Abstract
Drug resistance remains the key problem in cancer treatment. It is now accepted that each myeloma patient harbors multiple subclones and subclone dominance may change over time. The coexistence of multiple subclones with high or low chromosomal instability (CIN) signature causes heterogeneity and drug resistance with consequent disease relapse. In this study, using a tandem affinity purification–mass spectrometry (TAP-MS) technique, we found that NEK2, a CIN gene, was bound to the deubiquitinase USP7. Binding to USP7 prevented NEK2 ubiquitination resulting in NEK2 stabilization. Increased NEK2 kinase levels activated the canonical NF-κB signaling pathway through the PP1α/AKT axis. Newly diagnosed myeloma patients with activated NF-κB signaling through increased NEK2 activity had poorer event-free and overall survivals based on multiple independent clinical cohorts. We also found that NEK2 activated heparanase, a secreted enzyme, responsible for bone destruction in an NF-κB–dependent manner. Intriguingly, both NEK2 and USP7 inhibitors showed great efficacy in inhibiting myeloma cell growth and overcoming NEK2-induced and -acquired drug resistance in xenograft myeloma mouse models.
Keywords: Oncology
Keywords: B cells, Cancer
Introduction
Multiple myeloma is a plasma cell malignancy, characterized by organ dysfunction due to hypercalcemia, renal dysfunction, anemia, and/or bone destruction (CRAB) (1). Novel drugs, such as proteasome inhibitors and immunomodulatory drugs, combined with autologous stem cell transplantation (ASCT) have greatly improved patient outcome and led to complete remissions in the large majority of newly diagnosed patients with myeloma (2, 3). However, many patients still relapse, and after prolonged salvage treatment, the disease becomes resistant to every drug available and eventually death is inevitable. Of several genetic alterations, myeloma patients with 17p deletion, t(4;14), t(14;16), t(14;20), and 1q21 amplification show in general a more aggressive phenotype, associated with increased drug resistance (4–8). Myeloma cells typically produce a substantial amount of paraprotein and are heavily dependent on the ubiquitin-proteasome system (UPS), autophagy, and the unfolded protein response (UPR) to maintain cellular homeostasis (9). XBP1, a critical regulator of the UPR and a transcription factor, plays an important role in the response to bortezomib. Low expression or a mutation of XBP1 is associated with bortezomib resistance. Conversely, the spliced XBP1 isoform confers sensitivity to bortezomib (10). The deubiquitinase (DUB) USP7 can also alter drug resistance by modulating its targets, which include a range of oncogenes (e.g., MDM2) and tumor suppressors (e.g., PTEN). Increased USP7 activity leads to the stabilization or displacement of these targets, ultimately driving malignancy (11, 12). In myeloma, the USP7 inhibitor P5091 shows promise by specifically targeting myeloma cells and overcoming bortezomib resistance while not affecting the tumor microenvironment (13). We have shown that an increased chromosomal instability (CIN) signature is linked to drug resistance in myeloma using a systemic analysis of sequential myeloma samples treated at different stages during intensive treatment including autologous transplantation and posttransplant therapy (14). NEK2, a CIN gene, is the most significant gene associated with drug resistance and poor outcome based on almost 2,500 microarrays in myeloma and other cancers (14–16). Though we have demonstrated that NEK2 activates efflux drug pumps, the detailed mechanisms resulting in drug resistance conferred by NEK2 remain unknown.
NEK2 is a serine-threonine kinase originally found to have an important role in centrosome maintenance and cell division (17). In normal tissues, its expression is mostly limited to the testis and is highly regulated in a cell cycle–dependent manner. NEK2 plays a role in maintaining centrosome integrity by regulating the mitotic centrosome separation through reversible phosphorylation of its substrates C-NAP1, protein phosphatase 1 (PP1), NIP/centrobin, β-catenin, and SGO1 (18–23). However, in recent years our lab has found that NEK2 expression is progressively increased in myeloma cells of patients in complete remission, but with minimal residual disease, and in relapsed myeloma samples (14). High levels of NEK2 are linked to a poor event-free survival (EFS) and overall survival (OS) in myeloma and other cancers. High NEK2 also increases activation of oncogenes such as EZH2, AKT, and Wnt (14, 24, 25) in solid tumors, including breast cancer, lung cancer, ovarian cancer, etc. (20, 26, 27). Interestingly, NEK2 has not been found to have activating mutations, gene amplification, or a hypomethylated promoter in tumor cells, suggesting that the increased accumulation of NEK2 is the result of changes in transcription regulation and/or potentially protein stabilization. Since NEK2-overexpressing (NEK2-OE) cancer cells are more resistant to numerous standard antimyeloma drugs with different modes of action (e.g., bortezomib, doxorubicin, etoposide, and others) (14), it appears likely that NEK2 is activating numerous separate and perhaps overlapping pathways to promote drug resistance. Because proteasome inhibitors are probably the most powerful standard drugs in myeloma, we have focused our work on identifying the mechanisms related to NEK2-induced resistance to proteasome inhibitors in this study.
Here, we address the knowledge gap between the NEK2 accumulation and the mechanisms of drug resistance in myeloma. Using systemic biochemical in vitro and in vivo approaches, we uncovered NEK2 interacting proteins and determined the mechanism by which NEK2 is hijacking myeloma cells. We demonstrate the clinical relevance of our findings by correlating the downstream signaling pathway of NEK2 with patient outcome based on multiple independent clinical data sets. Furthermore, our work elucidates a NEK2 stabilization pathway by virtue of its interaction with the deubiquitinating enzyme USP7 that is also overexpressed in myeloma and other cancers (13). Using 2 distinct small-molecule compounds, we target NEK2 protein directly and indirectly in vivo and show that they are able to overcome bortezomib resistance and reduce tumor burden. Our findings should facilitate novel targeted approaches for myeloma therapy to prevent disease relapse.
Results
Tandem affinity purification–mass spectrometry identifies NEK2 binding to USP7.
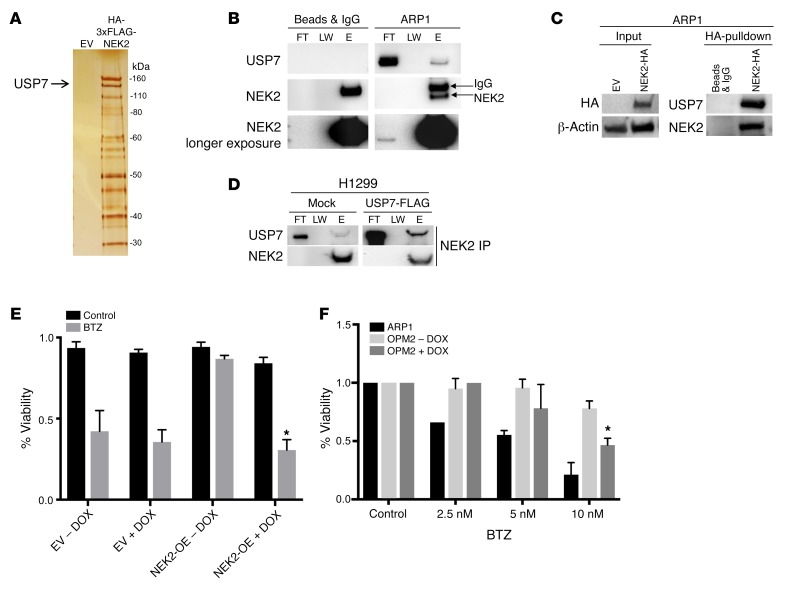
To better understand what pathways are involved in NEK2-mediated drug resistance, we decided to analyze its interacting partners. Tandem affinity purification–mass spectrometry (TAP-MS) using anti-HA and anti-FLAG antibodies was performed on ARP1 cells with stable expression of NEK2 tagged with HA-FLAG (3×). We found that over 60 proteins bound to NEK2 (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI98765DS1). We were particularly interested in the DUB USP7, because a USP7 inhibitor can induce apoptosis in myeloma cells resistant to bortezomib (13). To confirm the interaction between NEK2 and USP7, coimmunoprecipitation (Co-IP) of endogenous NEK2 in ARP1 cells was performed using anti-NEK2 antibodies; Western blotting was applied to detect the binding of USP7 with NEK2 protein (Figure 1B). NEK2 was also pulled down using an anti-HA antibody conjugated to dynabeads in NEK2-OE ARP1 cells. Endogenous NEK2 was pulled down in H1299 cells transfected with USP7-FLAG using NEK2 antibodies. Western blots showed that USP7 was bound to NEK2 in ARP1 (Figure 1C) and H1299 cells (Figure 1D), suggesting that the USP7 and NEK2 binding was not cell context specific. We thus hypothesized that USP7 might play a significant role in the NEK2-mediated drug resistance. USP7 is known to drive multiple pathways in various cancers and is being considered as a potential therapeutic target (13, 28). We then constructed a doxycycline-inducible USP7-shRNA vector, which was transduced into empty vector–containing (EV-transduced) or NEK2-OE myeloma cells, and tested the cells’ resistance to bortezomib. The viability of EV ARP1 cells showed a 60% reduction after bortezomib treatment (5 nM) for 48 hours, while NEK2-OE ARP1 cells were highly resistant to bortezomib (Figure 1E). However, knockdown of USP7 in the NEK2-OE ARP1 and OCI-MY5 (Supplemental Figure 1) cell lines led to less than 40% viability after bortezomib treatment, similar to the EV controls treated with bortezomib (Figure 1E). These results suggest that USP7 may be a novel NEK2-interacting protein and its expression is needed for the NEK2-mediated bortezomib resistance (14). To address sensitivity to bortezomib based on endogenous NEK2 levels, we treated ARP1 and OPM2 myeloma cell lines, which have low and high levels of NEK2 expression, respectively (Figure 2C), with 2.5, 5, and 10 nM bortezomib for 48 hours. We found that OPM2 cells were relatively resistant to bortezomib, while ARP1 cells were sensitive to bortezomib (Figure 1F). Furthermore, downregulation of NEK2 by NEK2-shRNA in the OPM2 cell line decreased its drug resistance (Figure 1F), suggesting that high NEK2 expression causes bortezomib resistance in myeloma cells.
Figure 1. USP7 interacts with NEK2.
(A) HEK293T cells were transfected with either empty vector (EV) or HA-FLAG(3×)-NEK2. Proteins binding to NEK2 were pulled down by tandem HA and FLAG antibodies and stained with silver prior to mass spectrometry. (B) ARP1 myeloma cells were lysed and NEK2 was immunoprecipitated using NEK2 antibodies. Western blots were probed with NEK2 and USP7 antibodies. FT, LW, and E represent flow through, last wash, and elution of the immunoprecipitation, respectively. (C) ARP1 myeloma cells were transduced with NEK2-HA plasmids. Transduced cells were lysed and NEK2 was immunoprecipitated using HA antibodies. Western blots were probed using NEK2 and USP7 antibodies. (D) H1299 cells were transfected with mock or USP7-FLAG overexpression vector. Endogenous NEK2 was immunoprecipitated and Western blots were analyzed using NEK2 and USP7 antibodies. (E) ARP1 myeloma cells transduced with EV + USP7-shRNA or NEK2-OE + USP7-shRNA were treated with doxycycline (DOX) or vehicle to suppress USP7 expression. After 72 hours, cells were treated with bortezomib (BTZ; 5 nM) for a further 24 hours and cell viability was measured using trypan blue stain. (F) OPM2 cells transduced with NEK2-shRNA were treated with DOX or vehicle to suppress NEK2 expression. ARP1 cells or OPM2 cells with or without silencing of NEK2 were treated with BTZ (2.5, 5, and 10 nM) for a further 24 hours and cell viability was measured using trypan blue stain. Viability experiments were performed in triplicate and a Student’s t test was performed and showed the significance at 10 nM with or without silencing of NEK2. *P < 0.05.
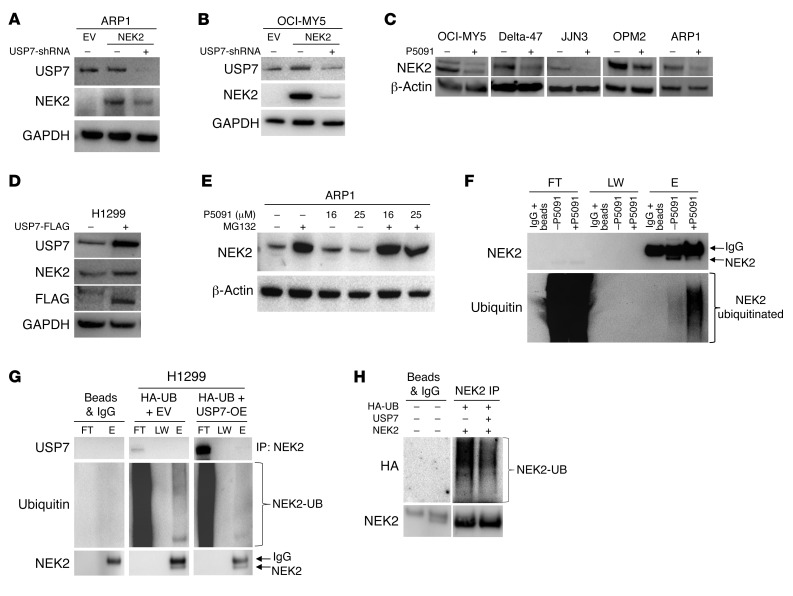
Figure 2. USP7 prevents ubiquitination of NEK2 protein.
(A and B) Knockdown of USP7 decreases NEK2 protein. ARP1 (A) and OCI-MY5 (B) myeloma cells were transfected with EV, NEK2, or NEK2 + USP7-shRNA. After 72-hour induction with doxycycline, cells were lysed. NEK2 and USP7 protein levels were analyzed by Western blot. (C) OCI-MY5, Delta-47, JJN3, OPM2, and ARP1 myeloma cell lines were treated with 16 μM P5091 for 24 hours. Cells were lysed and NEK2 levels analyzed by Western blot. (D) H1299 cells were transfected with mock or USP7-FLAG–overexpressing vectors, lysed, and NEK2 and USP7 levels were determined by Western blot. (E) ARP1 myeloma cells were treated with the proteasome inhibitor MG132 (10 μM) alone for 30 minutes or in combination with P5091 (16 and 25 μM) for an additional 5 hours. Cells were lysed and NEK2 levels were analyzed by Western blot. (F) OPM2 cells were treated with or without P5091 (25 μM for 2 hours) and protein was extracted with lysis buffer supplemented with NEM. Endogenous NEK2 was immunoprecipitated and analyzed by Western blot using NEK2 and ubiquitin antibodies. FT, LW, and E represent flow through, last wash, and elution of the immunoprecipitation, respectively. (G) H1299 cells were transfected with EV and HA-ubiquitin (HA-UB) or FLAG-USP7 and HA-UB. Cells were lysed and endogenous NEK2 was immunoprecipitated (IP) by NEK2 antibodies and ubiquitination levels were analyzed by Western blot. The higher-molecular-weight band is nonspecific IgG. (H) H1299 cells were transfected with NEK2-OE, HA-UB, and FLAG-USP7 or NEK2-OE and HA-UB. Cells were lysed and total NEK2 protein, including both endogenous and exogenous, was immunoprecipitated (IP) by anti-NEK2 antibodies and ubiquitination levels were analyzed using HA antibodies by Western blot.
NEK2 is stabilized by the DUB USP7.
USP7 is a deubiquitinating enzyme and a known stabilizer of numerous oncogenes. Since we found a NEK2-USP7 interaction, we hypothesized that USP7 might stabilize the NEK2 protein. To address this hypothesis, USP7 was knocked down by USP7-shRNA in the NEK2-OE myeloma cell lines ARP1 (Figure 2A) and OCI-MY5 (Figure 2B). Cell lysates were collected after 48 hours of doxycycline induction to inhibit USP7 expression. Western blotting was performed and showed that NEK2 protein was substantially depleted in NEK2-OE myeloma cells. To corroborate this finding, H1299 cells were transfected with the same USP7-shRNA vector and induced with doxycycline for 48 hours. Western blots showed that endogenous NEK2 levels were decreased (Supplemental Figure 2A). Five myeloma cell lines (OCI-MY5, Delta-47, JJN3, OPM2, and ARP1) were also treated with P5091, a USP7 inhibitor that selectively binds the USP7 active site and inhibits its activity, but not its expression (29). We found that P5091 depleted endogenous NEK2 protein after overnight treatment at 16 μM (Figure 2C). Because P5091 can target USP47, we treated ARP1 cells with P5091 overnight at 16 μM and analyzed the protein extract by Western blot. We found no detectable USP47 in ARP1 cells, while NEK2 protein was depleted by P5091 (Supplemental Figure 2B), suggesting that the NEK2 depletion is mediated by USP7 inhibition. We also examined NEK2 mRNA expression after P5091 treatment in myeloma cells. Briefly, ARP1 cells were treated with P5091 overnight at 16 μM followed by RNA extraction and quantitative PCR (qPCR) analysis. Using NEK2-specific primers, we found no significant changes in NEK2 mRNA levels after P5091 treatment (Supplemental Figure 2C). Next, the H1299 cells were transiently transfected with FLAG-USP7 vector for 48 hours. Results showed that endogenous NEK2 protein increased when USP7 was overexpressed (Figure 2D), supporting the notion that NEK2 protein is stabilized by USP7. Under normal conditions, NEK2 levels are tightly regulated throughout the cell cycle, mediating its depletion through the UPS (30). Thus, we tested if NEK2 depletion by P5091 was dependent on proteasome activity. ARP1 (Figure 2E), OCI-MY5 (Supplemental Figure 2D), and H1299 (Supplemental Figure 2E) cells were treated with the proteasome inhibitor MG132 (10 μM) alone for 30 minutes or in combination with P5091 (16 and 25 μM) for an additional 5 hours. Western blots showed that, although P5091 alone indeed depleted NEK2, treatment with MG132 blocked this effect, confirming that USP7 stabilizes NEK2 by preventing proteasomal degradation of NEK2. USP7 has been described mainly as a DUB. Therefore, we tested endogenous NEK2 ubiquitination in ARP1 cells treated with or without P5091 at a higher concentration (25 μM) for 2 hours to prevent significant depletion of endogenous NEK2 protein. Cells were lysed in the presence of N-ethylmaleimide (NEM) to prevent deubiquitination and endogenous NEK2 was immunoprecipitated using NEK2 antibodies. Western blots showed that the NEK2 ubiquitination smear was higher in cells treated with P5091 (Figure 2F). Additionally, we tested NEK2 ubiquitination in H1299 cells overexpressing USP7. H1299 cells were transfected with HA-ubiquitin (HA-UB) and EV or USP7-OE vectors. Endogenous NEK2 was immunoprecipitated using the same conditions described above. Western blots detected less ubiquitinated NEK2 protein in H1299 cells overexpressing USP7 (Figure 2G). Furthermore, we repeated the same experiment in H1299 cells with exogenous NEK2 expression. H1299 cells were transfected with HA-UB and NEK2 plus EV or USP7 vectors (Figure 2H). Transfection efficiency was analyzed by Western blot (Supplemental Figure 2F); endogenous and exogenous NEK2 was immunoprecipitated using NEK2 antibodies as shown in Figure 2F. Western blots detected less ubiquitinated NEK2 using HA antibodies (Figure 2H) in H1299 cells overexpressing USP7. Taken together, our data indicate that USP7 stabilizes NEK2 through deubiquitination of NEK2, and inhibition of USP7 with P5091 depletes NEK2 protein.
NEK2 activates the canonical NF-κB signaling pathway.
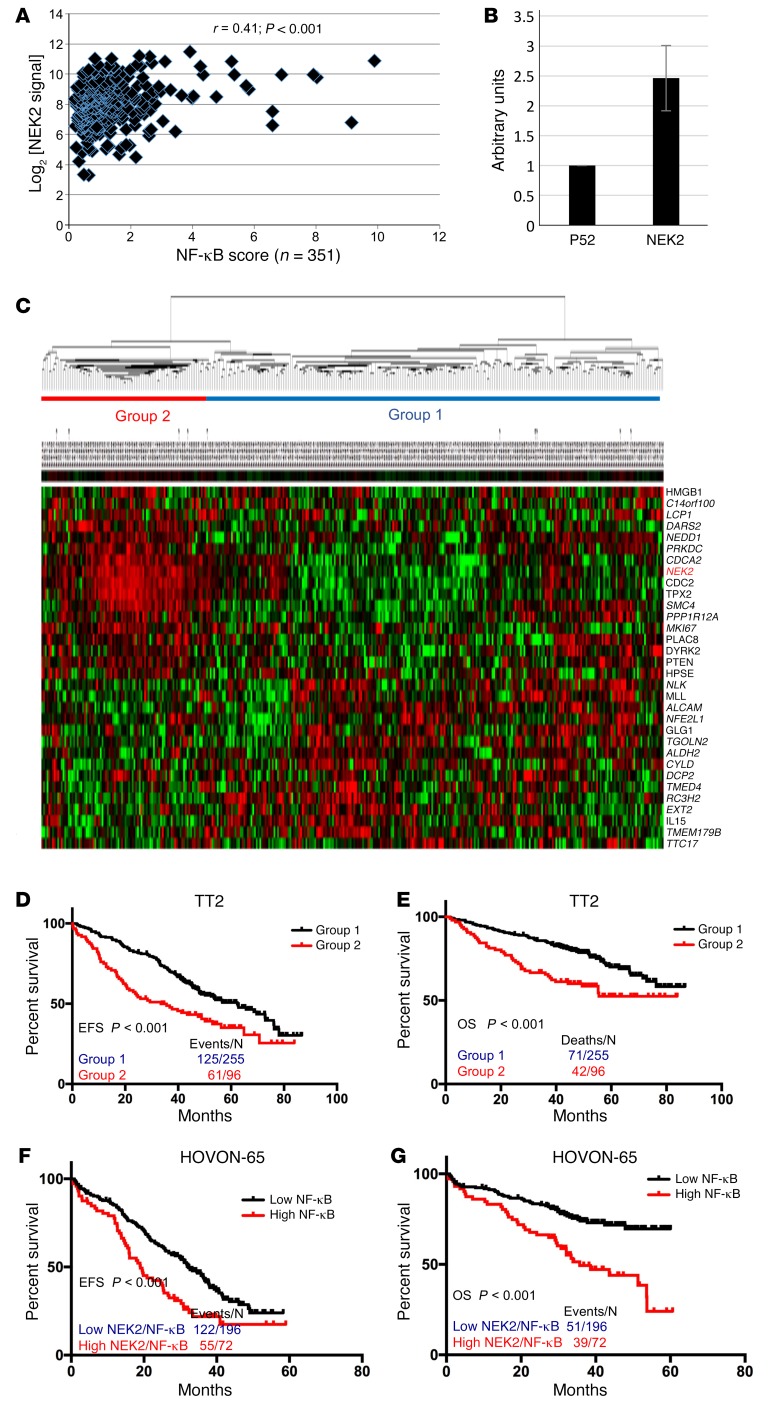
Our previous study indicated that about 44% of the genes (31 of 70) regulated by NEK2 are direct targets of the canonical NF-κB pathway (14). The p65 binding sites of these genes are listed in Supplemental Table 2, based on ENCODE at the University of California, Santa Cruz (UCSC). We have generated an NF-κB signaling score by averaging 11 NF-κB–targeted genes (31). In this study, the expression of NEK2 was correlated with the NF-κB signaling score using Affymetrix U133plus2.0 gene expression profiling (GEP) data from 351 purified bone marrow plasma cell populations of the Total Therapy 2 (TT2) cohort (32). Myeloma patients with high NEK2 showed a significantly increased NF-κB signaling score (Figure 3A; r = 0.41, P < 0.001). The myeloma cell lines ARP1 and OCI-MY5, overexpressing human NEK2 cDNA, showed a substantially higher expression of phosphorylated p65 (p-p65) protein than control cells (Supplemental Figure 3), supporting the concept that NEK2 activates the canonical NF-κB pathway in myeloma cells. To further confirm that increased p-p65 has functional significance, NF-κB transactivity in NEK2-OE and p52-OE H1299 cells (as a negative control, activator of noncanonical pathway; ref. 33) was determined by a dual-luciferase reporter assay. This assay was performed using a pNF-κB-luc construct containing 2 NF-κB binding elements in the promoter region, and a plasmid expressing Renilla luciferase as an internal control reporter. Transfected cells were exposed to the above 2 groups of NEK2-OE and p52-OE H1299 cells for 24 hours, and the reporter luciferase activity was determined and normalized to the Renilla luciferase activity using the Dual-Luciferase assay kit. As shown in Figure 3B, the luciferase activity was increased 2.5-fold in NEK2-OE H1299 cells when compared with the control cells.
Figure 3. NEK2 activates the canonical NF-κB signaling pathway.
(A) Affymetrix gene expression profiling data from 351 purified bone marrow plasma cell populations in the Total Therapy 2 (TT2) cohort was used to correlate the expression of NEK2 with the NF-κB signaling score. Pearson’s correlation was performed between log2 [NEK2 signal] and the NF-κB signaling score. (B) H1299 cells were transfected with luciferase vector under a p65-dependent promoter, an internal Renilla control vector, p52 as a negative control, or a NEK2-OE vector. Results show that NEK2 overexpression increased the luciferase signal 2.5-fold compared with p52. (C) An unsupervised hierarchical cluster analysis was used to classify 351 multiple myeloma samples using NEK2 and 31 NF-κB genes regulated by NEK2. (D–G) Kaplan-Meier analyses show event-free survival (EFS) and overall survival (OS) of multiple myeloma patients enrolled in TT2 (D and E) and HOVON-65 (F and G) cohorts. Each line represents different subgroups identified in C and is described in the figure and color coded.
To determine the clinical relevance of activation of the NF-κB signaling pathway by NEK2, the 31 NF-κB signaling genes regulated by NEK2 as well as NEK2 itself were applied in an unsupervised hierarchical cluster analysis of 351 TT2 samples. Two groups were identified and one (n = 96) contained myeloma samples with high expression of NEK2 and 16 other genes; while 15 genes, which dominated high expression in another subgroup, were negatively correlated with the expression of NEK2 (Figure 3C). Kaplan-Meier analyses were performed and showed that the group with higher expression of NEK2 and the 16 NF-κB genes had a significantly inferior EFS (Figure 3D, P < 0.001) and OS (Figure 3E, P < 0.001). We calculated a score for each sample using the 32 genes including NEK2 and evaluated patient outcome in an independent HOVON-65 cohort, which included 268 newly diagnosed myeloma samples (34). Consistent with our findings, the myeloma patients with an active NF-κB signaling showed a worse EFS (Figure 3F) and OS (Figure 3G) compared with those with an inactive NF-κB signaling.
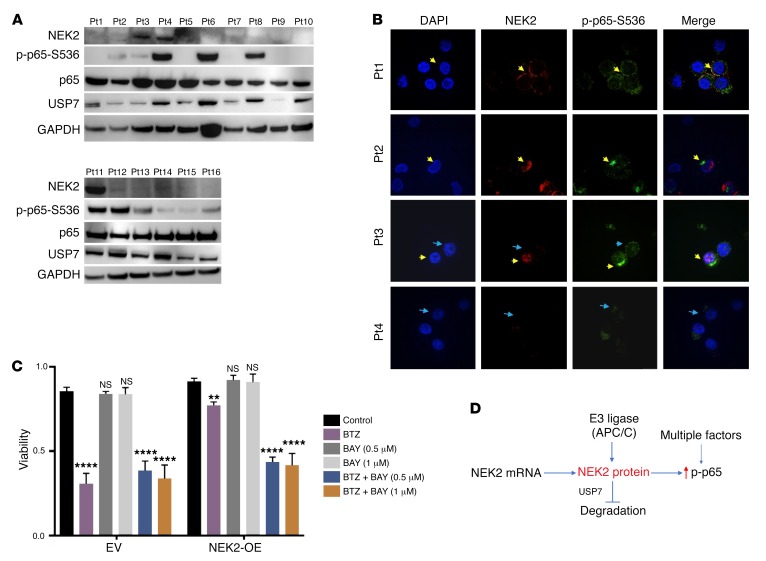
To further validate the physiological relevance of NEK2-mediated NF-κB activation, we analyzed primary myeloma samples to determine if high NEK2 expression activates the canonical NF-κB signaling pathway. Western blots from 16 primary myeloma samples showed that 3 samples (patients 3, 4, and 11) had strong NEK2 positivity (18.75%) (Figure 4A). This is consistent with the mRNA expression determined by GEP in that 15%–20% of myeloma patients had high expression of NEK2, which was associated with poor prognosis (14). As shown in Figure 4A, all 3 NEK2 strongly positive samples also showed p-p65 expression, but p-p65 was also expressed in some NEK2-negative myeloma samples, indicating that the canonical NF-κB pathway is also activated by alternative mechanisms. This is not surprising since NF-κB signaling is regulated by multiple mechanisms. USP7 protein was examined in these samples. We found that USP7 was always expressed in myeloma samples (Figure 4A), suggesting that NEK2 mRNA expressed in myeloma cells is translated to protein and can thus be stabilized by USP7. We next correlated NEK2 protein with p-p65 protein in patient samples using immunofluorescence. The results showed that in every cell with detectable NEK2, p-p65 activation was present (Figure 4B). In agreement with the results of Western blot, p-p65 was also detected in some cells in which NEK2 was not expressed (Figure 4B). Finally, to address the question whether inhibition of NF-κB could overcome NEK2-induced bortezomib resistance, NEK2-OE or EV ARP1 cells were incubated with the NF-κB inhibitor BAY11-7082 (BAY), at 0.5 and 1 μM alone or in combination with 5 nM bortezomib for 24 hours. While the NF-κB inhibitor did not kill NEK2-OE or EV ARP1 cells, bortezomib alone decreased cell viability from 90% to 30% in EV ARP1 cells but induced much less cell death in NEK2-OE ARP1 cells (Figure 4C). Though the combination of BAY with bortezomib did not further decrease cell viability in EV ARP1 cells compared with bortezomib alone, the cell death was significantly increased in NEK2-OE ARP1 cells, with viability of approximately 40% after the combination treatment (Figure 4C). Together, these data further support the notion that NEK2 activates the NF-κB signaling pathway and plays an important role in bortezomib resistance (Figure 4D).
Figure 4. NEK2 activates the canonical NF-κB signaling pathway in primary multiple myeloma samples.
(A) Primary myeloma samples from 16 patients (Pts) were lysed and analyzed by Western blot using NEK2, p-p65-S536, total p65 (p65), USP7, and GAPDH antibodies. (B) CD138-positive myeloma cells isolated from 4 primary myeloma patients were mounted on cytospin slides and analyzed by immunofluorescence using NEK2 and p-p65-S536 antibodies. DAPI staining was used to visualize nuclei. Yellow arrowheads indicate myeloma cells coexpressing NEK2 and p-p65-S536. Blue arrowheads show myeloma cells expressing p-p65-S536 with undetectable NEK2 levels. (C) EV and NEK2-OE ARP1 cells were treated with vehicle, BAY11-7082 (0.5 or 1.0 μM), and bortezomib (5 nM) alone or in combination. After 48 hours, cell viability was assessed by trypan blue staining and Dunnett’s method was used to calculate the multiplicity-adjusted P values for each treatment and control group pair. **P = 0.0023; ****P = 0.0001. NS, no significance. Experiment was performed in triplicate. (D) A model for NEK2 deubiquitination and stabilization by interacting with USP7. USP7 prevents E3 ligase APC/C (30) to ubiquitinate NEK2 resulting in its stabilization.
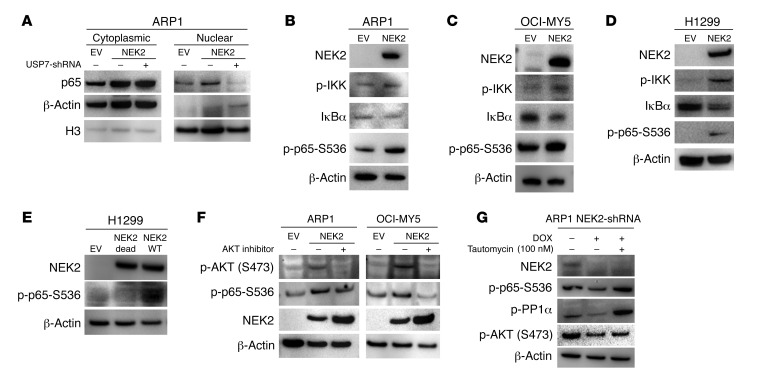
NEK2 activates NF-κB signaling via PP1α/AKT.
In addition to NF-κB–activating mutations that drive the pathway without the need of an extrinsic signal cascade, p65, the transcription factor of the NF-κB pathway, can be activated by numerous upstream signals in cancer cells, requiring IκB kinase α/β (IKKα/β) dimers (35, 36). To better understand the mechanism by which NEK2 activates NF-κB, ARP1 cells were transfected with either EV or NEK2-OE or NEK2-OE/USP7-shRNA as described in Figure 2A. Nuclear and cytoplasmic fractionations were performed in these isogenic myeloma cell lines to determine if p65 was stabilized in the nucleus of myeloma cells with high NEK2 expression. The p65 protein levels were substantially increased in both cytoplasmic and nuclear fractions in the NEK2-OE myeloma cells when compared with EV control cells (Figure 5A). Interestingly, the nuclear, but not the cytoplasmic accumulation of p65 was reversed after induction of USP7-shRNA (Figure 5A), suggesting that the stabilization of p65 is USP7 dependent. Since the IKKα/β dimer phosphorylates IκBα to facilitate p65 nuclear translocation (37, 38), ARP1, OCI-MY5, and H1299 cell lines with NEK2-OE or EV were also analyzed to test these regulators of canonical NF-κB by Western blot. Increased p-IKK, decreased IκBα, and higher levels of p-p65 at S536 were observed in NEK2-OE myeloma cell lines ARP1 (Figure 5B) and OCI-MY5 (Figure 5C), and the lung cancer cell line H1299 (Figure 5D). Because IκBα degradation by phosphorylation is required for the release of p65 into the nucleus, we also tested p-IκBα levels in NEK2-OE cancer cells. Briefly, H1299 cells were transiently transfected with EV or NEK2-OE plasmids for 24 hours and treated with MG132 to prevent protein degradation. Western blots showed that NEK2 overexpression increased phosphorylated levels of IκBα (Supplemental Figure 4A). To confirm that p65 activation was mediated by NEK2 activity, H1299 cells were transfected with EV, NEK2 wild-type cDNA (NEK2-WT), and the NEK2 K37R inactive mutant (NEK2-Dead). Western blots showed that p65 phosphorylation was increased in cells overexpressing NEK2-WT but did not change in cells expressing NEK2-Dead (Figure 5E). These data indicate that NEK2 acts as a potentially novel kinase activator of the canonical NF-κB pathway.
Figure 5. NEK2 activates NF-κB signaling via PP1α/AKT.
(A) USP7 was knocked down in ARP1 cells transduced with NEK2-OE after 72 hours induction with doxycycline (DOX). Nuclear and cytosolic fractionations were carried out. p65 levels were analyzed between EV and NEK2-OE with or without USP7 shRNA by Western blot. β-Actin and histone H3 (H3) were used as cytosolic and nuclear markers, respectively. (B–D) EV and NEK2-OE ARP1, OCI-MY5, and H1299 cells were lysed. NEK2, p65-S536 phosphorylation, IKK phosphorylation, and IκBα were analyzed by Western blot. (E) H1299 cells transiently transfected with EV or NEK2-OE (WT) or NEK2-K37R mutant (NEK2-Dead) were lysed, and NEK2 and p65-S536 phosphorylation was analyzed by Western blot. (F) ARP1 and OCI-MY5 cells transfected with EV or NEK2-OE were treated with vehicle or MK-2206 2HCl, an AKT inhibitor, for 30 minutes and then cells were lysed. p65-S536 phosphorylation was analyzed by Western blot. (G) NEK2-shRNA ARP1 cells were induced with DOX for 48 hours and then treated with tautomycin, a PP1α inhibitor, for another 24 hours. NEK2, p-p65-S536, p-PP1α, and p-AKT were analyzed by Western blot.
We have shown that NEK2 increases S473-AKT, a known activator of IKK in cancer cells (14). To further determine whether AKT mediates the activation of the canonical NF-κB pathway through NEK2, the NEK2-OE ARP1, OCI-MY5 (Figure 5F), as well as H1299 cell lines (Supplemental Figure 4B) were treated with MK-2206 2HCl (39), an AKT inhibitor, at 1 μM for 30 minutes. Western blots showed that MK-2206 2HCl suppressed NEK2-induced phosphorylation of p65 (Figure 5F), suggesting that AKT is responsible for the NEK2-driven canonical NF-κB activation. Based on our TAP-MS results and confirmation by a Co-IP experiment (data not shown), we were unable to demonstrate that NEK2 binds and activates AKT directly. However, PP1α, a phosphatase known to suppress AKT, has been shown to directly bind to NEK2 (14). Thus, we hypothesized that NEK2 phosphorylates and deactivates PP1α. Tautomycin, an established PP1α inhibitor, was used to treat ARP1 (Figure 5G) and H1299 (Supplemental Figure 4C) cells with knockdown of NEK2 by NEK2-shRNA. Tautomycin restored S473-AKT phosphorylation (Figure 5G), suggesting that the interaction between NEK2 and PP1α activates the AKT pathway resulting in increased nuclear p65.
NEK2 increases heparanase expression via NF-κB.
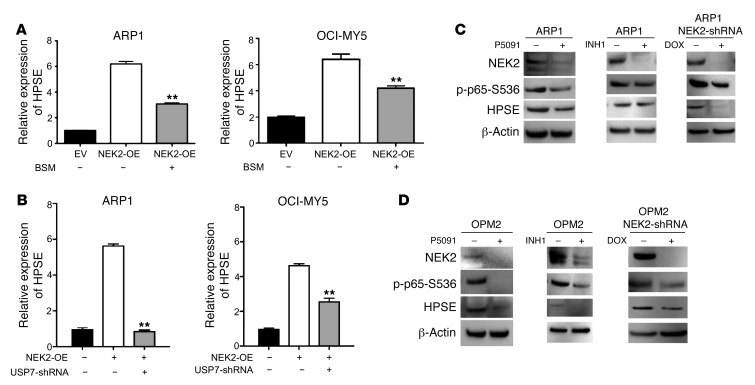
To determine whether nuclear p65 that is increased by NEK2 is functional as a transcription factor, we further evaluated the expression of a direct target in isogenic myeloma cell lines. Our group recently reported that NEK2 activates heparanase (HPSE), a secreted endoglycosidase responsible for bone destruction in myeloma patients through increased stimulation of osteoclast differentiation and that it is also a direct NF-κB target, as listed in Supplemental Table 2 (40). We found that inhibition of HPSE expression with small-molecule drugs inhibited osteoclast differentiation and osteolysis induced by NEK2 in vivo. However, the mechanism underlying the activation of HPSE by NEK2 has not been described. In this study, we verified that NEK2 expression was correlated strongly with HPSE in ARP1 and OCI-MY5 cells with NEK2-shRNA, EV, or NEK2-OE. Indeed, the expression of HPSE decreased when NEK2 was silenced and increased when NEK2 was overexpressed by GEP (Figure 6A). As shown in Figure 6B, the HPSE gene has RELA (p65) binding sites throughout the DNA sequence, suggesting it is a direct NF-κB target. To test this, chromatin immunoprecipitation (ChIP)-qPCR using antibodies against p65 was carried out in EV and NEK2-OE ARP1 cells. The results showed that NEK2-OE ARP1 cells significantly increased p65 occupancy in the HPSE gene (Figure 6C), corroborating the findings of the UCSC genome browser. To causally show that NEK2 drives HPSE through the canonical NF-κB pathway, we treated EV and NEK2-OE ARP1 cells with vehicle or the NF-κB inhibitor BAY. BAY prevented the increase in HPSE protein induced by NEK2 (Figure 6D). Furthermore, we used another NF-κB inhibitor, BSM-345541, which works by selectively targeting IKKβ, to treat NEK2-OE ARP1 and OCI-MY5 cells. Similar results were obtained and showed that BSM-345541 significantly blocked HPSE mRNA expression induced by NEK2 (Figure 7A), supporting the concept that the canonical NF-κB pathway mediates NEK2-induced HPSE activation. We have shown that NEK2 was stabilized by USP7. We then tested whether inhibition of USP7 could prevent NEK2-induced HPSE upregulation. Figure 7B demonstrates that NEK2-upregulated HPSE levels were reversed by USP7 silencing. To further confirm that NEK2 increases HPSE through NF-κB, parental ARP1 and OPM2 cells were treated with the USP7 inhibitor P5091 (overnight at 16 μM), the NEK2 inhibitor INH1 (for 24 hours at 25 μM), or knocked down by NEK2-shRNA. Western blots showed that suppressing endogenous NEK2 through P5091, INH1, or NEK2-shRNA resulted in decreased expression of p-p65-S536 and HPSE proteins (Figure 7, C and D). Taken together, these data support the notion that the canonical NF-κB pathway activated by NEK2 mediates bortezomib resistance and regulates the tumor microenvironment modulating HPSE protein expression.
Figure 6. NEK2 regulates heparanase expression via NF-κB.
(A) Total RNA from EV or NEK2-OE or NEK2-shRNA ARP1 cells were harvested for gene expression profiling; NEK2 and HPSE mRNA levels are presented as bar graphs of duplicate samples. (B) The map of p65 binding to the HPSE promoter was obtained from the UCSC genome browser ChIP-seq data (track name: GM12878+TNFa RELA). HSPE contains 2 RELA (p65) binding sites across its sequence. H3K4me3 and H3K27Ac, typical for promoter and activator binding elements, respectively, show strong peaks at the p65 binding site of the HPSE sequence (red box). (C) ChIP-qPCR was performed in control cell (EV) and NEK2-OE ARP1 myeloma cells using p65 antibodies. The DNA occupancy of the HPSE promoter was analyzed by qPCR and a Student’s t test was performed to assess the significance. *P < 0.05. (D) NEK2-OE ARP1 myeloma cells were treated with vehicle or the NF-κB inhibitor BAY11-7082 (BAY). Cell lysates from NEK2-OE ARP1 cells and the control EV ARP1 cells were probed with antibodies against NEK2, p-p65-S536, and HPSE by Western blot.
Figure 7. NEK2 regulates heparanase expression.
(A) EV and NEK2-OE ARP1 and OCI-MY5 cells were treated with vehicle or BSM-345541, and HSPE mRNA levels were analyzed by qRT-PCR. (B) HPSE mRNA levels were analyzed by qRT-PCR in EV, NEK2-OE, or NEK2-OE + USP7-shRNA ARP1 and OCI-MY5 myeloma cells. **P < 0.01 by Student’s t test. (C) ARP1 and (D) OPM2 cells were treated with P5091 (16 μM overnight), INH1 (25 μM for 24 hours), or NEK2-shRNA DOX (48 hours). Cells were lysed and proteins were analyzed by Western blot with NEK2, p-p65-S536, HPSE, and GAPDH antibodies.
INH1 prevents myeloma cell growth in vitro and in vivo.
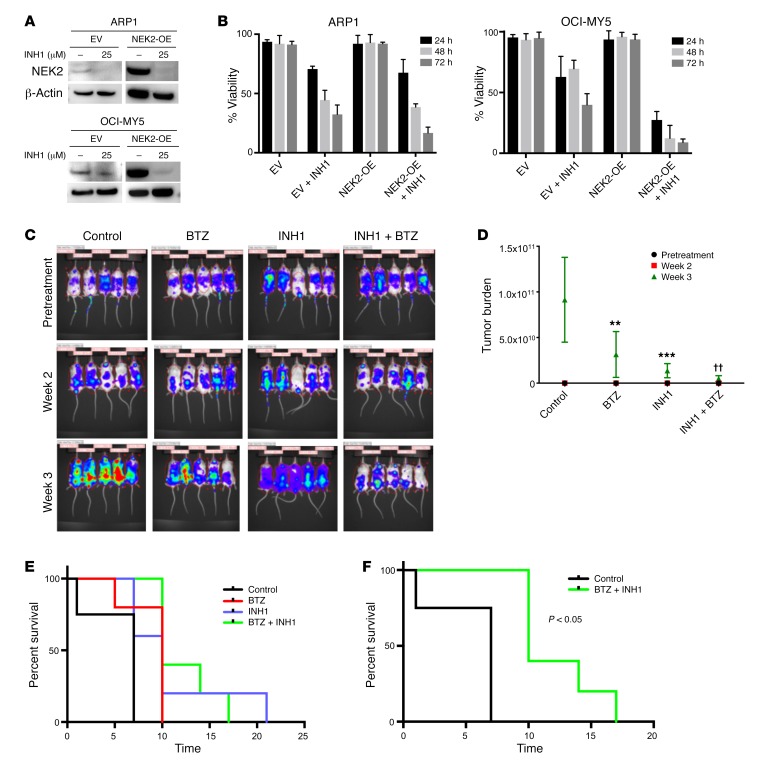
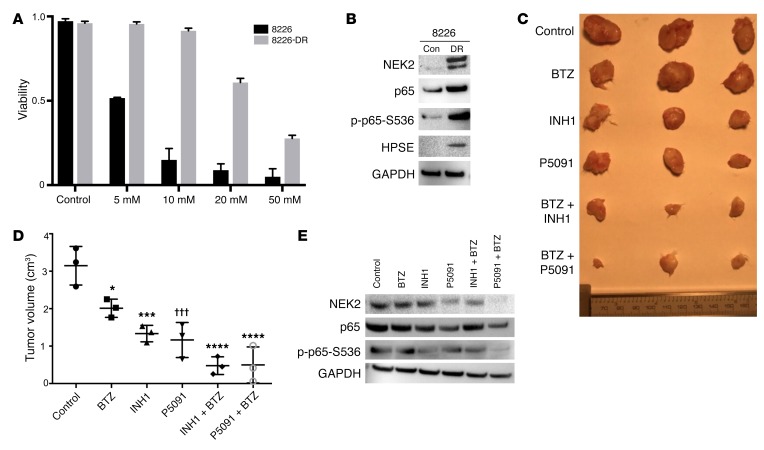
INH1 has been shown to selectively deplete NEK2 protein resulting in inhibition of cancer cell growth in both in vitro and in vivo models (41). In this study, ARP1 and OCI-MY5 myeloma cell lines with EV or NEK2-OE were treated with 25 μM INH1 for 24 hours. Western blots showed that both endogenous (EV) and overexpressed NEK2 (NEK2-OE) were substantially depleted after INH1 treatment (Figure 8A). We also tested whether INH1 was toxic to myeloma cells. Both ARP1 and OCI-MY5 myeloma cell lines with EV or NEK2-OE were treated with INH1 at 25 μM and cell viability was assessed every 24 hours for 3 days. INH1 killed both EV and NEK2-OE ARP1 and OCI-MY5 cells (Figure 8B). Interestingly, NEK2-OE ARP1 and OCI-MY5 cells, which were resistant to bortezomib, appeared more sensitive to INH1 than EV cells, showing a final viability of 20% and 10% after 3 days of INH1 treatment compared with 30% and 40% viability for the EV ARP1 and OCI-MY5 cell lines, respectively (P < 0.05). These results suggest that INH1 is an effective antimyeloma drug by blocking NEK2 gene function. An in vivo test of INH1 was performed in NOD-Rag1null mice by injecting 0.5 × 106 ARP1 cells per mouse, expressing luciferase to analyze tumor burden, through the tail vein, which allowed dissemination of cancer cells throughout the body. Mice were treated with (a) vehicle, (b) bortezomib (3 mg/kg, i.p., 2 times/week), (c) INH1 (100 mg/kg, i.p., 3 times/week), or (d) bortezomib + INH1 for 3 weeks. The tumor burden measured by luciferase intensity was significantly decreased at weeks 2 and 3 in mice treated with bortezomib or INH1 alone and additively decreased in the combination of bortezomib + INH1 when compared with the control (Figure 8, C and D). Kaplan-Meier survival analysis was performed for mice treated with these different drugs, and no significant difference was observed in mice treated with bortezomib or INH1 alone (Figure 8E), but the combination of bortezomib + INH1 significantly extended myeloma mouse survival compared with the control group (Figure 8F; P < 0.05).
Figure 8. INH1 inhibits myeloma growth in xenograft mice.
(A) EV or NEK2-OE ARP1 and OCI-MY5 myeloma cells were treated with 25 μM INH1 for 24 hours, lysed, and NEK2 levels were analyzed by Western blot. (B) EV or NEK2-OE ARP1 and OCI-MY5 myeloma cells were treated with 25 μM INH1 for 3 days. Cell viability was calculated every 24 hours using trypan blue staining. Viability experiments were performed in triplicate. (C) Approximately 0.5 × 106 ARP myeloma cells expressing luciferase were injected into NOD-Rag1null mice via the tail vein. One week after injection of ARP1 cells, mice were treated with (i) vehicle, (ii) bortezomib (BTZ; 3 mg/kg, i.p., 2 times/week), (iii) INH1 (100 mg/kg, i.p., 3 times/week), or (iv) BTZ + INH1. The tumor growth in the 4 groups of mice was evaluated using an in vivo imaging system. (D) Quantification of tumor burden was determined from C and Dunnett’s method was used to calculate the multiplicity-adjusted P values for each treatment and control group pair after 3 weeks of treatment. **P = 0.0096; ***P = 0.0008; ††P = 0.0015. (E) Kaplan-Meier curves showing survival of mice treated with BTZ, INH1, BTZ + INH1, or the control. (F) Kaplan-Meier curves showing that the combination of BTZ + INH1 significantly extended myeloma mouse survival compared with the control (P < 0.05).
Pharmacological inhibition of NEK2 or USP7 overcomes NEK2-induced bortezomib resistance.
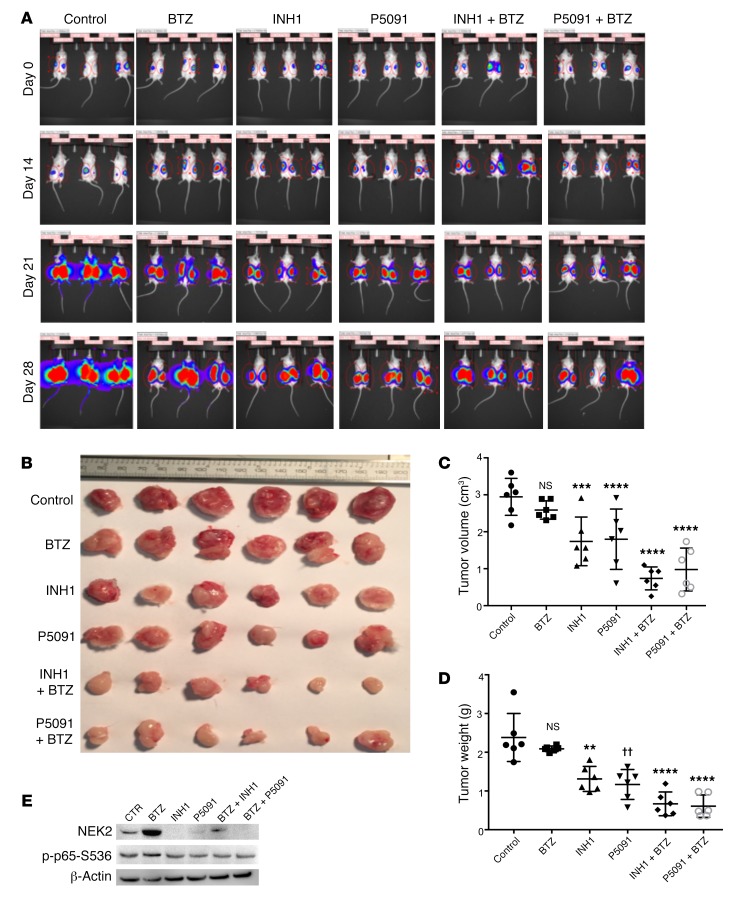
Because NEK2 confers drug resistance, we tested if NEK2 depletion, using P5091 or INH1, could overcome drug resistance in vivo using NEK2-OE ARP1 myeloma cells that are resistant to bortezomib. Approximately 0.5 × 106 ARP1 overexpressing NEK2 and luciferase were subcutaneously injected in both flanks of NOD-Rag1null mice. Mice were treated with (a) vehicle, (b) bortezomib (3 mg/kg, i.p., 2 times/week), (c) INH1 (100 mg/kg, i.p., 3 times/week), (d) P5091 (10 mg/kg, i.v., 2 times/week), (e) INH1 + bortezomib, or (f) P5091 + bortezomib for 4 weeks. No weight loss, used as toxicity side effect, was reported during the 4-week treatment (Supplemental Figure 5). As expected, bortezomib had very little effect on reducing tumor burden since NEK2-OE cells are highly resistant to this proteasome inhibitor (14). Inhibition of NEK2 with P5091 or INH1 alone had a strong antitumor effect and this effect was further increased after combination with bortezomib (Figure 9A). Tumors were harvested, photographed (Figure 9B), and analyzed for tumor size and volume at week 4. INH1 or P5091 alone and in combination with bortezomib resulted in a much smaller tumor size and weight (Figure 9, C and D). Tumor cells were isolated for Western blot. NEK2 levels were depleted in tumors treated with INH1, P5091, INH1 + bortezomib, or P5091 + bortezomib. Similarly, levels of p-p65-S536 were also suppressed in the tumor samples treated with INH1, P5091, INH1 + bortezomib, or P5091 + bortezomib, suggesting that these drugs target NEK2 and NF-κB in vivo (Figure 9E). Taken together, these data show that NEK2 inhibition by P5091 or INH1 may constitute a potential treatment for patients with advanced refractory myeloma who highly express NEK2 and are resistant to bortezomib.
Figure 9. INH1 and P5091 overcomes NEK2-induced bortezomib resistance.
(A) Approximately 0.5 × 106 NEK2-OE ARP1 cells expressing luciferase were injected subcutaneously in both left and right flanks of NOD-Rag1null mice. One week after injection of NEK2-OE cells, mice were treated with (i) vehicle, (ii) bortezomib (BTZ; 3 mg/kg, i.p., 2 times/week), (iii) INH1 (100 mg/kg, i.p., 3 times/week), (iv) P5091 (10 mg/kg, i.v., 2 times/week), (v) INH1 + BTZ, or (vii) P5091 + BTZ for 28 days. In vivo imaging showing the tumor growth in the different groups of mice before and after treatments at different time points. (B) Tumors from A were harvested and photographed. (C) Quantification of tumor volume from dissected tumors in B and Dunnett’s method was used to calculate the multiplicity-adjusted P values for each treatment and control group pair. ***P = 0.005; ****P = 0.0001. NS, no significance. (D) Quantification of tumor weight from dissected tumors in B and Dunnett’s method was used to calculate the multiplicity-adjusted P values for each treatment and control group pair. **P = 0.0032; ††P = 0.0052; ****P = 0.0001. NS, no significance. (E) Tumors from B were lysed and analyzed by Western blot using NEK2, p-p65-S536, and GAPDH antibodies.
Targeting NEK2 overcomes acquired drug resistance in vivo.
To correlate endogenous NEK2 levels with drug resistance, we examined the parental RPMI-8226 myeloma cell line (8226) and the RPMI-8226 drug-resistant cell line (8226-DR), which acquired drug resistance by prolonged exposure to a low dose of bortezomib. We first confirmed the sensitivity to bortezomib of these 2 cell lines. As shown in Figure 10A, the 8226-DR cells were markedly resistant to bortezomib, while 8226 cells were sensitive to bortezomib. As expected, the expression of NEK2, p-p65-S536, and HPSE was substantially increased, as determined by Western blot, in 8226-DR cells compared with parental 8226 cells (Figure 10B). An in vivo study was performed to determine whether targeting NEK2 could overcome acquired drug resistance. Approximately 0.5 × 106 8226-DR cells were subcutaneously injected into both flanks of NOD-Rag1null mice. Mice were treated with (a) vehicle, (b) bortezomib (3 mg/kg, i.p., 2 times/week), (c) INH1 (100 mg/kg, i.p., 3 times/week), (d) P5091 (10 mg/kg, i.v., 2 times/week), (e) INH1 + bortezomib, or (f) P5091 + bortezomib for 4 weeks. Results showed that bortezomib did not significantly change tumor volume. However, the combination of INH1 or P5091 with bortezomib significantly reduced tumor size (Figure 10, C and D). Western blots showed that the expression of NEK2 and p-p65-S536 was significantly decreased after treatment with P5091 or INH1 or in combination with bortezomib (Figure 10E). These data indicate that NEK2 may be a strong therapeutic target to overcome acquired drug resistance.
Figure 10. Targeting NEK2 overcomes acquired drug resistance in vivo.
(A) The parental RPMI-8226 (8226) and drug-resistant RPMI-8226 (8226-DR) myeloma cells were treated with different bortezomib (BTZ) concentrations. Cell viability was analyzed by trypan blue staining after 48 hours. Viability experiments were performed in triplicate. (B) Western blots showing the expression of NEK2, p65, p-p65-S536, HPSE, and GAPDH in 8226 and 8226-DR myeloma cells. (C) Images of representative dissected tumors from mice treated with (i) vehicle, (ii) BTZ, (iii) INH1, (iv) P5091, (v) INH1 + BTZ, or (vi) P5091 + BTZ for 28 days. (D) Tumor volume was quantified from dissected tumors in C and Sidak’s method was used to calculate the multiplicity-adjusted P values for active treatment groups versus control. *P = 0.0172; ***P = 0.0004; †††P = 0.0002; ****P < 0.0001. (E) Western blots showing the expression of NEK2, p65, p-p65-S536, and GAPDH from dissected tumors in C.
Discussion
Drug resistance remains the key problem in cancer treatment. The number of myeloma patients that eventually relapse and die due to drug-resistant subclone(s) is estimated at 12,000 annually (42–46). Specific molecular genetic lesions and tumor cell–stroma interactions influence the markedly variable clinical course and response to therapy (47). By applying interphase fluorescence in situ hybridization (FISH) and chromosomal microarrays, almost all myeloma patients have been found to be cytogenetically abnormal (48–56). Myeloma must therefore be characterized by significant genomic or CIN. Our recent study demonstrates that an increased CIN signature is associated with increased cell proliferation and drug resistance, and poor prognosis (14). High expression of NEK2, a key CIN gene, has been shown to induce premature splitting of centrosomes (57, 58), while suppression of NEK2 blocks spindle and chromosome segregation, resulting in accumulation of multiple centrosomes (59–61). These data support a model in which increased endogenous NEK2 protein levels promote centrosome separation by triggering dissolution of a putative intercentriolar linkage resulting in CIN in cancer cells. However, despite the advances described above, which were mostly made in normal cells, the biological functions of NEK2 in cancer in general and myeloma in particular are still poorly understood. Here, we aim at uncovering our knowledge gap about the observed NEK2 accumulation in cancer cells. We unveil what we believe to be a novel interaction that stabilizes NEK2 protein, allowing us to better understand why NEK2 protein levels are increased in myeloma, resulting in increased drug resistance.
We found that NEK2 activates the canonical NF-κB pathway based on the following observations: (a) NEK2 expression is positively correlated with increased NF-κB score in primary myeloma samples; (b) more than 40% of genes activated or inhibited by NEK2 are direct targets of p65; (c) the dual-luciferase reporter assay showed an increased transactivity of p65 in NEK2-OE cells; (d) ChIP-qPCR confirmed the functional activity of p65, which directly binds to the human genomic DNA sequences of HPSE and is upregulated in NEK2-OE cells; (e) importantly, we found that NEK2-positive myeloma cells showed elevated phosphorylated p65 in primary myeloma samples by both Western blot and immunofluorescence; and (f) overexpression of NEK2 decreases IκBα protein (an inhibitor of the classical NF-κΒ pathway), and increases nuclear p65. The IKKα inhibitor BAY11-7082 and the IKKβ inhibitor BMS-345541 can block NEK2-upregulated HPSE expression, further confirming that NEK2 regulates HPSE expression through activation of the IKK complex (IKKα/β/γ). The direct link between NEK2 and NF-κB function is potentially novel. The IKK complex can be activated directly by TAK1, TBK1, and AKT kinases or by K63- or M1-linked polyubiquitin chains, and leads to ubiquitination and proteasome degradation of IκBα in cancer cells (62). We have shown that NEK2 directly binds to PP1α (14), which is an inhibitor of the AKT pathway (63). Given that NEK2 activates AKT, and that the AKT pathway controls NF-κB activity, we evaluated whether activation of NF-κB by NEK2 is PP1α/AKT dependent. We found that NEK2 inhibits PP1α activity by its phosphorylation; this drives AKT phosphorylation at residue S473. Phosphorylated AKT cascades into the canonical NF-κB pathway by activating IKK, which degrades IκBα and releases p65, resulting in nuclear translocation. Though we demonstrate that the NEK2 kinase activates the canonical NF-κB signaling in a PP1α/AKT–dependent manner, 5 master transcription factors, i.e., NF-κB1/p105, NF-κB2/p100, RelA/p65, RelB, and RelC, are major members of the NF-κB family, and both canonical and noncanonical NF-κB pathways play a critical role in myeloma pathogenesis and progression (31, 64). Future work should determine whether NEK2 activates only the canonical NF-κB or also noncanonical NF-κB pathway in myeloma.
The advent of proteasome inhibitors has revolutionized myeloma treatment. Currently, bortezomib is used as an induction and maintenance therapy in conjunction with ASCT. However, bortezomib has dose-limiting toxicities and eventually myeloma cells become insensitive. A more targeted approach is required to overcome this resistance. Here we discovered that USP7, a DUB, is a NEK2-binding protein. We confirmed this USP7-NEK2 interaction in various cell lines and that this interaction leads to the deubiquitination and stabilization of NEK2. This stabilization mechanism provides an explanation for how NEK2 overcomes proteasomal degradation. NEK2 is not amplified or mutated, while UPS7 activity is increased in all myeloma cells. DUBs have been receiving attention due to their numerous roles in stabilizing oncogenes, displacing and deactivating tumor suppressors, and other activities. Within the 98 members of the DUB proteome, the USP family has a very prominent role in cancer-promoting proteins and has very differing amino acid sequences (65, 66). USP7, a member of the USP family, has many functions, such as displacing PTEN out of the nucleus, which is linked to more aggressive cancer behavior. Other functions include stabilization of MDM2, an E3 ligase for the tumor suppressor P53, and stabilization of the oncogenic TRAF6, which has been implicated in AKT activation (11, 12, 67, 68). High expression of USP7 is a predictor of poor prognosis in lung squamous cell carcinoma and large cell lymphoma. It promotes tumorigenesis in breast cancer and has been reported to be consistently overexpressed in myeloma (13, 69, 70). In this study, USP7-shRNA was shown to reverse these tumor-promoting effects, and P5091, a nonreversible USP7 inhibitor, showed strong cytotoxic effects on myeloma cells and was able to overcome NEK2-induced bortezomib resistance in in vitro and in vivo models, suggesting that this posttranslational modification also plays an important role in NEK2-related cell apoptosis, survival, and drug resistance in myeloma.
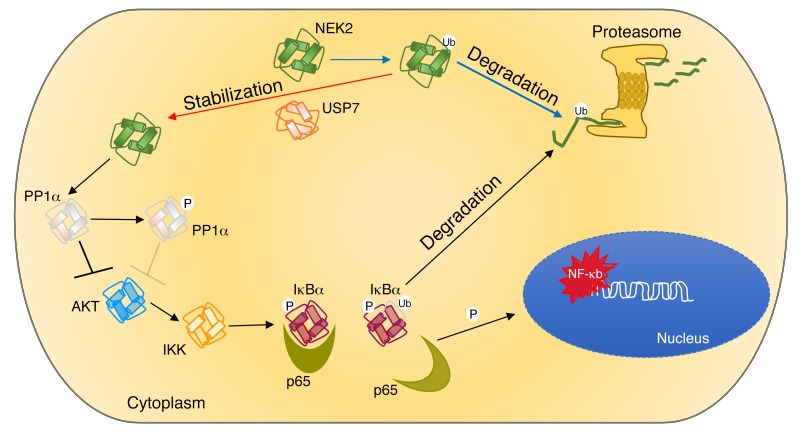
Under normal conditions NEK2 is present during most stages of the cell cycle except mitosis. Before mitosis, the KEN box near the C terminus is ubiquitinated and targeted for proteasomal degradation (30). We used INH1, another small-molecule drug that inhibits NEK2 binding to HEC1, which promotes its proteasomal degradation (41). As was the case with P5091, INH1 proved to be more cytotoxic for NEK2-OE cancer cells in vitro and in vivo. Based on these observations, we believe that NEK2 is being upregulated transcriptionally and escapes proteasomal degradation by increased deubiquitination. We found that NEK2 inhibitors and USP7 inhibitors had a similar effect on myeloma cells. Together, our data present a potentially novel mechanism by which the interaction of NEK2 with USP7 drives drug resistance in multiple myeloma. Figure 11 outlines every step of our research. Based on the results, we summarize that (a) USP7 leads to the deubiquitination and prevention of proteasomal degradation of NEK2, resulting in accumulation of this protein in myeloma cells. (b) The increased levels of NEK2 bind and phosphorylate PP1α, suppressing its phosphatase activity thus promoting AKT activation. And (c) AKT activation results in phosphorylation of the IKKα/β complex, triggering the canonical NF-κB cascade, which phosphorylates and targets IκBα for proteasomal degradation, releasing the sequestered p65 phosphorylated at S536 so that it can translocate to the nucleus, where it drives bortezomib resistance by activating its target genes. Our data also show that INH1 and P5091 can deplete NEK2 protein in vitro and in vivo, resulting in suppression of NEK2 downstream targets, thus overcoming bortezomib resistance. NEK2 activity is increased in the chemotherapy-surviving, minimal residual disease cells in complete remission. Therefore, we postulate that therapeutic targeting of NEK2, using either NEK2 or USP7 inhibitors, in patients with myeloma may be most effective as an adjuvant therapy when tumor mass is already extremely debulked.
Figure 11. Working model of the interaction between NEK2 and USP7.
USP7 binds to and stabilizes NEK2 by deubiquitination, allowing it to accumulate in myeloma cells. Accumulated NEK2 binds to and phosphorylates PP1α, resulting in loss its AKT-suppressing activity. Active AKT triggers the canonical NF-κB pathway by phosphorylating IKK, with subsequent phosphorylation and degradation of IκBα. p65 released from the complex with IκBα translocates into the nucleus, where it activates its target genes leading to drug resistance in myeloma. Ub, ubiquitin.
Methods
Cell culture.
Human myeloma cell lines (ARP1, OCI-MY5, and their derivative cell lines), a gift from John D. Shaughnessy Jr., University of Arkansas (Little Rock, Arkansas, USA), were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Invitrogen), penicillin (100 IU/ml), and streptomycin (100 μg/ml) in a humidified incubator at 37°C and 5% CO2/95% air. HEK293T and H1299 were cultured with DMEM (Invitrogen) with the same supplements.
Vectors and transfections.
NEK2 was subcloned into the pCDH vector under the CAG promoter and tagged with 3× FLAG and HA. The USP7-Flag vector was obtained from Addgene (plasmid 16655). Ubiquitin-HA was subcloned pcDNA3 (Addgene). The NEK2 K37R inactive mutant (NEK2-Dead) was a gift from Andrew M. Fry (University of Leicester, Leicester, United Kingdom). The shRNA sequence for USP7 is 5′-ATACAGTGTGTTCAAAGTA-3′ and was inserted into EcoRI and Xhol sites of the doxycycline-inducible expression vector pTRIPZ. Lentiviruses were packaged in HEK293T cells using VSV-G and psPAX2 helper vectors (Addgene) and calcium phosphate transfection (Promega). Transient transfections were done with Lipofectamine 2000 reagent according to the manufacturer’s guidelines (Invitrogen).
TAP-MS.
NEK2-bound proteins were sequentially purified using a FLAG HA Tandem Affinity Purification Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Eluted protein was run through SDS-PAGE and silver stained (Bio-Rad). Bands were excised and identified by high-resolution MS analysis at the proteomics core facility at the University of Iowa.
Cell lysis, immunoprecipitation, and Co-IP.
For total cell lysis, buffer containing 150 mM NaCl, 10 mM EDTA, 10 mM Tris pH 7.4, and 10% Triton X-100 was used and supplemented with Protease inhibitor cocktail (BioVision, Inc.) and DTT. For detection of phosphorylated proteins, antiphosphatase mini tablets (Thermo Fisher Scientific) were added to the lysis buffer. Cell pellets were resuspended in lysis buffer, vortexed vigorously for 15 seconds, and incubated on ice for 10 minutes, vortexed again, and centrifuged at 16,200 g for 10 minutes at 4°C. Lysed cell pellets were discarded and supernatant (protein lysate) was analyzed by Western blot or frozen at –80°C. For cell fractionations, a Nuclear/Cytosol Fractionation kit (BioVision, Inc.) was used according to the manufacturer’s protocol. For immunoprecipitations to probe ubiquitinated proteins, cells were lysed using regular lysis buffer described above supplemented with NEM (10 mM) to prevent deubiquitination. Lysates were collected and the same amounts of protein were incubated with 25 μl of protein G Dynabeads and 3 μl of anti-NEK2 antibody (Santa Cruz Biotechnology, D-8, sc-55601) overnight. Samples were washed 3 times with lysis buffer and eluted by rotating 20 minutes at room temperature with 350 mM DTT–supplemented sample buffer. Co-IPs were done using IP buffer (Thermo Fisher Scientific) supplemented with protease inhibitor cocktail. Samples were vortexed on low for 5 seconds, incubated on ice for 20 minutes, vortexed again, and centrifuged 16,200 g for 10 minutes.
Western blot.
Nanodrop (Thermo Fisher Scientific) was used to calculate cell lysate protein concentration and 100 μg of protein was used per well. Proteins were separated with NuPAGE 4% to 12% Bis-Tris Gel (NOVEX) at 200 V, then transferred to a nitrocellulose membrane for 1 hour at 400 mA at 4°C. The membrane was blocked for 20 minutes with 10% milk at 37°C or with 5% BSA overnight for phosphorylation antibodies. Antibodies against NEK2 (Santa Cruz Biotechnology, D-8, sc-55601), USP7 [Cell Signaling Technologies, HAUSP (D17C6) XP], GAPDH (Cell Signaling Technologies, 14C10), β-actin (Cell Signaling Technologies, D6A8), ubiquitin (Cell Signaling Technologies, 3933S), Flag (Sigma-Aldrich, monoclonal M2 F1804-200UG), IκBα (Cell Signaling Technologies, 9242S), phospho-IKK [Cell Signaling Technologies, p-IKK-α/β (S176/180), 16A6], p-p65-S536 [Cell Signaling Technologies, p-NF-κB p65 (S536), 93H1], HA (Cell Signaling Technologies, C29F4), and p65 [Cell Signaling Technologies, NF-κB p65 (D14E12) XP] were incubated overnight at a dilution of 1:1,000. Secondary rabbit antibody (ANASPEC, goat anti-rabbit IgG [H+L], HRP-conjugated) and secondary mouse antibody (Santa Cruz Biotechnology, goat anti-mouse IgG-HRP, sc-2005) were incubated for 1 hour at a concentration of 1: 10,000. For exposure, Immobilon Western HRP Substrate Peroxide Solution from Millipore was used. Imaging was done with a Bio-Rad ChemiDoc XRS+ with Image Lab Software.
Drug treatment conditions.
For shRNA induction, doxycycline was used at 100 ng/ml for 48 or 72 hours. P5091 (Selleckchem) was solubilized in DMSO at a concentration of 16 μM or 25 μM. MG132 (Selleckchem) was solubilized in DMSO and used at 10 μM for 5 hours. NF-κB inhibitor BSM-345541 (Selleckchem) was solubilized in DMSO and treated at 10 μM and BAY11-7082 (Selleckchem) was used at 0.5 or 1.0 μM. Bortezomib (Selleckchem) was used at 5 nM for 48 or 72 hours. INH1 (Selleckchem) was used at 25 μM. AKT inhibitor MK-2206 2HCl (Selleckchem) was used for 0.25 μM for 3 hours. Tautomycin (Selleckchem) was used at 100 nM for 24 hours. NEM was solubilized in ethanol and added to lysis buffer immediately before use at a concentration of 10 mM.
Cell viability.
Cell culture (10 μl) was mixed with 10 μl of trypan blue and samples were counted in a hemocytometer; translucent cells were counted as viable and blue-stained cells were counted as dead. Cell viability was calculated by dividing viable cells by total cell number; each sample was done in triplicate.
Luciferase-Renilla reporter assay.
NEK2, p52 (pCMV4-p52, Addgene) (negative control), luciferase (under p65-dependant promoter, pGL3-2×NF-κB-Luc), and Renilla (pRL-null, E2271; Promega) (internal control) vectors were transfected into H1299 cells using Lipofectamine 2000. After 24 hours, cells were harvested and a luciferase reporter assay was performed using Dual-Luciferase Reporter Assay System (Promega) under the manufacturer’s guidelines. Readings were done using a Berthold Detection Systems FB12 Luminometer with FB12 Sirius Software Version 1.5. All samples were done in triplicate.
ChIP.
The binding of the NF-κB p65 subunit to DNA in ARP1 was quantified by ChIP-qPCR. Briefly, chromatin (5 μg) from the ARP1 myeloma cell line was used in ChIP assay using antibodies (4 μg) against NF-κB p65 (Santa Cruz Biotechnology, sc-372). The ChIPed DNA fragments were quantified by Qubit DNA assay kit (Life Technologies), and the enrichment of DNA fragments that contain putative NF-κB binding sites in the gene promoter was quantified by qPCR containing DNA fragments using a Two-Color Real-Time PCR Detection System (Bio-Rad). The primers for HPSE used in the ChIP-qPCR assay are forward 5′-TGGAGAGCTGCCTGGAGATT-3′ and reverse 5′-AACCTCGAGCAGATGGCAGT-3′.
qPCR.
For quantitative analysis of gene expression, total RNA was isolated by RNeasy kit (Qiagen). Complementary DNA was synthesized using an Iscript reverse transcription kit according to the manufacturer’s instructions (Bio-Rad). Real-time qPCRs for human NEK2, HPSE, and β-actin were performed with SYBR Green Super Mixture Reagents (Bio-Rad) on the CFX Connect real-time system (Bio-Rad). PCR was initiated at 95°C for 3 minutes to hot-start the DNA polymerase and denature the template, and then 40 cycles consisting of denaturing at 95°C for 30 seconds, annealing and extension at 60°C for 30 seconds.
Immunofluorescence.
Bone marrow aspirates from human myeloma patients were sorted with CD138 magnetic beads and mounted onto cytospin slides for this study. Myeloma cells were fixed in 4% formaldehyde, and primary antibodies against NEK2 (Santa Cruz Biotechnology, D-8, sc-55601) and p-p65-S536 [Cell Signaling Technologies, p-NF-κB p65 (S536), 93H1] were added at a final dilution of 1:100. After overnight incubation of primary antibodies at 4°C, the secondary antibodies goat anti-mouse Alexa Flour-568 (Invitrogen) and goat anti-rabbit Alexa Flour 488 (Invitrogen) were added at a final dilution of 1:300 for 1 hour at room temperature. The slides were washed and mounted with DAPI. Images were obtained using a spinning disk confocal microscope (PerkinElmer UltraVIEW Vox).
Mice xenograft models.
For Figure 8, NOD.Cγ-Rag1 mice 6–8 weeks old (Jackson Laboratories) were injected with 0.5 million ARP1 + luciferase by tail vein injection to allow full-body dissemination. Mice were treated with one of the following: (a) bortezomib (3 mg/kg, i.p.), (b) INH1 (100 mg/kg, i.p.), (c) bortezomib (3 mg/kg, i.p.) + INH1 (100 mg/kg, i.p.), or (d) untreated control. Every 7 days for 3 weeks mice were imaged using a Carestream IN-Vivo MS FX Pro. Exposure time was kept constant in all images and tumor burden were analyzed by selecting areas around mice and quantifying pixel intensity for each mouse of each group. After stopping treatment, mice were kept alive for survival analysis. Mice were sacrificed when hind leg paralysis was evident.
For Figure 9, NOD.Cγ-Rag1 mice 6–8 weeks old (Jackson Laboratories) were injected subcutaneously with 0.5 million ARP1 NEK2-OE + luciferase into the left and right flanks. For 4 weeks, mice were treated with one of the following: (a) P5091 (10 mg/kg, i.v.), (b) INH1 (100 mg/kg, i.p.), (c) bortezomib (3 mg/kg, i.p.), (d) P5091 (10 mg/kg, i.v.) + bortezomib (3 mg/kg, i.p.), (e) INH1 (100 mg/kg, i.p.) + bortezomib (3 mg/kg, i.p.), or (f) untreated control. Every 7 days for 4 weeks the luciferase intensity of each mouse was imaged using the Carestream IN-Vivo MS FX Pro. After sacrificing, the mouse tumors were harvested, weighed, and measured with calipers to determine volume. Following the measurements, the tumors were stored at –80°C for later protein processing.
For Figure 10, NOD.Cγ-Rag1 mice 6–8 weeks old (Jackson Laboratories) were injected subcutaneously with 0.5 million 8226-DR into the left and right flanks. For 4 weeks, mice were treated with one of the following: (a) control, (b) bortezomib (3 mg/kg, i.p.), (c) INH1 (100 mg/kg, i.p.), (d) P5091 (10 mg/kg, i.v.), (e) INH1 (100 mg/kg, i.p.) + bortezomib (3 mg/kg, i.p.), or (f) P5091 (10 mg/kg, i.v.) + bortezomib (3 mg/kg, i.p.). After sacrificing, the mouse tumors were harvested to determine volume. Following the measurements, the tumors were stored at –80°C for later protein processing.
Microarray data sets.
Gene expression profiling data were obtained from previous studies and are available under the NCBI’s Gene Expression Omnibus under accession numbers GSE2658 (4, 6) and GSE19784 (34). For Figure 3D, we developed an NF-κB score for each sample obtained from the ratio of the average log2-scale expression of the 17 NEK2 positive genes (including NEK2) to 15 NEK2-negative genes. To validate the clinical relevance of NEK2/NF-κB signaling observed in the 351 TT2 samples, we analyzed the HOVON-65 cohort including 268 newly diagnosed myeloma samples. We present validation based on an independent cohort treated under a different treatment protocol in order to illustrate and provide strong supporting evidence for the association of the NEK2/NF-κB upregulated/downregulated mean ratio with EFS and OS.
Statistics.
Results are presented as average ± SD or as average ± SEM, as indicated in the figure legends. Statistical analysis was done using GraphPad Prism 6.05 and Prism 7.0. All other comparisons were analyzed by unpaired, 2-sided, independent Student’s t test, unless otherwise described in the figure legends. P < 0.05 was considered significant. In correlations of NEK2 and NF-κB score for patient survival, EFS and OS were determined using the Kaplan-Meier method, and the log-rank test was used for group comparison. Significance was set at P < 0.05.
Study approval.
Bone marrow aspirates were obtained from multiple myeloma patients. Written informed consent was obtained from all participants. The study was approved by the institutional State Key Laboratory of Experimental Hematology, Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Science & Peking Union Medical College, Tianjin, China. NOD.Cγ-Rag1 mice (Jackson Laboratories) were bred and maintained in compliance with the guidelines of the institutional animal care at the University of Iowa.
Author contributions
RFM and MH performed the experiments, collected and analyzed the data, generated the figures, and wrote and edited the manuscript. HB, ZG, and XZ performed experiments, collected and analyzed the data. HH provided technical counseling on experiments and edited the manuscript. IF supervised experiments, analyzed data, and edited the manuscript. YJ and LQ reviewed the data and edited the manuscript. FZ and GT designed and supervised this study, collected and analyzed data, wrote and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01CA152105 (to F. Zhan); the Multiple Myeloma Research Foundation (to F. Zhan); the International Myeloma Foundation (to F. Zhan); the America Society of Hematology (ASH) Bridge (to F. Zhan); the Leukemia and Lymphoma Society TRP (6094-12); the NIH Lymphoma Spore grant P50 CA097274; institutional start-up funds from the Department of Internal Medicine, Carver College of Medicine, University of Iowa (to F. Zhan); Cancer Center Support Grant NIH number P30 CA086862, and the Nature Science Foundation of China 81630007 (to L. Qiu).
Version 1. 06/04/2018
Electronic publication
Version 2. 07/02/2018
Print issue publication
Footnotes
ZG’s present address is: Children’s Institute at UT Southwestern, Dallas, Texas, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(7):2877–2893.https://doi.org/10.1172/JCI98765.
Contributor Information
Reinaldo Franqui-Machin, Email: reinaldo-franquimachin@uiowa.edu.
Mu Hao, Email: haomusg@163.com.
Hua Bai, Email: hua-bai@uiowa.edu.
Zhimin Gu, Email: Zhimin.Gu@UTSouthwestern.edu.
Xin Zhan, Email: xin-zhan@uiowa.edu.
Hasem Habelhah, Email: hasem-habelhah@uiowa.edu.
Yogesh Jethava, Email: yogesh-jethava@uiowa.edu.
Lugui Qiu, Email: qiulg@ihcams.ac.cn.
Ivana Frech, Email: ivana-frech@uiowa.edu.
Guido Tricot, Email: guido-tricot@uiowa.edu.
Fenghuang Zhan, Email: fenghuang-zhan@uiowa.edu.
References
- 1.Kumar SK, et al. Multiple myeloma. Nat Rev Dis Primers. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy PL, et al. Lenalidomide maintenance after autologous stem-cell transplantation in newly diagnosed multiple myeloma: a meta-analysis. J Clin Oncol. 2017;35(29):3279–3289. doi: 10.1200/JCO.2017.72.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreau P, Attal M, Facon T. Frontline therapy of multiple myeloma. Blood. 2015;125(20):3076–3084. doi: 10.1182/blood-2014-09-568915. [DOI] [PubMed] [Google Scholar]
- 4.Zhan F, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.[No authors listed] Erratum for the report: “Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells” by W. Qasim, et al. Sci Transl Med. 2017;9(377):aam9292. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 6.Shaughnessy JD, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 7.Hanamura I, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108(5):1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drach J, et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92(3):802–809. [PubMed] [Google Scholar]
- 9.Nikesitch N, Ling SC. Molecular mechanisms in multiple myeloma drug resistance. J Clin Pathol. 2016;69(2):97–101. doi: 10.1136/jclinpath-2015-203414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung-Hagesteijn C, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24(3):289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 12.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W, et al. ALDH1 activity identifies tumor-initiating cells and links to chromosomal instability signatures in multiple myeloma. Leukemia. 2014;28(5):1155–1158. doi: 10.1038/leu.2013.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Z, Xia J, Xu H, Frech I, Tricot G, Zhan F. NEK2 promotes aerobic glycolysis in multiple myeloma through regulating splicing of pyruvate kinase. J Hematol Oncol. 2017;10(1):17. doi: 10.1186/s13045-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17(2):470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, et al. Abnormal expression of Nek2 and β-catenin in breast carcinoma: clinicopathological correlations. Histopathology. 2011;59(4):631–642. doi: 10.1111/j.1365-2559.2011.03941.x. [DOI] [PubMed] [Google Scholar]
- 19.Jeong Y, Lee J, Kim K, Yoo JC, Rhee K. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J Cell Sci. 2007;120(pt 12):2106–2116. doi: 10.1242/jcs.03458. [DOI] [PubMed] [Google Scholar]
- 20.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237(2):155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Fu G, et al. Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 2007;17(7):608–618. doi: 10.1038/cr.2007.55. [DOI] [PubMed] [Google Scholar]
- 22.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141(7):1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahmanyar S, et al. β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22(1):91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, et al. Targeting NEK2 attenuates glioblastoma growth and radioresistance by destabilizing histone methyltransferase EZH2. J Clin Invest. 2017;127(8):3075–3089. doi: 10.1172/JCI89092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Velimezi G, et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol. 2013;15(8):967–977. doi: 10.1038/ncb2795. [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Franqui Machin R, Gu Z, Zhan F. Role of NEK2A in human cancer and its therapeutic potentials. Biomed Res Int. 2015;2015:862461. doi: 10.1155/2015/862461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;31(2):745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]
- 28.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Lill JR, Wertz IE. Toward understanding ubiquitin-modifying enzymes: from pharmacological targeting to proteomics. Trends Pharmacol Sci. 2014;35(4):187–207. doi: 10.1016/j.tips.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001;20(24):7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlogie B, et al. Thalidomide and hematopoietic-cell transplantation for multiple myeloma. N Engl J Med. 2006;354(10):1021–1030. doi: 10.1056/NEJMoa053583. [DOI] [PubMed] [Google Scholar]
- 33.Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21(1):71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broyl A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116(14):2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 35.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12(8):695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 36.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beg AA, Baldwin AS. The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7(11):2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 38.Shrum CK, Defrancisco D, Meffert MK. Stimulated nuclear translocation of NF-κB and shuttling differentially depend on dynein and the dynactin complex. Proc Natl Acad Sci U S A. 2009;106(8):2647–2652. doi: 10.1073/pnas.0806677106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa C, et al. Measurement of PIP3 levels reveals an unexpected role for p110β in early adaptive responses to p110α-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27(1):97–108. doi: 10.1016/j.ccell.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao M, et al. NEK2 induces osteoclast differentiation and bone destruction via heparanase in multiple myeloma. Leukemia. 2017;31(7):1648–1650. doi: 10.1038/leu.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu G, et al. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008;68(20):8393–8399. doi: 10.1158/0008-5472.CAN-08-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bataille R, Harousseau JL. Multiple myeloma. N Engl J Med. 1997;336(23):1657–1664. doi: 10.1056/NEJM199706053362307. [DOI] [PubMed] [Google Scholar]
- 43.Boccadoro M, Pileri A. Diagnosis, prognosis, and standard treatment of multiple myeloma. Hematol Oncol Clin North Am. 1997;11(1):111–131. doi: 10.1016/S0889-8588(05)70418-4. [DOI] [PubMed] [Google Scholar]
- 44.Barlogie B, et al. Standard chemotherapy compared with high-dose chemoradiotherapy for multiple myeloma: final results of phase III US Intergroup Trial S9321. J Clin Oncol. 2006;24(6):929–936. doi: 10.1200/JCO.2005.04.5807. [DOI] [PubMed] [Google Scholar]
- 45.Segeren CM, et al. Overall and event-free survival are not improved by the use of myeloablative therapy following intensified chemotherapy in previously untreated patients with multiple myeloma: a prospective randomized phase 3 study. Blood. 2003;101(6):2144–2151. doi: 10.1182/blood-2002-03-0889. [DOI] [PubMed] [Google Scholar]
- 46.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 47.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2(3):175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 48.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121(Pt 23):3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 49.Anguiano A, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27(25):4197–4203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bochtler T, et al. Evaluation of the cytogenetic aberration pattern in amyloid light chain amyloidosis as compared with monoclonal gammopathy of undetermined significance reveals common pathways of karyotypic instability. Blood. 2008;111(9):4700–4705. doi: 10.1182/blood-2007-11-122101. [DOI] [PubMed] [Google Scholar]
- 51.Ho PJ, Campbell LJ, Gibson J, Brown R, Joshua D. The biology and cytogenetics of multiple myeloma. Rev Clin Exp Hematol. 2002;6(3):276–300. doi: 10.1046/j.1468-0734.2002.00081.x. [DOI] [PubMed] [Google Scholar]
- 52.Calasanz MJ, et al. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997;18(2):84–93. doi: 10.1002/(SICI)1098-2264(199702)18:2<84::AID-GCC2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 53.Zandecki M, Laï JL, Facon T. Multiple myeloma: almost all patients are cytogenetically abnormal. Br J Haematol. 1996;94(2):217–227. doi: 10.1046/j.1365-2141.1996.d01-2939.x. [DOI] [PubMed] [Google Scholar]
- 54.Gabrea A, Leif Bergsagel P, Michael Kuehl W. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair (Amst) 2006;5(9–10):1225–1233. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance. Br J Haematol. 2006;134(6):573–589. doi: 10.1111/j.1365-2141.2006.06235.x. [DOI] [PubMed] [Google Scholar]
- 56.Iida S, Ueda R. Multistep tumorigenesis of multiple myeloma: its molecular delineation. Int J Hematol. 2003;77(3):207–212. doi: 10.1007/BF02983776. [DOI] [PubMed] [Google Scholar]
- 57.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci. 2002;115(pt 16):3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 58.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151(4):837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14(7):2876–2889. doi: 10.1091/mbc.e03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonn S, Khang I, Kim K, Rhee K. Suppression of Nek2A in mouse early embryos confirms its requirement for chromosome segregation. J Cell Sci. 2004;117(pt 23):5557–5566. doi: 10.1242/jcs.01476. [DOI] [PubMed] [Google Scholar]
- 61.Prigent C, Glover DM, Giet R. Drosophila Nek2 protein kinase knockdown leads to centrosome maturation defects while overexpression causes centrosome fragmentation and cytokinesis failure. Exp Cell Res. 2005;303(1):1–13. doi: 10.1016/j.yexcr.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 62.Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2(9):823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruvolo PP. Ceramide regulates cellular homeostasis via diverse stress signaling pathways. Leukemia. 2001;15(8):1153–1160. doi: 10.1038/sj.leu.2402197. [DOI] [PubMed] [Google Scholar]
- 64.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraile JM, Quesada V, Rodríguez D, Freije JM, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31(19):2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 66.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 67.Coombs N, et al. Helicobacter pylori affects the cellular deubiquitinase USP7 and ubiquitin-regulated components TRAF6 and the tumour suppressor p53. Int J Med Microbiol. 2011;301(3):213–224. doi: 10.1016/j.ijmm.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao GY, et al. USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour Biol. 2015;36(3):1721–1729. doi: 10.1007/s13277-014-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q, et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126(6):2205–2220. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.