Abstract
Background
Robust methods to culture primary airway epithelial cells were developed several decades ago and these cells provide the model of choice to investigate many diseases of the human lung. However, the molecular signature of cells from different regions of the airway epithelium has not been well characterized.
Methods
We utilize DNase-seq and RNA-seq to examine the molecular signatures of primary cells derived from human tracheal and bronchial tissues, as well as healthy and diseased (cystic fibrosis (CF)) donor lung tissue.
Results
Our data reveal an airway cell signature that is divergent from other epithelial cell types and from common airway epithelial cell lines. The differences between tracheal and bronchial cells are clearly evident as are common regulatory features. Only minor variation is seen between bronchial cells from healthy or CF donors.
Conclusions
These data are a valuable resource for functional genomics analysis of airway epithelial tissues in human disease.
1. Introduction
Major advances in understanding mechanisms of human lung disease have arisen through the development of methods to culture primary cells from the airway epithelium (1), (2). These cells may be differentiated on permeable supports to generate a polarized cell layer that at air-liquid interface are a robust model for investigating many aspects of airway epithelial function. Cells of nasal, tracheal and bronchial epithelial tissue are all amenable to these protocols, though the bronchial cells have been most extensively studied. Moreover, the latter are the desired endpoint of recent methods to differentiate human induced pluripotent stem cells (IPSCs) into airway epithelium (3, 4). Use of these primary cell cultures to study normal human airway epithelial biology (5) and dysfunction in diseases such as cystic fibrosis (CF), asthma (6) and chronic obstructive pulmonary disease (COPD) (7) among other disorders has provided important insights into disease mechanisms. The precise cellularity of each tracheal and bronchial culture will depend on many factors, including substantial donor-to-donor variation, both genetic and environmental, and specific culture protocols. Our goal was to compare the molecular signatures of primary human tracheal (HTE) and bronchial epithelial (HBE) cells to determine functional differences between them. We used genome-wide analysis of open chromatin, which is associated with active regulatory elements, combined with gene expression profiles of each cell type. The signature of HBE cells derived from healthy and CF donor lungs was also compared. The data revealed a clustering of open chromatin profiles in primary airway cells, which was distinct from that observed in lung epithelial cell lines and other epithelial cell types. Though there was substantial overlap between the tracheal and bronchial cells, distinct features of each cell population were evident and correlated well with divergent gene expression profiles. Also of note were the very limited differences between HBE and CFHBE primary cells, with only a few disease-related pathways identified.
2. Methods
2.1 HTE, HBE, CF HBE and NHBE cells
Human tracheae were collected post mortem from healthy donors and human tracheal epithelial (HTE) cells isolated and grown as described previously (1). Human bronchial epithelial (HBE) cells were obtained (at passage 1, P1) under protocol #03–1396 approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board (2). Informed consent was obtained from authorized representatives of all organ donors. Normal human bronchial epithelial (NHBE) cells were purchased from Lonza. Primary HTE, HBE and NHBE cells were grown on collagen-coated plastic in bronchial epithelial growth medium (BEGM, Lonza) (2). Cells were grown on plastic rather than permeable supports to provide adequate cell numbers for the DNAse-seq protocol and to ensure efficient DNase I digestion. See Supplemental Methods for detailed culture protocols.
2.2 DNase-seq and RNA-seq
DNase-seq and RNA-seq libraries were prepared according to standard protocols. See Supplemental Methods for detailed analysis methods.
3. Results and Discussion
3.1 DNase-seq in HTE, HBE and NHBE cells reveals substantial similarities between the open chromatin landscapes of the three cell types
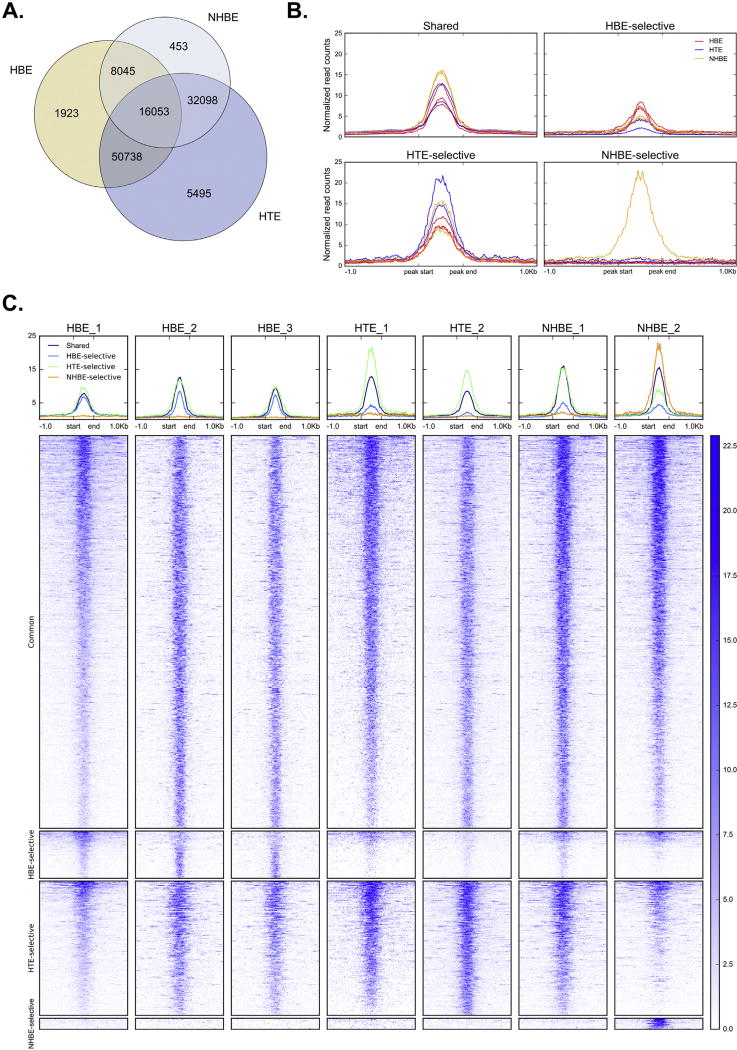
Regulatory elements within the genome are usually associated with regions of open chromatin, which can be readily detected by enzymatic or chemical methods. In order to compare open chromatin profiles in human bronchial (HBE) and tracheal (HTE) cells, we performed DNase-seq on three cell types: primary HBE cells cultured from three different donors, primary HTE cells from two different donors, and NHBE cells (Lonza CC-2541, a mixture of HBE and HTE cells) from two donor codes. After mapping reads to the human genome, we used the differential peak calling function of the MACS2 peak caller to identify subclasses of DNase-I hypersensitive sites (DHS) as follows: “common” DHS are present in all three (HBE/HTE/NHBE) cell types, “shared” DHS are present in pairs of samples, and “cell type selective” DHS are present in only one of the three cell types. A total of 56,649 peaks were seen in NHBE cells, 76,759 peaks were evident in HBE cells, and 104,384 peaks were called in HTE cells. Of these, 16,053 were common to all three of the cell types (Fig. 1A). Only a small percentage of peaks were cell type selective. NHBE showed 453 (0.8%) cell type-selective DHS, HBE had 1,923 (2.5%) such sites, and HTE 5,495 (5.3%). Pairwise comparison of cell type-selective and shared DHS revealed that HBE and HTE were the most similar, with high reciprocal overlaps of 87% (HBE) and 64% (HTE). In contrast, NHBE DHS appeared to represent a subset of HTE DHS, with 85% of NHBE DHS found in HTE, but only 46% of HTE DHS present in NHBE. NHBE and HBE were the least similar, with less than 50% of DHS in either cell type present in the other (HBE: 31%, NHBE: 43%). Since NHBE cells are established as mixture of tracheal and bronchial epithelial cells these profiles suggest that in the lot numbers that we analyzed cells of tracheal origin were predominant. Though the HBE cultures may have a small (1–5cm of trachea) contribution of tracheal cells these are a minor component of the population. Comparison of normalized DNase-seq signal across replicates (Fig. 1B–C) confirmed the cell type-specificity of DNase-I sensitivity at HTE- and HBE-selective DHS. The “NHBE-selective” DHS, however, were detectable exclusively in one NHBE replicate, while the other NHBE sample appeared most similar to the HTE profile.
Figure 1. Overlap between DHS in multiple human airway cell types.
A. Intersection of all DHS between HTE, HBE and Lonza NHBE. B–C. Average profiles (B) and heatmaps (C) of normalized DNase-seq signal at common and cell-type selective DHS ±1 kb.
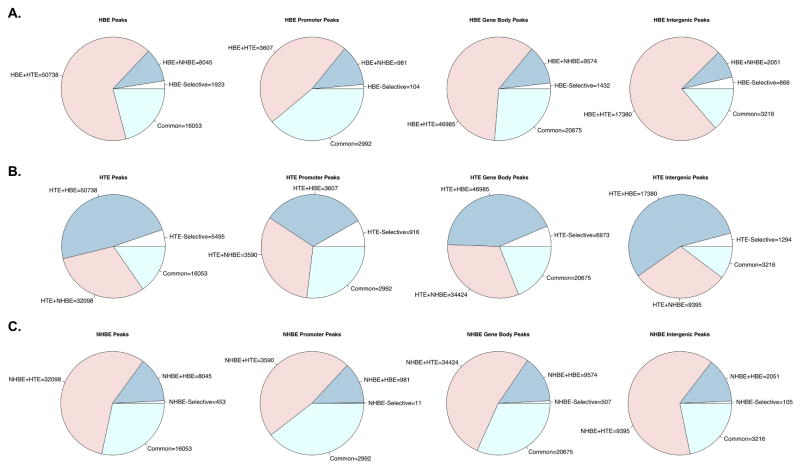
Next, we used gene annotations to classify DHS as either promoter-associated (within 2 kb upstream of an annotated transcription start site [TSS]), gene-body associated (between 20kb upstream and 20kb downstream of an annotated gene), or intergenic (at least 20kb from the nearest annotated gene). Pairwise comparison of cell type-selective and shared DHS in promoter regions, gene bodies and intergenic regions were largely consistent with the patterns observed when comparing all DHS (data not shown). Notably, nearly 50% reciprocal overlap was seen between NHBE and HBE DHS at promoters, in contrast to many fewer common sites in the overall and intergenic (HBE: 22%, NHBE: 36%) comparisons. This suggests that where the same genes are active in the 2 cultures types, they may often recruit different cis-regulatory elements to drive their promoters. Alternatively, more of the intergenic sites are not directly associated with the transcriptional program of the cells. To further compare the distribution of overlap between cell types, we next visualized the proportion of cell type-selective, shared (between 2 cell types) and common (seen in all 3 cell types) DHS by annotation in HBE (Fig. 2A), HTE (Fig. 2B) and NHBE (Fig. 2C) cells. In each cell type, the highest proportion of shared sites was found in the promoter-associated DHS (second column), supporting a hypothesis of a shared airway epithelial chromatin landscape at gene promoters. Promoters were also the location of the lowest proportion of cell type-selective DHS in both HBE and NHBE cells. In HTE cells, however, the highest fraction of HTE-selective sites was found at promoters. This difference likely reflects the lower proportion of promoter DHS shared between HTE and HBE relative to other annotation classes. Nonetheless, the most numerous class of promoter-associated HTE DHS overall remains those that are shared with HBE.
Figure 2. Distribution of peak classes in annotated human airway DHS.
Numbers of DHS by overlap with other cell types, sorted by annotations, in HBE (A), HTE (B) and Lonza NHBE (C).
3.2 Human primary airway cells cluster independently of other epithelial cells and unrelated cell types by DHS utilization
Next, we compared the open chromatin landscape of the primary human airway cells to DHS profiles of 1) airway cell lines that are commonly used for functional assays of airway epithelium, and 2) other human epithelial and non-epithelial cell types. Normalized read coverage was measured at all DHS in multiple airway cells types: the primary airway cells, an immortalized human bronchial epithelial cell line (16HBE14o-), two lung adenocarcinoma lines of different cellular origin (Calu3, bronchial and A549, alveolar type II), and primary HBE cultures from two donors with cystic fibrosis (CF-HBE). A more extensive comparison was done with other non-airway epithelial cell types (Caco2, colorectal adenocarcinoma, HepG2, hepatocellular carcinoma and HeLa-S3, cervical carcinoma) and non-epithelial cells (GM12878, lymphoblastoid, Human Umbilical Vein Endothelial Cells (HUVEC), endothelial, K562, erythroleukemia and FibroP, fibroblasts from 3 individuals with Parkinson’s disease) (8). See Supplemental Methods for data accession numbers.
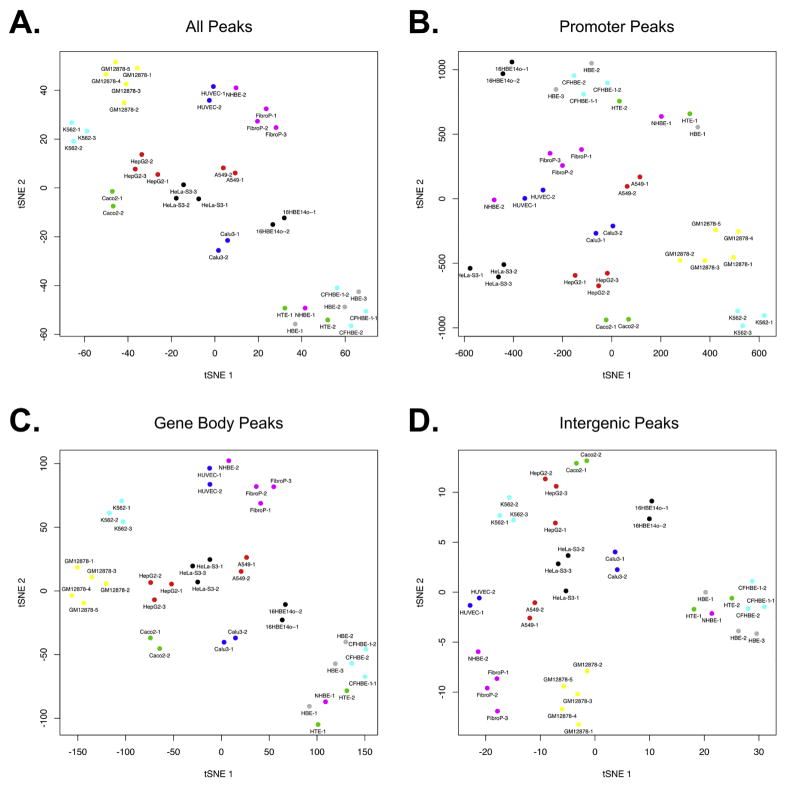
Visual clustering of all cell types by t-Distributed Stochastic Neighbor Embedding (tSNE), which enables the visualization of high-dimensionality data, is shown in Fig. 3. When all DHS are included (Fig. 3A), the human airway cultures cluster independently of the other cell types. An exception is one NHBE culture, which is located between biological replicas of HUVEC and FibroP samples, raising doubt about the cellularity of this commercial sample. Otherwise, the tSNE distribution of the airway cultures with respect to the other cell types was consistent with their cellular origin, with the human bronchial airway cell lines (16HBE14o- and Calu3) nearest and the more distantly related airway and other epithelial cell lines (A549, Caco2, HepG2 and HeLa-S3) slightly further away. When focusing on promoter-associated DHS (Fig. 3B), 16HBE14o- cells cluster together with the primary airway cultures, likely reflecting the normal bronchial epithelial origin of this cell line, which was immortalized with SV40 large T antigen. Inspection of the intergenic peaks (Fig. 3D) reinforces our previous observations (9, 10) that the majority of cell-specific regulatory elements lie in these regions. This is evident in the greater divergence in the tSNE plots of primary airway cells and all the long-term cell lines. For example, HeLa-S3 and Caco-2 non-airway epithelial cell lines are nearly as far from the primary airway cultures as the lymphoblastoid (GM12878) and erythroleukemia (K562) cell lines. It is also evident that clustering by gene-body associated (Fig. 3C) and intergenic (Fig. 3D) DHS is largely consistent with clustering by all DHS (Fig. 3A). Of note is the second NHBE culture (NHBE-2, Lonza), which clusters much closer to the other primary airway cells when considering only promoter DHS (Fig. 3B) than when considering all DHS (Fig. 3A). This suggests NHBE-2 shares active promoters with the other primary airway cultures, but not cell-type specific elements that we (9, 10), and others (11),(12) have shown to predominate in other genic and intergenic regions. This raises some questions about the precise differentiated state of this culture.
Figure 3. Clustering of human airway cultures by open chromatin.
tSNE dimensionality reduction plot showing relationships between 14 human cell types determined by all HBE-HTE-NHBE DHS (A), promoter DHS only (B), gene body DHS only (C) or intergenic DHS only (D).
3.3 Motif enrichment analysis reveals novel and known regulatory factors in primary airway epithelial cells
To reveal the transcriptional programs that could underlie the cell-specific signatures of open chromatin in HTE, HBE and NHBE cells we next measured transcription factor (TF) binding motif enrichment in our annotated DHS using Clover. The top 20 most significant cell-type specific overrepresented motifs in promoters and intergenic regions are summarized in Table 1 (full results in Supplementary table 1). Among promoter region DHS shared between the three cell types, nearly 300 TFs were over-represented, with more than 200 over-represented in shared intergenic DHS (2–10 kb from the nearest gene or > 20 kb away). Perhaps more informative were the relatively small number of enriched motifs that were found in cell type-selective DHS. Though shorter, these lists included several factors known to play a role in airway development and function.
Table 1.
Top 20 overrepresented motifs specific to cell-type selective promoter and intergenic DHS.
| Motifs overrepresented in: | |||
|---|---|---|---|
| Shared promoter DHS | HTE-selective promoter DHS only | HBE-selective promoter DHS only | NHBE-selective promoter DHS only |
| MA0741.1_KLF16 | MA0599.1_KLF5 | MA0501.1_MAF::NFE2 | MA0521.1_Tcf12 |
| MA0108.2_TBP | MA0039.2_Klf4 | MA0631.1_Six3 | MA0499.1_Myod1 |
| MA0517.1_STAT1::STAT2 | MA0746.1_SP3 | MA0067.1_Pax2 | MA0037.2_GATA3 |
| MA0095.2_YY1 | MA0152.1_NFATC2 | MA0056.1_MZF1 | MA0140.2_GATA1::TAL1 |
| MA0084.1_SRY | MA0747.1_SP8 | - | MA0050.2_IRF1 |
| MA0076.2_ELK4 | MA0477.1_FOSL1 | - | MA0766.1_GATA5 |
| MA0514.1_Sox3 | MA0478.1_FOSL2 | - | - |
| MA0062.2_Gabpa | MA0491.1_JUND | - | - |
| MA0601.1_Arid3b | MA0655.1_JDP2 | - | - |
| MA0624.1_NFATC1 | MA0099.2_FOS::JUN | - | - |
| MA0625.1_NFATC3 | MA0104.3_Mycn | - | - |
| MA0470.1_E2F4 | MA0058.3_MAX | - | - |
| MA0471.1_E2F6 | MA0647.1_GRHL1 | - | - |
| MA0645.1_ETV6 | MA0606.1_NFAT5 | - | - |
| MA0144.2_STAT3 | MA0742.1_Klf12 | - | - |
| MA0136.2_ELF5 | MA0825.1_MNT | - | - |
| MA0703.1_LMX1B | MA0472.2_EGR2 | - | - |
| MA0851.1_Foxj3 | - | - | - |
| MA0913.1_Hoxd9 | - | - | - |
| MA0910.1_Hoxd8 | - | - | - |
|
| |||
| Shared intergenic DHS | HTE-selective intergenic DHS only | HBE-selective intergenic DHS only | NHBE-selective intergenic DHS only |
|
| |||
| MA0478.1_FOSL2 | MA0528.1_ZNF263 | MA0486.2_HSF1 | MA0496.1_MAFK |
| MA0489.1_JUN(var.2) | MA0746.1_SP3 | MA0078.1_Sox17 | MA0472.2_EGR2 |
| MA0490.1_JUNB | MA0155.1_INSM1 | MA0770.1_HSF2 | MA0820.1_FIGLA |
| MA0491.1_JUND | MA0106.3_TP53 | MA0744.1_SCRT2 | MA0117.2_Mafb |
| MA0477.1_FOSL1 | MA0067.1_Pax2 | MA0869.1_Sox11 | MA0525.2_TP63 |
| MA0476.1_FOS | MA0113.3_NR3C1 | MA0083.3_SRF | MA0091.1_TAL1::TCF3 |
| MA0516.1_SP2 | MA0493.1_Klf1 | MA0795.1_SMAD3 | MA0732.1_EGR3 |
| MA0152.1_NFATC2 | MA0098.3_ETS1 | MA0025.1_NFIL3 | - |
| MA0599.1_KLF5 | MA0647.1_GRHL1 | MA0093.2_USF1 | - |
| MA0139.1_CTCF | MA0611.1_Dux | MA0526.1_USF2 | - |
| MA0099.2_FOS::JUN | MA0775.1_MEIS3 | MA0730.1_RARA(var.2) | - |
| MA0081.1_SPIB | MA0007.3_Ar | MA0639.1_DBP | - |
| MA0079.3_SP1 | MA0145.3_TFCP2 | MA0043.2_HLF | - |
| MA0056.1_MZF1 | MA0474.2_ERG | MA0843.1_TEF | - |
| MA0606.1_NFAT5 | MA0760.1_ERF | - | - |
| MA0462.1_BATF::JUN | MA0258.2_ESR2 | - | - |
| MA0500.1_Myog | MA0475.2_FLI1 | - | - |
| MA0442.1_SOX10 | MA0762.1_ETV2 | - | - |
| MA0624.1_NFATC1 | - | - | - |
| MA0499.1_Myod1 | - | - | - |
| MA0521.1_Tcf12 | - | - | - |
Over-represented motifs in primary airway cell promoter DHS
Notable in the list of HTE-selective peaks are the Kruppel-like factors KLF4 and KLF5, which are known to have a key role in early endoderm development (13). During ES cell differentiation in the mouse KLF4 expression inhibits endoderm differentiation while KLF5 inhibits mesodermal markers. Deregulation of KLF4 was also linked to repair processes in CF human airway epithelial cultures (14). Moreover, these factors have an extensive documented role in epithelial cancers (reviewed in (15)).
In NHBE selective peaks the IRF1 motif is significantly over-represented, which is of interest since this factor is a member of the interferon regulatory factor family, which has a key role in the immune response, immune cell development and regulation of cell growth and apoptosis (16), (17). Indeed, five of the six genes with NHBE-selective promoter DHS containing IRF1 motifs (MLLT1, Super Elongation complex subunit (MLLT11), H2.0-like homeobox (HLX), Zic Family Member 1 (ZIC1), Single-Minded Family BHLH Transcription Factor (SIM1) and Ubiquitin Specific Peptidase9, X-linked (USP9X) are annotated with gene ontology (GO) terms relating to development, cell growth and/or apoptosis. IRF1 is induced by interferon gamma (IFN-γ) and binds to the promoters of IFN-γ responsive genes, activating gene expression. IRF2, which is also induced by IFN-γ is synthesized after IRF1 and acts as and IRF1 agonist. We recently showed this pair of factors to regulate an airway-selective enhancer in the cystic fibrosis transmembrane conductance regulator CFTR gene (18). Another potentially interesting enriched motif is SIX homeobox 3 (SIX3) in HBE-selective promoters. SIX3 is well-studied in development, but its functions in normal epithelia have not been defined.
Overrepresented motifs in primary airway cell intergenic DHS
As mentioned above, data from many groups including ours (9, 10), (19) showed that the majority of cell type-selective DHS are located in intergenic regions. Hence the overrepresented TF motifs identified in these peaks are potentially the most informative in discerning the transcriptional network in individual cell types. Particularly interesting is the observation of overrepresentation of the grainy-head like transcription factor 1 (GRHL1) motif in HTE-selective peaks in intergenic (2–10kb and >20kb) sites. The 3 GRHL factors (GRHL1/2/3) all interact with the same motif (AACCGGTT) and are known to play a key role in epidermis development and differentiation (20),(21, 22),(23). All 3 factors are expressed in HTE and HBE cells though GRHL2 is best studied in the human airway epithelium, where it has a role in regulating cell adhesion, motility and differentiation (5). Also overrepresented in intergenic DHS (2–10kb) in HTE-selective sites are the motifs for several nuclear hormone receptors, the Androgen Receptor (AR), the Glucocorticoid Receptor, (GR, NR3C1) and Estrogen Receptor Beta (ER-β, ESR2). The AR observation is likely not an artifact of donor tissue gender as all HBE and HTE samples analyzed here were from males. In addition to these cell-type specific factors, many transcription factors with known roles in the airway appear in common DHS, including NF-κB (RELA) (24), Yin Yang 1 (YY1) (25), Homeobox A5 (HOXA5) (26) and several ETS transcription factors (ELF5, ETS1, ELF3) (27), (28), (29).
3.4 Gene expression is tightly correlated with nearby active regulatory elements identified by open chromatin
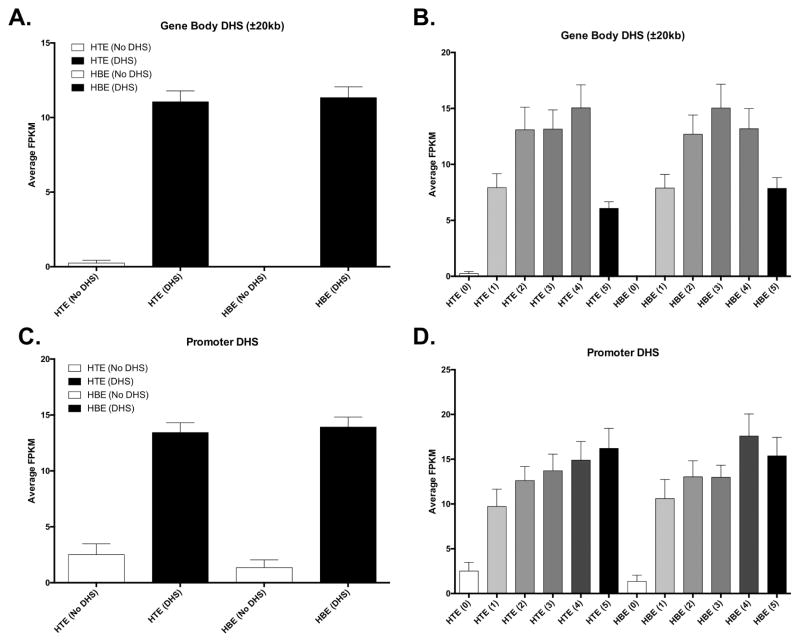
To determine if DHS presence and intensity were correlated with gene expression we compared RNA-seq data from HTE and HBE with our mapped DHS (Figure 4; GO term enrichment in Supplementary Table 2). We found that genes with no DHS nearby (no DHS between 20kb upstream and 20kb downstream) were rarely expressed in either HTE or HBE, while genes with nearby DHS were robustly detected (Fig. 4A). Next, we binned DHS by DNase-seq coverage into six bins (0–5; 0 = no reads; 5 = highest quintile). We found that the intensity of DHS hypersensitivity was positively correlated with the expression of nearby genes (Fig. 4B), although this trend did not extend to the most hypersensitive DHS (bin 5). Interestingly, we found that the lack of a promoter-associated DHS (within 2kb upstream of TSS) was less predictive of repressed gene expression (Fig. 4C) than the lack of a DHS anywhere near the locus, but the presence of a promoter-associated DHS was associated with higher average gene expression than a DHS in the general vicinity of a gene. The intensity of promoter-associated DHS was also correlated with gene expression, with the trend continuing to the most hypersensitive sites in this case (Fig. 4D).
Figure 4. Gene expression is tightly associated with open chromatin.
All plots: Average gene expression (FPKM) in HTE and HBE cultures (x-axis) in the indicated groups. (A) Genes with and without DHS in the gene body or 20kb up/downstream. (B) Genes with (1–5) and without (0) DHS in the gene body or 20kb up/downstream, with DHS binned by quintile intensity (1–5). (C) Genes with and without DHS in the promoter (2 kb). (D) Genes with (1–5) and without (0) DHS in the promoter (2 kb), with DHS binned by quintile intensity (1–5).
3.5 CF-HBE and WT-HBE
To assess the impact of cystic fibrosis on the open chromatin landscape of human bronchial epithelial cells, we compared RNA-seq and DNase-seq data sets from WT-HBE and CF-HBE cells from two different donors with CF. We found that both gene expression profiles and DHS were very similar between WT- and CF-HBEs, with only 116 differentially expressed genes detected (Supplementary Table 4) and >94% of all DHS shared between the two cell types (Fig. 5A). By DNase-seq, the CF-HBE open chromatin landscape appears to be a subset of the open chromatin observed in WT-HBE, with >99% of all DHS shared with WT-HBE. This is also true for promoter, intergenic and gene body DHS. In contrast, 4–6.5% of WT-HBE DHS are specific to WT-HBE. While this is a substantially larger proportion of sites than those unique to CF-HBE, this still reflects the high degree of similarity between the two cell types when compared to the pairwise comparisons between HTE, HBE and NHBE.
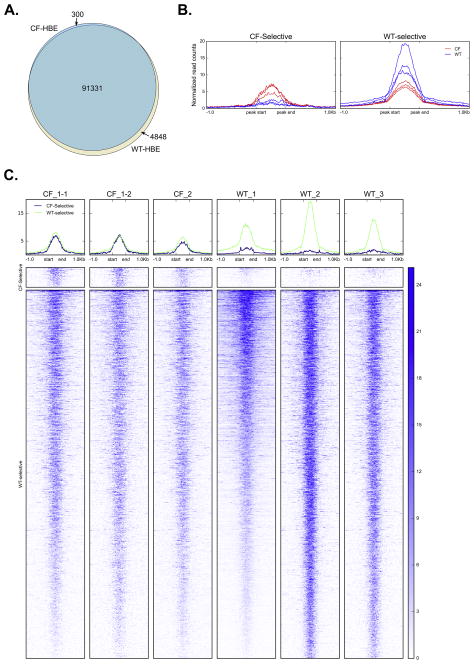
Figure 5. Overlap between DHS in CF and WT HBE.
A. Intersection of all DHS between CF-HBE and WT-HBE, including CF HBE technical replicates. B. Average profiles of normalized DNase-seq signal at WT- and CF-selective DHS ±1 kb. C. Heatmaps of DNase-seq signal at WT- and CF-selective DHS ±1 kb.
Despite the small number of unique sites in CF- and WT-HBE, the normalized DNase-seq signal (Fig. 5B–C) shows that these DHS are highly specific across replicates. The CF-selective sites are barely detectable in WT-HBE, while the WT-selective sites are significantly weaker in CF-HBE. To determine the functional relevance of these sites, we determined differential transcription factor (TF) binding motif over- and underrepresentation at CF-selective sites relative to WT-selective sites using Clover. The top 10 most significant over- and underrepresented motifs in CF-selective sites are summarized in Table 2 (full results in Supplementary Table 3). The most significantly enriched motifs at CF-selective sites are those associated with the AP-1 transcription factor (dimer composed of Jun, Fos or ATF). This factor is involved in regulation of inflammatory mediators, including IL-8, and has been reported to be dysregulated in CF airway epithelial cells (30). In contrast, the most significant underrepresented terms are ETS-family transcription factors, including E47-like factor 5 (ELF5) and Ets homologous factor (EHF), both of which were associated with CF lung disease severity in genome wide association studies (31). Moreover, we recently showed a pivotal role for EHF in epithelial dysfunction in lung disease (19, 32).
Table 2.
Top 10 over- and underrepresented motifs in CF-selective sites relative to WT-selective sites.
| Relative to WT-selective sites: | |
|---|---|
| Overrepresented in CF-selective sites | Underrepresented in CF-selective sites |
| MA0099.2_FOS::JUN | MA0473.2_ELF1 |
| MA0476.1_FOS | MA0028.2_ELK1 |
| MA0489.1_JUN(var.2) | MA0641.1_ELF4 |
| MA0490.1_JUNB | MA0131.2_HINFP |
| MA0477.1_FOSL1 | MA0759.1_ELK3 |
| MA0478.1_FOSL2 | MA0506.1_NRF1 |
| MA0491.1_JUND | MA0763.1_ETV3 |
| MA0462.1_BATF::JUN | MA0765.1_ETV5 |
| MA0655.1_JDP2 | MA0761.1_ETV1 |
| MA0442.1_SOX10 | MA0764.1_ETV4 |
In conclusion, we show the molecular signature of primary human tracheal and bronchial epithelial cells and describe the common and distinct features of these cell types utilizing standard culture protocols. Further experiments to include cells from additional healthy and CF organ donors could be an advantage, though the bioinformatics tools used here tend to eliminate variation that is not significant. Our data suggest that the open chromatin landscape is surprisingly consistent between lung tissue donors, since the majority of DHS are shared between HBE cells from WT individuals and CF patients. Moreover, there is very little cis-regulatory element divergence in HBE cells derived from patients with CF, when compared to those derived from healthy lungs.
3.6 Translational relevance
Many approaches to develop new therapies for human lung disease rely on cell-based assays. For example, high throughput pharmacological screens, gene therapy protocols and most recently gene editing methods rely on preclinical studies in human airway epithelial cells. The challenges of culturing and expanding primary cells leads to the use of surrogate cell line models. Our data show the detailed molecular signatures of each cell type. They illustrate the importance of using primary cells as models for the human airway. Moreover, the data resource generated will be of considerable use to investigators of lung health and disease.
Supplementary Material
Highlights.
Primary human bronchial and tracheal epithelial cells have unique molecular signatures that are distinct from commonly used airway cell lines.
Genome-wide analysis of open chromatin state in conjunction with global transcriptomics revealed a high-resolution profile for each cell type.
Only minor variation is seen between healthy and CF bronchial epithelial cells.
Acknowledgments
We thank Drs A Safa, L. Song and G E Crawford for DNAse-seq library making and sequencing and Dr P Faber and staff at the University of Chicago Genomics Core for RNA-seq.
All data were deposited into Gene Expression Omnibus (GEO) GSE with accession number GSE101993.
Funding sources.
This work was funded by the National Institutes of Health: R01HL094585, R01HD068901 and R01HL117843 (PI:A.H.); P30DK065988; also by the Cystic Fibrosis Foundation (Harris 11G0, 14P0, 16G0, 15XX0, 17XX0, Drumm 15R0).
Footnotes
Declaration of Interests: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davis PB, Silski CL, Kercsmar CM, Infeld M. Beta-adrenergic receptors on human tracheal epithelial cells in primary culture. Am J Physiol. 1990;258(1 Pt 1):C71–6. doi: 10.1152/ajpcell.1990.258.1.C71. [DOI] [PubMed] [Google Scholar]
- 2.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 3.Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nature biotechnology. 2012;30(9):876–82. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firth AL, Dargitz CT, Qualls SJ, Menon T, Wright R, Singer O, et al. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2014;111(17):E1723–30. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao X, Bali AS, Randell SH, Hogan BL. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol. 2015;211(3):669–82. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JA, Kim JH, Bi D, Mitchel JA, Qazvini NT, Tantisira K, et al. Unjamming and cell shape in the asthmatic airway epithelium. Nat Mater. 2015;14(10):1040–8. doi: 10.1038/nmat4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polosukhin VV, Cates JM, Lawson WE, Zaynagetdinov R, Milstone AP, Massion PP, et al. Bronchial secretory immunoglobulin a deficiency correlates with airway inflammation and progression of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;184(3):317–27. doi: 10.1164/rccm.201010-1629OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischof JM, Gillen AE, Song L, Gosalia N, London D, Furey TS, et al. A genome-wide analysis of open chromatin in human epididymis epithelial cells reveals candidate regulatory elements for genes coordinating epididymal function. Biol Reprod. 2013;89(4):104, 1–8. doi: 10.1095/biolreprod.113.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischof JM, Ott CJ, Leir SH, Gosalia N, Song L, London D, et al. A genome-wide analysis of open chromatin in human tracheal epithelial cells reveals novel candidate regulatory elements for lung function. Thorax. 2012;67(5):385–91. doi: 10.1136/thoraxjnl-2011-200880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome research. 2011;21(10):1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aksoy I, Giudice V, Delahaye E, Wianny F, Aubry M, Mure M, et al. Klf4 and Klf5 differentially inhibit mesoderm and endoderm differentiation in embryonic stem cells. Nature communications. 2014;5:3719. doi: 10.1038/ncomms4719. [DOI] [PubMed] [Google Scholar]
- 14.Crespin S, Bacchetta M, Bou Saab J, Tantilipikorn P, Bellec J, Dudez T, et al. Cx26 regulates proliferation of repairing basal airway epithelial cells. The international journal of biochemistry & cell biology. 2014;52:152–60. doi: 10.1016/j.biocel.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Limame R, Op de Beeck K, Lardon F, De Wever O, Pauwels P. Kruppel-like factors in cancer progression: three fingers on the steering wheel. Oncotarget. 2014;5(1):29–48. doi: 10.18632/oncotarget.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annual review of immunology. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annual review of immunology. 2008;26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Leir SH, Harris A. Immune Mediators Regulate CFTR Expression through a Bifunctional Airway-Selective Enhancer. Molecular and cellular biology. 2013;33(15):2843–53. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fossum SL, Mutolo MJ, Yang R, Dang H, O’Neal WK, Knowles MR, et al. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 2014;42(22):13588–98. doi: 10.1093/nar/gku1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308(5720):381–5. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- 21.Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308(5720):411–3. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 22.Wilanowski T, Caddy J, Ting SB, Hislop NR, Cerruti L, Auden A, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008;27(6):886–97. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boglev Y, Wilanowski T, Caddy J, Parekh V, Auden A, Darido C, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev Biol. 2011;349(2):512–22. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Hart LA, Krishnan VL, Adcock IM, Barnes PJ, Chung KF. Activation and Localization of Transcription Factor, Nuclear Factor-κ B, in Asthma. American Journal of Respiratory and Critical Care Medicine. 1998;158:1585–92. doi: 10.1164/ajrccm.158.5.9706116. [DOI] [PubMed] [Google Scholar]
- 25.Boucherat O, Landry-Truchon K, Bérubé-Simard F-A, Houde N, Beuret L, Lezmi G, et al. Epithelial inactivation of Yy1 abrogates lung branching morphogenesis. Development. 2015:142. doi: 10.1242/dev.120469. [DOI] [PubMed]
- 26.Boucherat O, Montaron S, Bérubé-Simard F-A, Aubin J, Philippidou P, Wellik DM, et al. Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2013:304. doi: 10.1152/ajplung.00006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzger DE, Xu Y, Shannon JM. Elf5 is an epithelium-specific, fibroblast growth factor–sensitive transcription factor in the embryonic lung. Developmental Dynamics. 2007;236:1175–92. doi: 10.1002/dvdy.21133. [DOI] [PubMed] [Google Scholar]
- 28.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho I-C. Ets-1 is a negative regulator of Th17 differentiation. The Journal of experimental medicine. 2007;204:2825–35. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver JR, Kushwah R, Wu J, Pan J, Cutz E, Yeger H, et al. Elf3 plays a role in regulating bronchiolar epithelial repair kinetics following Clara cell-specific injury. Lab Invest. 2011;91(10):1514–29. doi: 10.1038/labinvest.2011.100. [DOI] [PubMed] [Google Scholar]
- 30.Saadane A, Eastman J, Berger M, Bonfield TL. Parthenolide inhibits ERK and AP-1 which are dysregulated and contribute to excessive IL-8 expression and secretion in cystic fibrosis cells. J Inflamm (Lond) 2011;8:26. doi: 10.1186/1476-9255-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corvol H, Blackman SM, Boelle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nature communications. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fossum SL, Mutolo MJ, Tugores A, Ghosh S, Randell SH, Jones LC, et al. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J Biol Chem. 2017;292(26):10938–49. doi: 10.1074/jbc.M117.775304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.