Abstract
The biocontrol function of the repressor of cellulase expression I (ACE1) in Trichoderma atroviride was verified through constructing Δace1 mutant strain by Agrobacterium tumefaciens-mediated transformation. The activities of cell wall-degrading enzymes (cellulase, xylanase, chitinase, β-1,3-glucanase, and protease) in the supernatant of Δace1 mutant strain were distinctly higher than those of control strain, followed with the elevation of related genes transcript levels. Besides, the Δace1 mutant resulted in an elevating transcript level of xyr1, but no obvious change in the expression of cre1, which suggested that ACE1 was negative regulator of the xyr1 transcription, but not involved in cre1 transcription. On core polyketide synthases of four biosynthesis gene clusters for antibiotic secondary metabolites, only the transcription levels of encoding genes Try83179/TryH and Aza79482/AzaJ in Δace1 mutant strain were significantly higher than that in wild-type during antagonizing with pathogenic fungi Fusarium oxysporum and Rhizoctonia solani (with the inhibition rate of 30.7 and 19.8%, respectively). The biocontrol function of Δace1 mutant strain was remarkably enhanced. The results indicated that ACE1, indeed, acted as a repressor for cell wall-degrading enzymes and PKSs expression in T. atroviride, and the Δace1 mutant strain effectively made related enzymes activities improved with potential enhancement of biocontrol potency.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1314-z) contains supplementary material, which is available to authorized users.
Keywords: Trichoderma atroviride, ACE1, Biocontrol, Cell wall-degrading enzymes, Polyketide synthases
Introduction
Many species of the genus Trichoderma have been developed as biocontrol agents against plant diseases, such as Trichoderma reesei, T. virens, T. atroviride, and T. harazium. T. atroviride is qualified with prominent biocontrol efficiency deriving from its mycoparasitic activity. Mycoparasitism can assist Trichoderma spp. to antagonize plant pathogens by a combination of nutrient competition, secretion of extracellular hydrolytic enzymes, and antifungal secondary metabolites like diketopiperazines, sesquiterpenes, polyketides, peptaibols, and so on (Saxena et al. 2015; Oda et al. 2015). Since the fungal cell wall is composed of β-glucans, chitin, and mannan, the expression of large amounts of extracellular hydrolytic enzymes is seemed as the critical factor for cell wall degradation, such as β-glucanase, cellulase, and hemicellulase. Several genes coding for regulators of cell wall-degrading enzymes expression had been identified, including the carbon catabolite repressor CREI (Strauss et al. 1995), the repressor cellulase expression I (ACE1) (Saloheimo et al. 2000), the activator ACE1 (Schmoll and Kubicek 2003), and so on. To study the functions of ACE1 in T. reesei, the Δace1 mutant results in an increased expression of all the main cellulase and hemicellulase genes during growth on sophorose and cellulase (Aro et al. 2003). In addition, ACE1 could also inhibit xyr1 expression in d-xylose-induced cultures (Mach-Aigner et al. 2008). Cellulase production in T. reesei is subject to a variety of environmental and physiological conditions involving an intricate regulatory network with multiple transcription factors, such as xyr1, clr1, clr2, and so on. The mating type locus protein MAT1-2-1 is identified as an interacting partner for the key transcriptional activator xyr1 of T. reesei cellulase genes (Zheng et al. 2017). It is demonstrated that both clr1 and clr2 are required for the bulk of transcriptional response to cellulose in ascomycete fungi, including induction of all major cellulase and some major hemicellulase genes (Coradetti et al. 2012). Functional CLR1 is necessary for expression of clr2 and efficient cellobiose utilization. Phylogenetic analyses showed that CLR1 and CLR2 were conserved in the genomes of most filamentous ascomycete fungi capable of degrading cellulose.
Besides, the biocontrol effect of Trichoderma could also be characterized by a significant induction of transcripts encoding secondary metabolite-producing gene clusters, such as non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) (Baker et al. 2012; Mukherjee et al. 2012), which produced a considerable part of these metabolites that were reported to possess many activities, including antimicrobial, antifungal, antiviral, antioxidant, cytotoxic, nematicidal, and anti-inflammatory activities, etc (Osmanova et al. 2010).
Therefore, the potential role of ACE1 in the biocontrol of T. atroviride will be validated in Δace1 gene mutant model through the Agrobacterium tumefaciens-mediated transformation (ATMT) technology. The changes of enzyme activities, related encoding genes expression, and transcription levels of core PKSs of biosynthesis gene clusters will be addressed to testify potential mechanism.
Materials and methods
Strains, plasmid, and culture conditions
The wild-type fungus T. atroviride IMI206040, preserved in China General Microbiological Culture Collection Center (CGMCC NO. 10168), was adopted as the parent strain for the construction of the mutants. pCAMBIA1300 was customized as knock-out vector to construct the genetically modified strain. The plasmid contains a trpc promoter, an hph gene cloned from E. coli and a trpc terminator between its multiple cloning sites and bacterial resistance loci of kanamycin. Escherichia coli strain DH5α was also used for plasmid construction and grown on LB medium at 37 °C with an appropriate supplement of kanamycin (50 µg/ml). Agrobacterium tumefaciens strain AGL1 was applied in ATMT carrying the virulence genes essential for T-DNA transfer. AGL1 was routinely grown on Luria–Bertani (LB) containing kanamycin (100 µg/ml) and rifampicin (100 µg/ml) at 28 °C.
Construction of gene knock-out plasmid pC1300-Δace1
For the construction of the deletion cassette, primers for two DNA flanking fragments of ace1 (ace1-LF/ace1-LR and ace1-RF/ace1-RR) were initially designed using the nucleotide sequence of the T. atroviride ace1 gene (JGI protein identification [ID] 83090 [http://genome.jgi-psf.org/Triat2/Triat2.home.html]) and PCR-amplified from the genomic DNA of T. atroviride. The 1452-bp fragment ace1-up containing the 5′ flanking sequences of ace1 was digested by HindIII and BamHI, and cloned into pCAMBIA1300. Similarly, another 1224-bp fragment ace1-down containing the 3′ flanking sequences of ace1 was digested by KpnI and EcoRI. Subsequently, the recombinant plasmid pC1300-Δace1 containing the whole cassette ace1-up::hph::ace1-down for gene disruption was transferred into E. coli DH5α and cultured on plates containing kanamycin (50 µg/ml) at 37 °C overnight. Finally, three positive clones are chosen and verified by PCR and sequencing technology. All primers used in this paper were shown in Table S1.
Fungal transformations and analysis of transformants
The recombinant plasmid pC1300-Δace1 was then assembled into wide-type strain to generate the mutant Δace1 via ATMT (De Groot et al. 1998). First, the recombinant plasmid pC1300-Δace1 was introduced into A. tumefaciens AGL1 competent cells. The transformants were isolated from kanamycin-resistant colonies and verified by PCR specific amplification. Then, the transformation was achieved by co-cultivation of A. tumefaciens and T. atroviride conidia. A 100-µl aliquot of A. tumefaciens AGL1 cell suspension with an OD600 of 0.6–0.8 was mixed with 100 µl of a conidial suspension (5 * 106 ml− 1). The mixture was plated onto sterile cellophane discs including 200-µM acetosyringone as induction medium and then incubated at 25 °C for 36–48 h. Before the total discs were covered with white mycelia, the cellophane discs were carefully transferred to selective induction medium plates including 200-µg/ml hygromycin B to screen the transformed strains and 300-µg/ml timentin to inhibit the growth of (A) Tumefaciens AGL1. The possible mutant colonies would appear after dark incubation at 25 °C for 5–7 days. Each putative mutant was subsequently transferred to PDA medium containing 300-µg/ml timentin and 200-µg/ml hygromycin (B) Mitotic stability was tested by subculturing five generations on PDA medium without hygromycin B.
Screening of mutants
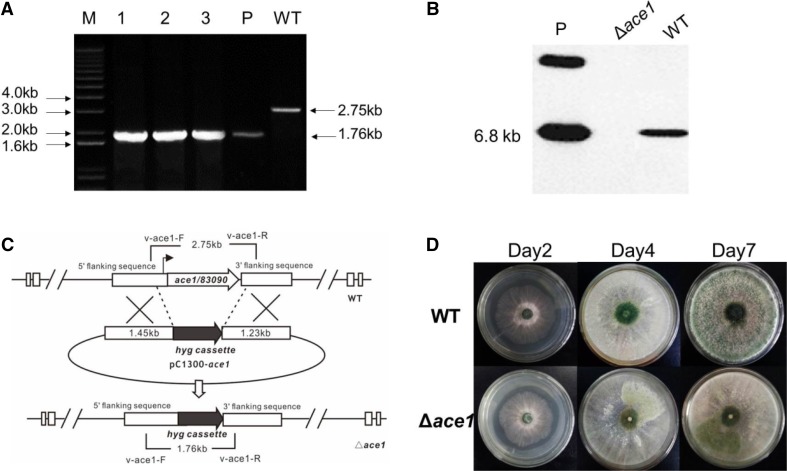
The ideal Δace1 knock-out transformants screened by real-time qPCR (Realplex 4 s, Eppendorf, USA). Primers v-ace1-F and v-ace1-R were designed to amplify the modified cassette. For a true mutant, the ORF of ace1 was replaced by the hph cassette (Fig. 2c); the PCR product (1.76 kb) was almost 1 kb smaller than wild-type strain (2.75 kb) (Fig. 2a). If there were additional T-DNA insertions in the mutants, both two PCR products would show in the gel (1.76 and 2.75 kb).
Fig. 2.
Construction of Δace1 mutant strain of Trichoderma atroviride in the sequence of pC1300:ace1-up::hyg::ace1-down. a Verification of the positive fragments of ace1 transformed; M, Marker 2000; 1–3: different mutant screened; P, plasmid positive control; WT: wide-type control; b Southern blot to verify ace1 was replaced with hybridization probe of ace1. WT wild-type strain, P plasmid, Δace1, ace1 knock-out mutant; c Mechanism of ATMT and primers design for vector pC1300-Δace1; d The phenotypes of WT and Δace1 after several days of cultivation on PDA plates
For further confirmation of the true mutants by southern blotting, amount of 40-µg DNA of transformants was digested with appropriate restriction enzyme HindIII and concentrated to 40 µl by ethanol. The DNA fragments were separated on 0.9% agarose gel. Subsequently, the gel was treated with 2.5% HCl (v/v) to break the DNA into smaller pieces and denatured with sodium hydroxide. After then, the DNA was transferred to a nylon membrane by suction 20 × SSC buffer. The probe (495-bp ace1 ORF, amplify with the primers ace1-F and ace1-R) labeling reactions and hybridization were carried out using Gene Images CDP-Star Detection Kit (Amersham, UK).
Enzyme activity assays
Briefly, the T. atroviride strain was cultured in liquid production medium consisted of peptone (0.1%), Urea (0.03%), KH2PO4 (0.2%), (NH4)2SO4 (0.14%), MgSO4·7H2O (0.03%), and CaCl2·6H2O (0.03%), and 1 ml of trace element solution (Fe2+, Mn2+, Zn2+, and Co2+), pH (5.4), for 3 days at 200 rpm. After incubating at 28 °C for 72 h, the culture filtrates were centrifuged at 3000 rpm for 10 min, and the supernatants were collected and used for enzyme assay. Enzyme activity assays of cell wall-degrading enzymes (cellulase, xylanase, chitinase, β-1,3-glucanase and protease) from the culture supernatants were performed by measuring the reduction of sugar from their respective substrates. Carboxymethylcellulase (CMC), xylan, laminarin, colloidal chitin, and gelatin were applied as substrates corresponding to the enzymes cellulase (Li et al. 2016), xylanase (Bailey et al. 1992), chitinase (Li et al. 2013a, b), β-1,3-glucanase (Noronha and Ulhoa 2000), and protease, respectively. Cellulase activity was determined in a reaction mixture (2.0 ml) containing 0.2 ml of the enzymes solution and 1.8 ml of a 1% (w/v) CMC solution prepared in sodium acetate buffer (50 mM, pH 5.0). The reaction mixture was incubated at 50 °C for 30 min, and the reducing sugar liberated in the reaction mixture was measured using the 3, 5-dinitrosalicylic acid (DNS) method. The reaction was stopped by adding 1.0 ml of 1.0-M sodium carbonate solution. Color development was measured at 405 nm. One unit (U) of enzyme activity was defined as the amount of enzyme required to liberate 1 mole of glucose per minute under the assay conditions. The enzyme activity is expressed as U·g− 1 (units per gram). The activities of xylanase were measured using DNS as well. At 50 °C and pH 5.0, 1 U of xylanase activity was defined as the amount of enzyme required to release 1-µmol xylose per minute. Xylose was used as standard for xylanase analysis. The protease activity was measured by incubating of 100 µl of culture filtrate with 400 µl of 50-mM Tris/HCl buffer (pH 7.5) containing 1.0% (w/v) gelatin at 37 °C for 4 h. The reaction was terminated by the addition of 0.4 ml of 10% (w/v) trichloroacetic acid and then centrifuged at 10,000×g for 10 min. After centrifugation, 850 µl of the supernatant was collected and mixed with 320 µl of 1-M NaOH, and A440 was measured. The absorbance changes at 440 nm were corrected using the nonenzymatic reaction (as measured under the same conditions in the absence of the culture filtrate). For measuring chitinase activity, a 100-ml fermentation medium (MgSO4·7H2O 0.6 g/L, FeSO4·7H2O 0.1 g/l, NH4NO3 3.0 g/L, KH2PO4 2.0 g/l, colloidal chitin 10% v/v, powdered chitin 2% w/w) was inoculated at 30 °C for 200 rpm in a shaking table. β-1,3-Glucanase activity was measured by mixing 125 µl of sample with 250 µl of 50-mmol/l acetate buffer (pH 5.0), containing 1% laminarin. The mixture was incubated at 45 °C for 30 min before termination of reaction. One unit of the enzyme activity was defined as the required amount of enzymes to catalyse µg of catabolized sugar per milliliter per minute under the respective reaction condition. All measurements were executed in triplicates.
Transcript analysis by quantitative PCR (qPCR)
The total RNAs of wild-type and Δace1 mutant strain under given conditions were extracted by TaKaRa MiniBEST Plant RNA Extraction Kit (Takara, Dalian, China). In addition, 500 ng of total RNA were subjected to reverse transcription using PrimeScript™ RT reagent Kit with gDNA Eraser Kit (Takara, Dalian, China) containing a blend of oligo (dT) and random primers. qPCRs were performed in SYBR Green® Realtime PCR Master Mix-Plus-Kit (TOYOBO, Shanghai, China), the primers were designed to give products of 180–240 bp with β-actin as a housekeeping control. Each reaction mixture contained 1-µl RT reaction product as template, 12.5-µl SYBR Premix Ex Taq, 1 µl of each primer (Table S1) of genes interested, and RNase-free water to a final volume of 25 µl. The PCR protocol consisted of 5 min of the initial denaturation at 95 °C followed by 40 cycles of 15 s at 95 °C, 10 s at 56 °C, and 20 s at 72 °C. A melting curve was performed after each run to check the specificity of PCR product. The qPCR experiments were delivered by Realplex 4 s, Eppendorf, USA.
The result was based on the calculated threshold cycle (CT) of each gene, which was normalized to the CT of β-actin amplified from each corresponding sample. The fold change was calculated according to the 2− ΔΔCT method (Livak and Schmittgen 2001).
Primary evaluation on antagonistic effect ofΔace1 mutant strain
To exhibit the biocontrol potency of Δace1 mutant and control strain, the new hyphae at the boundary of 3-day-old colony was selected as isolates for pot cultivation against pathogens Fusarium oxysporum and Rhizoctonia solani under artificially infestation conditions. Pots with isolates (5-mm diameter) of wild-type strain or pathogenic hyphae in each side of PDA plates were kept under greenhouse conditions at 28 °C for 5–7 days. The diameter of mycelia growth was measured and used to determine percentage of inhibition using the formula I = (C−T)/C*100%, where I represent percent inhibition; C radial growth of pathogen in the presence of control strains; T radial growth of pathogen in the presence of Δace1 mutant strain. Three replicates were done for each test pot.
Computational software and statistical analysis
The amino acid sequence of ACE1 was searched by BLAST and aligned by Clustal X 2.0.11. In addition, phylogeny analysis was achieved by MEGA5 after alignment by Clustal X2. Data analyses were performed using one-way analysis of variance (ANOVA), followed by Duncan Post hoc multiple comparison, using GraphPad Prism (version 5.0; San Diego, CA, USA). All data are presented as the mean standard error (SEM). Significance was set at P < 0.05 and P < 0.01. “a” means P < 0.05, and “b” means P < 0.01, compared with control group.
Results
Construction of the Δace1 mutant strain
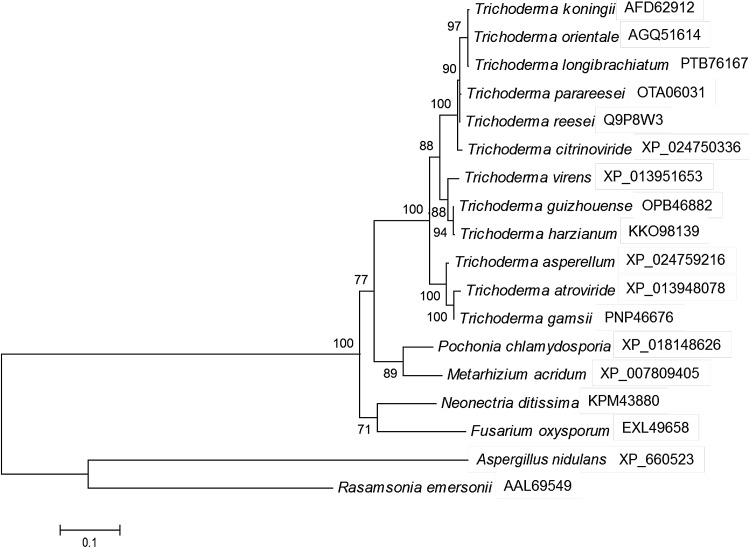
ACE1 is well known as a regulator binding to and activating the T. reesei cbh1 promoter, which was isolated by a yeast-based in vitro binding and gel mobility shift assays (Saloheimo et al. 2000). ACE1 contains three Cys2His2-type zinc fingers and is shown to bind in vitro to eight sites containing the core sequence 5′AGGCA scattered along the 1.15-kb cbh1 promoter. We used BLAST to search the T. atroviride protein database homologous to ACE1 in T. reesei (EGR51484.1). Result showed that there was a Cys2His2-type zinc-finger protein (ID 83090, NCBI Acession: XP_013948078, 85% identity), with a 2322-bp open reading frame (containing two introns) and coding a protein with 732 amino acid residues. Blast results also showed that the amino acid sequence of ACE1 in T. atroviride was identity to that in T. gamsii (98%), T. asperellum (97%), T. virens (88%), T. harzianum (88%), T. reesi (85%), Pochonia chlamydosporia (72%), and Aspergillus nidulans (37%) (Fig. 1). As to investigate the important antifungal role of ACE1 in T. atroviride, we first did the ace1 gene disruption by homologous recombination via ATMT.
Fig. 1.
Phylogeny analysis of ACE1 in T. atroviride and other strains. NCBI accession numbers of ACE1 in each strain were signed after strain name. It was the maximum-likelihood trees of ACE1 amino acid sequences in NCBI database after aligned by Clustal X. The number signed in branch represented the bootstrap value after 100 replications
After the PCR amplification, the knock-out vector was constructed by inserting the flanking fragments of ace1 to the multiple cloning site of plasmid pC1300. Then, the vector was applied in ATMT method to construct the ace1 knock-out mutant strain (Fig. 2a, b). In general, more than 20 transformants could grow in each plate [mix of 100 ul (A) tumefaciens AGL1 and 5*106-ml− 1 conidial suspension]. 36 transformants were transferred to PDA medium containing 300-µg/ml timentin and 200-µg/ml hygromycin (B). Three of them were later subjected to subculture twice and 100 single clones from these three transformants were tested by PCR amplification using primers v-ace1-F/R of to screen the mutant Δace1. The target Δace1 mutant showed only one 1.76-kb PCR product in gel, while the PCR product(2.75 kb) of wild type was larger than mutant (Fig. 2a). If there was additional T-DNA insertion in the mutant, both 1.76- and 2.75-kb product would be amplified at the same time. Southern bolt was applied to verify the idea mutant (Fig. 2b). Thereafter, the Δace1 mutant strain of T. atroviride in the sequence of pC1300:ace1-up::hyg::ace1-down was constructed successfully with no remarkable difference on dry weights of the mycelia compared with wild-type stain, but with a slight delay on sporulation (Fig. 2c, d). The genetic complementation experiment was also done to verify the function of ace1 (data not shown).
Activities of extracellular cell wall-degrading proteins and related encoding genes expression
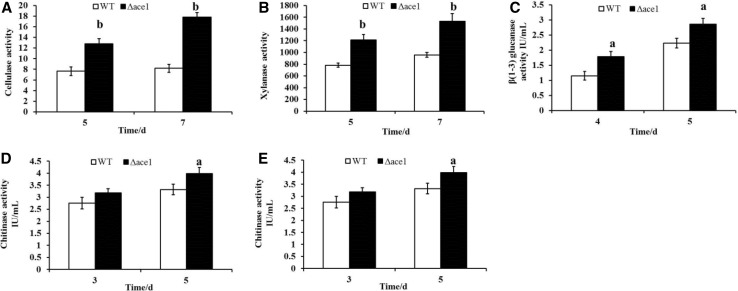
The activities of cell wall-degrading enzymes (cellulase, xylanase, chitinase, β-1,3-glucanase, and protease) between Δace1 mutant strain and control strain were evaluated after 3, 4, 5, and 7 days of growth on PDA. Carboxymethylcellulase (CMC), xylan, laminarin, colloidal chitin, and gelatin were applied as substrates for their activities assays, respectively. Besides, the total amount of secreted proteins in the culture supernatants was detected to be approximately 24.4 and 22.4% higher in Δace1 mutant strain than that in control strain after 5 and 7 day cultivation (5 days: 2.24 ± 0.02 vs. 1.80 ± 0.01; 7 days: 2.79 ± 0.02 vs. 2.28 ± 0.01, respectively). After 5 days of growth, the activities of cell wall-degrading enzymes (cellulase, xylanase, chitinase, β-1,3-glucanase, and protease) in the supernatant of Δace1 mutant strain were 1.7, 1.6, 1.1, 1.2, and 1.2 times higher than those of control strain (Fig. 3a–e) significantly (P < 0.05). In summary, the results indicated that the mutant of ace1 resulted in enhanced productivity of extracellular cell wall-degrading proteins in T. atroviride.
Fig. 3.
Gene mutant of ace1 enhanced activities of cell wall-degrading enzymes (a–e). a Cellulase activity; b xylanase activity; c β(1–3) glucanase activity; d chitinase activity; e protease activity. Data analyses were performed using one-way analysis of variance (ANOVA), followed by Duncan’s test, using GraphPad Prism 5 (San Diego, CA, USA). All data are presented as the mean standard error (SEM). “a” means P < 0.05, and “b” means P < 0.01, compared with control group
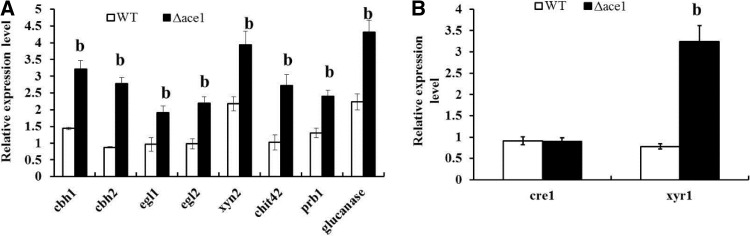
qRT-PCR was carried out to verify the related encoding gene expression (cbh1, cbh1, egl1, egl2, xyn2, chit42, prb1, and glucanase) of these cell wall-degrading enzymes. After 3 days of cultivation, the expressions of cbh1, cbh1, egl1, egl2, xyn2, chit42, prb1, and glucanase genes in Δace1 mutant strain were 2.2, 3.2, 2.0, 2.2, 1.8, 2.7, 1.8, and 1.9 times higher, respectively (P < 0.01, Fig. 4a). The regulator-encoding genes of cre1 and xyr1 were studied as well with β-actin as reference. Only the expression of xyr1 was impacted by the mutant of ace1 gene, which suggested that ACE1 might be a repressor for the transcription of the xyr1 gene (P < 0.01) but did not impact the expression of cre1 gene (Fig. 4b). These results provided good consistency with the relative results of enzyme activities.
Fig. 4.
Gene mutant of ace1 enhanced related genes expressions of cell wall-degrading enzymes (a) and their regulator-encoding genes (b). a Enzyme-related genes including cbh1, cbh1, egl1, egl2, xyn2, chit42, prb1, and glucanase were extracted after 5 days of cultivation. b Regulator-encoding genes of cre1 and xyr1 were studied. All genes expression was analyzed by RT-PCR with β-actin as reference. Data analyses were performed using one-way analysis of variance (ANOVA), followed by Duncan’s test, using GraphPad Prism 5 (San Diego, CA, USA). All data are presented as the mean standard error (SEM). “a” means P < 0.05, and “b” means P < 0.01, compared with control group
Expression of biosynthesis gene clusters for antibiotic secondary metabolites
We attempted to predict biosynthesis gene clusters of antibiotic secondary metabolites in T. atroviride genome (Trichoderma atroviride IMI 206040_v2.0) through the Antismath website (http://fungismash.secondarymetabolites.org/#!/start). Of all discovered 35 gene clusters, 13 cases were polyketide biosynthesis gene clusters and 6 cases for non-ribosomal peptides, which inferred that T. atroviride might have the great potential to produce microbial active chemical compounds. At last, four PKSs biosynthesis gene clusters were selected due to the high similarity to the reported gene clusters (Gao et al. 2011; Chen et al. 2017).
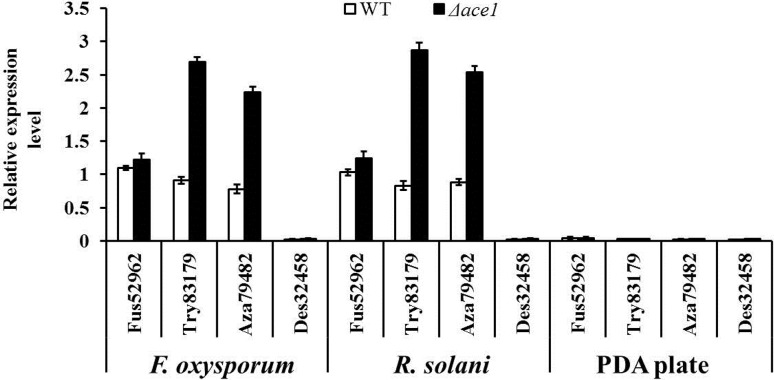
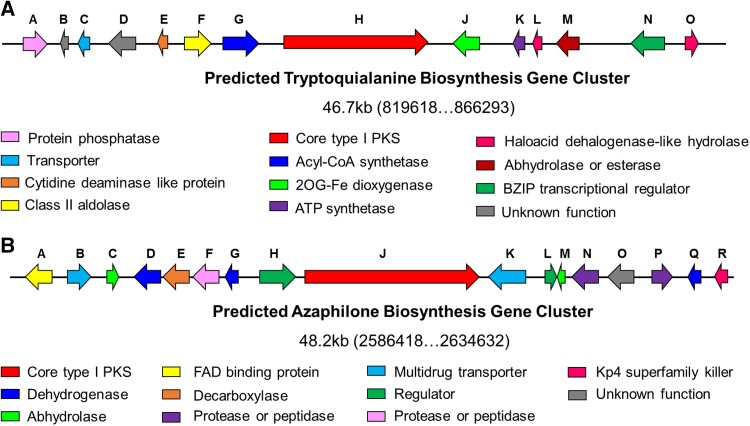
Subsequently, the transcription levels of these core PKSs in the hyphae cultivated on blank PDA plates or hyphae induced by pathogens F. oxysporum and R. solani were assessed. As shown in Fig. 5, four core PKSs were in a state of silence with few of transcription on blank PDA plates in both WT and Δace1 mutant strains. During antagonizing with pathogenic fungi, the transcription levels of Try83179/TryH and Aza79482/AzaJ in Δace1 mutant strain was prominently higher (F. oxysporum: 3.0 and 2.9 times; R. solani: 3.5 and 2.9 times, all P < 0.01) than that in wild-type one, which meant that the expressions of these two synthetase could be induced by pathogenic fungi, and ACE1 might inhibit the expression of these two genes to a certain extent. These two biosynthesis gene clusters were predicted and analyzed, as shown in Fig. 6 & Tables S2–S3. As typical PKSs synthesize gene clusters, both of them were composed of various enzymes related to the synthesis of secondary metabolites, regulator for genes transcription, transporters, and some other proteins. In addition, the transcription of synthetase in these clusters was mainly regulated by the regulator protein for transcription.
Fig. 5.
Transcription levels of four core PKS synthetases in the hyphae cultivated on blank PDA plates or hyphae induced by pathogens Fusarium oxysporum and Rhizoctonia solani. Data analyses were performed using one-way analysis of variance (ANOVA), followed by Duncan’s test, using GraphPad Prism 5 (San Diego, CA, USA). All data are presented as the mean standard error (SEM). “a” means P < 0.05, and “b” means P < 0.01, compared with control group
Fig. 6.
Predicted biosynthesis gene clusters of tryptoquialanine (a) and azaphilone (b)
The transcription of Des32458 gene in Δace1 mutant strain was also induced in the rising trend but with no significant difference after compared with the control strain. It inferred that Des32458 gene expression might not be regulated by ACE1. Besides, there was no obvious expression of Fus52962 in co-culture conditions, which indicated that it might not attend the antibiotic effect of T. atroviride at least on these two pathogens.
Δace1 mutants are more effective for biocontrol of plant pathogens
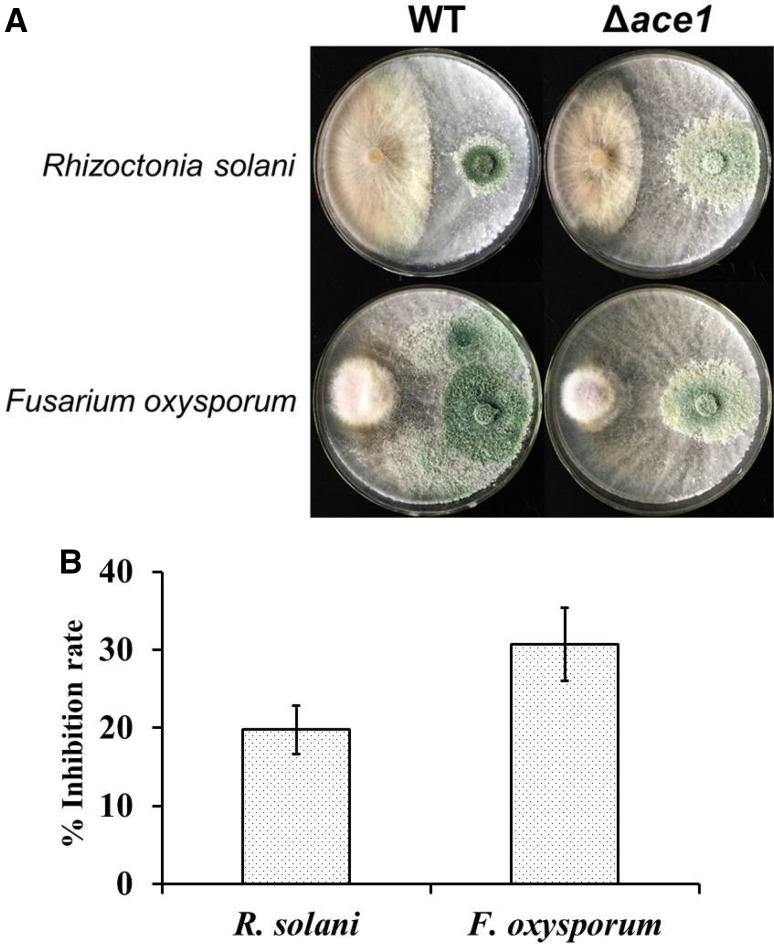
As mentioned above, not only the activities of cell wall-degrading enzymes could be enhanced by Δace1 mutant strain, but also the expressions of two core synthetases for antibiotic secondary metabolites were elevated in Δace1 mutant strain under the induction of pathogenic fungi. Both cell wall-degrading enzymes activities and antibiotic secondary metabolites were believed to be important factors for biocontrol. We compared the antagonism of both Δace1 mutant and WT strains against pathogenic fungi F. oxysporum and R. solani on PDA plates as shown in Fig. 7. After antagonizing with both F. oxysporum and R. solani, smaller clone with fewer mycelia was observed in Δace1 mutant strain. In contrast, the growth of WT strain was more vigorous with much more conidia, which meant that WT strain possessed less efficient on the resistance effect against these two pathogens. This result indicated that the repressor ACE1 indeed played important negative role in the biocontrol application of T. atroviride.
Fig. 7.
In vitro antagonistic effect (a) and related inhibition rate (b) of Δace1 mutant and control strain against the pathogens of F. oxysporum and R. solani
Discussion
The genus Trichoderma is well known for its opportunistic lifestyle, including saprotrophy, mycoparasitism, endophytism, and interactions with plants and animals. Trichoderma strains have become prominent biocontrol agents for their ability to antagonize other fungi and stimulate plant defences against phytopathogens (Schuster and Schmoll 2010). As biocontrol agents, secondary metabolite production in Trichoderma spp. is of particular importance. Trichoderma spp. are probably best known for their production of peptaibols, which are non-ribosomal peptides with antimicrobial and plant defence-stimulating activity (Viterbo et al. 2007), while relative studies on PKSs in Trichoderma are less. It is believed that PKS- and NRPS-encoding genes are linked to specific secondary metabolites (Bushley and Turgeon 2010; Chiang et al. 2010; Kubicek et al. 2011). From the results described above, the biocontrol potency of Δace1 mutant strain was remarkably improved at least partly via enhancing the activities of cell wall-degrading enzymes, and elevating the expressions of core PKSs of four biosynthesis gene clusters for antibiotic secondary metabolites.
The encoding genes regulators of cellulase and hemicellulase included carbon catabolite repressor (Strauss et al. 1995), the repressor ACE1 (Saloheimo et al. 2000), the activator ACE1I, the CCAAT-binding complexes, the activator xylanase regulator 1 (XYRI), and so on. It was reported that the expression of the main cellulase and xylanase genes was elevated in the Δace1 knock-out strain under sophorose- and cellulase-induced cultivations (Aro et al. 2003). The expression of xyr1 gene could also be repressed by ACE1 during growth on d-xylose (Mach-Aigner et al. 2008). Currently, a dimer of XYRI was believed to be the basis of the induction-specific complex for xyn1 expression, whilst ACE1 was deemed as a competitor of XYRI for one of the two binding elements in the xyn1 promoter. The functional ACE1 repressor complex consists of a single ACE1, XYRI, and ACE1-interacting protein.
As a kind of active filamentous fungi, Trichoderma could not only secrete a large number of cell wall-degrading enzymes, but also release a variety of antibacterial secondary metabolites to suppress or kill the competitors in ecological environment. In the application of industrial production, T. reesei is a major cellulase-degrading strain being used in cellulosic material hydrolysis for commercialization of cellulosic biofuels and biochemical (Li et al. 2014). Based on this study, it can be learnt that the knock-out of ace1 in T. atroviride significantly improve the activities of these related cell wall-degrading enzymes, which means that this approach may also provide a quotable experiences for its potential improvement on cellulosic material hydrolysis.
The current analysis of the three available Trichoderma genome sequences has indicated that the genomes of these organisms encode only a small catalogue of PKSs: T. atroviride and T. virens each encode 18, and T. reesei only 11 predicted PKSs (Kubicek et al. 2011; Martinez et al. 2008). Plenty of physiologically active fungal metabolites were discovered and reported, especially PKS-NRPS secondary metabolites, such as GKK1032 (including four components GKK1032A1, GKK1032A2, GKK1032A3, and GKK1032B) (Becker et al. 2012; Oikawa 2003), pyrrocidines A/B (He et al. 2002; Bigelis et al. 2006), hirsutellones A–F (Isaka et al. 2005, 2006), and so on, due to their significant antibiotic activities (Li et al. 2013a, b). GKK1032B was antibiotic at MIC = 20.8 µg/ml on Bacillus subtilis (Oikawa 2003). A potent antibiotic activity was detected for pyrrocidine A on Gram-positive bacteria including drug-resistant strains (piperacillin-resistant Staphylococcus and vancomycin-resistant Enterococcus strains) at MIC = 0.25–2 µg/ml. Pyrrocidine B was much less active. Hirsutellones A–C showed significant antimycobacterial activities against Mycobacterium tuberculosis H37Ra at MIC = 0.78 µg/ml, while hirsutellone D was active at 3.125 µg/ml (value for isoniazid: 0.06 µg/ml) (Li et al. 2013a, b). Hirsutellone F is a heterodimeric compound made of both hirsutellone A and hirsutellone B units. Its antimycobacterial activity was lower than that of the monomeric compounds, with an MIC value of 3.12 µg/ml against M. tuberculosis H37Ra. It was also active against the malarial parasite Plasmodium falciparum K1 with a moderate IC50 of 4.2 µg/ml (value for the monomers > 20 µg/ml) (Isaka et al. 2006).
Four biosynthesis gene clusters mentioned before for antibiotic secondary metabolites were in a state of silence with few of transcription on blank PDA plates in both WT and Δace1 mutant strains. It was assumed that those biosynthesis gene clusters might be induced to express only in special cultivation conditions to save energy and avoid material waste. There were only a few studies confusing on the changes of these biosynthesis gene clusters and related core PKSs in transcription level after relative gene mutant or silence (Mukherjee et al. 2012; Baker et al. 2012). In our results, during antagonizing with pathogenic fungi, the transcription levels of Try83179/TryH and Aza79482/AzaJ in Δace1 mutant strain were significantly higher (F. oxysporum: 3.0 and 2.9 times; R. solani: 3.5 and 2.9 times) than that in wild-type one, which meant that the expressions of these two synthetases could be induced by pathogenic fungi, and ACE1 might inhibit the expression of these two genes to a certain extent. On the other hand, it can be assured that Δace1 mutant is also beneficial for the synthesis of associated secondary metabolites (such as polyketides tryptoquialanine and azaphilone) and the improvement of biocontrol functions.
There are many limitations existed in this study. First, the enhanced activities of cell wall-degrading enzymes in Δace1 mutant strain were limited in a relatively low degree, which can be explained by the high baseline of associated enzymatic activities. Second, we only measured the transcription level of core PKSs, which was not a promise to the final expression of polyketides, since their biosynthesis was mediated by many other synthetases or regulators, such as haloacid dehalogenase-like hydrolase, abhydrolase, esterase, decarboxylase, and so on. At last, the further studies on many other core PKSs are warranted.
The biocontrol function of Δace1 mutant strain was remarkably enhanced under the conditions of co-culture with pathogens Fusarium oxysporum and Rhizoctonia solani. The results indicated that ACE1, indeed, acts as a repressor for cell wall-degrading enzymes expression in T. atroviride, and Δace1 effectively improved related enzymes activities with potential enhancement of biocontrol potency.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We really appreciate all the useful comment and support from our colleagues in Jiangxi University of Technology, China.
Funding
This work was supported by Jiangxi Provincial Science and Technology Project of Ministry of Education (Project No.: GJJ161137) and Natural Science Project of Jiangxi Science and Technology College (Project No.: 16ZRYB03).
Compliance with ethical standards
Conflict of interest
All authors declare that they have no competing interests.
Ethical statement
All procedures of this study were complied with local experimental operation specification. In addition, no relevant ethical statement was needed to be declared.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1314-z) contains supplementary material, which is available to authorized users.
References
- Aro N, Ilmén M, Saloheimo A, Penttilä M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23:257–270. doi: 10.1016/0168-1656(92)90074-J. [DOI] [Google Scholar]
- Baker SE, Perrone G, Richardson NM, Gallo A, Kubicek CP. Phylogenomic analysis of polyketide synthase-encoding genes in Trichoderma. Microbiology. 2012;158:147–154. doi: 10.1099/mic.0.053462-0. [DOI] [PubMed] [Google Scholar]
- Becker J, Liermann JC, Opatz T, Anke H, Thines E. GKK1032A2, a secondary metabolite from Penicillium sp. IBWF-029-96, inhibits conidial germination in the rice blast fungus Magnaporthe oryzae. J Antibiot (Tokyo) 2012;65:99–102. doi: 10.1038/ja.2011.114. [DOI] [PubMed] [Google Scholar]
- Bigelis R, He H, Yang HY, Chang LP, Greenstein M. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J Ind Microbiol Biotechnol. 2006;33:815–826. doi: 10.1007/s10295-006-0126-z. [DOI] [PubMed] [Google Scholar]
- Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zeng X, Yang YL, Xing YM, Zhang Q, Li JM, Ma K, Liu HW, Guo SX. Genomic and transcriptomic analyses reveal differential regulation of diverse terpenoid and polyketides secondary metabolites in Hericium erinaceus. Sci Rep. 2017;7:10151. doi: 10.1038/s41598-017-10376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Oakley BR, Keller NP, Wang CC. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl Microbiol Biotechnol. 2010;86:1719–1736. doi: 10.1007/s00253-010-2525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- Gao X, Chooi YH, Ames BD, Wang P, Walsh CT, Tang Y. Fungal indole alkaloid biosynthesis: genetic and biochemical investigation of the tryptoquialanine pathway in Penicillium aethiopicum. J Am Chem Soc. 2011;133:2729–2741. doi: 10.1021/ja1101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Yang HY, Bigelis R, Solum EH, Greenstein M, Carter G. Pyrrocidines A and B, new antibiotics produced by a filamentous fungus. Tetrahedron Lett. 2002;43:1633–1636. doi: 10.1016/S0040-4039(02)00099-0. [DOI] [Google Scholar]
- Isaka M, Rugseree N, Maithip P, Kongsaeree P, Prabpai S, Thebtaranonth Y. Hirsutellones A–E, antimycobacterial alkaloids from the insect pathogenic fungus Hirsutella nivea BCC 2594. Tetrahedron. 2005;61:5577–5583. doi: 10.1016/j.tet.2005.03.099. [DOI] [Google Scholar]
- Isaka M, Prathumpai W, Wongsa P, Tanticharoen M. Hirsutellone F, a dimer of antitubercular alkaloids from the seed fungus Trichoderma species BCC 7579. Org Lett. 2006;8:2815–2817. doi: 10.1021/ol060926x. [DOI] [PubMed] [Google Scholar]
- Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yang Z, Zhang RH, Zhang D, Chen S, Ma L. Effect of pH on cellulase production and morphology of Trichoderma reesei and the application in cellulosic material hydrolysis. J Biotechnol. 2013;168:470–477. doi: 10.1016/j.jbiotec.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Li XW, Ear A, Nay B. Hirsutellones and beyond: figuring out the biological and synthetic logics toward chemical complexity in fungal PKS–NRPS compounds. Natl Prod Rep. 2013;30:765–782. doi: 10.1039/c3np70016j. [DOI] [PubMed] [Google Scholar]
- Li Q, Ng WT, Wu JC. Isolation, characterization and application of a cellulose-degrading strain Neurospora crassa S1 from oil palm empty fruit bunch. Microb Cell Fact. 2014;13:157. doi: 10.1186/s12934-014-0157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun R, Yu J, Saravanakumar K, Chen J. Antagonistic and biocontrol potential of Trichoderma asperellum ZJSX5003 against the Maize Stalk Rot Pathogen Fusarium graminearum. Indian J Microbiol. 2016;56:318–327. doi: 10.1007/s12088-016-0581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl Environ Microbiol. 2008;74:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Buensanteai N, Moran-Diez ME, Druzhinina IS, Kenerley CM. Functional analysis of non-ribosomal peptide synthetases (NRPSs) in Trichoderma virens reveals a polyketide synthase (PKS)/NRPS hybrid enzyme involved in the induced systemic resistance response in maize. Microbiology. 2012;158:155–165. doi: 10.1099/mic.0.052159-0. [DOI] [PubMed] [Google Scholar]
- Noronha EF, Ulhoa CJ. Characterization of a 29-kDa beta-1,3-glucanase from Trichoderma harzianum. FEMS Microbiol Lett. 2000;183:119–123. doi: 10.1111/j.1574-6968.2000.tb08944.x. [DOI] [PubMed] [Google Scholar]
- Oda S, Kameda A, Okanan M, Sakakibara Y, Ohashi S. Discovery of secondary metabolites in an extractive liquid-surface immobilization system and its application to high-throughput interfacial screening of antibiotic-producing fungi. J Antibiot (Tokyo) 2015;68:691–697. doi: 10.1038/ja.2015.59. [DOI] [PubMed] [Google Scholar]
- Oikawa H. Biosynthesis of structurally unique fungal metabolite GKK1032A(2): indication of novel carbocyclic formation mechanism in polyketide biosynthesis. J Org Chem. 2003;68:3552–3557. doi: 10.1021/jo0267596. [DOI] [PubMed] [Google Scholar]
- Osmanova N, Schultze W, Ayoub N. Azaphilones: a class of fungal metabolites with diverse biological activities. Phytochem Rev. 2010;9:315–342. doi: 10.1007/s11101-010-9171-3. [DOI] [Google Scholar]
- Saloheimo A, Aro N, Ilmén M, Penttilä M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J Biol Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- Saxena A, Raghuwanshi R, Singh HB. Trichoderma species mediated differential tolerance against biotic stress of phytopathogens in Cicer arietinum L. J Basic Microbiol. 2015;55:195–206. doi: 10.1002/jobm.201400317. [DOI] [PubMed] [Google Scholar]
- Schmoll M, Kubicek CP. Regulation of Trichoderma cellulase formation: lessons in molecular biology from an industrial fungus. A review. Acta Microbiol Immunol Hung. 2003;50:125–145. doi: 10.1556/AMicr.50.2003.2-3.3. [DOI] [PubMed] [Google Scholar]
- Schuster A, Schmoll M. Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol. 2010;87:787–799. doi: 10.1007/s00253-010-2632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Mach RL, Zeilinger S, Hartler G, Stöffler G, Wolschek M, Kubicek CP. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 1995;376:103–107. doi: 10.1016/0014-5793(95)01255-5. [DOI] [PubMed] [Google Scholar]
- Viterbo A, Wiest A, Brotman Y, Chet I, Kenerley C. The 18mer peptaibols from Trichoderma virens elicit plant defence responses. Mol Plant Pathol. 2007;8:737–746. doi: 10.1111/j.1364-3703.2007.00430.x. [DOI] [PubMed] [Google Scholar]
- Zheng F, Cao Y, Wang L, Lv X, Meng X, Zhang W, Chen G, Liu W. The mating type locus protein MAT1-2-1 of Trichoderma reesei interacts with Xyr1 and regulates cellulase gene expression in response to light. Sci Rep. 2017;7:17346. doi: 10.1038/s41598-017-17439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.