Abstract
Exposure of protein modified surfaces to air may be necessary in several applications. For example, air contact may be inevitable during the implantation of biomedical devices, for analysis of protein modified surfaces, or for sensor applications. Protein coatings are very sensitive to dehydration and can undergo significant and irreversible alterations of their conformations upon exposure to air. With the use of two compatible solutes from extremophilic bacteria, ectoine and hydroxyectoine, the authors were able to preserve the activity of dried protein monolayers for up to >24 h. The protective effect can be explained by the preferred exclusion model; i.e., the solutes trap a thin water layer around the protein, retaining an aqueous environment and preventing unfolding of the protein. Horseradish peroxidase (HRP) immobilized on compact TiO2 was used as a model system. Structural differences between the compatible solute stabilized and unstabilized protein films, and between different solutes, were analyzed by static time-of-flight secondary ion mass spectrometry (ToF-SIMS). The biological activity difference observed in a colorimetric activity assay was correlated to changes in protein conformation by application of principal component analysis to the static ToF-SIMS data. Additionally, rehydration of the denatured HRP was observed in ToF-SIMS with an exposure of denatured protein coatings to ectoine and hydroxyectoine solutions.
I. INTRODUCTION
Proteins immobilized onto surfaces find application in protein chips, functional protein microarrays, biocatalysis, biosensors, and protein modified implant materials.1–4 In the latter, they promise a significantly better acceptance of the implant. Foreign body reactions and fibrosis can be decreased, and bone healing and implant ingrowth can be improved.5,6 Promising strategies include the “disguising” of the implant material with proteins recognized by the body as familiar7 or the immobilization of growth factors to induce tissue formation on the surface of the implant.8,9
Protein coatings on surfaces are extremely sensitive to drying.10 Hydrophobic domains of the protein will be exposed upon contact with air, and, consequently, the tertiary structure of the protein will be changed. The aim of this project was to significantly increase the shelf-life and stability of protein coated metal oxides. Xia et al. were able to preserve the protein activity and tertiary structure for short-term exposure to air when covering the protein modified surfaces with the sugar trehalose.11,12 The stabilization mechanism of the protein tertiary structure by trehalose was proposed to originate in the formation of hydrogen bonds to polar residues in the protein,13,14 and the sugar molecule was additionally shown to prevent amyloid formation of insulin or polyglutamine.15,16
In solution, it was shown that protein functionality can be preserved under harsh conditions to a great extent by addition of compatible solutes from extremophilic bacteria such as ectoine and hydroxyectoine.17–19 These substances sustain the protein tertiary structure even under hostile conditions such as extreme temperatures, urea, freeze-thaw treatments, and lyophilization. Compatible solutes are described as biologically inert molecules that do not interfere with the overall cellular functions even if they are present at high concentrations within the cytoplasm.20
Several theories are proposed to explain the protective function of the compatible solutes, among which the “preferential exclusion model” has gained considerable interest.21–23 This model describes that the molecules are excluded from the immediate hydration shell, as interaction of the solutes with the native protein is regarded as unfavorable. Unfolding free energy becomes greater in the presence of compatible solutes relative to that in their absence, as in the unfolded state a larger amount of protein surface area is in contact with the solute molecules. Consequently, minimization of the exposed protein surface and preferential hydration of the latter occurs, thereby enhancing the stability of the native conformation and thus making denaturation thermodynamically less favorable. The compatible solutes do not directly interact with the protein structure; i.e., they stabilize by not binding to the proteins but by trapping water between the protein and themselves. Recently, Wolkers et al. were able to show trehalose induced water retention at the biomolecule surface upon drying by infrared spectroscopy.24 Ectoines have already found application as cell protectants in skin care and as protein-free stabilizers of proteins and cells in life sciences.25
The protective properties of compatible solutes have, to date, been mainly investigated for protein solutions. The influence of compatible solutes on the protein conformation and functionality of immobilized protein coatings is the focus of this work. Horseradish peroxidase (HRP) adsorbed onto TiO2 was used as a model system, as titanium with a native or engineered TiO2 surface is a common medical implant material. The enzymatic activity of HRP can easily be quantified in a colorimetric reaction, where H2O2 is used as a substrate for the enzyme and the produced OH• radicals induce a color change in a dye molecule [e.g., 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS)]. The protein was immobilized via the linker molecule 1,1′-carbonyldiimidazole (CDI), as this linker provides a high protein coverage of TiO2.26
Surface analytical techniques based on mass spectrometry can provide insights into the protein structure at interfaces. Whereas techniques like hydrogen-deuterium exchange mass spectrometry provide information about protein conformation upon interactions with, e.g., ligands,27 time-of-flight secondary ion mass spectrometry (ToF-SIMS) was shown to be extremely valuable for the determination of the orientation of proteins adsorbed to solid supports. As the information depth of the technique (∼2 nm) is lower than the average protein diameter, conclusions about the orientation of the protein may be reached according to the amino acid pattern visible in the spectra.28,29 The spectra of the dry protein coatings were compared using principal component analysis (PCA). This data analysis method has been widely used for the interpretation of ToF-SIMS data, including analysis of protein samples.30 PCA can help to identify the major sources of spectral differences within a sample or between samples.31
II. EXPERIMENT
All chemicals were purchased from chemical suppliers and used without further purification. All organic solvents were of water-free grade.
A. Substrate preparation
Ti (100 nm) was sputtered onto a silica wafer in a Createc SP-P-US-6M-3Z UHV instrument, ultrasonically cleaned in ethanol and deionized (DI) water, and anodized at 20 V for 2 min in 1 mol/l H2SO4 (Merck), and a Pt sheet was used as a counter electrode. Anodization was carried out using a high-voltage potentiostat (Jaissle IMP 88-200 PC), connected to a digital multimeter (Keithley 2000) interfaced to a computer. The wafer was cut into pieces with an area of 0.5 cm2. All samples were rinsed with DI water after the anodization process and then cleaned by exposure to UV light for at least 30 min (while being completely immersed in DI water)32 to photocatalytically remove contaminations and subsequently dried under a stream of N2.
B. Protein adsorption and solute coating
Ectoine, hydroxyectoine, and trehalose will be referred to as “solutes” throughout this article.
HRP (Sigma-Aldrich) was immobilized onto the TiO2 substrate via the bioactive linker molecule CDI (Sigma-Aldrich). The linker was coupled to the oxide surface by assembly from a 25 mmol/l solution in CHCl3 (Sigma-Aldrich, purity >99.8%) at room temperature (RT, 20 °C) for 24 h. Samples were rinsed with chloroform and dried under a nitrogen stream. The CDI modified surfaces were immersed in a phosphate buffered saline (PBS) solution (pH 6.4) with a protein concentration of 100 μg/ml for 24 h at 4 °C. For one set of experiments, compatible solutes were added to the protein solution with a concentration of 0.5 mol/l. Samples were rinsed according to Foster et al.33 Subsequently, they were coated with 75 μl of a 26 mmol/l solute solution in DI water for 30 min, according to Xia et al.11 Finally, samples were spin-dried at 3000 rpm for 30 s and stored for the respective times at either RT or in a fridge (4 °C). To minimize artifacts by protein dissolution into the solute solution, references for the activity assay were prepared by coating the samples with 75 μl of DI water. For ToF-SIMS measurements, the reference samples were stored after the rinsing and spin-drying process, then coated directly prior to the measurements with 75 μl of the respective 26 mmol/l solute solution for 30 min, and spin-dried at 3000 rpm for 30 s. Additionally, some samples were spin-dried immediately after being covered with the solute solution.
Reference samples of pure solute molecules (“solute”) were prepared by evaporation of a droplet of the solute solution. The obtained references show a solute layer thick enough to attenuate the substrate signal in ToF-SIMS spectra.34
C. Activity assay
The protein coated samples were placed in a multiwell array containing 0.75 ml PBS solution (pH 6.4), 0.3% H2O2, and 0.05 mol/l ABTS (Fluka). H2O2 was added as a biological substrate for the enzyme and oxidizes the ABTS molecule subsequently, detectable by the intense green color of the stable ABTS• radical, which has absorption maxima at λ = 405 nm and λ = 747 nm.35 The intensity of the absorption of the differently stored samples was measured at λ = 747 nm on a UV-Vis spectrometer Lambda Bio XLS after 30 min incubation time at room temperature in the dark (to prevent any influence of the photocatalytically active TiO2 substrate). Absorption values were referenced to the absorption of the pure ABTS solution. Reference samples of pure TiO2 and solute coated TiO2 did not show any activity in the ABTS assays. Error bars depict the standard deviation of three measurements.
D. Surface analysis
Positive and negative static SIMS measurements were performed on a ToF.SIMS 5 spectrometer (ION-TOF, Münster). The primary analysis beam was a pulsed 25 keV Bi3+ liquid-metal ion beam. Spectra were recorded in the high mass resolution mode (m/Δm > 8000 at 29Si). The beam was electrodynamically bunched down to <1 ns to increase the mass resolution and rastered over a 100 × 100 μm2 area. The primary ion dose density was kept at 5 × 1011 ions/cm2, ensuring static conditions. Peaks were assigned using their accurate mass as well as their isotopic pattern. Poisson correction was used for integration of the signal intensities. Spectra were normalized to their total intensity and mass calibrated to CH3+, C2H3+, C3H5+, C4H9+, and C7H7+ signals for positive spectra and CH−, CH2−, C2−, CN−, S−, and CNO− for negative spectra. On each sample, five spectra were recorded, and samples were measured in triplicate. Spectra showing artifacts from ion beam instabilities (e.g., peak broadening) were removed from analysis.
PCA processing of the spectra was conducted with the “spectragui” software from the NESAC/BIO toolbox.36 The peak set used for PCA processing of the positive spectra is described in the supplementary material, Table S1 and Fig. S1.28,37–39,44 Peaks were normalized to the total intensity of the PCA peak set to eliminate any systematic differences in total secondary ion yield (absolute intensity) between spectra. The data set was also square root mean-centered to ensure that the differences in samples were due to variations around the means and not the variance of the means.40
The Research Collaboratory for Structural Bioinformatics Protein Data Base (RCSB PDB) database structure of 1HCH (Ref. 41) was used for structural evaluation of the denaturation of HRP, using the ngl viewer program.42
Optical images of the sample surface were recorded on a Nikon eclipse lv 150 microscope in incident light mode. Water contact angles were measured with droplets of 10 μl DI water and recorded with a Leica DFC295 digital camera using the leica application suite 3.7.0 software.
III. RESULTS AND DISCUSSION
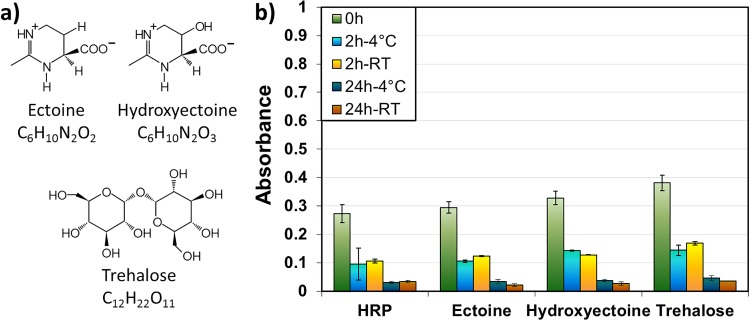
The aim of this study was to maximize the dry state lifetime of protein monolayers immobilized onto surfaces and to minimize their denaturation during sample storage. The protective properties of the compatible solutes ectoine and hydroxyectoine for stabilization of the dried protein coatings during air exposure were compared to the protective properties of the sugar trehalose. The structures of ectoine, hydroxyectoine, and trehalose are displayed in Fig. 1(a). HRP immobilized onto TiO2 surfaces via the bioactive linker molecule CDI was chosen as a model system. The enzymatic activity of the protein, used as a measure of its structural intactness, was investigated with a colorimetric activity assay (ABTS assay) for several protection strategies.
Fig. 1.
(a) Molecular structure of the compatible solutes ectoine, hydroxyectoine, and trehalose. (b) ABTS activity assay (UV-Vis spectroscopy) of HRP with solutes added during immobilization of the protein with different storage conditions.
Initially, a large excess of solutes (ectoine, hydroxyectoine, or trehalose) was added directly to the protein solution during the adsorption process, and samples were then rinsed according to the procedures in Foster et al.33 Figure 1(b) shows how the absorbance (λ = 747 nm, UV-Vis spectroscopy) measured for the enzymatic activity assay of HRP depends on the storage time and temperature. For each sample type, a freshly prepared sample (green) was compared to samples stored at 4 °C (blue) and room temperature (orange) for 2 and 24 h.
Slight improvements with solute addition were observed directly after the drying of the samples (0 h) and for short-term storage (2 h, both 4 °C and RT storage). However, all samples showed a comparable trend of loss of protein activity over storage time, with or without the addition of the small stabilizing molecules.
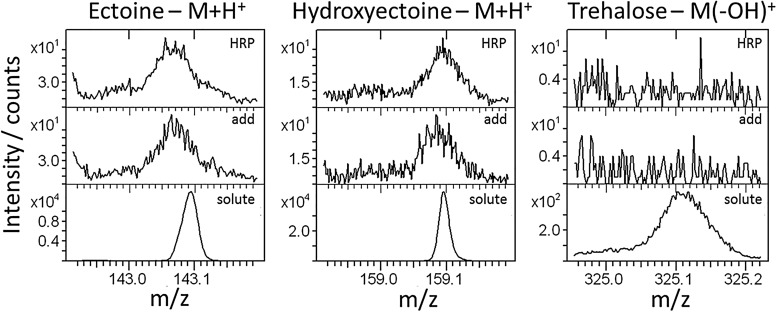
The samples were analyzed with ToF-SIMS after rinsing and drying. Figure 2 shows the major fragment of each solute for a solute-free sample (“HRP,” top), the protein layer adsorbed from solute-containing solution (“add,” middle), and an evaporated droplet of pure solute solution (“solute,” bottom).
Fig. 2.
Positive polarity ToF-SIMS spectra of the regions of the major signals of the solutes: ectoine and hydroxyectoine—molecular fragment plus H+; trehalose—molecular fragment minus OH−. Top: pure HRP; middle: solutes added to HRP solution during adsorption; bottom: pure solute drop-casted onto the substrate (individual y axes for each spectrum).
No clear signals originating from the small molecules could be detected on the protein surfaces with any of the solutes added to the protein solution; the regions for the major solute fragments are virtually identical to the ones of bare HRP. This was particularly surprising as a huge excess (∼50 000-fold amount) of solutes was used relative to the protein. Consequently, a strong interaction of the solutes and the HRP layer can be excluded. The absence of the solute molecules in the spectra is in agreement with the “preferential exclusion model”;21–23 i.e., the small molecules seem not to stabilize the protein by direct interaction, but a solute stabilized water shell may be encapsulating the protein. Assuming that the “preferential exclusion model” applies to the immobilized protein coatings, upon rinsing the protein sample the solutes would be washed away from the surface and consequently not exert any protective effects onto the dried protein monolayer. The latter effect is observed in the ABTS activity assay (cf. Fig. 1).
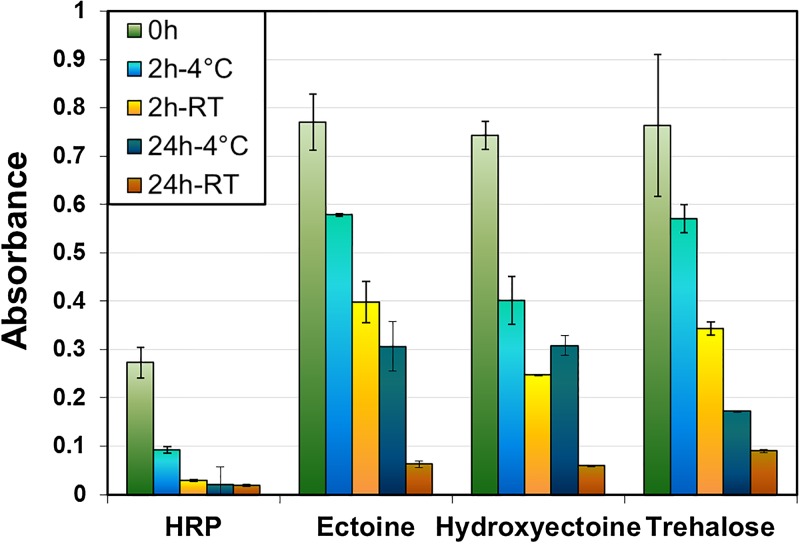
Xia et al. demonstrated the positive influence of spin-coated trehalose on the conformation stability of proteins. We used a similar approach for ectoine and hydroxyectoine. With this approach, solute fragments are clearly visible in the ToF-SIMS spectra; however, the protein signals are not obstructed (cf. S2 in the supplementary material44). Figure 3 shows the absorption values measured in the ABTS activity assay with and without the addition of solute coatings. For each sample type, a freshly prepared sample (green) was compared to samples stored at 4 °C (blue) and room temperature (orange) for 2 and 24 h.
Fig. 3.
ABTS activity assay (UV-Vis spectroscopy) of HRP coated with solutes directly after immobilization of the protein with different storage conditions.
Initial protein activity measured directly after adsorption and drying showed more than a doubling of the value observed for nonprotected HRP if the sample was coated with any of the suggested molecules. Even a very short drying step (“0 h” in Fig. 3; less than 1 min was needed for transfer from the spin coater into the test solution) seems to reduce the performance of protein coatings to a significant extent. This is most likely caused by a change in protein orientation and folding as soon as it comes in contact with air.
The enzymatic activity of a trehalose protected sample stored for 2 h at room temperature was comparable to the initial nonprotected activity, which is in good agreement with the results reported by Xia and Castner.12 Longer storage times at room temperature, however, were detrimental for the performance of the coatings, as only a small amount of HRP activity was retained. Storage at 4 °C clearly improved the durability of the dried coatings. The decrease in activity is significantly slower than at room temperature. For longer storage times at 4 °C, the compatible solutes ectoine and hydroxyectoine both outperform trehalose in their protective ability. The ABTS activity assay of the coated samples yielded absorbance values comparable to the initial nonprotected HRP coatings, even after 24 h of storage. These results indicate that immersion in solute solution prior to drying (however, after rinsing) protects and retains the functionality of immobilized protein coatings after drying. Compatible solute coatings allow storage of the modified surfaces for periods of several hours without loss of activity compared to nonprotected protein coatings.
To assess potential influences of the solute coatings on the bioavailability and bioactivity of the protein coatings, all coated samples were subjected to an additional rinsing step with DI H2O (S2 in the supplementary material44). The solute coatings do not form any strong interactions with the proteins and are removed after exposure to an aqueous environment, indicating that the protective coatings will be washed away in any aqueous or biological environment and will not obstruct the functionality of the protein coating.
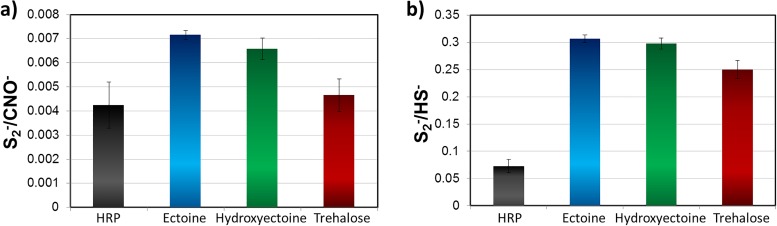
Changes in protein conformation were evaluated by ToF-SIMS. Protein denaturation is the consequence of the tertiary structure unfolding. Upon unfolding, disulfide bonds between two cysteine residues may become broken. It has been shown that the S2− signal in negative polarity ToF-SIMS spectra is an indicator for (heat induced) protein denaturation.43 Thus, the negative spectra of samples protected with the various solutes and stored for 24 h at 4 °C were evaluated (cf. Fig. 4). Comparing the S2− signal with the peptide backbone (CNO−), Fig. 4(a) shows the content of disulfide bonds within the part of the molecule accessible by ToF-SIMS. A clear increase of disulfide content is observed for ectoine and hydroxyectoine protected samples, indicating that the protein structure is intact to a higher degree than the unprotected and trehalose protected samples. This is in agreement with the results of the colorimetric activity assay (cf. Fig. 3). However, as the majority of cysteine residues are clustered on one end of the HRP molecule (RCSB PDB, 1HCH),38 the observations from Fig. 4(a) may also be explained by a differing preferential protein orientation for each solute, which could amount for the small deviation between trehalose protected and unprotected samples. Therefore, we additionally compared the S2− fragment to HS−, which is characteristic of nonbound thiol groups, i.e., broken disulfide bonds. The unprotected samples show a significantly lower amount of disulfide versus nonbound thiol compared to all protected coatings. The trend for the protected samples [Fig. 4(b)] is similar to the disulfide/peptide bond ratio; however, the difference between trehalose and (hydroxy-)ectoine protection is smaller. This indicates that (hydroxy-)ectoine coated HRP has a more intact tertiary structure than trehalose protected protein and all solute coatings preserve the disulfide bonds in HRP. Additionally, the orientation of the enzyme is likely to vary between the different protective coatings as well as unprotected samples.
Fig. 4.
Evaluation of the content of disulfide bridges within the tertiary structure for samples stored 24 h at 4°C: comparison of the disulfide (S2−) signal intensity to (a) the peptide backbone signal (CNO−) and (b) the HS− signal (free cysteine). Negative polarity ToF-SIMS spectra.
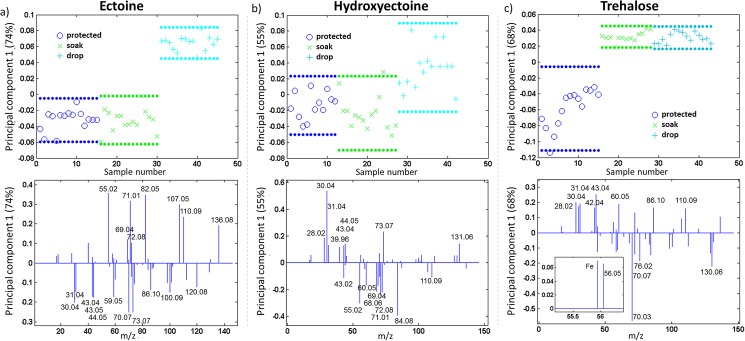
Changes in protein conformation upon denaturation were evaluated by the positive ToF-SIMS spectra. To compare the amino acid pattern of HRP protected with the three different solutes, all solute characteristic fragments were omitted from the respective peak lists (cf. S1 in the supplementary material44). For evaluation of the denaturation of the protein surfaces, sets of protected and unprotected samples were compared for each solute individually. The denatured samples were soaked in solute solution for 30 min directly prior to introduction into the ToF-SIMS chamber, as described previously.11,12 This treatment is necessary to minimize matrix effects and to prevent further vacuum induced denaturation of the uncoated protein. Matrix effects caused, e.g., by different kinds of solutes may influence the fragmentation and ionization probabilities of the underlying immobilized protein coating. HRP protected with solute coatings prior to storage was compared to unprotected samples that were covered with solutes by two differing methods after storage for 24 h at 4 °C, directly prior to analysis: the previously reported soaking method (“soak”) and drying of a droplet of solute solution on the sample by spin-drying immediately after contact (“drop”). PCA was used to visualize the differences between the protein coated samples. PCA scores and loadings from the first principal component obtained from the positive spectra are shown in Fig. 5. The scores describe the relationship between the samples (spectra) and are a projection of the original data points onto a given principal component axis. The loadings show which variables (peaks) are responsible for the separation seen in the scores plot. Together the scores and loadings represent a concise summary of the original data.29 PC1 separates the samples according to their largest differences in the sample set; a separate PCA evaluation was conducted for each solute.
Fig. 5.
Comparison of protected to unprotected samples coated with the solute prior to measurement by soaking or drop-coating: (a) ectoine, (b) hydroxyectoine, (c) trehalose (top: scores, bottom: loadings of PC1). All samples were stored for 24 h at 4 °C. The dotted lines in the score plots show the 95% confidence limits. Positive polarity ToF-SIMS spectra were analyzed.
While separation in the first principal component between (hydroxy-)ectoine protected and unprotected samples [scores, Figs. 5(a) and 5(b)] is dependent on the coating method, no changes between coating methods were observed for trehalose [scores, Fig. 5(c)]. With ectoine and hydroxyectoine, the major variation after 24 h storage at 4 °C for the protected and unprotected soaked samples lies within the samples and no clear separation is observed in the first principal component (PC).11,12 Further PCs did not separate the protected samples from the denatured ones coated by soaking (cf. S3 in the supplementary material44). This was surprising, as all other applied techniques indicate a change in the protein structure as samples turn more hydrophobic upon unprotected storage (optical microscopy and contact angle measurements, cf. S4 and S5 in the supplementary material44) and the enzymatic activity of an unprotected sample is significantly inferior to the enzymatic activities of solute protected HRP. The major difference in sample handling between the activity assay and the surface analysis was the additional coating step with a solute (“soak”).11,12 Consequently, the contact time between the denatured protein sample and (aqueous) solute solution was minimized by direct drying of the applied solute solution on the sample (“drop”). Subsequently, separation of protected and unprotected samples was possible in the case of ectoine and a trend was observed in the case of hydroxyectoine. This indicates that exposure of the protein to ectoine and hydroxyectoine solutions changes the protein orientation or conformation, whereas trehalose does not show this effect.
The corresponding loadings (bottom panels in Fig. 5) are discussed with respect to the 3D-protein structure of HRP (RCSB PDB, 1HCH).38 HRP has one acetate and two Ca2+ ligands, while the active site consists of a heme-porphyrin in the center of the protein. When looking at the distribution of hydrophilic and hydrophobic amino acids, no extended domains can be identified. However, all three histidine residues of the enzyme are located in close proximity to the central active site, with one histidine coordinated to the porphyrin iron atom. Four serine residues are also present in the heme pocket, i.e., in the center of the correctly folded enzyme. Tyrosine and cysteine residues are clustered in opposing regions of the protein. The single tryptophan residue (hydrophobic, but located between two polar amino acids: sequence Ser-Trp-Arg), arginine, lysine, proline, glutamic acid, and aspartic acid are prevalent on the surface of HRP. Two proline and several phenylalanine residues are located at the opening of the channel to the reactive center; the remaining phenylalanine is distributed throughout the entire structure.
Samples coated with ectoine [Fig. 5(a)] by the drop-coating method differ from protected and soaking-method coated samples by the following signals, loading positively in PC1 (scores above 0.1; assignments of fragments to amino acids in brackets): m/z 55.02, C3H3O+ (Tyr/ligand); m/z 69.04, C4H5O+ (Thr); m/z 71.01, C3H3O2+ (Ser); m/z 72.08, C4H10N+ (Val); m/z 82.05, C4H6N2+ (His); m/z 107.05, C7H7O+ (Tyr); m/z 110.09, C5H8N3+ (His, Arg); and m/z 136.08, C8H10NO+ (Tyr). The fragments loading negatively in PC1, i.e., predominantly contributing to protected samples (and the ones coated by the soaking method), are m/z 30.04, CH4N+ (Gly, Leu, and others); m/z 31.04, CH5N+ (amino acids); m/z 43.04, C2H5N+ (Ala, Leu, Ser); m/z 43.05, C3H7+ (carbohydrates); m/z 44.05, C2H6N+ (Ala, Asn, Leu); m/z 59.05, CH5N3+ (Arg); m/z 70.07, C4H8N+ (Pro, Val, Arg, Leu); m/z 73.07, C2H7N3+ (Arg); m/z 86.10, C5H12N+ (Ile, Leu); m/z 100.09, C4H10N3+ (Arg); and m/z 120.08, C8H10N+ (Phe). The major contributions to the drop-coated samples stem from amino acids located in the center of HRP, indicating an unfolding of the tertiary structure. The fragments corresponding to protected (and soaking-method coated unprotected) HRP are attributed mainly to amino acids located prevalently on the HRP surface.
Hydroxyectoine coated samples differ to a less significant extent [Fig. 5(b)], and a clear separation cannot be reached even with the drop-coating method. Signals corresponding slightly more to the drop-coated samples include m/z 28.02, CH2N+ (Leu + others); m/z 30.04, CH4N+ (Gly, Leu, and others); m/z 31.04, CH5N+ (amino acids); m/z 39.96, Ca+ (ligand); m/z 43.04, C2H5N+ (Ala, Leu, Ser); m/z 44.05, C2H6N+ (Ala, Asn, Leu); m/z 73.07, C2H7N3+ (Arg); and m/z 131.06, C9H7O+ (Phe). The negative loadings include m/z 43.02, C2H3O+ (ligand); m/z 55.02, C3H3O+ (Tyr/ligand); m/z 60.05, C2H6NO+ (Ser); m/z 68.06, C4H6NO+ (Pro, Lys); m/z 69.04, C4H5O+ (Thr); m/z 71.01, C3H3O2+ (Ser); m/z 72.08, C4H10N+ (Val); m/z 84.08, C4H6N+ (Pro, Lys); and m/z 110.09, C5H8N3+ (His, Arg). The overlap between sample types indicates that even a short exposure to hydroxyectoine can influence the protein conformation. The loadings suggest that the drop-coated samples contain even more of the outer shell components of HRP than the protected ones, indicating a fast reorientation process upon contact with hydroxyectoine solution.
Trehalose shows a clear distinction between protected and unprotected samples [Fig. 5(c)]. Moreover, the exposure time of unprotected (denatured) samples to trehalose prior to ToF-SIMS measurements does not influence the protein conformation. The scores of the trehalose protected samples show a larger spread than ectoine or hydroxyectoine protected ones, which may be related to the less homogeneous distribution of trehalose over the HRP surface (cf. S4 in the supplementary material44). The latter may also be related to the inferior protective ability observed in the activity assay for long term storage of the protein coatings. The positive loadings [above 0.1, Fig. 5(c) displays labeled loadings only above 0.15 for better visibility], characteristic to denatured HRP, include m/z 28.02, CH2N+ (Leu + others); m/z 30.04, CH4N+ (Gly, Leu, and others); m/z 31.04, CH5N+ (amino acids); m/z 42.04, C2H4N+ (Ala, Gly, His, Leu, Ser); m/z 43.04, C2H5N+ (Ala, Leu, Ser); m/z 43.05, C3H7+ (carbohydrates); m/z 60.05, C2H6NO+ (Ser); m/z 72.08, C4H10N+ (Val); m/z 86.10, C5H12N+ (Ile, Leu); m/z 110.09, C5H8N3+ (His, Arg); and m/z 136.08, C8H10NO+ (Tyr). Additionally, Fe+ loads positively on PC1 [inset in Fig. 5(c)]. The major fragments related to the protected protein are m/z 44.04, CH4N2+ (Arg); m/z 58.07, C3H8N+ (Glu); m/z 59.00, C2H3S+ (Cys); m/z 59.05, CH5N3+ (Arg); m/z 70.03, C3H4NO+ (Asn); m/z 70.07, C4H8N+ (Pro, Val, Arg, Leu); m/z 76.02, C2H6SN+ (Cys); m/z 84.04, C4H6NO+ (Gln, Glu); m/z 98.06, C4H4NO2+ (Asn); m/z 129.11, C5H13N4+ (Arg); and m/z 130.06, C9H8N+ (Trp). The unprotected samples show mainly fragments related to the HRP core, while the loadings of the protected ones correspond to amino acids located at the protein surface. Additionally, the domain with the cysteine cluster seems to be exposed in the protected samples, whereas the tyrosine rich region is related to the unprotected protein surface. This indicates that a change in protein orientation may additionally accompany the denaturation.
The contact time of unprotected HRP and (aqueous) ectoine solution has a significant influence on the amino acids exposed at the protein surface. This may be explained by rehydration and subsequent changes of the protein structure. Thus, additionally, the ABTS activity assay was performed on unprotected coatings stored for 24 h at 4 °C, both soaked in ectoine solution (30 or 60 min) and drop-coated with a solute (Fig. S6 in the supplementary material44). All coating methods led to an enzymatic activity that is roughly identical to (or even lower than) the solute-free sample. This indicates that the protein structure may change back to exposing the more hydrophilic domains upon immersion in aqueous ectoine or hydroxyectoine solution; however, the correct folding of the enzyme does not seem to be achieved.
The findings of this project show that ectoine and hydroxyectoine are most efficient for keeping a protein coated surface hydrated (hydrophilic amino acids are prevalent on the surface of the protein) and active. These solute molecules even induce a change in the folding of denatured proteins immobilized onto surfaces. Due to the latter effect, trehalose coatings are more well-suited for surface analytical investigation of protein denaturation, while the (hydroxy-)ectoine coatings are more promising for application in the dry storage of protein modified devices.
IV. CONCLUSION
In the present work, we investigate the stabilization of dried protein coatings on solid supports by the compatible solutes ectoine, hydroxyectoine, and the sugar trehalose. Horseradish peroxidase adsorbed to TiO2 surfaces was chosen as a model system. A colorimetric activity assay showed that the direct addition of the solutes into the protein solution did not stabilize the adsorbed protein coatings and no solute molecules could be detected on the surface by ToF-SIMS. This is consistent with the “preferred exclusion model”; i.e., the solutes seem to not directly interact with the proteins and are easily washed off.
Coating of the protein modified surfaces with solute molecules during the drying process increased the initial activity of the dried protein coating significantly and, consequently, is recommendable even for shortest periods of exposure to air. The activity of horseradish peroxidase after 24 h dry storage at 4 °C coated with ectoine and hydroxyectoine was comparable to freshly dried, unprotected protein surfaces. Compatible solutes are thus able to significantly extend the time frame for handling of protein coated surfaces exposed to air.
Ectoine and hydroxyectoine coated protein surfaces were furthermore shown to contain a higher amount of disulfide bonds than trehalose protected surfaces. Comparing the ToF-SIMS spectra of protected and unprotected HRP indicates that a rehydration of denatured protein is possible with ectoine and hydroxyectoine, whereas no change of the protein conformation was observed for trehalose coatings. A correct refolding of the denatured protein, however, could not be achieved.
In summary, trehalose coatings preserve the current state of orientation and conformation in protein coatings, which is important for, e.g., surface analytical investigation of protein denaturation. Hydroxyectoine and ectoine produce very homogeneous coatings on protein monolayers and hydrate the latter efficiently, allowing for longer periods of dry storage of protein coated surfaces.
ACKNOWLEDGMENTS
The authors thank Dan Graham, Ph.D., for developing the NESAC/BIO toolbox used in this study and the National Institutes of Health (NIH, Grant No. EB-002027) for funding and support of the toolbox development. The authors would like to thank the Deutsche Forschungsgemeinschaft (DFG) research unit FOR1718 FunCOS “Functional Molecular Structures on Complex Oxide Surfaces” for funding. They also thank Markus Licklederer, Friedrich-Alexander University of Erlangen, Germany, for the preparation of the sputtered TiO2 samples, Evangelia Anastasiou, Friedrich-Alexander University of Erlangen, Germany, for assistance in sample preparation, and Patrik Schmuki, Friedrich-Alexander University of Erlangen, Germany, for providing laboratory resources.
References
- 1.Berrade L., Garcia A. E., and Camarero J. A., Pharm. Res. 28, 1480 (2011). 10.1007/s11095-010-0325-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu S., Xie Z., Qian J., Blackshaw S., and Zhu H., WIREs Syst. Biol. Med. 3, 255 (2011). 10.1002/wsbm.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheldon R. A. and Pelt S., Chem. Soc. Rev. 42, 6223 (2013). 10.1039/c3cs60075k [DOI] [PubMed] [Google Scholar]
- 4.Sassolas A., Blum L. J., and Leca-Bouvier B. D., Biotechnol. Adv. 30, 489 (2012). 10.1016/j.biotechadv.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 5.Wang P. J., Thissen H., and Kingshott P., Acta Biomater. 45, 31 (2016). 10.1016/j.actbio.2016.08.054 [DOI] [PubMed] [Google Scholar]
- 6.Zhou D. and Ito Y., RSC Adv. 3, 11095 (2013). 10.1039/c3ra23313h [DOI] [Google Scholar]
- 7.Stachelek S. J. et al. , Biomaterials 32, 4317 (2011). 10.1016/j.biomaterials.2011.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X., Neoh K. G., Shi Z., Kang E.-T., Poh C., and Wang W., Biomaterials 31, 8854 (2010). 10.1016/j.biomaterials.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 9.Park J., Bauer S., Pittrof A., Killian M. S., Schmuki P., and von der Mark K., Small 8, 98 (2012). 10.1002/smll.201100790 [DOI] [PubMed] [Google Scholar]
- 10.Prestrelski S., Tedeschi N., Arakawa T., and Carpenter J. F., Biophys. J. 65, 661 (1993). 10.1016/S0006-3495(93)81120-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia N., May C. J., McArthur S. L., and Castner D. G., Langmuir 18, 4090 (2002). 10.1021/la020022u [DOI] [Google Scholar]
- 12.Xia N. and Castner D. G., J. Biomed. Mat. Res. 67A, 179 (2003). 10.1002/jbm.a.10063 [DOI] [PubMed] [Google Scholar]
- 13.Allison S. D., Chang B., Randolph T. W., and Carpenter J. F., Arch. Biochem. Biophys. 365, 289 (1999). 10.1006/abbi.1999.1175 [DOI] [PubMed] [Google Scholar]
- 14.Shi H., Tsai W.-B., Garrison M. D., Ferrari S., and Ratner B. D., Nature 398, 593 (1999). 10.1038/19267 [DOI] [PubMed] [Google Scholar]
- 15.Ha C. and Park C. B., Biotechnol. Bioeng. 90, 848 (2005). 10.1002/bit.20486 [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N. R., Doi H., Kurosawa M., Nekooki M., and Nukina N., Nat. Med. 10, 148 (2004). 10.1038/nm985 [DOI] [PubMed] [Google Scholar]
- 17.Lippert K. and Galinski E. A., Appl. Microbiol. Biotechnol. 37, 61 (1992). 10.1007/BF00174204 [DOI] [Google Scholar]
- 18.Barth S., Huhn M., Mattehey B., Klimka A., Galinski E. A., and Engert A., Appl. Environ. Microbiol. 66, 1572 (2000). 10.1128/AEM.66.4.1572-1579.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Göller K. and Galinski E. A., J. Mol. Catal. B Enzym. 7, 37 (1999). 10.1016/S1381-1177(99)00043-0 [DOI] [Google Scholar]
- 20.Brown A. D., Adv. Microb. Physiol. 17, 181 (1978). 10.1016/S0065-2911(08)60058-2 [DOI] [PubMed] [Google Scholar]
- 21.Ohtake S., Kita Y., and Arakawa T., Adv. Drug Del. Rev. 63, 1053 (2011). 10.1016/j.addr.2011.06.011 [DOI] [PubMed] [Google Scholar]
- 22.Arakawa T. and Timasheff S. N., Biophys. J. 47, 411 (1985). 10.1016/S0006-3495(85)83932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu I., Jindo Y., and Nagaoka M., J. Phys. Chem. B 111, 10231 (2007). 10.1021/jp068367z [DOI] [PubMed] [Google Scholar]
- 24.Wolkers W. F., Oldenhof H., and Glasmacher B., Cryobiology 61, 108 (2010). 10.1016/j.cryobiol.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 25.Lentzen G. and Schwarz T., Appl. Microbiol. Biotechnol. 72, 623 (2006). 10.1007/s00253-006-0553-9 [DOI] [PubMed] [Google Scholar]
- 26.Killian M. S. and Schmuki P., Surf. Interface Anal. 46, 193 (2014). 10.1002/sia.5497 [DOI] [Google Scholar]
- 27.Zhang J., Li J., Craig T. A., Kumar R., and Gross M. L., Biochemistry 56, 3523 (2017). 10.1021/acs.biochem.7b00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lhoest J. B., Detrait E., Aguilar P. B, and Bertrand P., J. Biomed. Mater. Res. 41, 95 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Tidwell C. D., Castner D. G., Golledge S. L., Ratner B. D., Meyer K., Hagenhoff B., and Benninghoven A., Surf. Interface Anal. 31, 724 (2001). 10.1002/sia.1101 [DOI] [Google Scholar]
- 30.Wagner M. S. and Castner D. G., Langmuir 17, 4649 (2001). 10.1021/la001209t [DOI] [Google Scholar]
- 31.Graham D. J. and Castner D. G., Biointerphases 7, 49 (2012). 10.1007/s13758-012-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cecilio de Oliveira Monteiro M., Schmuki P., and Killian M. S., Langmuir 33, 13913 (2017). 10.1021/acs.langmuir.7b03044 [DOI] [PubMed] [Google Scholar]
- 33.Foster R. N., Harrison E. T., and Castner D. G., Langmuir 32, 3207 (2016). 10.1021/acs.langmuir.5b04743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killian M. S., Gnichwitz J.-F., Hirsch A., Schmuki P., and Kunze J., Langmuir 26, 3531 (2010). 10.1021/la9032139 [DOI] [PubMed] [Google Scholar]
- 35.Thomas J. C., Pacholski C., and Sailor M. J., Lab Chip 6, 782 (2006). 10.1039/b516121e [DOI] [PubMed] [Google Scholar]
- 36.See: http://www.nb.uw.edu/mvsa/multivariate-surface-analysis-homepage.
- 37.Lhoest J.-B., Wagner M. S., Tidwell C. D., and Castner D. G., J. Biomed. Mater. Res. 57, 432 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Henry M. and Bertrand P., Surf. Interface Anal. 41, 105 (2009). 10.1002/sia.2993 [DOI] [Google Scholar]
- 39.Canavan H. E., Graham D. J., Cheng X., Ratner B. D., and Castner D. G., Langmuir 23, 50 (2007). 10.1021/la062330o [DOI] [PubMed] [Google Scholar]
- 40.Vandeginste B. G. M., Massart D. L., Buydens L. M. C., deJong S., Lewi P. J., and Smeyers-Verbeke J., Handbook of Chemometrics and Qualimetrics: Part B (Elsevier, Amsterdam, 1998). [Google Scholar]
- 41.Berglund G. I., Carlsson G. H., Smith A. T., Szöke H., Henriksen A., and Hajdu J., Nature 417, 463 (2002). 10.1038/417463a [DOI] [PubMed] [Google Scholar]
- 42.Rose A. S. and Hildebrand P. W., Nucleic Acids Res. 43, W576 (2015). 10.1093/nar/gkv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Killian M. S., Krebs H. M., and Schmuki P., Langmuir 27, 7510 (2011). 10.1021/la200704s [DOI] [PubMed] [Google Scholar]
- 44.See supplementary material at 10.1116/1.5031189E-BJIOBN-13-301806 for S1: Peak lists for positive polarity spectra; S2: solute and protein fragments visibility after coating and assessment of potential obstruction of bioactivity by solute coatings; S3: PCA comparing (hydroxy-)ectoine protected and unprotected HRP (soaking method only); S4: optical images of protected and unprotected samples after solute coating; S5: contact angle measurements of unprotected HRP; and S6: colorimetric assay of enzymatic activity of unprotected samples with solute addition. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at 10.1116/1.5031189E-BJIOBN-13-301806 for S1: Peak lists for positive polarity spectra; S2: solute and protein fragments visibility after coating and assessment of potential obstruction of bioactivity by solute coatings; S3: PCA comparing (hydroxy-)ectoine protected and unprotected HRP (soaking method only); S4: optical images of protected and unprotected samples after solute coating; S5: contact angle measurements of unprotected HRP; and S6: colorimetric assay of enzymatic activity of unprotected samples with solute addition. [DOI]