. For electron microscopy and microtubule nucleation analysis, the column was incubated overnight in buffer B plus 300 μg/ml GCP2 antigenic peptide. The elution was collected by passing 2 column volumes of buffer B through the column.
Electron Microscopy
Carbon-coated copper grids were glow discharged in air, and then 5 μl of purified γ-tubulin complex was applied to each grid for ∼2 min. Grids were rinsed in water, and then 5–10 μl of freshly filtered 1% uranyl acetate were applied for 30 s. Excess stain was wicked off with filter paper, and grids were allowed to dry at room temperature. Images were obtained on a Philips (Eindhoven, Netherlands) FEI Tecnai20 transmission electron microscope at 29,000× magnification with the use of a Gatan (Pleasanton, CA) 1024 × 1024 cooled CCD camera.
Microtubule Nucleation
Polymerization of microtubules was followed in a Lambda 20 spectrometer (Perkin Elmer-Cetus Instruments, Norwalk, CT) by turbidity at 350 nm, as described previously (Gaskin et al., 1974; Voter and Erickson, 1984), with the following modifications. Tubulin was purified from bovine brain by two cycles of polymerization/depolymerization followed by phosphocellulose chromatography (Mitchison and Kirschner, 1984). Before use in the turbidometric nucleation assay, the tubulin was cycled once more and centrifuged at 80,000 rpm (278,000 × g) in a TL100 rotor (Beckman Instruments, Fullerton, CA) for 20 min to remove microtubule “seeds.” To prevent polymerization of the tubulin, it was then kept on a 0°C ice-water slurry while reaction mixtures were prepared. Just before assaying, the tubulin was diluted into a mixture of γ-tubulin complex (femto- to nanomolar concentration range) and MgGly buffer (a 2× stock of buffer was used to generate a final concentration of 80 mM K-1,4-piperazinediethanesulfonic acid (PIPES), pH 6.8, 2 mM MgCl2, 1 mM EGTA, 3.4 M glycerol, 1 mM GTP). Nucleation was tested at a range of tubulin concentrations (3.9, 4.9, 5.9, 6.7, 7.8, and 9 μM). Each γ-tubulin complex-containing reaction was paired with a reaction containing the same amount of γ-tubulin complex elution buffer as a control. Reactions were quickly mixed and transferred to cuvettes on the ice-water slurry. The cuvettes were then placed in the spectrometer's nine-cell, Peltier changer, which was prewarmed to 37°C. Up to five reactions were followed simultaneously in the cell changer; the absorbance at 350 nm was recorded every 30 s for up to 90 min, depending on the experiment. The nucleation assays were repeated four times on different days with similar results.
To be certain that the turbidity curves represented the accumulation of bona fide microtubules, for one set of experiments, samples of each reaction at their polymerization plateau were removed, fixed for 3 min in 37°C 1% glutaraldehyde in BRB80 (80 mM K-PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA), diluted with 2 volumes of BRB80, and then sedimented through a 10% glycerol/1× BRB80 cushion onto electron microscopy grids. The presence of microtubules on the grids was confirmed by electron microscopy.
Mass Spectrometry Analysis
γ-Tubulin complex purified from 800 mg of 293 cell lysate (as described above) was concentrated by methanol/chloroform precipitation. Briefly, 800 μl of methanol were added to 200 μl of elution from the anti-GCP2 column, the solution was mixed, and 200 μl of chloroform were added, and the samples were mixed again. Water (600 μl) was added, and the solution was mixed by vortexing for 30 min and then centrifuged for 1 min in a microfuge at 14,000 × g. The upper layer was removed without disturbing the interphase, 600 μl of methanol were added to the interphase and lower phase, and the solution was mixed and then centrifuged for 5 min in a microfuge at 14,000 × g. The supernatant was removed and the pellet was dried. The pellet was resuspended in 40 μl of 2× pH 8.3 sample buffer (125 mM Tris, pH 8.3, 40% glycerol, 4% SDS, 0.05% bromophenol blue, 10 mM DTT) and heated for 5 min at 85°C. After returning to room temperature the sample was alkylated by adding 160 μg of iodoacetamide and incubating for 20 min in the dark.
The sample was separated by SDS-PAGE. The major Coomassie-stained bands were removed and subjected to in-gel digestion with trypsin. The masses of the resulting peptides were determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry and used for sequence database searches (O'Connell and Stults, 1997).
Cloning GCP5 and GCP6
Intron/exon boundaries in the genomic sequence of GCP6 were predicted with the use of GeneScan (www.genes.mit.edu/GENSCAN.html, Burge and Karlin, 1997). Theoretical masses of tryptic peptides were predicted with the use of FindPept (www.expasy.cbr.nrc.ca/tools/findpept.html). Total RNA was isolated from U2OS cells with the use of the RNAeasy kit (Qiagen). Reverse-transcription–polymerase chain reaction (RT-PCR) was performed with the use of either the Thermoscript RT-PCR system and Platinum Pfx polymerase (Life Technologies, BRL) or the Superscript One-Step RT-PCR system (Life Technologies, BRL). Clones were sequenced with the use of the BigDye system (Amersham Pharmacia Biotech).
RT-PCR of GCP6 was performed with the use of a primer 21 bp downstream of the start codon and a primer downstream of the stop codon to generate two nearly full-length GCP6 cDNAs from two separate RT-PCR reactions. These two clones, pTS1021 and pTS1031, contained multiple nucleotide changes compared with the genomic sequence. Four of the changes were shared between the two PCR products and expressed sequence tags (ESTs) and are assumed to be errors in the genomic sequence. In addition to these changes, each clone contained PCR-introduced mutations (three for pTS1021, seven for pTS1031). These mutations were eliminated by replacing the mutated sequences in pTS1021 with corresponding nonmutated sequence from pTS1031. The lack of mutations in this hybrid clone (pTS1032) was confirmed by sequencing the entire clone. Two lines of evidence suggest that the predicted GCP6 start codon is correct. First, this position corresponds to a methionine codon in a similar position in a zebrafish EST that has high sequence similarity to GCP6. Second, the 5′-untranslated region of GCP6 contains an in-frame stop codon that is upstream of the predicted start codon.
To make a full-length cDNA clone of GCP6, sense and antisense oligonucleotides corresponding to the extreme 5′-end of GCP6, plus 5 bp of the 5′-untranslated region were annealed and ligated to the 5′-end of pTS1032, resulting in pTS1106 (accession number AF272887). A full-length GCP5 cDNA was obtained by a combination of lambda cDNA library screening and RT-PCR. PCR primers from the 5′-end of the GCP5 sequence were used to amplify a 503-bp fragment from an EST clone (accession number AI863224, obtained from Research Genetics, Huntsville, AL). A radioactive probe derived from this fragment was used to screen 7 × 105 plaques from a lambda ZAP2 HeLa cell cDNA library (Stratagene, La Jolla, CA; Sambrook et al., 1989). The clone with the longest insert contained most of the gene, including the predicted start codon, preceded by in-frame stop codons. The position of the start codon corresponds to a methionine codon in a similar position in a zebrafish EST that has high sequence similarity to GCP5. RT-PCR was used to clone the 3′-end of GCP5 from U2OS cell RNA. The RT-PCR product was ligated to the library clone to form the full-length GCP5 cDNA (pTS1026; accession number AF272884). The entire clone was sequenced and found to match the GCP5 EST and genomic sequences except for two silent changes at bases 2022 and 2646.
Mammalian expression constructs for GCP5 and GCP6 were made by inserting the full-length coding sequences into derivatives of pCDNA I Neo (Invitrogen, San Diego, CA).
Antibodies
Peptides corresponding to residues 867–880 (CERLERLSAERSQKATPQ) of GCP2, residues 549–563 (CNSTRDFESIRLAHDH) of GCP4, residues 974–989 (TWRMESIEKMESDFKN) of GCP5, and residues 1764–1678 (CRQLQLFKHEMQHFVK) of GCP6 were synthesized (Research Genetics, Huntsville, AL). A cysteine residue was added to the NH2 terminus of the peptides to allow cross-linking to a carrier protein, and the COOH terminus was amidated. The peptides were coupled to keyhole limpet hemocyanin (Calbiochem, La Jolla, CA) with sulfo-succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (Pierce). Each of these coupled peptides was injected into two rabbits for antibody production (Covance, Richmond, CA).
Antibodies to GCP2 were affinity purified (Harlow and Lane, 1988) against the antigenic peptide immobilized on sulfolink coupling gel (Pierce). Antibodies to GCP4, GCP5, and GCP6 were affinity purified against TrpE-GCP4mh, GST-GCP5mh, and HisGCP6mycΔN fusion proteins, respectively. Plasmids encoding TrpE-GCP4mh (pTS1030), GST-GCP5mh (pTS1027), and HisGCP6mycΔN (pTS1126) were transformed into bacterial strains RR1 and BL21LysS, respectively. The expression of TrpE-GCP4mh was induced by the exclusion of tryptophan from the medium and the addition of 3β-indoleacrylic acid to 5 μg/ml. The expression of GST-GCP5mh and HisGCP6mycΔN was induced by the addition of isopropylthio-β-galactoside to 0.5 mM. All fusion proteins were insoluble and were isolated as inclusion bodies (Harlow and Lane, 1988). Inclusion bodies were resuspended in sample buffer and analyzed on a 10% SDS-polyacrylamide gel. The proteins were transferred to nitrocellulose and used to affinity purify the anti-GCP4, anti-GCP5, and anti-GCP6 antibodies (Harlow and Lane, 1988).
Protein Analysis
Cells were lysed as described above for purification of the γ-tubulin complex and then clarified by centrifugation for 10 min at 12,000 × g at 4°C for most experiments. Protein concentration was determined by the Bradford assay (Bio-Rad, Richmond, CA). Immunoblotting, sucrose gradients, and immunoprecipitations were performed as previously described (Murphy et al., 1998).
Microtubule-pelleting Assay
Bovine brain tubulin and rhodamine-conjugated tubulin were prepared as described by Hyman et al. (1991). Bovine brain tubulin was diluted to a final concentration of 2 mg/ml in BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA). Protease inhibitors (aprotinin, pepstatin, leupeptin, and PMSF), 1 mM GTP, and 1 mM DTT were added. Rhodamine-conjugated tubulin was added to the mixture to a final concentration of 0.2 mg/ml. The mixture was incubated with increasing amounts of taxol (Calbiochem-Novabiochem, San Diego, CA; 0.02 mM for 5 min, 0.2 mM for 5 min, 2 mM for 15 min) at 37°C. The resulting microtubules were examined by fluorescence microscopy. The average length of the microtubules was 5.2 ±2.4 μm (n = 77).
Cytoplasmic cell lysate was made from GCP6mh cells as described above except that cytochalasin B was added to a final concentration of 100 μg/ml. Lysates were prespun in a TLA 100.2 rotor (Beckman, Fullerton, CA) at 70,000 rpm for 10 min to remove protein aggregates. Microtubules were mixed with 1.5 mg of total lysate protein and incubated at 30°C for 30 min. Samples were centrifuged through a 1.5-ml cushion of 40% glycerol plus 1 mM GTP, aprotinin, pepstatin, leupeptin, and PMSF in BRB80. Taxol (10 mM) was added to cushions for samples with microtubules. The samples were spun at 50,000 rpm for 10 min at 30°C in a TLS-55 rotor (Beckman). Cushions were washed three times with water and aspirated, and pellets were resuspended in sample buffer by sonication. Samples were analyzed on 6 and 7.5% SDS-PAGE.
Immunofluorescence
Cells were prepared for immunofluorescence as previously described (Murphy et al., 1998). Primary antibodies were diluted as follows: anti-Myc (mouse monoclonal 9E10) diluted 1:250; anti-γ-tubulin (affinity-purified rabbit polyclonal EAD-2–4; Stearns and Kirschner, 1994) diluted 1:200; anti-γ-tubulin (mouse monoclonal GTU-88; Sigma) diluted 1:1000; anti-α-tubulin (rat monoclonal YL1/2, Sera-Lab, Westbury, New York) diluted 1:200, GCP6 antibody diluted 1:100. Secondary antibodies were from Jackson Immunoresearch Laboratories (West Grove, PA) and were diluted 1:200 or 1:100. Micrographs were taken with a cooled CCD camera (Princeton Instruments, Princeton, NJ) on an Axioskop microscope (Zeiss, Oberkochen, Germany).
RESULTS
Characterization of the Human γ-Tubulin Complex
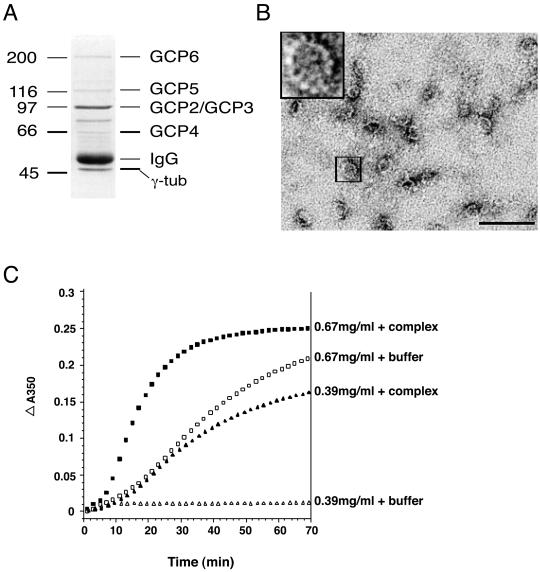
The γ-tubulin complex was purified from human 293 cells by a combination of biochemical steps and immunoaffinity with antibodies against GCP2, a previously defined component of the complex (Murphy et al., 1998). The protein composition of the purified material is shown in Figure 1A. The marked bands correspond to proteins that consistently copurified with γ-tubulin and the other known components of the complex. The components of the human γ-tubulin complex have molecular weights similar to those of γ-tubulin complexes from Xenopus (Zheng et al., 1995), Drosophila (Oegema et al., 1999), sheep (Detraves et al., 1997), pig (Fava et al., 1999), and mouse (Murphy et al., 1998) (Figure 1A).
Figure 1.
Characterization of the purified human γ-tubulin complex. (A) A Coomassie Blue-stained gel showing the proteins from the purified γ-tubulin (γ-tub) complex. The identities of the bands are indicated to the right. Size in kilodaltons is indicated to the left. (B) Negative stain electron microscopy of the purified complex. The human γ-tubulin complex is shaped like an open ring or spiral and has a diameter of 25 nm. Bar, 100 nm. (C) Microtubule nucleation by the complex was assayed by turbidity. Addition of γ-tubulin decreased microtubule polymerization lag time when compared with buffer control. Open symbols represent microtubule polymerization in the absence of the complex. Filled symbols represent microtubule polymerization in the presence of the complex. Two different concentrations of tubulin are shown: squares represent 0.67 mg/ml and triangles represent 0.39 mg/ml.
The γ-tubulin complexes from Xenopus (Zheng et al., 1995) and Drosophila (Oegema et al., 1999) have been shown to have a characteristic ring-shaped structure that mimics the end of a microtubule both in diameter and in subunit structure. To determine the morphology of the human γ-tubulin complex, it was examined by negative-stain electron microscopy (Figure 1B). We found that, like the Xenopus and Drosophila complexes, the human γ-tubulin complex is in the form of an open ring or spiral, with an average diameter of ∼25 nm and a defined subunit structure, with >10 but <15 subunits. A precise determination of the subunit structure will require more detailed analysis of the complex.
The γ-tubulin complex is necessary for microtubule nucleation at the centrosome (Felix et al., 1994; Stearns and Kirschner, 1994; Moritz et al., 1998) and is sufficient for microtubule nucleation in vitro (Zheng et al., 1995). We tested the ability of the purified human γ-tubulin complex to nucleate microtubules, with the use of a solution nucleation assay. A constant amount of the γ-tubulin complex was incubated with purified α-tubulin/β-tubulin heterodimer over a range of concentrations, and polymerization was assayed by turbidity over time. The human complex decreased the lag time for microtubule polymerization when compared with the buffer control and increased the total polymer formed (Figure 1C), indicating that it was active for microtubule nucleation.
Identification of the Components of the Human γ-Tubulin Complex
To identify the components of the human γ-tubulin complex, we performed tryptic digests of the major gel bands from the purified material and analyzed the resulting peptides by mass spectrometry. The peptide masses were searched against a virtual tryptic digest of the National Center for Biotechnology Information nonredundant protein database. Six bands, indicated in Figure 1A, were chosen for analysis based on their reproducible copurification with the use of several different methods and cell lines. The unmarked bands in Figure 1A did not reproducibly copurify and are assumed to be background proteins. Four of the copurifying bands were found to correspond to proteins previously shown to be components of the human γ-tubulin complex: γ-tubulin, GCP2, GCP3, and GCP4.
The peptide masses derived from the two remaining bands did not match any known proteins in the database. Because GCP2, GCP3, and GCP4 are related proteins, we reasoned that other proteins in the complex might also be related to these known components. We searched the human EST database and found a set of overlapping ESTs that defined a gene that was related to the other GCPs but was distinct from them. The corresponding genomic locus was identified as being on chromosome 22, and a putative cDNA was assembled from this sequence by computer analysis of intron/exon boundaries. We named the protein that would result from translation of this cDNA GCP6. The masses of theoretical tryptic fragments of this protein matched those from the band indicated as GCP6 in Figure 1A. With the use of this sequence information a full-length GCP6 cDNA (accession number AF272887) was cloned by RT-PCR of human mRNA. The GCP6 cDNA would encode a protein with a predicted molecular mass of 244 kDa.
We then used the sequences of GCP2, GCP3, GCP4, and GCP6 to identify a fifth member of this family in the EST database. Overlapping ESTs defined a gene located on chromosome 15. We named the protein that would result from the translation of this cDNA GCP5. Masses of theoretical tryptic digests of this protein match those from the band indicated as GCP5 (Figure 1A). A full-length GCP5 cDNA (accession number AF272884) was obtained by a combination of cDNA library screening and RT-PCR of human mRNA (see MATERIALS AND METHODS). The GCP5 cDNA would encode a protein with a predicted molecular mass of 118 kDa. No further members of this superfamily are present in the current version of the human genome sequence. Thus, aside from γ-tubulin, all known members of the human γ-tubulin complex are part of a unique protein superfamily, which we will refer to as the GCP superfamily.
The GCP Superfamily
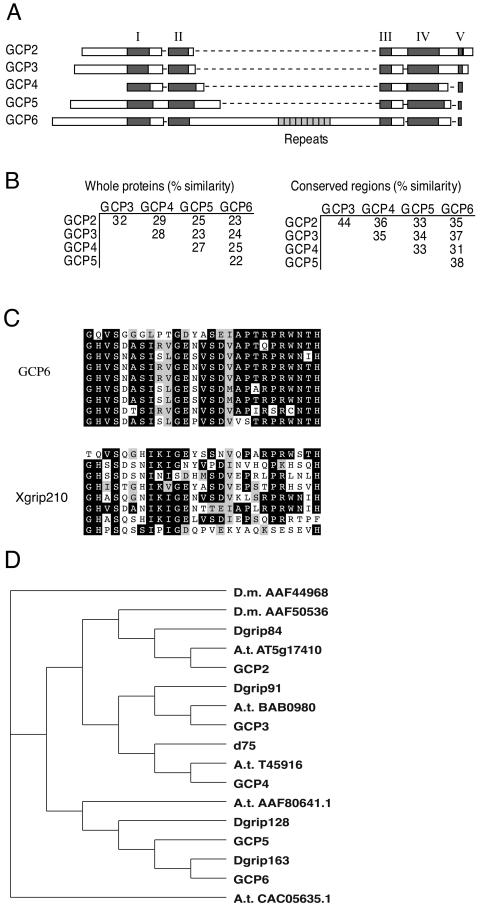
Comparison of the five human GCP protein sequences revealed that they share sequence similarity mainly in five regions (Figure 2A). Although the sequence similarity is low between full-length proteins (22–32%), the similarity within the five defined regions ranges from 31–44% (Figure 2B). The GCPs differ considerably in length and in the sequences outside of the regions defined above. At the extremes are GCP4, which is composed almost entirely of the defined regions, and GCP6, which has a long stretch of additional sequence between regions II and III (Figure 2A). Interestingly, this additional sequence in GCP6 contains nine tandem repeats of a 27-amino acid sequence (Figure 2C); these repeats bear no significant similarity to other known repeats.
Figure 2.
The GCPs are related to each other in five regions of conserved sequence. (A) Schematic diagram showing sequence conservation between the GCPs. Conserved regions are shown in dark gray. Unique regions are shown in white. The repeat region of GCP6 is shown in light gray. Gaps are indicated by dashed lines. (B) Percentage of sequence similarity between the GCPs. The amino acid sequences of the GCPs range from 22–32% similarity, whereas the conserved regions range from 31–44% similarity. (C) Alignment of human GCP6 and Xenopus Xgrip210 repeat regions. Black boxes indicate identical amino acids. Gray boxes indicate similar amino acids. (D) A phylogenetic tree of GCP homologues from human, D. melanogaster (D.m.), and A. thaliana (A.t.). The human GCP2 (accession number AF042379) and GCP3 (AF042378), GCP4 (AJ249677) and the Drosophila Dgrip84 (AF118379), Dgrip91 (AF118380), Dgrip128 (AJ291604), Dgrip163 (AJ291605), and d75p (AJ249678) sequences were previously published. GCP5 (AF272884) and GCP6 (AF272887) are described here, and the other proteins were identified by BLAST searches against the respective genome databases. The previously uncharacterized sequences are indicated as either D.m. or A.t., followed by the accession number. The repeat region of GCP6 was excluded in the comparison. The sequences were aligned with the use of ClustalW with a gap penalty of 50.
GCP2 and GCP3 are conserved in fungi, plants and animals, but GCP4, 5, and 6 appear to be both less widely distributed, and more divergent. GCP4, GCP5, and GCP6 are not found in the completed Saccharomyces cerevisiae and the current Schizosaccharomyces pombe genome databases. A frog orthologue of GCP6, named Xgrip210, has been described (Zhang et al., 2000), and there are multiple vertebrate ESTs corresponding to GCP4, GCP5, and GCP6, indicating general conservation of the structure of the γ-tubulin complex in vertebrates.
To further define the relationships among GCPs, we performed a phylogenetic analysis of homologous proteins from human, Drosophila melanogaster, and Arabidopsis thaliana (Figure 2D). There are seven GCP homologues in the Drosophila genome, and there are five in the Arabidopsis genome. Five of the Drosophila GCPs have been previously characterized: Dgrip84, Dgrip91 (Oegema et al., 1999), Dgrip128, Dgrip163 (Gunawardane et al., 2000), and d75p (Fava et al., 1999). Based on this analysis, the GCP2, GCP3, and GCP4 families are the most highly conserved within the GCP superfamily, and orthologues can be clearly identified in both Drosophila and Arabidopsis. Dgrip84 and A. thaliana 5G17410 are the predicted orthologues of GCP2, Dgrip91 and A. thaliana BAB0980 are the predicted orthologues of GCP3, and d75p and A. thaliana T45916 are the predicted orthologues of GCP4.
The identification of GCP5 and GCP6 orthologues is less definitive; however, pairwise alignments suggest that Dgrip128 and Dgrip163 are the Drosophila orthologues of GCP5 and GCP6, respectively. GCP5 has 18% sequence identity and 37% sequence similarity to Dgrip128 over the entire length of the proteins; GCP6 has 27% sequence identity and 48% sequence similarity to Dgrip163 in the most conserved C-terminal one-third of the proteins. The two remaining Arabidopsis GCP homologues are not sufficiently similar to either GCP5 or GCP6 to be definitively classified. Drosophila has two more GCPs than either human or Arabidopsis. D. melanogaster AAF0536 is most similar to GCP2, whereas D. melanogaster AAF44968 is not sufficiently similar to any of the GCPs to be classified. It will be interesting to see whether these extra Drosophila GCPs are redundant in function with other Drosophila GCPs or represent new functions.
The most striking feature of human GCP6 is the tandem repeat segment between regions II and III. These repeats are conserved in the GCP6 orthologues of other mammals and are also present in the Xenopus GCP6 orthologue Xgrip210 (Figure 2C), although they have diverged to such an extent that they went unnoticed in the original description of this protein (Zhang et al., 2000). Interestingly, the Drosophila GCP6 orthologue, Dgrip163, lacks this repeat region. Also we note that the reported start codon for the Xenopus GCP6 orthologue Xgrip210 (Zhang et al., 2000) corresponds to methionine at codon 72 in human GCP6 and is thus likely to be an internal methionine and not the true start codon.
GCP5 and GCP6 Are Components of the γ-Tubulin Complex
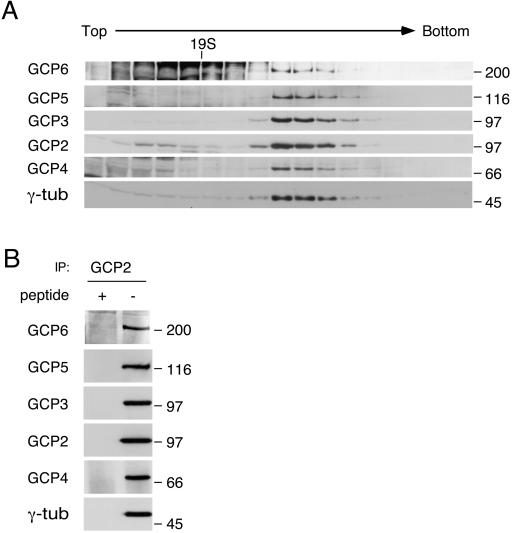
We confirmed that GCP5 and GCP6 are components of the γ-tubulin complex with the use of two methods. The γ-tubulin complex sediments as a 32S particle; if GCP5 and GCP6 are part of the γ-tubulin complex, they should cosediment with γ-tubulin. Antibodies against GCP5 and GCP6 were generated against peptides derived from the primary sequences. These antibodies recognized GCP5 and GCP6 proteins in the purified γ-tubulin complex, as well as bacterially expressed GCP5 and GCP6 fusion proteins. The antibodies were used to probe sucrose gradient fractions of human U2OS cell lysate. GCP5 and GCP6 cosedimented with γ-tubulin, GCP2, GCP3, and GCP4 (Figure 3A). Similarly, if GCP5 and GCP6 are components of the γ-tubulin complex, they should also coimmunoprecipitate with the other proteins in the complex. The γ-tubulin complex was immunoprecipitated from U2OS cell lysate with the use of anti-GCP2 antibody. In this and subsequent experiments, we used peptide competition as a test of specificity of the immunoprecipitation. The immunoprecipitates were probed with antibodies against γ-tubulin, GCP2, GCP3, GCP4, GCP5, and GCP6; each of these proteins coimmunoprecipitated with GCP2 (Figure 3B). Thus, GCP5 and GCP6 cosediment and coimmunoprecipitate with the other components of the γ-tubulin complex, indicating that they are components of the γ-tubulin complex.
Figure 3.
GCP5 and GCP6 cosediment and coimmunoprecipitate with γ-tubulin (γ-tub). (A) GCP5 and GCP6 from U20S cell lysate were examined by sucrose gradient sedimentation. Fractions from a 10–40% gradient were immunoblotted for γ-tubulin, GCP2, GCP3, GCP4, GCP5, and GCP6. The GCPs cosedimented with γ -tubulin. The anti-GCP6 antibody also recognizes nonspecific proteins in the lower molecular weight part of the gradient. The position of thyroglobulin (19S) is indicated. (B) U2OS cell lysate immunoprecipitated (IP) with anti-GCP2 antibody in the presence (+) and absence (−) of the antigenic peptide was subjected to immunoblot analysis with the use of antibodies against γ-tubulin, GCP2, GCP3, GCP4, GCP5, and GCP6. Size in kilodaltons is indicated to the right.
Each γ-Tubulin Complex Contains All Five GCPs
The γ-tubulin complex is unusual in having one unique component, γ-tubulin, and five related GCP components. We wished to determine whether all five of the GCPs are present in each γ-tubulin complex. Cell lines were generated that stably express individual GCP components with mh tags. Each of the epitope-tagged GCPs was found to cosediment on sucrose gradients with the γ-tubulin complex (Figure 4A). Interestingly, substantial amounts of the tagged proteins were also found in the lower molecular weight part of the gradient. The low molecular weight forms appeared to be proportional to the amount expressed, suggesting that excess protein cannot be incorporated into the complex. These epitope-tagged cell lines allowed us to distinguish the tagged protein from the endogenous protein and to immunoprecipitate each of the proteins specifically with the anti-myc antibody.
Figure 4.
(A) Cell lines stably expressing γ-tubulinmh, GCP2mh, GCP3mh, GCP4mh, GCP5mh, and GCP6mh were subjected to sucrose gradient sedimentation. Fractions were immunoblotted with anti-myc (myc) and anti-γ-tubulin (γ-tub) antibodies. Epitope-tagged proteins cosedimented with γ-tubulin. The position of thyroglobulin (19S) is indicated. (B) The γ-tubulin complex proteins were examined in pairwise combinations. Epitope-tagged proteins were immunoprecipitated with the anti-myc antibody in the presence (+) and absence (−) of the antigenic peptide. The immunoprecipitations were immunoblotted and probed with antibodies against other γ-tubulin complex proteins. Size in kilodaltons is indicated to the right.
To determine whether there are any components within the complex for which the presence of one excludes the presence of another, we examined the existence of each of the possible pairs of proteins within the complex. Tagged proteins were immunoprecipitated with the use of the myc antibody, and these immunoprecipitations were probed with antibodies against the other components. To ensure specificity, the proteins were immunoprecipitated in the presence and absence of competing antigenic peptide. Because all the γ-tubulin complex components could be identified in these pairwise combinations, no two components are mutually exclusive (Figure 4B). Given the discrete size of the γ-tubulin complex, this is most consistent with the complex containing all six components simultaneously.
Stoichiometry of the GCPs
We examined the stoichiometry of each of the components of the γ-tubulin complex with the use of the epitope-tagged cell lines described above. The experiment for each of the proteins was to immunoprecipitate the epitope-tagged version of the protein from the appropriate cell line and then to assay for the presence of the corresponding endogenous protein in the immunoprecipitate. If the endogenous protein is present, then there must be at least two copies of that protein in the γ-tubulin complex, whereas if the endogenous protein is not present, then there must be only a single copy of that protein in the complex. This technique was used previously to demonstrate that γ-tubulin and GCP3 are present in multiple copies within the γ-tubulin complex (Murphy et al., 1998).
We distinguished the tagged and endogenous proteins based on the mobility shift in SDS-PAGE gels caused by the tag. This shift was readily detected by Western blot analysis for GCP2mh, GCP4mh, and GCP5mh proteins (Figure 5). However, because of the large size of GCP6, no mobility shift could be detected for the GCP6mh protein. Immunoprecipitation of GCP2mh and GCP4mh with anti-myc antibodies resulted in the isolation of both tagged and endogenous proteins, indicating that GCP2 and GCP4 are present in multiple copies within the complex. Immunoprecipitation of GCP5mh with anti-myc antibodies resulted in the isolation of only the tagged protein, indicating that GCP5 is present in a single copy within the complex. An important control in this experiment is to immunoprecipitate the γ-tubulin complex independently of the epitope tag and demonstrate that both the tagged and endogenous proteins are detectable in the cell lines. We immunoprecipitated the tagged cell lines with the anti-GCP2 antibody and found that in all cases both tagged and endogenous proteins were present in the immunoprecipitate.
Figure 5.
GCP2 and GCP4 are present in multiple copies in the human γ-tubulin complex, whereas GCP5 is present in a single copy. Wild-type cells (wt) and cell lines stably expressing epitope-tagged GCPs were immunoprecipitated with anti-myc and anti-GCP2 antibodies. The immunoprecipitates (IP) were probed with antibodies against the endogenous protein corresponding to the protein tagged in that cell line. Immunoblots were also probed with antibodies against γ-tubulin or GCP2 as loading controls. There is less endogenous GCP4 on the blot because the GCP4mh cell line expresses higher levels of GCP4mh than the endogenous GCP4. Size in kilodaltons is indicated to the right.
GCP5 and GCP6 Localize to the Centrosome
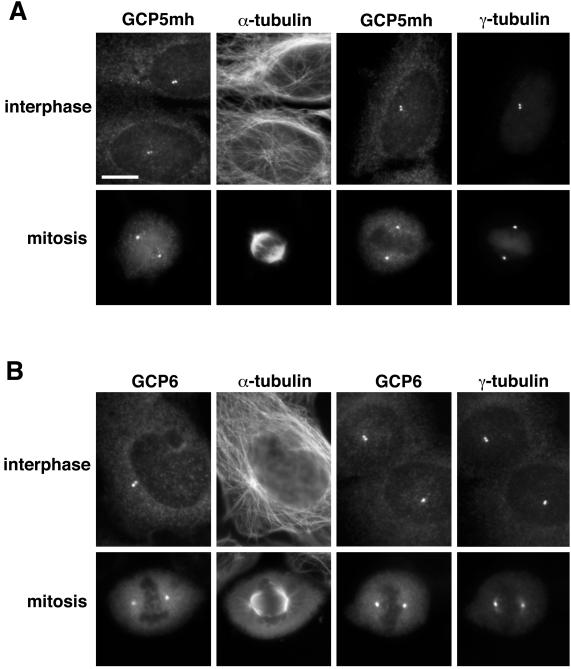
All known components of the γ-tubulin complex have been identified on the basis of their association with the cytoplasmic form of the complex; yet, the complex is believed to function at the centrosome. It is not clear whether there are differences in the composition of the cytoplasmic and centrosomal forms of the γ-tubulin complex. γ-Tubulin, GCP2, GCP3, and GCP4 have all been shown to localize to the centrosome, presumably as part of the complex. To determine whether GCP5 and GCP6 also localize to the centrosome, immunofluorescence microscopy was performed on human cells. For GCP6, both anti-GCP6 antibody on U2OS cells and anti-myc antibody on the GCP6mh cell line gave similar results. For GCP5, anti-myc immunofluorescence on the GCP5mh cell line was used because the anti-GCP5 antibody was not useful for immunofluorescence. Cells were double labeled with antibodies against either α-tubulin or γ-tubulin to visualize microtubules or the centrosome, respectively.
For both GCP5 and GCP6, staining was present at the foci of microtubule asters in interphase cells and at spindle poles of mitotic cells, colocalizing with γ-tubulin (Figure 6). This centrosomal staining was eliminated by competing antigenic peptide. In addition to the centrosome, γ-tubulin is detected in the midbody (Julian et al., 1993) and in the mitotic spindle, spreading from the poles (Lajoie et al., 1994). Interestingly, although GCP6 antibody stained the midbody of cells undergoing cytokinesis (data not shown), it did not stain the spindle microtubules of mitotic cells (Figure 6). This difference with γ-tubulin did not appear to be due to the intensity of labeling, because the centrosomal GCP6 signal was similar to that of γ-tubulin. We could not determine definitively whether GCP5 is present in the spindle or midbody because the anti-myc staining in the GCP5mh cell line was relatively weak. To determine whether GCP5 and GCP6 are core centrosomal components, we examined whether their localization to the centrosome is microtubule dependent. Cells were treated with nocodazole to depolymerize microtubules and labeled with anti–α-tubulin antibody to confirm microtubule depolymerization. For both GCP5 and GCP6, centrosomal staining remained in the absence of microtubules, indicating that they, like the other γ-tubulin complex proteins, are core centrosomal components.
Figure 6.
Localization of GCP5 and GCP6 by immunofluorescence. (A) Images of GCP5mh cell line stained with antibodies against the myc epitope, α-tubulin, and γ-tubulin. (B) Images of U2OS cells stained with antibodies against GCP6, α-tubulin, and γ-tubulin. Both GCP5mh and GCP6 colocalize with γ-tubulin at the centrosomes of both interphase and mitotic cells. Bar, 10 μm.
The Composition of Microtubule-bound γ-Tubulin Complex
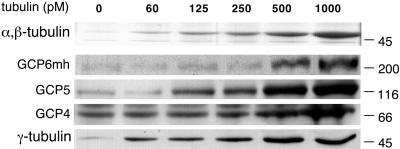
It is believed that γ-tubulin associates with the minus ends of microtubules as part of the entire γ-tubulin complex. This is supported by our previous observation that GCP2 and GCP3 from cell lysates associate with microtubules along with γ-tubulin (Murphy et al., 1998). To investigate whether GCP4, GCP5, and GCP6 also associate with microtubules, we performed a microtubule-pelleting assay. Varying amounts of taxol-stabilized microtubules were incubated with cell lysates and centrifuged through a glycerol cushion. The microtubule pellets were then assayed by Western blotting. γ-Tubulin, GCP2, GCP3, GCP4, GCP5, and GCP6 were all present in the pellets in an amount that increased proportionally with the amount of microtubules in the assay (Figure 7). This indicates that all the components of the γ-tubulin complex associate with microtubules and thus that the composition of the γ-tubulin complex does not change upon such association.
Figure 7.
GCP4, GCP5, and GCP6 copellet with taxol-stabilized microtubules. Equal amounts of GCP6mh cell lysates were incubated with increasing amounts of taxol-stabilized microtubules. Samples were centrifuged through a 10% glycerol cushion to pellet microtubules. Pellets were analyzed by SDS-PAGE and subsequent Coomassie Blue staining to examine α/β-tubulin and immunoblotting with antibodies against γ-tubulin, GCP4, GCP5, and myc. Size in kilodaltons is indicated to the right. Amount of microtubules in picomolar is indicated at the top.
DISCUSSION
The γ-tubulin complex is required for microtubule nucleation at the centrosome. To determine the composition of the human γ-tubulin complex, we purified the complex from cultured cells and performed mass spectrometry analysis of peptides derived from the constituent proteins. We found the four previously characterized members of the human γ-tubulin complex, γ-tubulin, GCP2, GCP3, and GCP4, and identified two new components, GCP5 and GCP6. Although the cytoplasmic form of the γ-tubulin complex was purified and characterized, all of the known components also localize with γ-tubulin to the centrosome in vivo and bind with γ-tubulin to microtubules in vitro. With the identification of GCP5 and GCP6, all of the major proteins in the human γ-tubulin complex have now been identified. Here we consider the implications of these results for γ-tubulin function.
Current ideas for the mechanism of γ-tubulin complex function are based in part on its morphology. Frog (Zheng et al., 1995), fly (Oegema et al., 1999), and human (this study) γ-tubulin complexes all share a characteristic ring, or spiral, shape and are ∼25 nm in diameter. This conservation of morphology strongly suggests that size and shape of the complex are important for function. Two models have been proposed for how the γ-tubulin complex nucleates microtubule polymerization. In the protofilament model, nucleation would occur by lateral interaction of α-tubulin/β-tubulin heterodimer subunits with a linear protofilament of γ-tubulin (Erickson and Stoffler, 1996). When not actively engaged with a microtubule, the linear γ-tubulin protofilament is proposed to adopt a ring conformation, similar to GDP-tubulin protofilaments from depolymerizing microtubules. In the template model, nucleation would occur by head-to-tail interaction of γ-tubulin with α-tubulin/β-tubulin heterodimers. Each of the 13 protofilaments of a typical microtubule would thus be templated directly by one of the repeated ring subunits of the γ-tubulin complex, (Zheng et al., 1995). Structural studies of the Drosophila (Moritz et al., 2000) and Xenopus (Keating and Borisy, 2000; Zhang et al., 2000) γ-tubulin complexes and their interaction with microtubules best fit the template model. Our results on the size and shape of the human γ-tubulin complex are also most consistent with the template model. It seems unlikely that the diameter and subunit structure of the γ-tubulin complex would be conserved to approximate those of the microtubule were it not for functional constraints.
γ-Tubulin, GCP2, and GCP3 are conserved in all eukaryotes and thus are likely to form a core unit of microtubule nucleation. However, neither γ-tubulin alone (Leguy et al., 2000) nor a γ-tubulin/GCP2/GCP3 small complex (Oegema et al., 1999) are able to nucleate microtubules efficiently, indicating that the other γ-tubulin complex components are important for nucleation. It is interesting that S. cerevisiae lacks GCP4, GCP5, and GCP6; it is likely that interactions with other proteins at the yeast spindle pole body serve the same function as these GCP components (reviewed by Jeng and Stearns, 1999). Although it is possible to clearly identify orthologues of γ-tubulin, GCP2, and GCP3 in all eukaryotes, the other GCPs are less well conserved. For example, the orthologue relationships of vertebrate GCP4, GCP5, and GCP6 to the GCP-like proteins in plants are not clear.
Why are so many related proteins found in the γ-tubulin complex? The GCP components share sequence similarity in restricted domains, and it seems likely that these conserved domains share common functions. This is the case for other complexes composed of related proteins, such as the CCT chaperonin complex (also known as the TCP-1 complex or TriC) and the RF-C DNA replication factor (also known as activator 1). CCT is composed of eight different subunits that shares conserved domains. The conserved domains of the CCT proteins are involved in the ATPase activity of each of the subunits and may also be involved in interactions between subunits (Kim et al., 1994). RF-C is composed of five subunits that shares eight regions of sequence similarity (reviewed by Mossi and Hubscher, 1998). These regions are involved in complex formation and substrate binding. By analogy, the regions of sequence similarity in the GCPs might have a role in folding of the proteins to the correct conformation or in GCP-GCP subunit interactions within the complex.
Although the GCPs are related, we suspect that they are not redundant in function. This is based on the finding that GCP2/Spc97p and GCP3/Spc98p are both essential proteins in yeast (Geissler et al., 1996; Knop et al., 1997) and that the proteins in the vertebrate γ-tubulin complexes are present in unique stoichiometries (Murphy et al., 1998; Zheng et al., 1995; this study). Again, this is similar to the CCT complex, in which each of the eight subunits is an essential protein in yeast. If the GCP components are not redundant, then what different functional properties might they possess? There are at least two large-scale interactions in which the γ-tubulin complex must take part. One is with the microtubule, which is likely to involve interaction of the γ-tubulin/GCP2/GCP3 subunits with tubulin heterodimers (see below) in a still ill- defined way. The other interaction is with the centrosome, or its equivalent in other organisms. The relatively low level of conservation of GCP4, GCP5, and GCP6 among eukaryotes suggests that they might be involved in this interaction; the organelles that carry out microtubule organization in eukaryotes are morphologically divergent, and thus the interacting proteins might be different.
The stoichiometry of the components within the γ-tubulin complex also suggests distinct functions for the individual components. Previous studies have assessed the stoichiometry of the components in the γ-tubulin complex by protein quantification, either by dye-binding in gels (Zheng et al., 1995; Oegema et al., 1999) or by immunoreactivity on Western blots (Fava et al., 1999). These studies revealed that γ-tubulin, GCP2, and GCP3 are the most abundant members of the complex. We took a complementary approach to determine whether individual components are present in single or multiple copies within the complex, relying on the ability to distinguish tagged and endogenous proteins in immunoprecipitates. Electron microscopy of the Drosophila γ-tubulin complex has shown that it consists of a ring of repeated subunits (Oegema et al., 1999), topped with an asymmetrical cap (Moritz et al., 2000). Based on the observations that γ-tubulin, GCP2, and GCP3 form a discrete subcomplex in yeast and Drosophila and that these proteins are the most abundant in the complex, it has been proposed that this γ-tubulin/GCP2/GCP3 subcomplex comprises the repeated subunit of the ring (Knop et al., 1997; Oegema et al., 1999). Consistent with this, we find that these three components are present in multiple copies (this study; Murphy et al., 1998). GCP5 is present in a single copy per complex, indicating that it must not be part of a repeated structure and is thus likely to be in the cap. GCP4, however, is present in multiple copies. If GCP4 resides in the cap, then it might interact with the repeated ring subunits. It is also possible that GCP4 resides within the repeated ring subunits themselves, although this seems less likely based on its absence from yeast.
The γ-tubulin complex is present in the cytoplasm and at the centrosome and is thought to be active for nucleation only at the centrosome in vivo. The function of the cytoplasmic γ-tubulin complex is not known, but one possibility is that the cytoplasmic form is a free pool that can be recruited to the centrosome to increase microtubule nucleation when needed, such as at mitosis. What difference(s), if any, exist between the cytoplasmic and centrosomal forms of the γ-tubulin complex? We have identified six proteins in the cytoplasmic form of the human γ-tubulin complex. Each of these proteins is also present at the centrosome and interacts with microtubules, presumably as part of the larger complex. Thus, the cytoplasmic form does not have any known subunits that are lost upon either interaction. Further characterization of the cytoplasmic and centrosomal forms will allow a determination of whether they differ in more subtle ways, such as modification of any of the constituent proteins.
ACKNOWLEDGMENTS
We thank Janos Demeter and Karine Piard-Ruster for critical reading of the manuscript and Paul Chang for analysis of the economics of phylogenetic analysis. This work was supported by a fellowship from the Searle Foundation and grant GM-52022 from the National Institutes of Health to T.S. S.M. was supported by grant PF-4198 from the American Cancer Society and by funding from the Stanford Cancer Biology Program.
Abbreviations used:
- CMV
cytomegalovirus
- DTT
dithiothreitol
- EST
expressed sequence tag
- GCP
gamma-tubulin complex protein
- mh
C-terminal myc epitope and 6× histidine tag
- PIPES
1,4-piperazinediethanesulfonic acid
- PMSF
phenylmethylsulfonyl fluoride
- RT-PCR
reverse-transcription–polymerase chain reaction
REFERENCES
- Bergen LG, Kuriyama R, Borisy GG. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Detraves C, Mazarguil H, Lajoie MI, Julian M, Raynaud MB, Wright M. Protein complexes containing gamma-tubulin are present in mammalian brain microtubule protein preparations. Cell Motil Cytoskeleton. 1997;36:179–189. doi: 10.1002/(SICI)1097-0169(1997)36:2<179::AID-CM7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M. Human 76p: a new member of the gamma tubulin associated protein family. J Cell Biol. 1999;147:857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix M-A, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin F, Cantor CR, Shelanski ML. Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J Mol Biol. 1974;89:737–755. doi: 10.1016/0022-2836(74)90048-5. [DOI] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J Cell Biol. 2000;151:1513–1524. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Heidemann SR, McIntosh JR. Visualization of the structural polarity of microtubules. Nature. 1980;286:517–519. doi: 10.1038/286517a0. [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Jeng R, Stearns T. Gamma-tubulin complexes: size does matter. Trends Cell Biol. 1999;9:339–342. doi: 10.1016/s0962-8924(99)01621-9. [DOI] [PubMed] [Google Scholar]
- Julian M, Tollon Y, Lajoie-Mazenc I, Moisand A, Mazarguil H, Puget A, Wright M. γ-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J Cell Sci. 1993;105:145–156. doi: 10.1242/jcs.105.1.145. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Kim S, Willison KR, Horwich AL. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie-Mazenc I, Tollon Y, Detraves C, Julian M, Moisand A, Gueth-Hallonet C, Debec A, Salles-Passador I, Puget A, Mazarguil H, et al. Recruitment of antigenic gamma-tubulin during mitosis in animal cells: presence of gamma-tubulin in the mitotic spindle. J Cell Sci. 1994;107:2825–2837. doi: 10.1242/jcs.107.10.2825. [DOI] [PubMed] [Google Scholar]
- Leguy R, Melki R, Pantaloni D, Carlier MF. Monomeric gamma-tubulin nucleates microtubules. J Biol Chem. 2000;275:21975–21980. doi: 10.1074/jbc.M000688200. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng YX. Xgrip109: a gamma-tubulin-associated protein with an essential role in gamma-tubulin ring complex (gamma-Turc) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–236. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossi R, Hubscher U. Clamping down on clamps and clamp loaders: the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Vinh DBN, Crawford DK, Davis TN. A genetic analysis of interactions with spc110p reveals distinct functions of spc97p and spc98p, components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- O'Connell KL, Stults JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997;18:349–359. doi: 10.1002/elps.1150180309. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent M, Barlat I, Langen H, Dargemont C. IkappaBalpha and IkappaBalpha/NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000;275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in-vitro. Proc Natl Acad Sci USA. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homolog of the yeast Spc98p and its association with gamma-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Bryna J, Bush DJ, Fujiwara K, Mooseker MS, Murphy DB, Snyder DH. Microtubules: evidence for 13 protofilaments. J Cell Biol. 1973;59:267–275. doi: 10.1083/jcb.59.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JM, Stearns T, Rieder CL, Palazzo RE. Centrosomes isolated from Spisula solidissima oocytes contain rings and an unusual stoichiometric ratio of alpha/beta tubulin. J Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voter WA, Erickson HP. The kinetics of microtubule assembly: evidence for a two-stage nucleation mechanism. J Biol Chem. 1984;259:10430–10438. [PubMed] [Google Scholar]
- Weber PJ, Eckhard CP, Gonser S, Otto H, Folkers G, Beck-Sickinger AG. On the role of thymopoietins in cell proliferation: immunochemical evidence for new members of the human thymopoietin family. Biol Chem. 1999;380:653–660. doi: 10.1515/BC.1999.081. [DOI] [PubMed] [Google Scholar]
- Weil CF, Oakley CE, Oakley BR. Isolation of mip (microtubule-interacting protein) mutations of Aspergillus nidulans. Mol Cell Biol. 1986;6:2963–2968. doi: 10.1128/mcb.6.8.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Keating TJ, Wilde A, Borisy GG, Zheng Y. The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J Cell Biol. 2000;151:1525–1536. doi: 10.1083/jcb.151.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. γ-Tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]