Abstract
NOTCH1 is a prevalent signaling pathway in T cell acute lymphoblastic leukemia (T-ALL), but crucial NOTCH1 downstream signals and target genes contributing to T-ALL pathogenesis cannot be retrospectively analyzed in patients and thus remain ill defined. This information is clinically relevant, as initiating lesions that lead to cell transformation and leukemia-initiating cell (LIC) activity are promising therapeutic targets against the major hurdle of T-ALL relapse. Here, we describe the generation in vivo of a human T cell leukemia that recapitulates T-ALL in patients, which arises de novo in immunodeficient mice reconstituted with human hematopoietic progenitors ectopically expressing active NOTCH1. This T-ALL model allowed us to identify CD44 as a direct NOTCH1 transcriptional target and to recognize CD44 overexpression as an early hallmark of preleukemic cells that engraft the BM and finally develop a clonal transplantable T-ALL that infiltrates lymphoid organs and brain. Notably, CD44 is shown to support crucial BM niche interactions necessary for LIC activity of human T-ALL xenografts and disease progression, highlighting the importance of the NOTCH1/CD44 axis in T-ALL pathogenesis. The observed therapeutic benefit of anti-CD44 antibody administration in xenotransplanted mice holds great promise for therapeutic purposes against T-ALL relapse.
Keywords: Hematology, Oncology
Keywords: Bone marrow, Leukemias, T cell development
Introduction
T cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy characterized by BM infiltration with immature lymphoblasts derived from transformed T cell precursors (1). A common cause of T-ALL is the dysregulation of signaling pathways that control T cell development in the thymus (2), such as NOTCH1, which is a key oncogenic player in human T-ALL (3). NOTCH1 is a member of the NOTCH family (NOTCH1–NOTCH4) of conserved transmembrane receptors that regulate survival, proliferation, and cell-fate decisions in different cell types, including T lymphocytes (4, 5). Binding of mammalian Notch receptors to Delta or Jagged ligands triggers a series of enzymatic cleavages by ADAM metalloproteases and γ-secretase that release the intracellular Notch domain (ICN). ICN translocates to the nucleus and forms a transcriptional complex with the DNA-binding protein CSL/RBPJ and coactivators of the mastermind-like (MAML) family (6). NOTCH1 plays a crucial role in T-ALL pathogenesis, as illustrated by the fact that over 60% of human T-ALLs contain gain-of-function NOTCH1 mutations that lead to ligand-independent NOTCH1 signaling (7).
Seminal studies have addressed the impact of dysregulated Notch signaling in T cell leukemogenesis using a well-defined murine model, wherein expression of constitutively active intracellular Notch1 (ICN1) in hematopoietic stem/progenitor cells (HSPCs) leads to extrathymic T cell development in the BM and results in a fatal murine T-ALL that resembles the human disease (8, 9). Weaker NOTCH1 alleles, such as those commonly found in human T-ALL, fail to initiate overt T cell leukemia in mice, but generate signals of sufficient strength to induce ectopic development and BM accumulation of aberrant CD4+CD8+ double-positive (DP) preleukemic T cells, which eventually cause an aggressive T-ALL in concert with other genetic alterations (10). Consequently, Notch-associated leukemogenesis is characterized by the initial interaction of preleukemic cells with the BM microenvironment, which may be a crucial step providing the supportive signals required for leukemia initiation (11). Therefore, either when the NOTCH1 mutation acts as the initial driving genetic event of T-ALL (12) or when oncogenic NOTCH1 complements other leukemogenic signals (10, 13), BM engraftment contributes to T-ALL pathogenesis. In both situations, NOTCH1 activation is an early hallmark of T cell leukemogenesis and a key regulator of leukemia-initiating cell (LIC) activity (13, 14), which suggests the participation of Notch signaling in the engraftment process. However, the nature of the initial signals that regulate BM engraftment of preleukemic cells and eventually contribute to the T-ALL transformation program remains largely unknown.
Recognizing the preleukemogenic events associated with aberrant NOTCH1 signaling must be clinically relevant, as it may lead to the identification of specific targets for developing improved therapeutic strategies to fight disease relapse, which is a main T-ALL clinical issue. However, retrospective analysis of human leukemic onset is unfeasible, and understanding the stepwise impact of NOTCH1 mutations on human T-ALL pathogenesis demands the availability of suitable in vivo models whose generation has been previously addressed, but has been more difficult than anticipated (15). Here, we have approached this aim using immunodeficient mice reconstituted with primary human cord blood (CB) HSPCs expressing constitutively active NOTCH1 and show the successful generation de novo of a clonal human leukemia that resembles T-ALL in patients. This T-ALL model has allowed the delineation of pathogenic events associated with the onset of the human disease. In particular, the identification of the adhesion molecule CD44 as a direct NOTCH1 target required for BM engraftment and LIC activity of human T-ALL xenografts highlights the importance of targeting the NOTCH1/CD44 axis in future therapeutic interventions.
Results
Oncogenic NOTCH1 drives BM engraftment and ectopic T cell development of human HSPCs and intrathymic precursors.
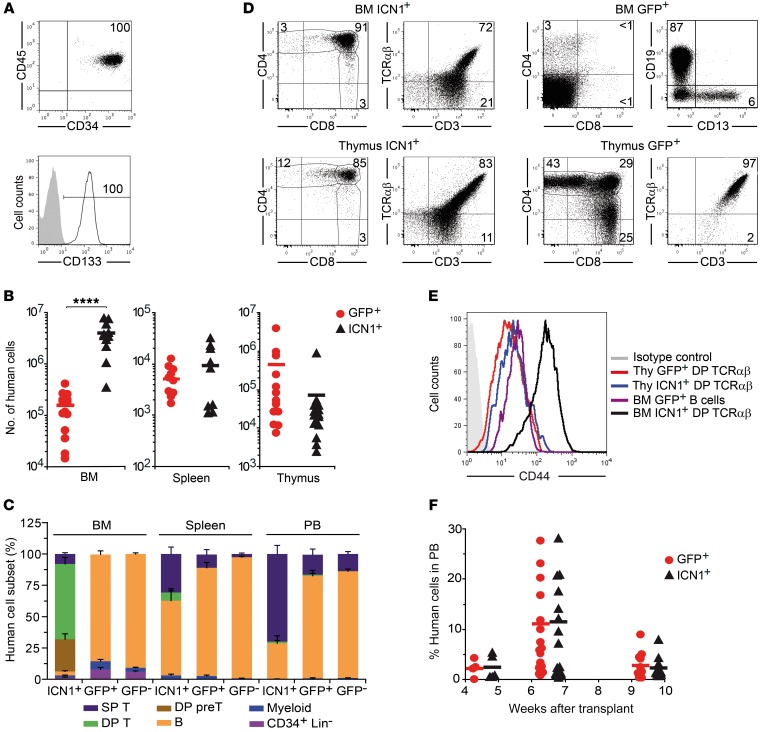
In an attempt to generate a human T-ALL in vivo, we took advantage of the oncogenic Notch1 approach, previously reported to induce an aggressive murine T-ALL (8, 9). To this end, CD34+CD133+ HSPCs isolated from human umbilical CB (Figure 1A) were transduced with a bicistronic retroviral vector encoding either constitutively active ICN1 and GFP as reporter or GFP alone, and cells were then transplanted into immunodeficient RAG-2–/– × γc–/– mice. Flow cytometry analysis of sequential BM aspirates revealed that enforced ICN1 expression significantly boosted human HSPC engraftment in mouse BM (25-fold at 9 weeks after transplant), but not in spleen and thymus (Figure 1B). Most BM grafting ICN1+ human cells displayed an aberrant CD4+CD8+ DP phenotype characteristic of immature thymocytes (>90% at 9 weeks after transplant; Figure 1, C and D) and showed unexpected high levels of the adhesion molecule CD44, specifically of the standard CD44 isoform (not shown), which is downregulated in conventional DP thymocytes (Figure 1E). A major proportion of ICN1+ DP ectopic T cells expressed the CD3–TCR-αβ complex, as did ICN1+ DP cells developing in the thymus, but about 25% displayed low CD3 expression and lacked both TCR-αβ (Figure 1D) and TCR-γδ (not shown), suggesting a differentiation blockade at the pre-TCR+ stage (16). In all animals, human ectopic T-lineage cells were exclusively derived from ICN1-transduced progenitors, whereas control GFP-transduced HSPCs differentiated mostly into CD19+ B lymphocytes in the BM and also produced a minor CD13+ myeloid subset, similarly to nontransduced HSPC controls (Figure 1, C and D). Importantly, ICN1+ T cells also reached the peripheral blood (PB) and spleen of transplanted mice, but essentially all of them displayed a CD4+ or CD8+ single-positive (SP) phenotype of conventional T cells, and very few showed the aberrant DP phenotype of their BM counterparts (Figure 1C). ICN1+ progenitors also produced B cells that colonized the PB and spleen, although at very reduced proportions compared with GFP+ controls (Figure 1C).
Figure 1. Constitutive NOTCH1 activation boosts BM engraftment of human HSPCs and promotes ectopic T cell development.
(A) Representative FACS phenotype of primary HSPCs from human CB (n = 5). (B) RAG-2–/– × γc–/– mice were transplanted with CD34+ CB HSPCs transduced with either retrovirus encoding ICN1 and GFP (ICN1; transduction efficiencies ± SEM: 21.0% ± 2.9%) or control retrovirus encoding GFP alone (GFP; 43.4% ± 8.2%). Shown are absolute numbers of human (CD45+) cells, normalized to 105 transduced input cells, in BM, spleen, and thymus at 9 weeks after transplant. (C) Percentages of human cells with the indicated cell phenotype recovered from the BM, spleen, and PB of mice shown in B. Percentages of cells derived from nontransduced (GFP–) HSPCs are also shown (SP T, CD4+ or CD8+ SP; DP T, CD4+CD8+ TCR-αβ+ DP; DP pre-T, CD4+CD8+ TCR-αβ–CD3lo; B, CD19+; Myeloid, CD33+CD13+; CD34+Lin–, CD34+ cells lacking lineage-specific markers). Mean values ± SEM are shown (n ≥ 4). (D) Representative phenotype of human cells derived from ICN1- or GFP-transduced HSPCs infiltrating the BM and thymus of mice shown in B (n ≥ 4). (E) Representative CD44 expression of human DP TCR-αβ+ and B cells from BM or thymus (Thy) of transplanted mice shown in B (n ≥ 6). (F) Percentages of human cells recovered at the indicated times after transplant from the PB of mice shown in B. Numbers were normalized to 25% of transduced input cells. Mean values from at least 4 independent experiments, with a combined total of 5 to 19 mice per group, are shown. ****P < 0.0001.
The observed engraftment of ICN1-transduced CB progenitors in RAG-2–/– × γc–/– mice contrasted with results from a previous study wherein no peripheral repopulation of NOD/SCID mice with ICN1+ human cells was observed (15). Thus, ICN1+ cells in our model could represent preleukemic cells able to eventually generate a human leukemia. However, such ICN1+ cells transiently engrafted the PB and showed repopulation kinetics indistinguishable from those of control GFP+ cells. Both reached maximal numbers at 7 weeks after transplant, but decreased significantly by 10 weeks (Figure 1F) and disappeared by 15 to 17 weeks (data not shown). Therefore, enforced NOTCH1 signaling in human HSPCs is able to promote early events associated with preleukemia generation and leukemia onset in mice, including ectopic generation of aberrant DP T cells (8); such putative preleukemic cells engrafted transiently in RAG-2–/– × γc–/– mice, with no evidence of leukemia generation, at least during the follow-up period of 17 weeks.
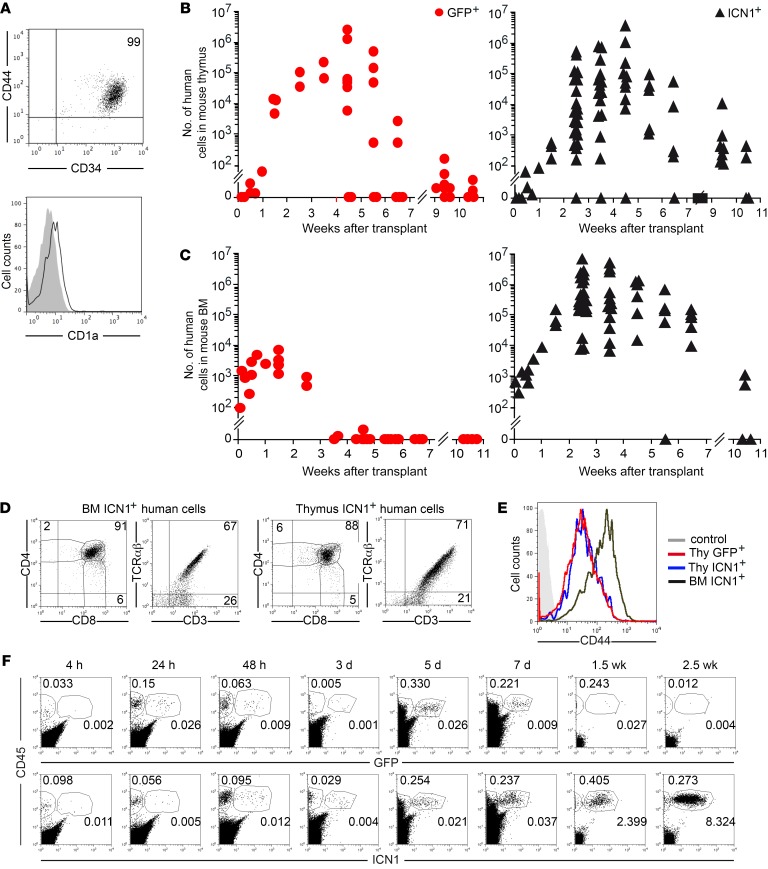
BM infiltration with transformed immature lymphoblasts derived from intrathymic T cell precursors is a common feature of T-ALL (1, 17). Therefore, thymus exit and exposure to a BM-supportive niche may be part of the T-ALL oncogenic program undertaken by preleukemic cells, as proposed for other hematologic neoplasias (11). To investigate whether NOTCH1 signaling contributes to that program, we analyzed BM seeding and engraftment potential of CD34+CD44+CD1a– early T cell precursors (ETPs) isolated from the human postnatal thymus (Figure 2A) and transplanted into RAG-2–/– × γc–/– mice upon transduction with either the ICN1 or the only-GFP vectors. We found that GFP-transduced ETPs reached the host thymus at 1 week after transplant, expanded in situ over 6 to 8 weeks, and disappeared after 10 weeks (Figure 2B), as observed with nontransduced thymocytes (not shown); however, they were unable to engraft the BM (Figure 2C). In contrast, ETPs expressing ICN1 recapitulated the behavior of ICN1+ HSPCs, as they efficiently engrafted the host BM for up to 10 to 12 weeks (Figure 2C) and also transiently repopulated the host thymus (Figure 2B). Importantly, as observed with HSPCs, ICN1+ ETPs generated ectopic DP cells in the BM, which were indistinguishable from their ICN1+ thymus counterparts in terms of CD4, CD8, and CD3–TCR-αβ expression (Figure 2D), but they aberrantly displayed upregulated levels of CD44 (Figure 2E). To assess whether ICN1 was able to improve cell engraftment by modulating homing into the BM, ICN1+ and GFP+ ETPs were next assayed in short-term homing assays, which showed that they displayed similar homing kinetics, as both reached the BM as early as at 4 hours after transplant and remained at equivalent numbers for up to 1 week (Figure 2F). However, GFP+ and nontransduced control ETPs dropped thereafter and became essentially undetectable by 2.5 weeks, while ICN1+ cell numbers increased steadily (up to 1000-fold) during the first 3 to 4 weeks after transplant (Figure 2, C and F). Therefore, active NOTCH1 does not markedly affect BM seeding of ETPs, but promotes their ectopic differentiation and expansion, likely contributing to the leukemogenic program.
Figure 2. Human thymic precursors overexpressing active NOTCH1 undergo BM engraftment and extrathymic differentiation.
(A) Representative phenotype of primary ETPs from human postnatal thymus (n = 20). (B and C) RAG-2–/– × γc–/– mice were transplanted with human ETPs transduced with ICN1 and GFP (19.3% ± 1.4% ICN1+) or GFP alone (28.0% ± 7.9% GFP+). Shown are absolute numbers of human cells recovered from thymus (B) and BM (C) at the indicated weeks after transplant. Numbers were normalized to 105 transduced input cells and represent combined results from 4 to 8 independent experiments, with a total of 35 to 75 mice per group. (D) Representative flow cytometry phenotype of human ICN1+ cells in BM and thymus of mice shown in B and C at 6 weeks after transplant (n = 4). (E) Representative CD44 expression of human DP TCR-αβ+ cells infiltrating the BM and thymus of mice shown in B and C (n ≥ 4). (F) Percentages of human CD45+ cells engrafting the BM of mice shown in B and C at the indicated times after transplant (n ≥4).
Aberrant CD44 expression is an early hallmark of mutant NOTCH1 signaling and extrathymic T cell development.
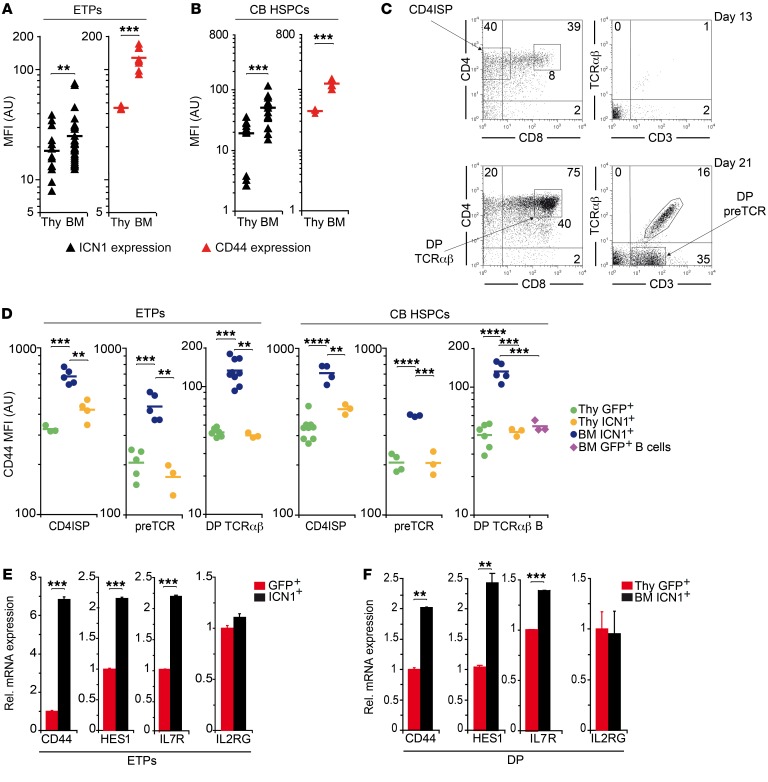
Upregulation of CD44 has been observed in preleukemic DP cells derived ectopically from murine BM progenitors expressing ICN1 (13, 17, 18), but a relationship between Notch activation and CD44 expression, although suggested, has not been studied in detail. Supporting a functional link between NOTCH1 activity and CD44 expression, quantitative FACS analysis revealed a tight correlation between CD44 surface expression and ICN1 transgene expression, assessed by GFP reporter levels, in the cellular progeny of human ICN1+ ETPs or CB HSPCs engrafting the thymus and BM of recipient mice (Figure 3, A and B), but such correlation was not observed in cells transduced only with GFP (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI92981DS1). Notably, the highest ICN1 and CD44 expression levels corresponded to T cells developing ectopically in the BM (Figure 3, A and B; and Supplemental Figure 1C), which included all sequential T cell maturation stages, i.e., CD4+CD3– immature SP (ISP), DP pre-TCR+, and DP CD3+TCR-αβ+ cells (Figure 3C), while their GFP+ control intrathymic counterparts showed downregulated levels of CD44 (Figure 3D). Quantitative gene expression analyses of ICN1-transduced ETPs revealed that CD44 upregulation was induced at the transcriptional level as early as at 36 hours after transduction, when Notch target genes HES1 and IL7R were concurrently upregulated, but the unrelated IL2RG gene remained unchanged (Figure 3E). Transcriptional upregulation of Notch targets was observed as well in ectopic ICN1+ DP CD44hi cells arising in the BM at 4 weeks after transplant, as compared with control DP thymocytes (Figure 3F). Collectively, these results provide evidence of a functional association between NOTCH1 activation and CD44 gene expression and indicate that CD44 upregulation is one of the earliest hallmarks of sustained NOTCH1 signaling, which is selectively associated with aberrant extrathymic T cell development.
Figure 3. CD44 upregulation is an early hallmark of aberrant NOTCH1 signaling and ectopic T cell development.
(A and B) GFP reporter and CD44 expression of ICN1+CD45+ human cells engrafting the thymus or BM of RAG-2–/– × γc–/– mice transplanted with either ETPs (A) or CB HSPCs (B) transduced with ICN1 and GFP. Cells were obtained at 2 to 6 and 9 weeks after transplant, respectively. Mean fluorescence intensity (MFI) combined results from 6 independent experiments, with a total of 3 to 30 mice per group, are shown. (C) Representative FACS phenotype of T-lineage cell subsets derived from ICN1-transduced human ETPs in the BM of RAG-2–/– × γc–/– mice shown in A at 13 and 21 days after transplant (CD4ISP, CD4+CD3-; DP TCR-αβ, CD4+CD8+CD3+TCR-αβ+; DP pre-TCR, CD4+CD8+CD3loTCR-αβ–). (D) CD44 expression levels of human CD4 ISP, DP pre-TCR, and DP TCR-αβ cells generated at 2, 3, and 6 weeks after transplant, respectively, in the BM and thymus of mice shown in A or at 9 weeks after transplant in the BM and thymus of mice shown in B. CD44 expression of control cells generated from either ETPs or CB HSPCs transduced with GFP (GFP+) in the thymus and of human B cells generated in the BM from GFP+ HSPCs is also shown. MFI ± SEM combined results from 6 (ETPs) and 3 (HSPCs) independent experiments, with a total of 3 to 9 mice per group are shown. Data were corrected by the FDR method. (E and F) Relative mRNA expression of CD44, HES1, IL7R, and IL2RG genes analyzed by qPCR and normalized to GAPDH in GFP- or ICN1-transduced human ETPs after 36 hours of transduction (C) or in DP thymocytes and BM ectopic DP cells derived from GFP- or ICN1-transduced human ETPs at 4 weeks after transplant. Data are shown as mean ± SEM of triplicates (n = 5). **P < 0.01; ***P < 0.001; ****P < 0.0001.
CD44 is a direct transcriptional target of Notch1 in T-ALL cells.
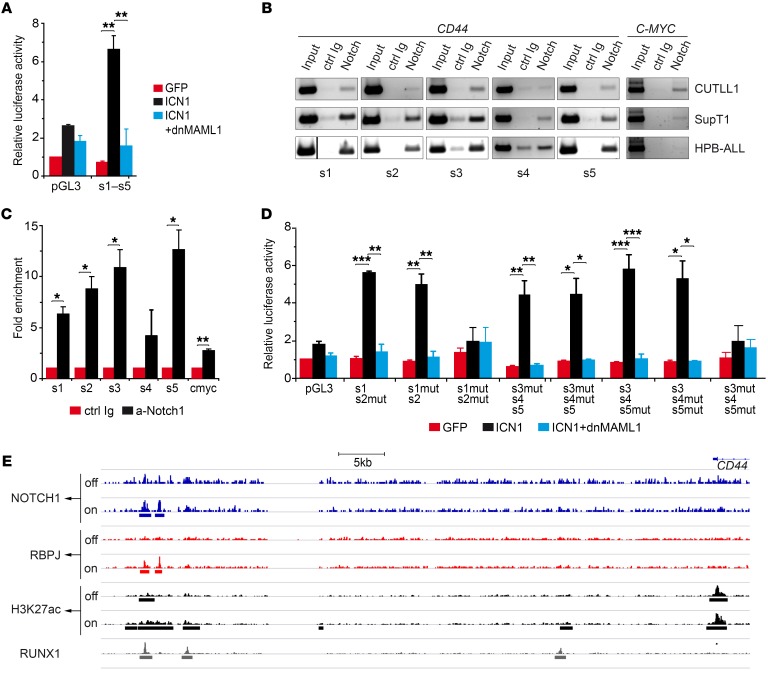
Supporting a direct role of NOTCH1 signaling in the induction of aberrant CD44 expression, enforced ICN1 expression induced CD44 upregulation at the clonal level in SupT1 and CUTLL1 T-ALL cell lines that display constitutive NOTCH1 activation and CD44 expression (19) (Supplemental Figure 2, A and B). Conversely, constitutive CD44 expression was downregulated upon transduction with a dominant negative form of the Notch coactivator MAML1 (dnMAML1) (Supplemental Figure 2A). These results were confirmed in mRNA expression assays showing that NOTCH1 signaling regulates CD44 as well as HES1 and IL7R transcriptional levels in a MAML1-dependent manner (Supplemental Figure 2C). Further experiments provided formal proof that CD44 is a direct NOTCH1 target. In particular, searching for putative CSL/RBPJ-binding sites in a human CD44 genomic sequence (Ensembl Gene database; https://uswest.ensembl.org:8443/sitedown.html) covering 5 kb upstream of the transcriptional start site (TSS) revealed 4 predicted CSL-binding sites at positions –4.7, –4.4, –3.8, and –3.5 kb (sites 1, 2, 3, and 5, respectively), which are conserved in nonhuman primates, and one at position –3.6 kb (site 4), also conserved in rodents. Reporter assays performed in SupT1 T-ALL cells transfected with a luciferase vector, which included the CD44 genomic region spanning the 5 putative CSL-binding sites (–4.8 to –3.5 kb), confirmed that ectopic ICN1 induced a significant transcriptional activity that was abolished by dnMAML1 (Figure 4A). Moreover, ChIP assays demonstrated that endogenous ICN1 expressed in CUTLL1, SupT1, and HPB-ALL T-ALL cell lines was associated in vivo with the CSL/RBPJ-binding motifs 1, 2, 3, and 5 in the CD44 gene and, as expected, also with the well-known NOTCH1 target MYC (20–22) (Figure 4, B and C). Site-directed mutagenesis confirmed the RBPJ-dependent autonomous functionality of those sites (Figure 4D). Therefore, CD44 is a CSL/MAML1-dependent direct NOTCH1 target in human T-ALL.
Figure 4. CD44 is a CSL/MAML-dependent direct target of NOTCH1.
(A) Luciferase reporter assays of SupT1 T-ALL cells cotransfected with a reporter construct containing a genomic CD44 region (–4.8 to –3.5 kb) spanning 5 putative CSL-binding sites along with ICN1-encoding retrovirus plus or minus retrovirus encoding dnMAML1 or with GFP-only control. Data are represented as mean fold induction ± SEM over luciferase activity of control cells cotransfected with an empty reporter vector (pGL3) and the GFP-only vector (n = 3). s1–s5, CSL-binding sites 1–5. (B) Representative ChIP assay performed either with anti-NOTCH1 Abs (anti-cleaved NOTCH1, for CUTLL1 and HPB-ALL, or anti–C-terminal NOTCH1, for SupT1) or with the corresponding control Ab. PCR amplification was done on input and immunoprecipitated DNA with primer pairs spanning each of the 5 CD44 CSL sites or the reported c-MYC CSL-binding site. The vertical dividing line separates lanes run noncontiguously on the same gel. (C) Quantitative data of ChIP assays in B. Results are shown as fold enrichment of anti-NOTCH1–immunoprecipitated DNA over DNA immunoprecipitated with control Abs. Bars represent mean values ± SEM (n = 3). Ctrl, control. (D) Luciferase reporter assay of SupT1 T-ALL cells cotransfected with a reporter vector containing the indicated combinations of WT and mutated (mut) sequences corresponding to the 5 putative CSL-binding sites identified in CD44, along with either retrovirus encoding ICN1 together or not with dnMAML1-encoding retrovirus or with GFP-only retrovirus as control. Data are represented as mean fold induction SEM over control cells as in A (n = 3). Data shown in A and D were corrected by FDR. (E) Chromatin landscapes upstream of the CD44 locus were derived from ChIP-seq raw data of human CUTLL1 cells under steady-state (basal), Notch-off, and Notch-on conditions (GEO GSE51800), previously described by Wang et al. (23). The presence of a 5′ distal enhancer containing a dynamic NOTCH1-PBPJ site and a RUNX1 site is shown. H3K27ac peaks associated with this dynamic NOTCH1 site are shown. *P < 0.05; **P < 0.01; ***P < 0.001.
A recent study by Aster’s group showed that direct NOTCH1 target genes in T-ALL could be transcriptionally regulated by dynamic NOTCH1 binding to H3K27ac-marked superenhancers that frequently overlap with RUNX1-binding sites (23). Importantly, based on their ChIP-sequencing (ChIP-seq) raw data (NCBI’s Gene Expression Omnibus database; GEO GSE51800), we identified 2 dynamic NOTCH1 sites located 55–60 kb upstream of the CD44 gene, near RUNX1 sites, which are strongly associated with H3K27ac marks sensitive to Notch signaling alterations (Figure 4E). Therefore, NOTCH1 may also activate CD44 gene expression through dynamic interactions with superenhancers, as shown for other NOTCH1 targets, such as IL7R (23).
Ectopic CD44hi DP T cells include preleukemic cells that generate an aggressive human T-ALL.
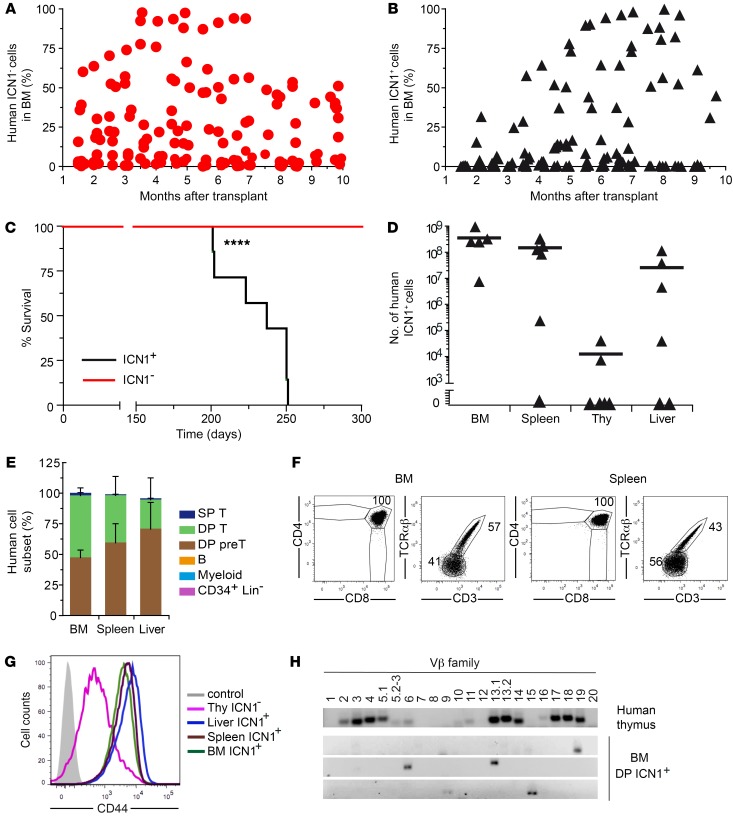
The finding that NOTCH1 signaling directly regulates CD44 transcription suggests that CD44 might be upregulated in preleukemic cells as part of the T-ALL oncogenic program. This possibility could not be confirmed in RAG-2–/– × γc–/– mice, wherein ICN1+ DP CD44hi aberrant cells engrafted transiently with no signs of leukemia progression (Figure 1F and Figure 2C). Importantly, these results concurred with the unexpected finding that control HSPCs also engrafted transiently in our model (Figure 1F), suggesting that the RAG-2–/– × γc–/– mouse strain is not optimal for assaying human hematopoietic engraftment and repopulation. To overcome this possible limitation, we moved to the NSG strain, which has been reported to support greater human engraftment in BM than other strains of mice (24), and assessed whether enforced ICN1 expression is leukemogenic for human HSPCs in NSG mice. Thus, ICN1-transduced (7.4% ± 1.7% ICN1+) human HSPCs were transplanted into 6- to 10-week-old NSG mice, and sequential BM aspirates were then analyzed by flow cytometry for the presence of ICN1+ and ICN1– human cells. We found that all NSG transplanted mice, in contrast with RAG-2–/– × γc–/– mice, supported multilineage (T, B, and myeloid cells) engraftment from control ICN1– human HSPCs during a follow-up period of 10 months (Figure 5A and data not shown) and that 50% simultaneously displayed long-term engraftment of ICN1+ cells (Figure 5B and Table 1). Strikingly, all animals with ICN1+ cell engraftment developed symptoms of disease and died of an aggressive T cell leukemia, with a median survival of 240 days (Figure 5C). Diseased mice showed up to 100-fold higher numbers of ICN1+ cells than ICN1– nontransduced cells in the BM (up to 109 in absolute numbers; Figure 5D), together with a high infiltration of peripheral organs, such as spleen (up to 3.4 × 108) and liver (up to 108), but thymus infiltration was lower, and PB infiltration was observed in 20% of mice (Table 1 and Figure 5D). This represents the first model, to our knowledge, of de novo generation of a human T-ALL in vivo.
Figure 5. Constitutive Notch1 signaling promotes an aggressive human T-ALL.
(A and B) CB HSPCs transduced with ICN1 and GFP (7.4% ± 1.7%) were transplanted into NSG mice. Combined data from 4 independent experiments, with a total of 18 mice, are shown as percentages of human cells derived from nontransduced ICN1– (A) or ICN1-transduced (B) HSPCs engrafting the mouse BM at the indicated months after transplant. (C) Kaplan-Meier survival curves of mice shown in A and B. (D) Absolute numbers of human ICN1+ cells infiltrating the BM, spleen, thymus, and liver of diseased mice shown in B at 7 months after transplant. Data were normalized to 105 transduced input cells. Mean values combined from 4 independent experiments with 5 to 6 mice/group are shown. (E) Mean percentages ± SEM of human ICN1+ cells of the indicated cell subsets, as defined in Figure 1C, infiltrating the BM, spleen, and liver of diseased mice shown in B (n ≥ 3). (F) Representative phenotype of human ICN1+ cells infiltrating the BM and spleen of diseased mice shown in B (n ≥ 7). (G) Representative CD44 expression of human DP T cells derived from ICN1-transduced or nontransduced CB HSPCs, infiltrating the indicated organs of diseased mice shown in B (n ≥ 7). (H) Reverse-transcriptase PCR (RT-PCR) analysis of the TCR-Vβ repertoire of 3 samples of human ICN1+ DP T cells infiltrating the BM of diseased mice shown in B (bottom panels) at 7 months after transplant (n = 8). The polyclonal repertoire of DP thymocytes isolated from human postnatal thymus is shown as control (upper panel). ****P < 0.0001.
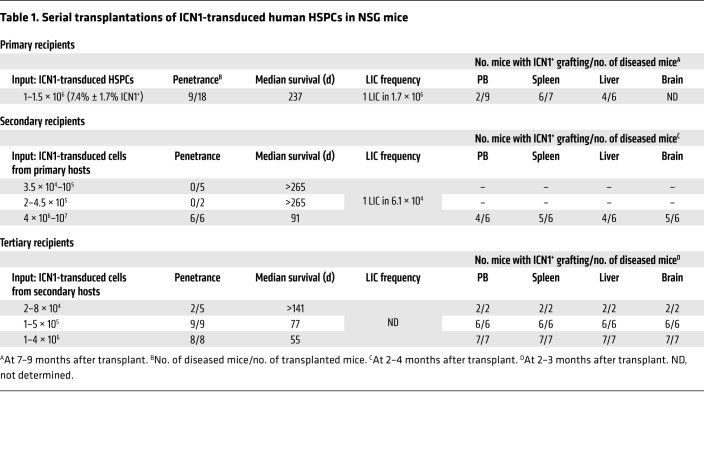
Table 1. Serial transplantations of ICN1-transduced human HSPCs in NSG mice.
Immunophenotypic analyses of sequential BM aspirates performed over a 9-month follow-up period revealed that engrafted ICN1+ cells in NSG mice were mostly DP T cells that expressed either the TCR-αβ or the pre-TCR, together with aberrantly high levels of CD44, and this phenotype was also observed in ICN1+ cells infiltrating the spleen and liver (Figure 5, E–G). Supporting their leukemogenic nature, BM ICN1+ DP cells displayed a restricted TCR-Vβ repertoire (Figure 5H) and, like spleen and liver ICN1+ cells (not shown), they were able to serially transfer an aggressive T-ALL disease to secondary and tertiary recipients (Table 1). Together, these data provide formal proof that the bulk of human ICN1+CD44hi DP T cells in the BM of NSG mice include T-ALL cells with LIC activity. Transplantation of these aberrant BM cells under limiting dilution conditions (102 to 107 cells/mouse) into secondary recipients revealed a LIC frequency of 1 in 1.7 × 106 ICN1+ BM cells, while LIC activity of leukemic cells arising in secondary recipients was increased by 28-fold, as revealed by tertiary transplants (Table 1 ). Increased LIC activity in sequential transplants resulted in a more aggressive disease with shorter latency, which correlated with an enhanced infiltration of leukemic DP CD44hi cells in blood, spleen, liver, and brain. Collectively, these data demonstrate that constitutive NOTCH1 signaling can induce a human aggressive leukemia in NSG mice that resembles T-ALL arising in patients.
CD44 is required for BM engraftment of NOTCH1-induced preleukemic cells and primary T-ALL cells.
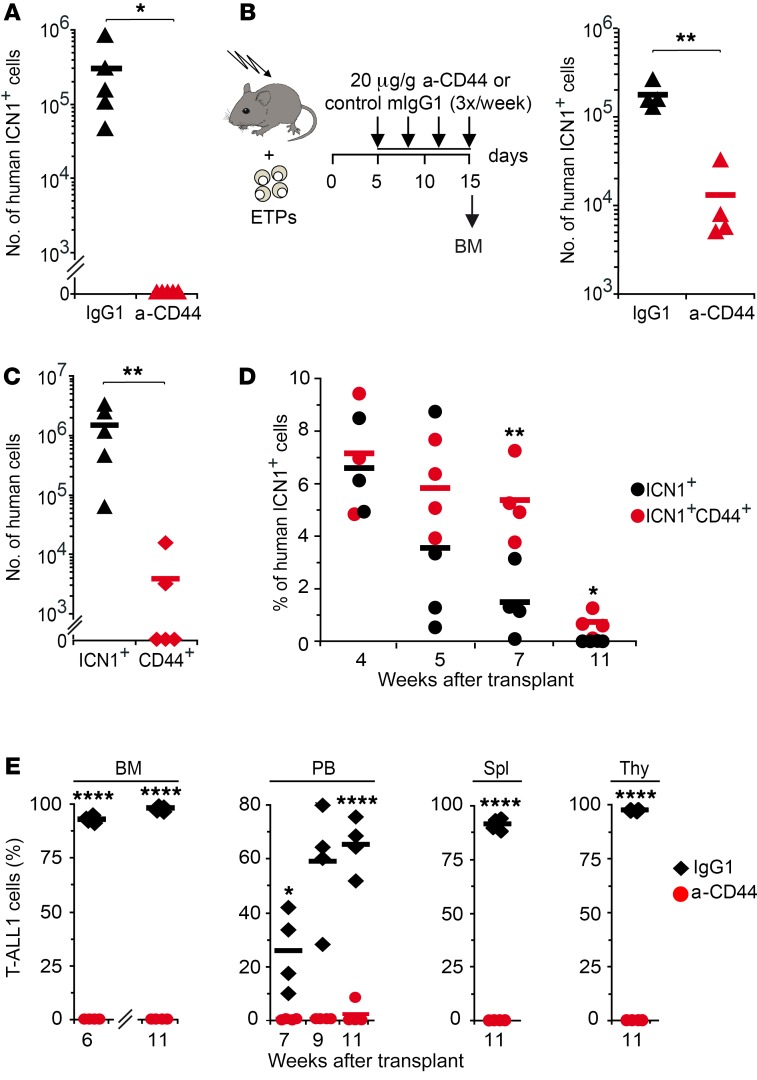
The role of CD44 as a critical mediator of interactions between normal HSPCs and the BM microenvironment has been known for many years (25, 26), and the expression of CD44 in some cancer-initiating cells has also been described more recently (27). Considering the critical role of the BM microenvironment in normal and neoplastic hematopoiesis (11), we sought to investigate the contribution of CD44 to NOTCH1-induced BM engraftment and leukemia progression. Thus, we first assessed the role of CD44 in BM engraftment of aberrant human cells derived from ICN1-transduced ETPs infused into Rag2–/– × γc–/– mice and found that cell treatment before transplantation with a neutralizing mAb that blocks CD44 interaction with its ligands (515, IgG1, BD Bioscience) prevented the engraftment and ectopic development of aberrant ICN1+ DP T cells in the BM, as compared with cells pretreated with an isotypic IgG1 control (Figure 6A). The participation of CD44 in BM engraftment was confirmed in a therapeutic assay wherein Rag2–/– × γc–/– mice transplanted with untreated ICN1-transduced ETPs were subjected to serial i.p. administration of the blocking anti-CD44 mAb HP2/9 (IgG1) (28, 29), starting at day 5 after transplant (Figure 6B), when ETPs have already seeded the host BM (Figure 2F and data not shown). We found a significant reduction (>100-fold) of ICN1+ T cell ectopic generation and expansion in anti-CD44–treated animals, compared with IgG1-treated controls (Figure 6B), indicating that NOTCH1 regulates the engraftment of aberrant preleukemic cells derived from human ETPs through its downstream target, CD44. Although CD44 overexpression was not sufficient by itself to promote cell engraftment in the absence of active NOTCH1 (Figure 6C), enforced CD44 expression improved and prolonged BM engraftment of ICN1+ cells (Figure 6D), suggesting a cooperative role of NOTCH1 and CD44. Based on these results, we hypothesized that NOTCH1-induced CD44 expression mediates the initial interaction of aberrant T cells with the BM microenvironment, further supporting the engraftment and expansion of preleukemic cells and likely the progression of the disease.
Figure 6. CD44 is essential for BM engraftment of ICN1-induced human preleukemic T cells and primary T-ALL cells.
(A) ICN1-transduced human ETPs were treated with either blocking anti-CD44 mAb (515; IgG1) or control IgG1 before transplantation into RAG-2–/– × γc–/– mice. Absolute ICN1+ cell numbers infiltrating the BM of 5 mice/group were analyzed at 3 weeks after transplant. (B) Schematic representation of experiment design (left): RAG-2–/– × γc–/– mice transplanted with ICN1-transduced ETPs received 3 weekly i.p. injections of either blocking anti-CD44 (HP2/9; IgG1) mAb or control IgG1, starting at day 5 after transplant. (Right) Absolute numbers of human ICN1+ cells infiltrating the BM of 4 mice/group analyzed at day 15 after transplant. (C) Human cell numbers infiltrating the BM of RAG-2–/– × γc–/– mice (5/group) after 3 weeks of transplant with ETPs transduced with either ICN1 or CD44. Data in A–C were normalized to 105 transduced input cells (n = 3). (D) Mean percentages ± SEM of human cells infiltrating the BM of RAG-2–/– × γc–/– mice (4/group) transplanted with ETPs cotransduced with ICN1 along with either a CD44-encoding or a control retrovirus (n = 4). (E) Cells obtained from the BM of a T-ALL patient (T-ALL1) were pretreated with either blocking anti-CD44 mAb 515 or control IgG1 and then transplanted into RAG-2–/– × γc–/– mice. Relative T-ALL1 cell numbers infiltrating the BM, PB, spleen, and thymus of 4 mice/group were analyzed at the indicated times after transplant (n = 3). *P < 0.05; **P < 0.01; ****P < 0.0001.
Besides CD44, other molecules, such as α4β1 integrin and the CXCR4 chemokine receptor, support the interaction of HSPCs with the BM niche (30–32) and could thus mediate the engraftment of aberrant DP T cells induced by active NOTCH1. However, we found no specific effect of ectopic ICN1 on CXCR4 or α4β1 expression levels on aberrant DP cells derived from ETPs or HSPCs (Supplemental Figure 3, A and B, and data not shown). In contrast, CXCR4 functional competence was consistently higher in ICN1+ DP cells than in DP controls, as revealed in an in vitro migration assay toward the CXCR4 ligand SDF-1 (CXCL12) (Supplemental Figure 3, C and D). Supporting the participation of CXCR4 in BM engraftment of human ICN1+ cells, treatment of transplanted RAG-2–/– × γc–/– mice with the CXCR4 antagonist AMD3100 (33) induced a robust mobilization of aberrant ICN1+ as well as control ICN1– cells from the BM to the circulation (Supplemental Figure 3, E and F). Therefore, NOTCH1 signaling may promote the interaction of human ICN1+ preleukemic cells with the supportive BM niche by inducing the expression of CD44 and also by regulating the activity of the CXCR4/SDF1 signaling axis.
It is currently believed that the microenvironmental context can enable and modify hematopoietic neoplasia (11). Therefore, we ought to investigate whether CD44 could also control the engraftment of T-ALL cells within the BM niche. To this end, primary leukemic cells that expressed active NOTCH1 and high CD44 levels (Supplemental Figure 4), obtained from a T-ALL patient (T-ALL1), were loaded with anti-CD44 mAb or control IgG1 prior to transplantation into Rag2–/– × γc–/– mice. Kinetic analyses showed that anti-CD44 pretreatment significantly impaired T-ALL engraftment and fully prevented leukemia burden and infiltration of thymus, spleen, and PB during a follow-up period of 11 weeks, as compared with IgG1-treated T-ALL cells (Figure 6E). These data indicate that CD44-mediated interactions of T-ALL cells with the BM microenvironment might control leukemic engraftment and further progression of the disease. Therefore, CD44 targeting may be a promising therapeutic strategy against T-ALL.
CD44 targeting prevents LIC function and progression in vivo of human T-ALL.
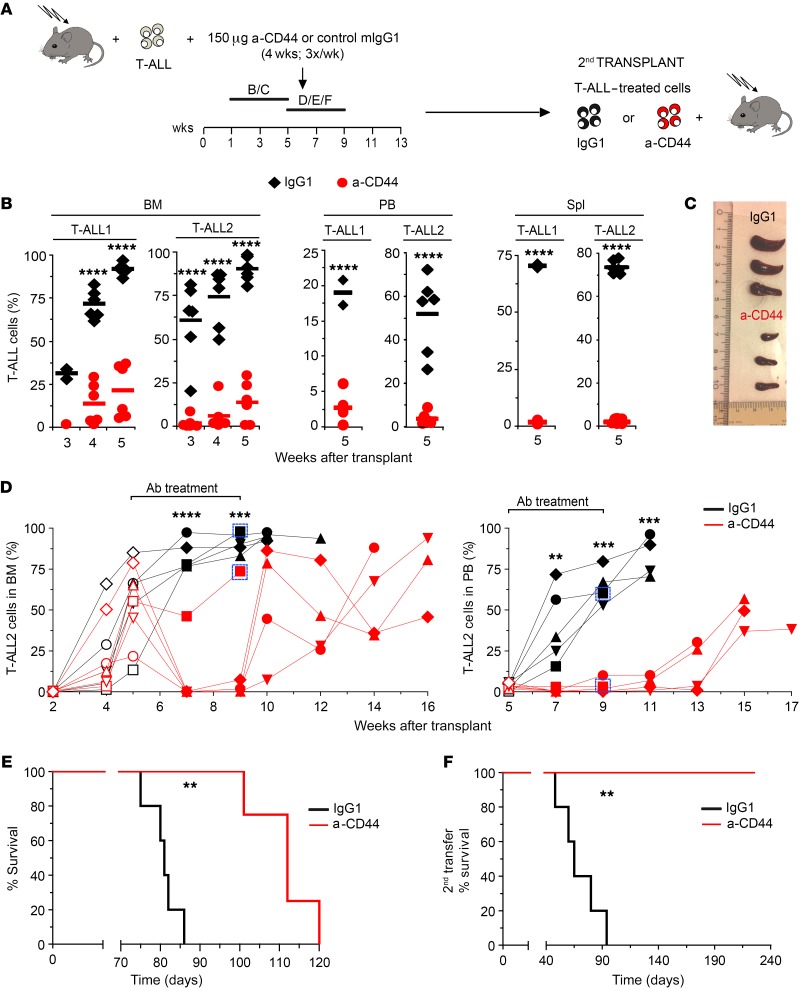
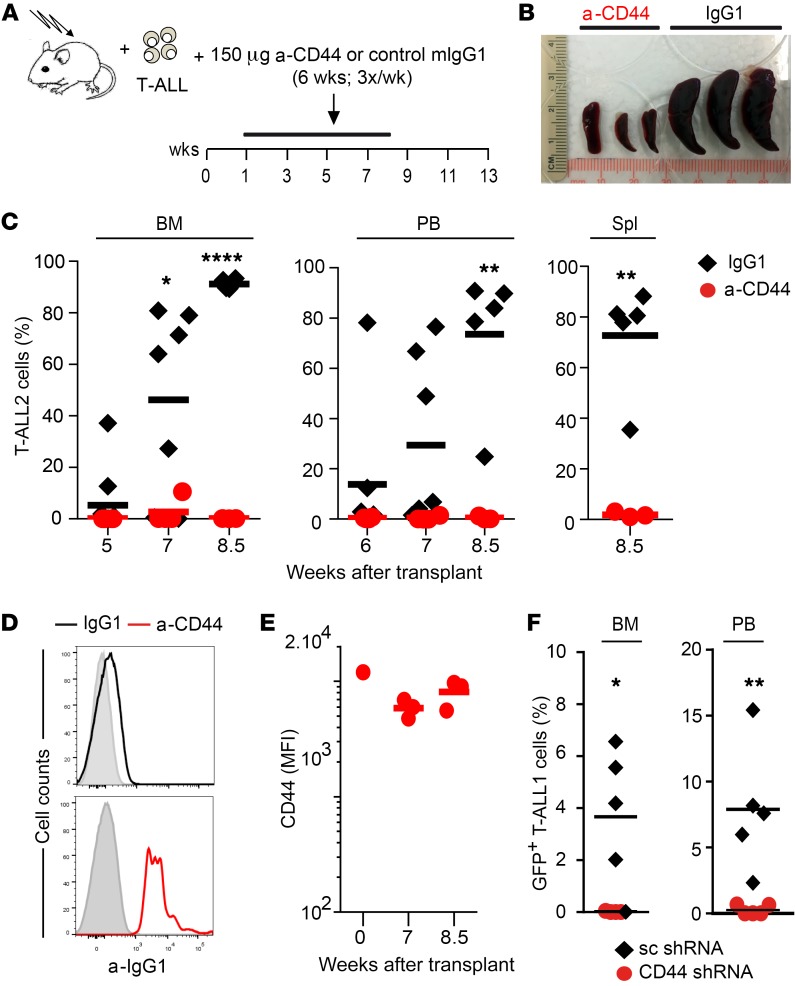
To directly explore the therapeutic potential of CD44 targeting, we next examined human T-ALL progression in mice subjected to anti-CD44 treatment once the disease was established. Two primary human T-ALLs with active NOTCH1 and distinct expression levels of CD44 (T-ALL1 or T-ALL2) (Supplemental Figure 4) were independently transplanted into Rag2–/– × γc–/– mice, which were then subjected to serial i.p. injections of either HP2/9 anti-CD44 mAb or control IgG1 during 4 weeks, starting at different times after transplant, when increasing proportions of T-ALL cells representative of disease severity had engrafted the BM (Figure 7A). In a first group of mice, treatment started at 1 week after transplant, when incipient BM engraftment of T-ALL cells was observed (3.8% ± 1.6%) and no PB infiltration was evident (not shown). Analysis of sequential BM aspirates of IgG1-treated mice revealed that T-ALL engraftment increased progressively to 95% or more at 5 weeks after transplant, when a significant leukemia burden was evident in PB and spleen (up to 60% and 75%, respectively) (Figure 7B). However, anti-CD44 administration from week 1 to week 5 after transplant significantly impaired T-ALL engraftment and resulted in a striking reduction of leukemic burden in PB and spleen, which correlated with a marked decrease of splenomegaly compared with that in controls (Figure 7, B and C), indicating that anti-CD44 treatment impairs human T-ALL progression.
Figure 7. CD44 targeting impairs LIC activity and progression in vivo of human T-ALL and prolongs survival.
(A) RAG-2–/– × γc–/– mice transplanted with primary human T-ALL1 or T-ALL2 cells (Supplemental Figure 4) received 3 weekly i.p. injections of either blocking anti-CD44 mAb (HP2/9) or control IgG1 during 4 weeks, starting at 1 week (B/C) or 5 weeks (D/E/F) after transplant. When indicated, T-ALL cells recovered from the BM of transplanted mice at the end of treatment were transplanted into secondary hosts. (B) Percentages of T-ALL1 and T-ALL2 cells infiltrating the BM, PB, and spleen of mice treated from weeks 1 to 5 after transplant as shown in A. Mean values from 3 independent experiments with a total of 13 to 18 mice for T-ALL and 3 to 8 mice for T-ALL2 are shown. (C) Image of representative spleens obtained at the end of treatment from mice shown in B. (D) Thorough analysis of anti-CD44 in vivo treatment showing percentages of human T-ALL2 cells infiltrating the BM (left) and PB (right) of 5 mice/group treated from weeks 5 to 9 after transplant as shown in A. Empty symbols represent cell percentages before the onset of Ab treatment. Boxes identify individual donors for secondary transplantations shown in F. (E) Kaplan-Meier survival curve of mice treated with anti-CD44 mAb (HP2/9) or control IgG1 in D. (F) Kaplan-Meier survival curve of secondary recipients transplanted with T-ALL2 cells (33 cells/mouse) obtained from the BM of individual donors represented as boxed in D. **P < 0.01; ***P < 0.001; ****P < 0.0001.
In a second group of mice, Ab treatment started when the disease was clearly established (at 5 weeks after transplant) and tumor burden reached up to 85% (55% ± 23%) of BM cells. In control IgG1-treated mice, T-ALL cells infiltrating the BM increased dramatically (up to 95%) during the next 2 weeks, and the incipient infiltration of PB observed at the onset of treatment resulted in a massive expansion (up to 70%). Despite the severity of the disease in those animals, anti-CD44 administration resulted in a significant drop of leukemia burden in the BM during the treatment period from weeks 5 to 9 after transplant, which paralleled an impaired T-ALL expansion in the PB, and this latter effect lasted for several weeks after the end of treatment (Figure 7D). As a consequence, the median survival of anti-CD44–treated mice was significantly prolonged (from 81 to 112 days) when compared with that of IgG1-treated controls (Figure 7E). Moreover, limiting dilution transplantation assays performed with T-ALL cells recovered from the BM of anti-CD44–treated or IgG1-treated mice provided formal proof that prolonged survival of the former correlated with a reduced frequency of leukemic cells with LIC activity. This correlation was observed even in poor responder mice, wherein anti-CD44 treatment was partially effective in terms of inhibition of leukemia progression. Indeed, LIC activity of CD44-targeted cells from a representative low responder (framed in Figure 7D) was 40-fold reduced compared with LIC frequency of T-ALL cells from an IgG1-treated control (from 1:569 to 1:14, respectively). Accordingly, survival of secondary recipients receiving CD44-targeted T-ALL cells was markedly prolonged (from 65 to more than 225 days) compared with that of recipients receiving T-ALL cells from IgG1-treated mice (Figure 7F). Therefore, in vivo administration of anti-CD44 impairs LIC function. Likewise, LIC function was diminished in T-ALL cells recovered from mice subjected to late anti-CD44 treatment, starting at 8 weeks after transplant, when a severe disease with massive BM engraftment (94% ± 4%) and evident PB infiltration (9% ± 2%) was developed. Accordingly, secondary hosts transplanted with such cells showed prolonged survival (from 61 to 100 days) compared with control mice (data not shown). Collectively, these data indicate that CD44 is essential to the LIC activity and progression in vivo of human T-ALL leukemic cells.
The clinical implications of these findings prompted us to investigate the potential mechanism underlying impaired T-ALL engraftment and progression following anti-CD44 treatment. We found that the HP2/9 anti-CD44 mAb mediated very poor complement-dependent cytotoxicity (CDC) against T-ALL cells in an in vitro assay (Supplemental Figure 5A), suggesting that CDC-induced killing of targeted T-ALL cells is not induced by HP2/9 in vivo. Accordingly, we found that few but detectable numbers of HP2/9 mAb–loaded T-ALL cells transplanted into Rag2–/– × γc–/– mice, identified by their CD7+CD5+ phenotype, were able to seed the mouse BM at very early times after transplant (5 hours, 24 hours, and 7 days) (Supplemental Figure 5B). More importantly, such T-ALL cells showed reactivity with a PE-coupled anti-mouse IgG1 Ab, indicating that the HP2/9 mAb persisted in vivo on the surface of essentially all transplanted cells (Supplemental Figure 5C), although HP2/9 binding seemed to decrease gradually over time (Supplemental Figure 5D). As a whole, these data suggest that anti-CD44 treatment impairs T-ALL engraftment in part by blocking the interaction of CD44 with its ligands expressed in the BM microenvironment, although a cytotoxic effect of the mAb in vivo cannot be formally excluded.
To confirm that in vivo treatment with the anti-CD44 HP2.9 mAb results in reduced T-ALL engraftment and progression by a CDC-independent mechanism, we extended our study to NSG mice, wherein complement function is impaired due to a 2 bp deletion in the coding region of the hemolytic complement gene (34). Notably, we found that HP2/9 mAb administration to NSG mice transplanted with T-ALL cells over 7 weeks, starting at 1 week after transplant (Figure 8A), reproduced the impaired progression of T-ALL observed in Rag2–/– × γc–/– mice and resulted in a striking reduction of BM engraftment and leukemia burden, with no signs of splenomegaly, as compared with that in IgG1-treated mice (Figure 8, B and C). These results confirm that anti-CD44 administration impairs leukemia progression in vivo by a CDC-independent mechanism. In addition, FACS analyses revealed that the very few T-ALL cells recovered from the BM of treated NSG mice displayed high reactivity with PE-coupled anti-IgG1, confirming the persistence of high levels of the HP2/9 mAb on the surface of T-ALL cells during the whole treatment period (Figure 8, D and E). We found no evidence of a cytotoxic effect of the HP2/9 mAb bound on the cell surface, as the HP2/9 mAb was unable to induce cell apoptosis or cell-cycle arrest of T-ALL cells, at least in vitro (Supplemental Figure 5, E and F). Therefore, we concluded that anti-CD44 mAb may impair T-ALL progression by blocking the ability of leukemic cells to interact with their supportive stromal niches. The interaction of CD44 with its major BM ligand hyaluronate (HA) (28) may provide the critical supportive signals that are impeded in the presence of anti-CD44; however, we found no evidence of a direct impact of the CD44-HA interaction on cell survival or proliferation of T-ALL cells, at least when they were cultured on HA-coated plates (Supplemental Figure 5G and data not shown). Despite the CD44 ligand involved, formal proof that CD44-mediated function is critical for human T-ALL propagation in vivo was directly provided in experiments showing that leukemia engraftment was significantly impaired in NSG mice transplanted with T-ALL cells in which CD44 expression was silenced by enforced expression of a CD44-specific shRNA, as compared with cells transduced with a control scramble shRNA (Figure 8F). Collectively, these results provide evidence that CD44 expression is crucial for human T-ALL maintenance and progression in vivo, suggesting that CD44 targeting is a promising approach for treating T-ALL relapse.
Figure 8. CD44-mediated interaction with the BM microenvironment is required for T-ALL cell leukemogenesis.
(A) NSG mice transplanted with primary T-ALL2 cells received 3 weekly i.p. injections of either blocking anti-CD44 mAb (HP2/9) or control IgG1 during 7 weeks, starting at 1 week after transplant. (B) Image of representative spleens obtained at the end of treatment from mice shown in A. (C) Percentages of T-ALL2 cells infiltrating the BM, PB, and spleen of 5 to 10 mice treated as shown in A. (D) Representative FACS analysis showing persistence of anti-CD44 HP2/9 mAb on the surface of T-ALL cells recovered from the BM of NSG mice represented in A at the end of treatment with either anti-CD44 mAb or IgG1. Bound HP2/9 mAb was detected by reactivity with PE-labeled anti-mouse IgG1 (n = 3). (E) FACS analysis showing levels of HP2/9 anti-CD44 mAb persisting on the surface of T-ALL2 cells that infiltrate the BM of anti-CD44–treated mice shown in A at the indicated weeks after transplant. Shown are MFI values from a total of 7 mice, determined as in D. (F) BM and PB infiltration of T-ALL1 cells transduced with a lentiviral vector encoding GFP and either CD44-specific shRNA (28% transduction efficiency) or a scramble control shRNA (36%). Data show percentages of GFP+ cells within infiltrating CD45+ T-ALL1 cells of 5 NSG mice/group at 11 weeks after transplant. *P < 0.05; **P < 0.01; ****P < 0.0001.
Discussion
The multistep oncogenic process that underlies human T-ALL pathogenesis is still poorly understood, mainly because retrospective studies are not feasible in patients and no models reproducing the earliest stages of the human disease are yet available. Here, we report an in vivo model of human T-ALL, which recapitulates the behavior of deregulated T cell progenitors in T-ALL patients and the generation de novo of a clonal human T-ALL. The model relies on oncogenic signaling provided by constitutive NOTCH1 activation and uncovers the earliest stages of the leukemogenic process that determines the engraftment of preleukemic T cells in the BM and the subsequent establishment and progression of the disease (1, 11). The clinical relevance of this model is strengthened by the demonstration that the CD44 adhesion molecule is a direct NOTCH1 transcriptional target that mediates crucial cell interactions with the BM microenvironment that result in preleukemic engraftment and further support T-ALL LIC activity and disease progression. Our study thus provides information that opens avenues for delineating specific targeting therapies for fighting T-ALL relapse.
Regardless of the particular oncogene driving T-ALL, NOTCH1 activation is known to be an early hallmark of T cell leukemogenesis and a key regulator of T-ALL LIC activity (13, 14). Accordingly, there is a high selective pressure for NOTCH1 activation in T-ALL (35). These findings concur with the fact that activating NOTCH1 mutations are present in more than 60% of human T-ALL cases (3, 7) and that many NOTCH1 targets are crucial regulators of T-ALL pathogenesis (19–22, 36, 37). Therefore, our model provides a valuable tool to decipher critical leukemogenic events that might be common to many subgroups of human T-ALL. Particularly, we show that CD44 upregulation may be one of the initial events that contribute to leukemogenesis in T-ALL patients. A link between Notch signaling and CD44 expression has been previously observed, though not formally established, in different cell types, including T cells (18, 38, 39). We now show that CD44 is a direct NOTCH1 target, whose transcription is regulated in T-ALL cells by MAML1/CSL-binding to its proximal promoter, but also through dynamic interactions with a superenhancer, which is common to other NOTCH1 targets involved in development and cancer, such as IL7R (23). Consequently, the important role that CD44 plays as a physiological mediator of HSPC anchoring within the BM niche during hematopoiesis can be extended to BM engraftment of aberrant T cell precursors (25–27). Considering that normal T cell progenitors downregulate CD44 as part of their maturation program (16), natural NOTCH1 mutations or improved NOTCH1 activation in patients may be a crucial event to induce CD44-mediated exposure of preleukemic cells to critical supportive signals within the BM niche, which will further promote their expansion and eventually their transformation. Supporting the contribution of long-lasting interactions between preleukemic cells and their surrounding BM niche to disease development, ectopic generation of aberrant nontumorigenic DP T cells showing upregulated levels of CD44 typically precedes malignant transformation induced by Notch1 mutations in mice (17, 18, 38–40). These results concur with our finding that NSG mice, which maintain long-term engraftment of human cells better than other mouse strains (24), supported the generation de novo of an aggressive human T-ALL with latency longer than 6 months, while Rag2–/– × γc–/– mice only supported short-term engraftment of aberrant ICN1+ DP T cells. Collectively, these data support our view that CD44 overexpression is an early hallmark of NOTCH1-associated T-ALL pathogenesis. However, the implication of CD44 in T-ALL pathogenesis has not been formally established. Using blocking anti-CD44 mAbs, we now provide evidence that CD44 critically controls the engraftment and expansion of human preleukemic T cells within the BM niche and the propagation of human T-ALL. The marked impact observed in vivo after treatment with the HP2/9 anti-CD44 mAb seems independent of cell loss due to CDC, as most mouse IgG1 mAbs are poor mediators of mouse complement activation (41) and CDC is not mediated in vitro by HP2/9. Moreover, HP2/9 administration also reduced leukemia burden in NSG mice, which lack CDC activity (34). We can also discard a direct cytotoxic effect of the HP2/9 mAb bound on the cell surface, at least in vitro. However, we cannot formally rule out the possibility that myeloid cells present in immunodeficient mice eliminate T-ALL cells through Ab-dependent cellular cytotoxicity or phagocytosis (42, 43), although it seems very unlikely considering that genetic silencing of CD44 was sufficient to impair T-ALL engraftment and progression in NSG mice. Therefore, our results suggest that blockage of CD44 by anti-CD44 mAbs may prevent T-ALL engraftment and progression by disrupting critical CD44–CD44 ligand–supportive interactions within the BM niche.
While the particular supportive signals provided in vivo by CD44 to T-ALL cells remain to be clarified, CD44 binding to its BM ligands may play a critical role. Particularly, HA influences most activities of CD44 (28, 44) and is required for normal BM hematopoiesis (45), seeming a good candidate to provide niche supportive signals to preleukemic and leukemic T-ALL cells. However, we found no evidence of a direct effect of HA on survival or proliferation of primary T-ALL cells, at least in vitro, suggesting that other CD44 ligands, such as collagen, laminin, fibronectin, or osteopontin, may fulfill cell-survival functions in vivo (44). Also, CD44 could function through functional regulation of additional molecules known to mediate HSPC interactions with the BM niche and cell survival, such as the CXCR4 chemokine receptor (30–32). Supporting this view, previous findings have shown that CXCR4 function can be regulated via CD44 binding to HA (46, 47), which concurs with our observation that ICN1+CD44hi DP cells have increased CXCR4-mediated function in vitro compared with normal DP thymocytes and can be mobilized in vivo in response to CXCR4 antagonists. Therefore, although NOTCH1 activation had no effect on surface expression levels of CXCR4 in our model, NOTCH1 signaling may contribute to T-ALL engraftment through the CXCR4/SDF1 signaling axis via CD44 induction. This possibility is further supported by the observation that CXCR4 is essential to the LIC activity of T-ALL cells, which were shown to be in direct contact with a vascular BM niche that expresses HA and produces SDF-1 (48, 49). Since HA is the major component of the BM extracellular matrix (28, 44), it is possible that the CD44-HA axis could ultimately control the highly dynamic interactions and promiscuous distribution of leukemia cells that migrate across the BM during T-ALL progression (50).
T-ALL cells with LIC activity are thought to be responsible for relapse following chemotherapy (51). Therefore, disruption of the specialized LIC niche is currently envisioned as a promising therapeutic strategy (11). We now propose that CD44 targeting may represent a valuable therapy against T-ALL relapse, as anti-CD44 administration markedly impairs LIC activity of an established T-ALL xenograft and results in prolonged survival. Given that the observed therapeutic effect correlated with persistence of the anti-CD44 mAb on the surface of T-ALL cells, we suggest that impaired T-ALL LIC activity results from a compromised interaction of CD44 with the LIC-supportive niche. However, CD44 expression levels do not directly correlate with LIC activity, as primary T-ALL1 and T-ALL2 cells with distinct CD44 expression displayed similar LIC frequencies (1:113 and 1:143, respectively), suggesting that, as previously reported, other factors, such as glycosylation, may regulate the binding activity of CD44 (reviewed in ref. 25).
Previous studies have highlighted the clinical significance of CD44 as a tumor-initiating marker in breast, colon, pancreatic, and liver cancers (25) and have demonstrated its contribution to the initiation of chronic and acute myeloid leukemias (52, 53). Our study now extends the pathological role of CD44 to initiation and progression of T-ALL. Accordingly, CD44 is among the top 5% of genes overexpressed in human T-ALL with active NOTCH1 (54), and its expression is a negative T-ALL prognostic factor associated with chemotherapy resistance (55). Therefore, the observed therapeutic benefit of anti-CD44 treatment, together with the proposal that novel therapeutic interventions aimed at avoiding chemo-resistance and disease relapse may not need to target specific BM stroma components, but rather the ability of T-ALL cells to interact stochastically with promiscuous BM microenvironment components (50), holds great promise for strategies that disrupt transient interactions of T-ALL cells with the supportive BM niche via CD44.
Methods
Human cell sample isolation and flow cytometry.
Thymus samples were removed during corrective cardiac surgery of patients aged 1 month to 4 years. ETPs were isolated as described (19) from thymocyte suspensions obtained by centrifugation on Ficoll-Hypaque (Lymphoprep, Axis-Shield PoC AS) using the Dynal CD34 Progenitor Cell Selection System (Life Technologies). HSPCs were obtained from Ficoll-Hypaque–purified CB samples by AutoMACS immunomagnetic sorting using the CD34 Progenitor Cell Isolation Kit (Miltenyi Biotec). Sorted populations were proved 99% or more CD34+ and negative for CD3, CD4, CD8, CD13, CD14, CD19, and CD56 lineage markers on reanalysis (Lin–). Primary T-ALL cells were Ficoll-Hypaque–purified from PB or BM patient samples obtained at the time of diagnosis. Flow cytometry mAbs included directly labeled mouse anti-human CD1a-PE (clone T6-RD1), CD4-PE-Cy5 (clone 13B8.2), CD13-PE-Cy5 (clone Immu.103.44), CD33-PE-Cy5 (clone D3HL60.251), CD34-PE (clone 581), CD34-PE-Cy5 (clone 581), CD34-APC (clone 581), and TCR-αβ–PE-Cy5 (clone IP26A) from Beckman Coulter; CD3-PE (clone UCHT1), CD3-APC (clone UCHT1), CD5-FITC (clone L17F12), CD7-FITC (clone 4H9), CD8-PE-Cy7 (clone RPA-T8), CD19-PE (clone HIB19), CD34-FITC (clone 581), CD44-FITC (clone G44-26), CD44-PE (clone G44-26), CD45-FITC (clone HI30), CD45-PE (clone HI30), CD45-APC (clone HI30), CD45-V450 (clone 2D1), and CD271-A647 (clone C40-1457) from BD Biosciences; CD8-PE (clone 3B5) and TCR-γδ-PE-Cy5 (clone 5A6.E9) from Invitrogen; CD133-PE (clone AC133/2) from Miltenyi Biotec and biotin-labeled anti-human TCR-αβ (clone IP26A) from eBiosciences. Goat anti-mouse IgG-APC (catalog 550826) was purchased from BD Biosciences; streptavidin-PE-Cy7 (catalog 25-4317-82) from eBiosciences; goat anti-mouse IgG1-A647 (catalog A-21240) from ThermoFisher; and streptavidin-PE (catalog 405203) and - APC (catalog 405207) from eBiosciences. Isotype-matched irrelevant Abs (BD Biosciences) were used to define background fluorescence. Flow cytometry was performed in a FACSCalibur or a FACSCanto II (BD Biosciences).
Retroviral and lentiviral constructs and cell transductions.
Retroviral bicistronic vectors included MigR1, encoding either ICN1 and GFP (MigR1-ICN1) or GFP alone (MigR1-GFP) or a dominant negative form of MAML1 fused to GFP (MigR1-dnMAML1) (9), and the pRV vector, encoding a truncated form of human NGFR (pRV-ΔNGFR) (Genetrix). cDNA encoding the full-length standard isoform of human CD44 (56) was subcloned into MigR1-GFP and pRV-ΔNGFR. Supernatant production and retroviral infections were performed as described (57). For CD44 gene silencing, CD44 shRNA (Mission TRCN000289233; Sigma-Aldrich) or scramble shRNA (Mission SHC002) was cloned into the pHRSIN-SFFVp-GFP lentiviral vector (58) and cotransfected into HEK-293 cells (ATCC CRL-1573) with the helper plasmids psPAX2 and pMD2.G (Addgene plasmids; catalog 12260 and 12259), provided by D. Trono. Supernatants collected after 48 hours of culture (58) were used for transduction of T-ALL cells that were precultured for 48 hours onto DLL4-expressing OP9 stromal cells (59) and then cultured for 24 hours onto RetroNectin-coated (Takara) plates (105 cells/ml) in the presence of rhIL7 (200 IU/ml; National Institute of Biological Standards and Controls [NIBSC]). Transduction efficiency was assessed by FACS analysis of GFP expression in 104 cells cultured for 3 days.
Cell cultures.
Mycoplasma-free SupT1, HPB-ALL, and CUTLL1 T-ALL cell lines (57, 60) were cultured in RPMI 1640 medium (Lonza) supplemented with 10% FCS. For NOTCH1-signaling inhibition, cells were cultured with 200 nM CompE (Enzo Biochem Inc.), and DMSO vehicle was used as control.
T-ALL generation, xenotransplantion, and LIC activity assays.
For human T-ALL generation, 3- to 5-day-old RAG-2–/– × γc–/– (61) or 6- to 10-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG; The Jackson Laboratory) mice were sublethally irradiated with 3.5 Gy or 1.5 Gy and subjected to intrahepatic (i.h.) or i.v. injection, respectively, with transduced human HSPCs or ETPs (1.0–1.5 × 106 cells/mice). LIC activity of human T-ALL cells arising de novo in the BM of NSG mice was assessed following their transplantation under limiting dilution conditions (from 102 to 106 cells/mouse) into secondary hosts. LIC frequency was calculated using ELDA software (http://bioinf.wehi.edu.au/software/elda/). For xenotransplantation assays, primary human T-ALL cells were transplanted by i.v. injection into 6- to 10-week-old RAG-2–/– × γc–/– or NSG mice. LIC activity of patient T-ALLs was calculated with the ELDA software upon limiting dilution transplantation (3 to 105 cells/mouse) into secondary hosts.
Anti-CD44 mAb pretreatment and in vivo administration.
For anti-CD44 pretreatment, ICN1-transduced ETPs or primary T-ALL cells were incubated at 4°C for 1 hour with 20 μg/ml of either blocking anti-CD44 mAb 515 (IgG1; BD Biosciences) or control mouse IgG1 prior to transplantation. When indicated, T-ALL cells were also pretreated with 100 μg/ml of anti-CD44 HP2/9 mAb (IgG1; 28, 29). For in vivo administration, xenotransplanted mice were serially i.p. injected (3 times/week) with 200 μl of PBS containing either blocking anti-CD44 mAb HP2/9 or IgG1 starting at different times after transplant. In vivo persistence of HP2/9 mAb on the surface of T-ALL cells was analyzed by FACS using a PE-labeled goat anti-mouse IgG1 Ab (Southern Biotech). Total BM cellularity was calculated considering that 2 femurs represented 10% of total BM.
TCR-Vβ repertoire analysis and real-time quantitative PCR.
Real-time quantitative PCR (qPCR) was performed as described (19) using TaqMan Gene Expression Assays (Applied Biosystems). To analyze T cell Vβ usage, cDNA from human DP cells was amplified by PCR using 22 different 5′ Vβ-specific primers and a common 3′ Cβ-specific primer, as previously described (62).
Luciferase reporter assays.
Human CD44 gene (ENSG00000026508) sequences of interest were amplified by conventional PCR and cloned into the pGL3-Promoter Luciferase Reporter Vector (Promega). Site-directed mutagenesis of potential CSL-binding sites (HTGGGAM to HGTACCM for sites 1, 2, 3, and 5; and TTCCCAC to TGTACCC for site 4; bold letters indicate mutations that were made) was performed using specific primers and PCR. Luciferase reporter assays were performed as described (19) using SupT1 cells cotransfected with the CD44 luciferase reporter vectors and either MigR1-GFP, MigR1-ICN1, or MigR1-ICN1 and MigR1-dnMAML1 plus the constitutively active Renilla reniformis luciferase-producing vector pRL-CMV (Promega).
ChIP assays.
To search for putative CSL/RBPJ-binding sites in human CD44, gene sequences (NCBI’s Gene database; ENSG00000026508) covering 5 kb upstream of the TSS were analyzed using MULAN software (http://mulan.dcode.org/). Sequences of 5 putative CSL/RBPJ-binding sites identified at positions –4.7, –4.4, –3.8, and –3.6 kb, and –3.5 kb (sites 1, 2, 3, 4, and 5, respectively) were as follows: site 1: CCTTGGGACT; site 2: TTCTGGGAAGC; site 3: TATGGGAAAG; site 4: GTTTCCCACAT; and site 5: GTTTGGGAAGG. ChIP assays were performed as described (19) using polyclonal Abs against either the C-terminal NOTCH1 domain (Santa Cruz Biotechnology Inc.) or cleaved NOTCH1 (Val1744) (Cell Signaling Technology). Goat or rabbit irrelevant Abs were used, respectively, as controls. Unbound chromatin (input) and immunoprecipitated DNA samples were analyzed by semiquantitative PCR with specific primers (Supplemental Table 1) amplifying the CSL-binding sites of CMYC or CD44 genes. Site-directed mutagenesis of potential CSL-binding sites (HTGGGAM to HGTACCM for sites 1, 2, 3, and 5; and TTCCCAC to TGTACCC for site 4) was performed using specific primers (Supplemental Table 2) and conventional PCR techniques.
Statistics.
Statistical significance was determined using 2-tailed Student’s t test. To account for multiple comparisons, data were corrected using the FDR method. Kaplan-Meier survival curves were compared using the log-rank test. In all cases, significance was defined as P < 0.05.
Study approval.
Experiments were conducted and human cell samples were obtained according to Declaration of Helsinki principles and to the procedures approved by the Spanish National Research Council Bioethics Committee. Written informed consent was received from participants before inclusion in the study. All animal studies were approved by the Dirección General de Consejería de Medio Ambiente, Administración Local y Ordenación del Territorio, Comunidad de Madrid, Madrid, Spain (Ref: PROEX 058/16).
Author contributions
MGP and MLT designed experiments and wrote the manuscript. MGP, PF, MM, CCM, JA, MJGL, AR, and CCM performed experiments. PGDM, CMC, and PM provided human samples and clinical experience. KW and HS provided support with the xenotransplantation model. CMC, DTS, and FSM provided critical reagents and helpful suggestions.
Supplementary Material
Acknowledgments
We thank J.C. Aster, C. Cabañas, A. Ferrando, M. Mellado, M.L. Gaspar, B. Andrés, A. Ursa and D.J.J. Waugh for providing useful reagents; M.A. Marcos for mouse transplantation advice; B. Alarcón, J.L. de la Pompa J. Fernández-Piqueras, S. Rodríguez de Córdoba, M. Ramírez and C. López-Larrea for helpful discussions and advice; Centro de Transfusión de la Comunidad de Madrid and the Pediatric Cardiosurgery Units from Ciudad Sanitaria La Paz (Madrid, Spain) for CB and thymus samples, respectively; and the Genomics and New Generation Sequencing Facility at the CBMSO for landscape chromatin analysis. This work was supported by funds from the Ministerio de Economía y Competitividad (MINECO) PLE-2009-0110, SAF2010-15106, SAF2013-44857-R, and SAF2016-75442-R (Agencia Estatal de Investigación/European Regional Development Fund, European Union), the Fundación Sandra Ibarra, the Fundación Asociación Española Contra el Cáncer (AECC CI13131229), the Instituto de Salud Carlos III (RTICC RD06/0014/1012), and the European Union Seventh Framework Programme (FP7/2007-20013) ThymiStem project (602587). Institutional grants from the Fundación Ramón Areces and Banco de Santander to the CBMSO are also acknowledged. MM and MJGL were supported by MINECO.
Version 1. 05/21/2018
Electronic publication
Version 2. 05/22/2018
Labels corrected in Figures 6, 7, and 8
Version 3. 07/02/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(7):2802–2818.https://doi.org/10.1172/JCI92981.
Contributor Information
Marina García-Peydró, Email: mgpeydro@cbm.csic.es.
Marta Mosquera, Email: mmosquera@cbm.csic.es.
María J. García-León, Email: mjgleon@cbm.csic.es.
Juan Alcain, Email: jalcain@cbm.csic.es.
Antonio Rodríguez, Email: a.rodriguez@uam.es.
Pablo Menéndez, Email: pmenendez@carrerasresearch.org.
Kees Weijer, Email: K.Weijer@amc.uva.nl.
Hergen Spits, Email: hergen.spits@amc.uva.nl.
Carlos Cuesta-Mateos, Email: carlos.cuesta@salud.madrid.org.
Cecilia Muñoz-Calleja, Email: cmunoz.hlpr@salud.madrid.org.
Francisco Sánchez-Madrid, Email: fsanchez.hlpr@salud.madrid.org.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 3.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223(2):262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/S1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 5.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/S1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 6.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 8.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20(20):7505–7515. doi: 10.1128/MCB.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scadden DT. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157(1):41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eguchi-Ishimae M, Eguchi M, Kempski H, Greaves M. NOTCH1 mutation can be an early, prenatal genetic event in T-ALL. Blood. 2008;111(1):376–378. doi: 10.1182/blood-2007-02-074690. [DOI] [PubMed] [Google Scholar]
- 13.Kindler T, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112(8):3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong F, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113(8):1730–1740. doi: 10.1182/blood-2008-02-138172. [DOI] [PubMed] [Google Scholar]
- 15.De Smedt M, et al. Active form of Notch imposes T cell fate in human progenitor cells. J Immunol. 2002;169(6):3021–3029. doi: 10.4049/jimmunol.169.6.3021. [DOI] [PubMed] [Google Scholar]
- 16.Trigueros C, Ramiro AR, Carrasco YR, de Yebenes VG, Albar JP, Toribio ML. Identification of a late stage of small noncycling pTalpha- pre-T cells as immediate precursors of T cell receptor alpha/beta+ thymocytes. J Exp Med. 1998;188(8):1401–1412. doi: 10.1084/jem.188.8.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allman D, et al. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med. 2001;194(1):99–106. doi: 10.1084/jem.194.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giambra V, et al. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-θ and reactive oxygen species. Nat Med. 2012;18(11):1693–1698. doi: 10.1038/nm.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-García S, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R{alpha} gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206(4):779–791. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26(21):8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci U S A. 2014;111(2):705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott SP, Eppert K, Lechman ER, Doedens M, Dick JE. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116(2):193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- 25.Ghaffari S, et al. CD44 isoforms in normal and leukemic hematopoiesis. Exp Hematol. 1999;27(6):978–993. doi: 10.1016/S0301-472X(99)00023-5. [DOI] [PubMed] [Google Scholar]
- 26.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106(6):1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 27.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 28.Liao HX, Lee DM, Levesque MC, Haynes BF. N-terminal and central regions of the human CD44 extracellular domain participate in cell surface hyaluronan binding. J Immunol. 1995;155(8):3938–3945. [PubMed] [Google Scholar]
- 29.de la Hera A, Acevedo A, Marston W, Sanchez-Madrid F. Function of CD44(Pgp-1) homing receptor in human T cell precursors. Int Immunol. 1989;1(6):598–604. doi: 10.1093/intimm/1.6.598. [DOI] [PubMed] [Google Scholar]
- 30.Méndez-Ferrer S, Frenette PS. Hematopoietic stem cell trafficking: regulated adhesion and attraction to bone marrow microenvironment. Ann N Y Acad Sci. 2007;1116:392–413. doi: 10.1196/annals.1402.086. [DOI] [PubMed] [Google Scholar]
- 31.Peled A, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 32.Lapidot T, Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16(10):1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- 33.Dar A, et al. Rapid mobilization of hematopoietic progenitors by AMD3100 and catecholamines is mediated by CXCR4-dependent SDF-1 release from bone marrow stromal cells. Leukemia. 2011;25(8):1286–1296. doi: 10.1038/leu.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma MK, et al. A novel hemolytic complement-sufficient NSG mouse model supports studies of complement-mediated antitumor activity in vivo. J Immunol Methods. 2017;446:47–53. doi: 10.1016/j.jim.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang MY, et al. High selective pressure for Notch1 mutations that induce Myc in T-cell acute lymphoblastic leukemia. Blood. 2016;128(18):2229–2240. doi: 10.1182/blood-2016-01-692855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi I, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113(8):1689–1698. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao L, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010;29(2):201–213. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13(1):73–84. doi: 10.1016/S1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205(12):2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuberger MS, Rajewsky K. Activation of mouse complement by monoclonal mouse antibodies. Eur J Immunol. 1981;11(12):1012–1016. doi: 10.1002/eji.1830111212. [DOI] [PubMed] [Google Scholar]
- 42.Koller CA, LoBuglio AF. Monocyte-mediated antibody-dependent cell-mediated cytotoxicity: the role of the metabolic burst. Blood. 1981;58(2):293–299. [PubMed] [Google Scholar]
- 43.Gül N, van Egmond M. Antibody-dependent phagocytosis of tumor cells by macrophages: a potent effector mechanism of monoclonal antibody therapy of cancer. Cancer Res. 2015;75(23):5008–5013. doi: 10.1158/0008-5472.CAN-15-1330. [DOI] [PubMed] [Google Scholar]
- 44.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4(7):528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 45.Goncharova V, et al. Hyaluronan expressed by the hematopoietic microenvironment is required for bone marrow hematopoiesis. J Biol Chem. 2012;287(30):25419–25433. doi: 10.1074/jbc.M112.376699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avigdor A, et al. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood. 2004;103(8):2981–2989. doi: 10.1182/blood-2003-10-3611. [DOI] [PubMed] [Google Scholar]
- 47.Fuchs K, Hippe A, Schmaus A, Homey B, Sleeman JP, Orian-Rousseau V. Opposing effects of high- and low-molecular weight hyaluronan on CXCL12-induced CXCR4 signaling depend on CD44. Cell Death Dis. 2013;4:e819. doi: 10.1038/cddis.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passaro D, et al. CXCR4 is required for leukemia-initiating cell activity in T cell acute lymphoblastic leukemia. Cancer Cell. 2015;27(6):769–779. doi: 10.1016/j.ccell.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Pitt LA, et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell. 2015;27(6):755–768. doi: 10.1016/j.ccell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawkins ED, et al. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature. 2016;538(7626):518–522. doi: 10.1038/nature19801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clappier E, et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med. 2011;208(4):653–661. doi: 10.1084/jem.20110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12(10):1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 53.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 54.Coustan-Smith E, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117(23):6267–6276. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoofd C, et al. CD44 promotes chemoresistance in T-ALL by increased drug efflux. Exp Hematol. 2016;44(3):166–71.e17. doi: 10.1016/j.exphem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Hill A, et al. Cortactin underpins CD44-promoted invasion and adhesion of breast cancer cells to bone marrow endothelial cells. Oncogene. 2006;25(45):6079–6091. doi: 10.1038/sj.onc.1209628. [DOI] [PubMed] [Google Scholar]
- 57.García-Peydró M, de Yébenes VG, Toribio ML. Sustained Notch1 signaling instructs the earliest human intrathymic precursors to adopt a gammadelta T-cell fate in fetal thymus organ culture. Blood. 2003;102(7):2444–2451. doi: 10.1182/blood-2002-10-3261. [DOI] [PubMed] [Google Scholar]
- 58.Garaulet G, et al. IL10 released by a new inflammation-regulated lentiviral system efficiently attenuates zymosan-induced arthritis. Mol Ther. 2013;21(1):119–130. doi: 10.1038/mt.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayllón V, et al. The Notch ligand DLL4 specifically marks human hematoendothelial progenitors and regulates their hematopoietic fate. Leukemia. 2015;29(8):1741–1753. doi: 10.1038/leu.2015.74. [DOI] [PubMed] [Google Scholar]
- 60.Palomero T, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 61.van Rijn RS, et al. A new xenograft model for graft-versus-host disease by intravenous transfer of human peripheral blood mononuclear cells in RAG2-/- gammac-/- double-mutant mice. Blood. 2003;102(7):2522–2531. doi: 10.1182/blood-2002-10-3241. [DOI] [PubMed] [Google Scholar]
- 62.Choi YW, Kotzin B, Herron L, Callahan J, Marrack P, Kappler J. Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc Natl Acad Sci U S A. 1989;86(22):8941–8945. doi: 10.1073/pnas.86.22.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.