Abstract
BACKGROUND
Downregulated sodium currents in heart failure (HF) has been linked to increased arrhythmic risk. Reduced expression of mRNA stabilizing protein ELAVL1/HuR may be responsible for the downregulation of sodium channel gene SCN5A mRNA.
OBJECTIVE
To investigate whether HuR regulates SCN5A mRNA expression and whether manipulation of HuR benefits arrhythmia control in HF.
METHODS
Quantitative real-time reverse-transcriptase PCR (qRT-PCR) were used to investigate the expression of SCN5A. Optical mapping of intact heart was adopted to study the effects of HuR on the conduction velocity and action potential upstroke in myocardial infarct (MI) HF mice after AAV-HuR injection.
RESULTS
HuR was associated with SCN5A mRNA in cardiomyocytes, and the expression of HuR was downregulated in failing hearts. The association of HuR and SCN5A mRNA protected SCN5A mRNA from decay. Injection of AAV-9 viral particles carrying HuR increased SCN5A expression in mouse heart tissues after MI. Optical mapping of intact heart demonstrated that overexpression of HuR improved action potential upstroke and conduction velocity in the infarct border zone, which reduced risks forming reentrant arrhythmia in MI.
CONCLUSIONS
Our data indicate that HuR is an important RNA binding protein in maintaining SCN5A mRNA abundance in cardiomyocytes. Reduced expression of HuR may be at least partially responsible for the downregulation of SCN5A mRNA expression in ischemic HF. Overexpression of HuR may rescue decreased SCN5A expression and reduce arrhythmia risk in HF. Increasing mRNA stability to raise ion channel currents may correct a fundamental defect in HF and represent a new paradigm in antiarrhythmic therapy.
Keywords: Cardiac sodium channel, RNA binding protein, mRNA decay, Arrhythmia, Heart failure
Introduction
Control of gene expression is a coordinated, multi-layered process, which includes active regulation of mRNA decay. The regulation of mRNA decay is a major mechanism by which mammalian cells control gene expression in response to changes in environmental conditions and cellular stress.1 mRNA decay mechanisms are involved in the regulation of the stability of certain mRNAs and the elimination of nonfunctional or otherwise defective transcripts. The half-life of mRNA is controlled by sequences or structures within the mRNA (cis-elements) and the trans-acting factors such as RNA-binding proteins (RBPs), which bind to these cis-elements. The association between specific cis-elements and selected RBPs can prevent mRNA degradation or lead to mRNA decay.
HuR (also known as ELAVL1) is one of the most prominent sequence-specific regulatory RNA binding proteins. Systemic analysis indicates that HuR coordinates gene expression outcomes at multiple interconnected steps of RNA processing.2 HuR binds adenylate-uridylate rich elements (AU-rich elements, AREs), one of the most well-characterized mRNA stability determinants in the 3′-untranslated region (3′-UTR),3 to regulate the stability of a wide range of mRNAs involved in key cellular processes.4
Voltage-gated cardiac sodium channels are essential for the myocyte membrane excitability. Among all the ionic currents involved in generating the cardiac action potential (AP), the cardiac sodium current (INa) plays a central role in the upstroke of the AP. The cardiac sodium channel consists of a large α subunit, encoded by SCN5A, and small β subunits. In patients, deletions or loss-of-function mutations of SCN5A have been associated with a wide range of arrhythmias.5 During common pathological conditions, such as myocardial ischemia and heart failure, decreased sodium current contributes to decreased upstroke velocity of the AP, decreased conduction velocity in heart tissue, and arrhythmic risk.6 Our previous results have shown that the expression levels of SCN5A mRNA and protein are decreased in failing hearts and that this decrease in current contributes to the increased risk of sudden death in heart failure (HF).7–9
Multiple mechanisms have been identified that contribute to the reduction in sodium current with myocardial pathologies. However, the molecular mechanisms underlying sodium current downregulation in the failing heart are far from clear. In this study, we identified a new post-transcriptional mechanism by which the expression of SCN5A mRNA is regulated. Our data described here suggest that HuR positively regulates SCN5A expression by protecting its mRNA against degradation. Overexpression of HuR rescues down-regulated SCN5A mRNA and reduces arrhythmia risk in ischemic cardiomyopathy.
Methods
Cell Culture and Transfection
Human fetal cardiomyocyte cell line RL14 (ATCC® RL-14, ATCC, Manassas, VA, USA) cells were grown in DMEM/F-12 nutrient mixture (GE Healthcare Life Sciences, Logan, UT) supplemented with 12.5% (v/v) fetal bovine serum (Sigma, St. Louis, MO), streptomycin, and penicillin. DNA plasmids were transfected into cultured cells using SuperFect (Qiagen, Valencia, CA) according to the manufacturer’s protocol.
Human left ventricular tissue acquisition
De-identified LV endocardium samples from patients with severe HF of ischemic (ICM; n=8) origin were collected at the time of LVAD implantation. Non-failing human LV endocardium samples (Control, n=8) were collected from donor hearts deemed not suitable for transplantation for technical or non-cardiac reasons from Mid-America Transplant Services.
Plasmid constructions
HA-fusion plasmids were generated by amplifying the coding region of HuR using PCR primers that included the HA sequences and subcloning into pcDNA3.1 or pTRIPO vectors to create a HA-tag at its N-terminus of HuR. pTRIPO was generated by replacing the AgeI–MluI fragment of pTRIPZ (Thermo Fisher Scientific, Waltham, MA) containing the turboRFP tag and the 5′mir30/3′mir30 sequences by an AgeI–HpaI–XhoI–EcoR1–MluI polylinker sequence to create the inducible lentiviral vector under the tet- on control system.10
Lenti-virus packaging and transduction
Three-plasmids (pTRIPO-HA-HuR, psPAX and pMD2.G) co-transfection into Lenti-X 293 cells (Clontech, Mountain View, CA) was used to make pseudotyped lenti virus as described using the manufacturer’s protocol. Plasmids psPAX2 (Addgene, plasmid # 12260, Cambridge, MA) and pMD2.G (Addgene, plasmid # 12259, Cambridge, MA) were gifts of Dr. Didier Trono. Meium containing pseudotyped lenti viral particles were collected and were concentrated using PEG-it™ Virus Precipitation Solution (System Biosciences, Palo Alto, CA 94303) and stored at −80°C.
Real-Time PCR quantification
Total RNA was isolated using the RNeasy Mini plus Kit (Qiagen, Valencia, CA) and was reverse transcribed into complementary DNA (cDNA) using SuperScript® VILO™ Master Mix (Thermo Fisher Scientific, Waltham, MA). Quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) was carried out using gene-specific primers, Fast SYBR® Green Master Mix and 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). Samples were run in triplicate and averaged. Gene expression levels were normalized to the level of β-actin. The primers for qRT-PCR were listed in supplementary table 1.
Western blotting
Equal amounts (30 μg) of proteins from heart tissues or cultured cells were separated on 4–10% mini-PROTEAN® TGX™ gels and transferred to Polyvinylidene difluoride membranes. The membranes were blotted with an anit-HuR (Santa Cruz Biotechnology, Santa Cruz, CA) antibody. An anti-β-actin antibody (Thermo Fisher Scientific, Waltham, MA) was used as a loading control for all studies. Proteins were visualized using Clarity™ Western ECL Blotting Substrate, and band intensities were detected by ChemiDoc MP imaging System and analyzed by Image Lab software (Biorad, Hercules, CA).
siRNA and Transfection
Silencer® select validated siRNA for HuR 5′-ACUUAUUCGGGAUAAAGUAtt-3′ and negative control siRNA were purchased from Life Technologies (Carlsbad, CA). siRNA was transfected into RL14 cells using Lipofectamine® RNAiMAX (Life Technologies, Carlsbad, CA) following the manufacturer’s protocol.
Electrophysiology
Sodium currents were measured using the whole-cell patch-clamp technique in voltage-clamp mode at room temperature as described before.11 Peak currents obtained during steps to −20 or −30 mV were used for comparison of the INa with or without HuR overexpression.
Mice
All animal protocols used in this study were approved by the IACUC (Institutional Animal Care and Use Committee) of Lifespan. Mice were housed in accordance with the regulations on mouse welfare and ethics of Lifespan and had free access to food and water. Twelve week old male C57BL/6 mice underwent either sham surgery or left anterior coronary artery ligation to induce myocardial infarction (MI). Two weeks after surgery, MI animals meeting inclusion criteria (ejection fraction < 45%) were randomized into control and treatment groups. AAV9 virial particles bearing either an empty vector or HuR were systemically injected into mice via the right jugular vein as previously described.12 Two weeks after viral injection, animals were sacrificed for cardiomyocytes isolation, tissue collection and Western blotting.
Optical Mapping
Isolated hearts were Langendorff-perfused with control Krebs-Henseleit buffer as described before.13 Hearts were stained with a voltage-sensitive dye, di-4-ANEPPS (Invitrogen, Carlsbad), delivered through a bubble trap, above the aortic cannula. ECG and perfusion pressure were continuously monitored (PowerLab, ADInstruments, Colorado Springs). The action potentials were recorded using a MiCAM Ultima-L CMOS camera (SciMedia, Japan) with high spatial (1×1 cm2 field of view, 100×100 μm2 pixel resolution) and temporal (2000 frames/s) resolution (emission filter >580nm). Data were filtered using a spatial Gaussian filter (5×5 pixel), and first derivatives (dF/dt) were calculated using a temporal polynomial filter (3rd order, 13 points). APD75 was measured between action potential upstroke and 75% recovery. Local conduction velocity (CV) was measured from calculating local gradient of activation time from 7×7 pixels, and the averaged value of CV was calculated from all the pixels. The action potentials were recorded from two locations, the infarct border zone (BZ) in the left ventricles and the remote zone in the anterior right ventricle area where infarct zone is minimally seen in the field of view. The vulnerability to VT was tested using 1) S1S2 stimulation protocol, 2) ramp pacing protocol in which CL was decreased by 5 ms until loss of 1:1 capture or induction of VT, 3) isoproterenol injection (140 nmol/L, 5 ml).
Results
HuR expression decreased in ischemic heart failure
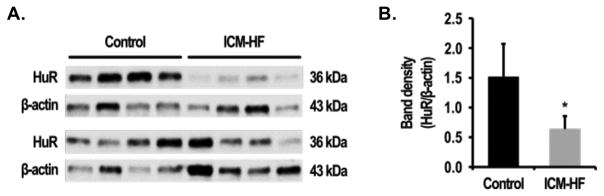
As HuR is the major Hu protein family member predominantly expressed in human heart (supplementary Figure 1), we tested whether the expression of HuR was altered in HF. Using left endocardium ventricular tissues from non-failing or ischemic failing human hearts, Western blot analysis revealed that the protein level of HuR decreased in the ischemic failing hearts (Figure 1A). Quantification of the ratios of intensity of HuR and β-actin showed that HuR was significantly lower in HF heart tissue samples (1.75±0.88 in vs. 0.57±0.17 respectively, P<0.05; Figure 1B).
Figure 1. HuR is reduced in human HF.
(A) Immunoblot analysis of HuR protein in normal control (n=8) and failing heart (n=8) tissue. (B) Quantification of HuR expression in control and failing hearts. Protein level was normalized by β-actin. Error bars represent mean ± SD. *P<0.05
HuR associated with SCN5A mRNA
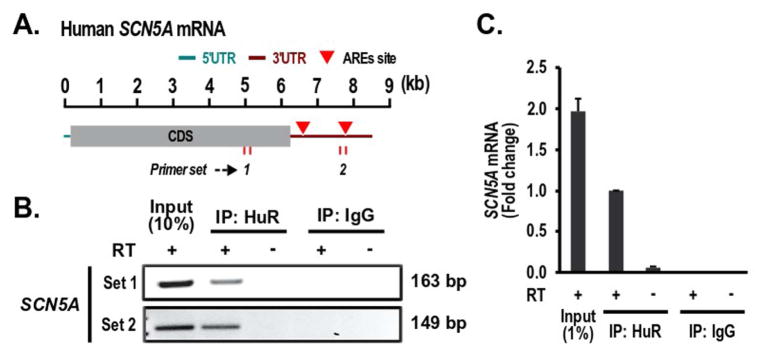
HuR action involves binding to mRNA AU rich elements (AREs). The association of SCN5A mRNA with HuR was examined by PCR analyses following reverse transcription of RNA isolated from the IP precipitates using the procedure described before.14 SCN5A PCR products were highly enriched in samples precipitated with anti-HuR antibody, while no SCN5A PCR product was detected in control IgG pellets. There was no SCN5A PCR product detected in samples precipitated with anti-HuR antibody but without reverse transcription (Figure 2B). Our qPCR results further confirmed the SCN5A mRNA enrichment in the samples treated with specific anti-HuR antibody (Figure 2C). These data suggest that HuR specifically associates with SCN5A mRNA.
Figure 2. HuR associates with SCN5A mRNA.
(A) Schematic representation of the SCN5A mRNA ARE sites in its 3′-UTR. The length of the mRNA is indicated by a kilo base (kb)-scale. (B) HuR binds to the SCN5A mRNA. After IP of RNA-protein complexes from cardiomyocytes using either anti-HuR antibody (Anti-HuR) or control IgG, RNA was isolated, and then, the cDNA was synthesized in a presence or absence of the reverse transcription enzyme. Two sets of primers to different regions of SCN5A were used to amplify SCN5A fragments. The PCR products were visualized with SYBR™ Safe DNA Gel Stain in agarose gels. (C) qRT-PCR was performed to confirm the results in (B).
HuR regulated SCN5A mRNA expression
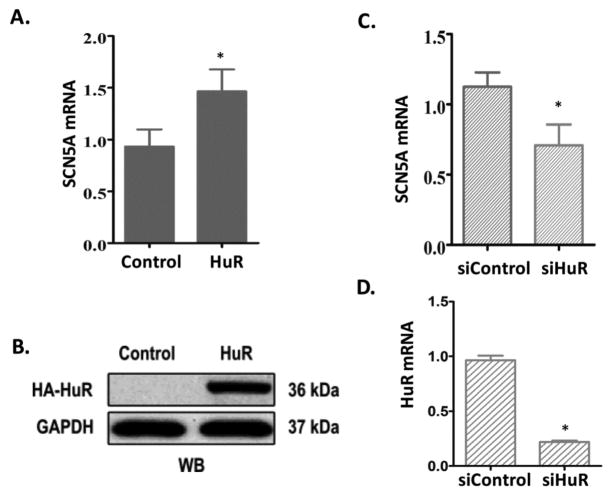
To test the physiological significance of the association of SCN5A mRNA and HuR, we examined the steady-state mRNA level of endogenous SCN5A in cardiomyocytes RL14 by manipulating the expression of HuR. As expected, overexpression of HuR increased the steady-state level of SCN5A mRNA by 57.3% (P < 0.05) compared with that in control group (Figure 3A). Western blot confirmed the overexpression of HA tagged HuR by anti-HA antibody (Figure 3B). On the other hand, in cardiomyocytes transfected with HuR siRNA (siHuR), the steady-state level of SCN5A mRNA decreased 36.98% (P<0.05) compared with in those transfected with scrambled siRNA (siCtrl; Figure 3C), and HuR mRNA decreased by 77.3% (P<0.05; Figure 3D). These results indicate that HuR positively regulates SCN5A mRNA expression.
Figure 3. HuR positively regulates SCN5A mRNA expression in cardiomyocytes.
(A) Ectopic overexpression of HuR increases the steady-state level of cardiac sodium channel SCN5A mRNA by qRT-PCR. (B) Immunoblot shows overexpressed HA-tagged HuR by anti-HA antibody. (C) Silencing of HuR by siRNA decreases the steady-state level of SCN5A mRNA by qRT-PCR. (D) Decreased HuR mRNA expression in cells transfected with siHuR by qRT-PCR. Levels of all mRNAs were normalized to β-actin. Error bars represent mean ± SD. *P<0.05.
HuR protected SCN5A mRNA from decay
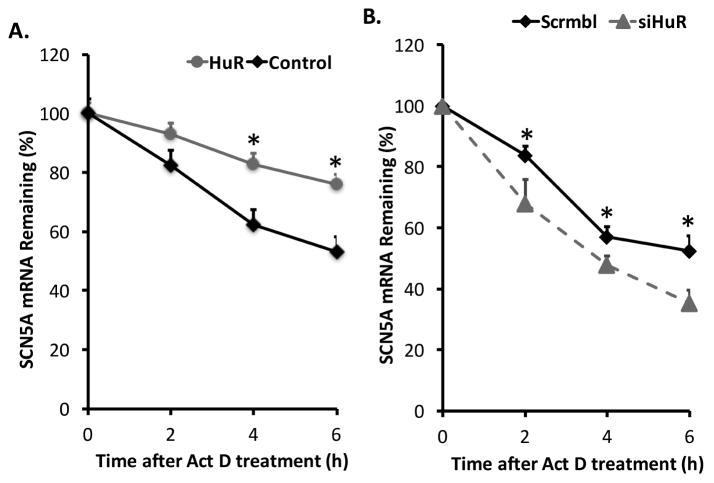
To further investigate the molecular pathway by which HuR regulates SCN5A mRNA expression, we established a doxycycline-inducible HuR overexpression system (Supplemental Figure S2A). Cardiomyocytes were infected with lenti-viral particles carrying an inducible HuR expression construct. Transcription was blocked by adding actinomycin D after 48 h of doxycycline induction. Then, cells were collected at different time points to isolate total RNA. Overexpression of HA-tagged HuR was confirmed by Western blot using anti-HA antibody (Supplemental Figure S2B). HuR stabilized SCN5A mRNA as indicated by the decreased SCN5A mRNA decay in the presence of HuR (Figure 4A). On the other hand, knock down of HuR by siRNA increased SCN5A mRNA degradation (Figure 4B). Therefore, the increase of the steady-state level of SCN5A when HuR was overexpressed was likely because of the increased stability of SCN5A mRNA mediated by HuR binding.
Figure 4. HuR protects SCN5A mRNA from decay.
(A) Overexpression of HuR increased SCN5A mRNA stability. Transcription of cells with the TetOn pTRIPz-HuR construct was blocked by addition of actinomycin D (Act D) (5 μg/mL) after 48 hours of doxycycline induction. Cells were harvested at different time points after Act D addition and the levels of SCN5A mRNA were measured using qRT-PCR. The graph shows the percentage of remaining SCN5A mRNA levels in relation to β-actin mRNA compared with the levels of normalized mRNA measured immediately before addition of Act D. The initial mRNA levels were set to 100%. (B) HuR-specific siRNA decreased the mRNA stability of SCN5A. Transcription was blocked with addition of Act D as described above 48 hours after transfection of scrambled (Scrmbl) or HuR-specific siRNA. The remaining SCN5A mRNA levels were measured as described above. Error bars represent mean ± SD. *P<0.05.
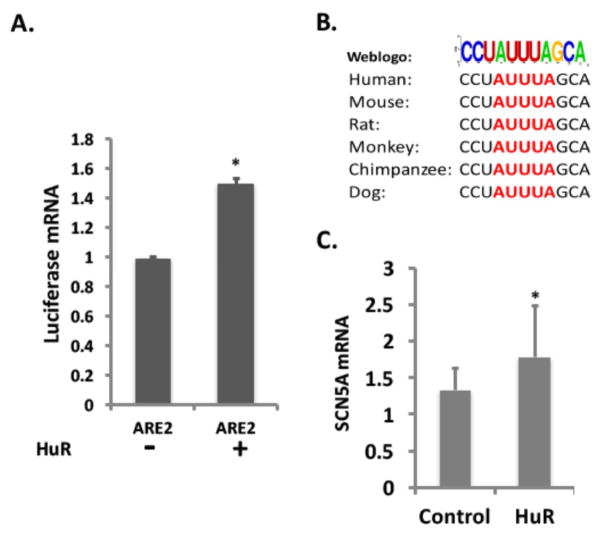
Computational predictions identified two HuR binding motifs in the 3′-UTR of the SCN5A mRNA (Figure 2A). We further investigated which copy was responsible for the SCN5A mRNA stabilizing effect. Both HuR binding motifs with surrounding sequences were individually cloned into the pGL3-promoter vector and transfected into cardiomyocytes with an inducible HuR overexpression construct. Luciferase mRNA levels were measured by RT-qPCR with or without doxycycline induction of HuR. Luciferase mRNA increased in the cells with the downstream HuR motif, which contained a canonical “AUUUA” sequence, while luciferase mRNA did not change upon HuR induction in the cells with the upstream HuR motif (Figure 5A and Supplemental Figure S3). These results indicated that the downstream HuR motif mediated the effects of HuR on SCN5A mRNA. Implying significance of the result across species, the downstream SCN5A ARE is well conserved in multiple species (Figure 5B). This functional ARE site was further confirmed by the fact that overexpression of HuR in mouse HL-1 cardiomyocytes increased mouse SCN5A mRNA expression (Figure 5C) since the mouse SCN5A mRNA harbors the downstream ARE only.
Figure 5. HuR stabilizes SCN5A mRNA through the downstream ARE.
(A) Cardiomyocytes with or without doxycycline induction were co-transfected with a pGL3-Promoter-ARE2 and a pEGFP vectors. Forty-eight hours after transfection, cells were harvested, and the abundance of luciferase mRNA was measured using qRT-PCR and normalized with EGFP mRNA. (B) Conservation of AREs (Red) in the SCN5A mRNA 3′UTR among species. (C) SCN5A mRNA expression levels in HL-1 mouse cardiomyocytes with or without HuR overexpression. Error bars represent mean ± SD. *P<0.05
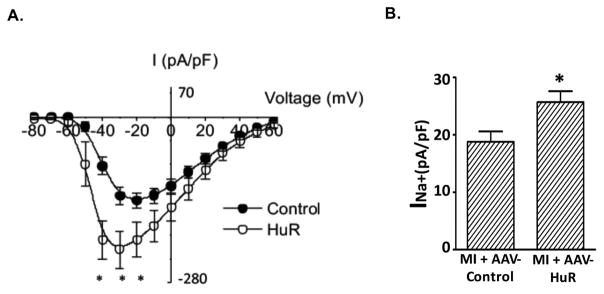
HuR overexpression increased sodium currents
To examine the role of HuR in regulating sodium currents, we cloned the functional HuR binding motif at the downstream of the SCN5A open reading frame in a GFP expression vector and transfected this construct into HEK293 cells with an inducible TetOn-HuR construct. With doxycycline-mediated HuR induction, peak sodium current increased by 67.8% (Figure 6A, P < 0.05). Overexpression of HuR also increased the peak current densities of Na+ currents at −30 mV in primary cardiomyocytes isolated from the mice received AAV-HuR injection (Figure 6B, P<0.05). Nevertheless, overexpression of HuR did not change the peak sodium current in cells transfected with SCN5A construct bearing the upstream ARE sequences (Supplemental Figure S4).
Figure 6. HuR increased sodium currents.
(A) Plasmids (pEGFP-C1-SCN5A-ORF-3′ARE2) were transfected into HEK293 cells with TetOn-HuR. Forty-eight hours after transfection, sodium currents were measured by patch clamp. Sodium currents are shown as the mean peak current ± SE. (*P<0.05). (B) Sodium currents were measured in primary cardiomyocytes isolated from mice treated with either AAV-control (n=14) or AAV-HuR (n=25). The peak current densities of Na+ currents at −30 mV are shown as mean + SE (*P<0.05).
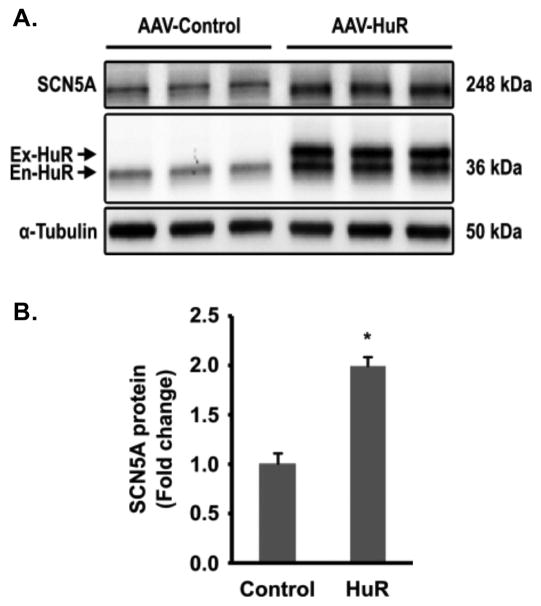
HuR overexpression increased cardiac sodium channel protein and reduced arrhythmia risk in ischemic failing hearts
To test the role of HuR in regulating SCN5A expression in vivo, we injected MI mice with either AAV-Control or AAV-HuR viral particles two weeks after coronary artery ligation and the confirmation of HF (ejection fraction < 45%). The expression of exogenous HuR was driven by a human cardiac specific α-MHC promoter. Two weeks after injection, mouse hearts were harvested to evaluate the expression of SCN5A and HuR by Western blot (Figure. 7A). Quantification of Western blots indicated that SCN5A protein increased in 119.2% in mice injected with AAV-HuR compared with in those injected with the AAV-Control vector (Figure 7B, P < 0.05). It is well known that HuR also positively regulates the expression of itself, 15, 16 which was also observed in this study (Figure 7A, En-HuR).
Figure 7. Overexpression of HuR increased the expression of SCN5A in vivo.
AAV-9 viral particles bearing either flag-tagged HuR or empty vector were injected into MI mouse 2 weeks after left anterior descending coronary artery ligation. Heart tissues were collected two weeks after injection, and Western blot was performed to examine the expression of SCN5A and HuR. (A) Representative Western blot for SCN5A and HuR in MI mice injected with control AAV (AAV-Control) or AAV bearing HuR (AAV-HuR); (B) Quantification of SCN5A expression in MI mice injected with AAV-Control (n=3) or AAV-HuR (n=3). Protein levels were normalized by α-tubulin. Ex-HuR: exogenous Flag-tagged HuR; En-HuR: endogenous HuR. Error bars represent mean ± SD. *P<0.05.
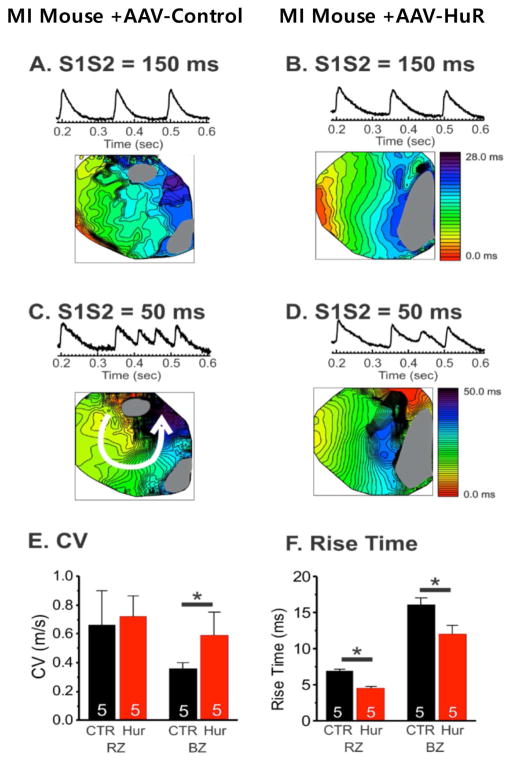
Optical mapping of 2-week post-AAV-Control or AAV-HuR injected hearts showed that HuR overexpression reduced conduction block and reentry formation under S1S2 stimulation from 150 ms pacing (S1S2=150 ms, Figure 8A & 8B) or from 50 ms pacing (S1S2=50 ms, Figure 8C & 8D) protocol (n=0/5 in HuR vs. n=4/5 in CTR, Figure 8C & 8D). Conduction velocity and action potential upstroke were markedly improved in the infarct boarder zone (Figure 8E and F), consistent with our Western blot results (Figure 7). Nevertheless, overexpression of HuR did not alter EF in MI mice (Supplemental Figure S5).
Figure 8. Cardiac optical mapping 2-weeks after either AAV-Control or AAV-HuR injection.
(A&B) Traces and activation maps from S1S2 = 150 ms pacing(C&D). S1S2 = 50 ms pacing caused reentry in MI AAV-control hearts but not in MI + AAV-HuR hearts (E&F). HuR overexpression (red) improved conduction velocity in the border zone (CL = 150 ms, CV = 0.58 ± 0.16 m/s in HuR vs.,0.36 ± 0.04 m/s in CTR, P<0.05) and action potential rise-time in the remote (4.5 ± 0.3 ms in HuR vs. 6.9 ± 0.2 ms in CTR) and border zones (12.0 ± 1.2 ms in HuR vs. 16.1 ± 1.0 ms in CTR). This was associated with protection against VT induction under multiple stimulation protocols. RZ: remote zone, BZ: border zone, RZ* p < 0.05.
Discussion
Arrhythmia is one of the major causes of mortality in HF patients. The increased arrhythmic risk is thought to be mediated in part by electrical remodeling, consisting of changes in cardiac ion channel currents. Most sarcolemmal ion channel currents are reduced in cardiomyopathy, and increasing ion channel currents can reduce arrhythmic risk.17 In HF, it is well documented that the expression of the SCN5A cardiac sodium channel gene decreases at both the protein and mRNA levels and that downregulation of cardiac sodium channels can contribute to conduction disturbances and arrhythmic risk.6, 8, 9, 18 Circulating white blood cell SCN5A mRNA levels correlate with those in heart, and assessing these levels is predictive of arrhythmic risk in HF.7
Although up or downregulation of the cardiac sodium channel current result in arrhythmic risk, few studies have addressed mechanisms controlling of SCN5A expression. To date, studies regarding SCN5A expression regulation have been mainly focused on transcriptional regulation. TBX5 and GATA4 are major transcription factors promoting SCN5A transcription while NFκB, TBX3 and Snail function as transcriptional SCN5A repressors.19–21 Recently, post-transcriptional regulation has emerged as important mechanisms in the regulation of eukaryotic gene expression. Previously, we have reported abnormal SCN5A pre-mRNA splicing leads to truncated non-functional SCN5A variants.8, 9
In addition to alternative pre-mRNA splicing, the stability of mRNA has emerged as a key post-transcriptional step in the regulation of eukaryotic gene expression. The RNA binding protein, HuR/ELAVL1, is a well-established posttranscriptional regulator, and it affects the fate of its target mRNAs by binding to specific AU- or U-rich elements in their 3′ UTRs.2, 22 In the present study, we discovered that SCN5A expression could be regulated by HuR through mRNA stability pathway. Our study suggests a novel molecular mechanism by which the expression of SCN5A is regulated via alterations in SCN5A mRNA stability.
The association of Hu proteins and mRNAs are cellular context dependent.23 In this case, HuR binding was specific to only one of the two putative AREs in the SCN5A mRNA, confirming that Hu proteins do not bind to and stabilize all ARE-containing mRNAs indiscriminately. The predominant site through which HuR stabilized SCN5A mRNA was the downstream ARE site containing a classic “AUUUA” motif. This SCN5A motif is conserved in many species. Overexpression of HuR in HL-1 mouse cardiomyocytes increased SCN5A mRNA expression further confirming this downstream putative ARE is the functional ARE, since this is the only putative ARE in the mouse SCN5A mRNA 3′UTR. The role of this ARE in cardiac sodium current regulation was confirmed by the fact that overexpression of HuR increased sodium current in HEK293 cells transfected by a SCN5A expression construct bearing the downstream ARE sequences.
HuR is downregulated in HF, reduced HuR can cause reduced SCN5A, and reduced expression of HuR has been shown to be responsible for the decreased mRNA stability,24, 25. Therefore, it seems likely that the reduced HuR is at least partially responsible for the downregulation of SCN5A and ultimately leads to increased risk of arrhythmia in HF. Our results demonstrated that overexpression of HuR in failing hearts could rescue the downregulation of SCN5A and improve arrhythmic risk after MI. These findings provide evidence that HuR plays a critical role in regulating the cardiac sodium channel.26 Since HuR is not specific for SCN5A, it is possible that the antiarrhythmic effect of overexpression involved additional HuR-dependent events. Nevertheless, this experiment establishes the possibility that raising ion channel levels could be a new antiarrhythmic strategy.
Conclusion
The mRNA stabilizing protein HuR positively regulated the cardiac sodium channel by protecting SCN5A mRNA from decay via association with the 3′-UTR. The expression of HuR decreased in ischemic heart failure. The overexpression of HuR reversed the downregulation of SCN5A in failing hearts and reduced arrhythmia risk. Therefore, HuR may contribute to HF-associated arrhythmia risk, and HuR overexpression may be a useful antiarrhythmic strategy.
Supplementary Material
Acknowledgments
We thank the Translational Cardiovascular Biobank and Repository at Washington University Medical School for assistance with human heart tissue samples.
Funding Sources
The present work was supported in part by National Institutes of Health grant R01 HL104025.
Appendix. Supplementary data
Supplementary data associated with this article can be found in the online version at
Footnotes
Disclosures
Dr. Dudley is the inventor of PCT/US2015/046162, Assessment of Heart Failure and Arrhythmic Risk Stratification by Measuring Circulating Hu Proteins. Other authors don’t have any conflict of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adjibade P, Mazroui R. Control of mRNA turnover: implication of cytoplasmic RNA granules. Semin Cell Dev Biol. 2014;34:15–23. doi: 10.1016/j.semcdb.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol LifeSci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srikantan S, Gorospe M. HuR function in disease. Front Biosci (Landmark Ed) 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Stat Med. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M, Gu L, Sulkin MS, Liu H, Jeong EM, Greener I, Xie A, Efimov IR, Dudley SC., Jr Mitochondrial dysfunction causing cardiac sodium channel downregulation in cardiomyopathy. J Mol Cell Cardiol. 2013;54:25–34. doi: 10.1016/j.yjmcc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao G, Brahmanandam V, Raicu M, Gu L, et al. Enhanced risk profiling of implanted defibrillator shocks with circulating SCN5A mRNA splicing variants: A pilot trial. J Am Coll Cardiol. 2014;63:2261–2269. doi: 10.1016/j.jacc.2014.02.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao G, Xie A, Huang SC, et al. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang LL, Pfahnl AE, Sanyal S, Jiao Z, Allen J, Banach K, Fahrenbach J, Weiss D, Taylor WR, Zafari AM, Dudley SC., Jr Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ Res. 2007;101:1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Sanyal S, Gao G, Gurung IS, Zhu X, Gaconnet G, Kerchner LJ, Shang LL, Huang CL, Grace A, London B, Dudley SC., Jr Cardiac Na+ current regulation by pyridine nucleotides. Circ Res. 2009;105:737–745. doi: 10.1161/CIRCRESAHA.109.197277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katwal AB, Konkalmatt PR, Piras BA, Hazarika S, Li SS, John Lye R, Sanders JM, Ferrante EA, Yan Z, Annex BH, French BA. Adeno-associated virus serotype 9 efficiently targets ischemic skeletal muscle following systemic delivery. Gene Ther. 2013;20:930–8. doi: 10.1038/gt.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziv O, Morales E, Song YK, Peng X, Odening KE, Buxton AE, Karma A, Koren G, Choi BR. Origin of complex behaviour of spatially discordant alternans in a transgenic rabbit model of type 2 long QT syndrome. J Physiol. 2009;587:4661–80. doi: 10.1113/jphysiol.2009.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–19. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govindaraju S, Lee BS. Adaptive and maladaptive expression of the mRNA regulatory protein HuR. World J Biol Chem. 2013;4:111–118. doi: 10.4331/wjbc.v4.i4.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zucal C, D’Agostino V, Loffredo R, Mantelli B, Thongon Natthakan, Lal P, Latorre E, Provenzani A. Targeting the multifaceted HuR protein, benefits and caveats. Curr Drug Targets. 2015;16:499–515. doi: 10.2174/1389450116666150223163632. [DOI] [PubMed] [Google Scholar]
- 17.Rutledge CA, Ng FS, Sulkin MS, Greener ID, Sergeyenko AM, Liu H, Gemel J, Beyer EC, Sovari AA, Efimov IR, Dudley SC. c-Src kinase inhibition reduces arrhythmia inducibility and connexin43 dysregulation after myocardial infarction. J Am Coll Cardiol. 2014;63:928–934. doi: 10.1016/j.jacc.2013.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borlak J, Thum T. Hallmarks of ion channel gene expression in end-stage heart failure. FASEB J. 2003;17:1592–608. doi: 10.1096/fj.02-0889com. [DOI] [PubMed] [Google Scholar]
- 19.Atack TC, Stroud DM, Watanabe H, Yang T, Hall L, Hipkens SB, Lowe JS, Leake B, Magnuson MA, Yang P, Roden DM. Informatic and functional approaches to identifying a regulatory region for the cardiac sodium channel. Circ Res. 2011;109:38–46. doi: 10.1161/CIRCRESAHA.110.235630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang LL, Sanyal S, Pfahnl AE, Jiao Z, Allen J, Liu H, Dudley SC., Jr NF-kB-dependent transcriptional regulation of the cardiac scn5a sodium channel by angiotensin II. Am J Physiol Cell Physiol. 2008;294:C372–9. doi: 10.1152/ajpcell.00186.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P, Christoffels VM. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–30. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akool el S, Kleinert H, Hamada FM, Abdelwahab MH, Forstermann U, Pfeilschifter J, Eberhardt W. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA-stabilizing factor HuR. Mol Cell Biol. 2003;23:4901–16. doi: 10.1128/MCB.23.14.4901-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin JS, Choi HE, Seo S, Choi JH, Baek NI, Lee KT. Berberine Decreased Inducible Nitric Oxide Synthase mRNA Stability through Negative Regulation of Human Antigen R in Lipopolysaccharide-Induced Macrophages. J Pharmacol Exp Ther. 2016;358:3–13. doi: 10.1124/jpet.115.231043. [DOI] [PubMed] [Google Scholar]
- 26.Eberhardt W, Doller A, Akool el S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol Ther. 2007;114:56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.