Abstract
Rationale and Objectives
We evaluate utilizing convolutional neural networks (CNNs) to optimally fuse parenchymal complexity measurements generated by texture analysis into discriminative meta-features relevant for breast cancer risk prediction.
Materials and Methods
With IRB approval and HIPAA compliance, we retrospectively analyzed “For Processing” contralateral digital mammograms (GE Healthcare 2000D/DS) from 106 women with unilateral invasive breast cancer and 318 age-matched controls. We coupled established texture features (histogram, co-occurrence, run-length, structural), extracted using a previously validated lattice-based strategy, with a multi-channel CNN into a hybrid framework in which a multitude of texture feature maps are reduced to meta-features predicting the case or control status. We evaluated the framework in a randomized split-sample setting, using the area under the curve (AUC) of the receiver operating characteristic (ROC) to assess case-control discriminatory capacity. We also compared to CNNs directly fed with mammographic images, as well as to conventional texture analysis, where texture feature maps are summarized via simple statistical measures which are then used as inputs to a logistic regression model.
Results
Strong case-control discriminatory capacity was demonstrated on the basis of the meta-features generated by the hybrid framework (AUC=0.90), outperforming both CNNs applied directly to raw image data (AUC=0.63, p<0.05) and conventional texture analysis (AUC=0.79, p<0.05).
Conclusion
Our results suggest that informative interactions between patterns exist in texture feature maps derived from mammographic images, and can be extracted and summarized via a multi-channel CNN architecture toward leveraging the associations of textural measurements to breast cancer risk.
Keywords: digital mammography, breast cancer risk, convolutional neural network, parenchymal texture
Introduction
The stratification of breast cancer risk levels is becoming increasingly important and is rapidly evolving beyond the “one-size-fits-all” approach in breast cancer screening to personalized regimens tailored by individual risk profiling1, 2. Starting from the pioneering work of Wolfe3, studies have consistently shown an association of the breast parenchymal complexity (i.e., the distribution of fatty and dense tissues) on breast images with levels of breast cancer risk. In particular, full-field digital mammography, which is routinely used for breast cancer screening4, has demonstrated substantial potential in providing novel quantitative imaging biomarkers related to breast cancer risk. Mammographic density is one of the strongest risk factors for breast cancer5, 6, while studies increasingly support significant associations of breast cancer risk with mammographic texture descriptors7–9, which reflect more refined, localized characteristics of the breast parenchymal pattern.
In early studies investigating the role of mammographic texture in breast cancer risk assessment10–14, textural measurements have been estimated within a single region of interest (ROI) in the breast. In an attempt to provide more granular texture estimates, more recent studies have proposed sampling the parenchymal tissue through the entire breast for subsequent texture analysis15–17. For instance, in a recently proposed lattice-based strategy15, each texture descriptor is calculated within multiple non-overlapping local square ROIs through the breast and texture measurements are then averaged over the breast regions sampled by the lattice. In a preliminary case-control evaluation15, the lattice-based texture features were shown to outperform state-of-the-art features extracted from the retroareolar or central breast region, thereby suggesting that enhanced capture of the heterogeneity in the parenchymal texture within the breast may also improve the associations of texture measures with breast cancer risk. However, by averaging regional texture values, important information about the overall parenchymal tissue complexity might be still missed and, therefore, an improved fusion approach, which retains richer information about texture variability over the breast, might leverage the potential of such granular texture measurements provided by multiple ROIs.
Convolutional neural networks (CNNs) and “Deep Learning” technology18, 19 are able to automatically generate hierarchical representations useful for a particular learning task via neural networks with multiple hidden layers. In the last few years, CNNs have demonstrated a remarkable impact on medical image analysis20, 21 and they are now gaining increasing attention in applications with digital mammography, including primarily mass or lesion detection and characterization22–25. However, their potential in breast cancer risk prediction remains largely unexplored with only a few studies having used CNNs to relate mammographic patterns to breast cancer risk26, 27 or breast density28. Moreover, although CNNs have been primarily applied to raw imaging data, they may also have value as efficient feature-fusion techniques, especially in cases that conventional feature summarization approaches may have limitations.
In this study, we employ a multi-channel CNN architecture to fuse parenchymal texture feature maps into discriminative meta-features relevant for breast cancer risk prediction. The rationale is that CNNs can also be used as a high-level approach to effectively summarize high-dimensional patterns resulting from handcrafted features such as texture descriptors, for which conventional feature summarization and/or dimensionality reduction approaches may be limited in capturing the available rich information. Our hypothesis, therefore, is that by capturing sparse, subtle interactions between localized patterns present in texture feature maps derived from mammographic images, a multitude of textural measurements can be optimally reduced to meta-features capable of improving associations with breast cancer. We assess our hypothesis by evaluating these meta-features in a case-control study with digital mammograms, and we also compare their performance to the performances of conventional texture analysis and CNNs when fed directly with raw mammographic images.
Materials & Methods
Study Dataset
In this IRB-approved, HIPAA-compliant study under a waiver of consent, we retrospectively analyzed a case-control dataset of raw (i.e., ‘FOR PROCESSING’) mediolateral-oblique (MLO)-view digital mammograms previously reported15, 29. Briefly, the cases included women diagnosed with biopsy-proven unilateral primary invasive breast cancer (n = 106) at age 40 years or older, recruited by a previously-completed, IRB-approved, multimodality breast imaging trial (2002–2006; NIH CAXXXXX), in which all women had provided informed consent. For cases the contralateral images were analyzed (i.e., images from the unaffected breast) as a surrogate for inherent breast tissue properties associated with high breast cancer risk, as done in prior case-control studies of mammographic pattern analysis15, 29–31. Controls were randomly selected from the eligible population of women seen for routine screening over the closest possible overlapping time period at the same institution, and had negative breast cancer screening and confirmed negative one-year follow-up. Controls were age-matched to cases on 5-year intervals at a 3:1 ratio (n = 318), yielding a total sample size of 424.
Image Acquisition
All images were acquired using either a GE Senographe 2000D or Senographe DS full-field digital mammography (FFDM) system (image size: 2294×1914 pixels; image spatial resolution: 10 pixels/mm in both dimensions). Prior to further analysis, images were log-transformed, then inverted, and, finally, intensity-normalized by a z-score transformation within the breast region which was automatically segmented in each image using the publicly available “Laboratory for Individualized Breast Radiodensity Assessment” (LIBRA)29, 32. These common preprocessing steps help to alleviate differences between studies via intensity histogram alignment, while also maintaining the overall pattern of the breast parenchyma17. Additionally, we flipped horizontally the images of right MLO views so that the breast is always on the same side of the image.
Revealing Meta-features of Breast Parenchymal Complexity
We developed a hybrid framework which coupled state-of-the-art texture analysis with CNNs (Fig. 1). In the first step which follows the previously validated lattice-based strategy for parenchymal texture analysis15, a regular lattice was virtually overlaid on the mammographic image and a set of texture descriptors were computed on 6.3×6.3 mm2 (63×63 pixels2) local square ROIs centered on each lattice point within the breast. For each image, this step generated a set of 36×30 pixels2 texture feature maps (each pixel corresponds to an ROI defined by the lattice), one for each texture descriptor, that represent the spatial distribution of the corresponding textural measurements as sampled by the lattice over the entire breast.
Fig. 1.
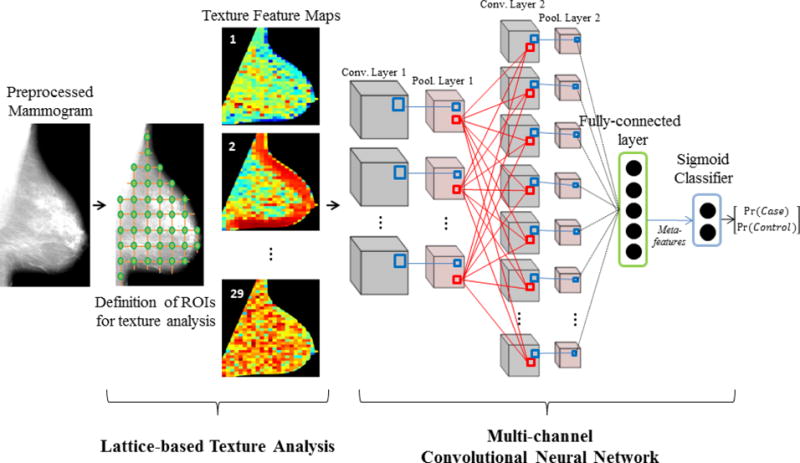
Hybrid framework workflow: Employing multi-channel convolutional neural networks to fuse texture feature maps into case-control discriminative meta-features.
Our texture feature set included a total of 29 established texture descriptors (Table 1 and Appendix for detailed feature descriptions), including gray-level histogram, co-occurrence, run-length, and structural features, all of which have been previously used for mammographic pattern analysis and breast cancer risk assessment7. Briefly, gray-level histogram features are common first-order statistics which describe the distribution of gray-level intensity. The co-occurrence features also consider the spatial relationships of pixel intensities in specified directions and are based on the gray-level co-occurrence matrix (GLCM) which encodes the relative frequency of neighboring intensity values. Run-length features capture the coarseness of texture in specified directions by measuring strings of consecutive pixels which have the same gray-level intensity along specific linear orientations. Finally, structural features capture the architectural composition of the parenchyma by characterizing the tissue complexity, the directionality of flow-like structures in the breast, and intensity variations between central and neighboring pixels.
Table 1.
Parenchymal texture feature maps (TFMs) extracted from each digital mammogram of the study dataset.
| Gray-level Histogram | |
|---|---|
| TFM1 | 5th Percentile |
| TFM2 | 5th Mean |
| TFM3 | 95th Percentile |
| TFM4 | 95th Mean |
| TFM5 | Entropy |
| TFM6 | Kurtosis |
| TFM7 | Max |
| TFM8 | Mean |
| TFM9 | Min |
| TFM10 | Sigma |
| TFM11 | Skewness |
| TFM12 | Sum |
|
| |
| Co-occurrence | |
|
| |
| TFM13 | Contrast |
| TFM14 | Correlation |
| TFM15 | Homogeneity |
| TFM16 | Energy |
| TFM17 | Entropy |
| TFM18 | Inverse Difference Moment |
| TFM19 | Cluster Shade |
|
| |
| Run-length | |
|
| |
| TFM20 | Short Run Emphasis |
| TFM21 | Long Run Emphasis |
| TFM22 | Gray Level Non-uniformity |
| TFM23 | Run Length Non-uniformity |
| TFM24 | Run Percentage |
| TFM25 | Low Gray Level Run Emphasis |
| TFM26 | High Gray Level Run Emphasis |
|
| |
| Structural | |
|
| |
| TFM27 | Edge-enhancing index |
| TFM28 | Box-Counting Fractal Dimension |
| TFM29 | Local Binary Pattern |
The set of 29 texture feature maps generated for each mammogram were then fed as separate “channels” of breast parenchymal complexity into a multi-channel CNN with a LeNet-like design, a pioneering CNN architecture suitable for small input images33. The multi-channel CNN consisted of two hidden convolutional/down-sampling layers (2×2 max-pool) and each convolutional layer contained 10 convolutional maps with 5×5 and 4×3 kernels for the first and second hidden layers, respectively. The second hidden layer fed into a fully-connected layer with five nodes, which in turn outputs five meta-features used as inputs to a final logistic regression layer (LR) predicting the case or control status. The hyperbolic tangent activation function was utilized in all but the LR layer, which utilized the sigmoid activation.
The multi-channel CNN was trained via training (n=200) and validation (n=100) sets, with the final model tested on 124 women; membership assignment into each set was carried out by stratified random sampling. Training occurred by sequentially feeding the CNN with the training image data (forward pass) and updating the weights (backward pass) via stochastic gradient descent (batch-size = 1 and learning rate = 0.01) so as to minimize the difference between the LR classification and the ground-truth case-control status34. Then, the trained model was applied to the validation set and the validation error was measured as the mean difference between the LR outputs and the ground-truth information. To better train our model while also avoiding over-fitting, this process was repeated 200 times (epochs) maximum, until the validation error got higher than it was in the previous epoch35. The final trained CNN was applied to the “unseen” images of the test set, via a single forward pass of all the test image data and the case-control discriminatory capacity was assessed via the area under the curve (AUC) of the receiver operator characteristic (ROC).
The multi-channel CNN was implemented using Theano v.0.8.2 on Python v.2.7 and parameters involved in the CNN architecture and training were empirically set.
Comparative Evaluation
To investigate the potential advantage of the proposed hybrid framework, we compared with the case-control discriminatory capacity of 1) conventional texture feature analysis and 2) CNNs applied directly to the original mammographic images (Fig. 2). DeLong’s tests36 were used to test for AUC differences between our hybrid framework and each of these two approaches.
Fig. 2.
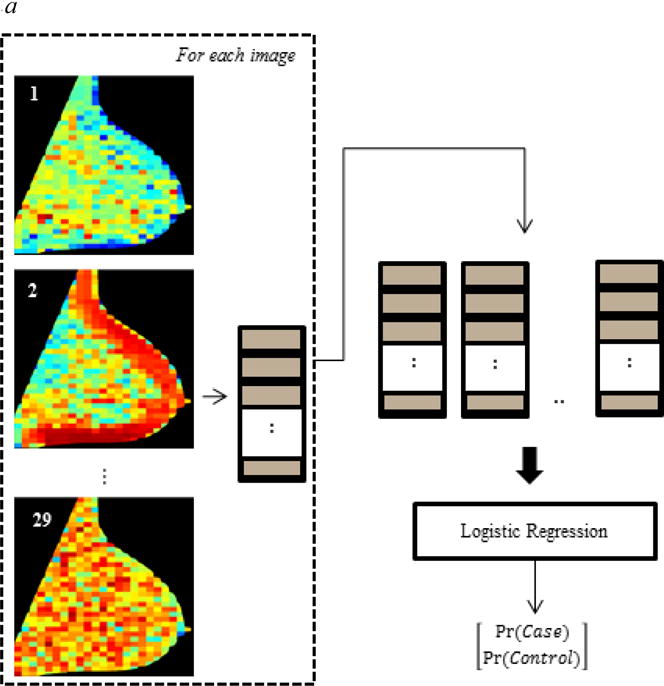

Design of comparative evaluation experiments: Evaluating the case-control discriminatory capacity of (a) conventional texture analysis and (b) convolutional neural networks applied directly to the original images.
Briefly, to evaluate conventional parenchymal texture analysis, each lattice-based texture feature map was summarized by the mean and the standard deviation of the corresponding feature across all the ROIs defined by the lattice15, thereby generating a 58-element feature vector for each breast (Fig. 2 (a)). To build a logistic regression model (similar in principle to the final layer of the CNN) while also limiting potential over-fitting, the texture feature vectors of subjects belonging to the training and validation sets were processed with elastic net regression37, a regularized technique suitable for strongly correlated features which assigns zero coefficients to weak covariates during model construction. The model was then fed with the texture feature vectors of the test set to obtain an estimate of its discriminatory capacity (AUC) which was compared to the AUC of the hybrid framework.
To assess the performance of CNNs when fed directly with the mammographic images, we developed a single-channel CNN with input the pre-processed image downscaled by a factor of four, as typically done in the field to address the computational issues of handling full-resolution images28. Our network was developed using the Lasagne library (Release 0.2.dev1)38 built on top of Theano, with an overall architecture as shown in Fig. 2 (b). The CNN consisted of five hidden convolutional/down-sampling layers, feeding into a pair of fully-connected layers and a logistic regression classifier. The parameters of the network were initialized using the recipe of Glorot & Bengio39 and, similarly to the multi-channel CNN, the CNN was trained and tested via distinct training, validation and test phases, now using the pre-processed mammograms of the same training, validation and test sets, respectively.
Results
Strong linear separability of cancer cases from controls was demonstrated on the basis of the five meta-features generated by the proposed hybrid framework. Figure 3 shows the predicted probabilities, along with the ground-truth labels of the test set. The corresponding case-control classification performance was AUC = 0.90 (95% CI: 0.82–0.98), with sensitivity and specificity equal to 0.81 and 0.98, respectively.
Fig. 3.
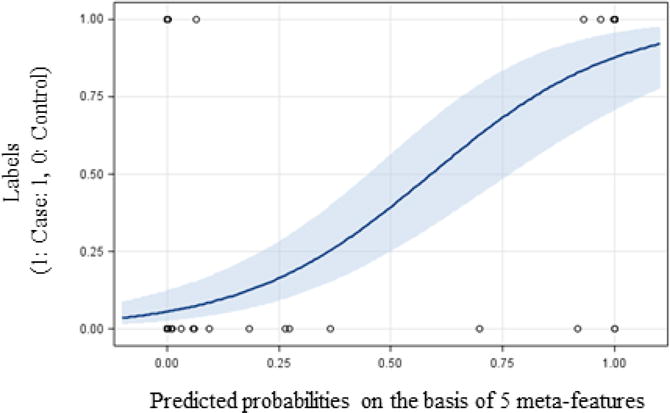
Case-control classification outcomes of the hybrid framework: Probabilities (with 95% confidence limits) of test images to belong to a cancer case as predicted by the hybrid approach versus corresponding ground-truth labels (1: Case, 0: Control).
When the texture feature maps were summarized into simple statistical measures (i.e., average and standard deviation), the discriminative performance was significantly lower (DeLong’s test, p = 0.03). Specifically, 12 texture features were selected by elastic net regression for inclusion in the logistic regression model, including one gray-level histogram feature, four co-occurrence, four run-length, and three structural features; among these, eight represented mean values and four the variability in the texture feature maps (Table 2). With these features, the lattice-based strategy demonstrated an AUC = 0.79 (95% CI: 0.69-0.89). The hybrid framework outperformed also the single-channel CNN (DeLong’s test, p = 0.0004) which showed an AUC = 0.63 (95% CI: 0.57–0.81) in the same case-control dataset (Fig. 4).
Table 2.
Texture features selected by elastic net regression. For each feature, the logistic regression coefficient (b), the p-value and 95% confidence interval (CI) for b are provided.
| b | p-value | 95% CI | |
|---|---|---|---|
| TFM13_mean | −0.59 | 0.013 | [−1.05 −0.12] |
| TFM17_mean | 0.03 | 0.897 | [−0.44 0.50] |
| TFM19_mean | −0.69 | 0.001 | [−1.08 −0.29] |
| TFM22_mean | −1.31 | 0.395 | [−4.34 1.71] |
| TFM23_mean | 0.76 | 0.582 | [−1.96 3.48] |
| TFM24_mean | −0.14 | 0.602 | [−0.64 0.37] |
| TFM28_mean | 1.07 | 0.437 | [−1.62 3.75] |
| TFM29_mean | 0.05 | 0.944 | [−1.34 1.44] |
| TFM11_std | 0.24 | 0.642 | [−0.79 1.28] |
| TFM15_std | 0.52 | 0.031 | [0.05 1.00] |
| TFM22_std | 0.14 | 0.483 | [−0.25 0.53] |
| TFM28_std | −0.54 | 0.002 | [−0.87 −0.20] |
Fig. 4.
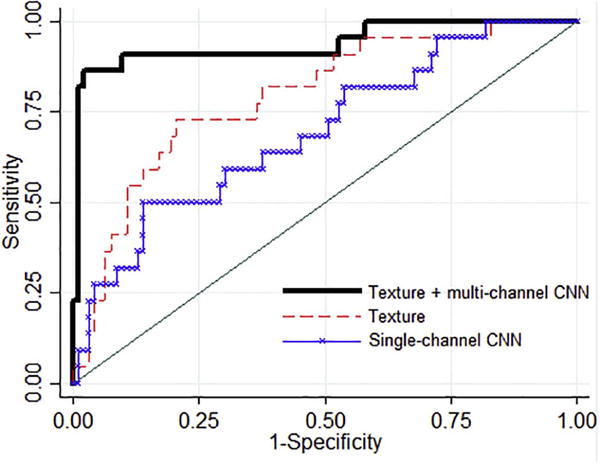
Comparative evaluation results: The hybrid approach,, i.e., texture analysis followed by multi-channel CNNs, (AUC = 0.90) compared to conventional parenchymal texture analysis (AUC = 0.79) or single-channel CNNs applied directly to the original images (AUC = 0.63). CNNs: convolutional neural networks.
Discussion
We assessed the feasibility of a texture-feature fusion approach, utilizing a CNN architecture able to capture sparse, subtle, yet relevant interactions between localized patterns present in texture feature maps derived from mammographic images. This approach may also be interpreted as a methodology for non-linear feature reduction, in which an entire ensemble of parenchymal texture feature maps is reduced down to a small number of meta-features which may serve as a basis for separating cases from controls via a simple linear classifier such as the logistic regression classifier implemented in the last layer of our CNN architecture.
Our hybrid framework demonstrated a promising case-control classification performance. Moreover, it outperformed standard texture analysis (AUC = 0.90 vs. AUC = 0.79), providing preliminary evidence that coupling texture analysis with this CNN-based texture-feature fusion approach can leverage textural features in breast cancer risk assessment. Additionally, the proposed hybrid framework also outperformed CNNs fed directly with raw mammograms. Our results from the single-channel CNN are in line with the few reported similar analyses with raw mammographic images26, in which comparable case-control classification performance (AUC = 0.57 – 0.61) was demonstrated by features learned with a multilayer convolutional architecture in multiscale patches of film and digital mammograms. However, given the relatively small size of our dataset, and considering that no data augmentation was applied in our study which might increase the performance of CNNs especially when deeper networks are used, a more extensive comparison between our hybrid approach and CNNs fed directly with raw image data is warranted.
Limitations of our study must also be noted. First, we evaluated our framework using as “case” images the contralateral mammograms of cancer cases, rather than prior unaffected screening mammograms of women in whom breast cancer was later detected. This is a common first-step approach in this field7, based on the premise that the contralateral unaffected breasts of cancer cases share inherent breast tissue properties that may predispose to a higher breast cancer risk. In future studies, we aim to test our hybrid framework on larger datasets in which prior negative screening mammograms for cancer cases will be analyzed. Larger datasets, combined with data augmentation techniques to further increase the volume of training data, may also allow us to develop deeper networks and to boost the performance of our CNNs, as well as to consider established demographic and clinical risk factors (e.g., body-mass-index, parity, family history of breast cancer) in our logistic regression models. Moreover, this study used only images from two FFDM devices, both of which were from the same vendor. Considering the reported substantial differences in textural measurements across image acquisition settings17, FFDM representations, and vendors40, in our future analyses we will also test the performance of our method by incorporating data from multiple FFDM vendors. Additionally, because of limited computational resources, we had to heavily rely on our experience in the choice of learning CNN hyper-parameters. Given the promising results obtained, in a future step, we will perform a systematic search for optimal hyper-parameters following grid or random search optimization41, which might further improve the performance of our CNNs.
In conclusion, this work introduced a texture feature fusion method, implemented via convolutional neural networks, designed for the detection of interactions among a multitude of hand-designed features, and optimized for the assessment of breast cancer risk. Our results suggest that informative interactions between localized patterns exist in feature maps derived from mammographic images, and can be extracted and summarized via a multi-channel CNN architecture toward leveraging textural measurements in breast cancer risk prediction. Upon further optimization and validation, such tools might be useful in routine breast cancer screening to provide each woman a breast cancer risk score, as well as to visualize parenchymal complexity patterns and breast locations associated with increased levels of breast cancer risk, towards assisting radiologists’ recommendations for supplemental breast cancer screening or breast cancer prevention treatments.
Acknowledgments
Funding Information:
Support for this project was provided by the Susan G. Komen for the Cure® Breast Cancer Foundation [PDF17479714] and the National Cancer Institute at the National Institutes of Health [Population-based Research Optimizing Screening through Personalized Regimens (PROSPR) Network (U54CA163313), an R01 Research Project (2R01CA161749-05), and a Resource-Related Research Project–Cooperative Agreement (1U24CA189523-01A1)].
Appendix
Descriptions of texture features
Gray-level histogram features are 13 well-established first-order statistics42, which were calculated from the gray-level intensity histogram of the image using 128 histogram bins17, 40.
The co-occurrence features reflect the spatial relationship between pixels and summarize the information encoded by the gray-level co-occurrence matrix (GLCM)43 which corresponds to the relative frequency with which two neighboring pixels, one with gray level i and the other with gray level j, occur in the image. Such matrices are a function of the distance (d) and the orientation (ϑ) between the neighboring pixels. In this study, the M × M GLCM matrices were estimated using M = 128 gray levels to balance computational precision with efficiency, d = 11 pixels17, 40, and the co-occurrence features were computed by averaging over four orientations.
Run-length features capture the coarseness of a texture in specified directions44, 45. A run is defined as a string of consecutive pixels with the same gray-level intensity along a specific linear orientation. Fine textures tend to contain more short runs with similar gray-level intensities, while coarse textures have more long runs with different gray-level intensities. Similarly to the GLCM, a M × N run-length matrix is defined, representing the number of runs with pixels of gray-level intensity equal to i and length of run equal to j along a specific orientation. In correspondence with the co-occurrence features, the run-length statistics were estimated for M = 128 gray levels, N = 11 pixels17, 40, and averaged over four polar-grid-driven orientations.
The structural features include (a) the edge-enhancing index, which is based on edge enhancing diffusion and it describes the directionality of flow-like structures within the image46, (b) the local binary pattern (LBP), which captures intensity variations between central and neighboring pixels47 and (c) the fractal dimension (FD), which reflects the degree of complexity and was estimated using the box-counting method48.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest Statement
Dr. Conant reports membership on the Hologic, Inc., Scientific Advisory Board. The other authors have no relevant conflicts of interest to disclose.
References
- 1.Onega T, Beaber EF, Sprague BL, Barlow WE, Haas JS, Tosteson AN, et al. Breast cancer screening in an era of personalized regimens: A conceptual model and National Cancer Institute initiative for risk-based and preference-based approaches at a population level. Cancer. 2014;120(19):2955–64. doi: 10.1002/cncr.28771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siu AL. Screening for breast cancer: US Preventive Services Task Force recommendation statement. Annals of internal medicine. 2016 doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 3.Wolfe JN. Breast patterns as an index for developing breast cancer. Am J Roentgenol. 1976;126:1130–7. doi: 10.2214/ajr.126.6.1130. [DOI] [PubMed] [Google Scholar]
- 4.McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical Diagnosis and Management of Breast Cancer. Journal of Nuclear Medicine. 2016;57(Supplement 1):9S–16S. doi: 10.2967/jnumed.115.157834. [DOI] [PubMed] [Google Scholar]
- 5.Ng K-H, Lau S. Vision 20/20: Mammographic breast density and its clinical applications. Medical Physics. 2015;42(12):7059–77. doi: 10.1118/1.4935141. [DOI] [PubMed] [Google Scholar]
- 6.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiology Biomarkers & Prevention. 2006;15(6):1159–69. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 7.Gastounioti A, Conant EF, Kontos D. Beyond breast density: a review on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Research. 2016;18(1):91. doi: 10.1186/s13058-016-0755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Brentnall AR, Cuzick J, Harkness EF, Evans DG, Astley S. A novel and fully automated mammographic texture analysis for risk prediction: results from two case-control studies. Breast Cancer Research. 2017;19(1):114. doi: 10.1186/s13058-017-0906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkel RR, Nielsen M, Petersen K, Lillholm M, Nielsen MB, Lynge E, et al. Mammographic density and structural features can individually and jointly contribute to breast cancer risk assessment in mammography screening: a case–control study. BMC cancer. 2016;16(1):414. doi: 10.1186/s12885-016-2450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Giger ML, Lan L, Janardanan J, Sennett CA. Comparative analysis of image-based phenotypes of mammographic density and parenchymal patterns in distinguishing between BRCA1/2 cases, unilateral cancer cases, and controls. Journal of Medical Imaging. 2014;1(3):031009. doi: 10.1117/1.JMI.1.3.031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häberle L, Wagner F, Fasching PA, Jud SM, Heusinger K, Loehberg CR, et al. Characterizing mammographic images by using generic texture features. Breast Cancer Res. 2012;14(2):R59. doi: 10.1186/bcr3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manduca A, Carston MJ, Heine JJ, Scott CG, Pankratz VS, Brandt KR, et al. Texture features from mammographic images and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(3):837–45. doi: 10.1158/1055-9965.EPI-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Giger ML, Olopade OI, Margolis A, Lan L, Chinander MR. Computerized Texture Analysis of Mammographic Parenchymal Patterns of Digitized Mammograms. Academic Radiology. 2005;12:863–73. doi: 10.1016/j.acra.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 14.Torres-Mejia G, De Stavola B, Allen DS, Perez-Gavilan JJ, Ferreira JM, Fentiman IS, et al. Mammographic features and subsequent risk of breast cancer: a comparison of qualitative and quantitative evaluations in the Guernsey prospective studies. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1052–9. doi: 10.1158/1055-9965.EPI-04-0717. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y, Keller BM, Ray S, Wang Y, Conant EF, Gee JC, et al. Parenchymal texture analysis in digital mammography: A fully automated pipeline for breast cancer risk assessment. Medical Physics. 2015;42(7):4149–60. doi: 10.1118/1.4921996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun W, Tseng T-LB, Qian W, Zhang J, Saltzstein EC, Zheng B, et al. Using multiscale texture and density features for near-term breast cancer risk analysis. Medical Physics. 2015;42(6):2853–62. doi: 10.1118/1.4919772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller BM, Oustimov A, Wang Y, Chen J, Acciavatti RJ, Zheng Y, et al. Parenchymal texture analysis in digital mammography: robust texture feature identification and equivalence across devices. Journal of Medical Imaging. 2015;2(2):024501. doi: 10.1117/1.JMI.2.2.024501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 19.Bengio Y. Learning deep architectures for AI. Foundations and trends® in Machine Learning. 2009;2(1):1–127. [Google Scholar]
- 20.Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging. 2016;35(5):1285–98. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. arXiv preprint arXiv: 170205747. 2017 doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: Deep convolutional neural network with transfer learning from mammography. Medical Physics. 2016;43(12):6654–66. doi: 10.1118/1.4967345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, et al. Large scale deep learning for computer aided detection of mammographic lesions. Medical Image Analysis. 2017;35:303–12. doi: 10.1016/j.media.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of Medical Imaging. 2016;3(3):034501. doi: 10.1117/1.JMI.3.3.034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samala RK, Chan H-P, Hadjiiski LM, Helvie MA, Cha K, Richter C. Multi-task transfer learning deep convolutional neural network: application to computer-aided diagnosis of breast cancer on mammograms. Physics in medicine and biology. 2017 doi: 10.1088/1361-6560/aa93d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallenberg M, Petersen K, Nielsen M, Ng A, Diao P, Igel C, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Transactions on Medical Imaging. 2016;35(5):1322–31. doi: 10.1109/TMI.2016.2532122. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Giger ML, Huynh BQ, Antropova NO, editors. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. SPIE; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geras KJ, Wolfson S, Kim S, Moy L, Cho K. High-Resolution Breast Cancer Screening with Multi-View Deep Convolutional Neural Networks. arXiv preprint arXiv: 170307047. 2017 [Google Scholar]
- 29.Keller BM, Chen J, Daye D, Conant EF, Kontos D. Preliminary evaluation of the publicly available Laboratory for Breast Radiodensity Assessment (LIBRA) software tool: comparison of fully automated area and volumetric density measures in a case–control study with digital mammography. Breast Cancer Research. 2015;17(1):1–17. doi: 10.1186/s13058-015-0626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Moschidis E, Taylor C, Astley S. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014. Springer; 2014. Breast cancer risk analysis based on a novel segmentation framework for digital mammograms; pp. 536–43. [DOI] [PubMed] [Google Scholar]
- 31.Eng A, Gallant Z, Shepherd J, McCormack V, Li J, Dowsett M, et al. Digital mammographic density and breast cancer risk: a case-control study of six alternative density assessment methods. Breast Cancer Res. 2014;16(5):439. doi: 10.1186/s13058-014-0439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller BM, Nathan DL, Wang Y, Zheng Y, Gee JC, Conant EF, et al. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Medical Physics. 2012;39(8):4903–17. doi: 10.1118/1.4736530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86(11):2278–324. [Google Scholar]
- 34.LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, et al. Backpropagation applied to handwritten zip code recognition. Neural computation. 1989;1(4):541–51. [Google Scholar]
- 35.Prechelt L. Early stopping—but when? Neural Networks: Tricks of the Trade. Springer; 2012. pp. 53–67. [Google Scholar]
- 36.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988:837–45. [PubMed] [Google Scholar]
- 37.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67(2):301–20. [Google Scholar]
- 38.Lasagne: a lightweight library to build and train neural networks in Theano. Available from: https://lasagne.readthedocs.io/en/latest/
- 39.Glorot X, Bengio Y, editors. Aistats. 2010. Understanding the difficulty of training deep feedforward neural networks. [Google Scholar]
- 40.Gastounioti A, Oustimov A, Keller BM, Pantalone L, Hsieh M-K, Conant EF, et al. Breast parenchymal patterns in processed versus raw digital mammograms: A large population study toward assessing differences in quantitative measures across image representations. Medical Physics. 2016;43(11):5862–77. doi: 10.1118/1.4963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergstra J, Bengio Y. Random search for hyper-parameter optimization. The Journal of Machine Learning Research. 2012;13(1):281–305. [Google Scholar]
- 42.Materka A, Strzelecki M. Technical university of lodz, institute of electronics, COST B11 report. Brussels: 1998. Texture analysis methods–a review; pp. 9–11. [Google Scholar]
- 43.Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. Systems, Man and Cybernetics, IEEE Transactions on. 1973;(6):610–21. [Google Scholar]
- 44.Galloway MM. Texture analysis using gray level run lengths. Computer graphics and image processing. 1975;4(2):172–9. [Google Scholar]
- 45.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognition Letters. 1990;11(6):415–9. [Google Scholar]
- 46.Weickert J. Coherence-enhancing diffusion filtering. International Journal of Computer Vision. 1999;31(2-3):111–27. [Google Scholar]
- 47.Ojala T, Pietikäinen M, Mäenpää T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2002;24(7):971–87. [Google Scholar]
- 48.Caldwell CB, Stapleton SJ, Holdsworth DW, Jong RA, Weiser WJ, Cooke G, et al. Characterisation of mammographic parenchymal pattern by fractal dimension. Physics in Medicine & Biology. 1990;35(2):235–47. doi: 10.1088/0031-9155/35/2/004. [DOI] [PubMed] [Google Scholar]


