Abstract
Spinal muscular atrophy (SMA), a degenerative motor neuron (MN) disease, caused by loss of functional survival of motor neuron (SMN) protein due to SMN1 gene mutations, is a leading cause of infant mortality. Increasing SMN levels ameliorates the disease phenotype and is unanimously accepted as a therapeutic approach for patients with SMA. The ubiquitin/proteasome system is known to regulate SMN protein levels; however, whether autophagy controls SMN levels remains poorly explored. Here, we show that SMN protein is degraded by autophagy. Pharmacological and genetic inhibition of autophagy increases SMN levels, while induction of autophagy decreases these levels. SMN degradation occurs via its interaction with the autophagy adapter p62 (also known as SQSTM1). We also show that SMA neurons display reduced autophagosome clearance, increased p62 and ubiquitinated proteins levels, and hyperactivated mTORC1 signaling. Importantly, reducing p62 levels markedly increases SMN and its binding partner gemin2, promotes MN survival, and extends lifespan in fly and mouse SMA models, revealing p62 as a potential new therapeutic target for the treatment of SMA.
Keywords: Neuroscience, Stem cells
Keywords: Autophagy, Molecular biology, Neurodegeneration
Introduction
Spinal muscular atrophy (SMA) is an early-onset, autosomal recessive neuromuscular disease caused by low levels of survival motor neuron (SMN) protein (1). SMA is primarily characterized by the degeneration of motor neurons (MNs) of the spinal cord and skeletal muscle atrophy. However, recent studies have revealed a wide range o f pathologies in multiple peripheral tissues in humans and mice (2). Because of a gene duplication event, humans harbor 0–6 copies of SMN2, a paralog to SMN1. A single nucleotide difference between SMN1 and SMN2 leads to alternative splicing of SMN2 (3, 4). While SMN1 primarily produces a full-length SMN (SMN-FL) transcript, approximately 90% of the SMN2 gene product lacks exon 7 (SMNΔ7), a highly unstable form of the protein (5, 6). SMN2 is a major SMA disease modifier, with higher SMN2 copy numbers correlating with delayed disease onset and better prognosis. SMN is ubiquitously expressed, localized in the cytoplasm and nuclear foci, termed gems, and plays an essential housekeeping role in the biogenesis of spliceosomal small nuclear ribonucleoprotein particles (snRNPs) (7) and axonal transport of mRNA (8). A large body of evidence suggests that increasing SMN levels repairs the disease phenotype. Although a number of compounds that increase SMN levels by stimulating its transcription or altering the SMN2 splicing pattern have been identified (9–14), SMN is also regulated at the level of protein degradation. We and others have shown that SMN protein is degraded by the ubiquitin-proteasome system (UPS) (15–20). However, the role of macroautophagy (hereafter referred to as autophagy) in regulating SMN protein levels has not been investigated. Autophagy is a highly conserved intracellular degradative pathway that eliminates cell components to maintain cellular homeostasis (21). While the proteasome degrades short-lived proteins, autophagy turns over damaged, redundant, and aggregated proteins as well as cellular organelles in bulk but also in a selective manner through a number of proteins, called autophagy receptors, that specifically recognize certain cargo and deliver it to autophagosomes (APs) for degradation (22, 23). The role of autophagy in neurodegenerative disorders has been extensively studied (24–28). However, its contribution to the pathophysiology of SMA remains unknown.
Here, we demonstrate that SMN protein levels are regulated by autophagy. We show that lysosomal inhibition increases the levels of total SMN protein, ubiquitinated SMN, SMN-binding partner gemin2, and the number of nuclear gems as well as levels of partially functional SMNΔ7. We also show that SMN degradation is mediated by its ubiquitination and interaction with the autophagy receptor p62 (also known as SQSTM1). Our study unravels a highly activated mTOR pathway in SMA-affected cells that contributes to the deficient AP clearance and subsequent accumulation of ubiquitinated proteins and p62. Importantly, depletion of p62 levels markedly rescues MN death in vitro and extends the lifespan of SMA fly and mouse models. Taken together, our results point to new therapeutic possibilities for the treatment of SMA by dampening autophagy-dependent SMN degradation.
Results
Autophagy regulates SMN protein levels.
Restoring SMN protein levels above a certain threshold is the most common approach to treating SMA. While SMN protein levels are controlled at transcriptional and posttranscriptional levels, in the past few years, therapeutic strategies have focused primarily on the transcriptional regulation of SMN. It is thought that SMN degradation occurs mainly via the UPS (15–20). However, since SMN is part of large multi-protein complexes including the SMN complex, formed by SMN and gemins (2–8) (key components of spliceosomes) (7), stress granules (29), and axonal granules involved in axonal transport (8), we wondered whether SMN degradation occurred in bulk as part of these large protein complexes via autophagy.
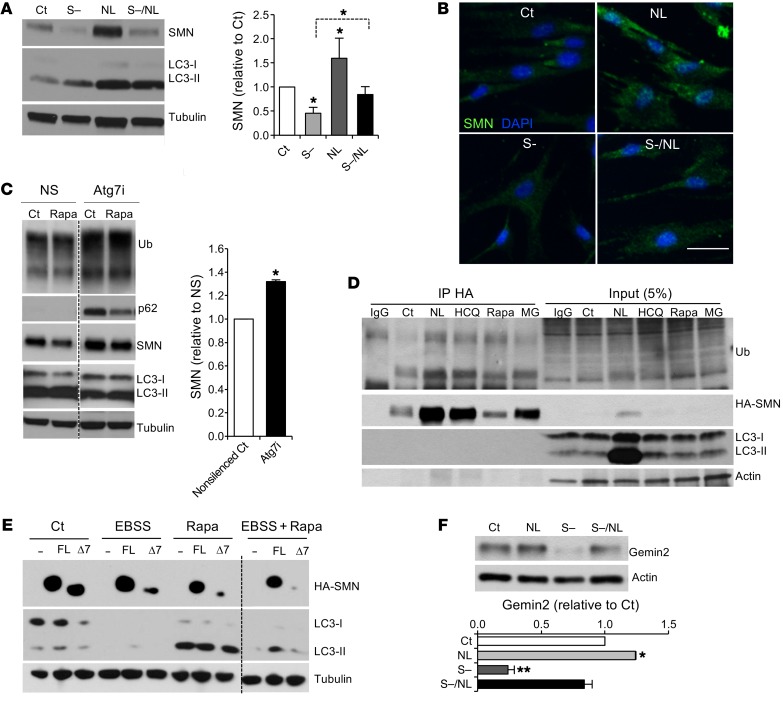
To determine whether autophagy regulates SMN degradation, we first exposed human fibroblasts to the best-known autophagy inducer, starvation, by depriving them of serum and blocked lysosomal degradation by exposing cells to the lysosomal inhibitors ammonium chloride and leupeptin (NL). We found that, not only did SMN levels decrease with serum starvation, but, importantly, SMN levels were elevated when autophagy activity was blocked (Figure 1, A and B), indicating that SMN is degraded via autophagy. We also sought to determine whether autophagy controls SMN protein levels in MNs, the most severely affected cell type in the disease. Given that autophagy is not efficiently triggered by starvation in neurons (21), we used shRNA delivery to reduce the expression of Atg7, a crucial gene required for autophagosome formation, in mouse embryonic stem cell–derived (ESC-derived) MNs. As expected, we found that MNs with reduced Atg7 showed increased levels of the autophagy substrate p62, accumulation of ubiquitinated proteins, as well as reduced levels of the AP marker LC3-II (Figure 1C). As anticipated, we also noted an increase in the levels of SMN when compared with levels in control cultures (Figure 1C), confirming that autophagy meditates SMN degradation. It is traditionally accepted that, upon ubiquitination, SMN degradation occurs solely via the UPS. Our results led us to explore whether ubiquitinated SMN is also degraded by autophagy. In fact, when HA-SMN-FL–expressing HEK293T cells were treated with modulators of autophagy (the inhibitors NL and weak base hydroxychloroquine [HCQ] and a well-known activator of autophagy, rapamycin) or the proteasome inhibitor MG132 and subjected to IP using an anti-HA antibody, we detected an accumulation of ubiquitinated SMN upon both proteasomal and lysosomal blockage (Figure 1D).
Figure 1. Autophagy regulates SMN protein levels.
(A) Representative immunoblot and quantification of protein lysates from human control fibroblasts after treatment with lysosomal inhibitors (NL), serum starvation (S–), or a combination of both (S–/NL) for 24 hours (n = 5 independent experiments). (B) Representative image of immunostaining against SMN (green) on human control fibroblasts after the indicated treatments (nuclei are labeled with DAPI, blue). Scale bar: 50 μm. (C) Representative immunoblot and quantification of protein lysates from WT mouse ESC–derived MNs infected with lentivirus carrying shRNA against Atg7 (Atg7i) or empty control (NS) for 7 days and treated for the last 24 hours of the culture with rapamycin or control media (results are expressed relative to NS control cells; n = 3 independent experiments). (D) Representative IP from HEK293T lysates transfected with HA-SMN plasmid and treated with lysosomal inhibitors (NL and HCQ), rapamycin, or the proteasome inhibitor MG132 (MG) for 24 hours and immunoblotted against ubiquitin, HA, LC3, and actin. (E) Representative immunoblot from HEK293T lysates transfected with empty vector (–) or the HA-tagged versions of SMN-FL or SMNΔ7 (Δ7) and cultured in control or amino acid–free media (EBSS), rapamycin, or both for 24 hours. (F) Representative immunoblot and quantification from human control fibroblasts after a 24-hour treatment with serum-free media, NL, or both, showing the levels of the SMN-binding partner gemin2 (member of the SMN complex) (n = 3 independent experiments). *P < 0.05 and **P < 0.01, by 2-tailed t test. All results are shown as the mean ± SEM. Ct, control; Rapa, rapamycin; Ub, ubiquitin.
Given these findings, we next determined whether the truncated form of SMN protein, SMNΔ7, is also an autophagy substrate. Cells from patients with SMA express higher levels of SMNΔ7 than SMN-FL. Although SMNΔ7 is highly unstable, it retains some function, at least in the presence of SMN-FL (5, 6, 30). While it has been reported that SMNΔ7 is quickly degraded by the proteasome (16), its oligomerization with SMN-FL increases its stability (30). Because SMNΔ7 binds to SMN-FL, we postulated that SMNΔ7 might also be degraded by autophagy. To answer this question, we transfected HEK293T cells with HA-tagged forms of both proteins and induced autophagy by subjecting the cells to amino acid starvation (EBSS media), rapamycin, or both. Indeed, induction of autophagy led to decreased levels of SMN-FL and SMNΔ7, which were more pronounced upon combining both autophagy-inducing stimuli (Figure 1E), demonstrating that SMNΔ7 protein levels are also under autophagy control. Interestingly, the levels of gemin2, an SMN-binding protein and essential spliceosome component, were also decreased upon autophagy induction (with serum starvation), and increased upon lysosomal inhibition (NL) (Figure 1F). Taken together, these data demonstrate that autophagy degrades ubiquitinated SMN and other components of the SMN complex, such as gemin2, which in turn suggests that the pool of SMN degraded by autophagy is the one associated with SMN complexes.
SMN degradation by autophagy is mediated by p62.
Autophagy is not only an in-bulk catabolic process, it also selectively targets specific substrates for degradation. Several autophagy cargo recognition proteins, or autophagy receptors, have been identified, including p62, NBR1, optineurin, NDP52, and NIX (22, 23). They share key domains that allow binding to autophagic cargo for their delivery for lysosomal degradation. Through their ubiquitin-binding domain (UBA motif), these receptors bind to polyubiquitinated cargo, and via their LC3-binding domain (LC3-interacting region [LIR] motif), they interact with LC3, which coats the inner membrane of autophagosomes (22, 31–33). These proteins are also autophagy substrates and, hence, serve as markers for autophagy flux.
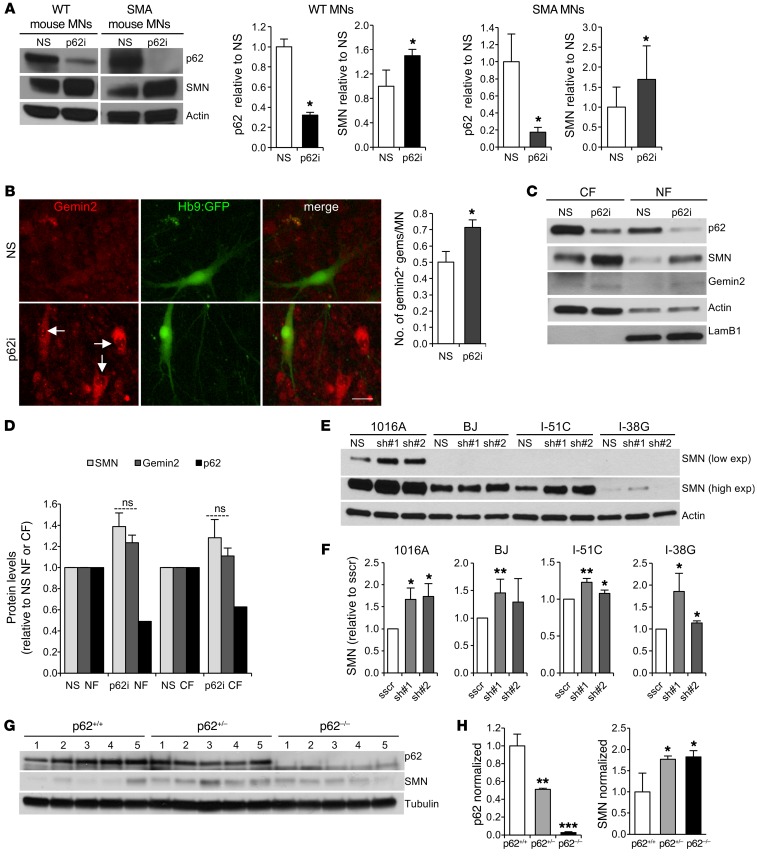
To investigate whether autophagy receptors mediate SMN degradation, we focused first on the best-characterized autophagy receptor, p62. Depletion of p62 levels with shRNAs markedly increased SMN protein levels in WT mouse MNs (Figure 2A), without increasing SMN mRNA expression levels (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI95231DS1). These results led us to explore whether the same was true in SMA MNs. We obtained ESCs from a severe SMA mouse model, SMNΔ7, in which Smn has been deleted, and 2 copies of the human SMN2 transgene and 2 copies of the human SMN2 transgene lacking exon 7 have been inserted into the genome (Smn–/– SMN2+/+ SMNΔ7+/+ Hb9::GFP) (30). SMN protein levels in both ESCs and MNs derived from these mice were reduced by approximately 70% when compared with levels in WT controls (Supplemental Figure 1, B and C). Knockdown of p62 in SMNΔ7 MNs led to increases in SMN protein levels similar to those detected in WT MNs (Figure 2A).
Figure 2. SMN degradation by autophagy is specifically mediated by p62.
(A) Representative immunoblot of protein lysates from WT and SMA mouse ESC–derived MNs 7 days after infection with the nonsilencing control (NS) or shRNA against p62 (p62i). Graph shows quantification of p62 and SMN protein levels (results are expressed relative to the nonsilenced control cells; n = 5 independent experiments). (B) Immunostaining against gemin2 and quantification of the number of gemin2+ nuclear gems in SMA mouse ESC–derived MNs. Scale bar: 25 μm (results are expressed relative to NS control cells (n = 3 independent experiments; 600 MNs per condition were counted). (C and D) Representative immunoblot and quantification of subcellular fractionation 3T3 protein lysates 6 days after infection with lentivirus carrying shRNA against p62 or NS control (ns, not significant). CF, cytoplasmic fraction; NF, nuclear fraction. (E) Representative immunoblot of human iPSC–derived MN lysates from healthy control cells (1016A and BJ) and SMA patients with different disease severities (type II, I-51C; type I, I-38G). MNs were infected with a lentivirus expressing NS shRNA control or p62 shRNA and lysed 7 days after infection. exp, expression. (F) Quantification of SMN and p62 protein levels. Results are expressed relative to scramble-silenced (sscr) cells (n = 7, 6, 4, and 3 independent experiments, from left to right). (G and H) Representative immunoblot and quantification of protein lysates obtained from p62–/–, heterozygotic (p62+/–), or WT (p62+/–) adult mouse brains (results are expressed relative to p62+/+ mice; n = 5 mice per group). *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed t test. All results are shown as the mean ± SEM.
We also studied the number of nuclear gems. Gems are multiprotein nuclear structures composed of SMN molecules, gemins2–7 and snRNPs involved in the transcription and processing of many types of nuclear RNAs (7). A reduction of their number per cell is an SMA hallmark. Interestingly, p62 downregulation led to an increase in the number of gemin2+ and SMN+ nuclear gems in SMA MNs and also in other neurons in the culture (non-MN population) (Figure 2B and Supplemental Figure 1D). This finding led us to explore whether the effect of p62 depletion on increases in SMN protein levels is more marked in the nucleus than in the cytoplasm. However, cellular fractionation assays of p62-knockdown NIH3T3 cells that also exhibited significantly higher SMN levels in total lysates when compared with the nonsilenced control (NS) (Supplemental Figure 1E) showed similar increases in SMN and gemin2 levels in both cellular fractions, which indicates that stabilization of these proteins occurs in both the nucleus and cytoplasm (Figure 2, C and D).
We extended these results by showing that p62 regulates SMN protein levels in human MNs. We derived MNs from induced pluripotent stem cells (iPSCs) from healthy control and SMA patients affected by different disease severities — a severe line (type II, I-51C) and a very severe line (type I, I-38G) — that we have previously described (34, 35). These iPSCs and the derived MNs express amounts of SMN that reflect their disease severities (35). Similar to what we observed in mouse MNs, depletion of p62 levels in human MNs also led to an increase in SMN protein levels (Figure 2, E and F, and Supplemental Figure 1F). To verify the importance of p62 as a mediator of SMN degradation, we studied SMN protein levels in brains from adult p62-null (p62–/–) mice. Confirming our results, brains from heterozygous and p62–/– mice showed markedly increased SMN protein levels when compared with levels in littermate controls (Figure 2, G and H). Together, these data indicate that p62 regulates SMN and gemin2 protein levels in all cell types studied, including human control and SMA iPSC–derived MNs, and in mice.
p62 interacts with SMN.
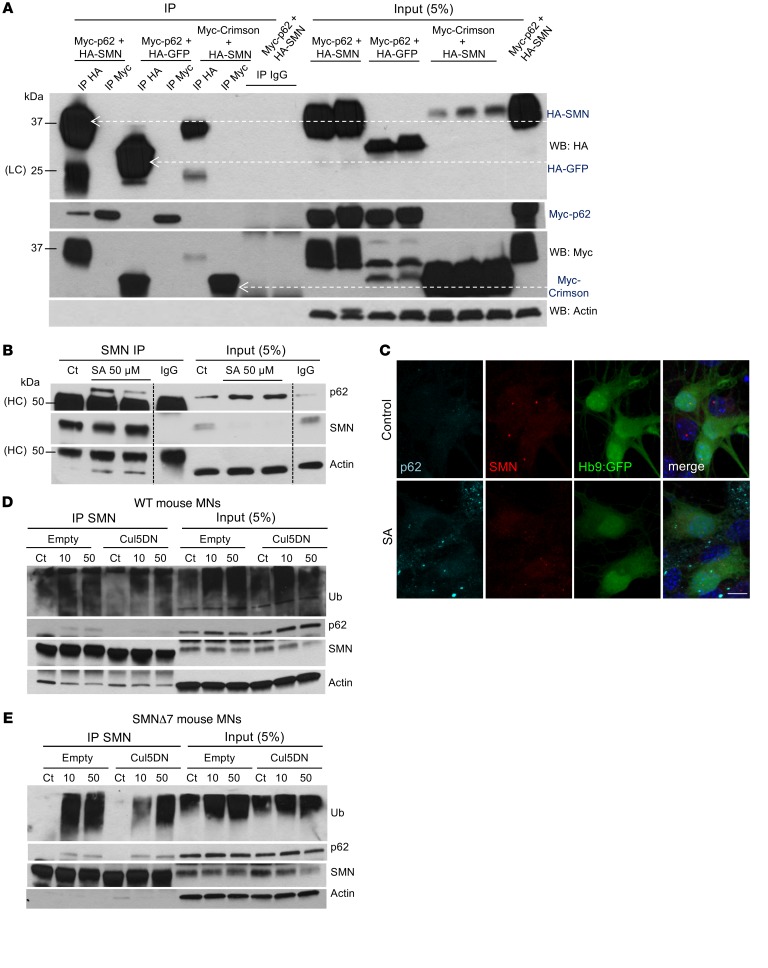
Given our findings, we next explored how p62 functions as an autophagy receptor to target SMN for degradation. We performed co-IP analysis after transfecting HEK293T cells with HA-SMN and Myc-p62 or various combinations of plasmids used as negative controls (HA-GFP plus Myc-p62 and HA-SMN plus Myc-E2-crimson, a far-red fluorescent protein). We found that, while Myc-p62 IP did not pull down HA-SMN, HA-SMN did coimmunoprecipitate with Myc-p62 (Figure 3A). Co-IPs of HA-GFP with Myc-p62 and of HA-SMN with Myc-E2-crimson were negative, as we expected (Figure 3A). These results indicate that SMN and p62 indeed interact with one another. To confirm these data, we performed IP of endogenous proteins in mouse MNs. Interestingly we did not detect interaction between SMN and p62 under basal conditions. However, binding of SMN to p62 was detected when we induced the incorporation of SMN into protein complexes. It has been reported that besides being distributed throughout the cytoplasm and localizing to the nucleus when assembled into gems, SMN can also cluster in cytoplasmic structures called stress granules (SGs) in response to oxidative stress (29). SGs are mainly composed of RNA-binding proteins such as Tia1, TIAR, FUS, and SMN, among others (29, 36), and mRNA, and their main function is to prevent the translation of mRNA during cellular stress (29). To test whether p62 interacts with SMN in SGs, we induced oxidative stress in HEK293T cells transfected with combinations of the constructs described above by exposing them to 10 and 50 μM sodium arsenite (NaAsO, referred to hereafter as SA) for 6 hours, a commonly used method to induce SG formation. We observed that HA-SMN strongly coimmunoprecipitated with Myc-p62 upon SG formation (Supplemental Figure 2A).
Figure 3. p62 interacts with SMN.
(A) HA and Myc representative IP from HEK293T lysates transfected with the indicated plasmids and immunoblotted against HA, Myc, and p62. WB, Western blot. (B) Representative IP of endogenous SMN protein from mouse SMA MN lysates treated with control or SA for 6 hours and immunoblotted against p62, SMN, LC3, and actin. (C) Representative image from mouse SMA ESC–derived MNs cultured in control conditions or with SA and immunostained against p62 (cyan) and SMN (red) (MNs express Hb9:GFP, and nuclei are stained with DAPI, blue). Scale bar: 10 μm. (D) Representative IP of endogenous SMN protein from WT and SMA mouse MNs. (E) MN lysates infected with lentivirus expressing the DN form of Cul5 for 5 days compared with empty vector–infected cells. MN cultures were treated with control media or SA (10 or 50 μM) for the last 6 hours of the culture. Membranes were immunoblotted against ubiquitin, p62, SMN, and actin. HC, heavy IgG chain; LC, light IgG chain.
To explore the reproducibility of these findings with endogenous proteins, we subjected mouse SMA MNs to similar analyses. Importantly, upon treatment with SA, endogenous SMN coimmunoprecipitated with p62 (Figure 3B). Interestingly, in WT MNs, exposure to SA resulted in a lower ratio of immunoprecipitated SMN to SMN in total lysates when compared with controls (Supplemental Figure 2B). By contrast, in SMA MNs, this difference was not as pronounced, indicating that upon SA-induced SG formation, the pool of SMN present in SGs (and therefore not available in protein lysates because of their incorporation into large aggregates and not extracted by a mild lysis detergent) was possibly larger in WT cells than in SMA MNs. These results agree with those of previous studies reporting that SMN-deficient cells have impaired SG formation, which increases their susceptibility to death (37). Second, we noted that co-IP of SMN with p62 was more efficient in SMA MNs (Figure 3B) than in WT MNs (Supplemental Figure 2B) and that localization of p62 in aggregates upon SA treatment was more evident in SMA MNs (Figure 3C and Supplemental Figure 2C). Therefore, the incorporation of SMN in SGs, or SG formation itself, might be impaired in SMA cells; however, the interaction between SMN and p62 is enhanced in the diseased cells.
Recently published data from our laboratory suggest that SMN is ubiquitinated by Cullin5-E3 ubiquitin ligase (Cul5), targeting it for degradation, and that overexpression of a dominant-negative (DN) form of Cul5 stabilizes SMN (35). We postulated that ubiquitinated SMN could at least be a fraction of the SMN protein that is recognized by p62 and degraded via autophagy. To test this possibility, we infected mouse SMA and WT ESC–derived MNs with lentiviruses carrying Cul5DN or a control empty vector, and we exposed them to SA to induce the localization of SMN in stress granules. We performed SMN IP and, as expected, we observed that for both types of MNs, there was a reduction in the amount of pulled-down, ubiquitinated SMN after Cul5DN expression (Figure 3, D and E). As a result, in WT MNs, the interaction of SMN with p62 was reduced in Cul5DN-expressing MNs compared with that seen in the empty vector–treated MNs (Figure 3D). Interestingly, and in agreement with our previous results, the binding between SMN and p62 was still maintained after Cul5DN overexpression in SMA MNs. This again suggests that in the diseased cells, the fraction of p62 that binds to SMN, at least under SG-formation conditions, is larger, or p62-SMN interaction stronger, than in WT cells (Figure 3E). Taken together, these data suggest that p62 interacts readily with the pool of SMN that is assembled in protein complexes. Furthermore, these data indicate that preventing SMN ubiquitination reduces its ability to bind p62.
SMN deficiency leads to autophagy failure and accumulation of p62/ubiquitinated proteins.
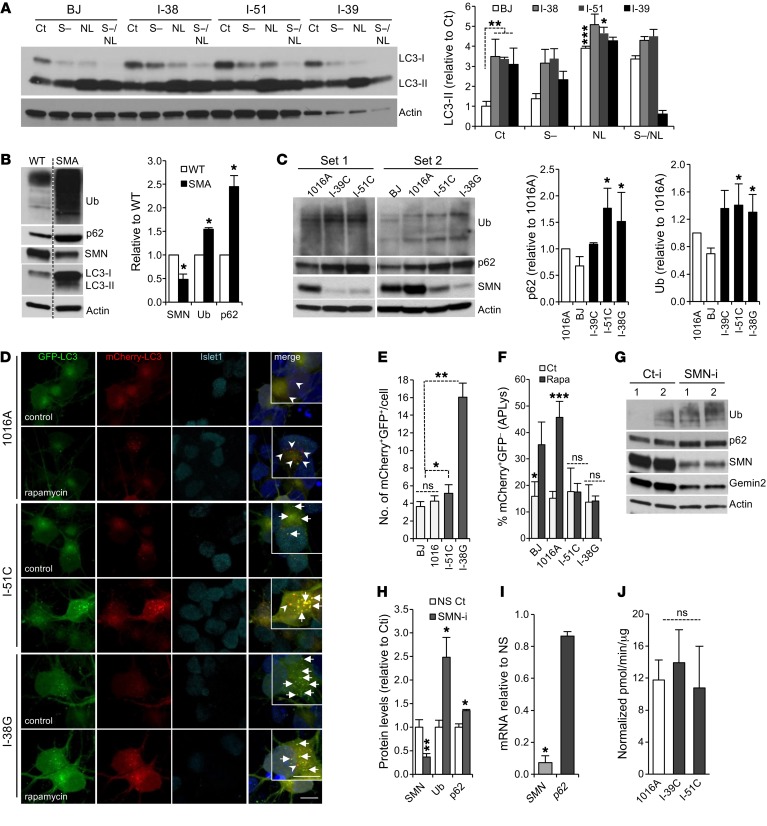
Autophagy dysfunction has been widely described in multiple neurodegenerative diseases; however, its participation in the pathophysiology of SMA remains largely unexplored. Thus, we decided to investigate autophagy function in SMA cells. First, fibroblasts from patients with different SMA severities (type I, I-38; type II, I-51; and type III, I-39) showed higher levels of LC3-I and LC3-II than did control fibroblasts (Figure 4A and Supplemental Figure 3A), suggesting that AP formation remains intact in SMA. To determine whether the increase in LC3-II represents an increase in AP formation and not a block in its fusion with lysosomes or its degradation, we studied autophagy flux by measuring LC3-II turnover in the presence and absence of lysosomal inhibitors. If autophagy flux occurs properly, the amount of LC3-II should be higher in the presence of lysosomal inhibitors (38). In SMA fibroblasts, we observed that the increase in LC3-II following lysosomal blockage under control and starvation conditions was reduced when compared with control fibroblasts (Figure 4A), indicating a defect in autophagy flux. This was accompanied by increased p62 protein levels in SMA fibroblasts — a hallmark of autophagy blockage (Supplemental Figure 3B). These data suggest that SMN deficiency impairs autophagy in human fibroblasts.
Figure 4. Autophagy activity is impaired upon SMN deficiency.
(A) Representative immunoblot to determine autophagy flux from human control and SMA fibroblast protein lysates. Cells were treated in the presence or absence of serum (S–) for 24 hours, with or without lysosomal inhibitors (NL) for the last 4 hours of the culture. Graph shows the quantification of LC3-II levels. (B) Representative immunoblot of protein lysates from WT and SMA mouse ESC–derived MNs. Graph shows quantification (results are expressed relative to WT MNs; n = 3 independent experiments). (C) Representative immunoblot from human iPSC–derived MN lysates from control cells (1016A and BJ) and SMA patients with different disease severities (type III, I-39C; type II, I-51C; type I, I-38G). Graphs show quantifications (results are expressed relative to 1016A control MNs; n = 5 independent experiments). (D) Representative confocal images of human control and SMA iPSC–derived MNs infected with mCherry-GFP-LC3 lentivirus. MNs were fixed 8 days after infection. Magnifications are shown in the insets. Scale bars: 10 μm. (E and F) Graphs show the quantification of APs (yellow, marked by arrows) and the percentage of APLys (red, marked by arrowheads). n = 146, 44, 114, and 86 transduced MNs for BJ, 1016A I-51C, and I-38G cells, respectively. (G) Representative immunoblot of HEK293T cells transfected with SMN RNAi (SMN-i) or control RNAi (Ct-i) for 3 days (duplicates are shown). (H) Quantification of protein levels (results are expressed relative to NS control cells; n = 3 independent experiments). (I) qRT-PCR showing p62 mRNA expression upon SMN RNAi–mediated knockdown in HEK293T cells. Gene expression is indicated as the fold change of 2-ΔΔCt with respect to actin, normalized to RNAi control cells (results are expressed relative to NS control cells; n = 2 independent experiments). (J) Quantification of the chymotrypsin-like activity in control versus SMA human MNs. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed t test. All results are shown as the mean ± SEM.
In order to validate these results, we explored whether a similar dysfunction in autophagy occurs in SMA MNs. Indeed, murine SMA MNs showed higher levels of p62 (while mRNA expression remained unchanged, Supplemental Figure 3C) and ubiquitinated proteins compared with WT MNs and higher levels of LC3-I and LC3-II (Figure 4B), which correlated with increased LC3 immunostaining in MNs in culture (Supplemental Figure 3D). We confirmed these results in human iPSC–derived MNs that showed higher levels of p62 and ubiquitinated proteins in SMA compared with healthy MNs (Figure 4C and Supplemental Figure 3E). To investigate how relevant these results from in vitro MNs are to the disease in vivo, we immunostained cervical spinal cord sections from SMNΔ7 mice and WT littermates for p62 and ubiquitin. In line with our in vitro results, we observed that Hb9:GFP+ ventral MNs from P10 SMNΔ7 mice showed increased p62 and ubiquitin staining compared with that seen in WT MNs (Supplemental Figure 3F). These results demonstrate that autophagy is impaired in both mouse and human MNs.
Autophagy can be dysregulated at several steps, and in the context of neurodegenerative diseases, a growing number of studies have reported that alterations at each of these steps can lead to autophagy failure (39). Our results indicate that AP clearance is partially impaired in SMA cells. To further explore this possibility, we studied autophagy flux by using an mCherry-GFP-LC3 construct. This allowed us to distinguish APs (both green and red giving rise to yellow puncta) from autophagolysosomes (APLys, red) since fusion with acidic lysosomes leads to loss of GFP fluorescence at low pH and unmasking of the mCherry fluorescence (40). Consequently, we infected human iPSC–derived MNs with lentiviruses expressing this construct and analyzed the presence of APs (yellow) and APLys (red) by confocal microscopy. SMA MN cultures had an increased number of APs under basal conditions (Figure 4, D and E) and a reduced increase in the percentage of APLys upon autophagy induction with rapamycin when compared with WT MN cultures (Figure 4, D and F), suggesting that the formation of APs is not impaired in SMA MNs, but rather their clearance.
We next questioned whether the defect in autophagy activity detected in SMA cells was an indirect consequence of long-term SMN deficiency or whether, by contrast, acute reduction in SMN levels per se could lead to autophagy impairment. To answer this question, and since MNs have poor transfection efficiency, we used HEK293T cells to silence SMN1 with siRNA and determined the levels of p62 and ubiquitination by Western blotting. We observed that both markers were increased after acute loss of SMN when compared with controls (Figure 4, G and H), while p62 mRNA expression levels remained unchanged (Figure 4I), indicating that p62 accumulation was nontranscriptional and occurred in essence from reduced turnover. Finally, to rule out the possibility that UPS activity dysfunction was responsible for the accumulation of p62 and ubiquitin in SMA MNs, we quantified chymotrypsin-like activity in protein lysates from control and SMA iPSC–derived MNs and observed that at least this activity associated with the proteasome complex was not reduced in diseased cells (Figure 4J). Together, these data indicate that the lack of SMN leads to an impairment of autophagy function that in turn results in accumulation of p62/ubiquitinated proteins in MNs and non-MNs. However, autophagy activity is not completely blocked, and we postulated that it can still sequester p62-SMN protein complexes for degradation.
SMN deficiency leads to mTORC1 activation.
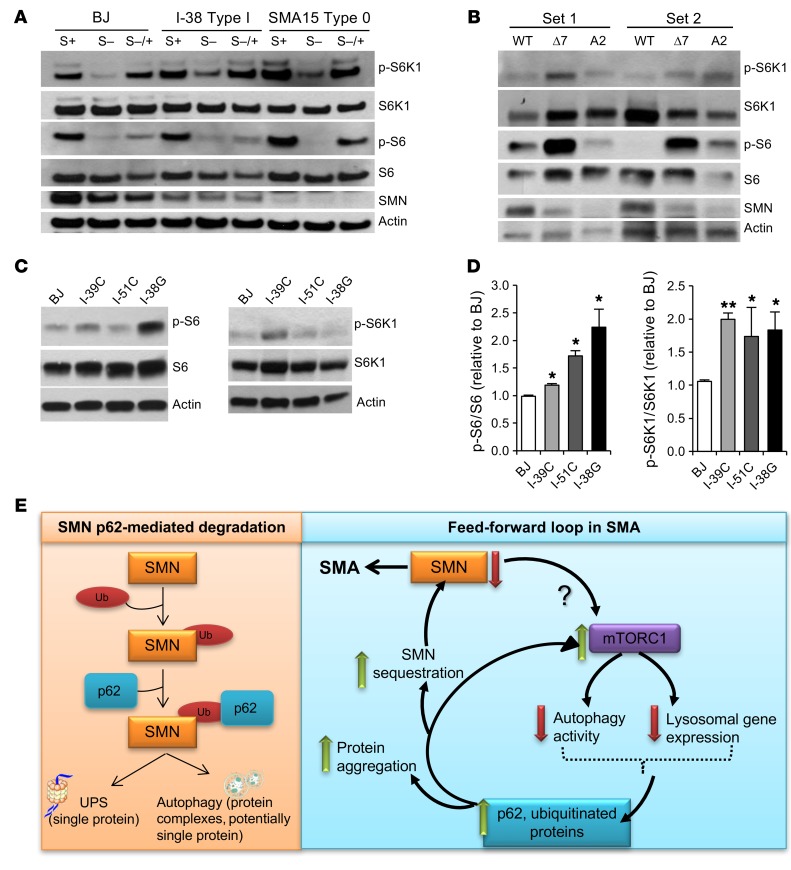
We next sought to explore how deficiency in SMN protein levels led to the autophagy malfunction that eventually resulted in the build-up of protein aggregates or toxic species (24, 41–43). It has been reported that p62, besides serving as an autophagy cargo adaptor, also mediates mTOR activation by promoting its localization to the lysosomal membrane (44, 45). The protein kinase mTOR is the master regulator of cellular metabolism and growth and is part of 2 protein complexes, mTORC1 and mTORC2 (46). When active, mTORC1 suppresses autophagy at different levels (47–49). Given the accumulation of p62 that we and others have observed in SMA cells (50, 51) and the associated autophagy activity dysfunction, we sought to determine whether decreased SMN levels affect mTOR activity. To address this question, we compared healthy control human fibroblasts with fibroblasts from patients affected by the most severe forms of the disease, SMA type I and type 0, with the latter type carrying only 1 SMN2 copy. We deprived the fibroblasts of serum overnight and then incubated them with full media for 30 minutes to measure mTOR activity. We observed that not only did the control-treated SMA fibroblasts show increased phosphorylated levels of the mTOR downstream targets S6K1 and S6 when compared with healthy fibroblasts (Figure 5A and Supplemental Figure 4A), but the decrease in their phosphorylation upon starvation was less pronounced than in control cells, suggesting a higher basal mTOR activity in the diseased fibroblasts. Next, we investigated whether mTOR activity was also enhanced in SMA mouse and human MNs of different severities and obtained similar results, i.e., higher levels of phosphorylated S6K1 (p-S6K1) and p-S6 compared with healthy controls (Figure 5B, Supplemental Figure 4B, and Figure 5, C and D, respectively). These data indicate that SMN deficiency results in mTOR pathway activation. It has been previously reported that p62 activates mTOR (44, 45). We postulate that these results, together with the elevated p62 levels observed in SMA-affected cells, suggest that the increase in mTORC1 activity in SMA leads to the accumulation of p62 protein, which in turn activates mTOR and results in a feed-forward cycle that results in further autophagy dysfunction and SMN sequestration (Figure 5E).
Figure 5. SMN deficiency results in mTOR activation, which could contribute to the autophagy impairment observed in SMA cells.
(A) Representative immunoblot from human healthy control and SMA types I and 0 fibroblast lysates. Cells were cultured in the presence of 10% serum (S+), deprived of serum (S–) for 16 hours, or deprived of serum and then incubated for an additional 30 minutes with serum (S–/+). Phosphorylated and total levels of the mTOR targets S6K1 and S6 were measured. Quantifications are shown in Supplemental Figure 4A. (B) Representative immunoblot of mouse WT and SMA (SMNΔ7 and A2) MN lysates to measure mTOR pathway activity. Quantifications are shown on Supplemental Figure 4B. (C) Representative immunoblot from human healthy control and SMA MN lysates to measure mTOR pathway activity. (D) Quantifications of p-S6/S6 and p-S6K1/S6K1 ratios from human healthy control and SMA MN lysates. *P < 0.05 and **P < 0.01, by 2-tailed t test (n = 6 independent experiments). Data indicate the mean ± SEM, expressed relative to BJ healthy control MNs. (E) Scheme of the interplay between low SMN-p62-mTOR–defective autophagy.
Reducing p62 levels promotes MN survival in vitro and increases the lifespan of fly and mouse SMA models.
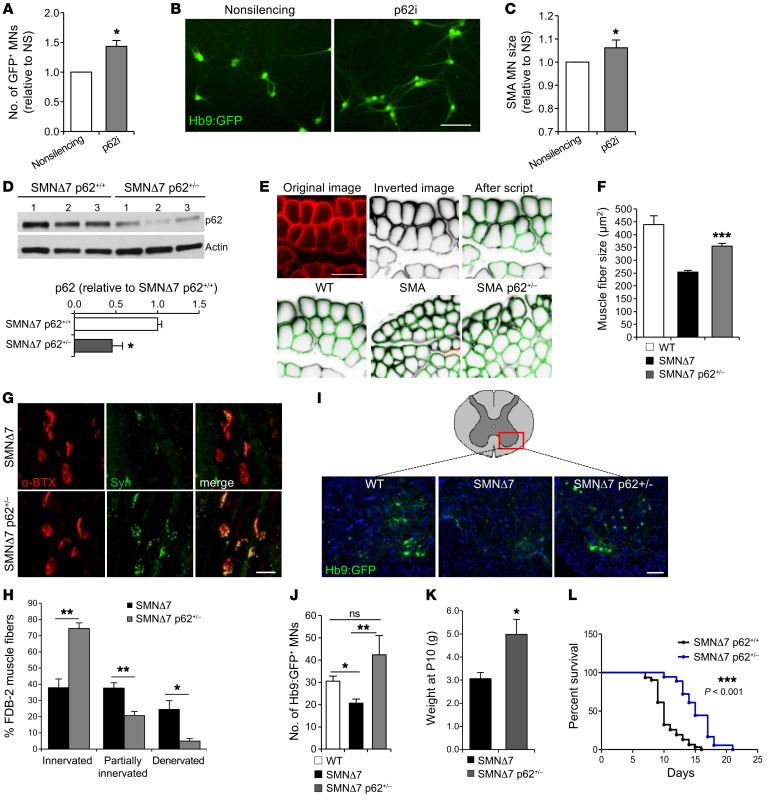
Our data show that a reduction of p62 protein levels leads to notable increases in SMN and other proteins of the SMN complex including gemin2. Consequently, we explored whether this stabilization of SMN and its binding partners is associated with cytoprotection of SMA cells. We have previously shown that higher SMN levels directly correlate with better survival of both WT and SMA MNs under any kind of stress (34, 35). Thus, we decided to investigate the effect on MN survival of a p62-mediated increase in SMN. We knocked down p62 by lentiviral infection in mouse SMA MNs and quantified the number of surviving Hb9:GFP+ MNs 10 days later. p62 knockdown resulted in a 50% increase in MN survival compared with cultures infected with a scramble control virus (NS) (Figure 6, A and B). In addition, p62-knockdown SMA MNs had a larger soma size, an indication of better overall neuronal health (Figure 6C). We also explored whether reducing p62 levels in WT MNs could reduce the basal rates of death due to culture-intrinsic stress. Indeed, we observed that WT MNs also had better survival and a larger soma size when p62 was knocked down compared with the NS-treated MNs (Supplemental Figure 5, A–C).
Figure 6. p62 knockdown promotes MN survival in vitro and ameliorates the disease phenotype of SMA models in vivo.
(A) Quantification of Hb9:GFP+ SMA mouse ESC–derived MNs 9 days after infection with p62 shRNA lentivirus compared with the NS-infected cells (results are expressed relative to NS-treated MNs; n = 6 independent experiments) and (B) representative image. Scale bar: 50 μm. (C) Quantification of SMA MN soma size after p62 knockdown (n = 4,100 NS; n = 5,500 p62i MNs measured). (D) Representative immunoblot and quantification of spinal cord protein lysates of SMNΔ7 mice and SMNΔ7 hemizygotes for p62 (n = 5 SMNΔ7 p62+/+ mice, n = 3 SMNΔ7 p62+/– mice). (E) Representative images of cryosectioned TA muscles. Top: Original image showing laminin immunostaining (red), signal inverted by Columbus script, and individual muscle fibers recognized by the script, highlighted in green. Bottom: TA sections from WT, SMNΔ7, and SMNΔ7 p62+/– P10 mice. Scale bar: 50 μm. (F) Quantification of TA fiber size (n = 5 WT mice, n = 4 SMNΔ7 mice, n = 4 SMNΔ7 p62+/– mice). (G) Immunofluorescence images of SMNΔ7 and SMNΔ7 p62+/– FDB-2 P10 muscles showing neuromuscular junctions immunostained for nerve terminals (anti-synaptophysin [Syn], green) and endplates (α-bungarotoxin [α-BTX], red). Scale bar: 20 μm. (H) Quantification of fully innervated, partially innervated, and fully denervated endplates (n = 4 SMNΔ7 mice, n = 4 SMNΔ7 p62+/– mice). (I) Representative images of cervical spinal cord cryosections from P10 WT, SMNΔ7, and SMNΔ7 p62+/– mice showing Hb9:GFP+ MNs (nuclei are stained with DAPI; central canals are outlined with dotted circles). Scale bar: 100 μm. (J) Average number of MNs per section (n = 10 WT mice, n = 9 SMNΔ7 mice, n = 4 SMNΔ7 p62+/– mice). (K) Weight measurements of P10 mice (n = 12 SMNΔ7 mice, n = 8 SMNΔ7 p62+/– mice). (L) Kaplan-Meier plot showing survival curves for SMNΔ7 p62+/+ mice (black line) and SMNΔ7 p62+/– (blue line) mice. n = 31 SMNΔ7 p62+/+ mice; n = 18 SMNΔ7 p62+/– mice. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed t test (A, C, D, F, H, J, and K) and Mantel-Cox and Gehan-Breslow-Wilcoxon tests (L). All results are shown as the mean ± SEM.
These results prompted us to investigate whether reducing p62 levels caused an amelioration of the SMA disease phenotype in vivo. We first studied a fly model of SMA, in which the expression of an RNAi targeting Smn is activated only in adult flies using a ubiquitous tubulin-Gal4 driver, which is under the control of the temperature-sensitive GAL80 protein (Gal80ts). Larvae were raised at 18°C, when tubulin-Gal4 is inactive, to prevent SMN reduction from having developmental effects, and adult flies were then switched to 29.5°C to drive expression of the RNAi. We used the actin gene-switch–inducible (actin-GS–inducible) Gal4-UAS expression system (52, 53), in which UAS RNAi expression is driven by Gal4 when flies are fed mifepristone (RU486). As expected, RNAi against a control gene (Contr-i) did not affect lifespan (Supplemental Figure 6, A and B), and Smn RNAi (Smn-i) decreased Smn mRNA levels by 70% (Supplemental Figure 6A). We did not expect that Smn RNAi would produce a strong decrease in lifespan, given that its expression is only activated during adult life. Indeed, we observed a small, although significant, reduction of the mean lifespan when it was coexpressed with the control RNAi (Supplemental Figure 6, A and C). Next, we analyzed the effects of reducing p62 expression on SMN protein levels and found that it resulted in enhanced SMN protein expression (Supplemental Figure 6, D and E), without affecting Smn mRNA expression (Supplemental Figure 6F). In addition, we tested whether p62 downregulation affected the lifespan of Smn-deficient flies by coexpressing Smn-i with p62-i. Importantly, while reducing p62 levels alone did not affect lifespan (Supplemental Figure 6, G and H), the flies expressing Smn-i and p62-i showed a complete rescue of the Smn RNAi–induced shortened lifespan (Supplemental Figure 6, I and J). These results indicate that decreasing p62 levels increases SMN protein in vivo and rescues the shortened lifespan of SMN-deficient flies.
We next sought to explore whether these observations could be reproduced in the SMA SMNΔ7 mouse, which in our colony survived an average of 10 days. Given than p62-KO mice are fertile and relatively healthy until they reach a mature age, when they develop obesity and bone and metabolic problems (54), we aimed to measure the disease manifestations in p62-KO SMNΔ7 mice. However, breeding SMNΔ7 p62-KO mice resulted in embryonic lethality of SMNΔ7 pups (data not shown), presumably because p62 is a scaffold protein with multiple modules that interact with other key signaling proteins (55). Instead, we crossed SMNΔ7 mice with p62-KO mice in order to obtain SMNΔ7 p62 heterozygotic mice (smn–/– hSMN2+/+ hSMNΔ7+/+ Hb9:GFP+ p62+/– or just SMNΔ7 p62+/– for simplicity). We confirmed that p62 protein levels in spinal cords from SMNΔ7 p62+/– mice were indeed reduced by half (Figure 6D) and then studied several aspects of the mouse SMA disease phenotype. We first used an unbiased, automated method to measure muscle fiber size of tibialis anterior (TA) muscle cryosections at the disease end stage (P10). We found that SMNΔ7 p62+/– TA muscle fibers had a significant increase in size compared with TA muscle fibers from SMNΔ7 mice (homozygous for p62) (Figure 6, E and F). Next, we evaluated the innervation status of the flexor digitorum brevis 2 (FDB-2) appendicular muscle, which is reported to show strong denervation in SMNΔ7 mice (56). Fully innervated endplates were defined by the complete overlap of presynaptic nerve terminals and immunostained with synaptophysin, and postsynaptic motor endplate acetylcholine receptors were labeled with α-bungarotoxin. Partially denervated endplates were identified by the partial overlap of pre- and postsynaptic labeling, and fully denervated endplates were devoid of any presynaptic labeling (Supplemental Figure 5D). Quantification of the percentage of denervated endplates revealed a robust amelioration of this pathologic phenotype in SMNΔ7 p62 mice. While in P10 SMNΔ7 muscles, only 40% of the endplates were fully innervated (Figure 6, G and H), nearly 80% of the endplates in SMNΔ7 p62 muscles were fully innervated and only approximately 5% were denervated (Figure 6, G and H). Additionally, we analyzed the spinal MN loss characteristically seen in this SMA mouse model (30) and observed that, while SMNΔ7 mice at P10 displayed a 50% reduction in the number of MNs, SMNΔ7 p62+/– P10 mice had similar numbers of ventral MNs compared with WT mice (Figure 6, I and J). This notable improvement in muscle, MNs numbers, and muscle innervation correlated with a significant increase in the body weight of SMNΔ7 p62+/– mice compared with that of SMNΔ7 mice (Figure 6K). Importantly, the rescue of multiple SMA-associated phenotypes translated to a 50% increase in the mean lifespan (5 more days) (Figure 6L). These data validate our in vitro results and confirm that p62 plays an important role in SMA pathology. Additionally, this work shows that reducing p62 levels, which are aberrantly increased in the disease, could represent a new strategy for developing SMA therapeutics.
Discussion
In this study, we have identified the autophagy receptor p62 as a regulator of SMN protein levels and discovered a molecular mechanism that explains the autophagy dysfunction affecting SMA cells. Our data reveal that mTOR overactivation causes such a dysfunction in SMA MNs, resulting in the accumulation of ubiquitinated proteins and p62. We suggest that abnormally high p62 levels aggravate the autophagy impairment and constitute the basis for the buildup of toxic species that increase susceptibility to cell death.
SMA is an early-onset neuromuscular disease associated with loss-of-function mutations or complete loss of the SMN1 gene, resulting in very low SMN protein levels. Therefore, it is commonly accepted that approaches aimed at increasing SMN levels will have therapeutic value in treating the disease. Several of these approaches have recently entered clinical trials after showing promising results in in vitro and in vivo models (57), with one, Spinraza (nusinersen), having been recently approved. However, most of these approaches focus on modifying the expression or splicing of the SMN2 gene or on directly replacing the SMN1 gene via viral delivery. SMN is also controlled at the posttranscriptional level, but to date the approaches targeting this aspect of SMN biology have not achieved therapeutic success. Therefore, one main focus of our work has been to identify new pathways that mediate SMN protein degradation, with a view toward stabilizing the protein and therefore increasing its levels. It is known that SMN forms part of protein complexes (gems, SGs, complexes along axons) whose disassembly and degradation mechanisms are unclear. Since the UPS, previously shown to regulate SMN levels, only targets single proteins, we focused on the other major cellular catabolic pathway, the autophagy-lysosomal system, which is in charge of degrading protein complexes, protein aggregates, and organelles. We postulated that autophagy could also regulate SMN protein levels by at least targeting the fraction that is localized to protein complexes. With this study, we found that, indeed, autophagy degrades SMN — not only SMN-FL, but also the most abundantly transcribed form of SMN in SMA patients, SMNΔ7, which is partially functional, and therefore blocking its degradation may be quite relevant for slowing disease progression. Our data also show that the autophagy receptor p62 is a mediator of selective SMN degradation. SMN and p62 interact, and this interaction is partially blocked when SMN ubiquitination is inhibited. We also demonstrate that reducing p62 levels leads to a significant enhancement not only of SMN, but also of some of its main binding partners, such as gemin2 and the number of SMN+, gemin2+ nuclear gems. Importantly, such increases had functional consequences, since they improved the survival of SMA MNs in vitro, abolished the shortening effect on fly lifespan induced by Smn-i, and notably improved multiple aspects of the disease pathology of SMNΔ7 mice, including increasing their lifespan by 50%.
One recently published study also implicated autophagy in the control of SMN protein levels (51). This work, however, showed that inducing autophagy increases SMN levels, while at the same time, it claimed that SMA cells exhibit increased autophagy. Although, there are differences between our studies, both of them demonstrated the critical role of autophagy in SMA pathogenesis, and our study, using multiple cell types and models, further uncovers the mechanistic control of SMN degradation by selective autophagy via the molecular adaptor p62. Additional studies will be required to understand the discrepancies between our findings and the previously published results.
For many years, autophagy was considered a bulk degradation process induced by nutrient deprivation and lacking specificity. However, almost 20 years ago, selective autophagy was reported in yeast (58), and since then, multiple studies have confirmed that autophagy is a more selective process than originally thought (59). Indeed, p62 and other proteins with similar amino acid domains, such as NBR1, optineurin, NDP52, and others, have been shown to function as autophagy cargo recognition proteins (31, 32). A hallmark of most neurodegenerative diseases is the accumulation of misfolded and aggregation-prone proteins and damaged organelles (39, 60). Numerous studies have reported the importance of autophagy receptors in delivering protein aggregates and organelles to APs for degradation (22). To date, the presence of these toxic species in SMA had not been reported. However, recent studies have identified intracellular trafficking problems that affect mitochondria (61), the endosomal-lysosomal system (62, 63), and axonal transport, growth, or morphology (26, 64–66). An impairment of the last steps of the autophagic process could well be another consequence of SMN deficiency, but such an autophagy dysfunction could also be the source of those cellular defects, similar to what has been reported for other neurological disorders. These studies, together with the data presented in this work, suggest that SMA shares more neurodegenerative disease hallmarks than previously recognized.
Since autophagy, through p62, plays a role in mediating SMN degradation, and since most neurodegenerative disorders involve defects in autophagy activity, we investigated whether this pathway was functioning properly in SMA. Indeed, we observed that all SMA cells analyzed — human fibroblasts as well as mouse and human iPSC–derived MNs — had higher amounts of LC3-I and LC3-II, which correlates with increased number of APs and impaired AP turnover (Figure 4). We observed that this defect was accompanied by increased levels of p62 and ubiquitinated proteins and that an acute decrease in SMN protein levels produced the same results. Our findings agree with those of recent studies that reported autophagy alterations upon SMN downregulation in cell lines (50, 67, 68) and answer the previously unresolved question of why autophagy is dysfunctional in SMA and explain the nature of the interplay between SMN and autophagy activity. We postulate that, as a consequence of SMN deficiency–induced autophagy impairment, ubiquitinated proteins and p62 accumulate. This is likely to increase the appearance of misfolded and aggregated proteins, leading to additional problems in cellular trafficking and AP clearance. This hypothesis is in line with a previous study from Komatsu and colleagues demonstrating that p62 plays an important role in inclusion body formation when autophagy is deficient (69). Another possible consequence of enhanced p62 protein levels is overactivation of mTORC1, which would in turn further interfere with autophagy activity (70, 71). There is also some disagreement as to the role that mTOR plays in SMA. On the one hand, several studies in SMA mice suggest that mTOR is downregulated and that increasing its activity can promote MN survival and enhance lifespan (72–74). On the other hand, another study showed that SMA type III muscle shows overexpression of mTOR (75). It is therefore possible that the activity of mTOR in SMA differs between different tissues or even cell populations. The increased mTOR activity that we detected in SMA cells could be a cellular response to limit further accumulation of APs and aggregation-prone material, but at the same time, it could be blocking autophagy reactivation via its inhibitory effect on TFEB, the master transcriptional regulator of lysosomal and autophagy genes (76, 77). Further studies are needed to understand whether the observed alterations of autophagy/lysosomal function and the mTORC1 signaling pathway in SMA MNs are direct, and maybe independent, consequences of SMN loss of function, or the indirect result of accumulative events triggered by the deficiency of the protein.
In summary, in this study, we have identified a pathway that controls SMN levels in cells including MNs, a finding that may uncover new avenues for identifying potential targets for therapeutic intervention. Furthermore, we have revealed an autophagy dysfunction in SMA MNs that is possibly caused by mTOR overactivation, resulting in the accumulation of ubiquitinated proteins and p62. p62 is a key protein that links autophagy, mTORC1, ubiquitination, and, now, SMN protein levels. We postulate that abnormally high p62 levels may aggravate autophagy impairment and constitute the basis for the buildup of toxic species that enhance susceptibility to cell death. Understanding the molecular mechanisms that control this loop, as well as whether compromised autophagy contributes to SMA progression, will be essential for the future development of therapeutics that act on this pathway to prevent neuronal degeneration.
Methods
Mouse colonies.
FVB mice were obtained from Charles River Laboratories. The original breeding pair of heterozygous SMNΔ7 mice (Smn+/– hSMN2+/+ hSMNΔ7+/+) on a FVB background and the Hb9:GFP mice on a C57Bl/6 background were provided by The Jackson Laboratory and backcrossed with mice on a FVB background for more than 10 generations. p62 SQSTM1–/– mice on a C57Bl/6 background were provided by Toru Yanagawa (University of Tsukuba, Tsukuba, Japan) and backcrossed with mice a FVB background for more than 7 generations.
Spinal MN and muscle histology.
The spinal cord and TA muscles were dissected from WT, SMNΔ7, and SMNΔ7 p62+/– mice euthanized at P10 and were fixed in 4% paraformaldehyde in PBS overnight. The spinal cords were processed as previously described (35), and images were captured with an LSM 700 Inverted Confocal Microscope (Leica). For evaluation of the innervation state of the FDB-2 muscle, hind paws were fixed in 4% paraformaldehyde in PBS overnight and then rinsed in PBS. The FDB-2 muscles were dissected, permeabilized in 0.5% Triton X-100, and blocked in 5% normal goat serum (NGS) under constant shaking for 1 hour, and then incubated with anti-synaptophysin antibody for 24 hours at 4°C. Samples were subsequently washed with several changes of PBS with shaking for 6 hours, incubated with secondary antibody overnight at 4°C, and then washed with PBS with shaking for 6 hours. Finally, samples were incubated with α-bungarotoxin for 1 hour at room temperature to label acetylcholine receptors (AChRs), washed again for 1 hour, and mounted and imaged as described above.
Mouse MN differentiation.
Mouse WT SMN+/+ SMN2+/+ Hb9:GFP and SMA SMN–/– SMN2+/+ Hb9:GFP, and SMN–/– SMN2+/+ SMNΔ7 Hb9:GFP ES cells were maintained and differentiated as described previously (35). Briefly, ESCs were differentiated into MNs using an embryoid body method with treatments of retinoic acid (Sigma-Aldrich) and a hedgehog agonist (Curis, Inc.). After 7 days, MNs were dissociated and plated onto poly-ornithine coated plates (PerkinElmer).
Human MN differentiation.
The iPSCs were grown on Matrigel-coated dishes (BD Biosciences), cultured with mTeSR (STEMCELL Technologies), and split using Gentle Cell Dissociation Reagent (STEMCELL Technologies) until the initiation of the MN differentiation protocol, when they were dissociated with Accutase (STEMCELL Technologies) and cultured in mTeSR as embryoid bodies in ultra-low attachment dishes (Corning). LDN (1 μM, Stemgent) and SB 431542 (10 μM, Stemgent) were added 1 day later (day 1) to induce neural differentiation and media gradually changed to knockout serum replacement and DMEM media (Life Technologies, Thermo Fisher Scientific). Retinoic acid (1 μM , Sigma-Aldrich) and BDNF (10 ng/ml) were added 2 days later (day 3), and 1 μM smoothened agonist 1.3 (EMD Millipore) was added 5 days later (day 8). DAPT (2.5 μM, R&D Systems) was added on day 10 along with, 2 μM cytosine arabinoside (Ara-c, Sigma-Aldrich) to eliminate progenitors and dividing cells. From day 5 to day 10, media were gradually changed to DMEM/F12 supplemented with 2% B27 and 1% N2 (Life Technologies, Thermo Fisher Scientific). After 14 days of differentiation, the cultures were dissociated with papain/DNase solution (Worthington) and plated in 50 μg/ml poly-lysin (EMD Millipore), 10 μg/ml laminin (Life Technologies, Thermo Fisher Scientific) and 10 μg/ml fibronectin-coated (Corning) plates (Greiner). The medium used was Neurobasal containing 2% B27 and 1% N2, supplemented with Glutamax (Life Technologies, Thermo Fisher Scientific), NEAA (EMD Millipore), 20% glucose, 0.2 μM ascorbic acid (Sigma-Aldrich), and 20 ng/ml of BDNF, GDNF, and CNTF (R&D Systems).
Immunocytochemistry and image analysis.
Cells were fixed in 4% paraformaldehyde (PFA) at room temperature for 20 minutes. Following standard protocols, cells were immunostained with primary antibodies followed by fluorescently labeled secondary antibodies (Life Technologies, Thermo Fisher Scientific) as previously described (35). For quantification of mCherry-LC3+ APLys and mCherry-GFP-LC3+ APs, human iPSC–derived MNs were cultured in 12-mm glass coverslips and images captured using a ×63 objective on an LSM 700 Inverted Confocal Microscope (Leica). Quantification was performed on images of 40 to 100 lentivirus-transduced neurons per line and per treatment, with maximum projection of all Z-stack sections using ImageJ software (NIH). Three independent experiments were performed. TA muscle fiber size was measured in images taken from random visual fields using a ×20 objective on a Nikon Eclipse Ti microscope. Subsequent image quantification was automatically performed using the PerkinElmer Columbus Image Data Storage and Analysis System. The muscle fibers (2,000–3,000 per mouse) were measured. Innervation of FDB-2 muscles was quantified in images taken with a ×20 objective on a LSM 700 inverted confocal microscope. The innervation status of postsynaptic endplates was determined according to the extent to which the endplate overlaid with presynaptic components. Fully innervated endplates were defined by the complete overlap of presynaptic (i.e., synaptophysin) and postsynaptic (α-bungarotoxin) labeling. Partially denervated endplates were those with only part of the endplate being covered by presynaptic labeling. Fully denervated endplates were devoid of any presynaptic labeling. Endplates (300–700 per mouse) were analyzed from randomly selected visual fields. The quantification was performed by an investigator blinded to the muscle genotypes.
Chemicals, antibodies, and plasmids.
For mammalian cell experiments, the following antibodies were used: rat HA and mouse β-actin (Cell Signaling Technology, 3700S); rabbit β-tubulin (Abcam, ab6046); rabbit islet 1 (Abcam, ab109517); SQSTM1/p62 (Abcam, ab56416); rabbit gemin2 (Abcam, ab150383); rabbit lamin B1 (Abcam, ab16048); mouse SMN (BD Biosciences, 610647); rabbit ubiquitin (Santa Cruz Biotechnology, SC-9133); mouse c-Myc (Novus Biologicals, NB600-302); rabbit LC3 (2775); rabbit p-P70S6K1 (Thr389, Cell Signaling Technology, 9234S); rabbit P70S6K1 (Cell Signaling Technology, 2708S); rabbit p-S6 (Ser235/236, Cell Signaling Technology, 2211s); rabbit S6 (Cell Signaling Technology, 2217S); synaptophysin (Synaptic Systems, 101002); and laminin (Sigma-Aldrich, L9393). For NMJ analysis, Alexa Fluor 555–conjugated α-bungarotoxin (Life Technologies, Thermo Fisher Scientific, B35451) was used. For fly experiments, SMN (gift of Spyros Artavanis-Tsakonas, Harvard Medical School, Boston, Massachusetts, USA, and Gregory Matera, University of North Carolina, Chapel Hill, North Carolina, USA) and tubulin (Sigma-Aldrich, T5168) antibodies were used. cDNAs encoding human HA-tagged SMN-FL protein or a truncated SMN protein carrying a deletion of exon 7 (Δ7) were subcloned into the pEF-DEST51 vector using the Gateway cloning system (Life Technologies, Thermo Fisher Scientific). Myc-p62 plasmid was provided by María T. Diaz-Meco (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, California, USA), and HA-GFP and Myc-E2-crimson plasmids were purchased from Addgene (plasmid numbers 22612 and 38770, respectively).
Transfection and lentiviral transduction.
For overexpression of SMN-FL and SMN-Δ7 proteins, HEK293T cells were transfected with Lipofectamine 2000 (Life Technologies, Thermo Fisher Scientific) with the HA-tagged forms of the proteins following the manufacturer’s instructions. A pool of RNAi (20 nM) against p62 (SQSTM1) or SMN (Santa Cruz Biotechnology, sc-29679 and sc-36510, respectively) or control RNAi (QIAGEN, 1027280) was transfected into HEK293T cells and mouse MNs (Santa Cruz Biotechnology, sc-29828) using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies, Thermo Fisher Scientific) following the manufacturers’ instructions. Cells were exposed to the silencing mix for 6 hours, following which silencing medium was replaced with fresh complete media. Lentiviral shRNA plasmids targeting Atg7 and a nonsilencing control, as well as mCherry-GFP-LC3 constructs were provided by Ana M. Cuervo (Albert Einstein College of Medicine, New York, New York, USA) and previously published (40, 78). Plasmids encoding the DN form of human Cul5 in PHAGE vector and empty PHAGE vector were purchased from Addgene (plasmid numbers 41916 and 44012, respectively) and used for lentiviral production. Vectors encoding short hairpins against mouse (TRCN0000238133 and TRCN0000098615) and human (TRCN0000430110 and TRCN0000007234) p62 (SQSTM1) were purchased from Sigma-Aldrich. Lentiviral stocks were prepared by Lipofectamine transfection of these vectors and the packaging vectors pMDLg/pRRE, pRSV-Rev, and pMD2.VSVG into HEK293T cells. Supernatants were collected over a 72-hour period, titered, and used for infection. The efficiency of infection at 72 hours, determined by the percentage of GFP+ cells or the percentage of cells surviving 1 μM puromycin treatment, depending on the vector, exceeded 90% for all constructs.
Cell culture treatments.
For serum or amino acid removal, cells were washed with PBS followed by culture in DMEM or EBSS for 24 hours, respectively. The lysosomal inhibitors leupeptin (Fisher BioReagents, Thermo Fisher Scientific), ammonium chloride (Sigma-Aldrich) and HCQ (Sigma-Aldrich) were used at 100 μM, 20 mM, and 10 μM, respectively, for 4 to 6 hours. Rapamycin (R&D Systems) was used at 200 nM and MG132 (Sigma-Aldrich) at 100 nM. For SG formation induction, cells were exposed to 10 or 50 μM sodium arsenate for 6 hours. Details on IP, cellular fractionation, and immunoblot analysis are provided in the Supplemental Experimental Procedures.
Chymotrypsin-like activity measurement.
Human iPSC–derived MNs were washed in PBS, homogenized with 0.5% NP-40, and centrifuged for 15 minutes at 4°C at 16,200 g. Supernatant were collected and the assay performed using the Proteasome Activity Assay Kit (Abcam, ab107921) according to the manufacturer’s instructions.
Reverse transcription PCR.
Total RNA was extracted with the TRIzol Reagent (Life Technologies, Thermo Fisher Scientific), followed by DNase digestion using RQ1 RNase-Free DNase (Promega). Total RNA was reverse transcribed with the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative reverse transcription PCR (qRT-PCR) was performed with iQ SYBR Green Supermix (Bio-Rad) and a CFX96 Real-Time PCR Detection System (Bio-Rad). Relative quantitations of mRNA levels were determined using the comparative Ct method. For mammalian cell experiments, 18s and actin were used as housekeeping genes, and for fly experiments, RpL32 and α-tubulin 84B were used as a normalization reference. The following mouse primers were used: SMN, forward, 5′-AAGGCACAGCCAGAAGAAAA-3′, SMN, reverse, 5′-TCACAGGTCGGGGAAAGTAG-3′; p62, forward, 5′-GCTGCCCTAACCCACATCT-3′, p62, reverse, 5′-CGCCTTCATCCGAGAAAC-3′; and IL6, forward, 5′-AGTTGCCTTCTTGGGACTGA-3′, IL6, reverse, 5′-TCCACGATTTCCCAGAGAAC-3′. The following human primers were used: SMN, forward, 5′-AACATCAAGCCCAAATCTGC-3′, SMN, reverse, 5′-TGGTCCAGAAGGAAATGGAG-3′; and p62, forward, 5′-AGAACGTTGGGGAGAGTGTG-3′, p62, reverse, 5′-GCGATCTTCCTCATCTGCTC-3′. The following primers were used for fly: Smn, forward, 5′-AGCGGAAAGAAAAAGACACC-3′, Smn, reverse, 5′-TCGAAAAGATGTGATGTGATGA-3′; ref(2)P(p62), forward, 5′-AATCGAGCTGTATCTTTTCCAGG-3′,ref(2)P(p62), reverse, 5′-AACGTGCATATTGCTCTCGCA-3′.
Fly lifespan analysis.
For survival analysis, flies were collected within 24 hours from eclosion, sorted by sex under light CO2 anesthesia, and reared at standard density (20–25 flies per vial) on cornmeal, soy flour, and yeast fly food at 25°C and 60% humidity and a 12-hour light/12-hour dark cycle. Flies were transferred to fresh vials every 2 days and dead flies counted. RU486 (Mifepristone) dissolved in ethanol was administered in the media at a final concentration of 150 μg/ml. The efficiency of Smn and p62 RNAi lines was tested using the ubiquitous actin5c-Gal4 driver. As a control, we used RNAi against the white gene, responsible for the eye color that we and others have showed does not affect lifespan when it is mutated or suppressed with RNAi (79). The following RNAi lines were used: control RNAi (HMS00017), Smn-i (JF02057), p62-551i (HMS00551), and p62-938i (HMS00938).
Statistics.
Statistical significance was determined using a 2-tailed Student’s t test for groups of 2 and by ANOVA for groups of 3 or more. A CI of 95% was used for all comparisons. The statistical significance for the Kaplan-Meier analysis was determined by Mantel-Cox and Wilcoxon tests. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal studies were reviewed and approved by the IACUC of Harvard University and performed in accordance with institutional and federal guidelines. The Pediatric Neuromuscular Clinical Research Network recruited SMA patients and collected the fibroblasts later obtained by the Rubin laboratory for the derivation of iPS cell lines. Tissue collections for all human subjects who donated tissue used in this research were conducted with consent under protocols that were approved by the Columbia University IRB. Subsequent use of the SMA patients’ samples in the Rubin laboratory at Harvard University was reviewed by the Harvard University Committee on the Use of Human Subjects (CUHS). The generation of iPSC lines by the Harvard iPSC Core was conducted in a fee-for-service capacity. The information regarding both the SMA iPSC lines and the healthy control cell lines BJ and 1016A has been previously published (35).
See complete unedited blots in the supplemental material.
Author contributions
NRM and AP conceived the study. NRM, AP, and TG designed the study methodology. NRM and AP performed formal analysis. NRM, AP, RMG, and TG conducted the studies. EMN provided research assistance. NRM, AP, NP, and LLR provided resources. NRM wrote the original draft of the manuscript. NRM, AP, TG, NP, RS, and LLR reviewed and edited the manuscript. NRM and LLR were responsible for funding acquisition.
Supplementary Material
Acknowledgments
We thank the Pediatric Neuromuscular Clinical Research Network for recruiting SMA patients and the Harvard Stem Cell Institute iPSC Core Facility for generating the iPSCs from SMA patients’ fibroblasts. We thank Catherine McGillivary and Diane Faria (Department of Stem Cell and Regenerative Biology, Histology Core, Harvard University, Cambridge, Massachusetts, USA) for histology assistance. We thank Patricia Boya (Department of Cellular and Molecular Biology, Centro de Investigaciones Biológicas, CSIC) for critically reviewing this manuscript. We are grateful to Lance S. Davidow for image analysis support and J. LaLonde(Department of Stem Cell and Regenerative Biology, Harvard University, and Harvard Stem Cell Institute, Cambridge, Massachusetts, USA) for editorial assistance. This work was supported by the Muscular Dystrophy Association (MDA376743, to NRM); the SMA Foundation (to LLR); the National Institute of Neurological Disorders and Stroke (NINDS) (P01NS066888, to LLR); and the National Institute of General Medical Sciences (R01-AG043517, to RS and R01-GM084947, to NP). AP is supported by a LAM Foundation Fellowship Award (LAM00105E01-15).
Version 1. 04/19/2018
In-Press Preview
Version 2. 06/11/2018
Electronic publication
Version 3. 07/02/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(7):3008–3023.https://doi.org/10.1172/JCI95231.
Contributor Information
Natalia Rodriguez-Muela, Email: nrodriguezmuela@fas.harvard.edu.
Andrey Parkhitko, Email: Andrey_Parkhitko@hms.harvard.edu.
Tobias Grass, Email: tobiasgrass@fas.harvard.edu.
Rebecca M. Gibbs, Email: rgibbs@g.harvard.edu.
Erika M. Norabuena, Email: emnorab@gmail.com.
Norbert Perrimon, Email: perrimon@receptor.med.harvard.edu.
Rajat Singh, Email: rajat.singh@einstein.yu.edu.
Lee L. Rubin, Email: lee_rubin@harvard.edu.
References
- 1.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Iascone DM, Henderson CE, Lee JC. Spinal muscular atrophy: from tissue specificity to therapeutic strategies. F1000Prime Rep. 2015;7:04. doi: 10.12703/P7-04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 5.Lorson CL, et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19(1):63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 6.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24(5):438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296(1):51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Fallini C, et al. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol. 2014;74(3):319–332. doi: 10.1002/dneu.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim SR, Hertel KJ. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3’ splice site pairing. J Biol Chem. 2001;276(48):45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 10.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82(4):834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29(24):7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman EY, et al. Morpholino antisense oligonucleotides targeting intronic repressor Element1 improve phenotype in SMA mouse models. Hum Mol Genet. 2014;23(18):4832–4845. doi: 10.1093/hmg/ddu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickson A, Osman E, Lorson CL. A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum Gene Ther. 2008;19(11):1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naryshkin NA, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345(6197):688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 15.Kwon DY, Motley WW, Fischbeck KH, Burnett BG. Increasing expression and decreasing degradation of SMN ameliorate the spinal muscular atrophy phenotype in mice. Hum Mol Genet. 2011;20(18):3667–3677. doi: 10.1093/hmg/ddr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29(5):1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makhortova NR, et al. A screen for regulators of survival of motor neuron protein levels. Nat Chem Biol. 2011;7(8):544–552. doi: 10.1038/nchembio.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon DY, et al. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell. 2013;24(12):1863–1871. doi: 10.1091/mbc.e13-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han KJ, et al. Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J Biol Chem. 2012;287(52):43741–43752. doi: 10.1074/jbc.M112.372318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8(24):4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Xu Z, Yang L, Xu S, Zhang Z, Cao Y. The receptor proteins: pivotal roles in selective autophagy. Acta Biochim Biophys Sin (Shanghai) 2015;47(8):571–580. doi: 10.1093/abbs/gmv055. [DOI] [PubMed] [Google Scholar]
- 23.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. 2016;428(9 Pt A):1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 25.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104(36):14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 28.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285(5):3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 31.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 32.Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 33.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 34.Ng SY, et al. Genome-wide RNA-Seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell. 2015;17(5):569–584. doi: 10.1016/j.stem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Muela N, et al. Single-cell analysis of SMN reveals its broader role in neuromuscular disease. Cell Rep. 2017;18(6):1484–1498. doi: 10.1016/j.celrep.2017.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572(1-3):69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Zou T, Yang X, Pan D, Huang J, Sahin M, Zhou J. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol. 2011;31(4):541–550. doi: 10.1007/s10571-011-9647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest. 2015;125(1):65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 42.Boya P, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25(3):1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(4):517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44(1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15(6):555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 47.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.e08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Custer SK, Androphy EJ. Autophagy dysregulation in cell culture and animals models of spinal muscular atrophy. Mol Cell Neurosci. 2014;61:133–140. doi: 10.1016/j.mcn.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Periyakaruppiah A, de la Fuente S, Arumugam S, Bahí N, Garcera A, Soler RM. Autophagy modulators regulate survival motor neuron protein stability in motoneurons. Exp Neurol. 2016;283(Pt A):287–297. doi: 10.1016/j.expneurol.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 52.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98(22):12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98(22):12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez A, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(3):211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282(24):4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 56.Ling KK, Gibbs RM, Feng Z, Ko CP. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2012;21(1):185–195. doi: 10.1093/hmg/ddr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faravelli I, Nizzardo M, Comi GP, Corti S. Spinal muscular atrophy--recent therapeutic advances for an old challenge. Nat Rev Neurol. 2015;11(6):351–359. doi: 10.1038/nrneurol.2015.77. [DOI] [PubMed] [Google Scholar]
- 58.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112(Pt 22):4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 59.Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4(8):1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 60.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13(7):805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller N, Shi H, Zelikovich AS, Ma YC. Motor neuron mitochondrial dysfunction in spinal muscular atrophy. Hum Mol Genet. 2016;25(16):3395–3406. doi: 10.1093/hmg/ddw262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimitriadi M, et al. Decreased function of survival motor neuron protein impairs endocytic pathways. Proc Natl Acad Sci U S A. 2016;113(30):E4377–E4386. doi: 10.1073/pnas.1600015113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hosseinibarkooie S, et al. The power of human protective modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am J Hum Genet. 2016;99(3):647–665. doi: 10.1016/j.ajhg.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang QJ, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26(31):8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman LG, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J Neurosci. 2012;32(22):7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodríguez-Muela N, Germain F, Mariño G, Fitze PS, Boya P. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell Death Differ. 2012;19(1):162–169. doi: 10.1038/cdd.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcera A, Bahi N, Periyakaruppiah A, Arumugam S, Soler RM. Survival motor neuron protein reduction deregulates autophagy in spinal cord motoneurons in vitro. Cell Death Dis. 2013;4:e686. doi: 10.1038/cddis.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piras A, et al. Inhibition of autophagy delays motoneuron degeneration and extends lifespan in a mouse model of spinal muscular atrophy. Cell Death Dis. 2017;8(12):3223. doi: 10.1038/s41419-017-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 70.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 71.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51(3):283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ning K, et al. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum Mol Genet. 2010;19(16):3159–3168. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 73.Tseng YT, Chen CS, Jong YJ, Chang FR, Lo YC. Loganin possesses neuroprotective properties, restores SMN protein and activates protein synthesis positive regulator Akt/mTOR in experimental models of spinal muscular atrophy. Pharmacol Res. 2016;111:58–75. doi: 10.1016/j.phrs.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 74.Kye MJ, et al. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet. 2014;23(23):6318–6331. doi: 10.1093/hmg/ddu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Millino C, et al. Different atrophy-hypertrophy transcription pathways in muscles affected by severe and mild spinal muscular atrophy. BMC Med. 2009;7:14. doi: 10.1186/1741-7015-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 77.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodríguez-Muela N, et al. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;12(3):478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parkhitko AA, Binari R, Zhang N, Asara JM, Demontis F, Perrimon N. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 2016;30(12):1409–1422. doi: 10.1101/gad.282277.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.