Abstract
Background
Due to rapid expansion of clinical genetic testing, an increasing number of genetic variants of undetermined significance are being identified in children with unclear diagnostic value. Variants found in genes associated with heritable channelopathies, such as long QT syndrome (LQTS), are particularly difficult to interpret given the risk of sudden cardiac death associated with pathologic mutations. Objective: To determine whether variants in LQTS-associated genes from whole exome sequencing (WES) represent disease-associated biomarkers or background genetic “noise.”
Methods
WES variants from Baylor Genetics Laboratories were obtained for 17 LQTS-associated genes. Rare variants from healthy controls were obtained from the GnomAD database. LQTS case variants were extracted from literature. Amino acid-level mapping and signal-to-noise calculations were conducted. Clinical history and diagnostic studies were analyzed for WES subjects evaluated at our institution.
Results
Variants in LQTS case-associated genes were present in 38.3% of 7,244 WES probands. There was a similar frequency of variants in the WES and healthy cohorts for LQTS1-3 (11.2 and 12.9%, respectively) and LQTS4-17 (27.1 and 38.4%, respectively). WES variants preferentially localized to amino acids altered in control individuals compared to cases. Based on amino acid-level analysis, WES-identified variants are indistinguishable from healthy background variation while LQTS1 and 2 case-identified variants localized to clear pathologic “hot spots.” No individuals who underwent clinical evaluation had clinical suspicion for LQTS.
Conclusions
The prevalence of incidentally identified LQTS-associated variants is ~38% among WES tests. These variants most likely represent benign healthy background genetic variation rather than disease associated mutations.
Keywords: genetic testing, genetics, long QT syndrome, mutation, variant of undetermined significance, whole exome sequencing
INTRODUCTION
Congenital long QT syndrome (LQTS) is characterized by delayed cardiac repolarization in the absence of an underlying syndrome or structural heart disease which can manifest as a prolonged QT interval on a resting electrocardiogram (ECG). This repolarization defect does not typically have a direct pathologic impact on the heart unless the patient is exposed to triggers such as physical exertion, emotion, auditory stimuli, QT prolonging drugs, or during the postpartum period which electrically destabilizes the heart leading to the potentially lethal dysrhythmia of torsades de pointes (TdP).1 This places the individual at risk of syncope, seizures, and sudden cardiac death (SCD). A relatively common arrhythmia syndrome, LQTS affects as many as 1 in 2,000 persons.2, 3 A prototypical cardiac channelopathy, heritable mutations in genes encoding cardiac channelopathies are believed to be the cause of LQTS.
It has been traditionally held that 65–75% of LQTS cases are due to mutations in 1 of 3 major genes: KCNQ1-encoded IKs potassium channel (Kv7.1, LQTS1),4 KCNH2-encoded IKr potassium channel (Kv11.1, LQTS2),5 and SCN5A-encoded INa sodium channel (Nav1.5, LQTS3).6, 7 These ion channels play key roles in modulating the cardiac action potential, and LQTS-associated genetic defects in these channels delay cellular repolarization. In addition to these 3 major genes, over a dozen others have been identified as rare causes of LQTS. To date, hundreds of mutations have been identified in 17 LQTS-susceptibility genes; however, genes associated with LQTS4 through 17 are rare among documented LQTS probands.8 Due to the clear link between heritable mutations in these 17 genes and the pathogenesis of LQTS, a clinical “gene panel” genetic test has been developed. Recent advances in genetic sequencing technology via whole exome sequencing (WES) has made genetic interrogation of a larger number of genes achievable with increasing utilization in the clinical setting among patients with a heterogeneous, or otherwise unclear, phenotypic presentation. While expansive genetic sequencing tests have provided an increased ability to detect pathologic genetic variation, it has also drastically increased the identification of incidentally identified variants of unclear diagnostic significance (VUS).
To address this, we compiled genetic variants found in LQTS-associated genes reported in one of the world’s largest cohorts of clinical WES testing. We then compared this WES cohort to a cohort of genetic variants identified in subjects diagnosed with LQTS and ostensibly healthy individuals. We find remarkable similarity between WES cohort variants and healthy genetic variation and no evidence of LQTS in individuals with a variant who underwent clinical evaluation. We conclude that incidentally identified variants likely represent healthy background variation.
METHODS
Genes and Gene Variants
Genes and gene variants are detailed in the Supplemental Methods.
Study Cohorts
This IRB-approved study included 3 cohorts and has been previously described9 and is fully detailed in Supplemental Methods.
Whole Exome Sequencing
Sequencing was done as previously described10 and detailed in Supplemental Methods.
Nomenclature
Variant nomenclature was done as previously described9 and detailed in Supplemental Methods.
Primary Sequence Comparison and Statistics
Primary sequence comparison and statistics were performed as previously described9 and detailed in Supplemental Methods.
RESULTS
Prevalence of incidentally identified LQTS1-17 variants in a large WES cohort
A large cohort of clinical WES genetic test referrals was interrogated to establish the background variance in LQTS1-17 variants found incidentally in next-generation sequencing referrals. The overall WES cohort included 7,938 total individuals with 7,244 probands. Among probands, the median age of genetic testing was 6.1 years with 54.0% were males, 45.2% females, and 0.8% fetal samples (Figure 1A and Supplemental Table 1). Individuals undergoing WES testing were referred from various institutions with approximately 90% originating within the United States and approximately 50% from the state of Texas.
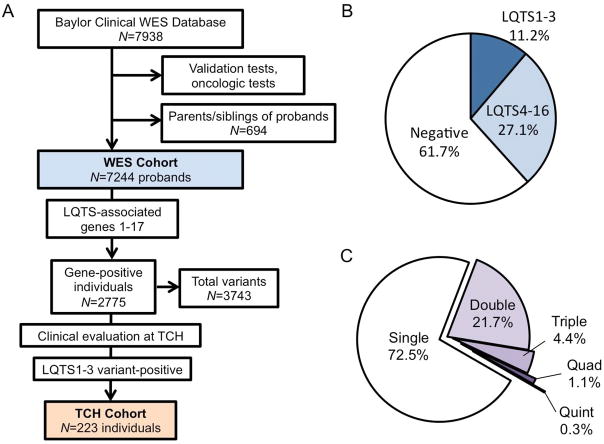
Figure 1.
A, Schematic of the study methodology. Synonymous variants, known polymorphisms, and variants interpreted as benign were excluded creating a proband-based WES cohort (light blue fill). Subset analysis of individuals seen a TCH with a variant in a LQTS1-3 gene (tan fill) was conducted. B, Pie chart of WES cohort demonstrating individuals with no LQTS-associated gene variants (negative, white fill) and individuals who are variant positive for LQTS1-3 (dark blue fill) and LQTS4-17 (light blue fill). C, Pie chart of WES variant-positive individuals depicting the number of variants hosted by a single proband.
LQTS1-17 variants were identified and deemed either “likely pathogenic” or a VUS by WES testing in 2775 individuals (38.8% [37.3–40.0]) with a total of 3743 unique variants identified. Variants localizing to the major genes commonly associated with LQTS, LQTS1-3, were identified in 813 individuals (11.2% [10.5–12.0] of WES probands). There were 1962 individuals (27.1% [26.1–28.1]) hosting variants localizing to LQTS4-17 genes without a LQTS1-3 variant (Figure 1B). Among variant-positive individuals, the majority hosted a single variant (2012, 72.5% [70.8–74.2] among variant-positive probands), while 603 (21.7% [20.2–23.3]) hosted 2 variants, 122 (4.4% [3.6–5.2]) hosted 3, 31 (1.1% [0.8–1.6]) hosted 4, and 7 individuals hosted 5 unique variants (0.3% [0.1–0.5]). These results are summarized in Figure 1C. Taken together, these results suggest that LQTS-associated variants are incidentally identified in a significant proportion of WES tests and are predominantly VUSs found in LQTS4-17-associated genes.
Incidentally identified LQTS-associated variants have a similar gene frequency to healthy individuals and not LQTS cases
To compare the frequency of incidentally identified WES variants to known cases of LQTS, a LQTS case cohort was compiled from a number of international cohort-based studies including Splawski et al (N=262 individuals),11 Tester et al (N=541),7 Hedley et al (N =44),12 Al-Hassnan et al (N=56),13 Lieve et al (N=855),14 and Napolitano et al (N=430)15 for a total of 2188 LQTS cases. Among these, 992 (45.3% [43.2–47.5]) were positive for a likely disease-associated variant in a LQTS1-3-associated gene. This was approximately 4-fold higher than the WES cohort frequency of 11.2% (P<0.0001). While comprehensive genotyping data for rare causes of LQTS was not available for all studies, LQTS4-17 mutation-positive probands were rare and had a yield of approximately (1.9% [1.3–2.5]) among LQTS cases.
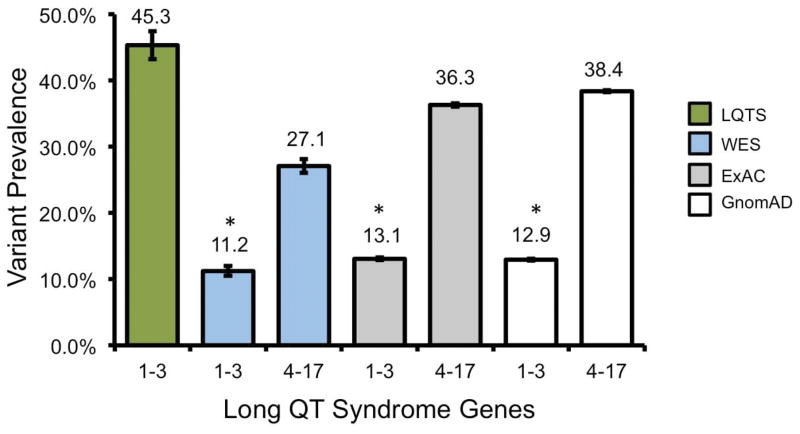
Given the marked disparity between the frequency of variants in LQTS1-3 genes among LQTS cases and among incidentally identified individuals, we next calculated the frequencies of rare LQTS-associated gene variants among ostensibly healthy individuals from the GnomAD cohort. There was a statistically significant, although modest, difference between the prevalence of WES variants in LQTS1-3 genes and the GnomAD control cohort (11.2% [10.5–12.0] versus 12.9% [12.8–13.1], respectively (P<0.0001). Furthermore, there was a significant difference between the prevalence of incidentally identified variants in LTQS4-17 genes and rare variants in the control cohort. These results are summarized in Figure 2. This was consistent with the findings utilizing the ExAC database (Supplemental Results). Taken together, these results suggest that approximately 13% of ostensibly healthy individuals will have a rare variant found in LQTS1-3 genes which is modestly different than the rate of incidentally identified variants found in WES testing. In comparison, genes associated with rare causes of LQTS are common with a rate of approximately 38% in healthy individuals and 27% in WES testing.
Figure 2.
Variant prevalence for LQTS, WES, and control cohorts. Bar graph of the frequency of LQTS-associated gene variants among individuals with LQTS (green fill), WES (light blue fill) and ostensible healthy control individuals (ExAC, light gray and GnomAD, dark gray fill). Error bars denote 95% CI. *, P<0.0001
LQTS-associated WES variants localize to genes that are rare causes of LQTS and reflect healthy background prevalence
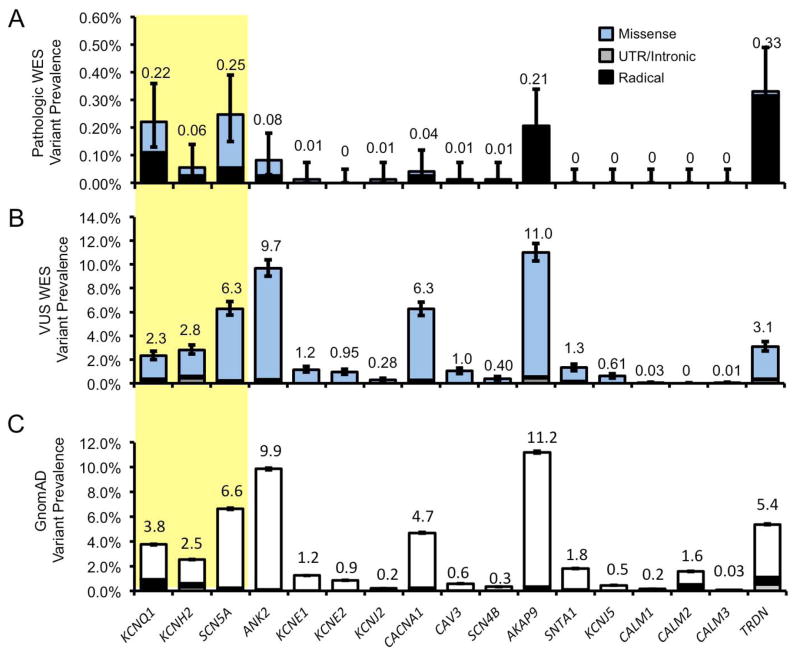
To determine the gene-specific prevalence of variants in LQTS-associated genes in next generation sequencing, we next stratified variant prevalence by genetic loci. Within the WES cohort, we found a small number of individuals with a “likely pathologic” variant while the vast majority hosted VUSs. These VUSs predominantly localized to a small number of LQTS-associated genes which are traditionally held to be rare causes of LQTS. These include AKAP9 (11.0% [10.3–11.8] of all WES referrals) and ANK2 (9.7% [9.0–10.4%]). The two most common genetic causes of LQTS, KCNQ1 and KCNH2, had a relatively low frequency of WES identified VUSs with a frequency of 2.3% [2.0–2.7] and 2.8% [2.4–3.2], respectively. These results are summarized in Figure 3A and B (further detailed in Supplemental Results). Overall, this subset analysis demonstrates that WES-identified variants tended to be VUSs and localized to genes which are rare causes of LQTS, while genes commonly associated with LQTS pathogenicity had a relatively low rate of incidentally identified WES variants.
Figure 3.
WES and control cohort gene-specific variant prevalence. A, Bar graph of WES variants (blue fill) deemed “likely pathogenic” at time of genetic testing for each LQTS-associated gene. B, Variants deemed variants of undetermined significance (VUS). C, Rare variants among GnomAD control cohort (white fill). Missense (blue/white), intronic and untranslated regions (UTR, gray), and radical mutations (black) are noted. Error bars denoted 95% CI.
We next compared the gene-specific variant prevalence within these cohorts to determine what degree of similarity was reflected at the individual gene level. As with WES variants, control variants were predominantly missense (46.8% [46.6–47.0] variant frequency) and rarely radical (3.2% [3.1–3.2]) in any LQTS-associated gene. These rare variants were found to predominantly localize to LQTS genes with a higher prevalence of WES-identified variants and were rare causes of LQTS. Healthy rare variants were found most commonly in AKAP9 (11.2% [11.1-1.3] variant frequency) and. ANK2 (9.9% [9.7–10.0]). These results are summarized in Figure 3C. Among all LQTS genes, there were widely congruent gene frequencies between the WES and control individuals and reinforce the remarkable similarity between the prevalence of incidentally identified WES variants and rare variants found in healthy individuals.
Given the notable similarity between WES and healthy variants, we evaluated the possibility that the variation in variant frequency was directly related to coding sequence length. ANK2, the gene locus with the highest frequency of rare variants among the WES (20.1%, relative frequency) and GnomAD (19.2%) cohorts demonstrated a nearly identical relative coding nucleotide length of 23.2%. Similarly, AKAP9 comprised 23.0% of the relative primary sequence length, 23.1% of the WES variants, 21.8% of the GnomAD variants. Overall, we found that relative protein length between LQTS1-17 gene products correlated tightly with relative frequency of WES and GnomAD rare variants (Supplemental Figure 1).
Gene-specific signal-to-noise for LQTS-associated genes
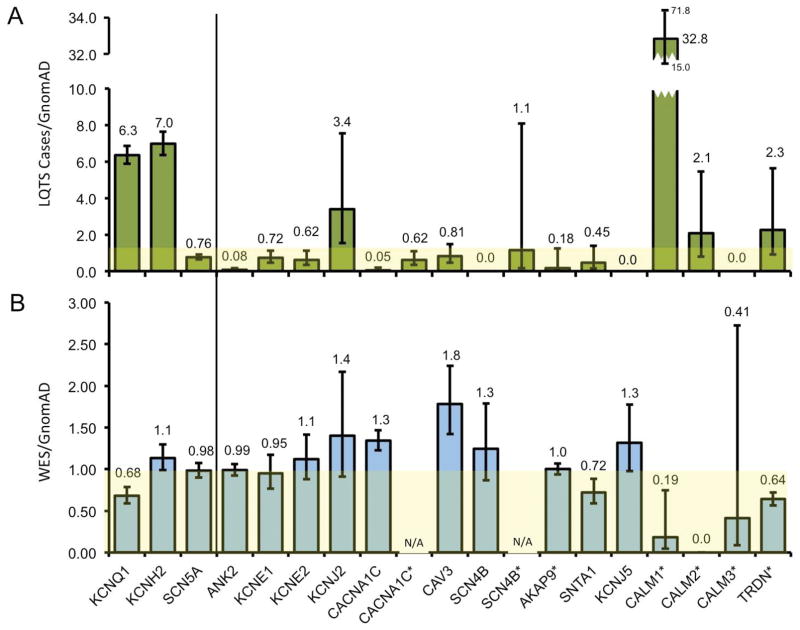
Given the similarity of LQTS gene frequency in WES variants and the background genetic variation noted in healthy individuals, we next calculated gene-specific “signal-to-noise” ratios by normalizing the frequency of variants in LQTS case and WES cohorts, respectively, against the background rate of variants in healthy individuals. In keeping with previous studies, the genes with the highest ratios were KCNQ1 (6.3 [5.9–6.9]) and KCNH2 (7.0 [6.3–7.6]), 2 of the 3 canonical causes of LQTS. SCN5A demonstrated a ratio of 0.76 [0.64–0.92]. Interestingly, KCNJ2 (3.4 [1.5–7.6]) also demonstrated a relatively high signal to noise ratio. Among genes only evaluated in cohorts of LQTS-positive, genotype-negative individuals, only CALM1 (32.8 [14.9–71.9]) demonstrated a signal to noise ratio greater than 2. These results are summarized in Figure 4A. These results were similar with an expected higher signal-to-noise ratio when a lower GnomAD MAF threshold of <0.001 was utilized (Supplemental Results).
Figure 4.
Gene-specific signal-to-noise frequencies. A, Bar graph of the frequency of LQTS/GnomAD-associated gene variants in the LQTS case cohort (green fill). B, Bar graph of the frequency of WES cohort/GnomAD-associated gene variants (blue fill). Yellow background denotes signal-to-noise threshold of 1. * Indicates a phenotype-positive genotype-negative proband sample. Error bars denote 95% CI.
With the high signal-to-noise ratio among truly pathologic variants in canonical LQTS-associated genes, we next examined WES-variants. Only CAV3 demonstrated a signal-to-noise ratio greater than 1 (1.8% [1.4–2.2]). This was driven by a high recurrence WES variant CAV3-T78M that made up 50% of all CAV3 variants within the WES cohort. This variant was found in the GnomAD cohort with a variant MAF of 0.0027. These results are summarized in Figure 4B. Overall, these results suggest that incidentally identified WES variants rare rise above the frequency of rare background variation on an individual gene basis.
Variant co-localization between cohorts
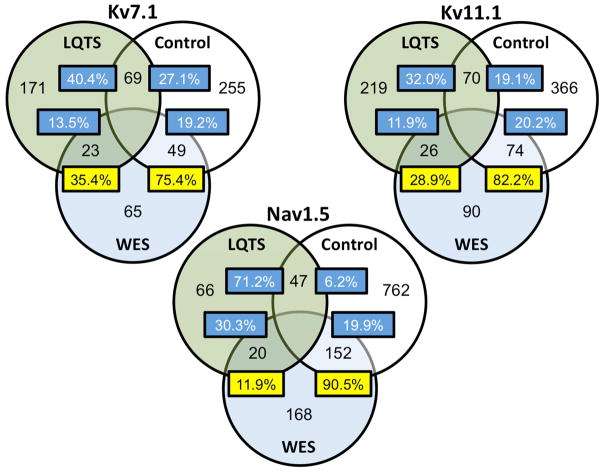
To determine whether variants identified among the clinical WES cohort more frequently altered amino acids affected by control or case variants, we mapped the variants along the primary sequence of Kv7.1, Kv11.1, and Nav1.5. There was relatively low degree of overlap in variant location between WES and LQTS cases, including 35.4% [23.9–48.2] for Kv7.1, 28.9% [19.8–39.4] for Kv11.1, and 11.9% [7.4–17.8] for Nav1.5. In comparison, there was a markedly higher frequency of variant overlap among WES and control residues. Specifically, 75.4% [63.1–85.2] for Kv7.1, 82.2% [72.7–89.5] for Kv11.1, and 90.4% [86.0–94.5] for Nav1.5. Overall, WES cohort variants in Kv7.1 were 3-fold more likely to co-localize to residues found in controls, 4-fold more likely in Kv11.1, and 13-fold more likely in Nav1.5 (P<0.0001). Findings are demonstrated in Figure 5.
Figure 5.
Variant position overlap between LQTS, WES, and control cohorts. Venn diagram of the co-localization of variants to residues shared between the LQTS (green fill), control (white fill), and WES (blue fill) cohorts. The numbers within the circles demonstrate number of unique amino acid residues that host variants/mutations within each cohort. Numbers within overlapping portions of the circles reflect the number of residues with variants shared between respective cohorts. The proportion of shared variants, out of the cohort total, is noted within each box.
Amino acid-level signal-to-noise calculations among Kv7.1, Kv11.1, and Nav1.5
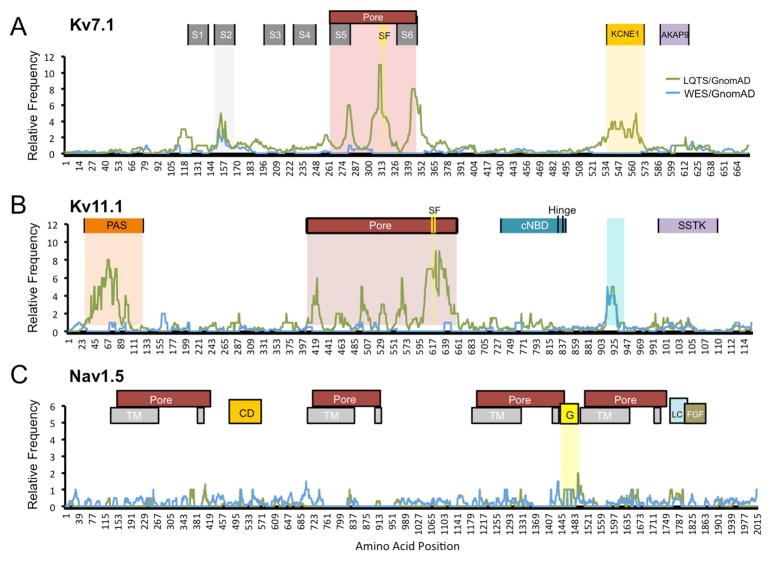
Given the amino-acid level similarities between WES and control variants, and previous evidence that LQTS-associated mutations localize to pathologic hotspots within the ion channel, we next conducted amino-acid level signal-to-noise comparisons.16 Interestingly, for Kv7.1, signal-to-noise ratios for disease-associated variants peaked in the second transmembrane domain, the pore region, and the KCNE1-binding site. Within the pore region, three distinct peaks were seen localizing to the S5 and S6 transmembrane domains of the pore with a large peak immediately N-terminal to the Kv7.1 ion selectivity filter. There were no such peaks when WES variants were normalized to controls. In Kv11.1, large LQTS disease-associated peaks were noted at the Per-Arnt-Sim domain and the pore domain. Within the pore domain, the region immediately C-terminal to the selectivity filter represented the highest signal within the peptide sequence. Interestingly, we observed a high signal-to-noise peak localizing to Kv11.1 residues 912–930 for both LQTS and WES variants. This region has no known functional domain. Finally, for Nav1.5, there were no significant signal-to-noise peaks for either LQTS or WES variants. These findings are depicted in Figure 6 with individual variant mapping in Supplemental Figures 2–4. Overall, these findings suggest that there are functional domains of Kv7.1 and Kv11.1 with a higher likelihood of hosting LQTS-associated pathologic mutations while WES variants do not.
Figure 6.
Amino-acid level signal-to-noise variant frequency for the three major gene products associated with LQTS. Frequency of LQTS (green) and WES (blue) cohort variant frequency versus amino acid position for Kv7.1 (A), Kv11.1 (B), and Nav1.5 (C). AKAP9, AKAP9-protein binding; CD, cytoplasmic; cNBD, C-terminal nucleotide binding; FGF, FGF13 binding; G, inactivation gate; KCNE1, KCNE1-protein binding; LC, L-type Ca2+ channel binding; PAS, Per-Arnt-Sim; SF, selectivity filter; S1–S6, transmembrane; SSTK, SSTK-interacting protein; TM, transmembrane; TSSK6-activating co-chaperone protein binding domains.
Indication for WES testing and variant yield among institutional referrals
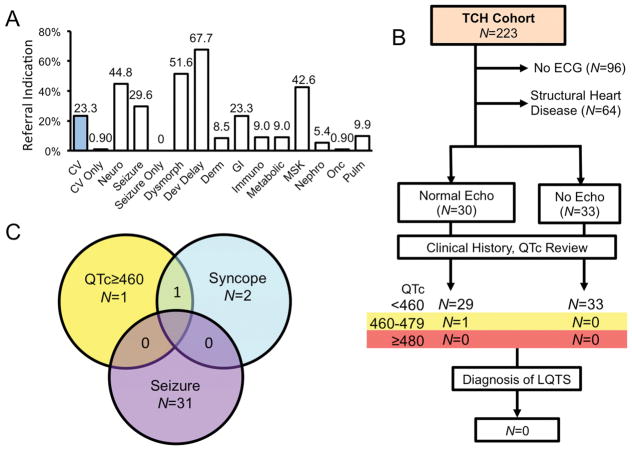
To evaluate whether children referred for WES genetic testing demonstrated pre-WES testing suspicion for LQTS, or post-test diagnosis of LQTS, we next evaluated the clinical records of WES referrals from our institution (TCH). A total of 223 unique probands were identified who were referred for WES testing from TCH and who hosted either “likely pathologic” or VUSs in one of the LQTS1-3 genes. The median age of this cohort was 5.2 [1.8–8.6] years at the time of genetic testing. Reflective of the WES cohort as a whole, most individuals hosted missense VUSs. Further, the indication for WES testing referral was rarely only cardiac in nature (0.9% [0.11–3.20], Figure 7, Supplemental Results, and Supplemental Table 2).
Figure 7.
Pre- and post-test clinical suspicion for LQTS among TCH WES referrals. A, Bar graph of the frequency of indication by organ systems for WES referral. Individuals referred for cardiovascular (CV, blue fill). B, Schematic of the TCH cohort evaluation. Individuals seen at TCH with an ECG and no structural heart disease were included. C, Venn diagram of associated symptoms among individuals with QTc>460 (borderline) on at least 1 ECG.
Clinical evaluation of institutional referrals with variants in LQTS genes
To determine whether any individual with incidentally identified variants in LQTS-associated genes have clinical suspicion for LQTS, clinical records from institutional referrals were reviewed. The vast majority of children demonstrated no personal or family history of arrhythmic disease, syncope, seizures, or sudden death. Given the link between multiple LQTS-associated variants and severe LQTS, individuals with 4 or more variants within the WES Cohort were separately evaluated. Two of these carried borderline prolonged QTc intervals of 467ms and 461ms, respectively; both were noted to have significant congenital anomalies and had congenitally malformed hearts. No individuals were found to have QTcs >480ms, evidence of ventricular arrhythmias, suspected channelopathy or LQTS (Figure 7 and Supplemental Results). Taken together, these results suggest that incidentally identified LQTS1-3-associated variants are not associated with QT prolongation or LQTS diagnoses. In the absence of a positive family history of LQTS or in the setting of refractory epilepsy, these variants are unlikely to represent disease susceptibility.
DISCUSSION
While early studies of LQTS gene variation in ostensibly healthy individuals have estimated that ~5% of ostensibly healthy individuals will host a rare variant in a LQTS-associated gene16, the American College of Medical Genetics and Genomics (ACMG) recommends the reporting of any “known pathologic” variant associated with LQTS.17 Recently, a curated list of disease-associated genes associated with diseases that present during childhood were proposed by the BabySeq project.18 A reflection of rapidly increasing fetal and neonatal WES testing, the authors put forward a list of gene loci associated with clinically actionable, and thus reportable, variants. All major LQTS-associated genes are designated as clinically actionable and, thus, reportable. While it is clear that broad genetic sequencing has diagnostic value, these studies do not address the large number of incidentally identified rare variants with unclear diagnostic value.
Not unique to LQTS, several studies have demonstrated a significant rate of rare background variation in other channelopathic and cardiomyopathic diseases among healthy individuals.19–21 We have found that ~6% of ostensibly healthy individuals will host rare variants in either of the two genes associated with the channelopathy catecholaminergic polymorphic ventricular tachycardia and, ~9% of individuals undergoing clinical WES testing will host a “likely pathogenic” variant or VUS.9 Here, we identify a background rate of rare variants in LQTS1-3 genes to be ~13% among individuals comprising the GnomAD database. A significantly higher ~38% of individuals hosted a variant in one of the LQTS4-17 genes, predominantly localizing to AKAP9 and ANK2. Similarly, incidentally identified variants from the WES cohort were noted to comprise similar prevalence with the prevalence of LQTS1-3 in pathologic cases was ~45%. This, combined with a high signal-to-noise ratio in the common genes associated with LQTS (KCNQ1 and KCNH2), suggests that incidentally identified variation is likely background genetic noise.
Our data suggests that a pathologic variant in KCNQ1 or KCNH2 connotes a higher chance for a post-test probability of LQTS in the context of pre-test clinical suspicion. This reflects the probabilistic approach to diagnosis established by Bayes Theorem which determines the probability of a future event is related to prior knowledge of the conditions associated with it. As such, identification of an incidentally identified variant in a gene with a low signal-to-noise ratio would hold less diagnostic value. Further, an incidentally identified variant in the absence of concerning family history or clinical suspicion for LQTS, is unlikely to represent monogenic markers of disease even if localizing to a LQTS-associated gene. We acknowledge the significant challenge inherent in variant pathogenicity predication and believe that further study utilizing tools, such as machine learning and artificial intelligence, has a role in continuing to refine predictive modeling. Due to these factors, care must be given in the interpretation of incidentally identified VUSs.
In addition to the gene locus itself, the diagnostic interpretation of a VUS should involve variant location within the gene sequence, particularly with variants in KCNQ1-encoded Kv7.1 and KCNH2-encoded Kv11.1. Our finding of variable signal-to-noise at the amino acid level supports a number of previous studies suggesting that the biophysical impact of potassium channel mutations may be largely dependent on location. This so-called “intragenic risk” is well established in Kv7.1 whereby LQTS-associated, loss-of-function mutations in the pore domain selectivity filter (S1–S4) are linked to a high risk of TdP and SCD22, 23 and mutations with the PAS domain of Kv11.1 are associated with channel trafficking defects and severe LQTS.24, 25 In addition to these known functional domains, we report a significant signal-to-noise peak in Kv11.1 residues 912-930. To our knowledge, this region of Kv11.1 has no identified functional domain or regulatory role. Given the critical functionality of the other domains identified by our analysis, we hypothesize that localization of disease-associated mutations normalized to rare background variation may be a tool for identifying biophysically and physiologically relevant protein domains.
The clinical diagnosis of LQTS can be a challenge, even in the setting of high clinical suspicion for the disease. While many individuals are diagnosed in childhood, the average of diagnosis is often in the second or third decade of life.7 Due to the referral bias of our institution, the age of genetic testing in the TCH cohort was ~5–6 years. It is notable that ~40% of LQTS probands had a personal history of syncope or a family history of LQTS while this was largely absent in the TCH cohort.7 While our evaluation for clinical signs, symptoms, or history of LQTS was retrospective in nature, we cannot exclude the possibility of a clinically occult LQTS diagnosis or a diagnosis later in life.
CONCLUSIONS
Incidentally identified LQTS-associated gene variants are common in children undergoing whole exome sequencing, associated with over 1/3 of all tests. Based on amino acid-level signal-to-noise analysis, WES-identified variants are indistinguishable from healthy background variation while LQTS cased-identified variants localized to clear pathologic “hot spots” within KCNQ1-encoded Kv7.1 and KCNH2-encoded Kv11.1.
Supplementary Material
Acknowledgments
FUNDING SOURCES
APL is supported by the National Institutes of Health K08-HL136839 and L40-HL129273, the Pediatric and Congenital Electrophysiology Society Paul C. Gillette Award, pilot grant funding from the Baylor College of Medicine Department of Pediatrics, and the McCrae Foundation.
Footnotes
DISCLOSURES
JAR and YY receive salary support from Baylor Miraca Genetics Laboratories. All other authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Landstrom AP, Tester DJ, Ackerman MJ. Role of genetic testing for sudden death predisposing heart conditions in athletes. 2011 [Google Scholar]
- 2.Schwartz PJ, Stramba-Badiale M, Crotti L, et al. Prevalence of the Congenital Long-QT Syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kass RS, Moss AJ. Long QT syndrome: novel insights into the mechanisms of cardiac arrhythmias. J Clin Invest. 2003;112:810–815. doi: 10.1172/JCI19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Curran ME, Splawski I, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 5.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 7.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Landstrom AP, Dailey-Schwartz AL, Rosenfeld JA, Yang Y, McLean MJ, Miyake CY, Valdes SO, Fan Y, Allen HD, Penny DJ, Kim JJ. Interpreting Incidentally Identified Variants in Genes Associated With Catecholaminergic Polymorphic Ventricular Tachycardia in a Large Cohort of Clinical Whole-Exome Genetic Test Referrals. Circulation Arrhythmia and electrophysiology. 2017:10. doi: 10.1161/CIRCEP.116.004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 12.Hedley PL, Durrheim GA, Hendricks F, Goosen A, Jespersgaard C, Stovring B, Pham TT, Christiansen M, Brink PA, Corfield VA. Long QT syndrome in South Africa: the results of comprehensive genetic screening. Cardiovascular journal of Africa. 2013;24:231–237. doi: 10.5830/CVJA-2013-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hassnan ZN, Al-Fayyadh M, Al-Ghamdi B, et al. Heart Rhythm. 2017. Clinical profile and mutation spectrum of long QT syndrome in Saudi Arabia: The impact of consanguinity. [DOI] [PubMed] [Google Scholar]
- 14.Lieve KV, Williams L, Daly A, Richard G, Bale S, Macaya D, Chung WK. Results of genetic testing in 855 consecutive unrelated patients referred for long QT syndrome in a clinical laboratory. Genet Test Mol Biomarkers. 2013;17:553–561. doi: 10.1089/gtmb.2012.0118. [DOI] [PubMed] [Google Scholar]
- 15.Napolitano C, Priori SG, Schwartz PJ, Bloise R, Ronchetti E, Nastoli J, Bottelli G, Cerrone M, Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 16.Kapa S, Tester DJ, Salisbury BA, Harris-Kerr C, Pungliya MS, Alders M, Wilde AA, Ackerman MJ. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards SAN, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehn HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. 2015 doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceyhan-Birsoy O, Machini K, Lebo MS, Yu TW, Agrawal PB, Parad RB, Holm IA, McGuire A, Green RC, Beggs AH, Rehm HL. A curated gene list for reporting results of newborn genomic sequencing. Genet Med. 2017;19:809–818. doi: 10.1038/gim.2016.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landstrom AP, Ackerman MJ. The Achilles’ heel of cardiovascular genetic testing: distinguishing pathogenic mutations from background genetic noise. Clin Pharmacol Ther. 2011;90:496–499. doi: 10.1038/clpt.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapplinger JD, Landstrom AP, Bos JM, Salisbury BA, Callis TE, Ackerman MJ. Distinguishing hypertrophic cardiomyopathy-associated mutations from background genetic noise. J Cardiovasc Transl Res. 2014;7:347–361. doi: 10.1007/s12265-014-9542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapplinger JD, Landstrom AP, Salisbury BA, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57:2317–2327. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess DE, Bartos DC, Reloj AR, et al. High-risk long QT syndrome mutations in the Kv7. 1 (KCNQ1) pore disrupt the molecular basis for rapid K(+) permeation. Biochemistry. 2012;51:9076–9085. doi: 10.1021/bi3009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jons C, Moss AJ, Lopes CM, et al. Mutations in conserved amino acids in the KCNQ1 channel and risk of cardiac events in type-1 long-QT syndrome. J Cardiovasc Electrophysiol. 2009;20:859–865. doi: 10.1111/j.1540-8167.2009.01455.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999;274:10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 25.Ke Y, Ng CA, Hunter MJ, Mann SA, Heide J, Hill AP, Vandenberg JI. Trafficking defects in PAS domain mutant Kv11. 1 channels: roles of reduced domain stability and altered domain-domain interactions. Biochem J. 2013;454:69–77. doi: 10.1042/BJ20130328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.