Abstract
Objective
Recent studies demonstrate that infection with the Human Immunodeficiency Virus-1 (HIV) is associated with accelerated aging effects in adults according to a highly accurate epigenetic biomarker of aging known as epigenetic clock. However, it not yet known whether epigenetic age acceleration occurs as early as adolescence in perinatally HIV-infected (PHIV+) youth.
Design
Observational study of PHIV and HIV-uninfected adolescents enrolled in the Cape Town Adolescent Antiretroviral Cohort (CTAAC) Study.
Methods
The Illumina EPIC array was used to generate blood DNA methylation data from 204 PHIV and 44 age-matched, uninfected (HIV-) adolescents aged 9 to 12 years old. The epigenetic clock software and method was used to estimate two measures of epigenetic age acceleration. Each participant completed a comprehensive neuropsychological test battery upon enrolment to CTAAC.
Results
HIV is associated with biologically older blood in PHIV+ adolescents according to both measures of epigenetic age acceleration. One of the measures, extrinsic epigenetic age acceleration, is negatively correlated with measures of cognitive functioning (executive functioning, working memory, processing speed).
Conclusions
Overall, our results indicate that epigenetic age acceleration in blood can be observed in PHIV+ adolescents and that these epigenetic changes accompany poorer cognitive functioning.
Keywords: HIV, DNA methylation, biomarker, aging, perinatal HIV, epigenetic clock
INTRODUCTION
Childhood HIV infection and its consequences on development remain a significant public health issue in several countries. There are an estimated 2.3 million children under the age of 15 years living with HIV/AIDS in sub-Sahara Africa, with 330,000 in South Africa (SA) alone. A growing number of perinatally HIV-infected (PHIV+) children in SA are surviving into adolescence, due to earlier diagnosis and initiation of effective antiretroviral therapies (ARTs). However, as elsewhere, PHIV+ adolescents face unique health issues, particularly related to neurocognitive and neurobiological complications resulting from HIV that will adversely hinder their transition into adult roles. HIV infection leads to changes in the structure and function of the central nervous system, which has significant consequences on academic performance and psychosocial functioning that compound over the lifespan[1–4]. Recent data from our lab indicates a pattern of damaged neuronal microstructure, smaller gray matter volumes, reduced cortical surface area and decreased gyrification in SA adolescents living with HIV and suggests abnormal neurodevelopment in perinatally infected adolescents[5]. The South African Department of Health first line antiretroviral (ART) regimen for children older than 3 years and more than 10kg is Abacavir (ABC) + lamivudine (3TC) + efavirenz (EFV). First-line treatment failure may be a key factor associated with CNS damage in perinatally infected adolescents.
The complexity of the problem presents unique public health challenges, including how to identify those youths that are at risk for neurocognitive complications or who already have as-of-yet undetectable neurobiological aberrations due to HIV, and how to intervene effectively so that they can realize their full potential. Yet despite the obvious public health emergency this represents, the complex interaction these factors have on neurocognitive and neurophysiological functioning during this important life phase remain poorly understood. Indeed, recent findings that asymptomatic older PHIV+ children considered “slow progressors” have measurable neurocognitive deficits that correlate with diffusion tensor imaging aberrations[6] is alarming. Therefore, it is important to understand the neurocognitive and neurodevelopmental issues in this group to assist in their transition to adulthood roles, and to open up avenues for helpful interventions.
Epigenetic regulation is an essential aspect of neurodevelopment and cell differentiation and is affected by environmental factors, thus making it a potent mechanism for understanding environmental effects on neurobehavioral and neurophysiological phenotypes[7] from early developmental aberrations into neurodegenerative diseases late in life[8, 9]. Because epigenetic mechanisms such as DNA methylation (DNAm) act as an interface between the environment and the genome, it is especially relevant when discussing neurocognitive problems in PHIV+ children, for whom a substantial environmental component is likely involved. We, and others, have shown that host DNAm is disrupted by HIV infection[10–14]. Specifically, we recently found evidence that HIV infection is associated with accelerated epigenetic aging based on the epigenetic clock[15] in both brain and peripheral blood mononuclear cells[10]. That is, brains of adults infected with HIV demonstrated age acceleration of 7.4 years compared to uninfected controls, and 5.2 years in peripheral blood mononuclear cells, findings that have been validated by other groups[12–14]. Furthermore, we have found that accelerated epigenetic aging can be observed in brain tissue samples of adults diagnosed pre-mortem with HIV-associated neurocognitive disorders (HAND)[16]. As such, the epigenetic clock method lends itself for detecting accelerated biological aging effects due to HIV infection in adult cohorts, and for determining whether accelerated epigenetic aging has significant health consequences. However, it is not yet known whether accelerated epigenetic aging effects are already detectable in adolescents and, if so, whether these epigenetic changes correlate with measures of cognitive performance, immune status and ART treatment regimen
In the current study, we apply the epigenetic clock to a cohort of South African adolescents in order to determine if perinatally acquired HIV infection is associated with accelerated aging in children and adolescents, and if so, whether or not it is also associated with neurocognitive impairment, current CD4, viral load and ART treatment line. Our hypotheses were that PHIV infection would be associated with accelerated aging, and that accelerated aging would be associated with neurocognitive impairment.
MATERIALS AND METHODS
This is a cross-sectional analysis of enrollment data from a substudy of the Cape Town Adolescent Antiretroviral Cohort (CTAAC), a longitudinal study of PHIV+ adolescents on ART in Cape Town, South Africa. The broader CTAAC study seeks to investigate markers of chronic disease processes and progression in five key areas (general adolescent development, neurocognitive function, pulmonary disease, cardiovascular function, and musculoskeletal disease) over a 48 month period. For the CTAAC substudy, PHIV+ adolescents were recruited from four routine ART services across Cape Town, and were eligible to participate if they were aged 9–12 years, had been on ART for longer than 6 months, and knew their HIV-status. Controls were recruited from the same communities and schools attended by the adolescents living with HIV in an attempt to control for household income and quality of education. Controls were confirmed to be HIV negative, and were frequency matched to PHIV+ participants on age, sex and ethnicity. Controls were excluded if they had known pre-existing disease or if informed consent and assent was not obtainable. All youth screened for the control cohort underwent rapid HIV testing prior to enrolment to confirm negative status. Exclusion criteria for both groups were: 1) an uncontrolled medical condition, such as diabetes mellitus, epilepsy, or tuberculosis (TB) requiring hospital admission; 2) an identified CNS condition, including meningitis (TB or bacterial), cerebrovascular accident, lymphoma, history of head injury with loss of consciousness greater than 5 minutes or any radiological evidence of skull fracture, history of perinatal complications including hypoxic ischemic encephalopathy or neonatal jaundice requiring exchange transfusion, or neurodevelopmental disorder not attributed to HIV. All participants in the study completed neuroimaging studies, although this data is not presented here. A clinical neuroradiologist examined all the MRI scans to look for evidence of other secondary causes of CNS effects (including congenital infections such as CMV). The primary caregiver of each adolescent provided written informed consent prior to participation, and child assent was obtained. The study was approved by the University of Cape Town’s Faculty of Health Sciences Human Research Ethics Committee and the University of California Los Angeles Institutional Review Board.
Data and samples were obtained from 204 PHIV+ and 44 HIV- CTAAC participants between 2013 and 2015. Baseline health and sociodemographic questionnaires were administered in order to obtain general health information, past history and data on ancestry, and treatment. ART regimen (first/second/third line) and date of initiation of ART were abstracted from routine care records.
Neurocognitive Functioning
Each participant completed a comprehensive neuropsychological test battery administered by trained research assistants in their home language. The neuropsychological test battery was comprised of a number of individual neuropsychological tests previously described in detail[17]. We conducted Cronbach’s alpha tests on various combinations of neuropsychological tests in order to determine the statistical strength of each cognitive domain. Internal consistency was determined among the neuropsychological tests within each domain. The threshold for acceptable internal consistency was a Cronbach’s α ≥ 0.7, which was found for general intellectual functioning, attention, working memory, visual memory, verbal memory, motor coordination, processing speed and executive function. Composite cognitive domain scores were calculated by averaging the scores of the all tests that comprised each domain, so that a single score for each domain was determined for each participant. In addition, an overall cognitive impairment score was determined for each participant[17].
Blood Collection and Processing
Blood samples were drawn at enrolment. DNA extraction from blood samples was accomplished using the QIAsymphony DSP DNA Midi kit and protocol. DNA was quantified using BioDrop (Whitehead Scientific, South Africa) and normalized to a concentration of 5–10ng/ul. All samples were tracked using a Laboratory Information Management System (LIMS; Freezerworks, USA).
DNA Methylation
DNA methylation analysis was performed with the Illumina Infinium MethylationEPIC BeadChip, which measures bisulfite-conversion-based, single-CpG resolution DNAm levels at 866,836 CpG sites in the human genome. The standard protocol of Illumina methylation assays quantifies methylation levels by the β value using the ratio of intensities between methylated (signal A) and un-methylated (signal B) alleles. Specifically, the β value is calculated from the intensity of the methylated (M corresponding to signal A) and un-methylated (U corresponding to signal B) alleles, as the ratio of fluorescent signals β = Max(M,0)/[Max(M,0) + Max(U,0) + 100]. Thus, β values range from 0 (completely un-methylated) to 1 (completely methylated)[18]. We used the noob normalization method[19], which is implemented in the "minfi" R package[20].
DNA methylation age and the epigenetic clock
Chronological age has been shown to have a profound effect on DNA methylation levels[21–24]. As a result, several highly accurate epigenetic biomarkers of chronological age have been proposed[15, 25–28]. These biomarkers use weighted averages of methylation levels at specific CpG sites to produce estimates of age (in units of years), referred to as "DNA methylation age" (DNAm age) or "epigenetic age". Recent studies support the idea that these measures are at least passive biomarkers of biological age. For instance, the epigenetic age of blood has been found to be predictive of all-cause mortality[29–32], frailty[33], lung cancer[34], and cognitive and physical functioning[35], while the blood of the offspring of Italian semi-supercentenarians (i.e. participants aged 105 or older) was shown to have a lower epigenetic age than that of age-matched controls[36]. Further, the utility of the epigenetic clock method using various tissues and organs has been demonstrated in applications surrounding Alzheimer's disease[37], centenarian status[36, 38], development[39], Down syndrome[40], frailty[33], HIV infection[10], Huntington's disease[41], obesity[42], menopause[43], osteoarthritis[44], and Parkinson's disease[45].
DNAm age (also referred to as epigenetic age) was calculated from human blood samples profiled with the Illumina Infinium EPIC array platform. We considered two different age estimators described in the following. First, we used the multi-tissue DNAm age estimator from Horvath (2013)[15], which is defined as a prediction method of age based on the DNAm levels of 353 CpGs. Predicted age, referred to as DNAm age, correlates with chronological age in sorted cell types (CD4+ T cells, monocytes, B cells, glial cells, neurons), tissues, and organs, including: whole blood, brain, breast, kidney, liver, lung, saliva[15]. Second, we applied the Hannum measure of DNAm age based on 71 CpGs, which was developed using DNA methylation data from blood[27].
Despite high correlations, epigenetic age estimates can deviate substantially from chronological age at the individual level. After adjusting DNAm age for chronological age, one arrives at measures of epigenetic age acceleration. Here we focus on two widely used measures of epigenetic age acceleration denoted by AgeAccelerationResidual and EEAA, respectively. By definition, both measures are independent of chronological age (at the time of blood draw). For both measures of age acceleration, positive (negative) values indicate that the blood sample is older (younger) than expected based on chronological age. The mathematical definition of these measures is briefly reviewed here. AgeAccelerationResidual is defined as the (raw) residual resulting from regressing the multi-tissue DNAm age estimate on chronological age. By definition, AgeAccelerationResidual is independent of chronological age. Extrinsic epigenetic age acceleration (EEAA) can be interpreted as an enhanced version of the Hannum measure of DNAm age estimator because it up-weights the contributions of age-related blood cell counts[32, 46].
In a recent large scale meta-analysis involving over 13 thousand subjects from 13 cohorts, we have shown that both AgeAccelerationResidual and EEAA are predictive of mortality, independent of chronological age, even after adjusting for additional risk factors, and within the racial/ethnic groups that we examined (Caucasians, Hispanics, African Americans)[32]. The EEAA measure is attractive because a) it exhibited the strongest predictive association with all-cause mortality[32] and b) it revealed the strongest association with cognitive functioning measures in our study. AgeAccelerationResidual is attractive because a) it is less confounded by changes in blood cell composition, b) it applies to all tissue samples, and c) it is particularly accurate in adolescents, and d) we previously used it in our study of HIV+ adults[10, 15].
Both AgeAccelerationResidual and EEAA were computed using the epigenetic clock software (http://labs.genetics.ucla.edu/horvath/dnamage/) where they are denoted as AgeAccelerationResidual and BioAge4HAStaticAdjAge, respectively.
While AgeAccelerationResidual is relatively robust with respect to changes in blood cell composition, EEAA capitalizes on age related changes in blood cell types and captures aspects of immunosenescence[32].
The epigenetic clock method and software applies to data generated using any Illumina platform (including the EPIC array). Missing CpG probes were automatically imputed by the software. Mathematical details and software tutorials for the epigenetic clock can be found in the Additional files of[15]. An online age calculator can be found at our webpage: http://labs.genetics.ucla.edu/horvath/dnamage/.
Statistical Analyses
Pearson correlation tests were used for correlating continuous variables (e.g. DNAmAge and chronological age). A non-parametric group comparison test (Kruskal Wallis test) was used to relate an ordinal outcome to a categorical variable (e.g. HIV group status).
RESULTS
Two hundred and four (204) HIV infected adolescents and 44 frequency-matched controls were enrolled. Demographic characteristics were similar between HIV+ adolescents and controls, Table 1. HIV infected adolescents were more likely to have had repeated grades at school (p=0.02). Most HIV-infected adolescents were on first line ART (79.9%), with a mean CD4 count of 955 cells/mm3, median viral load of <LOD (level of detection) copies/mL and mean age of initiation of ART of 3.41 years. There were significant differences in cognitive ability between the two groups in the following domains: general intellectual functioning, executive functioning, attention, working memory, verbal memory, visual memory, visual spatial ability, language and processing speed (see Table 1).
Table 1.
Baseline demographic and clinical characteristics of the CTAAC cohort
| Variable | HIV- infected (N = 204) |
Controls (N = 44) |
t | p |
|---|---|---|---|---|
| Age in years: Mean (SD) | 10.38 (0.88) | 10.38 (1.09) | −.01 | .99 |
| Gender: Male/Female | 100/104 | 20/24 | .43 | .67 |
| Ethnicity: Black African/Other | 187/17 | 44/0 | −1.99 | .05 |
| Home language: isiXhosa/Other | 182/20 | 42/2 | 1.12 | .26 |
| Education in years: Mean (SD) | 3.20 (1.13) | 3.39 (1.35) | .96 | .34 |
| Repeated grades: YES (%) | 121 (59.3) | 18 (40.9) | −2.24 | .02* |
| Low household income: (%)a | 199 (98.5) | 44 (100) | −.72 | .47 |
| ART regimen: First/Second/Third/Missing | 163/32/8/1 | N/A | N/A | N/A |
| Duration of ART: mean years (SD) | 7.18 (2.46) | N/A | N/A | N/A |
| Age of initiation of ART: mean years (SD) | 3.41 (2.53) | N/A | N/A | N/A |
| Current EFV treatment: YES (%) | 106 (52.0) | N/A | N/A | N/A |
| Viral load: median (IQR) (copies/mL) | 0.0 (40.0) | N/A | N/A | N/A |
| 1.0 | ||||
| CD4 count: mean (SD) (cells/mm3) | 955.12 (504.96) | N/A | N/A | N/A |
| Cognitive domains: Z-score means (SD) | ||||
| General intellectual function | −.55 (.78) | .00 (.79) | 4.12 | .00* |
| Executive function | −.45 (.62) | −.01 (.59) | 4.15 | .00* |
| Motor coordination | .03 (.99) | .00 (.91) | −.18 | .86 |
| Attention | −.39 (.92) | −.00 (.92) | 2.55 | .01* |
| Working memory | −.46 (.61) | −.00 (.63) | 4.40 | .00* |
| Visual spatial | −.35 (.89) | −.00 (1.00) | 2.31 | .02* |
| Visual memory | −.49 (.87) | −.00 (.98) | 3.31 | .00* |
| Language | −.38 (.102) | −.00 (1.00) | 2.21 | .03* |
| Verbal memory | −.52 (1.26) | .00 (.84) | 3.37 | .00* |
| Processing speed | −.55 (.63) | .00 (.68) | 5.13 | .00* |
HIV+ adolescents exhibit epigenetic age acceleration
The high accuracy of the epigenetic clock method is revealed by the surprisingly high correlation between DNAm age and chronological age (r=0.36, p=5.3E-9) despite the narrow age range (adolescents are aged between 9–12 years). HIV+ adolescents exhibit increased levels of AgeAccelerationResidual and EEAA compared to HIV- controls (Figure 1A,B). Increased epigenetic age acceleration can even be observed in HIV+ individuals whose viral load is not detectable (VL < 50 copies/mL).
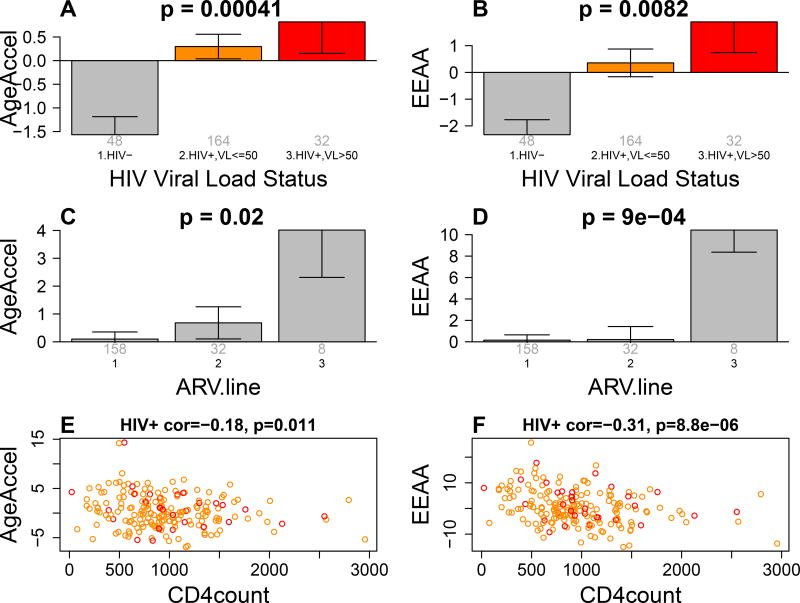
Figure 1. Epigenetic age acceleration versus HIV status.
A,B) The x axis reports 3 groups: HIV negative controls, HIV+ adolescents with non-detectable viral load (orange bar), and HIV+ individuals with detectable viral load (red bar). A) The y-axis reports the mean age acceleration residual based on Horvath 2013. B) Extrinsic epigenetic age acceleration (y-axis). The title of the bar plots reports the Kruskal Wallis p value.
C,D) ARV treatment failure (x-axis) versus C) AgeAccelerationResidual and D) EEAA. The group with ARV.line=3 corresponds to HIV+ adolescents who failed the ARV treatment repeatedly.
E,F) Observed CD4+ T cell count versus epigenetic age acceleration in HIV+ adolescents. Points are colored by HIV viral load status (red=detectable virus).
We observe significant associations between HIV status and epigenetic age acceleration even after removing the 17 adolescents of mixed ancestry from the analysis (p=3.7E-7 for AgeAccelerationResidual and p=1.4E-6 for EEAA). EEAA and to a lesser extent AgeAccelerationResidual exhibit significant negative correlations with current CD4 T cell counts in HIV+ adolescents.
Our analysis of ART line (first/second/third) (variable ARV.line) reveals that adolescents who are on second or third line ART treatment are more likely to have accelerated ageing (Figure 1C,D). Both AgeAccelerationResidual and EEAA remain significantly associated with current CD4 T cell counts and ARV.line even after adjusting for viral load, educational status, ethnicity, and other potential confounders (Table 2). Epigenetic age acceleration exhibits substantial heterogeneity in HIV+ adolescents. In HIV+ adolescents, age at ART initiation nor ART duration were significantly correlated with measures of epigenetic age acceleration nor with cognitive assessments. Similarly, household income was not correlated with epigenetic age acceleration in HIV+ adolescents. However, CD4+ T cell counts was negatively correlated with epigenetic age acceleration in HIV+ adolescents (Figure 1). A multivariate model analysis reveals that HIV viral load, current CD4 T cell count, ethnicity, ARV duration, ARV.line, and education explain only 8.8 percent of the variable in AgeAcceleration but 17.3 percent of the variation of EEAA in HIV+ adolescents (Table 2).
Table 2.
Virologic, treatment, and education factors associated with age acceleration measures
| Outcome: AgeAccelerationResidual. R^2=0.088 |
Outcome: EEAA. R.square=0.173 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Covariate | Coef | Std. Error |
P- value |
Coef | Std. Error |
P- value |
|
| ||||||
| Detectable.HIV.viral.load (VL > 50) | 3.71E-01 | 6.56E-01 | 5.73E-01 | 1.50E+00 | 1.22E+00 | 2.22E-01 |
| Current CD4 T count | −1.14E-03 | 5.19E-04 | 2.97E-02 | −3.38E-03 | 9.69E-04 | 6.03E-04 |
| Race (Coloured) | 3.11E-01 | 8.84E-01 | 7.26E-01 | −3.24E+00 | 1.65E+00 | 5.11E-02 |
| ARV duration | −5.53E-02 | 9.94E-02 | 5.79E-01 | −2.27E-01 | 1.86E-01 | 2.23E-01 |
| ARV line | 1.16E+00 | 4.90E-01 | 1.90E-02 | 2.31E+00 | 9.15E-01 | 1.23E-02 |
| Education (Highest Grade) | −2.99E-01 | 2.23E-01 | 1.81E-01 | −8.39E-01 | 4.16E-01 | 4.50E-02 |
| Repeated Grade (Yes) | −7.44E-01 | 5.02E-01 | 1.40E-01 | −2.61E-01 | 9.38E-01 | 7.81E-01 |
Extrinsic epigenetic age acceleration relates to cognitive functioning measures
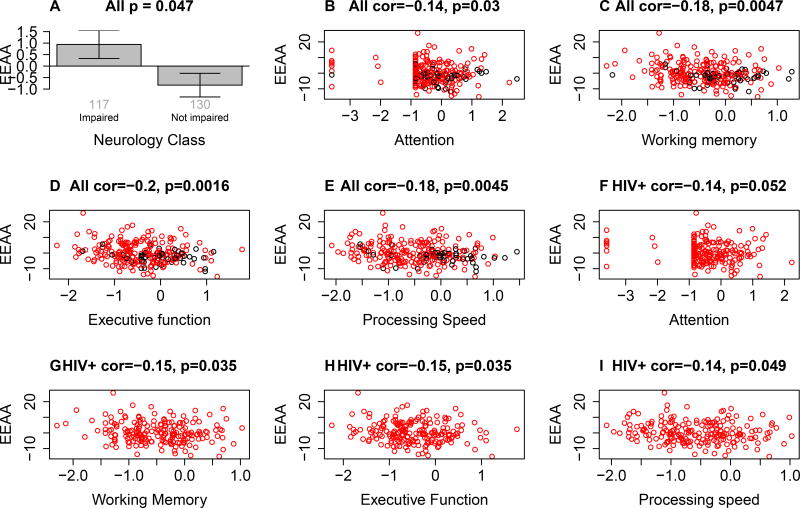
We correlated both measures of epigenetic age acceleration with cognitive functioning measures but found significant correlations only for the extrinsic measure of epigenetic age acceleration: EEAA is significantly associated with overall cognitive impairment status (Figure 2A), and more specifically with the following cognitive domains: attention, working memory, executive functioning, and processing speed (Figure 2B–E). This association does not reflect confounding by HIV status since we continue to observe significant associations between EEAA and working memory, executive functioning, and processing speed (Figure 2F–I) after restricting the analysis to HIV+ individuals. These findings suggest that an epigenetically older immune system is associated with lower cognitive functioning. A multivariate linear model analysis reveals that both HIV infection and EEAA are independently associated with cognitive performance measures: executive functioning, working memory, and processing speed (Model 1 in Table 3).
Figure 2. Cognitive functioning versus epigenetic age acceleration of blood.
EEAA versus (A) neurology class (x-axis), (B) attention, (C) working memory, (D) executive functioning E) processing speed in all adolescents (both HIV+ and HIV-). Points are colored by HIV status (red=HIV+, black=HIV-). (F–i) reports analogous findings for HIV+ adolescents, i.e. HIV- controls were omitted. Overall, increased epigenetic age acceleration is associated with impaired cognitive functioning. The title of panel A reports a Kruskal Wallis test p-value while the remaining panels report Pearson correlation test p values.
Table 3.
Multivariate linear regression model of measures of cognitive functioning.
| Outcome (dependent variables) |
Model 1 | |||
|---|---|---|---|---|
| Executive Function | Covariate | Coef | SE | P-value |
| Age | −1.44E-3 | 4.61E-2 | 9.75E-1 | |
| HIV+ | −3.88E-1 | 1.07E-1 | 3.34E-4 | |
| EEAA | −1.72E-2 | 6.72E-3 | 1.10E-2 | |
| Working memory | Covariate | Coef | SE | P-value |
| Age | −4.81E-2 | 4.30E-2 | 2.65E-1 | |
| HIV+ | −3.84E-1 | 9.95E-2 | 1.48E-4 | |
| EEAA | −1.38E-2 | 6.29E-3 | 2.93E-2 | |
| Processing speed | Covariate | Coef | SE | P-value |
| Age | −1.10E-1 | 4.64E-2 | 1.84E-2 | |
| HIV+ | −4.58E-1 | 1.08E-1 | 3.07E-5 | |
| EEAA | −1.53E-2 | 6.81E-3 | 2.55E-2 | |
| Attention | Covariate | Coef | SE | P-value |
| Age | 3.23E-2 | 6.37E-2 | 6.13E-1 | |
| HIV+ | −2.50E-1 | 1.47E-1 | 9.08E-2 | |
| EEAA | −1.77E-2 | 9.30E-3 | 5.87E-2 |
Rows correspond to different multivariate models whose outcome variable (dependent variable) can be found in the first column. Columns report the regression coefficients, standard errors, and Wald test p-value. Model 1 contains three covariates: age at the time of the blood draw. HIV status, and extrinsic epigenetic age acceleration.
Multivariate regression model analysis revealed that EEAA remained negatively associated with executive functioning (p=0.023), working memory (p=0.049) and processing speed (p=0.024) even after adjusting for HIV status, educational level (assessed by highest grade, and repeated grade), ethnicity, and household income.
DISCUSSION
This is the first study to show that a) perinatally acquired HIV infection is associated with epigenetic age acceleration in blood, and b) extrinsic epigenetic age acceleration is associated with lower cognitive functioning in these adolescents.
We found significant differences between the HIV-infected participants and demographic-matched controls in general intellectual functioning, executive functioning, attention, working memory, verbal memory, visual memory, visual spatial ability, language and processing speed. Based on the results of a recent meta-analysis, large effects (ESE >= 0.8) of HIV status, suggest greater impairment in HIV-infected children and adolescents for the following domains (ranked from largest to smallest effect size): working memory, processing speed, executive function, and visual memory[47]. Small ESEs were observed for visual spatial ability, attention, language, general intellectual functioning, motor coordination and verbal memory. A study conducted in South Africa found that even asymptomatic HIV infected children performed poorly on cognitive measures, and once again, most notably on tests of general intellectual functioning, visual spatial ability, visual memory and semantic fluency (i.e.: executive functioning)[6]. Executive functioning emerges as one of the more prominent domains to be affected in this population. In a previous study conducted in Cape Town, HIV-infected youth were found to perform significantly poorer in executive function tasks, particularly in processing speed, cognitive flexibility and inhibition[17]. Gaining insight into the factors contributing to cognitive impairment is essential, as subtle impairments may progress to more pronounced complications that will influence future intellectual performance, job opportunities, and community participation of HIV-infected youth. As with recent findings in HIV+ adults[48], the current study suggests that epigenetic age acceleration may be a contributing factor to poorer cognitive function in adolescents living with HIV.
We previously reported ambiguous results regarding the correlation between HIV viral load and epigenetic age acceleration in HIV+ adults: while a positive correlation could be observed in one data set, it could not replicated in another[10]. In the current data set, we do not observe a significant association between viral load and epigenetic age acceleration in HIV+ adolescents which might reflect the low number of HIV+ individuals with detectable viral load (n=32). Our analysis of ART regimen reveals that adolescents who are on second or third line ART treatment are more likely to have accelerated ageing. Second line ART, increased viral load, low CD4 counts and poor cognitive function has been associated with poor white matter integrity in youth living with HIV in South Africa[49]. While we have data on how many in the cohort have had to switch to second or third line ART, we do not have adequate information as to why the switch had to occur. Whether this was due to side effects or virological failure is unknown.
Our study cannot exclude the possibility that the observed epigenetic aging effects in HIV+ adolescents reflect changes in cell composition because our methylation data were generated from unfractionated blood. EEAA and AgeAccelerationResidual are highly correlated (r=0.59, p < 2.2e-16) but these measures have different properties. By construction, EEAA relates to cell abundance measures that are known to change with age (e.g. naive CD8T cells or conversely exhausted CD8T cells). Further, EEAA exhibits stronger negative correlations with CD4 T cell counts (r=−0.31,p=1.2×10−5) than AgeAccelerationResidual (r=−0.19,p=0.0092, Figure 1). EEAA is attractive since it captures aspects of immunosenescence whereas AgeAccelerationResidual is attractive since it is less confounded by differences in cell composition[32, 50]. DNAm age of whole blood probably reflects properties of hematopoietic stem cells in the bone marrow because a) the majority of cells in whole blood are neutrophils and b) different blood cell types (neutrophils, B cells, T cells, monocytes) have roughly the same age[10]. Epigenetic age acceleration effects due to HIV have been observed in sorted blood cell[13]. Future research should carefully study which blood cell types exhibit cell intrinsic epigenetic age acceleration effects due to HIV infection.
One would perhaps not expect DNAm age of blood to be associated with measures of cognitive functioning in young adolescents. However, other studies in HIV-uninfected older adults reported associations between epigenetic age acceleration of blood (based on the multi-tissue DNAm age) and white matter integrity in the brain[51, 52] and various measures of cognitive or memory functioning in older individuals[35, 53].
Limitations of this study include the cross sectional design, but longitudinal follow-up is underway to better understand the impact of epigenetic age acceleration on cognition in later adolescence. The control group is also small in comparison- to the participant group, but was nevertheless well-matched. Although other congenital infections and incidental CNS abnormalities were excluded as far as possible on history, clinical examination and on clinical review of the MRI scans, it remains a possibility that there may be some overlapping effects of undiagnosed conditions such as congenital CMV. A further limitation is the lack of objective measures of early life environmental exposures (e.g., psychological trauma) that may be more common in HIV-infected youths. Future research should focus on delineating specific factors that may account for accelerated aging in PHIV+ youth.
Conclusion
To our knowledge, this is the first cohort study to examine the impact of epigenetic age acceleration in adolescents living with HIV and the association with cognitive function. The study took place in South Africa, one of the countries most affected by HIV/AIDS with the highest rate of new HIV infections in the world. Despite a mean age of initiation of ART treatment of 3.4 years and usage for more than 7 years, considerable cognitive impairment is apparent. Longitudinal follow-up of our cohort will be crucial for determining the impact of epigenetic aging in adolescents living with HIV. Correlates of epigenetic aging need to be elucidated, including associations with measures of viral load, treatment duration, concurrent infections, and ART prescription and treatment adherence, in order to understand their impact on the long-term development of neurocognitive disorders in pediatric HIV.
Supplementary Material
Acknowledgments
Financial support
This study was funded primarily by R21MH107327-01 (AJL and JH)
SH was primarily supported by R21AG046954-01A1 and U34AG051425-01
Funding for CTAAC provided by R01-HD074051 (HJZ)
HJZ and DS are supported by the South African Medical Research Council
Footnotes
Potential conflicts of interest
All authors: No reported conflicts.
Author contributions
AJL, JH, DJS, and SH conceived of the study. HZ is PI of the CTAAC study, from which most of the data were derived. SH carried out the statistical analysis. JH, SH, and AJL wrote the first draft of the article. The remaining authors conceived of and aided with the CTAAC study, including collection of the DNA samples and phenotypic data. DL and MK generated the DNA methylation data. All authors helped in the interpretation of the findings and the write up of the article.
References
- 1.Ruel TD, Boivin MJ, Boal HE, Bangirana P, Charlebois E, Havlir DV, et al. Neurocognitive and motor deficits in HIV-infected Ugandan children with high CD4 cell counts. Clin Infect Dis. 2012;54(7):1001–1009. doi: 10.1093/cid/cir1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willen EJ. Neurocognitive outcomes in pediatric HIV. Mental retardation and developmental disabilities research reviews. 2006;12(3):223–228. doi: 10.1002/mrdd.20112. [DOI] [PubMed] [Google Scholar]
- 3.Shanbhag MC, Rutstein RM, Zaoutis T, Zhao H, Chao D, Radcliffe J. Neurocognitive functioning in pediatric human immunodeficiency virus infection: effects of combined therapy. Archives of pediatrics & adolescent medicine. 2005;159(7):651–656. doi: 10.1001/archpedi.159.7.651. [DOI] [PubMed] [Google Scholar]
- 4.Robertson K, Liner J, Hakim J, Sankale JL, Grant I, Letendre S, et al. NeuroAIDS in Africa. J Neurovirol. 2010;16(3):189–202. doi: 10.3109/13550284.2010.489597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoare J, Fouche JP, Phillips N, Joska JA, Paul R, Donald KA, et al. White matter micro-structural changes in ART-naive and ART-treated children and adolescents infected with HIV in South Africa. AIDS. 2015;29(14):1793–1801. doi: 10.1097/QAD.0000000000000766. [DOI] [PubMed] [Google Scholar]
- 6.Hoare J, Fouche JP, Spottiswoode B, Donald K, Philipps N, Bezuidenhout H, et al. A diffusion tensor imaging and neurocognitive study of HIV-positive children who are HAART-naive "slow progressors". J Neurovirol. 2012;18(3):205–212. doi: 10.1007/s13365-012-0099-9. [DOI] [PubMed] [Google Scholar]
- 7.Gapp K, Woldemichael BT, Bohacek J, Mansuy IM. Epigenetic regulation in neurodevelopment and neurodegenerative diseases. Neuroscience. 2014;264:99–111. doi: 10.1016/j.neuroscience.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 8.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. The Lancet Neurology. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 9.Rangasamy S, D'Mello SR, Narayanan V. Epigenetics, autism spectrum, and neurodevelopmental disorders. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2013;10(4):742–756. doi: 10.1007/s13311-013-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. The Journal of infectious diseases. 2015 doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickabaugh TM, Baxter RM, Sehl M, Sinsheimer JS, Hultin PM, Hultin LE, et al. Acceleration of age-associated methylation patterns in HIV-1-infected adults. PLoS One. 2015;10(3):e0119201. doi: 10.1371/journal.pone.0119201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulias K, Lieberman J, Greer EL. An Epigenetic Clock Measures Accelerated Aging in Treated HIV Infection. Mol Cell. 2016;62(2):153–155. doi: 10.1016/j.molcel.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross AM. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62 doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung JM, Fishbane N, Jones M, Morin A, Xu S, Liu JC, et al. Longitudinal study of surrogate aging measures during human immunodeficiency virus seroconversion. Aging (Albany NY) 2017;9(3):687–705. doi: 10.18632/aging.101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine AJ, Quach A, Moore DJ, Achim CL, Soontornniyomkij V, Masliah E, et al. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2016;22(3):366–375. doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoare J, Phillips N, Joska JA, Paul R, Donald KA, Stein DJ, et al. Applying the HIV-associated neurocognitive disorder diagnostic criteria to HIV-infected youth. Neurology. 2016;87(1):86–93. doi: 10.1212/WNL.0000000000002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunning M, Barbosa-Morais N, Lynch A, Tavare S, Ritchie M. Statistical issues in the analysis of Illumina data. BMC Bioinformatics. 2008;9(1):85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Research. 2013;41(7):e90–e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakyan VK. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome research. 2010;20 doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teschendorff AE. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research. 2010;20 doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S, Zhang Y, Langfelder P, Kahn R, Boks M, van Eijk K, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung M, Pfeifer GP. Aging and DNA methylation. BMC biology. 2015;13(1):1–8. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, et al. Epigenetic predictor of age. PLoS One. 2011;6 doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11(6):1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 27.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Q, Weidner CI, Costa IG, Marioni RE, Ferreira MR, Deary IJ, et al. DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy. Aging (Albany NY) 2016;8(2):394–401. doi: 10.18632/aging.100908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni R, Shah S, McRae A, Chen B, Colicino E, Harris S, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16(1):25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2015 doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perna L, Zhang Y, Mons U, Holleczek B, Saum K-U, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clinical Epigenetics. 2016;8(1):1–7. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8 doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women's health initiative. Aging (Albany NY) 2015;7(9):690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44 doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Blasio AM, Delledonne M, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 2015;7 doi: 10.18632/aging.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging (Albany NY) 2015;7(12):1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath S, Mah V, Lu AT, Woo JS, Choi OW, Jasinska AJ, et al. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY) 2015;7(5):294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker RF, Liu JS, Peters BA, Ritz BR, Wu T, Ophoff RA, et al. Epigenetic age analysis of children who seem to evade aging. Aging (Albany NY) 2015;7(5):334–339. doi: 10.18632/aging.100744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horvath S, Garagnani P, Bacalini M, Pirazzini C, Salvioli S, Gentilini D, et al. Accelerated Epigenetic Aging in Down Syndrome. Aging Cell. 2015;14(1) doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, et al. Huntington's disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 2016;8(7):1485–1512. doi: 10.18632/aging.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine ME, Lu AT, Chen BH, Hernandez DG, Singleton AB, Ferrucci L, et al. Menopause accelerates biological aging. Proc Natl Acad Sci U S A. 2016;113(33):9327–9332. doi: 10.1073/pnas.1604558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vidal L, Lopez-Golan Y, Rego-Perez I, Horvath S, Blanco FJ, Riancho JA, et al. Specific increase of methylation age in osteoarthritis cartilage. Osteoarthritis and Cartilage. 2016;24:S63. [Google Scholar]
- 45.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany NY) 2015;7(12):1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips N, Amos T, Kuo C, Hoare J, Ipser J, Thomas KG, et al. HIV-Associated Cognitive Impairment in Perinatally Infected Children: A Meta-analysis. Pediatrics. 2016;138(5) doi: 10.1542/peds.2016-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine AJ, Quach A, Moore DJ, Achim CL, Soontornniyomkij V, Masliah E, et al. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2015 doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoare J, Fouche JP, Phillips N, Joska JA, Donald KA, Thomas K, et al. Clinical associations of white matter damage in cART-treated HIV-positive children in South Africa. J Neurovirol. 2015;21(2):120–128. doi: 10.1007/s13365-014-0311-1. [DOI] [PubMed] [Google Scholar]
- 50.Chen BH, Carty CL, Kimura M, Kark JD, Chen W, Li S, et al. Leukocyte telomere length, T cell composition and DNA methylation age. Aging (Albany NY) 2017;9(9):1983–1995. doi: 10.18632/aging.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hodgson K, Carless MA, Kulkarni H, Curran JE, Sprooten E, Knowles EE, et al. Epigenetic Age Acceleration Assessed with Human White-Matter Images. The Journal of Neuroscience. 2017;37(18):4735. doi: 10.1523/JNEUROSCI.0177-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raina A, Zhao X, Grove ML, Bressler J, Gottesman RF, Guan W, et al. Cerebral white matter hyperintensities on MRI and acceleration of epigenetic aging: the atherosclerosis risk in communities study. Clinical Epigenetics. 2017;9(1):21. doi: 10.1186/s13148-016-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degerman S, Josefsson M, Nordin Adolfsson A, Wennstedt S, Landfors M, Haider Z, et al. Maintained memory in aging is associated with young epigenetic age. Neurobiology of Aging. 2017;55:167–171. doi: 10.1016/j.neurobiolaging.2017.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.